Abstract
An expansion of polyglutamines in the N terminus of huntingtin causes Huntington's disease (HD) and results in the accrual of mutant protein in the nucleus and cytoplasm of affected neurons. How mutant huntingtin causes neurons to die is unclear, but some recent observations suggest that an autophagic process may occur. We showed previously that huntingtin markedly accumulates in endosomal–lysosomal organelles of affected HD neurons and, when exogenously expressed in clonal striatal neurons, huntingtin appears in cytoplasmic vacuoles causing cells to shrink. Here we show that the huntingtin-enriched cytoplasmic vacuoles formed in vitro internalized the lysosomal enzyme cathepsin D in proportion to the polyglutamine-length in huntingtin. Huntingtin-labeled vacuoles displayed the ultrastructural features of early and late autophagosomes (autolysosomes), had little or no overlap with ubiquitin, proteasome, and heat shock protein 70/heat shock cognate 70 immunoreactivities, and altered the arrangement of Golgi membranes, mitochondria, and nuclear membranes. Neurons with excess cytoplasmic huntingtin also exhibited increased tubulation of endosomal membranes. Exogenously expressed human full-length wild-type and mutant huntingtin codistributed with endogenous mouse huntingtin in soluble and membrane fractions, whereas human N-terminal huntingtin products were found only in membrane fractions that contained lysosomal organelles. We speculate that mutant huntingtin accumulation in HD activates the endosomal–lysosomal system, which contributes to huntingtin proteolysis and to an autophagic process of cell death.
Keywords: Huntington's disease, autophagy, lysosomes, endosome tubulation, cathepsin D, N-terminal huntingtin, huntingtin proteolysis
Huntingtin is a protein of unknown function, is enriched in neurons, and resides mainly in the cytoplasm. An expanded polyglutamine tract in the N terminus of huntingtin causes Huntington's disease (HD), a neurological disorder associated with the selective loss of striatal and cortical neurons (Vonsattel and DiFiglia, 1998). Neurons affected in HD accumulate mutant huntingtin in the nucleus and cytoplasm (DiFiglia et al., 1997; Sapp et al., 1997;Gutekunst et al., 1998). Although the cause of protein accumulation in HD is unknown, experimental models developed in mice and in cell cultures have demonstrated that an excess of mutant huntingtin, especially the N-terminal region, in the nucleus or cytoplasm can cause cellular dysfunction or cell death (Hackam et al., 1998; Saudou et al., 1998; Peters et al., 1999). Features of apoptosis have been reported in some of these models, but the process of cell death in HD still remains unclear.
We showed that, in dying neurons in the HD brain, huntingtin aberrantly accumulated in perinuclear regions and in numerous punctate cytoplasmic structures that resembled endosomal–lysosomal organelles (Sapp et al., 1997). These results may be important in understanding how mutant huntingtin induces cell death because the endosomal–lysosomal–vacuolar pathway has been tied to the handling of other disease proteins, such as prions and Aβ peptide 1–42 (Taraboulos et al., 1992; Cataldo et al., 1996), and to cell death by autophagy, a process whereby cells remove cytosolic proteins and organelles and degrade themselves from within. Autophagy may precede and coexist with apoptosis, can be induced by apoptotic stimuli in the presence of caspase inhibitors (Xue et al., 1999), and may contribute to cell death in neurons through the regulation of lysosomal proteases cathepsin B and D (Ohsawa et al., 1998).
We described recently an in vitro model of HD using a clonal mouse striatal cell line transiently transfected with human huntingtin (Kim et al., 1999a). The exogenous wild-type and mutant huntingtins accumulated diffusely in the cytoplasm and formed cytoplasmic vacuoles or nuclear and cytoplasmic inclusions (mutant huntingtin only). Cells with cytoplasmic vacuoles became shrunken, whereas cells with inclusions did not. Like inclusions, the vacuoles localized N-terminal fragments of huntingtin. Previously, we observed that endogenous wild-type and mutant huntingtin associate with endosomes in primary fibroblasts (Velier et al., 1998) and that huntingtin accumulates in lysosomal-like organelles in the HD brain (Sapp et al., 1997). In view of these results, we undertook biochemical, immunohistochemical, and electron microscopy studies to further characterize vacuoles that accumulate exogenous huntingtin and other effects of huntingtin expression in the cytoplasm of clonal striatal cells. We found that the vacuoles associated with exogenous huntingtin incorporated the lysosomal enzyme cathepsin D in proportion to polyglutamine length in huntingtin and had the ultrastructural features of autophagosomes. N-terminal fragments of huntingtin were mainly found in membrane fractions that contained vacuoles. Huntingtin expression also induced extensive tubulation of endosomal membranes. We speculate that the endosomal–lysosomal system is the main path for huntingtin degradation and proteolysis and that autophagy contributes to cell death in HD.
MATERIALS AND METHODS
Antibodies. Antisera used in this study were directed against the following peptides or proteins: huntingtin (Ab1) (DiFiglia et al., 1995); FLAG (clone M5) and β-tubulin isotype III (monoclonals; Sigma, St. Louis, MO); calnexin C terminus (polyclonal; Stressgen, Victoria, British Columbia, Canada); GM130 and syntaxin 6 (monoclonals; Transduction Laboratories, Lexington, KY); transferrin receptor (monoclonal; Zymed Laboratories, San Francisco, CA); histone (monoclonal; Boehringer Mannheim, Indianapolis, IN); ubiquitin (polyclonal; Dako, Carpinteria, CA); C2 subunit of proteasome and the whole complex (polyclonals) (Mengual et al., 1996); cathepsin D [d23, made in sheep, gift of Dr. Ann Cataldo (Nathan S. Kline Institute for Psychiatric Research, Orangeburg, NY and New York University, New York, NY) (Cataldo et al., 1996); polyclonal Ab-2 (Oncogene, Cambridge, MA)]; rab7 (goat polyclonal; Santa Cruz Biotechnology, Santa Cruz, CA); and heat shock protein 70/heat shock cognate 70 (HSC70) (HSP70/HSC70) and lysosome-associated membrane protein-2 (LAMP2) (monoclonals; Stressgen). Secondary antibodies included Bodipy FL anti-mouse IgG (Molecular Probes, Eugene, OR), Cy3 anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA), and Texas Red anti-sheep IgG (Vector Laboratories, Burlingame, CA).
Expression constructs. The construction of expression plasmids used in this study has been described previously (Kim et al., 1999a). Briefly, two series of constructs, FH3221and FH9774, were made using pCDNA3 (Invitrogen, Carlsbad, CA) in which the sequence for FLAG was followed in frame with the first 3221 base pairs or 9774 base pairs of wild-type huntingtin cDNA. The FH constructs were modified to have variable CAG repeat lengths (18, 46, or 100) and are designated FH3221-18, FH3221-46, and FH3221-100 and FH9774-18, FH9774-46, and FH9774-100. In addition, we subcloned the first 3221 base pairs of the huntingtin cDNA with 18 or 100 glutamines into pCDNA3 without the FLAG tag, and these constructs are designated H3221-18 and H3221-100. The plasmid containing the cDNA for the c-Jun N-terminal protein kinase (JNK)-interacting protein 1 (JIP1) (Yasuda et al., 1999) preceded by a FLAG tag was a generous gift of Dr. R. J. Davis (Howard Hughes Medical Institute, University of Massachusetts Medical School, Worcester, MA). All expression plasmids contained the cytomegalovirus promoter.
Cell culture and transfections. Clonal striatal cells (X57) were produced by somatic cell fusion of embryonic day 18 mouse striatal neurons with neuroblastoma cells (N18TG2) (Wainwright et al., 1995). Cells were cultured in DMEM with high glucose (4.5 gm/l) supplemented with 10% fetal bovine serum and 50 U/ml penicillin–streptomycin at 37°C, with 5% CO2. All culture reagents were obtained from Life Technologies (Grand Island, NY). Cells were transiently transfected using the activated dendrimer Superfect Transfection Reagent (Qiagen, Valencia, CA). Cells were grown to 80% confluency in 100 mm tissue culture dishes, washed once with serum-free medium (DMEM), and then incubated with a mixture of 120 μl of Superfect reagent and 25 μg of DNA in 3 ml of complete medium at 37°C for 3 hr. Transfection medium was removed and replaced with normal growth medium. For immunocytochemistry, cells were trypsinized and plated on to uncoated glass coverslips directly after transfection. Mock transfections included the expression vector with no insert or the vector with FLAG sequence.
Subcellular fractionation and Western blot analysis. Cells were washed three times with ice-cold PBS and then scraped in 1 ml of homogenization buffer (in mm: 10 triethanolamine, 10 acetic acid, 250 sucrose, 1 EDTA, and 1 mmDTT, pH 7.4) with protease inhibitors (complete, mini-EDTA-free; Boehringer Mannheim). Cells were passed 15 times through a 26.5 gauge needle and monitored by light microscopy until 95% disruption was achieved. Crude homogenates (CH) were centrifuged at 2000 × g at 4°C for 10 min to obtain a crude pellet (P1). The low-speed supernatant (S1) was centrifuged at 100,000 ×g for 1 hr at 4°C to obtain the high-speed pellet (P2) and the high speed supernatant (S2). All pellets were washed twice in homogenization buffer and resuspended in a small volume of the same buffer. Equal amounts of protein from each fraction were analyzed by SDS-PAGE on 10% acrylamide low-bis (0.05%) gels using aN,N′-diallyltartardiamide (DATD) stacking gel, and Western blot analysis was performed as described previously (DiFiglia et al., 1995). Antibody dilutions for Western blotting were as follows: Huntingtin, Ab1, 0.5 μg/ml; calnexin, 1:1000; GM130, 1:250; proteasome (C2 subunit), 1:1000; transferrin receptor, 1:1000; histone, 5 μg/ml; cathepsin D (Ab-2), 2.5 μg/ml; HSP70/HSC70, 1 μg/ml; and β-tubulin, isotype III, 1:1000.
Isolation of nuclei. Cells were disrupted as described above, except the homogenization buffer (in mm:20 Tricine-NaOH, 250 sucrose, 25 KCl, and 5 MgCl2, pH 7.8 plus protease inhibitors) was changed to favor the isolation of intact nuclei. Homogenization was monitored by light microscopy and care was taken not to disrupt nuclei. Homogenates were centrifuged at 2000 × g at 4°C for 10 min. The supernatant (S1) was removed, the crude pellet was washed once, and nuclei were reisolated by centrifugation and resuspended in 1 ml of buffer (P1). This crude nuclear fraction was brought to 25% iodixanol (Optiprep; Accurate Chemicals, Westbury, NY) and layered on a discontinuous iodixanol gradient (30%, 35%). Gradients were centrifuged in a SW41 swing bucket rotor at 10,000 × gfor 20 min at 4°C. The first and second layers were collected separately. The nuclei at the 30%/35% interface were collected, and the final layer was collected. Equal volumes from each fraction were analyzed by SDS-PAGE and Western blot as described above.
Immunocytochemistry and confocal microscopy. Cells were grown on uncoated glass coverslips for the indicated times after transfection and washed twice with PBS containing 1 mm CaCl2 and 1 mm MgCl2. Cells were then fixed for 20 min with 4% paraformaldehyde in PBS containing calcium and magnesium ions. Subsequent washes were done with PBS without Mg2+/Ca2+. Cells were washed twice, permeablized with 0.2% Triton X-100 in PBS for 30 min, and then incubated in blocking solution (4% normal goat serum in PBS) for 1 hr at room temperature. Fixed cells were incubated in primary antibodies diluted in blocking solution overnight at 4°C. Antibodies were used at the following dilutions: Ab1, 0.5 μg/ml; Ab585, 1:500; M5, 10 μg/ml; anti-ubiquitin, 1:100; anti-GM130, 1:50; anti-cathepsin D (Oncogene), 1:500; anti-transferrin receptor, 4 μg/ml; HSP70/HSC70, 5 μg/ml; and anti-proteasome (whole complex), 1:500. Cells were then incubated with secondary antibodies (1:500) for 2 hr at room temperature, washed five times with PBS, and then dehydrated step-wise and mounted in Cytoseal 60 (Stephens Scientific, Riverdale, NJ). Individual images for each excitation wavelength (488 and 568 nm) were obtained using a Bio-Rad (Hercules, CA) 1024 laser confocal microscope through a 100× objective with oil immersion and merged in Adobe Systems (Salinas, CA) Photoshop to determine colocalization. Densitometry of cathepsin D labeling was performed using Sigma Scan Pro (Jandel Scientific, San Rafael, CA). For each labeled cell, the average signal intensity for cathepsin D in the region occupied by FLAG-positive vacuoles was determined. The average signal intensity for cathepsin D in an equivalent area of a nearby FLAG-negative cell was obtained and subtracted from the average intensity obtained in the FLAG-positive cell. Student's ttest was used to compare the mean corrected average signal intensities.
Electron microscopy. Cells were plated and transfected in plastic tissue culture dishes, allowed to grow, fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.1% Triton X-100 for 5 min, and blocked 1 hr in 4% normal goat serum. Fixed cells were incubated overnight at 4°C with the monoclonal antibody M5 (10 μg/ml) in blocking solution. The primary antibody was detected using an ABC kit (Vector Laboratories) with DAB or with gold-labeled (5 nm) Protein A (Goldmark Biochemicals, Phillipsburg, NJ). Cells were post-fixed with 2.5% glutaraldehyde, treated with 1% osmium and 1% uranyl acetate, sequentially dehydrated through 50, 70, 90, and 100% ethanol, and embedded using an ethanol soluble Epon mix (Lx112; LADD). The plastic from the dish was broken away from the embedded cells, which were thin sectioned and viewed using a JEOL 100CX electron microscope. The P1 pellet was prepared using the method described above for isolation of nuclei but without further purification over a discontinuous gradient. The pellet was washed three times with PBS with partial resuspension and centrifugation at 1500 × g and then fixed with 4% paraformaldehyde and 2.5% glutaraldehyde in PBS for 30 min. After several washes, the material was collected by centrifugation, treated en block with 1% osmium and 1% uranyl acetate, dehydrated, embedded, thin sectioned, and examined as above.
RESULTS
Morphology, distribution, and time course of appearance of dispersed and perinuclear vacuoles
In cells examined within 24 hr after transfection of truncated or full-length FLAG-huntingtin cDNAs, the cytoplasmic staining detected with FLAG antibody in many cells was reticular and consisted of very small discrete tubular structures (Fig.1a, shown for truncated huntingtin). The immunoreactive vacuoles that appeared in a small proportion of cells had irregular shapes and sizes and occurred in the cell bodies and proximal and distal portions of neurites (Fig.1b,c, shown for truncated huntingtin). Some vacuoles had a ring-like appearance (Fig.1d,e,f,g, shown for truncated and full-length huntingtin). In some cells, coalescence of vacuoles in the perinuclear region formed a single large complex (Fig. 1d,e,g, shown for truncated and full-length huntingtin). Vacuoles were also detected using Ab585, an antibody to an internal site in huntingtin (Fig. 1h). Expression of an untagged huntingtin cDNA produced vacuoles detectable with Ab1, an antibody to theN terminus of huntingtin (Fig. 1i, shown for truncated huntingtin). The vacuole formation was specific to huntingtin expression, because overexpression of an unrelated protein, JIP1 (Yasuda et al., 1999), bearing a FLAG tag at the N terminus produced diffuse cytoplasmic labeling but no vacuoles (Fig.1h). In a previous study, we showed that N-terminal huntingtin fragments were localized to vacuoles 24–48 hr after transfection. We performed an experiment to see whether the appearance of dispersed and perinuclear vacuoles coincided with the generation of N-terminal fragments, which appeared at 9 hr and were maximal at 24 hr after transfection in Western blots (Kim et al., 1999a). Cells were examined at 5, 7, 9, and 24 hr after transfection of the FLAG mutant huntingtin construct FH3221-100. At 5 and 7 hr, FLAG staining was cytoplasmic in 100% of labeled cells. At 9 hr, 7.6% of labeled cells had dispersed vacuoles. At 24 hr, 4.4% of labeled cells had dispersed vacuoles, and 11.8% displayed perinuclear vacuoles. (Additional experiments showed the same results and revealed that the perinuclear vacuoles could be detected as early as 18 hr with FH3221-100.) Critically, as found in our previous study (Kim et al., 1999a), few cells with nuclear or cytoplasmic inclusions were found in the first 24 hr, accounting for only ∼0.4% of the labeled cells at 24 hr. [Cells with inclusions are maximal 4–6 d after transfection (Kim et al., 1999a).] Thus, the formation of vacuoles and not inclusions coincided with the production of N-terminal huntingtin fragments.
Fig. 1.
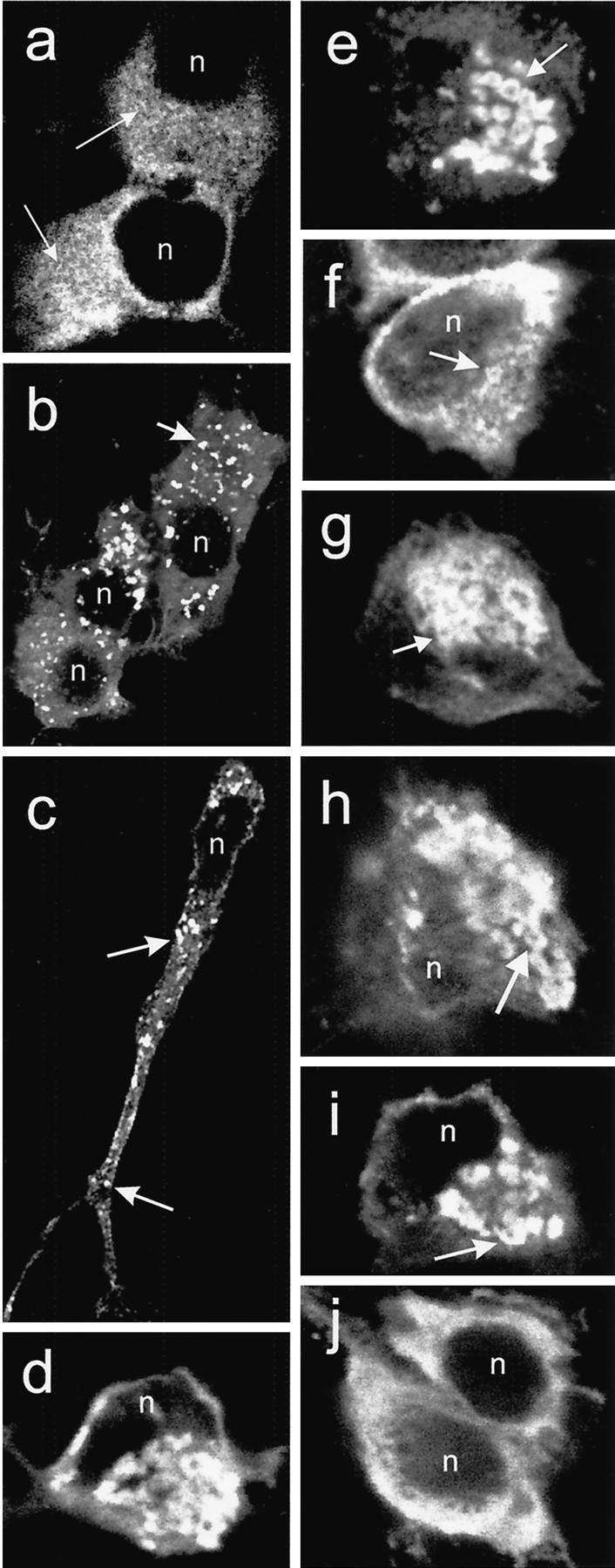
Overexpressed truncated FLAG-huntingtin in transfected clonal striatal cells, detected with a FLAG antibody, is localized in the cytoplasm in a meshwork of fine tubules (arrows) and punctate structures (a), in dispersed vacuoles throughout cell bodies (b, c) and neurites (c,arrows), and in vacuoles coalesced in the perinuclear region (d, e, arrow). Full-length FLAG-tagged huntingtin with a normal (f) or expanded (g) repeat also produce FLAG-positive vacuoles (arrows). Vacuolar staining can be obtained using huntingtin antibody Ab585 that recognizes an internal epitope of huntingtin (h,arrow). Huntingtin antibody Ab1 made to the N terminus detects vacuoles in cells expressing untagged
Molecular characterization of huntingtin-positive vacuoles
To characterize the FLAG-huntingtin vacuoles, we colabeled cells with a variety of subcellular markers and analyzed them by confocal microscopy. FLAG-positive vacuoles were immunoreactive for cathepsin D, a marker for lysosomes (Fig. 2), using two anti-cathepsin D antisera. Typically, in the double-labeled vacuoles, a rim of FLAG immunostaining surrounded a core of cathepsin D immunoreactivity with some overlap at the interface of the two markers. Relative to the well defined rim of FLAG labeling, the internal cathepsin D staining had a diffuse amorphous boundary. Because the lumen of the vacuole was accessible to cathepsin D antibody, it is unlikely that the absence of FLAG immunoreactivity in the core of the vacuole is attributable to incomplete access of FLAG antibody. FLAG-huntingtin and cathepsin D were localized in dispersed vacuoles (Fig. 2, top row) and perinuclear vacuoles of intact cells (Fig. 2, middle rows), and within condensed vacuoles that appeared in cell fragments (Fig. 2, bottom row). Overall, cathepsin D labeling was increased in FLAG-positive cells that had vacuoles compared with untransfected cells in the same field. The average signal intensity of cathepsin D within vacuoles (see Materials and Methods) was significantly greater in cells expressing FH3221-100 (n = 17 cells;p < 0.0001, for unpaired samples) compared with cells expressing FH3221-18 (n = 13 cells) or FH3221-46 (n = 15 cells) (Fig. 2, middle). FLAG-huntingtin vacuoles were also occasionally labeled for transferrin receptor, a marker of the endosomal–recycling system (data not shown). This agreed with our previous observations in human control and HD fibroblasts that endogenous wild-type and mutant huntingtin associated with endosomal membranes (Velier et al., 1998). We attempted additional characterization of the vacuoles with antisera to the late endosomal markers rab7 and the lysosomal membrane marker LAMP2, but the antisera did not produce staining in our cells. Vacuoles did not label with the endoplasmic reticulum (ER) marker calnexin, the cis/medial Golgi marker GM130 (Fig. 3a), or with thetrans-Golgi marker syntaxin 6 (data not shown). In some cells with extensive huntingtin-positive perinuclear vacuoles, Golgi membranes labeled with GM130 antisera were displaced from a perinuclear position (Fig. 3a, middle), and/or labeling was reduced in extent (Fig. 3a, bottom) compared with cells with diffuse huntingtin expression (Fig. 3a,top). Thus, huntingtin-enriched vacuoles were endosomal–lysosomal organelles and could affect Golgi organization.
Fig. 2.
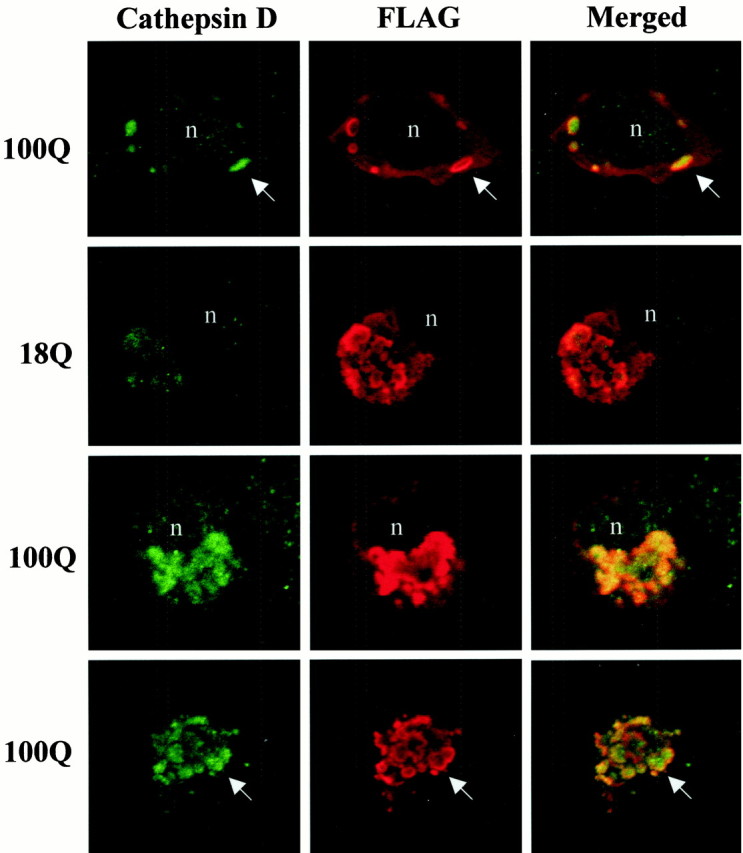
FLAG-huntingtin-immunoreactive vacuoles contain cathepsin D, and accumulation of cathepsin D is polyglutamine length-dependent. Confocal immunofluorescence microscopy of cells transfected with FH3221-18 or FH3221-100 then double immunostained after 24 hr for cathepsin D (green) and FLAG (red). Top three rows show intact cells, and bottom row shows a cell fragment. Dispersed vacuoles are in the cell in top row. Condensed vacuoles appear in the cells in the two middle rows. Note that FLAG-huntingtin immunoreactivity is present mainly along the periphery of the vacuole, and cathepsin D is inside the vacuole (arrows). The intensity of cathepsin D labeling in FLAG-immunoreactive vacuoles increases with polyglutamine expansion (compare two middle rows). Merged images onright show cathepsin D in green, FLAG inred, and the overlap in yellow.n, Nucleus.
Fig. 3.
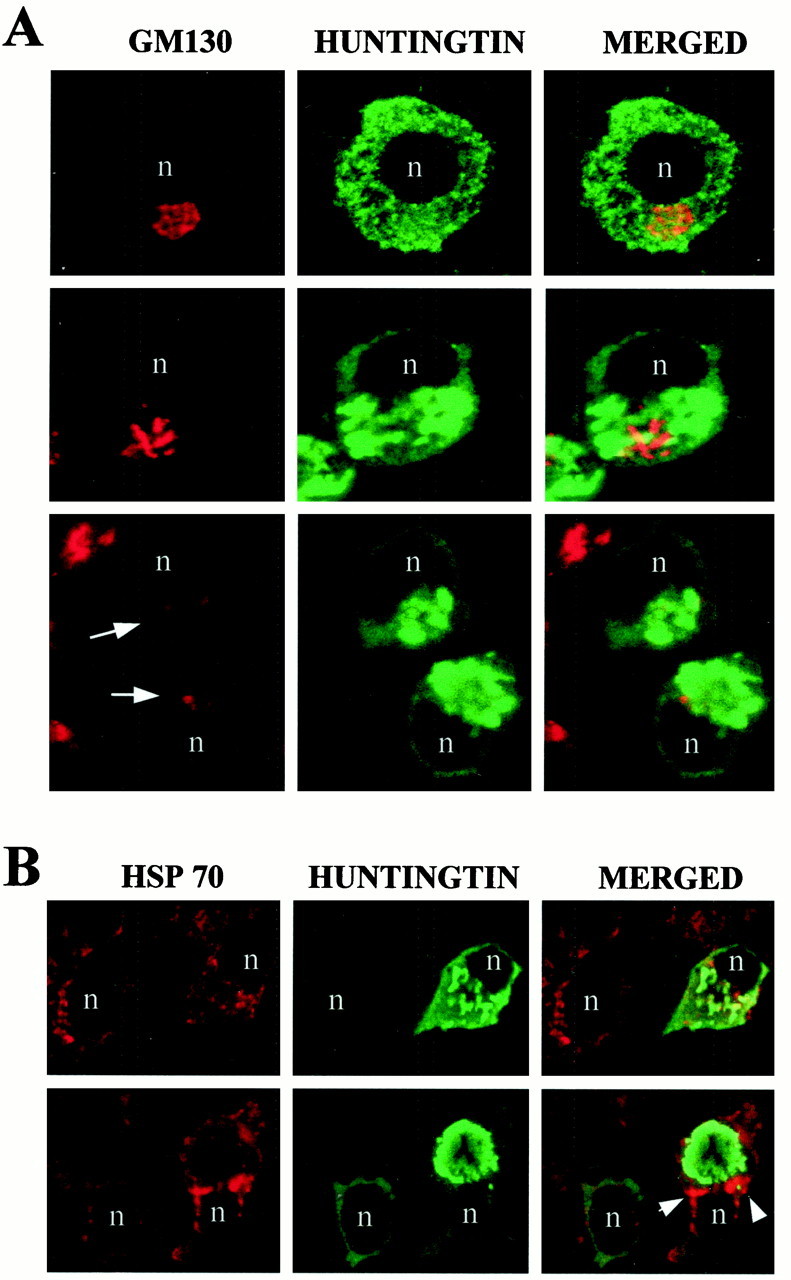
Distribution of Golgi labeling (A) and HSP70/HSC70 (B) in clonal striatal cells expressing exogenous huntingtin.A, Confocal immunofluorescence microscopy of cells transfected with FH3221-100 and double immunostained after 20 hr for the Golgi marker GM130 (red) and for huntingtin with Ab1 (green). A cell with diffuse huntingtin expression but lacking vacuoles shows perinuclear position of the Golgi (top row). Cells containing huntingtin-positive vacuoles show the Golgi displaced from the perinuclear region (middle row) or loss of immunoreactivity for GM130 (bottom row,arrows). Cells with reduced Golgi staining were imaged at the cross-sectional plane containing the highest level of GM130 immunoreactivity. B, HSP70/HSC70 localization in cells expressing FH3221-100 and double-labeled after 24 hr for HSP70/HSC70 (red) and for huntingtin with Ab1 (green). HSP70/HSC70 immunoreactivity is not enriched in huntingtin-positive vacuoles (top row). The punctate cytoplasmic staining for HSP70/HSC70, which may be mitochondria, is the same in huntingtin-positive and huntingtin-negative cells. One cell with a large huntingtin-positive vacuole shows increased HSP70/HSC70 immunoreactivity in the cytoplasm surrounding the vacuole (arrows), which may be within clustered mitochondria, but is not within the vacuole (bottom row). Merged images on right show GM130 (A) or HSP70/HSC70 (B) inred, huntingtin in green, and the overlap in yellow. n, Nucleus.
Immunoreactivity for ubiquitin, proteasome, and HSP70 has been found in the nuclear inclusions formed by mutant huntingtin or other polyglutamine disease proteins (Davies et al., 1997; DiFiglia et al., 1997; Paulson et al., 1997; Cummings et al., 1998; Chai et al., 1999). We investigated whether ubiquitin, proteasome, and HSP70 were also associated with huntingtin-immunoreactive vacuoles. Ubiquitin labeling was not present in huntingtin-positive vacuoles, and its localization in the cytoplasm was similar in FLAG-negative and FLAG-positive cells (results not shown). Overall, similar results were obtained using an antibody against the whole proteasome complex (data not shown). In some cells, however, proteasome labeling was increased at the borders of condensed perinuclear vacuoles and rarely within the vacuoles (data not shown). Immunoreactivity for HSP70/HSC70 in the cytoplasm was not different in most cells expressing exogenous huntingtin compared with cells expressing only endogenous huntingtin (Fig. 3b,top). However, in some cells with very large huntingtin-labeled vacuoles, HSP70/HSC70 staining was markedly increased in the cytoplasm surrounding the vacuoles (Fig.3b, bottom). The increase may be attributable to vacuole-induced displacement and accumulation of organelles, especially mitochondria that contain HSP70 family members (Kang et al., 1990) (see below).
Ultrastructure of cells expressing FLAG-huntingtin
To identify the subcellular structures associated with the accumulation of exogenous huntingtin, we analyzed by electron microscopy clonal striatal cells transfected with FLAG-huntingtin constructs containing 18, 46, and 100 glutamine repeats. Cells were fixed at 20 or 24 hr after transfection, labeled with FLAG antisera, and identified by immunoperoxidase (Fig.4) or immunogold (Fig.5) labeling. In general, labeled cells displayed marked enfolding of the nuclear membrane (Figs.4b,d, 5f) and perinuclear clustering and disruption of mitochondria. Labeled cells also contained a diverse group of autophagic vacuoles (Dunn, 1990a; Jia et al., 1997), lysosome-like bodies, and tubulovesicular structures, which were not seen in unlabeled cells or in cells transfected with a FLAG-only construct (Fig. 4a). The vacuoles and lysosome-like bodies were present throughout the cytoplasm and/or concentrated in perinuclear regions, similar to the distribution of dispersed and perinuclear FLAG-huntingtin-positive vacuoles seen by immunofluorescence. Immunoperoxidase label was more abundant in the cytoplasm of cells with fewer vacuoles than in cells with more vacuoles. The vacuoles had a variety of characteristics, including single- or double-limiting membranes (Figs.4c,d,e,5a,c), electron-lucent cores (Fig.4d), internal electron-dense vesicles (Figs.4c,d,e, 5a), whorls of internal membranes (myelin bodies; finger-print profiles) (Fig. 4f), and occasionally internalized mitochondria and portions of cytoplasm. Some of the lysosome-like bodies had radiating filaments and resembled Lewy bodies (Fig. 5b). Immunoperoxidase label was diffusely present in the cytoplasm (Fig.4b) and on the limiting membranes of autophagic vacuoles (Fig. 4c) but was hard to discern inside vacuoles and lysosome-like bodies because of their electron-dense quality (Fig.4b). In immunogold-labeled cells, gold particles were seen within and around late autophagic vacuoles and the lysosome-like dense bodies with radiating filaments (Fig.5a,b). Early autophagosomes that were found in the extracellular space adjacent to labeled cells also contained gold particles within the lumen and on the limiting membranes of the vacuoles (Fig. 5c). Gold label was more frequent in early autophagosomes (Fig. 5c) than late autophagic vacuoles (Fig. 5a). The latter may reflect a loss of epitope recognition attributable to digestion by cathepsins and other proteases. Gold particles were also dispersed in the matrix of the cytoplasm, at plasma membranes, especially sites of clathrin-coated pit invagination (Fig. 5d), and on the internal and limiting membranes of endosomes and multivesicular organelles (Fig.5e).
Fig. 4.
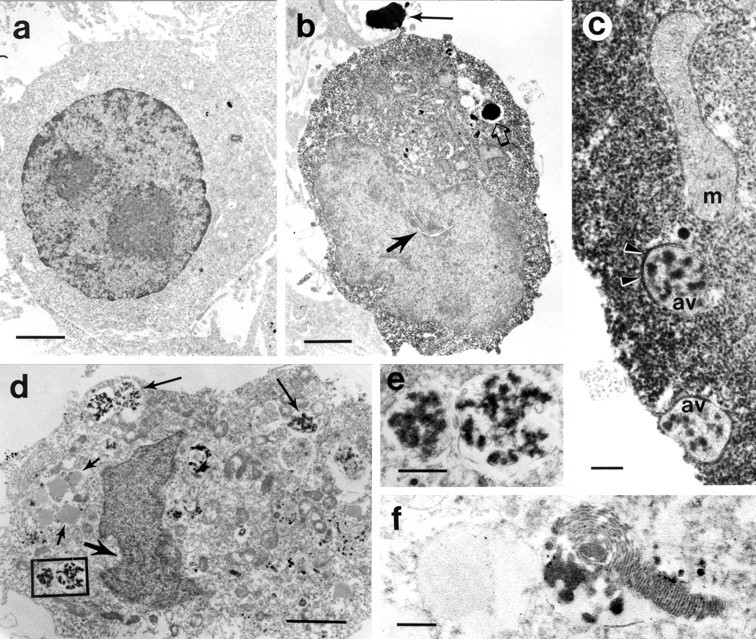
Ultrastructure of transfected clonal striatal cells. Transiently transfected cells were immunostained for FLAG and processed using the immunoperoxidase method. a, Electron micrograph of a typical control cell fixed 20 hr after transfection with the expression vector alone shows an unindented nucleus and normal distribution of organelles with no immunoperoxidase label. Electron micrographs from FLAG-positive cells are shown in b–f. Cells were fixed and immunostained 20 hr after transfection with FH3221-100 (b), FH3221-46 (d–f), and FH3221-18 (c). b, Intense immunoreactivity is present throughout the cytoplasm. Large lysosome-like bodies are present in the cytoplasm (open arrow) and outside the cell (long filled arrow). The nucleus is unlabeled and indented (short filled arrow). c, Peripheral portion of a cell body contains intense immunoperoxidase staining and two early autophagic vacuoles (av), which have peroxidase labeling on the double-limiting membranes (arrowheads). The nearby mitochondria (m) is unlabeled, and its inner membranes are disrupted. d, Cell with indented nucleus (short large arrow), numerous early autophagic vacuoles (small arrows), and late autophagic vacuoles (long arrows). This cell had only sparse immunoperoxidase label remaining in scattered regions of the cytoplasm. e, Two late autophagic vacuoles from the boxed region ind are filled with electron-dense vesicles.f, A vacuole containing a membrane whorl (also called myelin body or fingerprint profile) within an immunoperoxidase-labeled cell. Electron-dense tubulovesicular structures are present within the vacuole. Scale bars: a, b,d, 2 μm; c, f, 0.2 μm;e, 0.5 μm.
Fig. 5.
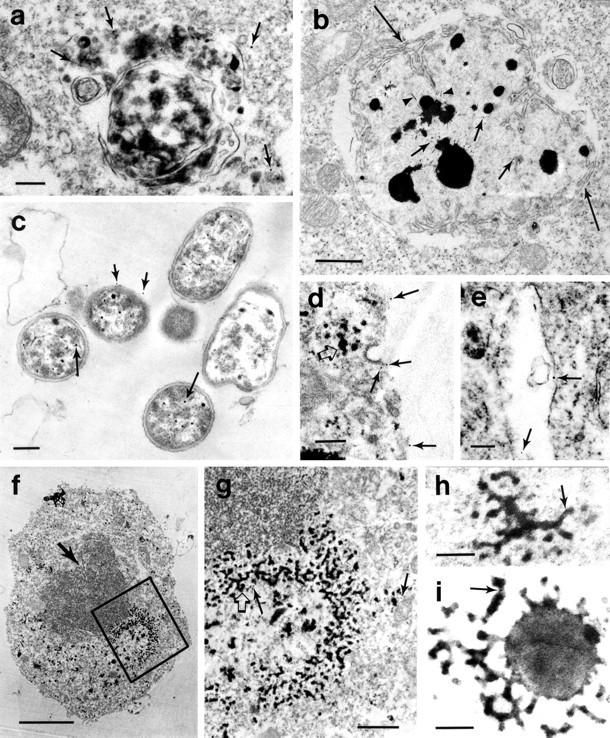
Immunogold labeling of FLAG-huntingtin in clonal striatal cells. Cells were transfected with FH3221-46 and then fixed and stained after 20 hr (a, b,d, f, g, h) or transfected with FH3221-100 and then fixed and stained after 24 hr (c, e, i).a, Gold deposits appear within autophagosomes (top arrows) and in the nearby cytoplasm (bottomright arrow). b, Lysosome-like bodies with radiating filaments (arrowheads) are enveloped with cisternae and limiting membranes (long arrows). Gold particles are scattered within the filamentous matrix of the organelle (short arrows). Mitochondria with disrupted cisternae are to theleft of the organelle. c, Extracellular early autophagosomes with double membranes show gold particles in the lumen (long arrows) and along the cytoplasmic face of the outer limiting membrane (short arrows).d, Gold label (arrows) appears along the cell surface and at clathrin-coated vesicles and invaginations.e, Gold deposits (arrows) appear on the limiting membrane and within the lumen of an early endosomal–multivesicular organelle. f, Tubulovesicular network composed of electron-dense vesicles and tubules appears in the perinuclear region. Nucleus is indented (arrow).g, Higher magnification of boxed region in f. Immunogold is present in the cytoplasm (short arrows), and electron-dense segments of tubules are clearly visible (open arrow). h,i, Gold particles (arrows) are attached or near electron-dense tubulovesicular organelles. h, A tubulovesicular body in the peripheral cytoplasm. i, A tubulovesicular body with ballooned segment located in the extracellular space. Gold particle is on tubule (arrow). Note that some tubules are electron-lucent and electron-dense, whereas the vesicles are electron-dense. Scale bars: a–e,h, i, 0.2 μm; f, 2 μm,g, 0.5 μm.
Tubulovesicular bodies that appeared in immunoperoxidase- and immunogold-labeled cells were present throughout the cytoplasm, near plasma membranes, in perinuclear locations, and in the extracellular space. Their widespread intracellular distribution corresponds to the reticulum of tubules observed with FLAG-huntingtin immunofluorescence (Fig. 1a). In immunogold-labeled cells, gold particles were near or adjacent to the tubulovesicular bodies (Fig.5f,g,h,i). The size of the tubules and the budding–fusing vesicles connected to them was ∼30–50 nm. The vesicles, which appeared to be uncoated, were electron-dense, whereas the tubules were a mixture of electron-dense and electron-lucent (Fig.5h,i). Hypertrophy in some tubules gave them a ballooned appearance (Fig. 5i).
Western blot analysis of the subcellular distribution of expressed full-length huntingtin, truncated huntingtin, and N-terminal huntingtin fragments
Subcellular fractions of cells transfected with full-length huntingtin were examined by SDS-PAGE and Western blot (Fig.6). Fraction P1 is a heterogenous mixture containing nuclei, ER remnants, large sorting–recycling organelles, and some cytoskeletal elements. Fraction S2 is expected to contain soluble elements of the cytosol, and fraction P2 should contain membranous organelles, including ER, Golgi, endosomes and small vesicles, lysosomes, mitochondria, and large particles such as ribosomes and proteasomes. Mutant full-length human huntingtin and endogenous mouse huntingtin fractionated with similar relative distributions (Fig. 6b) and were both weakly present in P1 and strong in S2 and P2 (Fig. 6). In contrast, the N-terminal huntingtin fragments of 80–90 kDa in size, which were derived from the overexpressed mutant huntingtin, were detectable in P1 and P2 and were absent from S2 (Fig. 6b). [These cleaved N-terminal products were also absent in mock transfected cells (Fig.6c).] The P1 pellet represents a small fraction (<10%) of the total cellular protein; therefore, based on signal intensity of the Western blot that was derived from gels loaded with equal amounts of protein from each fraction, most of the N-terminal 80–90 kDa fragments were in the membrane-enriched P2 fraction. The 80–90 kDa fragments are of special interest because they are the size expected by cleavage at caspase sites in mutant huntingtin (Goldberg et al., 1996; Wellington et al., 1998) and are inhibited by the caspase inhibitorN-benzyloxycarbonyl-val-ala-asp-fluoromethyl ketone (Z-VAD-FMK) (Kim et al., 1999a). In addition to their presence in intact cells, we found that N-terminal products of 80–90 kDa had markedly accumulated in the protein extracts collected from the growth medium and washes of transfected cells, indicating that the 80–90 kDa products were in “dead” cells or cell fragments that had been sloughed off (Fig. 6b, Debris). Inclusion of the stacking gel in the transfer showed that all of the exogenously expressed human mutant huntingtin was resolved in our SDS-PAGE system. The results were identical with expression of wild-type huntingtin, except that the N-terminal products were smaller (60–70 kDa) because of the normal polyglutamine repeat length. All results were confirmed in two sets of additional experiments.
Fig. 6.
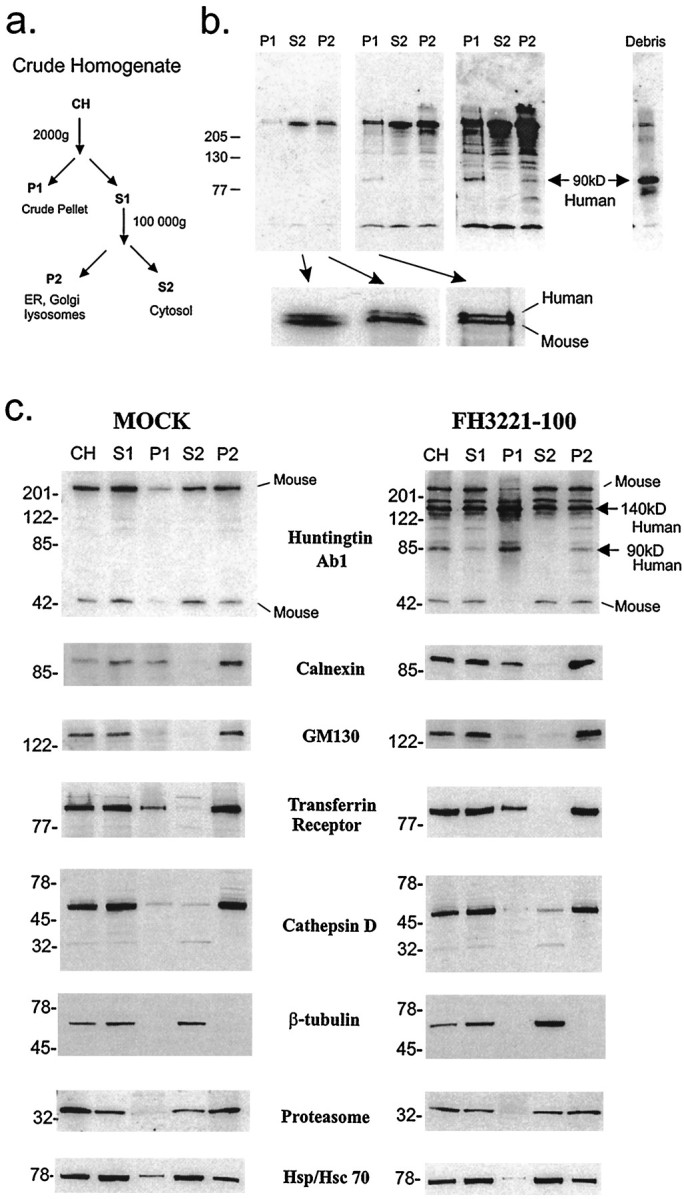
Biochemical analysis of subcellular fractions from cells expressing full-length mutant huntingtin or truncated mutant huntingtin. a, Scheme for subcellular fractionation using differential centrifugation with expected distribution of organelles. CH, Crude homogenate; S1, 2000 × g supernatant; P1, 2000 × g pellet; S2, 100,000 ×g supernatant; P2, 100,000 ×g pellet. b, Western blot shown at different exposures was probed with anti-huntingtin antisera Ab1. Protein fractions were from cells expressing FH9774-100. Cells were collected 24 hr after transfection. Twenty-five micrograms of protein were loaded per lane. Mutant huntingtin (Human) appears slightly above the normal endogenous (Mouse) protein in P1, S2, and P2. Bottom panels show enlargements of the two full-length bands from S2, P1 (longer exposure), and P2. N-terminal fragments are evident at longer exposures. The 90 kDa fragment derived from overexpressed mutant huntingtin is present in P1 and P2 but not S2, and abundantly in cellular debris recovered from the growth medium and washes that were pooled, centrifuged, and resuspended in 100 μl of homogenization buffer, 3 μg loaded. The stacking gel was included in the transfer to nitrocellulose and shows no protein. Apparent molecular weight is indicated in kilodaltons. c, Analysis of truncated huntingtin and its N-terminal huntingtin fragments from cells transfected with plasmid encoding FLAG only (MOCK) or transfected with FH3221-100. Cells were collected 24 hr after transfection. Fifteen micrograms of protein were loaded per lane. The identity and purity of the fractions was assessed with various markers: calnexin for the ER, GM130 for the cis/medial Golgi, transferrin receptor for endosomal–recycling compartments, and cathepsin D for lysosomes. β-tubulin was used to detect the presence of cytosolic constituents. Proteasome and HSP70 distribution are also shown. Apparent molecular weight is indicated in kilodaltons on the left. In blots probed with Ab1, endogenous full-length huntingtin (Mouse) is at the top, and a cleaved endogenous huntingtin fragment migrates at 50 kDa (Mouse). The expressed truncated huntingtin protein runs at the expected size of ∼140 kDa (top right arrow,Human) and as a modified form at ∼175 kDa. The 140 and 175 kDa proteins are present in S1, P1, S2, and P2, whereas the 90 kDa N-terminal product (bottom right arrow,Human) is present in P1 and P2 but not S2.
Subcellular fractions were also isolated from mouse striatal cells that expressed a FLAG-tagged truncated huntingtin or FLAG-only (Fig.6c). Transfection of FH3221-100 produced a fusion protein of the expected size of ∼140 kDa in the same fractions as endogenous huntingtin (Fig. 6c). In fraction P1, the level of the 140 kDa protein was stronger than the level of endogenous huntingtin or full-length FLAG-huntingtin (Fig. 6). The N-terminal huntingtin fragments (especially the 80 and 90 kDa fragments) fractionated similarly to the fragments produced from the full-length protein and were present in P1 and P2 and absent from S2 (Fig. 6c). There was an identical distribution of huntingtin and its N-terminal products seen with expression of truncated wild-type (18 glutamines) or mutant huntingtin with 46 glutamines (results not shown). The identity and purity of the fractions were confirmed by the detection of marker proteins, including calnexin for the ER, GM130 for the cis/medial Golgi (Nakamura et al., 1995), transferrin receptor for the endosomal–recycling system, and cathepsin D for lysosomes, the C2 subunit of the proteasome, and HSP70/HSC70. Results confirmed that the major cellular components had fractionated as expected and did not change whether cells expressed FLAG only or the wild-type or mutant huntingtin proteins. It is noteworthy that the proteasome did not distribute to fraction P1, suggesting that the N-terminal fragments accumulated in fraction P1 were not irreversibly associated with the proteasome. Importantly, the biochemical data were consistent with the immunofluorescence and electron microscopy findings; a significant portion of the expressed huntingtin and the N-terminal huntingtin fragments were associated with a fraction (P2) containing membrane-bounded cytoplasmic organelles. Mutant huntingtin expressed in human neuroblastoma cell line SY5Y also cofractionated with endogenous human huntingtin and showed a pattern of subcellular distribution similar to that observed in the mouse clonal striatal cells (data not shown).
Although the total yield of protein (and hence N-terminal fragments) obtained from fraction P1 is much less compared with other fractions, we determined whether the N-terminal huntingtin fragments found in the P1 fraction were in the nucleus or were part of a structure in the cytoplasm. Nuclei were isolated from fraction P1 on a discontinuous iodixanol density gradient (Fig.7a) and identified in fraction 3 using histone as a marker for nuclei (Fig. 7b). The 140 kDa truncated huntingtin protein was weakly detected in fraction 3, but the N-terminal products including the prominent 90 kDa product were not found in fraction 3 (Fig. 7b). This result showed that N-terminal huntingtin products derived from truncated huntingtin had segregated with a cytoplasmic component of the P1 fraction.
Fig. 7.
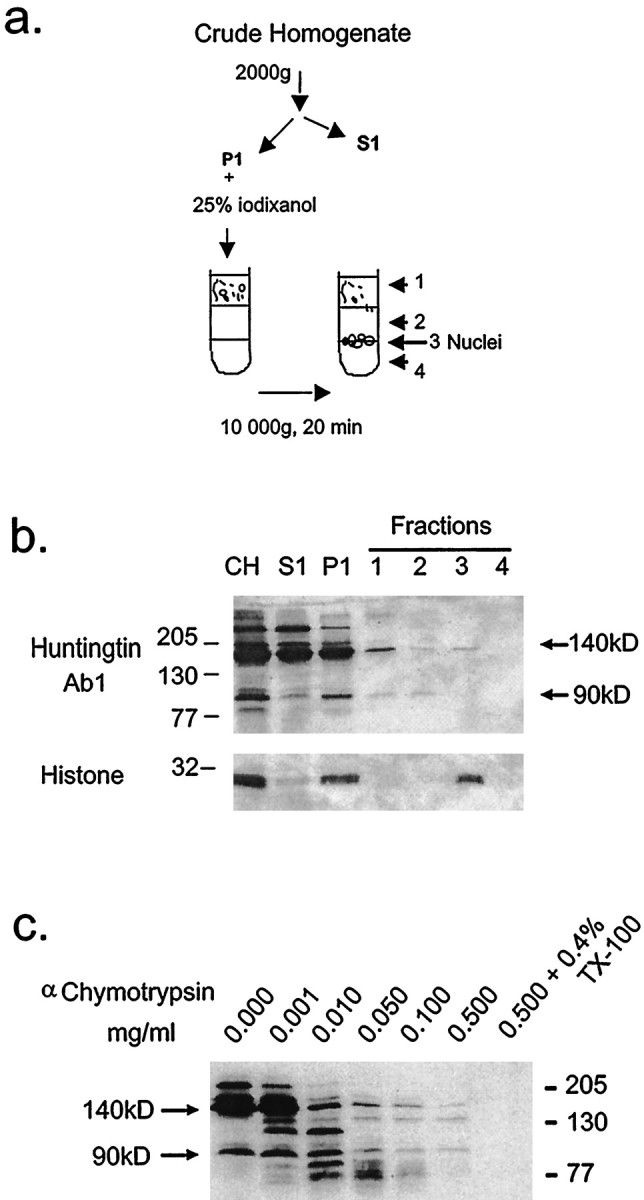
Biochemical analysis of P1 fraction and α-chymotrypsin digestion from cells transfected with FH3221-100. a, Scheme of nuclear isolation. The low speed pellet (P1) was fractionated on a discontinuous iodixanol density gradient as described in Methods and Materials. Fractions are as follows: CH, crude homogenate; S1, 2000 × gsupernatant; P1, 2000 × g pellet; fraction 1, 25% iodixanol step; fraction 2, 30% iodixanol step; fraction 3, 30%/35% interface; fraction 4, 35% iodixanol step. b, Western blots of fractions probed with anti-huntingtin antisera Ab1. Twenty microliters from each fraction was loaded per lane. Nuclear fraction (fraction 3) was identified using histone as a marker and contained the 140 kDa truncated huntingtin but not the N-terminal 90 kDa fragment. Apparent molecular weight is indicated in kilodaltons on theleft. c, Western blots of equal volumes of crude homogenates (CH). The crude homogenate was treated with varied amounts of α-chymotrypsin for 30 min on ice, stopped with 2 μl of 200 mm PMSF, and then analyzed by Western blot.
Electron microscopy of fraction P1
To determine which cytoplasmic components in the P1 fraction might be associated with N-terminal huntingtin fragments, we examined P1 fractions obtained from cells expressing FH3221-18, FH3221-46, and FH3221-100 or FLAG only by electron microscopy. We found electron-dense lysosome-like bodies and tubulovesicular profiles around intact nuclei (Fig.8a) in the fractions from cells expressing wild-type and mutant huntingtin but not in cells with FLAG only. The lysosome-like dense bodies were surrounded by vesiculated tubules and one or two limiting membranes and groups of cisternae (Fig. 8b). These results suggested that the N-terminal huntingtin products in the P1 fraction were associated with dense membranous organelles.
Fig. 8.
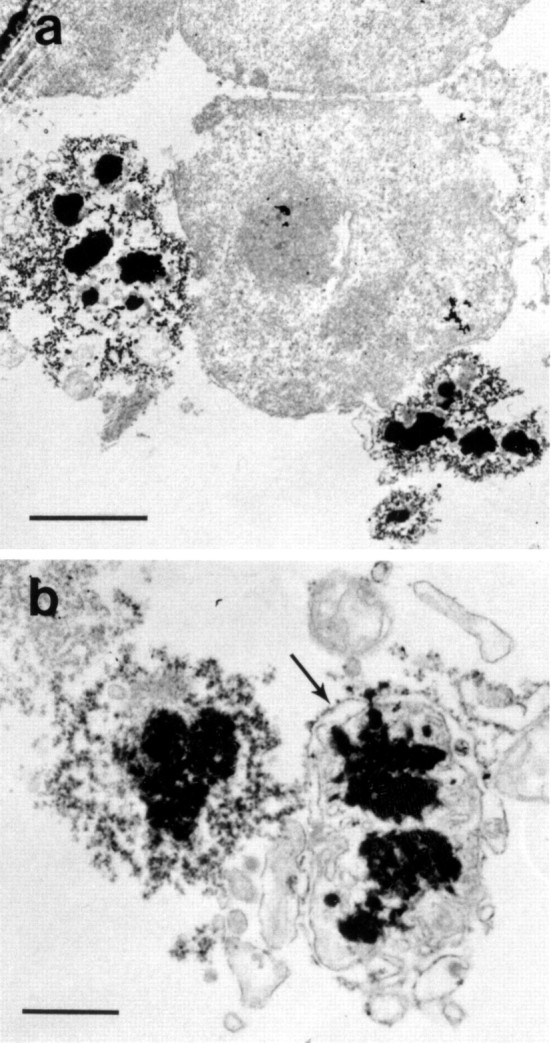
Electron microscopy of P1 pellet. Clonal striatal cells were transfected with FH3221-100. The P1 pellet was prepared 24 hr later and processed for electron microscopy.a, An intact nucleus is isolated with groups of lysosome-like bodies. b, Examples of lysosome-like dense bodies surrounded by electron-dense tubulovesicular structures and ER membranes (right, arrow). The lysosome-like dense bodies were absent from the P1 pellets of mock-transfected cells. Scale bars: a, 2 μm;b, 0.5 μm.
Proteolysis of huntingtin and its N-terminal products
Some of the expressed huntingtin and most of its N-terminal fragments had segregated to fractions with membrane-bound organelles (fractions P1 and P2). To determine whether the expressed huntingtin was on the outside of membrane-bound organelles or internalized to them, we treated homogenates from cells expressing FH3221-100 with increasing concentrations of α-chymotrypsin. Huntingtin and its N-terminal products were almost entirely susceptible to cleavage by α-chymotrypsin (Fig.7c), indicating that most of the protein was on the cytosolic surface of membranous organelles or free in the cytoplasm. A small amount of the remaining huntingtin succumbed to degradation with the addition of a mild detergent (0.4% Triton X-100), suggesting that a portion of the expressed huntingtin was enclosed within a membrane.
DISCUSSION
This study found that the cytoplasmic accumulation of human wild-type or mutant huntingtin in clonal mouse striatal cells induces activation of the endosomal–lysosomal system and autophagy. Increased endosomal–lysosomal activity may have occurred to remove excess protein, in response to abnormal direct effects of huntingtin on membrane morphology, and/or because wild-type and mutant huntingtin normally associate with endosomes (Velier et al., 1998). Our results help to explain the preponderance of endosomal–lysosomal-like organelles and tubulovesicular structures that label for huntingtin in the HD brain compared with the control brain (Sapp et al., 1997) and the numerous multivesicular bodies, endosomes, and lysosomes observed in biopsy tissue from HD patients examined in the 1970s (Tellez-Nagel et al., 1975; Roizin et al., 1979). In our study, transfected cells that developed huntingtin-positive vacuoles became shrunken and developed nuclear invaginations, which are features seen in the HD brain (Tellez-Nagel et al., 1975; Roizin et al., 1979), in HD transgenic mice (Davies et al., 1997), and in cells undergoing autophagy (Hornung et al., 1989; Ohsawa et al., 1998).
We show that cytoplasmic vacuoles that accumulate exogenous wild-type and mutant huntingtin (Kim et al., 1999a) are autophagosomes. Early autophagic vacuoles or autophagosomes form when cytosolic constituents, which have been sequestered into endosomes, become enveloped by rough or smooth ER membranes. The autophagosome matures to a degradative vacuole or autolysosome by acquiring a single-limiting membrane and lysosomal enzymes and acid hydrolases (Dunn, 1990a,b), which make them electron-dense. Some autolysosomes may present as sacs of membrane whorls or multivesicular bodies and can be extruded from cells (Jia et al., 1997) as we also observed. Many of the huntingtin-positive vacuoles had a single-limiting membrane, were multivesicular with electron-dense vesicles, and contained cathepsin D. Cathepsin D labeling was significantly more intense in vacuoles that expressed huntingtin with the largest polyglutamine expansion (100 glutamines). If increasing levels of cathepsin D are an index of vacuole maturation, then expression of mutant huntingtin may accelerate vacuole formation. This finding could explain why there are more cell fragments with vacuoles after expression of highly expanded mutant huntingtin than wild-type huntingtin (Kim et al., 1999a). Unlike inclusions, huntingtin-positive vacuoles expressed little or no immunoreactivity for ubiquitin, proteasome, or HSP70, which are involved in the removal of misfolded proteins (Buchner, 1996; Lee and Goldberg, 1998). Huntingtin may associate with these proteins at other subcellular sites, however.
Autophagy regulates normal cell growth and differentiation through the degradation of cytosolic proteins (Dunn, 1990a; Hollenbeck, 1993; Liang et al., 1999) but can also be activated by apoptotic stimuli (Xue et al., 1999). Ohsawa and colleagues (1998) found that PC12 cells deprived of serum or NGF showed a rapid rise in autophagosomes and cathepsin D-labeled granules in the early stages of apoptosis and suggested that autophagy and cathepsin D might be early positive regulators of apoptosis. Basal levels of autophagy can increase the sensitivity of some cells to apoptotic stimuli (Jia et al., 1997). This could be a basis for the selective vulnerability of some neuronal populations to neurodegeneration in HD. The signaling pathways that regulate autophagy in neurons are poorly understood but in yeast use a nonubiquitin protein conjugation system (Mizushima et al., 1998) and have biochemical overlap with mammalian proteins involved in secretion and endocytosis (Gerrard et al., 2000; Yoshimori et al., 2000). The overproduction of autophagosomes in a disease state, such as HD, could also interfere with normal membrane trafficking events in the cell, depleting membrane receptor proteins from the cell surface and draining lipid resources. Displacement of Golgi membranes and mitochondria by vacuoles could affect the function of these organelles (Dudani et al., 1990; Lippincott-Schwartz et al., 1991; Evtodienko et al., 1996; Xue et al., 1999). Increased endosome–lysosome activity has been implicated in the pathology of other neurodegenerative diseases, including Alzheimer's disease (Cataldo et al., 1996) and prion diseases (Boellaard et al., 1991; Laszlo et al., 1992; Taraboulos et al., 1992), and the coexistence of features of autophagy and apoptosis has been seen in substantia nigra neurons in Parkinson's disease (Anglade et al., 1997). Vacuoles have been observed in cells expressing other polyglutamine containing disease proteins, but their role in pathogenesis is unclear (Clark et al., 1997; Paulson et al., 1997).
Biochemical analysis showed that at least half of the total expressed huntingtin and most of the N-terminal huntingtin fragments were present in membrane fractions in which endosomes and lysosomes segregate. These data were consistent with immunofluorescence, immunogold, and α-chymotrysin digestion assays, suggesting that huntingtin was on or near the limiting membranes of autophagic vacuoles. The high concentration of N-terminal huntingtin fragments associated with autolysosomes suggests that lysosomal proteases could be involved in huntingtin proteolysis. The integrity of vacuoles could be compromised by excess huntingtin and release proteases in a manner similar to that seen in neuroblastoma cells that have taken up toxic Aβ1–42 peptide (Yang et al., 1998). Lysosomes treated with a membrane-permeabilizing agent release proteases and activate caspase 3 (Ishisaka et al., 1998), a proapoptotic caspase known to cleave huntingtin near its N terminus (Goldberg et al., 1996; Wellington et al., 1998). Cathepsin B activates caspase 3 at a neutral pH, compatible with a function in the cytosol or on membrane surfaces (Schotte et al., 1998). Caspases and cathepsin B are both sensitive to relatively low concentrations of the broad-acting caspase inhibitor Z-VAD-FMK (Schotte et al., 1998). This raises the possibility that the blockade of huntingtin proteolysis by Z-VAD-FMK (Kim et al., 1999a) involves inhibition of cathepsins. We speculate that, in HD neurons, lysosomes involved in mutant huntingtin degradation release proteases that activate caspase 3 and result in cleavage of N-terminal huntingtin.
The marked tubulation of membranes seen in cells expressing either wild-type or mutant huntingtin was unexpected and may provide clues to a membrane-associated function of huntingtin. The appearance and distribution of the tubulovesicular structures suggested that they are early and recycling endosomes (Stoorvogel et al., 1996; Prekeris et al., 1999). In human fibroblasts, huntingtin associates with endosomal membranes containing clathrin (Velier et al., 1998). The tubulation induced by huntingtin expression could affect vesicle budding, fusion, or transport of endosomes. Tubulation and altered function of endosomes occurs in cells treated with the toxin brefeldin A (Damke et al., 1991;Lippincott-Schwartz et al., 1991; Tooze and Hollinshead, 1992;Stoorvogel et al., 1996), which blocks assembly of coat proteins (clathrin, β-cop) onto membranes (Donaldson et al., 1991; de Figueiredo and Brown, 1995) and in cells overexpressing suppressor of K+ transport growth defect 1 (SKD1), a protein involved in endosome transport (Yoshimori et al., 2000). We showed previously that, in clonal striatal cells treated with forskolin or a D1 receptor agonist, endogenous huntingtin was recruited with clathrin to plasma membranes and localized to clathrin-coated vesicles, supporting a role for huntingtin in endocytosis (Kim et al., 1999b). In the present study, we found immunogold labeling for the expressed FLAG-huntingtin on clathrin-coated vesicles at plasma membranes. Huntingtin interacts with proteins thought to be involved in endocytosis, including huntington interacting protein 1 (HIP1) (Kalchman et al., 1997; Wesp et al., 1997), α-adaptin C (Faber et al., 1998), and SH3GL3 (Sittler et al., 1998). Another huntingtin binding partner, huntingtin-associated protein 1 (HAP1), localizes to membrane-bound organelles, tubulovesicular bodies, and budding vesicles (Martin et al., 1999) and may mediate vesicle transport by microtubules (Li et al., 1998). HIP1, SH3GL3, and HAP1 interact differently with wild-type and mutant huntingtin (Li et al., 1995; Kalchman et al., 1997; Sittler et al., 1998). Collectively, these observations suggest that the accumulation of mutant huntingtin in HD induces tubulation and other changes in endosomal membranes that could affect neuronal function.
Despite the high levels of expression achieved with transient transfection, the subcellular distributions of exogenous wild-type and mutant huntingtins were remarkably similar to each other and to the endogenous huntingtin by subcellular fractionation and Western blot analysis. These findings are consistent with data from HD brain showing that the mutant protein is transported and expressed similarly to wild-type in neurons (Aronin et al., 1995). We examined an early post-transfection period during which cells expressed mutant protein mainly in the cytoplasm and rarely had nuclear inclusions or evidence of SDS-insoluble products on Western blot. These results suggest that mutant huntingtin could accumulate in the cytoplasm and cause affects on membrane trafficking without mistargeting or aggregating.
In conclusion, the expression of wild-type or mutant huntingtin in the cytoplasm of clonal striatal cells caused a variety of effects on the endosomal–lysosomal system, including autophagy and membrane tubulation. We speculate that these changes contribute to the marked cellular dysfunction that occurs in HD neurons. Our data suggest that the endosomal–lysosomal pathway is the main pathway for removal of excess huntingtin and that lysosomal activity may regulate the cleavage of N-terminal fragments that later aggregate in nuclear and cytoplasmic inclusions of HD neurons. Although relatively little huntingtin reached the nucleus in our cells, it may have had some effects on cell function (Saudou et al., 1998). Nevertheless, our results strongly suggest that a significant contribution to neuropathology in HD involves an autophagic process that is induced by the cytoplasmic accumulation of mutant huntingtin.
Footnotes
This work was supported by National Institutes of Health Grants NS16367 and NS35711 (to M.D.), NS38194 (to N.A.), T32-AG00222 (K.B.K.), and a grant from the Huntington's Disease Society of America (to M.D.). We thank Mr. Lawrence Cherkas for his assistance with the photography, Yun J. Kim for his assistance with confocal microscopy, Kristy Brown for advise with electron microscopy, and Yumei Wang and Zheng-Hong Qin for help with the studies involving cathepsin D localization.
Correspondence should be addressed to Dr. Marian DiFiglia, Department of Neurology, Massachusetts General Hospital East, 149 13th Street, Room 6604, Charlestown, MA 02129. E-mail:difiglia@helix.mgh.harvard.edu.
REFERENCES
- 1.Anglade P, Vyas S, Javoy-Agid F, Herrero MT, Michel PP, Marquez J, Mouatt-Prigent A, Ruberg M, Hirsch EC, Agid Y. Apoptosis and autophagy in nigral neurons of patients with Parkinson's disease. Histol Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- 2.Aronin N, Chase K, Young C, Sapp E, Schwarz C, Matta N, Kornreich R, Landwehrmeyer B, Bird E, Beal MF, Vonsattel JP, Smith T, Carraway R, Boyce FM, Young AB, Penney JB, DiFiglia M. CAG expansion affects the expression of mutant huntingtin in the Huntington's disease brain. Neuron. 1995;15:1193–1201. doi: 10.1016/0896-6273(95)90106-x. [DOI] [PubMed] [Google Scholar]
- 3.Boellaard JW, Kao M, Schlote W, Diringer H. Neuronal autophagy in experimental scrapie. Acta Neuropathol (Berl) 1991;82:225–228. doi: 10.1007/BF00294449. [DOI] [PubMed] [Google Scholar]
- 4.Buchner J. Supervising the fold: functional principles of molecular chaperones. FASEB J. 1996;10:10–19. [PubMed] [Google Scholar]
- 5.Cataldo AM, Hamilton DJ, Barnett JL, Paskevich PA, Nixon RA. Properties of the endosomal–lysosomal system in the human central nervous system: disturbances mark most neurons in populations at risk to degenerate in Alzheimer's disease. J Neurosci. 1996;16:186–199. doi: 10.1523/JNEUROSCI.16-01-00186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chai Y, Koppenhafer SL, Bonini NM, Paulson HL. Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J Neurosci. 1999;19:10338–10347. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark HB, Burright EN, Yunis WS, Larson S, Wilcox C, Hartman B, Matilla A, Zoghbi HY, Orr HT. Purkinje cell expression of a mutant allele of SCA1 in transgenic mice leads to disparate effects on motor behaviors, followed by a progressive cerebellar dysfunction and histological alterations. J Neurosci. 1997;17:7385–7395. doi: 10.1523/JNEUROSCI.17-19-07385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummings CJ, Mancini MA, Antalffy B, DeFranco DB, Orr HT, Zoghbi HY. Chaperone suppression of ataxin-1 aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 9.Damke H, Klumperman J, von Figura K, Braulke T. Effects of brefeldin A on the endocytic route. J Biol Chem. 1991;266:24829–24833. [PubMed] [Google Scholar]
- 10.Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 11.de Figueiredo P, Brown WJ. Role for calmodulin in organelle membrane tubulation. Mol Biol Cell. 1995;6:871–887. doi: 10.1091/mbc.6.7.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiFiglia M, Sapp E, Chase K, Schwarz C, Meloni A, Young C, Martin E, Vonsattel JP, Carraway R, Reeves SA, Boyce FM, Aronin N. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 13.DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 14.Donaldson JG, Lippincott-Schwartz J, Klausner RD. Guanine nucleotides modulate the effects of brefeldin A in semipermeable cells: regulation of the association of a 110-kD peripheral membrane protein with the Golgi apparatus. J Cell Biol. 1991;112:579–588. doi: 10.1083/jcb.112.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudani AK, Austin RC, Venner TJ, Gupta RS. Effects of anti-mitotic and anti-mitochondrial agents on the cellular distribution of microtubules and mitochondria. Cytobios. 1990;63:95–108. [PubMed] [Google Scholar]
- 16.Dunn WA., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990a;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn WA., Jr Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J Cell Biol. 1990b;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evtodienko YV, Teplova VV, Sidash SS, Ichas F, Mazat JP. Microtubule-active drugs suppress the closure of the permeability transition pore in tumor mitochondria. FEBS Letters. 1996;393:86–88. doi: 10.1016/0014-5793(96)00875-7. [DOI] [PubMed] [Google Scholar]
- 19.Faber PW, Barnes GT, Srinidhi F, Chen J, Gusella JF, MacDonald M. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 20.Gerrard SR, Bryant NJ, Stevens TH. VSP21 controls entry of endocytosed and biosynthetic proteins into the yeast prevacuolar compartment. Mol Cell Biol. 2000;11:613–626. doi: 10.1091/mbc.11.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg YP, Nicholson DW, Rasper DM, Kalchman MA, Koide HB, Graham RK, Bromm M, Kazemi-Esfarjani P, Thornberry NA, Vaillancourt JP, Hayden MR. Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the glutamine tract. Nat Genet. 1996;13:442–449. doi: 10.1038/ng0896-442. [DOI] [PubMed] [Google Scholar]
- 22.Gutekunst CA, Li SH, Yi H, Ferrante RJ, Li XJ, Hersch SM. The cellular and subcellular localization of huntingtin-associated protein 1 (HAP1): comparison with huntingtin in rat and human. J Neurosci. 1998;18:7674–7686. doi: 10.1523/JNEUROSCI.18-19-07674.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hackam AS, Sinjaraja R, Wellington CL, Metzler M, McCutcheon K, Zhang T, Kalchman M, Hayden MR. The influence of huntingtin protein size on nuclear localization and cellular toxicity. J Cell Biol. 1998;141:1097–1105. doi: 10.1083/jcb.141.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollenbeck PJ. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J Cell Biol. 1993;121:305–315. doi: 10.1083/jcb.121.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hornung JP, Koppel A, Clarke PGH. Endocytosis and autophagy in dying neurons: an ultrastructural study in chick embryos. J Comp Neurol. 1989;283:425–437. doi: 10.1002/cne.902830310. [DOI] [PubMed] [Google Scholar]
- 26.Ishisaka R, Utsumi T, Yabuki M, Kanno T, Furuno T, Inoue M, Utsumi K. Activation of caspase-3-like protease by digitonin-treated lysosomes. FEBS Lett. 1998;435:233–236. doi: 10.1016/s0014-5793(98)01080-1. [DOI] [PubMed] [Google Scholar]
- 27.Jia L, Dourmashkin RR, Allen PD. Inhibition of autophagy abrogates tumour necrosis factor alpha induced apoptosis in human T lymphoblastic leukaemic cells. Br J Haematol. 1997;98:673–685. doi: 10.1046/j.1365-2141.1997.2623081.x. [DOI] [PubMed] [Google Scholar]
- 28.Kalchman MA, Koide HB, McCutcheon K, Graham RK, Nichol K, Nishiyama K, Kazemi-Esfarjani P, Lynn FC, Wellington C, Metzler M, Goldberg YP, Kanazawa I, Gietz RD, Hayden MR. HIP1, a human homologue of S. cerevisiae Sla2p, interacts with membrane associated huntingtin in the brain. Nat Genet. 1997;16:44–53. doi: 10.1038/ng0597-44. [DOI] [PubMed] [Google Scholar]
- 29.Kang P-J, Ostermann J, Shilling J, Neupert W, Craig EA, Pfanner N. Requirement of hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- 30.Kim M, Lee HS, Laforet G, McIntyre C, Martin EJ, Chang P, Kim TW, Williams M, Reddy PH, Tagle D, Boyce FM, Won L, Heller A, Aronin N, DiFiglia M. Mutant huntingtin expression in clonal striatal cells: dissociation of inclusion formation and neuronal survival by caspase inhibition. J Neurosci. 1999a;19:964–973. doi: 10.1523/JNEUROSCI.19-03-00964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim M, Velier J, Chase K, Laforet G, Kalchman MA, Hayden MR, Won L, Heller A, Aronin N, DiFiglia M. Forskolin and dopamine D1 receptor activation increase huntingtin's association with endosomes in immortalized neuronal cells of striatal origin. Neuroscience. 1999b;89:1159–1167. doi: 10.1016/s0306-4522(98)00400-x. [DOI] [PubMed] [Google Scholar]
- 32.Laszlo L, Lowe J, Self T, Kenward N, Landon M, McBride T, Farquhar C, McConnell I, Brown J, Hope J, Mayers RJ. Lysosomes as key organelles in the pathogenesis of prion encephalopathies. J Pathol. 1992;166:333–341. doi: 10.1002/path.1711660404. [DOI] [PubMed] [Google Scholar]
- 33.Lee DHL, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 34.Li SH, Gutekunst CA, Hersch SM, Li XJ. Interaction of huntingtin-associated protein with dynactin P150Glued. J Neurosci. 1998;18:1261–1269. doi: 10.1523/JNEUROSCI.18-04-01261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li XJ, Li SH, Sharp AH, Nucifora FC, Jr, Schilling G, Lanahan A, Worley P, Snyder SH, Ross CA. A huntingtin-associated protein enriched in brain with implications for pathology. Nature. 1995;378:398–402. doi: 10.1038/378398a0. [DOI] [PubMed] [Google Scholar]
- 36.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 37.Lippincott-Schwartz J, Yuan L, Tipper C, Amherdt M, Orci L, Klausner RD. Brefeldin A's effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell. 1991;67:601–616. doi: 10.1016/0092-8674(91)90534-6. [DOI] [PubMed] [Google Scholar]
- 38.Martin EJ, Kim M, Velier J, Sapp E, Lee H, Laforet G, Won L, Chase K, Bhide P, Heller A, Aronin N, DiFiglia M. Analysis of huntingtin-associated protein 1 in mouse brain and immortalized striatal neurons. J Comp Neurol. 1999;403:421–430. [PubMed] [Google Scholar]
- 39.Mengual E, Arizti P, Rodrigo J, Gimenez-Amaya JM, Castaño JG. Immunohistochemical distribution and electron microscopic subcellular localization of the proteasome in the rat CNS. J Neurosci. 1996;16:6331–6341. doi: 10.1523/JNEUROSCI.16-20-06331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohsawa Y, Isahara K, Kanamori S, Shibata M, Kametaka S, Gotow T, Watanabe T, Kominami E, Uchiyama Y. An ultrastructural and immunohistochemical study of PC12 cells during apoptosis induced by serum deprivation with special reference to autophagy and lysosomal cathepsins. Arch Histol Cytol. 1998;61:395–403. doi: 10.1679/aohc.61.395. [DOI] [PubMed] [Google Scholar]
- 43.Paulson HL, Perez MK, Trottier Y, Trojanowski JQ, Subramony SH, Das SS, Vig P, Mandel JL, Fischbeck KH, Pittman RN. Intranuclear inclusions in expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 44.Peters MF, Nucifora FC, Jr, Kushi J, Seaman HC, Cooper JK, Herring WJ, Dawson VL, Dawson TM, Ross CA. Nuclear targeting of mutant huntingtin increases toxicity. Mol Cell Neurosci. 1999;14:121–128. doi: 10.1006/mcne.1999.0773. [DOI] [PubMed] [Google Scholar]
- 45.Prekeris R, Folleti DL, Scheller RH. Dynamics of tubulovesicular recycling endosomes in hippocampal neurons. J Neurosci. 1999;19:10324–10327. doi: 10.1523/JNEUROSCI.19-23-10324.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roizin L, Stellar S, Liu JC. Neuronal nuclear-cytoplasmic changes in Huntington's chorea: electron microscope investigations. In: Chase TN, Wexler NS, Barbeau A, editors. Advances in neurology, Vol 23. Raven; New York: 1979. pp. 95–122. [Google Scholar]
- 47.Sapp E, Schwarz C, Chase K, Bhide PG, Young AB, Penney J, Vonsattel JP, Aronin N, DiFiglia M. Huntingtin localization in brains of normal and Huntington's disease patients. Ann Neurol. 1997;42:604–612. doi: 10.1002/ana.410420411. [DOI] [PubMed] [Google Scholar]
- 48.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 49.Schotte P, Van Criekinge W, Van deCraen M, Van Loo G, Desmedt M, Grooten J, Cornelissen M, De Ridder L, Vandekerckhove J, Friers W, Vandenabeele P, Beyaert R. Cathepsin B-mediated activation of the proinflammatory caspase-11. Biochem Biophys Res Commun. 1998;251:379–387. doi: 10.1006/bbrc.1998.9425. [DOI] [PubMed] [Google Scholar]
- 50.Sittler A, Walter S, Wedemeyer N, Hasenbank R, Scherzinger E, Eickhoff H, Bates GP, Lehrach H, Wanker EE. SH3GL3 associates with the huntingtin Exon 1 protein and promotes the formation of polygln-containing protein aggregates. Mol Cell. 1998;2:427–436. doi: 10.1016/s1097-2765(00)80142-2. [DOI] [PubMed] [Google Scholar]
- 51.Stoorvogel W, Oorschot V, Geuze HJ. A novel class of clathrin-coated vesicles budding from endosomes. J Cell Biol. 1996;132:21–33. doi: 10.1083/jcb.132.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taraboulos A, Raeber AJ, Borchelt DR, Serban D, Prusiner SB. Synthesis and trafficking of prion proteins in cultured cells. Mol Biol Cell. 1992;3:851–863. doi: 10.1091/mbc.3.8.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tellez-Nagel I, Johnson AB, Terry RD. Studies on brain biopsies of patients with Huntington's chorea. J Neuropathol Exp Neurol. 1975;33:308–332. doi: 10.1097/00005072-197404000-00008. [DOI] [PubMed] [Google Scholar]
- 54.Tooze J, Hollinshead M. In AtT20 and HeLa cells brefeldin A induces the fusion of tubular endosomes and changes their distribution and some of their endocytic properties. J Cell Biol. 1992;118:813–830. doi: 10.1083/jcb.118.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Velier J, Kim M, Schwarz C, Kim TW, Sapp E, Chase K, Aronin N, DiFiglia M. Wild-type and mutant huntingtins function in vesicle trafficking in the secretory and endocytic pathways. Exp Neurol. 1998;152:34–40. doi: 10.1006/exnr.1998.6832. [DOI] [PubMed] [Google Scholar]
- 56.Vonsattel JPG, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 57.Wainwright MS, Perry BD, Won LA, O'Malley KL, Wang WY, Ehrlich ME, Heller A. Immortalized murine striatal neuronal cell lines expressing dopamine receptors and cholinergic properties. J Neurosci. 1995;15:676–688. doi: 10.1523/JNEUROSCI.15-01-00676.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wellington CL, Ellerby LM, Hackam AS, Margolis RL, Trifiro MA, Singaraja R, McCutcheon K, Salvesen GS, Propp SS, Bromm M, Rowland KJ, Zhang T, Rasper D, Roy S, Thornberry N, Pinsky L, Kakizuka A, Ross CA, Nicholson DW, Bredesen DE, Hayden MR. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J Biol Chem. 1998;273:9158–9167. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- 59.Wesp A, Hicke L, Palecek J, Lombardi R, Aust T, Munn AL, Riezman H. End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:2291–2306. doi: 10.1091/mbc.8.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xue L, Fletcher GC, Tolkovsky AM. Autophagy is activated by apoptotic signalling in sympathetic neurons: an alternative mechanism of death execution. Mol Cell Neurosci. 1999;14:180–198. doi: 10.1006/mcne.1999.0780. [DOI] [PubMed] [Google Scholar]
- 61.Yang AJ, Chandswangbhuvana D, Margol L, Glabe CG. Loss of early endosomal/lysosomal impermeability is an early event in amyloid A beta1–42 pathogenesis. J Neurosci Res. 1998;52:691–698. doi: 10.1002/(SICI)1097-4547(19980615)52:6<691::AID-JNR8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 62.Yasuda J, Whitmarsh AJ, Cavanagh J, Sharma M, Davis RJ. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol Cell Biol. 1999;19:7245–7254. doi: 10.1128/mcb.19.10.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshimori T, Yamagata F, Yamamoto A, Mizushima N, Kabeya Y, Nara A, Miwako I, Ohashi M, Ohsumi M, Ohsumi Y. The mouse SKD1, a homologue of yeast vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol Biol Cell. 2000;11:747–763. doi: 10.1091/mbc.11.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]


