Abstract
PSD-95, DLG, ZO-1 (PDZ) domain-mediated protein interactions have been shown to play important roles in the regulation of glutamate receptor function at excitatory synapses. Recent studies demonstrating the rapid regulation of AMPA receptor function during synaptic plasticity have suggested that AMPA receptor interaction with PDZ domain-containing proteins may be dynamically modulated. Here we show that PKC phosphorylation of the AMPA receptor GluR2 subunit differentially modulates its interaction with the PDZ domain-containing proteins GRIP1 and PICK1. The serine residue [serine-880 (Ser880)] in the GluR2 C-terminal sequence (IE SVKI) critical for PDZ domain binding is a substrate of PKC and is phosphorylated in vivo. In vitro binding and coimmunoprecipitation studies show that phosphorylation of serine-880 within the GluR2 PDZ ligand significantly decreases GluR2 binding to GRIP1 but not to PICK1. Immunostaining of cultured hippocampal neurons demonstrates that the Ser880-phosphorylated GluR2 subunits are enriched and colocalized with PICK1 in the dendrites, with very little staining observed at excitatory synapses. Interestingly, PKC activation in neurons increases the Ser880 phosphorylation of GluR2 subunits and recruits PICK1 to excitatory synapses. Moreover, PKC stimulation in neurons results in rapid internalization of surface GluR2 subunits. These results suggest that GluR2 phosphorylation of serine-880 may be important in the regulation of the AMPA receptor internalization during synaptic plasticity.
Keywords: GRIP1, PICK1, AMPA receptor, phosphorylation, PKC, PDZ domain
Glutamate receptors mediate excitatory synaptic transmission in the CNS and are critical for most neuronal processes, including synaptic plasticity, neuronal development, and several neurological and psychiatric disorders (Sheng and Kim, 1996; Kim and Huganir, 1999). Two major ionotropic receptors at glutamatergic synapses are AMPA and NMDA receptors, which are heteromeric complexes of homologous subunits (GluR1–4 for AMPA receptors and NR1, NR2A-D for NMDA receptors) that differentially combine to form various receptor subtypes (Choi, 1988; Bliss and Collingridge, 1993; Seeburg, 1993; Hollmann and Heinemann, 1994). AMPA receptors mediate rapid excitatory synaptic transmission, whereas NMDA receptors are essential for the induction of activity-dependent synaptic plasticity underlying learning and memory (Choi, 1988; Bliss and Collingridge, 1993; Seeburg, 1993; Hollmann and Heinemann, 1994). Both AMPA and NMDA receptors are highly concentrated at the postsynaptic membrane of excitatory synapses where the C-terminal tails of their subunits associate with cytoskeletal and signaling molecules by PSD-95, DLG, ZO-1 (PDZ) domain-mediated interaction (Sheng and Kim, 1996; Kim and Huganir, 1999). PDZ domains are modular protein–protein interaction motifs that specifically bind the C termini of membrane or membrane-associated proteins (Woods and Bryant, 1991; Cho et al., 1992;Kistner et al., 1993; Kennedy, 1995). The NR2 subunits of NMDA receptors specifically interact with the PDZ domains of PSD-95/SAP90 family proteins through their C-terminal sequence (IESDV) (Kim et al., 1995; Kornau et al., 1995). In contrast, the C termini sequence (ESVKI) of the GluR2 and GluR3 subunits of AMPA receptors bind to specific PDZ domains in several proteins including GRIP1 and -2 (glutamate receptor interacting protein), ABP (AMPA receptor binding protein), and PICK1 (protein interacting with C kinase 1) (Dong et al., 1997, 1999;Srivastava et al., 1998; Xia et al., 1999).
PDZ domain-containing proteins have been suggested to regulate synaptic targeting of glutamate receptors and K+channels at synapses (Sheng and Kim, 1996; Kim and Huganir, 1999). PSD-95 colocalizes with NMDA receptors at excitatory synapses and can induce clustering of NMDA receptors and Shaker K+ channels in heterologous cells (Kim et al., 1995, 1996). Genetic studies in Drosophila have shown that the PSD-95-related protein Disks Large (DLG) is critical for the synaptic clustering of Shaker-type K+channels (Tejedor et al., 1997). Similarly, PICK1 has been shown to induce clustering of AMPA receptors in heterologous expression systems (Xia et al., 1999). Moreover, overexpression of the C-terminal domain of the GluR2 subunit results in the loss of synaptic AMPA receptor clusters in cultured neurons (Dong et al., 1997). PDZ domain-containing proteins have also been suggested to regulate synaptic signaling pathways. The PSD95/SAP90 family proteins interact with several proteins involved in signal transduction, including SynGAP, a neuronal RasGTPase-activating protein and neuronal nitric oxide synthase (nNOS) (Brenman et al., 1996; Chen et al., 1998; Kim et al., 1998). The targeted deletion of the PSD-95 gene in mice appears to disrupt downstream signaling from the NMDA receptor without affecting the synaptic localization of NMDA receptors (Migaud et al., 1998). GRIP1 and -2 interact with several signaling proteins, including EPH receptors, ephrins, and GRASP1 (GRIP-associated proteins 1), a novel neuronal rasGEF (Ye et al., 2000). Moreover, PICK1 interacts with PKCα. Thus PDZ domain-containing proteins play several functional roles and may serve as adaptor proteins that couple receptors to the synaptic cytoskeleton as well as downstream signal transduction cascades.
Recent studies have demonstrated that AMPA receptor function can be rapidly regulated during several forms of synaptic plasticity including long-term potentiation (LTP) and long-term depression (LTD) (Isaac et al., 1995; Liao et al., 1995; Carroll et al., 1999; Kim and Huganir, 1999; Liao et al., 1999; Shi et al., 1999). These results suggest that AMPA receptor–PDZ domain interactions may be dynamically regulated. We have previously proposed that the serine within the C-terminal PDZ ligand (IESVKI) of the GluR2 and GluR3 subunits may be a potential phosphorylation site to regulate AMPA receptor binding to PDZ domains (Dong et al., 1997). Here we show that phosphorylation of Ser880 within the GluR2 PDZ ligand differentially regulates GluR2 interaction with GRIP1 and PICK1. PKC activation in neurons increases Ser880 phosphorylation and induces internalization of GluR2 subunits. These results suggest that the regulation of GluR2 interaction with PDZ domain-containing proteins by phosphorylation may modulate surface expression of AMPA receptors during synaptic plasticity.
MATERIALS AND METHODS
Generation and characterization of anti-GluR2-pS880.Anti-GluR2-pS880 antibody was raised against the synthetic peptide LVYGIESVKIA corresponding to amino acids 873–883 of GluR2, with phosphoserine included at the Ser-880. Anti-GluR2-pS880 antibodies were affinity-purified from sera by sequential chromatography of the Affi-Gel (Bio-Rad) columns covalently linked to unphosphorylated and Ser880-phosphorylated GluR2 peptides. Antibody characterization was performed on the 3-week-old high-density cortical cultures isolated from 18-d-old embryonic rats (Goslin and Banker, 1991) or the HEK293T cells expressing GluR2 or GluR2 S880A, in which serine-880 is mutated to alanine. The cells were harvested in a hypoosmotic buffer (2 mm HEPES, 5 mm EDTA, 1 μm okadaic acid, 50 mm NaF, 10 mm sodium pyrophosphate, 1 mm sodium orthovanadate) with protease inhibitor cocktail (PIC: 2 μg/ml aprotinin, 1 μg/ml leupeptin, 2 μg/ml antipain, 10 μg/ml benzamide, 1 mm phenylmethylsulfonylfluoride). The homogenates were centrifuged at 14000 × g for 10 min at 4°C. The membrane fraction was resuspended in SDS sample buffer, loaded onto SDS-PAGE gels, transferred to PVDF membrane (Immobilon-P membrane, Millipore, Bedford, MA), and analyzed by immunoblotting with anti-GluR2-pS880 and anti-GluR2 C-terminal or anti-GluR2 N-terminal antibodies (Chemicon, Temecula, CA). To test the specificity of anti-GluR2-pS880, the antibodies were preabsorbed with either unphosphorylated or Ser880-phosphorylated GluR2 peptides at a concentration of 0.2 μg/ml, or PVDF membrane was treated with λ−phosphatase for 30 min before immunoblotting. Immunoprecipitation from rat brain membrane preparations (P2) were performed according to the procedures described by Luo et al. (1997) with modifications as described (Kim et al., 1996). The immunoprecipitates were eluted with SDS-PAGE sample buffer and subjected to immunoblot analysis.
PKC phosphorylation of GluR2. The human embryonic kidney (HEK)293T cells expressing GluR2 and the 3-week-old high-density cortical cultures were treated with either control solution or kinase activators [20 μm forskolin or 0.2–1 μmphorbol 12-myristate 13-acetate (TPA)] for 15 min. The membrane fraction was then analyzed by quantitative immunoblotting with anti-GluR2-pS880 and anti-GluR2 C-terminal or anti-GluR2 N-terminal antibodies (Chemicon). The immunoblots were visualized by enhanced chemifluorescence (ECF) development (Amersham Pharmacia, Arlington Heights, IL) and quantified on a Storm Imaging System and ImageQuant software (Molecular Dynamics). To quantify the relative degree of GluR2 phosphorylation, we calculated the ratio of the intensity of GluR2 labeling with the phosphorylation site-specific antibody over the intensity of labeling with C-terminal phosphorylation-independent GluR2 antibody. Then the phosphorylation ratio of control samples was taken as 100%, and the phosphorylation ratio of TPA- or forskolin-treated samples was normalized to the ratio of control samples to obtain percentage phosphorylation. For in vitro phosphorylation of GluR2 fusion protein, bacterial glutathione S-transferase (GST) fusion proteins containing the last 50 amino acids of GluR2 C terminus or GluR2 mutants (S880A or S863A) were constructed and purified as described previously (Roche et al., 1996). The phosphorylation reaction was performed with 1 μg of fusion protein, 50 μm ATP, and 0.1 μg of PKC for 30 min at 30°C in a 100 μl total volume (10 mm HEPES, pH 7, 10 mmMgCl2, 1 mmCaCl2, 50 mg/ml phosphatidylserine, and 5 mg/ml diolein). Reactions were quenched by the addition of 50 μl of 3 × SDS-PAGE sample buffer, and the fusion proteins were analyzed by immunoblotting with anti-GluR2-pS880 and anti-GluR2C-terminal antibodies.
Transfection of HEK293T cells and interaction studies. The cDNAs subcloned into the vectors pBKCMV (full-length GRIP1), pRK5 (GluR2 or GluR2S880E), or pRK5 with an N-terminal myc-tag (PICK1; amino acids 1–386) were transfected into HEK293T cells using calcium phosphate coprecipitation as described previously (Dong et al., 1997). For in vitro binding studies, the cells transfected with GRIP1 or PICK1 cDNAs were harvested in 1% Triton X-100 in ice-cold immunoprecipitation buffer (25 mm Tris-HCP with 100 mmNaCl, 5 mm EDTA, 5 mm EGTA, 1 μm OKA, 50 mm NaF, 1 mm sodium vanadate, and PIC). The lysates were then incubated with either unphosphorylated or Ser880-phosphorylated GluR2 peptide-conjugated affinity resin at 4°C for 1 hr. The column was prepared by coupling 2 mg/ml peptides with 2 ml of activated AffiGel-10 resin. The bound proteins were washed five times with the same buffer, eluted by SDS sample buffer, and analyzed by immunoblotting with either anti-GRIP1 or anti-PICK1 antibodies. For coimmunoprecipitation of GluR2 with GRIP1 or PICK1, the transfected HEK293T cells were treated for 15 min with either control solution or 1 μm TPA at 37°C and were solubilized in ice-cold immunoprecipitation buffer with 2% Triton X-100. Coimmunoprecipitation was performed using anti-GRIP1 antibody or anti-myc (myc-PICK1) antibody, respectively, according to procedures as described previously (Dong et al., 1997; Xia et al., 1999). For immunocytochemistry, PICK1, Narp, and either GluR2 or mutant GluR2S880E cDNAs were triply transfected into HEK293T cells, which were grown on coverslips coated with 0.2% gelatin. The cells were fixed and triple-stained with anti-N-terminal GluR2, anti-PICK1, and anti-Narp antibodies as described previously (O'Brien et al., 1999; Xia et al., 1999). For triple transfection with GRIP1, Narp, and either GluR2 or mutant GluR2S880E, the cells were stained with anti-N-terminal GluR2, anti-GRIP1, and anti-Narp antibodies.
Yeast cotransformation assay for protein interaction. Yeast assays were performed as described previously (Dong et al., 1997; Xia et al., 1999), using the PJ69 strain harboring HIS3, ADE2, and β-galactosidase as reporter genes. The cDNAs subcloned into the vectors pPC97 (the final 50 amino acids of wild-type GluR2 or mutant GluR2 S880E), pPC86 (PICK1; amino acids 1–386 or GRIP1; PDZ4–6) were transformed into PJ69 cells. The positive clones were selected for their growth in quadruple minus plates (Leu-, Trp-, His-, Ade-) and assayed for β-galactosidase activity with X-gal as a substrate.
Immunocytochemistry in hippocampal neurons and data analysis. Low-density hippocampal cultures from 18-d-old embryonic rats were prepared as described (Goslin and Banker, 1991). Three-week old neurons on coverslips were treated with either control solution or 200 nm TPA for 15 min and then fixed and stained as described previously (Liao et al., 1999). The anti-GluR2-pS880 antibodies were directly conjugated with Cy3TM (Amersham Life Science; red stain). Total GluR2 was visualized using anti-N-terminal GluR2 antibodies (Chemicon). Synapse was visualized by anti-synaptophysin or anti-N-terminal NR1 antibodies. Images of the pyramidal neurons were taken with a digital camera (Princeton Instruments) using identical exposure times to visualize the difference of fluorescence intensity. The images were analyzed with MetaMorph Imaging System (Universal Imaging Co.). Synaptic clusters of Ser880-phosphorylated GluR2 were counted with MetaMorph after setting a threshold of fluorescent intensity that was similar to the dendritic shaft. Only the GRIP1 and PICK1 clusters that colocalized with either synaptophysin or synaptic GluR2 were considered as synaptic and were counted manually. The number of clusters was normalized with the dendritic length after all dendritic branches in an image were manually traced and measured.
Biotinylation. For steady-state biotinylation, the 3-week-old high-density cultured cortical neurons were treated with either control or 1 μm, TPA for 15 min at 37°C to stimulate PKC. Cultures were cooled on ice, washed two times with ice-cold artificial CSF (ACSF) containing (in mm): 125 NaCl, 2.5 KCl, 25 NaHCO3, 1 NaH2PO4, 10 dextrose, 2.5 CaCl2, 1.25 MgCl2, and 5% CO2, and then incubated with ACSF containing 1 mg/ml Sulfo-NHS-LC-Biotin (Pierce Chemical Company, Rockford, IL) for 20 min on ice. Unreacted biotinylation reagent was washed once with ice-cold ACSF and quenched by two successive 20 min washes in ACSF containing 100 mm glycine, followed by two washes in ice-cold TBS (50 mm Tris, pH 7.5, 150 mm NaCl). Cultures were harvested in modified RIPA buffer (1% Triton X-100, 0.5% SDS, 0.5% deoxycholic acid, 50 mm NaPO4, 150 mm NaCl, 2 mm EDTA, 50 mm NaF, 10 mm sodium pyrophosphate, 1 mm sodium orthovanadate, and PIC). The homogenates were centrifuged at 14,000 × g for 15 min at 4°C. The resulting supernatant was incubated with 100 μl of 50% NeutraAvidin agarose (Pierce Chemical Company) for 3 hr at 4°C. After the NeutraAvidin agarose was washed five times with RIPA buffer, bound proteins were eluted with SDS sample buffer by boiling for 15 min. Total protein and isolated biotinylated proteins were analyzed by quantitative immunoblotting with anti-GluR2 C-terminal and anti-NR2B C-terminal antibodies. For the internalization assay, cortical neurons were first labeled with biotin on ice and then treated with 1 μm TPA for 15 min at 37°C to allow internalization. Cultures were cooled on ice, and the biotins on the surface were then stripped with stripping buffer (50 mm glutathione, 75 mm NaCl, and 75 mm NaOH, 10% FBS, pH 8.5–9.0). The neurons were solubilized in RIPA buffer, and the biotinylated GluR2 subunits were analyzed by immunoblotting with anti-GluR2-C-terminal and anti-R2-pS880 antibodies. All of the immunoblots were visualized by ECF development (Amersham Pharmacia) and quantified on a Storm Imaging System (Molecular Dynamics).
Data analysis. All data are reported as mean ± SE. Sample size n refers to the number of images processed in immunocytochemistry and the number of dishes analyzed in biotinylation as well as kinase activation experiments. Group paired ttest for kinase activation experiment was used to test the difference between the control and testing groups, whereas for immunocytochemistry and biotinylation experiments, Student's t test was used (*p < 0.05, **p < 0.01, ***p < 0.001).
RESULTS
The GluR2 subunit C-terminal PDZ ligand is phosphorylated at serine-880 in vivo
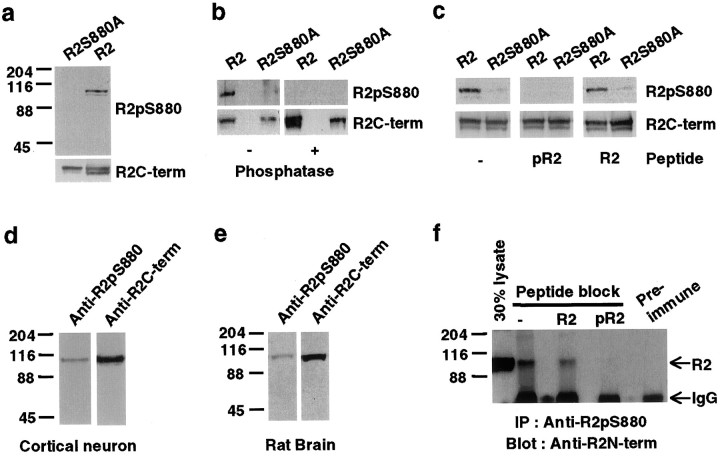
To determine whether the serine residue (Ser880) within the GluR2 PDZ ligand was phosphorylated in vivo, we generated a phosphorylation site-specific antibody against a synthetic phosphopeptide (containing a phosphorylated Ser880) corresponding to the last 10 amino acids of GluR2. The resulting antibody, anti-GluR2-pS880, was characterized in HEK293T cells transfected with wild-type GluR2 or mutant GluR2 (R2S880A), in which Ser880 was mutated to alanine. The anti-GluR2-pS880 antibody detected a single protein of 105 kDa, the predicted molecular weight of the GluR2 subunit, in GluR2-transfected HEK293T cells. Mutation of Ser880 to alanine eliminated immunorecognition of GluR2 by anti-GluR2-pS880, demonstrating the specificity of the antibody (Fig.1a). Dephosphorylation of GluR2 with λ−phosphatase before immunoblotting abolished immunorecognition by the anti-GluR2-pS880 antibody demonstrating the phosphorylation dependence of the antibody (Fig. 1b). In addition, preabsorption of the antibodies before immunoblotting with 0.2 μg/ml of Ser880-phosphorylated GluR2 C-terminal peptide (pR2) blocked the recognition of GluR2 by anti-GluR2-pS880, whereas preabsorption with the unphosphorylated peptide (R2) had no effect (Fig. 1c).
Fig. 1.
The GluR2 subunit C-terminal PDZ ligand is phosphorylated on Ser880. a–c, The phosphorylation site-specific anti-GluR2-pS880 antibody was characterized on immunoblots of HEK293T cells expressing GluR2. The expression of GluR2 was analyzed by using a phosphorylation-independent anti-GluR2-C-terminal antibody (Anti-R2C-term). a, Anti-GluR2-pS880 antibody recognized wild-type GluR2 but not the mutant GluR2 in which serine-880 was mutated to alanine.b, Anti-GluR2-pS880 no longer recognized GluR2 when the PVDF membrane was treated with λ-phosphatase. c, Preabsorption of the anti-GluR2-pS880 antibody with Ser880-phosphorylated R2 peptide (pR2) but not unphosphorylated peptide (R2) blocked GluR2 recognition by anti-GluR2-pS880. d, e, Western Blot for GluR2-pS880 in homogenates of cortical culture neurons (d) and rat brain (e).f, Preabsorption of the anti-GluR2-pS880 antibody with Ser880-phosphorylated R2 peptide (pR2) but not unphosphorylated peptide (R2) blocked immunoprecipitation of GluR2 with the anti-GluR2-pS880.
We examined whether GluR2 was phosphorylated at serine-880 in cultured neurons and rat brain. The anti-GluR2-pS880 antibody recognized a single 105 kDa protein in both cultured cortical neurons and rat brain homogenates (Fig. 1d,e). In addition, GluR2 was immunoprecipitated with the anti-GluR2-pS880 antibodies from detergent extracts of rat brain membranes. Preabsorption of the antibodies with the Ser880-phosphorylated peptide (pR2) but not the unphosphorylated peptide (R2) blocked the immunoprecipitation of GluR2, demonstrating that the anti-GluR2-pS880 antibody immunoprecipitated GluR2 in a phosphorylation-specific manner (Fig.1f). Quantification of immunoprecipitation showed that ∼5% of the total GluR2 in rat brain was phosphorylated at serine-880, whereas ∼30% of the total GluR2 was phosphorylated at serine-880 in 3-week-old cultured cortical neurons (data not shown). These results suggest a significant level of phosphorylation of GluR2 on Ser880 in vivo.
PKC phosphorylates serine-880 of GluR2
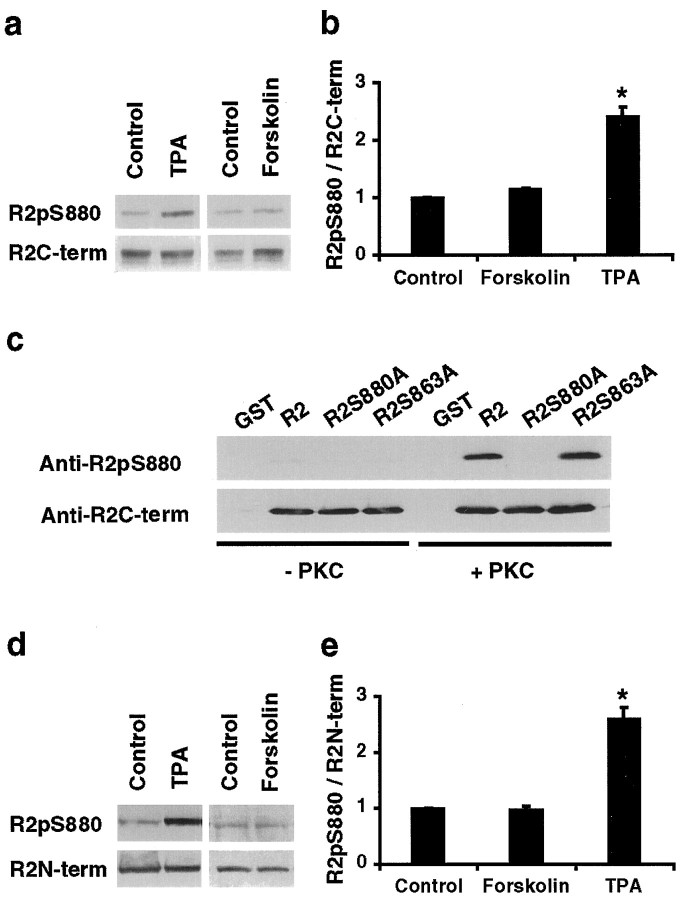
The amino acid sequence surrounding Ser880 (ESVKI) is a PKC consensus site (S/T-X-K/R) indicating that PKC may phosphorylate this site. To determine whether PKC phosphorylates Ser880, we used the phosphorylation site-specific antibody to see whether PKC activators increased Ser880 phosphorylation in GluR2-transfected HEK293T cells. To quantify the relative degree of GluR2 phosphorylation, we calculated the ratio of the intensity of GluR2 labeling with the phosphorylation site-specific antibody over the intensity of labeling with C-terminal phosphorylation-independent GluR2 antibody. Treatment of HEK293T cells with 1 μm phorbol ester (TPA) to activate PKC increased the relative phosphorylation at serine-880 (240 ± 15% of control, n = 4; p < 0.05) (Fig.2a, b). In contrast, no significant change in Ser880 phosphorylation was observed with 20 μm forskolin, a PKA activator (110 ± 2% of control; p > 0.05) (Fig.2a, b).
Fig. 2.
PKC phosphorylates serine-880 of GluR2 in vitro and in vivo. a, Western blot for GluR2-pS880 in transfected HEK293T cells that were treated with control solution (Control), 1 μm phorbol 12-myristate 13-acetate (TPA), or 20 μm forskolin for 15 min. Total amount of GluR2 was detected by anti-GluR2-C-terminal antibody. b, The ratios of intensity of the signal (intensity of anti-GluR2-pS880 antibody labeling/intensity of anti-GluR2 C-terminal antibody labeling) were calculated and normalized to the control cells. PKC stimulation by TPA significantly increases Ser880 phosphorylation of GluR2.c, Immunoblot analysis of the in vitrophosphorylation reaction of purified GST fusion proteins corresponding to the C termini of GluR2, GluR2S880A, and GluR2S863A with purified PKC. PKC directly phosphorylates GluR2 but not GluR2S880A mutant fusion proteins. d, Western blot for GluR2-pS880 in 3-week-old, high-density cortical culture neurons, which were treated with control solution (Control), 200 nm TPA, or 20 μm forskolin for 15 min. Total amount of GluR2 was detected by anti-GluR2-N-terminal antibody (Chemicon).e, The ratios of intensity of the signals (intensity of anti-GluR2-pS880 antibody labeling/intensity of anti-GluR2 N-terminal antibody labeling) were calculated and normalized to the control neurons. PKC stimulation in neurons significantly increases Ser880 phosphorylation of GluR2.
To confirm that serine-880 is directly phosphorylated by PKC, we performed in vitro phosphorylation reactions using purified fusion proteins corresponding to the C terminus of GluR2 and purified rat brain PKC. Immunoblot analysis with anti-GluR2-pS880 demonstrated that the fusion protein of the GluR2 C terminus was phosphorylated at serine-880 by PKC in vitro (Fig. 2c). Mutation of serine-880 to alanine completely eliminated this phosphorylation, whereas the mutation of serine-863, another GluR2 PKC phosphorylation site (B. J. McDonald, A. L. Mammen, H. J. Chung, and R. L. Huganir, unpublished data), had no effect on Ser880 phosphorylation (Fig. 2c). These results demonstrate that PKC directly phosphorylates serine-880 in vitro and provide evidence that PKC directly phosphorylates this site in vivo.
To determine whether PKC can phosphorylate GluR2 on serine-880 in neurons, we examined the modulation of Ser880 phosphorylation in cortical neuronal cultures. GluR2 is basally phosphorylated at serine-880, but the phosphorylation was significantly increased by treatment of neurons with 0.2 μm TPA (260 ± 2% of control, n = 12; p < 0.05) (Fig.2d,e). In contrast, 20 μmforskolin had little effect on phosphorylation of serine-880 (98 ± 4% of control; n = 5, p > 0.05) (Fig. 2d,e). Preabsorption of the anti-GluR2-pS880 antibody with the Ser880-phosphorylated GluR2 peptide, but not with the equivalent unphosphorylated peptides, abolished the antibody recognition of both basal and phorbol ester-stimulated GluR2 (data not shown). These results strongly suggest that PKC phosphorylates serine-880 of GluR2 in neurons.
Phosphorylation of GluR2 PDZ ligand differentially regulates its interaction with GRIP1 and PICK1
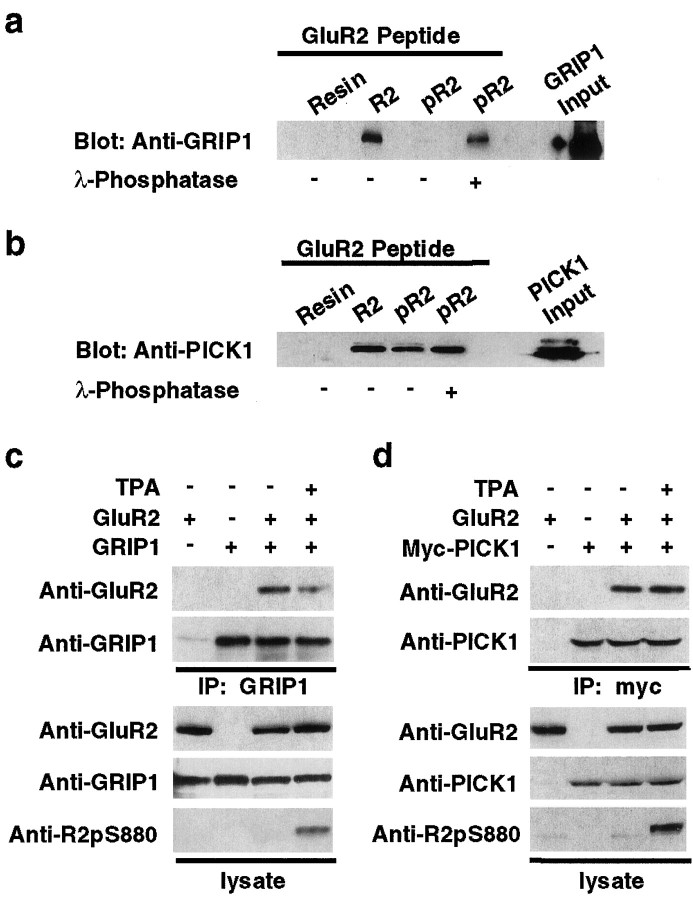
To investigate whether phosphorylation of serine-880 in the GluR2 C-terminal PDZ ligand can regulate the interaction of GluR2 with GRIP1 and PICK1, we analyzed the binding of GRIP1 or PICK1 to Ser880-phosphorylated (pR2) or unphosphorylated GluR2 C-terminal peptides (R2) in vitro. HEK293T cell lysates expressing GRIP1 or PICK1 were incubated with the GluR2 peptides-conjugated affinity columns, and then bound proteins were eluted and analyzed by immunoblot analysis. When the GluR2 C-terminal peptide was phosphorylated at Ser880, GRIP1 no longer interacted with the GluR2 peptide (Fig. 3a). However, GRIP1 interaction with the GluR2 C termini could be recovered when the phosphorylated peptide was dephosphorylated by λ-phosphatase treatment before GRIP1 binding (Fig. 3a). In contrast, PICK1 bound to both the unphosphorylated and phosphorylated GluR2 peptide (Fig. 3b). These results demonstrate that phosphorylation of the GluR2 PDZ ligand differentially regulated binding to the PDZ domains of PICK1 and GRIP1 in vitro.
Fig. 3.
Ser880 phosphorylation of GluR2 C terminus differentially regulates the interaction of GluR2 with GRIP1 and PICK1.a, b, In vitro binding of GRIP1 (a) or PICK1 (b) with Ser880-phosphorylated GluR2 peptides. Extracts of HEK293T cells expressing GRIP1 or PICK1 were incubated with Ser880-phosphorylated R2 peptides (pR2) or unphosphorylated peptides (R2) immobilized on Affigel resins, and bound GRIP1 or PICK1 was detected by immunoblotting. a, GRIP1 did not interact with pR2 peptide, whereas phosphatase treatment of pR2 peptide recovered GRIP1 binding. b, PICK1 binds to both pR2 and R2 peptide. c, d, Coimmunoprecipitation of GluR2 with GRIP1 (c) or PICK1 (d) from transfected HEK293T cells, with or without 1 μm TPA treatment. Increase in Ser880 phosphorylation of GluR2 C terminus by PKC activation attenuated GRIP1 interaction with GluR2 (c) but had no effect on the binding affinity of PICK1 to GluR2 (d).
We then examined whether PKC activation in HEK293T cells could modulate the interaction of GluR2 with the PDZ domains of GRIP1 in vivo. Phorbol ester treatment increased Ser880 phosphorylation of GluR2 and decreased the association of GluR2 with GRIP1 as measured by coimmunoprecipitation of GluR2 with GRIP1 (Fig. 3c). These results suggest that the increase in Ser880 phosphorylation by PKC significantly impaired the interaction between GluR2 and GRIP1. In contrast, coimmunoprecipitation of GluR2 with PICK1 was unaffected by TPA treatment, demonstrating that PICK1 binds to both Ser880-phosphorylated and unphosphorylated GluR2 in vivo(Fig. 3d).
Replacement of serine-880 with glutamate disrupts GluR2 interaction with GRIP1 but not PICK1
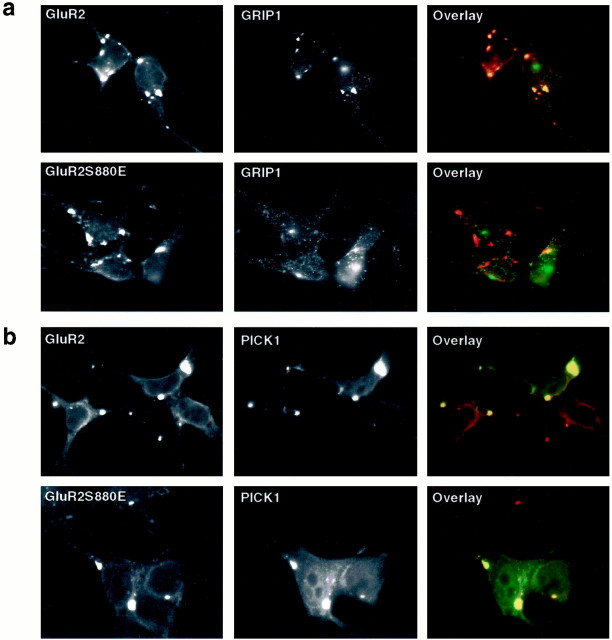
We further investigated the role of Ser880 phosphorylation on the interaction of the GluR2 C terminus with GRIP1 or PICK1 by examining the targeting of GRIP1 or PICK1 to GluR2 clusters in a heterologous expression system. We have shown previously by immunostaining that the extracellular neuronal immediate early gene Narp can induce the clustering of GluR2 subunits into receptor-rich patches when coexpressed in HEK293T cells (O'Brien et al., 1999). Interestingly, triple transfection of HEK293T cells with Narp, GRIP1, and GluR2 resulted in the recruitment of GRIP1 to the Narp-induced GluR2 clusters. However, GRIP1 was not recruited to the Narp-induced mutant GluR2 clusters (GluR2S880E) in which serine-880 was replaced with a glutamate residue to mimic the negative charge of phosphorylated serine-880 (Fig. 4a). Thus, the GluR2 mutant subunit (S880E) appeared not to interact with GRIP1 in HEK293T cells. In contrast, PICK1 was recruited to both the Narp-induced wild-type GluR2 and GluR2S880E clusters, suggesting that PICK1 could interact with both the wild-type and the GluR2S880E mutant, which mimicked the negative charge of Ser880 phosphorylation (Fig.4b). Thus, the replacement of serine-880 with glutamate disrupted the association of GRIP1, but not PICK1, with GluR2 clustersin vivo.
Fig. 4.
S880E mutation of GluR2 disrupts GRIP1 interaction with GluR2 but not PICK1. a, Immunostaining of HEK293T cells that were transfected with Narp, GRIP1, and GluR2 or GluR2S880E mutant. The transfected cells were double-labeled with the mouse anti-GluR2-Nterm (red) and anti-GRIP1 antibodies (green). Narp induces clustering of GluR2, and GRIP1 coclusters with GluR2; however, S880E mutation disrupts GRIP1 coclustering with GluR2. b, Immunostaining of HEK293T cells that were transfected with Narp, PICK1, and GluR2 or GluR2S880E mutant. The transfected cells were double-labeled with the mouse anti-GluR2-Nterm (red) and anti-PICK1 antibodies (green). S880E mutation did not disrupt coclustering of PICK1 and GluR2.
Similarly, in the yeast two-hybrid system, the wild-type GluR2 C terminus interacted well with GRIP1, whereas the GluR2S880E mutant failed to interact. In contrast, the mutant GluR2 S880E C terminus interacted with PICK1 as well as the wild-type GluR2 (data not shown). Moreover, we also performed a yeast two-hybrid screen of a hippocampal cDNA library with the mutant GluR2S880E C terminus as bait to search for other proteins that may bind to the Ser880-phosphorylated GluR2 subunits. Interestingly, under conditions in which we normally isolate many GRIP1 cDNAs using the wild-type GluR2 C terminus as bait, the only positive interacting clone isolated in this nonbiased screen was PICK1 (data not shown). These results demonstrate that the replacement of serine-880 with glutamate (S880E) greatly attenuated the interaction of GluR2 C terminus with GRIP1, but retained its interaction with PICK1 in yeast two-hybrid assays.
Neuronal localization of Ser880-phosphorylated GluR2 subunits
We examined the subcellular localization of Ser880-phosphorylated GluR2 in relation to the distribution of total GluR2 subunit in rat hippocampal neurons using immunocytochemical techniques. The anti-GluR2-pS880 antibodies demonstrated that most of the Ser880-phosphorylated GluR2 was localized in the dendritic shafts as small clusters, with only a low level of immunostaining detected at spiny synapses (Fig. 5a). This is in contrast to the distribution of the total population of GluR2 detected using an N-terminal antibody, which showed high levels of clustered GluR2 (Fig. 5a) that colocalize with synaptic markers on dendritic spines (Liao et al., 1999) (data not shown). A low level of GluR2 N-terminal staining also colocalized with Ser880-phosphorylated GluR2 in the dendritic shafts (Fig.5a). Preabsorption of anti-GluR2-pS880 antibodies with its antigen blocked the immunostaining showing the specificity of the antibody (data not shown). To confirm that Ser880-phosphorylated GluR2 is not localized at synapses, we double-labeled hippocampal neurons with a synaptic marker, anti-synaptophysin monoclonal antibody. No colocalization of the Ser880-phosphorylated GluR2 was observed with synaptophysin (Fig. 5c). In addition, other synaptic proteins, NR1 subunits, did not colocalize with the Ser880-phosphorylated GluR2 subunits at excitatory synapses (Fig.5e). These results suggest that Ser880 phosphorylation may be involved in synaptic targeting of GluR2 subunits.
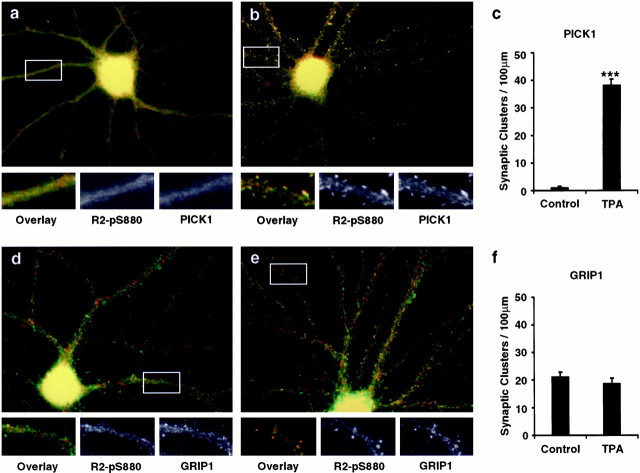
Fig. 5.
Localization of GluR2-pS880 in cultured hippocampal neurons. a, b, Three-week-old low-density hippocampal culture neurons were double-labeled with the rabbit anti-GluR2-pS880 (red) and the mouse anti-GluR2-Nterm antibodies (green).a, In control neurons, GluR2-pS880 colocalizes with N-terminal GluR2 staining in dendrites but not with synaptic GluR2 at synapses. b, In the neurons treated with 200 nm TPA for 15 min, PKC activation dramatically increased the staining of Ser880-phosphorylated GluR2 in the spine heads as well as in the dendrites. c, d, Neurons were double-labeled with the rabbit anti-GluR2-pS880 (red) and the mouse anti-synaptophysin antibodies (green). c, GluR2-pS880 does not colocalize with synaptophysin. d, In the neurons treated with 200 nm TPA for 15 min, PKC activation increases the colocalization of Ser880-phosphorylated GluR2 with synaptophysin at synapses. e, f, Neurons were double-labeled with the rabbit anti-GluR2-pS880 (red) and the mouse anti-NR1 N-terminal antibodies (green). e, GluR2-pS880 does not colocalize with NR1. f, In the neurons treated with 200 nm TPA for 15 min, PKC activation increases the colocalization of Ser880-phosphorylated GluR2 with NR1 at synapses.g, Statistical analysis of the effect of PKC activation on the number of synaptic GluR2-pS880 clusters per 100 μmdendrite. TPA treatment significantly increased synaptic GluR2-pS880 clusters (n = 36 for control andn = 40 for TPA, t test,p < 0.001). However, there was no change in the total number of synaptic GluR2 clusters (n = 15 for control and n = 20 for TPA, t test,p > 0.05) and synaptic NR1 clusters (n = 10 for control and n = 10 for TPA, t test, p > 0.05).
PKC activation increases synaptic Ser880-phosphorylated GluR2 as well as synaptic targeting of PICK1
Interestingly, activation of PKC by treatment of the neurons with 0.2 μm phorbol esters dramatically increased the phosphorylation of GluR2 in the spine heads or in the spine necks, a thread-like extrusion from the dendritic shafts, as well as in the dendritic shafts (Fig. 5b). The appearance of synaptic Ser880-phosphorylated GluR2 subunits was observed as early as 5 min after TPA treatment (data not shown). In addition, double staining of neurons with anti-GluR2-pS880 and anti-synaptophysin or anti-NR1 antibodies showed colocalization of the Ser880-phosphorylated GluR2 subunits with synaptophysin and NR1 at spiny synapses after TPA treatment (Fig. 5d,f). PKC activation dramatically increased the number of synaptic clusters of Ser880-phosphorylated GluR2 [control = 1 ± 0.1 per 100 μm dendrite (n = 36); TPA = 41 ± 1.0 per 100 μm dendrite (n = 40); p < 0.001] (Fig. 5g). In contrast, the total number of GluR2 synaptic clusters, which were detected by the phosphorylation-independent anti-GluR2 N-terminal antibodies, did not change significantly [control = 44 ± 1.5 per 100 μm dendrite (n = 15); TPA = 42 ± 1.6 per 100 μm dendrite (n = 20); p > 0.05] (Fig.5g). Similarly, there was no statistically significant change in the total number of NR1 synaptic clusters [control = 44 ± 2.0 per 100 μm dendrite (n = 10); TPA = 45 ± 3.7 per 100 μm dendrite (n = 10);p > 0.05] (Fig. 5g).
Because Ser880-phosphorylated GluR2 binds to PICK1 but not to GRIP1, we also examined the subcellular localization of phosphorylated GluR2 in relation to the distribution of GRIP1 and PICK1 in rat hippocampal neurons using immunocytochemical techniques. PICK1 colocalized with Ser880-phosphorylated GluR2 in the dendrites, but under these conditions, very little PICK1 was observed at the spiny synapses (Fig.6a). This is inconsistent with our previous results where we saw that PICK1 was localized to synapses (Xia et al., 1999). However, we have recently found that the synaptic localization of PICK1 is quite variable, depending on culture conditions, and is regulated by PKC. As shown in Figure 6b, PKC activation caused a dramatic redistribution of PICK1 with the Ser880-phosphorylated GluR2 subunits to dendritic spines (Fig.6b). Phorbol ester treatment significantly increased the number of synaptic PICK1 clusters [control = 1 ± 0.4 per 100 μm dendrite (n = 16); TPA = 38 ± 2.2 per 100 μm dendrite (n = 15); p < 0.001] (Fig. 6c). These results demonstrate that PKC activation increases the phosphorylation of synaptic GluR2 and induces a redistribution of the level of PICK1 at excitatory synapses.
Fig. 6.
PKC activation dramatically increases the synaptic GluR2-pS880 and PICK1 in cultured hippocampal neurons.a–c, Three-week-old low-density hippocampal culture neurons were double-labeled with anti-PICK1 antibodies (green) and the anti-GluR2-pS880 antibodies (red). a, In control neurons, PICK1 colocalizes with GluR2-pS880 in dendrites. b, In the neurons treated with 200 nm TPA for 15 min, PKC activation increases synaptic PICK1 staining. c, Statistical analysis of the effect of PKC activation on number of synaptic PICK1 clusters per 100 μm dendrite. PKC activation significantly increased the synaptic PICK1 clusters (n = 16 for control and n = 15 for TPA, t test, p < 0.001).d–f, Neurons were double-labeled with anti-GRIP1 antibodies (green) and the anti-GluR2-pS880 antibodies (red). d, In control neurons, GRIP1 does not colocalize well with GluR2-pS880 in both dendrites and spines. e, In the neurons treated with 200 nm TPA for 15 min, no significant change in the synaptic GRIP1 distribution was observed. f, Statistical analysis of the effect of PKC activation on number of synaptic GRIP1 clusters per 100 μm dendrite. PKC activation moderately decreased GRIP1 clusters at synapse (n = 7 for control andn = 8 for TPA, t test,p > 0.05).
Previous studies have shown that GRIP1 colocalizes with total GluR2 at some synapses (Dong et al., 1997, 1999). However, double-labeling neurons with antibodies against GRIP1 and Ser880-phosphorylated GluR2 showed that GRIP1 did not colocalize well with Ser880-phosphorylated GluR2 both in the dendrites and the spiny synapses (Fig.6d). PKC activation had little effect on the subcellular localization of GRIP1, causing colocalization of GRIP1 with some Ser880-phosphorylated GluR2 subunits at spiny synapses (Fig.6e). Quantification shows a small but not significant decrease in the number of synaptic GRIP1 clusters after PKC activation [control = 21 ± 1.6 per 100 μm dendrite (n = 7); TPA = 18 ± 1.8 per 100 μm dendrite (n = 8); p > 0.05] (Fig.6f).
PKC activation induces internalization of GluR2
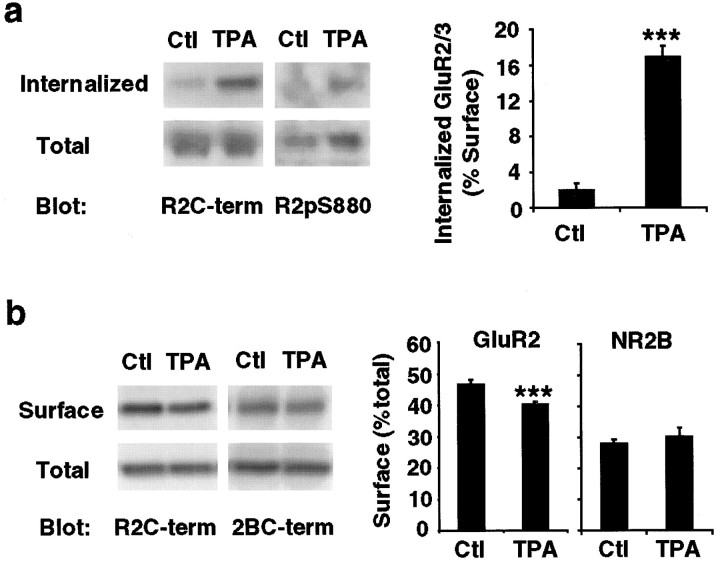
The Ser880 phosphorylation of synaptic GluR2 as well as PICK1 mobilization to synapses after PKC activation may be involved in surface membrane trafficking of AMPA receptors. To test this hypothesis, we measured the internalized surface GluR2 subunits after TPA treatment in cortical neuron cultures using a reversible surface biotinylation technique. Neurons were first labeled with biotin at 4°C, followed by the TPA treatment for 15 min at 37°C to allow membrane trafficking. Neurons were put back at 4°C to stop membrane trafficking, and the biotin remaining on the surface was cleaved by treatment with the reducing agent glutathione. At this step, only the internalized surface proteins were protected from the reducing agent and remained biotinylated. The biotinylated internalized surface molecules were then purified on avidin-conjugated resin and analyzed by quantitative immunoblot analysis using anti-GluR2-C-terminal antibodies. The amount of the surface GluR2, which was internalized after PKC stimulation, was quantified by comparing the amounts of biotinylated protein before and after glutathione reduction and presented as the percentage of the amount of total surface GluR2. In this analysis, we found that PKC activation dramatically increased the level of internalized surface GluR2 subunits (control = 2.0 ± 0.7%, n = 8; TPA = 17.0 ± 1.1%,n = 8, t test: p < 0.001) (Fig. 7a). In addition, the immunoblot analysis using anti-GluR2-pS880 antibody demonstrated that the internalized GluR2 subunits after TPA stimulation were phosphorylated at serine-880. These results suggest that GluR2 phosphorylation at serine-880 may regulate the internalization rate of GluR2.
Fig. 7.
PKC activation induces internalization of GluR2 in cultured cortical neurons. a, Internalization assay was performed in 3-week-old high-density cultured cortical neurons to examine the internalized GluR2 subunits after TPA treatment. The biotinylated, internalized GluR2 subunits were analyzed by immunoblotting with anti-GluR2-C-terminal and anti-GluR2-pS880 antibodies. The internalized GluR2 subunits were quantified as the percentage of total surface GluR2 subunits (n = 8 for both control and TPA, t test; p< 0.001). PKC activation induces rapid internalization of surface GluR2 subunits. b, Surface biotinylation was performed in cortical neurons after TPA treatment to examine the steady-state level of total surface GluR2 subunits. The biotinylated GluR2 subunits were analyzed by immunoblotting with anti-GluR2-C-terminal antibody. The steady-state level of surface GluR2 subunits was quantified as the percentage of total GluR2 subunits (n = 8 for both control and TPA, t test; p < 0.001). PKC stimulation results in a decrease in total level of surface GluR2 subunits but not surface NR2B subunits of NMDA receptors (n = 8 for both control and TPA, ttest; p > 0.05).
To test whether PKC activation decreased the surface expression of GluR2 subunits, we measured the steady-state level of surface GluR2 subunits after PKC activation in cortical neuron cultures using a surface biotinylation technique. After treating neurons with control solution or TPA for 15 min, the surface molecules were labeled with biotin at 4°C, purified on avidin-conjugated resin, and analyzed by quantitative immunoblot analysis using anti-GluR2-C-terminal and anti-NR2B-C-terminal antibodies. The amount of the surface protein was quantified by comparing the amount of biotinylated protein with the total protein amount and presented as the percentage of the total protein. PKC stimulation caused a small yet statistically significant decrease in the level of total surface GluR2 subunits (control = 47 ± 1.2%, n = 8; TPA = 40 ± 0.6%,n = 8, t test: p < 0.001) (Fig. 7b). In contrast, the surface level of NR2B subunits was not affected by TPA treatment, demonstrating that TPA-induced internalization is specific to GluR2 subunits (control = 28 ± 1.3%, n = 8; TPA = 30 ± 2.6%,n = 8, t test: p > 0.05) (Fig. 7b). These results suggest that an increase in GluR2 phosphorylation at serine-880 after PKC activation may be associated with internalization of surface GluR2 subunits.
DISCUSSION
PDZ domains are ubiquitous protein modules that mediate protein interactions through their binding to the C termini of target proteins (Sheng and Kim, 1996; Kim and Huganir, 1999). The postsynaptic density, a prominent structure at excitatory synapses, contains several PDZ domain-containing proteins that appear to be important for the synaptic targeting and signal transduction of glutamate receptors (Sheng and Kim, 1996; Kim and Huganir, 1999). Glutamate receptor function at synapses has recently been shown to be rapidly regulated, suggesting that dynamic regulatory mechanisms may exist to modulate the association of glutamate receptors with synaptic proteins (Isaac et al., 1995; Liao et al., 1995, 1999; Carroll et al., 1999; Shi et al., 1999). The C-terminal intracellular region of glutamate receptors contains the major domains for interaction with intracellular proteins but also contains the major sites of protein phosphorylation that have been shown to modulate ion channel properties (McGlade-McCulloh et al., 1993; Wang et al., 1994; Roche et al., 1996; Mammen et al., 1997). In this study, we investigated whether protein phosphorylation of AMPA receptor subunits may also regulate their interaction with PDZ domain-containing proteins. We demonstrated that the PDZ domain interacting region of GluR2 is phosphorylated at serine-880 in vivo. We also showed that serine-880 is phosphorylated by PKCin vitro and that this Ser880 phosphorylation is increasedin vivo by phorbol ester treatment. These results suggest that PKC phosphorylates this site in vivo, although we cannot exclude the possibility that other downstream kinases activated by PKC stimulation may phosphorylate this site in vivo.
Phosphorylation of Ser880 of GluR2 differentially regulates the interaction of the GluR2 C terminus with the PDZ domains of GRIP1 and PICK1. Interestingly, phosphorylation of Ser880 disrupts GluR2 binding to GRIP1, whereas both phosphorylated and unphosphorylated GluR2 can interact with PICK1. While this work was in progress, Matsuda et al. (1999) also found that GluR2 is phosphorylated on Ser880 by PKC and that this phosphorylation inhibits its interaction with GRIP1 (Matsuda et al., 1999). The differential effect of GluR2 phosphorylation on binding to GRIP1 and PICK1 may arise from the different structure of the GRIP1 and PICK1 PDZ domains. The PDZ domain of GRIP1 that interacts with GluR2 (PDZ5) is a type2 PDZ domain, which selectively binds to ligands with hydrophobic amino acids at the -2 residue such as the GluR2 C terminus (ES VKI) (Dong et al., 1997; Songyang et al., 1997). In contrast, the PDZ domain of PICK1 is a type 1b PDZ domain, which binds to C-terminal PDZ ligands with a typical T/SXV motif, such as occurs in the C termini of PKCα, in which a hydroxyl group at position -2 is important for binding to PDZ domains (Songyang et al., 1997; Staudinger et al., 1997). PICK1, however, also binds to the C termini of GluR2, EPH receptors and the ephrins, which are type 2 PDZ ligands (Torres et al., 1998; Xia et al., 1999). These results demonstrate that the PICK1 PDZ domain is less selective than PDZ5 in GRIP1 and may account for the binding of PICK1 to GluR2 regardless of the state of Ser880 phosphorylation.
Immunostaining of cultured hippocampal neurons with the Ser880 phosphorylation-specific antibody showed that under control conditions Ser880-phosphorylated GluR2 was enriched and colocalized with PICK1 in dendritic shafts with very little staining observed at synapses containing nonphosphorylated GluR2. This is different from our previous results where we saw some synaptic localization of PICK1 in control neurons (Xia et al., 1999). The variability in synaptic PICK1 staining may be attributable to culture conditions and may reflect PKC activation in the neurons. Because our data suggest that synaptic localization of PICK1 is dynamic and regulated by PKC, both conditions (some or no synaptic localization of PICK1 in control neurons) may coexist in vivo. On the contrary, GRIP1 did not colocalize well with phosphorylated GluR2 and was found colocalized with total GluR2 at some spiny synapses. Interestingly, treatment of neurons with PKC activators dramatically increased the Ser880 phosphorylation of GluR2, both in the dendrites and at synapses, as well as synaptic targeting of PICK1. In contrast, the number of GRIP1 synaptic clusters decreased slightly. These observations are consistent with the differential interaction of GRIP1 and PICK1 with the phosphorylated and unphosphorylated GluR2 and suggest that Ser880 phosphorylation of GluR2 may regulate the molecular composition of synapses. No significant change in GRIP1 distribution after PKC activation suggests that GRIP1 may be securely anchored to the postsynaptic density by interacting with synaptic cytoskeletal proteins through its multiple PDZ domains.
Recent data have indicated that the interaction of the C termini of GluR2 with PDZ domain-containing proteins is important for synaptic targeting of AMPA receptors. Overexpression of the GluR2 C-terminal PDZ ligand inhibits the synaptic clustering of AMPA receptors in cultured neurons, indicating that PDZ domain-mediated interactions of GluR2 are critical for synaptic targeting of AMPA receptors (Dong et al., 1997). These data suggested that the modulation of the interaction of GluR2 with GRIP1 and PICK1 by phosphorylation might regulate the membrane trafficking of AMPA receptors. In fact, activation of PKC in cortical culture neurons increases Ser880 phosphorylation of GluR2 and induces internalization of the surface GluR2 subunits, resulting in the reduction of the total level of surface GluR2. Interestingly, PKC activation also induced dramatic mobilization of PICK1 to synapse, suggesting that PICK1 interaction with Ser880-phosphorylated GluR2 may be involved in the internalization of AMPA receptors.
Changes in the synaptic localization of AMPA receptors have recently been suggested to be important in many forms of synaptic plasticity, including LTP and LTD (Carroll et al., 1999; Kim and Huganir, 1999;Liao et al., 1999; Shi et al., 1999). Induction of LTP in hippocampal slices has been shown to induce a rapid redistribution of AMPA receptors to synaptic spines (Shi et al., 1999), whereas LTD induction in neuronal cultures decreases the surface expression of AMPA receptors (Carroll et al., 1999). There is now extensive evidence that PKC activation is required for LTD in cerebellar Purkinje cells (Linden, 1994; De Zeeuw et al., 1998), and recent studies have provided evidence that cerebellar LTD may be mediated by AMPA receptor endocytosis (Wang and Linden, 2000). In addition, metabotropic glutamate receptor-dependent LTD in the CA1 region of the hippocampus (Stanton et al., 1991; Bolshakov and Siegelbaum, 1994; Yang et al., 1994) has been shown to require PKC activation (Oliet et al., 1997; Otani and Connor, 1998; Wang et al., 1998). Our results suggest that GluR2 phosphorylation at serine-880 may regulate the internalization of AMPA receptors during several forms of synaptic plasticity by modulating GluR2 interaction with PDZ domain-containing proteins.
The finding that PKC activation regulates GluR2 internalization is surprising because most previous studies have suggested that PKC potentiates AMPA receptor function possibly through the phosphorylation of Ser831 on the GluR1 subunit (Roche et al., 1996; Carroll et al., 1998). In addition, PKC activation has recently been shown to be required for 5HT-induced activation of silent spinal sensory synapses by a mechanism dependent on GluR2 interaction with GRIP (Li et al., 1999). However, as mentioned above, PKC activation appears to regulate AMPA receptor internalization during LTD in cerebellar Purkinje cells. The differences between these various systems is not clear; however, it is possible that the subunit composition of AMPA receptors and the expression level of specific receptor-associated proteins at the synapses in these systems may be important in this differential regulation of AMPA receptor function. For example, in sensory neurons the interaction between GluR2 and GRIP, but not PICK1, is thought to be involved in synaptic activation, whereas our results suggest that the interaction of GluR2 and PICK1 in cortical neurons is involved in receptor internalization. In addition, in cerebellar Purkinje cells, AMPA receptors consist of GluR2/3 heteromers and do not contain significant levels of the GluR1 subunit that may be required for the AMPA receptor potentiation by PKC. The molecular mechanisms by which PKC affects AMPA receptor function is obviously very complex, and further experiments will be necessary to dissect the interaction of these processes.
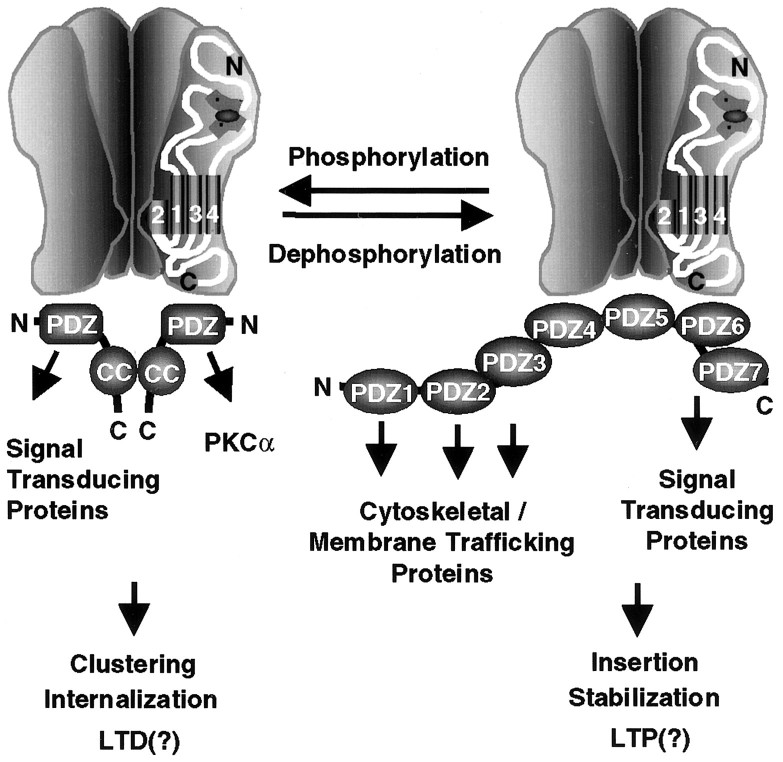
Finally, the differential association of the AMPA receptor with GRIP1 and PICK1 may also regulate the coupling of the receptor to distinct protein complexes involved in signal transduction (Kim and Huganir, 1999). GRIP1 contains seven PDZ domains, each of which interacts independently with various proteins, including the EPH receptor tyrosine kinases and their ligands the ephrins (Kim and Huganir, 1999) as well as GRASP1 (GRIP-associated protein 1), a novel neuronal rasGEF (Ye et al., 2000). Although PICK1 contains only a single PDZ domain, it can multimerize and link AMPA receptors to other proteins, including PKCα (Kim and Huganir, 1999). Thus phosphorylation of Ser880 may regulate the formation of AMPA receptor signal transduction complexes at excitatory synapses in addition to regulating AMPA receptor membrane trafficking (Fig. 8).
Fig. 8.
Model for modulation of the macromolecular AMPA receptor protein complex by phosphorylation of GluR2 PDZ ligand. PKC phosphorylation of Ser880 in GluR2 C-terminal PDZ ligand differentially regulates its interaction with PDZ domains of GRIP and PICK1. This differential regulation of PDZ domain-mediated interaction may modulate surface expression of AMPA receptors as well as the composition of the AMPA receptor complex and the subsequent downstream signaling cascades at excitatory synapses.
Footnotes
This work was supported by the Howard Hughes Medical Institute and the National Institutes of Health (R.L.H.). We thank C. Doherty and J. Bernhardt for technical support.
Correspondence should be addressed to Dr. Richard L. Huganir, Department of Neuroscience, Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, 904A PCTB, 725 N. Wolfe Street, Baltimore, MD 21205. E-mail: rhuganir@jhmi.edu.
REFERENCES
- 1.Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 2.Bolshakov VY, Siegelbaum SA. Postsynaptic induction and presynaptic expression of hippocampal long-term depression. Science. 1994;264:1148–1152. doi: 10.1126/science.7909958. [DOI] [PubMed] [Google Scholar]
- 3.Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, Froehner SC, Bredt DS. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 4.Carroll RC, Nicoll RA, Malenka RC. Effects of PKA and PKC on miniature excitatory postsynaptic currents in CA1 pyramidal cells. J Neurophysiol. 1998;80:2797–2800. doi: 10.1152/jn.1998.80.5.2797. [DOI] [PubMed] [Google Scholar]
- 5.Carroll RC, Lissin DV, von Zastrow M, Nicoll RA, Malenka RC. Rapid redistribution of glutamate receptors contributes to long-term depression in hippocampal cultures. Nat Neurosci. 1999;2:454–460. doi: 10.1038/8123. [DOI] [PubMed] [Google Scholar]
- 6.Chen HJ, Rojas-Soto M, Oguni A, Kennedy MB. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron. 1998;20:895–904. doi: 10.1016/s0896-6273(00)80471-7. [DOI] [PubMed] [Google Scholar]
- 7.Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 8.Choi DW. Glutamate neurotoxicity and diseases of the nervous system. Neuron. 1988;1:623–634. doi: 10.1016/0896-6273(88)90162-6. [DOI] [PubMed] [Google Scholar]
- 9.De Zeeuw CI, Hansel C, Bian F, Koekkoek SK, van Alphen AM, Linden DJ, Oberdick J. Expression of a protein kinase C inhibitor in Purkinje cells blocks cerebellar LTD and adaptation of the vestibulo-ocular reflex. Neuron. 1998;20:495–508. doi: 10.1016/s0896-6273(00)80990-3. [DOI] [PubMed] [Google Scholar]
- 10.Dong H, O'Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature. 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 11.Dong H, Zhang P, Song I, Petralia RS, Liao D, Huganir RL. Characterization of the glutamate receptor-interacting proteins GRIP1 and GRIP2. J Neurosci. 1999;19:6930–6941. doi: 10.1523/JNEUROSCI.19-16-06930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goslin K, Banker G. Culturing nerve cells. MIT; London: 1991. [Google Scholar]
- 13.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 14.Isaac JT, Nicoll RA, Malenka RC. Evidence for silent synapses: implications for the expression of LTP. Neuron. 1995;15:427–434. doi: 10.1016/0896-6273(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy MB. Origin of PDZ (DHR, GLGF) domains. Trends Biochem Sci. 1995;20:350. doi: 10.1016/s0968-0004(00)89074-x. [DOI] [PubMed] [Google Scholar]
- 16.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 17.Kim E, Cho KO, Rothschild A, Sheng M. Heteromultimerization and NMDA receptor-clustering activity of Chapsyn-110, a member of the PSD-95 family of proteins. Neuron. 1996;17:103–113. doi: 10.1016/s0896-6273(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 18.Kim JH, Huganir RL. Organization and regulation of proteins at synapses. Curr Opin Cell Biol. 1999;11:248–254. doi: 10.1016/s0955-0674(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 19.Kim JH, Liao D, Lau LF, Huganir RL. SynGAP: a synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron. 1998;20:683–691. doi: 10.1016/s0896-6273(00)81008-9. [DOI] [PubMed] [Google Scholar]
- 20.Kistner U, Wenzel BM, Veh RW, Cases-Langhoff C, Garner AM, Appeltauer U, Voss B, Gundelfinger ED, Garner CC. SAP90, a rat presynaptic protein related to the product of the Drosophila tumor suppressor gene dlg-A. J Biol Chem. 1993;268:4580–4583. [PubMed] [Google Scholar]
- 21.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 22.Li P, Kerchner GA, Sala C, Wei F, Huettner JE, Sheng M, Zhuo M. AMPA receptor-PDZ interactions in facilitation of spinal sensory synapses. Nat Neurosci. 1999;2:972–977. doi: 10.1038/14771. [DOI] [PubMed] [Google Scholar]
- 23.Liao D, Hessler NA, Malinow R. Activation of postsynaptically silent synapses during pairing-induced LTP in CA1 region of hippocampal slice. Nature. 1995;375:400–404. doi: 10.1038/375400a0. [DOI] [PubMed] [Google Scholar]
- 24.Liao D, Zhang X, O'Brien R, Ehlers MD, Huganir RL. Morphological detection of postsynaptic silent synapses in developing hippocampal neurons. Nature Neuroscience. 1999;2:37–43. doi: 10.1038/4540. [DOI] [PubMed] [Google Scholar]
- 25.Linden DJ. Long-term synaptic depression in the mammalian brain. Neuron. 1994;12:457–472. doi: 10.1016/0896-6273(94)90205-4. [DOI] [PubMed] [Google Scholar]
- 26.Luo J, Wang Y, Yasuda RP, Dunah AW, Wolfe BW. The majority of N-methyl-d-aspartate receptor complexes in adult rat cerebral cortex contain at least three different subunits (NR1/NR2A/NR2B). Mol Pharmacol. 1997;51:79–86. doi: 10.1124/mol.51.1.79. [DOI] [PubMed] [Google Scholar]
- 27.Mammen AL, Kameyama K, Roche KW, Huganir RL. Phosphorylation of the alpha-amino-3-hydroxy-5-methylisoxazole4-propionic acid receptor GluR1 subunit by calcium/calmodulin-dependent kinase II. J Biol Chem. 1997;272:32528–32533. doi: 10.1074/jbc.272.51.32528. [DOI] [PubMed] [Google Scholar]
- 28.Matsuda S, Mikawa S, Hirai H. Phosphorylation of serine-880 in GluR2 by protein kinase C prevents its C terminus from binding with glutamate receptor-interacting protein. J Neurochem. 1999;73:1765–1768. doi: 10.1046/j.1471-4159.1999.731765.x. [DOI] [PubMed] [Google Scholar]
- 29.McGlade-McCulloh E, Yamamoto H, Tan S-E, Brickey DA, Soderling TR. Phosphorylation and regulation of glutamate receptors by calcium/calmodulin-dependent protein kinase II. Nature. 1993;362:640–642. doi: 10.1038/362640a0. [DOI] [PubMed] [Google Scholar]
- 30.Migaud M, Charlesworth P, Dempster M, Webster LC, Watabe AM, Makhinson M, He Y, Ramsay MF, Morris RG, Morrison JH, O'Dell TJ, Grant SG. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature. 1998;396:433–439. doi: 10.1038/24790. [DOI] [PubMed] [Google Scholar]
- 31.O'Brien RJ, Xu D, Petralia RS, Steward O, Huganir RL, Worley P. Synaptic clustering of AMPA receptors by the extracellular immediate gene product NARP. Neuron. 1999;23:309–323. doi: 10.1016/s0896-6273(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 32.Oliet SH, Malenka RC, Nicoll RA. Two distinct forms of long-term depression coexist in CA1 hippocampal pyramidal cells. Neuron. 1997;18:969–982. doi: 10.1016/s0896-6273(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 33.Otani S, Connor JA. Requirement of rapid Ca2+ entry and synaptic activation of metabotropic glutamate receptors for the induction of long-term depression in adult rat hippocampus. J Physiol (Lond) 1998;511:761–770. doi: 10.1111/j.1469-7793.1998.761bg.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roche KW, O'Brien RJ, Mammen AL, Bernhardt J, Huganir RL. Characterization of multiple phosphorylation sites on the AMPA receptor GluR1 subunit. Neuron. 1996;16:1179–1188. doi: 10.1016/s0896-6273(00)80144-0. [DOI] [PubMed] [Google Scholar]
- 35.Seeburg PH. The TINS/TiPS Lecture. The molecular biology of mammalian glutamate receptor channels. Trends Neurosci. 1993;16:359–365. doi: 10.1016/0166-2236(93)90093-2. [DOI] [PubMed] [Google Scholar]
- 36.Sheng M, Kim E. Ion channel associated proteins. Curr Opin Neurobiol. 1996;6:602–608. doi: 10.1016/s0959-4388(96)80091-2. [DOI] [PubMed] [Google Scholar]
- 37.Shi SH, Hayashi Y, Petralia RS, Zaman SH, Wenthold RJ, Svoboda K, Malinow R. Rapid spine delivery and redistribution of AMPA receptors after synaptic NMDA receptor activation. Science. 1999;284:1811–1816. doi: 10.1126/science.284.5421.1811. [DOI] [PubMed] [Google Scholar]
- 38.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 39.Srivastava S, Osten P, Vilim FS, Khatri L, Inman G, States B, Daly C, DeSouza S, Abagyan R, Valtschanoff JG, Weinberg RJ, Ziff EB. Novel anchorage of GluR2/3 to the postsynaptic density by the AMPA receptor-binding protein ABP. Neuron. 1998;21:581–591. doi: 10.1016/s0896-6273(00)80568-1. [DOI] [PubMed] [Google Scholar]
- 40.Stanton PK, Chattarji S, Sejnowski TJ. 2-Amino-3-phosphono-propionic acid, an inhibitor of glutamate-stimulated phosphoinositide turnover, blocks induction of homosynaptic long-term depression, but not potentiation, in rat hippocampus. Neurosci Lett. 1991;127:61–66. doi: 10.1016/0304-3940(91)90895-z. [DOI] [PubMed] [Google Scholar]
- 41.Staudinger J, Lu J, Olson EN. Specific interaction of the PDZ domain protein PICK1 with the COOH terminus of protein kinase C-alpha. J Biol Chem. 1997;272:32019–32024. doi: 10.1074/jbc.272.51.32019. [DOI] [PubMed] [Google Scholar]
- 42.Tejedor FJ, Bokhari A, Rogero O, Gorczyca M, Zhang J, Kim E, Sheng M, Budnik V. Essential role for dlg in synaptic clustering of Shaker K+ channels in vivo. J Neurosci. 1997;17:152–159. doi: 10.1523/JNEUROSCI.17-01-00152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torres R, Firestein BL, Dong H, Staudinger J, Olson EN, Huganir RL, Bredt DS, Gale NW, Yancopoulos GD. PDZ proteins bind, cluster, and synaptically colocalize with Eph receptors and their ephrin ligands. Neuron. 1998;21:1453–1463. doi: 10.1016/s0896-6273(00)80663-7. [DOI] [PubMed] [Google Scholar]
- 44.Wang L-Y, Dudek EM, Browning MD, MacDonald JF. Modulation of AMPA/kainate receptors in cultured murine hippocampal neurones by protein kinase C. J Physiol (Lond) 1994;475:431–437. doi: 10.1113/jphysiol.1994.sp020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Wu J, Rowan MJ, Anwyl R. Role of protein kinase C in the induction of homosynaptic long-term depression by brief low frequency stimulation in the dentate gyrus of the rat hippocampus in vitro. J Physiol (Lond) 1998;513:467–475. doi: 10.1111/j.1469-7793.1998.467bb.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang YT, Linden DJ. Expression of cerebellar long-term depression requires postsynaptic clathrin-mediated endocytosis. Neuron. 2000;25:635–647. doi: 10.1016/s0896-6273(00)81066-1. [DOI] [PubMed] [Google Scholar]
- 47.Woods DF, Bryant PJ. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 48.Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 49.Yang XD, Connor JA, Faber DS. Weak excitation and simultaneous inhibition induce long-term depression in hippocampal CA1 neurons. J Neurophysiol. 1994;71:1586–1590. doi: 10.1152/jn.1994.71.4.1586. [DOI] [PubMed] [Google Scholar]
- 50.Ye B, Liao D, Zhang X, Zhang P, Dong H, Huganir RL. GRASPI: a neuronal rasGEF associated with the AMPA receptor/GRIP complex. Neuron. 2000;26:603–617. doi: 10.1016/s0896-6273(00)81198-8. [DOI] [PubMed] [Google Scholar]