Abstract
The synaptic localization, clustering, and immobilization of neurotransmitter receptors and ion channels play important roles in synapse formation and synaptic transmission. Although several proteins have been identified that interact with AMPA receptors and that may regulate their synaptic targeting, little is known about the interaction of AMPA receptors with the cytoskeleton. In studies examining the interaction of the AMPA receptor GluR1 subunit with neuronal proteins, we determined that GluR1 interacts with the 4.1G and 4.1N proteins, homologs of the erythrocyte membrane cytoskeletal protein 4.1. Using the yeast two-hybrid system and a heterologous cell system, we demonstrated that both 4.1G and 4.1N bind to a membrane proximal region of the GluR1 C terminus, and that a region within the C-terminal domain of 4.1G or 4.1N is sufficient to mediate the interaction. We also found that 4.1N can associate with GluR1 in vivo and colocalizes with AMPA receptors at excitatory synapses. Disruption of the interaction of GluR1 with 4.1N or disruption of actin filaments decreased the surface expression of GluR1 in heterologous cells. Moreover, disruption of actin filaments in cultured cortical neurons dramatically reduced the level of surface AMPA receptors. These results suggest that protein 4.1N may link AMPA receptors to the actin cytoskeleton.
Keywords: GluR1, protein 4.1, cytoskeleton, synaptic, AMPA receptor, yeast two-hybrid
Glutamate is the major excitatory neurotransmitter in the CNS. Glutamate receptors consist of three subclasses, namely AMPA, kainate, and NMDA receptors, based on their physiological and pharmacological properties (Hollmann and Heinemann, 1994). These receptors are heteromeric complexes of homologous subunits, which combine to form a variety of receptor subtypes. The synaptic targeting, clustering, and immobilization of glutamate receptors are crucial for efficient excitatory synaptic transmission. Recent studies have identified a variety of proteins that may play a role in the aggregation and immobilization of neurotransmitter receptors (Sheng and Kim, 1996; Kim and Huganir, 1999). For example, the PSD95/DIg/Z0-1 (PDZ) domain containing-protein postsynaptic density 95 (PSD-95)/synapse-associated protein 90 (SAP90) and its family members SAP102 and PSD-93/Chapsyn-110 have been shown to physically associate with the C termini of the NMDA receptor subunits and may be involved in the synaptic localization of NMDA receptors (Sheng and Kim, 1996; Kornau et al., 1997; O'Brien et al., 1998a). Genetic studies inDrosophila have revealed that the discs-large (DLG) protein, a PSD-95 related protein, is essential for the synaptic clustering of Shaker-type K+ channels (Tejedor et al., 1997). The C termini of the AMPA receptors have also been shown to interact with several PDZ domain-containing proteins such as glutamate receptor interacting protein (GRIP)/AMPA binding protein (ABP) (Dong et al., 1997; Srivastava et al., 1998), protein interacting with C-kinase (PICK1) (Xia et al., 1999), and SAP97 (Leonard et al., 1998) via their C-terminal T/SXV motifs. These interactions are thought to be important for the sorting and synaptic expression of these receptors.
The actin cytoskeleton has also been found to be critical for the immobilization of these receptors (Allison et al., 1998; Kim and Lisman, 1999). Recent studies in hippocampal neurons in culture have shown that disruption of F-actin decreased the number of clusters of NMDA and AMPA receptors on dendritic spines, suggesting the immobilization of these receptors depends on the integrity of the F-actin network (Allison et al., 1998). This observation was further supported by electrophysiological studies (Kim and Lisman, 1999;Krucker et al., 2000), which indicated that dynamic actin filaments are important for basal synaptic transmission as well as induction and maintenance of long-term potentiation. In addition, several cytoskeletal proteins have been shown to associate with NMDA receptors. α-Actinin-2, a member of the spectrin–dystrophin family of actin-binding proteins, binds to the C termini of both the NR1 and NR2B subunits of the NMDA receptor (Wyszynski et al., 1997). The neurofilament light chain and Yotiao, a novel cytoskeletal protein with a coiled-coil structure, have also been found to interact with the C terminus of the NR1 subunit (Ehlers et al., 1998; Lin et al., 1998). However, the direct or indirect interaction of cytoskeletal proteins with AMPA receptors has not been reported. Here we used the yeast two-hybrid system to identify that proteins 4.1G and 4.1N, homologs of the erythrocyte membrane cytoskeletal protein 4.1, can interact with the GluR1 subunit of the AMPA receptor. Protein 4.1, originally identified in red blood cells and called red blood cell protein 4.1 (4.1R), is critical for the organization and maintenance of the spectrin–actin cytoskeleton and for the attachment of the cytoskeleton to the cell membrane through interaction with integral membrane proteins such as glycophorin C and Band 3 (Tyler et al., 1979; Anderson and Lovrien, 1984; Pasternack et al., 1985). The erythrocyte 4.1R has a 30 kDa N-terminal domain that interacts with glycophorin C, calmodulin, and p55 (Tanaka et al., 1991; Hemming et al., 1994; Marfatia et al., 1994, 1995), a 16 kDa domain critical for membrane association, a 10 kDa domain containing the binding site for spectrin and actin complexes, and a 22–24 kDa C-terminal domain (CTD) whose function is for the most part unknown. 4.1G is ubiquitously expressed in cells, whereas 4.1N is enriched in neurons (Walensky et al., 1999). Both 4.1G and 4.1N share homology to 4.1R at the 10, 16, and 22–24 kDa domains but are distinct at their N-terminal domains. Our results show that both 4.1G and 4.1N bind to a membrane proximal region in the C terminus of GluR1, and a consensus region within the CTDs of 4.1G and 4.1N is sufficient to mediate the binding. Protein 4.1N also associates with GluR1 in vivo and colocalizes with AMPA receptors in excitatory synapses. Moreover, we found that GluR1 truncation mutants lacking the membrane proximal region capable of 4.1 binding have decreased plasma membrane expression. Overexpression of the CTDs of 4.1 or disruption of the F-action network also resulted in reduced GluR1 surface expression. These data suggest that protein 4.1G and 4.1N may serve to cross-link AMPA receptors to the actin cytoskeleton at excitatory synapses.
MATERIALS AND METHODS
Yeast two-hybrid screening and interaction assay. The cDNA corresponding to the C-terminal tail (the last 82 amino acids, 8.5 kDa) of rat GluR1 was amplified by PCR and subcloned in-frame into theSalI–BglII site of the pPC97 yeast vector containing the GAL4 DNA-binding domain (Chevray and Nathans, 1992). This bait plasmid was then transformed into yeast strain PJ69-4A (James et al., 1996). Yeast two-hybrid screening (Fields and Song, 1989) was performed using a random-primed cDNA library from rat hippocampus (Dong et al., 1997) subcloned into theSalI–NotI site of the pPC86 vector containing the GAL4 activation domain (Chevray and Nathans, 1992). Positive clones were selected on plates lacking leucine, tryptophan, and adenine and confirmed by growth on quadruple minus plates (Leu−, Trp−, Ade−, and His−) with 2 mm 3-aminotriazole and by liquid assay for β-galactosidase activity (Reynolds and Lundblad, 1989).
The yeast two-hybrid system was also used to investigate the interaction between protein 4.1G and GluR1. The C-terminal domain of GluR1 with various truncations was subcloned into the pPC97 vector, and the CTD of rat 4.1G (see Fig. 4B) was subcloned into the pPC86 vector. Combinations of these constructs (one in pPC97 and the other in pPC86) were co-transformed into PJ69-4A yeast cells and selected on Leu− and Trp− plates for double transformants, which were further plated on quadruple minus plates (Leu−, Trp−, Ade−, and His−) with 2 mm 3-aminotriazole to test for interaction. Any interaction detected was also confirmed by liquid assay for β-galactosidase activity. Vector pPC97 or pPC86 with no insert was used in control experiments.
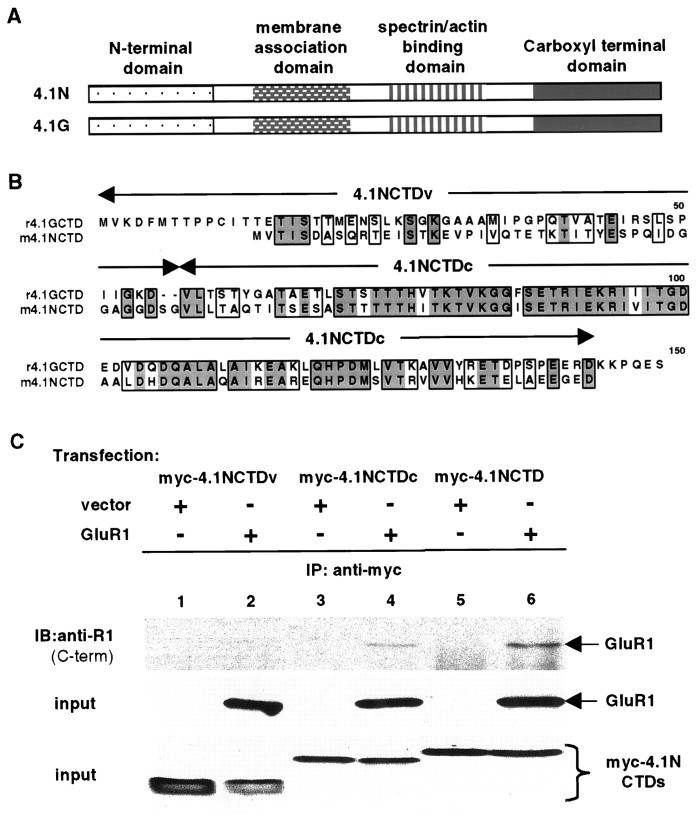
Fig. 4.
A consensus region within 4.1N is sufficient for interaction with GluR1. A, Schematic diagram of domain structures of 4.1N and 4.1G. The two proteins have significant homology at the membrane association domain, spectrin–actin binding domain, and CTD but have little homology at the N-terminal domain.B, Sequence alignment of CTDs of 4.1N and 4.1G. Identical amino acid residues are boxed anddarkly shaded; conserved residues areboxed and lightly shaded. The alignment shows that CTDs of the two proteins contain a variable N-terminal region (4.1NCTDv) and a highly conserved C-terminal region (4.1NCTDc). C, Association of the consensus region of 4.1NCTD with GluR1. A myc-tagged variable region (myc-4.1NCTDv; lanes 1, 2) or consensus region (myc-4.1NCTDc; lanes 3, 4) of 4.1NCTD or the entire 4.1NCTD (myc-4.1NCTD;lanes 5, 6) was co-transfected with vector (lanes 1, 3, 5) or with GluR1 (lanes 2, 4, 6) into 293T cells. Immunoprecipitations were performed on solubilized cell lysates using anti-myc antibody followed by SDS-PAGE and immunoblotting with anti-GluR1 antibody (top panel). The presence of GluR1 (middle panel) and myc-tagged proteins (bottom panel) in the input for immunoprecipitation was confirmed by Western blot using anti-GluR1 C-terminal and anti-myc antibodies, respectively.
Cell cultures and transfection. HEK 293T cells and COS cells were maintained in minimal essential medium (MEM; Life Technologies, Gaithersburg, MD) with 10% fetal bovine serum (Life Technologies) and 0.5% l-glutamine. The cDNA of the full-length rat GluR1 or its various mutants or the full-length protein 4.1N was subcloned into the mammalian expression vector pRK5. cDNAs were also subcloned into the mammalian expression vector pRK5 bearing a myc or hemagglutinin tag upstream of the cloning site where indicated. Five to 10 μg of each cDNA were used to transfect HEK 293T cells, and additional pRK5 vector DNA was added when necessary to equalize the total amount of DNA transfected. Transfection was done by calcium phosphate coprecipitation, as described (Blackstone et al., 1992a). After 36–48 hr, the cells were harvested and solubilized in immunoprecipitation buffer (in mm: 5 EDTA, 5 EGTA, 1 Na3VO4, 10 Na pyrophosphate, and 50 NaF in PBS, pH 7.4) with 1% Triton X-100 at 4°C for 1 hr. Cell lysates were centrifuged at 100,000 ×g for 10 min at 4°C, and the supernatant was either immediately used for immunoprecipitation or stored at −80°C.
Neuronal cultures. Low-density hippocampal neurons were cultured following the standard procedure described by Goslin and Banker (1991). Sprague Dawley rats of embryonic day 17 were used to culture cortical neurons as described with minor modification (Ghosh and Greenberg, 1995). Cells were maintained in MEM supplemented with 5% heat-inactivated horse serum.
Preparation of P2 fraction from brain tissue. Brain tissue from male Sprague Dawley rats, 4–6 weeks old, was homogenized in 10 volumes of buffered sucrose (0.32 m sucrose in 4 mm HEPES, pH 7.4, with protease inhibitors antipain, chymotrypsin, leupeptin, and Trasylol, and 0.1 mm PMSF) in a glass–Teflon homogenizer. The homogenate was centrifuged at 800 × g for 10 min, the supernatant was then subjected to another centrifugation at 9000 × g for 15 min. The supernatant from the second centrifugation, the crude synaptosomal fraction (P2), was either immediately used for immunoprecipitation or stored at −80°C. For immunoprecipitation, the P2 fraction was first solubilized in 1% sodium deoxycholate at 36°C for 30 min, followed by adding 0.1 volume of 1% Triton X-100 in 50 mm Tris-Cl, pH 9.0, and the preparation was centrifuged for 10 min at 100,000 × g (Luo et al., 1997). The supernatant was then used for immunoprecipitation.
Deletion mutagenesis of GluR1. Deletion mutants of GluR1 were generated by PCR-based site-directed mutagenesis (Quick Change; Stratagene, La Jolla, CA) according to the instructions of the manufacturer. Throughout this paper, unless otherwise specified, R1*875, R1*823, R1*812, and R1*807 were C-terminal deletions from amino acids 875, 823, 812, and 807, respectively as numbered by Hollmann et al. (1989), in which the first amino acid starts after the signal peptide sequence. Deletion constructs were confirmed by sequencing, and the expression of the mutants was verified by Western blot.
Coimmunoprecipitation and immunoblotting.Immunoprecipitation was performed as previously described (Lau et al., 1996; Luo et al., 1997) with modifications. For each reaction, ∼250 μg of solubilized HEK 293T cell lysate or solubilized rat brain P2 preparation was first incubated with 25 μl of 1:1 slurry of protein A/Sepharose CL-4B (Amersham Pharmacia Biotech, Arlington Heights, IL; prepared in PBS, pH 7.4) at 4°C for 1 hr to clarify any nonspecific binding to protein A from the lysate. At the same time, 5–10 μg of affinity-purified polyclonal antibodies, 1 μl of unpurified rabbit serum containing specific polyclonal antibodies, or 1 μl of mouse ascites containing specific monoclonal antibodies was preincubated with 25 μl of 1:1 slurry of protein A/Sepharose for 1 hr, and the protein A-antibody complex was spun down at 3000 rpm for 2 min. The clarified supernatants of the lysates were then added to the antibody-bound protein A beads, and the mixture was incubated for 2 hr at 4°C. After immunoprecipitation, the complex was spun down and washed once with 1% Triton X-100 in immunoprecipitation buffer (IPB), once with 1% Triton X-100 in IPB containing additional 0.5 m NaCl, and finally once with IPB. The proteins were eluted by Laemmli sample buffer (Laemmli, 1970) and were separated by SDS-PAGE. The gels were transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA), and the membranes were immunoblotted as described previously (Lau et al., 1996). Control coimmunoprecipitation reactions were performed by using preimmune serum or by preabsorption of the antibodies with their immunogenic peptide at a concentration of 50 μg/ml. The antibodies used in the experiments have been previously described: anti-GluR1 C-terminal antibody (Blackstone et al., 1992b), anti-GluR1 N-terminal antibody (Mammen et al., 1997a), and anti-4.1N antibody (Walensky et al., 1999). All coimmunoprecipitations were performed at least three times with similar results.
Cell surface biotinylation. COS cells were used in cell surface biotinylation analyses. Twenty-four hours after transfection, cells were biotinylated with 1 mg/ml sulfo-NHS-S-S-biotin (Pierce, Rockford, IL) for 20 min at 4°C, using previously described methods with minor modifications (Mammen et al., 1997b). To precipitate biotinylated proteins, supernatants of cell lysate were mixed with UltraLink immobilized neutravidin beads (Pierce) and rotated for 2 hr at 4°C. The beads were washed five times with IPB and then eluted with SDS-PAGE sample buffer supplemented with 50 mm DTT for 1 hr at 37°C. Protein biotinylation was efficiently reversed by DTT in this procedure. Both total and biotinylated proteins were resolved by SDS-PAGE, transferred to PVDF membranes, and probed with an anti-GluR1 N-terminal antibody. Alkaline phosphatase-conjugated anti-rabbit IgG (Pierce) was used as secondary antibody to visualize the specific signals by chemifluorescence (ECF) substrate (Amersham Pharmacia Biotech). The membranes were dried between filter papers and scanned in Storm 860 (Molecular Dynamics, Sunnyvale, CA) at 700–900 V. The scanned digital images were quantitated using ImageQuant software (Molecular Dynamics). The amount of GluR1 surface expression was analyzed by determining the relative ratio of biotinylated GluR1 to total GluR1. By checking the ratio of the control samples loaded at different concentrations, we found that the ECF method gave a better linear signal than conventional chemiluminescence (ECL) detection.
RESULTS
Identification of interaction between GluR1 and protein 4.1G using the yeast two-hybrid system
To identify proteins interacting with the cytosolic tail of GluR1, the last 82 amino acids of GluR1 were used as bait in a yeast two-hybrid screen against a randomly primed rat brain hippocampal cDNA library. Among 100 million independent colonies screened, only 9 positive clones were isolated, which were all homologous to the erythrocyte cytoskeletal protein 4.1R. The amino acid sequence of four of the clones were identical to the CTD of protein 4.1G, a novel homolog of erythrocyte protein 4.1 (Walensky et al., 1998). The other five positive clones were also homologous to the CTD of protein 4.1, but no identical match with any cloned member of the protein 4.1 family of proteins was found. On the basis of sequence homology, these positives could be splice variants or homologs of protein 4.1G.
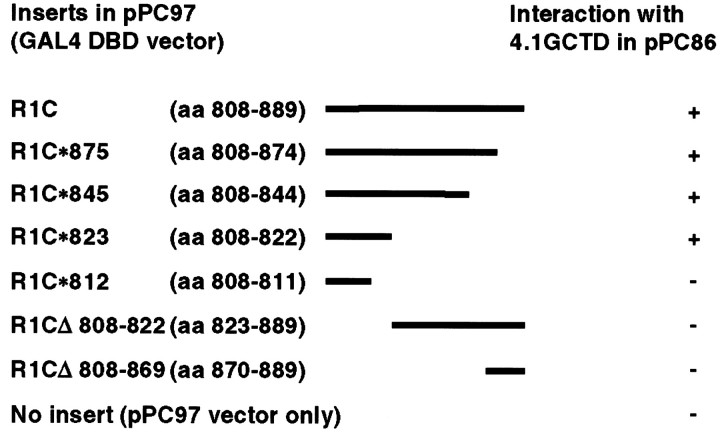
To map the interaction site on GluR1, we used the yeast two-hybrid interaction assay and serial deletions of GluR1 C terminus. As shown in Figure 1, deleting the PDZ domain ligand contained in the last 10 amino acids of GluR1 did not abolish its interaction with protein 4.1G. Deletion constructs of GluR1 up to amino acid 823 of GluR1 still interacted with protein 4.1G, whereas deletion after amino acid 812 abolished the interaction (Fig. 1). Control experiments using a vector that only expresses the GAL4 DNA binding domain did not interact (Fig. 1). These results suggest that the membrane proximal region between amino acids 812 and 823 in the C terminus of GluR1 is important for the interaction with protein 4.1G.
Fig. 1.
Yeast two-hybrid analysis of GluR1 and protein 4.1G interaction. The cytosolic tail of GluR1 and its various deletions were subcloned into yeast vector pPC97 containing the GAL4 DNA binding domain. The CTD of protein 4.1G was subcloned into pPC86 containing the GAL4 activation domain and co-transformed with GluR1 constructs into yeast. Double transformants were selected and scored for growth on plates lacking leucine, tryptophan, adenine, and histidine and for lacZ activity. Those that tested positive for interaction are designated +; those that tested negative are designated −.
Association of GluR1 with protein 4.1N and 4.1G in transfected HEK 293T cells
To confirm that GluR1 interacts with 4.1G, we examined whether they formed a complex in transfected HEK 293T cells. HEK 293T cells were transiently transfected with a construct encoding the full-length GluR1 with and without a construct encoding the CTD of protein 4.1G (4.1GCTD). After 2 d the cells were harvested and solubilized with 1% Triton X-100, and the cell lysates were subjected to immunoprecipitation using an anti-GluR1 antibody. As shown in Figure2A, immunoprecipitation of protein 4.1GCTD from co-transfected 293T cell lysates resulted in the specific co-immunoprecipitation of GluR1 (lane 3). The recent isolation of protein 4.1N (Walensky et al., 1999), a novel neuronal homolog of protein 4.1 highly related to 4.1G, and identification of its association with the postsynaptic density and localization at dendritic spines (Walensky et al., 1999) raise the possibility that GluR1 may also interact with 4.1N. Co-immunoprecipitation experiments from HEK293 cells transfected with GluR1 and 4.1N showed that, similar to 4.1G, 4.1N associates with GluR1 in this system (Fig. 2B).
Fig. 2.
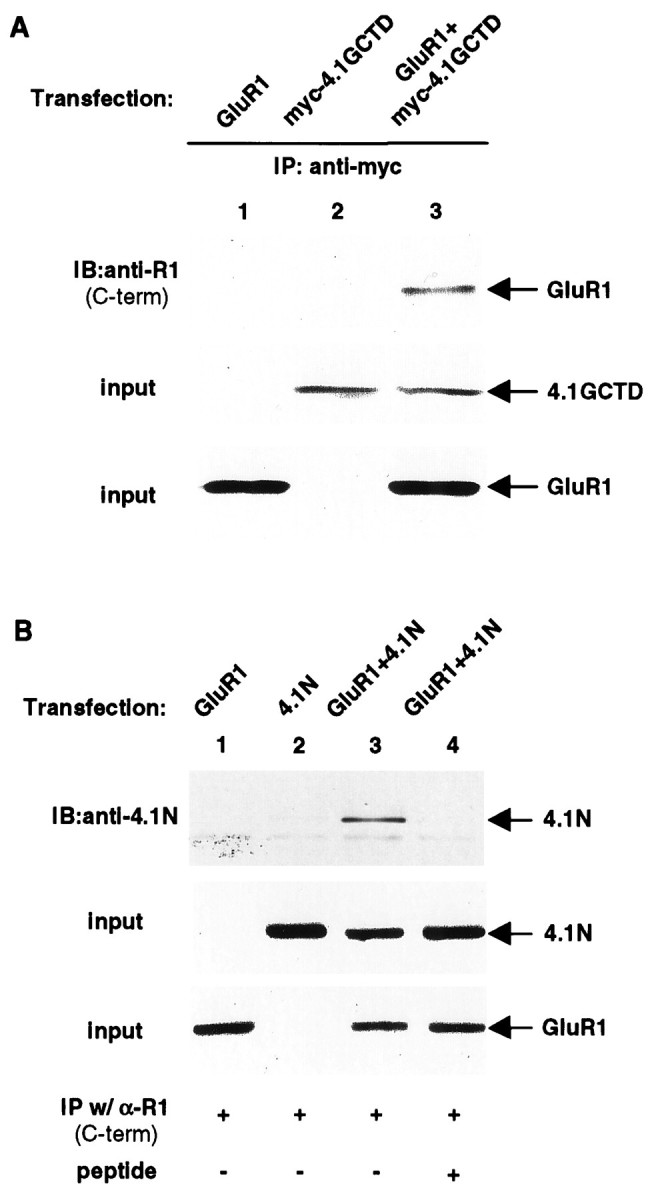
Association of GluR1 with 4.1G and 4.1N in HEK 293T cells. A. Coimmunoprecipitation of 4.1GCTD with GluR1 from 293T cells. Full-length GluR1 and myc-tagged 4.1GCTD were transfected into 293T cells either individually (lanes 1, 2) or together (lane 3). The transfected cells were solubilized with 1% Triton X-100, and the solubilized cell lysates were immunoprecipitated with anti-myc antibody. The immunoprecipitates were resolved on SDS-PAGE followed by immunoblotting with anti-GluR1 C-terminal antibody (top panel). The presence of myc-tagged 4.1GCTD (middle panel) and GluR1 (bottom panel) in the input for immunoprecipitation was confirmed by immunoblotting with anti-myc and anti-GluR1 C-terminal antibodies, respectively. IP, Immunoprecipitation; IB, immunoblot; same for other legends. B, Coimmunoprecipitation of 4.1N with GluR1 from 293T cells. Full-length GluR1 and full-length 4.1N were transfected into 293T cells either individually (lanes 1, 2) or combined (lanes 3, 4). The immunoprecipitation from cell lysates is the same as inA, except here the anti-GluR1 C-terminal antibody is used. The immunoprecipitates were resolved on SDS-PAGE and probed with anti-4.1N antibody (top panel). As a control experiment in lane 4, the anti-GluR1 C-terminal antibody was preincubated with its specific antigen before immunoprecipitation (see Materials and Methods). The presence of 4.1N (middle panel) and GluR1 (bottom panel) in the input for immunoprecipitation was confirmed by Western blot using anti-4.1N and anti-GluR1 antibodies, respectively.
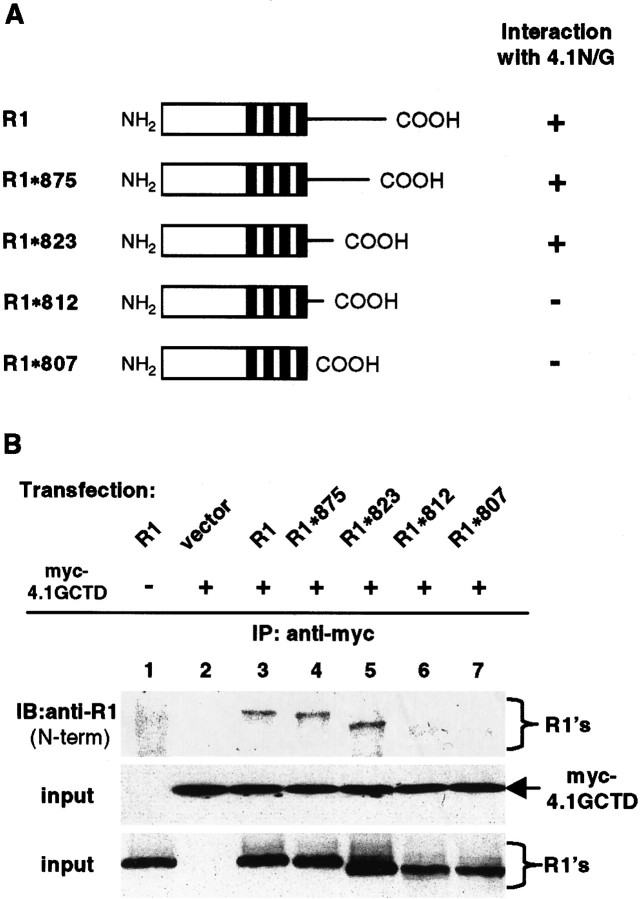
To confirm the yeast two-hybrid data that indicated that the membrane proximal region between amino acids 812 and 823 in the GluR1 C terminus is important for interaction, we generated C-terminal deletion mutants of GluR1 and tested their ability to associate with protein 4.1G/4.1N in transfected HEK 293T cells. Deletion of the C terminus of GluR1 after amino acid 823 (R1*823) did not disrupt its coimmunoprecipitation with 4.1G (Fig. 3B) or 4.1N (data not shown), whereas deletion of GluR1 at amino acid 812 (R1*812) or 807 (R1*807) eliminated the co-immunoprecipitation with both 4.1G (Fig. 3B) and 4.1N (data not shown). These results agree with the yeast two-hybrid analysis and provide additional evidence that the membrane proximal region from amino acid 812 to 823 is important for the binding of GluR1 to 4.1G and N.
Fig. 3.
Requirement of a membrane proximal region for association of GluR1 with proteins 4.1G and 4.1N. A, Schematic diagram of GluR1 deletion constructs used inB. B, The membrane proximal region in the GluR1 C terminus is required for binding to protein 4.1G. Full-length and C-terminal deletions of GluR1 were transfected with 4.1GCTD, and then solubilized cell lysates were immunoprecipitated with anti-myc antibody. The immunoprecipitates were subjected to SDS-PAGE and Western blot with anti-GluR1 N-terminal antibody (top panel). The presence of myc-tagged 4.1GCTD (middle panel) and GluR1 and its deletions (bottom panel) in the input for immunoprecipitation was confirmed by Western blot using anti-myc and anti-GluR1 N-terminal antibodies, respectively.
The CTDs of protein 4.1G and 4.1N contain a highly conserved C-terminal region and a variable N-terminal region (Fig.4B). To test whether this conserved region is responsible for the interaction with GluR1, we generated constructs containing the variable and conserved regions of 4.1NCTD (Fig. 4B, 4.1NCTDv, 4.1NCTDc) and tested their interaction with GluR1 in 293T cells. As shown in Figure4C, the conserved but not the variable region of 4.1NCTD interacted with GluR1 (lanes 4, 2, respectively). Similar results were obtained for 4.1GCTD (data not shown).
Protein 4.1N associates with AMPA receptors in vivoand colocalizes with AMPA receptors in excitatory synapses
To see whether AMPA receptors can associate with 4.1N in vivo, we performed coimmunoprecipitation experiments from brain lysates. Briefly, rat brain membrane preparations were solubilized with 1% deoxycholate, and then the AMPA receptors were immunoprecipitated with an antibody against the N terminus of the GluR2 subunit. GluR1 antibodies were not used in this experiment, because the anti-N-terminal GluR2 antibody gives more efficient immunoprecipitation of AMPA receptors in these extracts. As shown in Figure5A, protein 4.1N specifically co-immunoprecipitated with the AMPA receptor complex. Preabsorption of the antibody with antigen before the immunoprecipitation abolished the coimmunoprecipitation (Fig. 5A, lane 3), and PSD-95, a protein known to associate with NMDA receptors, did not coimmunoprecipitate. We were unable to perform coimmunoprecipitation experiments to examine for the association of 4.1G and GluR1 from brain membrane preparations, because we do not have a specific anti-4.1G antibody.
Fig. 5.
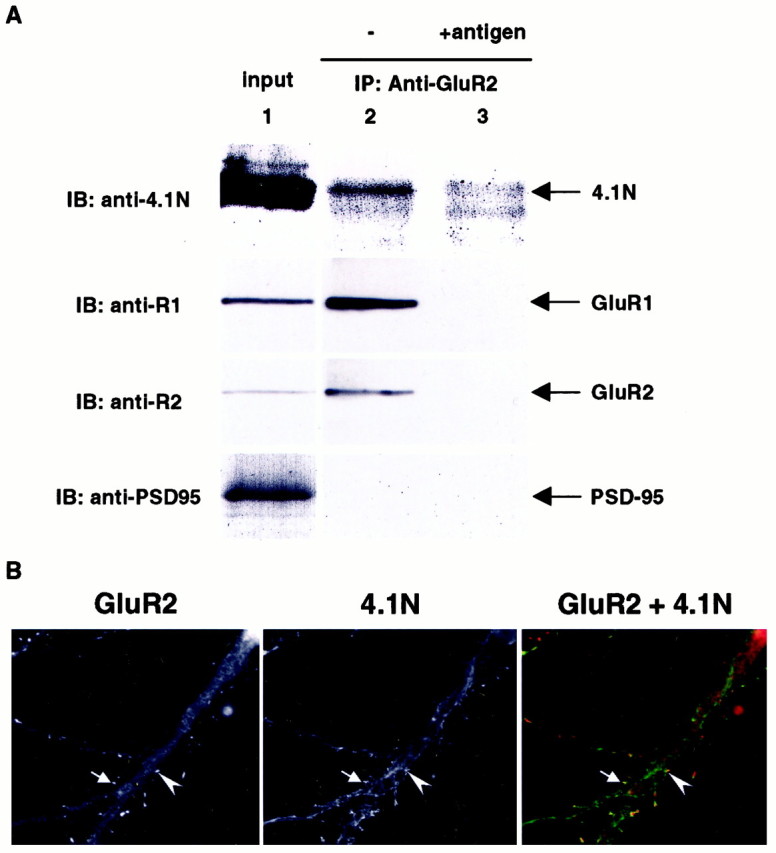
Protein 4.1N interacts with GluR1 in vivo and colocalizes with AMPA receptor complex in excitatory synapses. A, Rat brain membrane preparation (P2) was solubilized with 1% deoxycholate and immunoprecipitated with either anti-GluR2 N-terminal antibody (lane 2) or the same antibody with antigen preabsorption (lane 3). The immunoprecipitated complex and the input (lane 1) were resolved by SDS-PAGE and probed with anti-4.1N (top panel), anti-GluR1 C-terminal (upper middle panel), anti-GluR2 C-terminal (lower middle panel), and PSD-95 (bottom panel) antibodies. B, Primary hippocampal neurons were double-stained with monoclonal anti-GluR2 antibody (Chemicon, Temecula, CA) and polyclonal anti-4.1N antibody as primary antibodies, followed by rhodamine-conjugated anti-mouse IgG and FITC-conjugated anti-rabbit IgG as secondary antibodies. The staining was visualized and digitized using a fluorescent microscope (Zeiss, Thornwood, NY) with a digital camera controlled by the Metamorph program (Universal Imaging, West Chester, PA). Arrows and arrowheadsindicate the area of colocalization.
Previous studies have shown that 4.1N can be detected in PSD fractions and is localized at excitatory synapses (Walensky et al., 1999). We therefore investigated whether 4.1N colocalized with AMPA receptors in neurons. Figure 5B shows the staining of primary hippocampal neuronal cultures using an anti-GluR2 antibody to label AMPA receptors and an anti-4.1N antibody. The dendritic staining pattern of 4.1N is reminiscent of cytoskeleton structure and is enriched in a number of dendritic spines that colocalize with GluR2 (arrows).
Deletion of 4.1 binding region attenuates GluR1 plasma membrane expression in heterologous cells
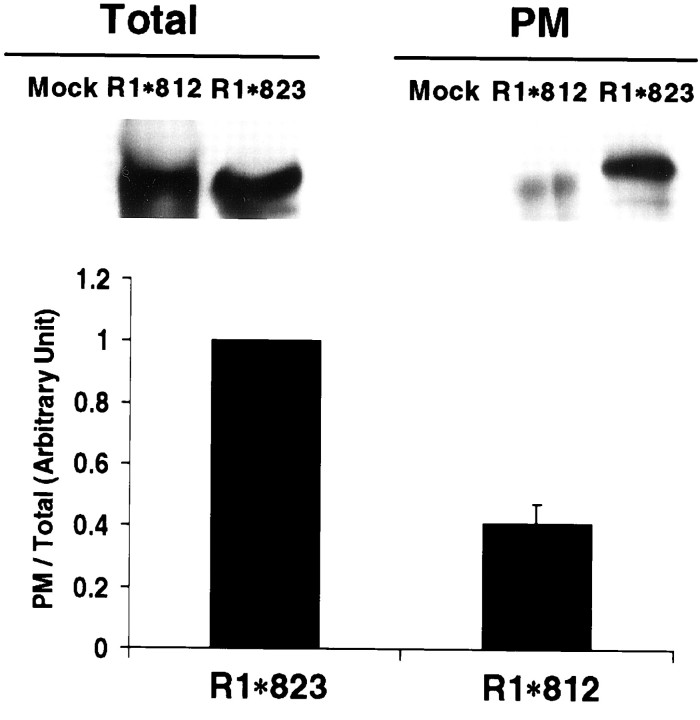
It has been well documented that the protein 4.1 family of proteins functions in membrane protein–cytoskeletal interactions (Bennett and Gilligan, 1993). Postsynaptic spines are enriched with F-actin, which directly contacts the PSD (Harris and Kater, 1994). Maintenance of synaptic transmembrane proteins often requires their cytoskeleton attachment. We therefore explored whether disruption of the interaction of GluR1 with the endogenous 4.1G in heterologous cells would affect the membrane trafficking of GluR1. GluR1 mutants with (R1*823) or without (R1*812) the membrane proximal region required for 4.1G/4.1N association were expressed in heterologous cells, and the plasma membrane expression of GluR1 was analyzed using cell surface biotinylation and quantitative immunoblotting techniques (Fig.6). GluR1-transfected cells were treated with a water-soluble, membrane-impermeable derivative of biotin to label proteins on the cell surface. Biotinylated molecules were then isolated by neutravidin beads, and a comparison of the steady-state levels of surface wild-type (WT) and mutant GluR1 subunits was analyzed by SDS-PAGE and quantitative ECF immunoblot analyses. The overall expression of the two mutant GluR1 subunits was similar, and both proteins were associated with intracellular and plasma membrane fractions, similar to WT GluR1 (data not shown). In contrast, the levels of surface expression of the two mutant receptors detected using biotinylation techniques were significantly different (Fig. 6). The GluR1 subunit capable of binding 4.1N and 4.1G (R1*823) was efficiently expressed on the surface, in sharp contrast to the GluR1 mutant (R1*812), which does not interact with 4.1. Deletion of the whole C-terminal domain (R1*807) rendered a similar result to R1*812 (data not shown). These results suggest that the interaction of GluR1 with endogenous 4.1G in COS cells may regulate the plasma membrane expression of GluR1.
Fig. 6.
Deletion of the GluR1 4.1 binding region reduces surface expression of GluR1. The membrane proximal region in the GluR1 C terminus is important for receptor surface expression. COS cells were transfected with pRK5 (Mock), GluR1*812 (R1*812), or GluR1*823 (R1*823). Surface expression of GluR1 was determined by biotinylation, as described in Materials and Methods. Samples from total cell lysates (Total) and biotinylated samples (PM) were analyzed for GluR1. For quantitation, the signals from biotinylated samples were divided by the total GluR1 signals to obtain the ratio of surface expression for GluR1*823 (R1*823) and GluR1*812 (R1*812). The ratio of surface expression of GluR1*812 was normalized to that of GluR1*823 to obtain the relative value of surface expression. n = 3. The error bar indicates SEM.
Actin filaments are required for maintenance of surface GluR1 in heterologous cells
Previous studies have suggested that the cortical cytoskeleton is involved in synaptic targeting of AMPA receptors (Allison et al., 1998;Kim and Lisman, 1999). To determine whether the actin cytoskeleton, possibly through a 4.1 association, regulated the surface expression of GluR1, we analyzed the effect of cytoskeletal disrupting agent latrunculin A (an actin polymerization inhibitor) on the plasma membrane distribution of wild-type and mutant GluR1 subunits (Fig.7). COS cells transfected with wild-type GluR1 or the two GluR1 mutants with and without the 4.1 binding region (R1*823 and R1*812) were treated with latrunculin A for 2 hr. Interestingly, latrunculin A drastically reduced surface expression of WT GluR1 (Fig. 7A) and the GluR1 mutant (R1*823) that is capable of 4.1 binding (Fig. 7B) but had no effect on the GluR1 mutant (R1*812) that is unable to bind 4.1 (Fig. 7B). The total amount of GluR1 was not affected by latrunculin A treatment. These results indicate that the 4.1 binding region is important for the latrunculin inhibition of GluR1 surface expression and suggest that 4.1 is important for the attachment of GluR1 with the actin cytoskeleton.
Fig. 7.
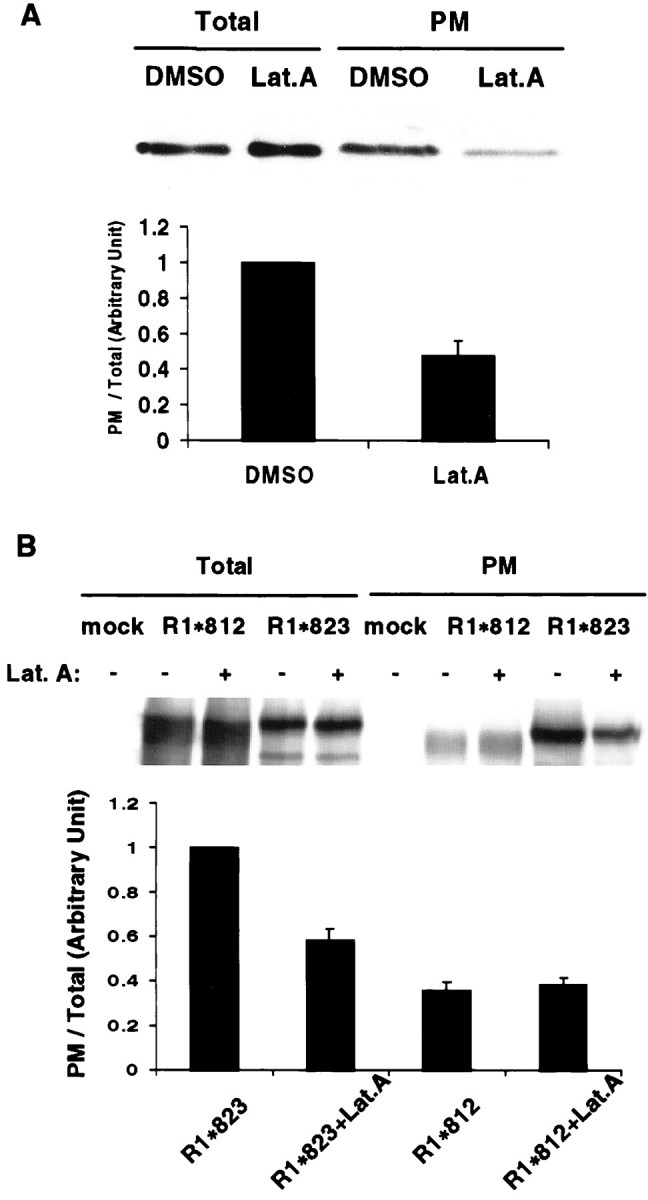
F-actin regulates GluR1 surface expression in COS cells. A, Disruption of F-actin inhibits surface expression of GluR1 in COS cells. COS cells were transiently transected with WT GluR1. Twenty-four hours after the transfection, either DMSO or a concentrated DMSO stock of latrunculin A (Lat.A) was added to the culture medium. Final concentration of latrunculin A is 5 μm. Cells were further incubated at 37°C for 2 hr before cell surface biotinylation. Samples from cell lysates (Total) and biotinylated samples (PM) were analyzed for GluR1 using an anti-GluR1 N-terminal antibody. For quantitation, the biotinylated samples were normalized to total GluR1 signals to obtain the ratio of surface expression for the control and latrunculin-treated cells. The ratio of surface expression of the latrunculin-treated cells was normalized to that of control cells to express the relative surface expression. n = 3. The error bar indicates SEM.B. The C-terminal membrane proximal region of GluR1 is important for latrunculin inhibition of surface expression. COS cells transfected with pRK5 (Mock), R1*812, or R1*823 were treated with either DMSO (Lat.A −) or latrunculin A (Lat.A +), as described in Materials and Methods. For quantitation, the signals from biotinylated samples were normalized to the total GluR1 signal to obtain the ratio of surface expression. The ratio of the surface expression was then normalized to that of R1*823 without latrunculin. R1*823,R1*823+Lat.A, R1*823 without or with latrunculin A, respectively;R1*812,R1*812+Lat.A, R1*812 without or with latrunculin A, respectively. n = 3. Error bars indicate SEM.
GluR1 surface expression is attenuated by overexpression of the CTDs of 4.1
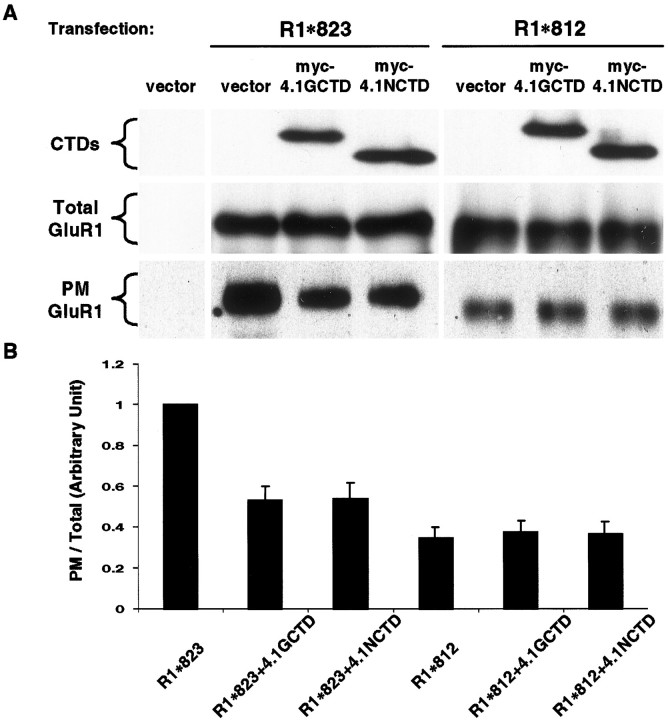
The CTDs of proteins 4.1N and 4.1G were used to further explore the specific effect of 4.1 homologs on F-actin-dependent surface anchoring of GluR1. Because the CTDs are sufficient for association with GluR1 but do not contain the domain for spectrin and actin binding (Fig. 4A), we postulated that they could serve as “dominant negative” constructs to examine the role of 4.1 in GluR1 surface expression. CTDs or the vector were cotransfected with different GluR1 constructs in COS cells, and the steady-state levels of surface GluR1 were analyzed by biotinylation techniques. Overexpression of either 4.1N or 4.1G CTD specifically attenuated the surface expression of the R1*823 mutant containing the C-terminal 4.1 binding region but not the R1*812 mutant lacking this region (Fig.8). These results indicate that the CTDs may disrupt the interaction of GluR1 with endogenous 4.1 and support the idea that F-actin and 4.1N association affects GluR1 surface expression. Combined with previous actin disruption studies, our data suggest that 4.1N and 4.1G serve as a link between GluR1 and cortical cytoskeleton that is required for GluR1 plasma membrane stability.
Fig. 8.
Overexpression of CTDs of 4.1 attenuated GluR1 surface expression. COS cells were cotransfected using R1*812 or R1*823, together with pRK5 (vector), 4.1GCTD (myc-4.1GCTD), or 4.1NCTD (myc-4.1NCTD). Both CTDs were myc-tagged. Transfected cells were incubated at 37°C for 24 hr, followed by cell surface biotinylation, as described in Materials and Methods. A, Immunoblots. Top panel, Total cell lysates blotted with anti-myc antibody;middle panel, total cell lysates blotted with anti-GluR1 N-terminal antibody; bottom panel, biotinylated samples blotted with anti-GluR1 N-terminal antibody. B, Quantitation of surface expression. The signals from biotinylated samples were normalized to the total GluR1 signal to obtain the ratio of surface expression and then normalized to the surface expression of GluR1*823 to obtain the relative value of surface expression.n = 3. Error bars indicate SEM.
Actin filaments are required for maintenance of surface GluR1 in cortical neurons
Latrunculin A was also used to investigate the importance of the actin cytoskeleton in the surface expression of AMPA receptors in neurons (Fig. 9A). Rat cortical neurons were cultured in vitro for 3 weeks and treated with 5 μm latrunculin A for 2 hr (Allison et al., 1998). The biotinylation assay was then used to quantify surface expression of GluR1. As seen in the COS cells, latrunculin A inhibited the surface expression of GluR1 in neurons. These results suggest that surface expression of GluR1 in neurons may also be regulated by a 4.1-mediated interaction with the actin cytoskeleton.
Fig. 9.
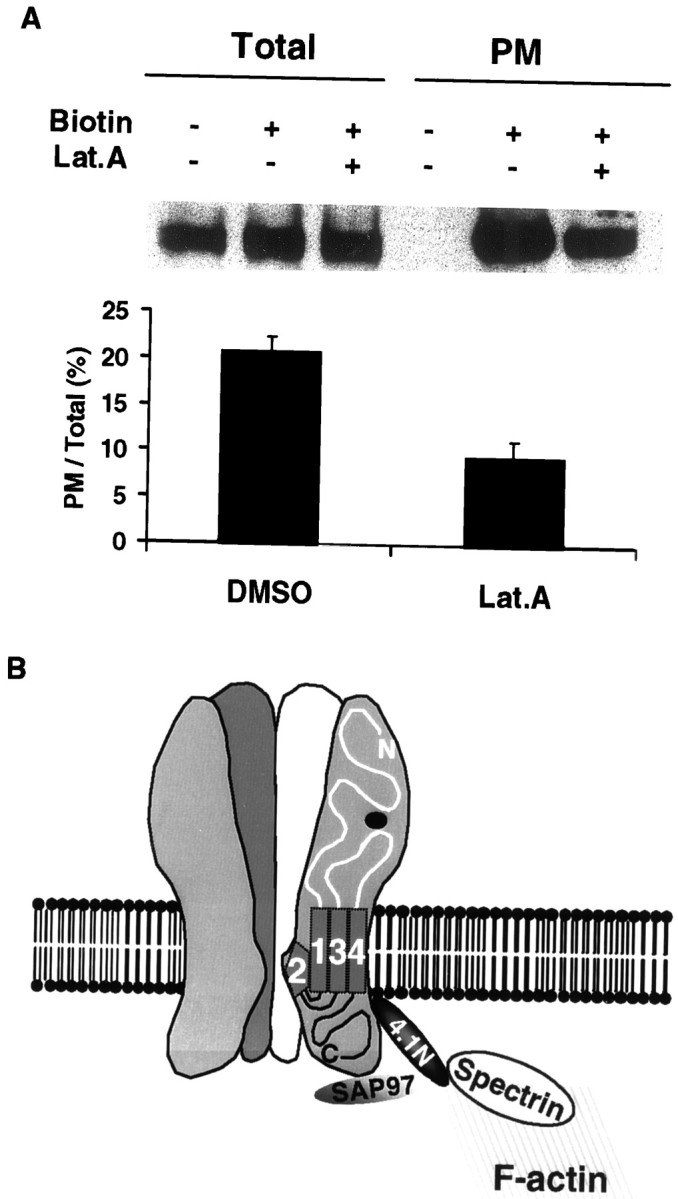
Actin cytoskeleton is required for synaptic maintenance of AMPA receptors. A, Actin polymerization is required for synaptic maintenance of AMPA receptors. Cultured rat hippocampal neurons were treated with either DMSO (Lat.A−) or latrunculin A (Lat.A +) followed by cell surface biotinylation (Biotin +, −) as described in Materials and Methods. Samples from cell lysates (Total) and biotinylated samples (PM) were analyzed for GluR1 via anti-GluR1 antibody. Samples of immunoblots (top panel) and a summary of quantitation (bottom panel) are presented. For quantitation, the signals from biotinylated samples were normalized to total GluR1 signals to obtain the ratio of surface expression [PM/Total (%)].DMSO, Lat.A, GluR1 without or with latrunculin A, respectively. n = 4. Error bars indicate SEM.B, Schematic model of AMPA receptor–actin cytoskeleton cross-linking by 4.1N. The interaction of the GluR1 subunit with protein 4.1N and SAP97 may link surface AMPA receptors to the cortical actin cytoskeleton network underneath the synaptic plasma membrane and PSD.
DISCUSSION
Excitatory synapses on dendritic spines are enriched in actin filaments, which are oriented longitudinally in the neck and form a lattice in the spine head (Fifkova and Delay, 1982; Matus et al., 1982;Cohen et al., 1985; Harris and Kater, 1994). Previous studies have shown that the actin network is important for the synaptic localization and immobilization of glutamate receptors (Allison et al., 1998). In this study we have shown that the AMPA receptor subunit GluR1 interacts with the actin-associated proteins 4.1N and 4.1G. Proteins 4.1N and 4.1G bind to GluR1 in vitro, using both the yeast two-hybrid system and a mammalian heterologous expression system. In addition, we show that 4.1N colocalizes with AMPA receptors in excitatory synapses in hippocampal neurons in culture and is associated with GluR1 in vivo in rat brain. Deletion analysis demonstrated that a stretch of 11 amino acids in the membrane proximal region in the GluR1 C terminus is required for 4.1N binding, whereas a consensus region within the CTDs of 4.1G and 4.1N is sufficient to mediate the interaction. In exploring the functional significance of this interaction, we found that GluR1 truncation mutants that lack the 4.1 binding region have reduced surface expression in heterologous cells. Moreover, overexpression of the CTDs of 4.1N and 4.1G, which should disrupt the interaction of GluR1 with endogenous 4.1G, attenuated GluR1 surface expression. Disruption of the F-actin network with latrunculin A also resulted in decreased plasma membrane GluR1 both in heterologous cells and in cultured neurons. These results suggest that protein 4.1N may play a functional role in the anchoring of AMPA receptors to the actin cytoskeleton and stabilizing the surface expression of the receptors. Recent studies have shown that the membrane proximal 39 amino acids of the GluR1 C terminus appear to be important for its sorting to dendrites (Ruberti and Dotti, 2000), suggesting that binding of protein 4.1 may be involved in this sorting process.
Recent studies have provided evidence that a network of PDZ domain-containing proteins may also play an important role in the cellular targeting of glutamate receptors in the CNS (Sheng and Kim, 1996; O'Brien et al., 1998a; Kim and Huganir, 1999). The PDZ domain-containing proteins PSD-95, SAP102, and SAP93 directly interact with NMDA receptor subunits, whereas the PDZ domain-containing proteins GRIP/ABP, PICK1, and SAP97 directly bind to AMPA receptor subunits. PDZ domain-containing proteins in non-neuronal cells have also been shown to be important as organizers of the cortical cytoskeletal network (Fanning and Anderson, 1996). For example, erythrocyte protein 4.1 interacts with the membrane protein p55, a palmitoylated erythrocyte membrane protein with a single PDZ domain and glycophorin C, and these interactions may serve to couple glycophorin C to the actin cytoskeleton (Marfatia et al., 1994; Marfatia et al., 1995). In addition, human CASK, a homolog of the Caenorhabditis elegans PDZ-containing protein LIN-2, has been shown to bind protein 4.1 (Cohen et al., 1998). Moreover, recently it has been reported that the PDZ domain-containing protein SAP97/hDlg, a member of the PSD-95 family, binds to 4.1 in epithelial cells (Lue et al., 1994). Interestingly, SAP97 is also localized to excitatory synapses and has recently been shown to specifically bind to the C-terminal PDZ ligand of GluR1. These results suggest that SAP97 and 4.1 may form a trimeric complex with GluR1 and cooperate in the regulation of the synaptic localization and immobilization of AMPA receptors (Fig.9B).
Various recent studies have indicated that the level of functional surface synaptic AMPA receptors can be modulated by activity-dependent mechanisms (Isaac et al., 1995; Liao et al., 1995, 1999; Rao and Craig, 1997; Lissin et al., 1998, 1999; O'Brien et al., 1998b; Turrigiano et al., 1998; Carroll et al., 1999; Shi et al., 1999). Both electrophysiological and morphological studies have indicated that chronic and rapid changes in synaptic activity can regulate receptor surface expression providing homeostatic and acute mechanisms for controlling synaptic efficiency. Chronic increases or decreases in synaptic activity decrease or increase, respectively, the levels of AMPA receptor surface expression (Rao and Craig, 1997; O'Brien et al., 1998b; Turrigiano et al., 1998). Moreover, patterns of synaptic firing that can result in rapid changes in synaptic transmission, such as those that induce long-term potentiation and long-term depression, can result in rapid changes in the distribution of AMPA receptors (Isaac et al., 1995; Liao et al., 1995, 1999; Rao and Craig, 1997; Lissin et al., 1998, 1999; O'Brien et al., 1998b; Turrigiano et al., 1998; Carroll et al., 1999; Shi et al., 1999). It is possible that the association of GluR1 with the membrane cytoskeleton via 4.1N may be regulated and play a role in these processes. The C-terminal domain of GluR1 contains the major sites for AMPA receptor phosphorylation (Roche et al., 1996;Mammen et al., 1997b, 1999), and phosphorylation of this region may regulate 4.1 binding. Moreover, several studies in erythrocytes have shown that phosphorylation of protein 4.1 regulates its interaction with the actin cytoskeleton and membrane proteins (Pinder et al., 1995). Future investigation of the dynamic regulation of AMPA receptor targeting will facilitate our understanding of the potential role of protein 4.1 in synaptic plasticity.
Footnotes
This work was supported by the Howard Hughes Medical Institute and National Institutes of Health. We thank Jeff Bernhardt for the making of GluR1 mutants, Xiaoqun Zhang for providing rat primary hippocampal cultures, and Carol Doherty for making and purifying anti-GluR1 antibodies.
L.S. and F.L. contributed equally to this work.
Correspondence should be addressed to Dr. Richard L. Huganir, Howard Hughes Medical Institute, Department of Neuroscience, The Johns Hopkins University School of Medicine, 725 North Wolfe Street, PCTB 904, Baltimore, MD 21205. E-mail: rhuganir@jhmi.edu.
Dr. Walensky's present address: Department of Neurology, Children's Hospital, 300 Longwood Street, Boston, MA 02115.
REFERENCES
- 1.Allison DW, Gelfand VI, Spector I, Craig AM. Role of actin in anchoring postsynaptic receptors in cultured hippocampal neurons: differential attachment of NMDA versus AMPA receptors. J Neurosci. 1998;18:2423–2436. doi: 10.1523/JNEUROSCI.18-07-02423.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson RA, Lovrien RE. Glycophorin is linked by band 4.1 protein to the human erythrocyte membrane skeleton. Nature. 1984;307:655–658. doi: 10.1038/307655a0. [DOI] [PubMed] [Google Scholar]
- 3.Bennett V, Gilligan DM. The spectrin-based membrane skeleton and micron-scale organization of the plasma membrane. Annu Rev Cell Biol. 1993;9:27–66. doi: 10.1146/annurev.cb.09.110193.000331. [DOI] [PubMed] [Google Scholar]
- 4.Blackstone CD, Levey AI, Martin LJ, Price DL, Huganir RL. Immunological detection of glutamate receptor subtypes in human central nervous system. Ann Neurol. 1992a;31:680–683. doi: 10.1002/ana.410310620. [DOI] [PubMed] [Google Scholar]
- 5.Blackstone CD, Moss SJ, Martin LJ, Levey AI, Price DL, Huganir RL. Biochemical characterization and localization of a non-N-methyl-D-aspartate glutamate receptor in rat brain. J Neurochem. 1992b;58:1118–1126. doi: 10.1111/j.1471-4159.1992.tb09370.x. [DOI] [PubMed] [Google Scholar]
- 6.Carroll RC, Lissin DV, von Zastrow M, Nicoll RA, Malenka RC. Rapid redistribution of glutamate receptors contributes to long-term depression in hippocampal cultures. Nat Neurosci. 1999;2:454–460. doi: 10.1038/8123. [DOI] [PubMed] [Google Scholar]
- 7.Chevray PM, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen AR, Woods DF, Marfatia SM, Walther Z, Chishti AH, Anderson JM, Wood DF. Human CASK/LIN-2 binds syndecan-2 and protein 4.1 and localizes to the basolateral membrane of epithelial cells. J Cell Biol. 1998;142:129–138. doi: 10.1083/jcb.142.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen RS, Chung SK, Pfaff DW. Immunocytochemical localization of actin in dendritic spines of the cerebral cortex using colloidal gold as a probe. Cell Mol Neurobiol. 1985;5:271–284. doi: 10.1007/BF00711012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong H, O'Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature. 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 11.Ehlers MD, Fung ET, O'Brien RJ, Huganir RL. Splice variant-specific interaction of the NMDA receptor subunit NR1 with neuronal intermediate filaments. J Neurosci. 1998;18:720–730. doi: 10.1523/JNEUROSCI.18-02-00720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fanning AS, Anderson JM. Protein-protein interactions: PDZ domain networks. Curr Biol. 1996;6:1385–1388. doi: 10.1016/s0960-9822(96)00737-3. [DOI] [PubMed] [Google Scholar]
- 13.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 14.Fifkova E, Delay RJ. Cytoplasmic actin in neuronal processes as a possible mediator of synaptic plasticity. J Cell Biol. 1982;95:345–350. doi: 10.1083/jcb.95.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh A, Greenberg ME. Distinct roles for bFGF and NT-3 in the regulation of cortical neurogenesis. Neuron. 1995;15:89–103. doi: 10.1016/0896-6273(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 16.Goslin K, Banker G. Rat hippocampal neurons in low-density culture. In: Banker G, Goslin K, editors. Culturing nerve cells. MIT; Cambridge, MA: 1991. pp. 251–281. [Google Scholar]
- 17.Harris KM, Kater SB. Dendritic spines: cellular specializations imparting both stability and flexibility to synaptic function. Annu Rev Neurosci. 1994;17:341–371. doi: 10.1146/annurev.ne.17.030194.002013. [DOI] [PubMed] [Google Scholar]
- 18.Hemming NJ, Anstee DJ, Mawby WJ, Reid ME, Tanner MJ. Localization of the protein 4.1-binding site on human erythrocyte glycophorins C and D. Biochem J. 1994;299:191–196. doi: 10.1042/bj2990191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 20.Hollmann M, O'Shea-Greenfield A, Rogers SW, Heinemann S. Cloning by functional expression of a member of the glutamate receptor family. Nature. 1989;342:643–648. doi: 10.1038/342643a0. [DOI] [PubMed] [Google Scholar]
- 21.Isaac JT, Nicoll RA, Malenka RC. Evidence for silent synapses: implications for the expression of LTP. Neuron. 1995;15:427–434. doi: 10.1016/0896-6273(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 22.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two- hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim CH, Lisman JE. A role of actin filament in synaptic transmission and long-term potentiation. J Neurosci. 1999;19:4314–4324. doi: 10.1523/JNEUROSCI.19-11-04314.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JH, Huganir RL. Organization and regulation of proteins at synapses. Curr Opin Cell Biol. 1999;11:248–254. doi: 10.1016/s0955-0674(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 25.Kornau HC, Seeburg PH, Kennedy MB. Interaction of ion channels and receptors with PDZ domain proteins. Curr Opin Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- 26.Krucker T, Siggins GR, Halpain S. Dynamic actin filaments are required for stable long-term potentiation (LTP) in area CA1 of the hippocampus. Proc Natl Acad Sci USA. 2000;97:6856–6861. doi: 10.1073/pnas.100139797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Lau LF, Mammen A, Ehlers MD, Kindler S, Chung WJ, Garner CC, Huganir RL. Interaction of the N-methyl-d-aspartate receptor complex with a novel synapse-associated protein, SAP102. J Biol Chem. 1996;271:21622–21628. doi: 10.1074/jbc.271.35.21622. [DOI] [PubMed] [Google Scholar]
- 29.Leonard AS, Davare MA, Horne MC, Garner CC, Hell JW. SAP97 is associated with the alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR1 subunit. J Biol Chem. 1998;273:19518–19524. doi: 10.1074/jbc.273.31.19518. [DOI] [PubMed] [Google Scholar]
- 30.Liao D, Hessler NA, Malinow R. Activation of postsynaptically silent synapses during pairing-induced LTP in CA1 region of hippocampal slice. Nature. 1995;375:400–404. doi: 10.1038/375400a0. [DOI] [PubMed] [Google Scholar]
- 31.Liao D, Zhang X, O'Brien R, Ehlers MD, Huganir RL. Regulation of morphological postsynaptic silent synapses in developing hippocampal neurons. Nat Neurosci. 1999;2:37–43. doi: 10.1038/4540. [DOI] [PubMed] [Google Scholar]
- 32.Lin JW, Wyszynski M, Madhavan R, Sealock R, Kim JU, Sheng M. Yotiao: A novel protein of neuromuscular junction and brain that interacts with specific splice variants of NMDA receptor subunit NR1. J Neurosci. 1998;18:2017–2027. doi: 10.1523/JNEUROSCI.18-06-02017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lissin DV, Gomperts SN, Carroll RC, Christine CW, Kalman D, Kitamura M, Hardy S, Nicoll RA, Malenka RC, von Zastrow M. Activity differentially regulates the surface expression of synaptic AMPA and NMDA glutamate receptors. Proc Natl Acad Sci USA. 1998;95:7097–7102. doi: 10.1073/pnas.95.12.7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lissin DV, Carroll RC, Nicoll RA, Malenka RC, von Zastrow M. Rapid, activation-induced redistribution of ionotropic glutamate receptors in cultured hippocampal neurons. J Neurosci. 1999;19:1263–1272. doi: 10.1523/JNEUROSCI.19-04-01263.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lue RA, Marfatia SM, Branton D, Chishti AH. Cloning and characterization of hdlg: the human homologue of the Drosophila discs large tumor suppressor binds to protein 4.1. Proc Natl Acad Sci USA. 1994;91:9818–9822. doi: 10.1073/pnas.91.21.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo J, Wang Y, Yasuda RP, Dunah AW, Wolfe BW. The majority of N-methyl-d-aspartate receptor complexes in adult rat cerebral cortex contain at least three different subunits (NR1/NR2A/NR2B). Mol Pharmacol. 1997;51:79–86. doi: 10.1124/mol.51.1.79. [DOI] [PubMed] [Google Scholar]
- 37.Mammen AL, Huganir RL, O'Brien RJ. Redistribution and stabilization of cell surface glutamate receptors during synapse formation. J Neurosci. 1997a;17:7351–7358. doi: 10.1523/JNEUROSCI.17-19-07351.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mammen AL, Kameyama K, Roche KW, Huganir RL. Phosphorylation of the alpha-amino-3-hydroxy-5-methylisoxazole 4-propionic acid receptor GluR1 subunit by calcium/calmodulin-dependent kinase II. J Biol Chem. 1997b;272:32528–32533. doi: 10.1074/jbc.272.51.32528. [DOI] [PubMed] [Google Scholar]
- 39.Mammen AL, Kamboj S, Huganir RL. Protein phosphorylation of ligand-gated ion channels. Methods Enzymol. 1999;294:353–370. doi: 10.1016/s0076-6879(99)94022-3. [DOI] [PubMed] [Google Scholar]
- 40.Marfatia SM, Lue RA, Branton D, Chishti AH. In vitro binding studies suggest a membrane-associated complex between erythroid p55, protein 4.1, and glycophorin C. J Biol Chem. 1994;269:8631–8634. [PubMed] [Google Scholar]
- 41.Marfatia SM, Leu RA, Branton D, Chishti AH. Identification of the protein 4.1 binding interface on glycophorin C and p55, a homologue of the Drosophila discs-large tumor suppressor protein. J Biol Chem. 1995;270:715–719. doi: 10.1074/jbc.270.2.715. [DOI] [PubMed] [Google Scholar]
- 42.Matus A, Ackermann M, Pehling G, Byers HR, Fujiwara K. High actin concentrations in brain dendritic spines and postsynaptic densities. Proc Natl Acad Sci USA. 1982;79:7590–7594. doi: 10.1073/pnas.79.23.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Brien RJ, Lau LF, Huganir RL. Molecular mechanisms of glutamate receptor clustering at excitatory synapses. Curr Opin Neurobiol. 1998a;8:364–369. doi: 10.1016/s0959-4388(98)80062-7. [DOI] [PubMed] [Google Scholar]
- 44.O'Brien RJ, Kamboj S, Ehlers MD, Rosen KR, Fischbach GD, Huganir RL. Activity-dependent modulation of synaptic AMPA receptor accumulation. Neuron. 1998b;21:1067–1078. doi: 10.1016/s0896-6273(00)80624-8. [DOI] [PubMed] [Google Scholar]
- 45.Pasternack GR, Anderson RA, Leto TL, Marchesi VT. Interactions between protein 4.1 and band 3. An alternative binding site for an element of the membrane skeleton. J Biol Chem. 1985;260:3676–3683. [PubMed] [Google Scholar]
- 46.Pinder JC, Gardner B, Gratzer WB. Interaction of protein 4.1 with the red cell membrane: effects of phosphorylation by protein kinase C. Biochem Biophys Res Commun. 1995;210:478–482. doi: 10.1006/bbrc.1995.1685. [DOI] [PubMed] [Google Scholar]
- 47.Rao A, Craig M. Activity regulates the synaptic localization of the NMDA receptor in hippocampal neurons. Neuron. 1997;19:801–812. doi: 10.1016/s0896-6273(00)80962-9. [DOI] [PubMed] [Google Scholar]
- 48.Reynolds A, Lundblad V. Yeast vectors and assay for expression of cloned genes. In: Aubusel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. Wiley; New York: 1989. pp. 13.6.1–13.6.4. [DOI] [PubMed] [Google Scholar]
- 49.Roche KW, O'Brien RJ, Mammen AL, Bernhardt J, Huganir RL. Characterization of multiple phosphorylation sites on the AMPA receptor GluR1 subunit. Neuron. 1996;16:1179–1188. doi: 10.1016/s0896-6273(00)80144-0. [DOI] [PubMed] [Google Scholar]
- 50. Ruberti F, Dotti CG. Involvement of the proximal C terminus of the AMPA receptor subunit GluR1 in dendritic sorting (in process citation). J Neurosci 20 2000. RC78 (1–5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheng M, Kim E. Ion channel associated proteins. Curr Opin Neurobiol. 1996;6:602–608. doi: 10.1016/s0959-4388(96)80091-2. [DOI] [PubMed] [Google Scholar]
- 52.Shi SH, Hayashi Y, Petralia RS, Zaman SH, Wenthold RJ, Svoboda K, Malinow R. Rapid spine delivery and redistribution of AMPA receptors after synaptic NMDA receptor activation. Science. 1999;284:1811–1816. doi: 10.1126/science.284.5421.1811. [DOI] [PubMed] [Google Scholar]
- 53.Srivastava S, Osten P, Vilim FS, Khatri L, Inman G, States B, Daly C, DeSouza S, Abagyan R, Valtschanoff JG, Weinberg RJ, Ziff EB. Novel anchorage of GluR2/3 to the postsynaptic density by the AMPA receptor-binding protein ABP. Neuron. 1998;21:581–591. doi: 10.1016/s0896-6273(00)80568-1. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka T, Kadowaki K, Lazarides E, Sobue K. Ca2(+)-dependent regulation of the spectrin/actin interaction by calmodulin and protein 4.1. J Biol Chem. 1991;266:1134–1140. [PubMed] [Google Scholar]
- 55.Tejedor FJ, Bokhari A, Rogero O, Gorczyca M, Zhang J, Kim E, Sheng M, Budnik V. Essential role for dlg in synaptic clustering of Shaker K+ channels in vivo. J Neurosci. 1997;17:152–159. doi: 10.1523/JNEUROSCI.17-01-00152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- 57.Tyler JM, Hargreaves WR, Branton D. Purification of two spectrin-binding proteins: biochemical and electron microscopic evidence for site-specific reassociation between spectrin and bands 2. 1 and 4 1. Proc Natl Acad Sci USA. 1979;76:5192–5196. doi: 10.1073/pnas.76.10.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walensky LD, Gascard P, Fields ME, Blackshaw S, Conboy JG, Mohandas N, Snyder SH. The 13-kD FK506 binding protein, FKBP13, interacts with a novel homologue of the erythrocyte membrane cytoskeletal protein 4.1. J Cell Biol. 1998;141:143–153. doi: 10.1083/jcb.141.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walensky LD, Blackshaw S, Liao D, Watkins CC, Weier HU, Parra M, Huganir RL, Conboy JG, Mohandas N, Snyder SH. A novel neuron-enriched homolog of the erythrocyte membrane cytoskeletal protein 4.1. J Neurosci. 1999;19:6457–6467. doi: 10.1523/JNEUROSCI.19-15-06457.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyszynski M, Lin J, Rao A, Nigh E, Beggs AH, Craig AM, Sheng M. Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature. 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 61.Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]