Abstract
The expression of α1-antichymotrypsin (ACT) is significantly enhanced in affected brain regions in Alzheimer's disease. This serine proteinase inhibitor specifically colocalizes with filamentous β-amyloid deposits and recently has been shown to influence both formation and destabilization of β-amyloid fibrils. In the brain, ACT is expressed in astrocytes, and interleukin-1 (IL-1), tumor necrosis factor α (TNF), oncostatin M (OSM), and IL-6/soluble IL-6 receptor complexes control synthesis of this inhibitor. Here, we characterize a molecular mechanism responsible for both IL-1 and TNF-induced expression of ACT gene in astrocytes. We identify the 5′ distal IL-1/TNF-responsive enhancer of the ACT gene located 13 kb upstream of the transcription start site. This 413-bp-long enhancer contains three elements, two of which bind nuclear factor kB (NF-kB) and one that binds activating protein 1 (AP-1). All of these elements contribute to the full responsiveness of the ACT gene to both cytokines, as determined by deletion and mutational analysis. The 5′ NF-kB high-affinity binding site and AP-1 element contribute most to the enhancement of gene transcription in response to TNF and IL-1. In addition, we demonstrate that the 5′ untranslated region of the ACT mRNA does not contribute to cytokine-mediated activation. Finally, we find that overexpression of the NF-kB inhibitor (IkB) totally inhibits any activation mediated by the newly identified IL-1/TNF enhancer of the ACT gene.
Keywords: α1-antichymotrypsin, Alzheimer's disease, IL-1, TNF, regulation, transcription, enhancer, NF-kB, AP-1
Alzheimer's disease (AD), the most common degenerative disorder of the CNS, is characterized by cerebral degeneration, neuronal cell death, and accumulation of deposits in the affected areas of brain (Selkoe, 1991). These filamentous deposits contain polymers of a 40–42 amino acid β-amyloid peptide (Aβ) released by action of specific proteinases (Selkoe, 1991; Sinha et al., 1999) from a transmembrane protein, referred to as β-amyloid precursor protein (APP). However, deposition of Aβ by itself is not sufficient to produce the AD clinical syndrome. Among the additional mechanisms implicated in the development of AD, inflammation-related factors are involved in a number of key steps in the pathological cascade, especially in the formation of neuritic plaques (Eikelenboom et al., 1994). Initially, immunohistological studies have shown the presence of complement factors in neuritic plaques. Complement activation, in turn, is believed to participate in the activation of microglial cells and the stimulation of synthesis of pro-inflammatory cytokines, including interleukin-1 (IL-1), tumor necrosis factor (TNF), and IL-6. This may initiate the beginning of a vicious cycle leading to further amplification of Aβ deposition, because pro-inflammatory cytokines have been shown to directly upregulate APP expression (Donnelly et al., 1990) as well as indirectly influence the balance between Aβ and so-called Aβ-associated proteins, including α1-antichymotrypsin (ACT) (Abraham et al., 1988; Snow et al., 1988; Namba et al., 1991).
ACT is a member of the serine proteinase inhibitor (serpin) family (Potempa et al., 1994). In vitro, it has been shown to either stimulate formation or destabilize already preformed fibrillar forms of Aβ, depending on the ratio of ACT and Aβ (Fraser et al., 1993; Ma et al., 1994). Recently, it has been demonstrated that Aβ inserts into two β sheets of ACT, which apparently leads to transformation of the latter protein from inhibitor to substrate (Janciauskiene et al., 1998). Thus, this interaction could result in lower levels of functional inhibitor, leading to uncontrolled proteolysis by an enzyme normally inhibited by ACT. Although the identity of a hypothetical target proteinase(s), normally inhibited by ACT in the brain, is still not known, this serpin has recently been shown to inhibit degradation of Aβ (Yamin et al., 1999).
Astrocytes have been shown to be the major source of ACT in affected brain regions in AD (Abraham et al., 1988). For this reason, a state of cerebral “acute phase,” similar to that found in liver, as a response to neuronal degeneration and accumulation of deposits has been proposed (Vandenabeele and Fiers, 1991). Proinflammatory cytokines from the IL-1 and IL-6 families have been suggested to mediate this response and upregulate expression of the ACT gene. In fact, IL-1, TNF, and recently, OSM have been shown to regulate ACT expression in astrocytes, whereas IL-6 was ineffective because of the lack of functional IL-6 receptors (Das and Potter, 1995; Kordula et al., 1998). In addition, regulatory elements that mediate responses to OSM have been identified in the promoter region of the ACT gene (Kordula et al., 1998). However, the mechanisms of IL-1- and TNF-induced activation of this gene have so far remained unclear, and an understanding of these processes is a prerequisite to any attempt to control ACT expression as an approach to future therapy.
In this report we characterize a molecular mechanism responsible for upregulation of ACT expression in primary human astrocytes by IL-1 and TNF.
MATERIALS AND METHODS
Cell culture. Human cortical astrocyte cultures were established exactly as described previously (Kordula et al., 1998). Cells were cultured in DMEM supplemented with 10% fetal calf serum, antibiotics, sodium pyruvate, and nonessential amino acids.
Cytokines and cell stimulation. Cells were stimulated with 25 ng/ml OSM (R&D Systems, Minneapolis, MN), 5 ng/ml IL-1 (a gift from Immunex Corp., Seattle, WA), or 10 ng/ml TNFα (a gift of Cutter Laboratories, Berkeley, CA). Dexamethasone (Dex) (1 μm; Sigma, St. Louis, MO) was also added to enhance cytokine action.
RNA preparation and Northern blot analysis. Total RNA was prepared using the phenol extraction method (Rose-John et al., 1988). Samples of RNA (5 μg) were subjected to formaldehyde gel electrophoresis using standard procedures (Sambrook et al., 1989) and transferred to Hybond-N membranes (Pharmacia Biotech, Little Chalfont, UK) according to the manufacturer's instructions. The filters were prehybridized at 68°C for 3 hr in 10% dextran sulfate, 1m sodium chloride, and 1% SDS and hybridized in the same solution with a 1.4 kb EcoRI–EcoRI fragment of ACT cDNA (a gift of Dr. H. Rubin, University of Pennsylvania) labeled by random priming (Feinberg and Vogelstein, 1983). After the hybridization, nonspecifically bound radioactivity was removed by washing in 2× SSC at room temperature, followed by two washes in 2× SSC and 1% SDS at 68°C for 20 min.
Synthetic oligonucleotides. The following oligonucleotides were synthesized to generate pStACTCAT: 5′-CAAGCTTGGATCCACTAGTAGATCTT-3′ and 5′-CTAGAAGATCTACTAGTGGATCCAAGCTTGCATG-3′. A pair of primers PR-INTL (5′-GGGTCTCCATGGGGCTGCCTCG-3′) and PR-INTEXR (5′-GGTACCATGGTCTCCATTCTCAACTCT-3′) was used in the PCR to obtain a DNA fragment containing the first intron of the ACT gene (ex-in-ex). Primers PRBglII-246 (5′-ATGAAGATCTAATAAGCAGATAAA AAC-3′) and either PRNcoI (5′-TCTCCATGGTCAACTCTGCCTCAGGGAGCTGGATGTAG-3′) or PRNcoII (5′-TCTCCATGGTCAACTCTCAGGGAGCTGGATGTAG-3′) were used in the PCR to incorporate two variants of the untranslated region (UTR) of the ACT mRNA in the front of the CAT gene (un1 and un2, respectively). Mutants containing point mutations in the NF-kB(5′), NF-kB(3′), and AP-1 elements were generated by PCR using Pwo polymerase (Roche, Indianapolis, IN) and the following primers: 5′-ACAGGGATCCCTGCAGAGATGCGGGAA GTCTAGAGGACAGCAGGAAAGTC-3′ (mut5′), 5′-GCTAGGATCCCCAGGAGCAAAGCTTCCTAGAGCCGGACCCTC-3′ (mut3′), 5′-ACTGTGGAATTCAGCTCCTCTGCAGTG-3′ (mutAP-1t), and 5′-GAGCTGAATTCCACAGTTTTGTCTGG-3′ (mutAP-1b). Primers 5′-GCTAGGATCCCCAGGAGCAAAGTCC-3′ (E1), 5′-GTGGGGATCCCAGATAATGAGTAAC-3′ (E2), 5′-ACAGGGATCCCTGCAGAGATGCG-3′ (E3), and 5′-TGCAGGATCCCAGACAAAACTGTG-3′ (E4) were used to obtain PCR products E1E2, E1E4, and E3E4. All oligonucleotides used for gel retardation assays were designed to contain single-stranded 5′ overhangs of four bases at both ends after annealing (Table1).
Table 1.
Oligonucleotides used in gel retardation assays
| Name | Sequence |
|---|---|
| 5′-GATCTATGCGGGAAGTCCCAGGGA-3′ | |
| 5′ NF-kB | 3′-ATACGCCCTTCAGGGTCCCTCTAG-5′ |
| 5′-GATCTCTAGGGGACTTTGCTCCTGA-3′ | |
| 3′ NF-kB | 3′-AGATCCCCTGAAACGAGGACTCTAG-5′ |
| 5′-GATCTGAGCTGACTCACACA-3′ | |
| AP-1 | 3′-ACTCGACTGAGTGTGTCTAG-5′ |
Library screening and plasmid construction. A human genomic library (from a HT1080 fibrosarcoma cell line) was obtained from Stratagene (La Jolla, CA). Phages 4.5 × 106 were screened using a (−352 to +25) PCR fragment of the ACT promoter. A single phage harboring a DNA fragment containing the first exon, first intron, and 14719-bp-long 5′ flanking region of the ACT gene was isolated. Plasmids pACT-3573CAT, pACT-244CAT, ptkCATΔEH, pSPI-3(-148)CAT, pUCACT, and prT-61 were described previously (Bugno et al., 1995; Kordula and Travis, 1996;Kordula et al., 1998). Expression plasmid pRSVIkB was a gift from Dr. K. Brand (Munich, Germany). Plasmid pStACTCAT was generated by an insertion of a double-stranded oligonucleotide, described above, intoSphI/XbaI sites of pACT-244CAT. Plasmids p'd'ACTtkCAT, p'e'ACTtkCAT, p'f'ACTtkCAT, and p'g'ACTtkCAT were constructed by insertion of BamHI–BamHI fragments (see Fig. 2, d, e, f,g) into the BamHI site of ptkCATΔEH. Plasmids p'a'StCAT, p'b'StCAT, p'c'StCAT, p'd'StCAT, p'e'StCAT, p'f'StCAT, and p'g'ACTtkCAT were constructed by insertion ofBamHI–BamHI fragments (a,b, c, d, e, f,g) into the BamHI site of pStACTCAT. The plasmid p-2431ACT(ex-in-ex)CAT was generated as follows. TheBamHI–NcoI fragment from pUCACT was cloned intoBglII–NcoI sites of the pCAT3-promoter (Promega, Madison, WI). The obtained plasmid p(B-N)CAT was digested withNcoI, and the NcoI-digested PCR product (ex-in-ex) was inserted. Plasmids p-244un1CAT and p-244un2CAT were constructed by insertion ofBglII–NcoI-digested PCR products (un1 and un2) into BglII–NcoI sites of pCAT-promoter. Plasmids pΔBglIIACTCAT, pΔHindIIIACTCAT, and pΔSphIACTCAT were the deletion mutants of p'a'StCAT from which the BglII–BglII,HindIII–HindIII, orSphI–SphI fragments were removed. Plasmids pSSCAT, prSSCAT, p1EECAT and p2EECAT were obtained by cloningSphI–SphI or EcoRV–EcoRV fragments from p'a'StCAT into either the SphI orBamHI/blunt site of pStACTCAT. Plasmids pER-SCAT, pee1CAT, and pee2CAT were generated by insertion ofEcoRI/blunt–SacI/blunt orEcoO109I/blunt–EcoO109I/blunt fragments from p1EECAT into the BamHI/blunt site of pStACTCAT. Plasmid pdelee1EECAT derives from p1EECAT from which the EcoO109I–EcoO109I fragment was deleted. Plasmids pΔACTCAT, pΔ5ACTCAT, and pΔ3ACTCAT were constructed by insertion of BamHI-digested E1E2, E1E3, and E3E4 PCR products into the BamHI site of pStACTCAT. Plasmids p(mut5′)ACTCAT, p(mut3′)ACTCAT, p(dm)ACTCAT, p(mutAP)ACTCAT, p(mut5′+AP)ACTCAT, p(mut3′+AP)ACTCAT, and p(muttriple)ACTCAT, analogous to pΔ5ACTCAT but with introduced point mutations in the 5′NF-kB, 3′NF-kB, or AP-1 elements, were generated by insertion of BamHI-digested PCR products into the BamHI site of pStACTCAT. Plasmids pSPI-3(IL-1enh)CAT, p(IL-1enh)CAT, pTIMP-1(IL-1enh)CAT, and p2x(IL-1enh)StACTCAT were constructed by insertion of the BamHI-digested E3E1 product into the BamHI site of pSPI-3(-148)CAT, ptkCATΔEH, prT-61CAT, or pStACTCAT. All constructs were sequenced on both strands.
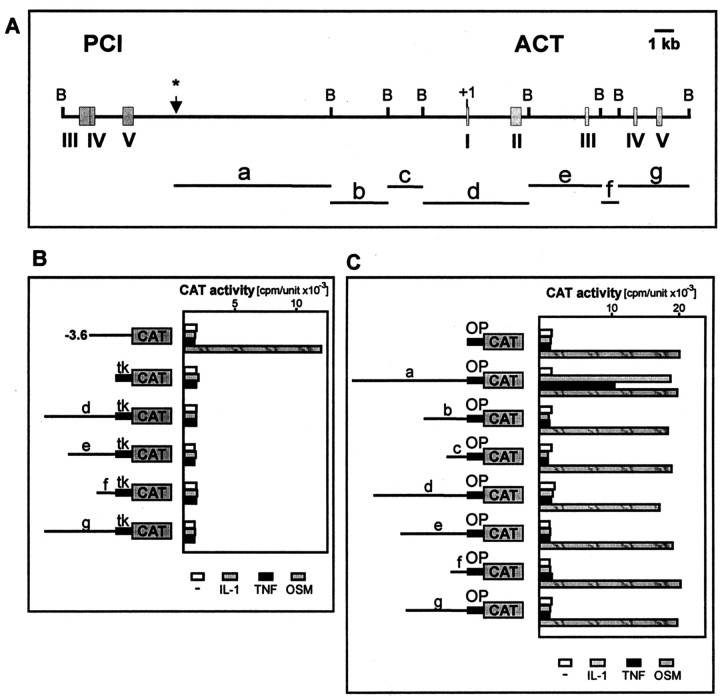
Fig. 2.
Localization of the IL-1/TNF response element of the ACT gene. A, Structure of the DNA fragment containing the ACT gene and its 5′ flanking region (GenBank accession no. AL049839). Exons coding for protein C inhibitor (PCI) and antichymotrypsin (ACT) are indicated by numbers.B marks restriction sites for BamHI.Asterisk indicates the end of the DNA fragment harbored by phage. BamHI–BamHI fragments used for construction of reporter plasmids are marked by letters (a–g). B, Human astrocytes were transfected with plasmids pact-3573CAT, ptkCATDEH, p'd'ACTtkCAT, p'e'ACTtkCAT, p'f'ACTtkCAT, or p'g'ACTtkCAT, and β-galactosidase expression vector as internal control for transfection efficiency. One day after transfection, cells were stimulated with the indicated cytokines, cultured for another 24 hr, and harvested. CAT activities were normalized to β-galactosidase activities (cpm/unit × 10−3). tkindicates thymidine kinase minimal promoter from ptkCATDEH.C, Human astrocytes were transfected with pStACTCAT, p'a'StCAT, p'b'StCAT, p'c'StCAT, p'd'StCAT, p'e'StCAT, p'f'StCAT, p'g'StCAT, and pCH110. Cells were treated with cytokines and harvested as described above. CAT activities were normalized to β-galactosidase activities (cpm/unit × 10−3). OP indicates 244-bp-long ACT promoter containing elements mediating response to OSM but not to IL-1 or TNF.
Transient transfections. Cells were transfected in 12-well clusters using FuGENE6 reagent (Roche, Indianapolis, IN), according to the supplier's instructions. Plasmids (200 ng of the reporter CAT plasmid and 100 ng of pCH110) and 0.6 μl of FuGENE6 diluted in 50 μl of serum-free medium were used for one well containing cells growing in 500 μl of culture medium. One day after transfection cells were stimulated, cultured another 24 hr, and harvested. Protein extracts were prepared by freeze-thawing (Gorman, 1985), and protein concentration was determined by the BCA method (Sigma). Chloramphenicol acetyltransferase (CAT) and β-galactosidase assays were performed as described (Delegeane et al., 1987; Seed and Sheen, 1988). CAT activities are normalized to the internal control β-galactosidase activity and are means ± SEM (three to six determinations).
Nuclear extract preparation and gel retardation assays.Nuclear extracts were prepared as described (Baeuerle and Baltimore, 1988). Double-stranded DNA fragments were labeled by filling in 5′ protruding ends with Klenow enzyme using [α32P]dCTP (3000 Ci/mmol). Gel retardation assays were performed according to published procedures (Fried and Crothers, 1981; Sawadogo et al., 1988). Nuclear extracts (5 μm) and ∼10 fmol (10,000 cpm) of probe were used. The polyclonal anti-NF-kB p65 antiserum (H-286) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
RESULTS
IL-1 and TNF upregulate expression of ACT mRNA in primary human astrocytes
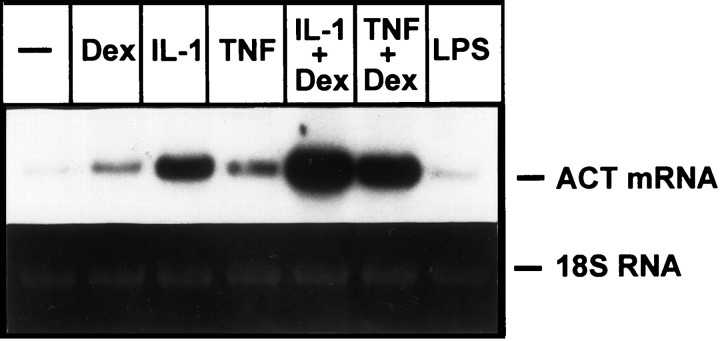
IL-1, TNF, and OSM have previously been shown to regulate ACT expression in both astrocytes and astrocytoma cells (Das and Potter, 1995; Lieb et al., 1996; Kordula et al., 1998). In astrocytes the magnitude of ACT stimulation by these cytokines was comparable, suggesting that all three cytokines should be considered as potential regulators of ACT expression in the brain under inflammatory conditions. To measure the effect of IL-1 and TNF on ACT expression in our astrocyte preparations, we stimulated these cells with cytokine in either the presence or absence of Dex, a synthetic glucocorticoid known to enhance cytokine action in hepatic cells. Figure 1 shows that although control astrocytes express barely detectable amounts of ACT mRNA, cytokine treatment results in substantial upregulation of ACT mRNA expression. This cytokine-activated synthesis of ACT mRNA was enhanced by Dex, although the glucocorticoid, by itself, had little effect. For comparison we also stimulated cells with LPS; however, this compound did not influence the production of ACT mRNA.
Fig. 1.
Expression of ACT mRNA in human astrocytes. Human astrocytes were treated with IL-1 (5 ng/ml), TNFα (10 ng/ml), or LPS (1 μg/ml). RNA was isolated after 18 hr and subjected to Northern blot analysis using ACT cDNA as a probe. Bottom panelshows 18S RNA stained with ethidium bromide on the membrane.
Identification of the DNA fragment containing IL-1/TNF response element(s)
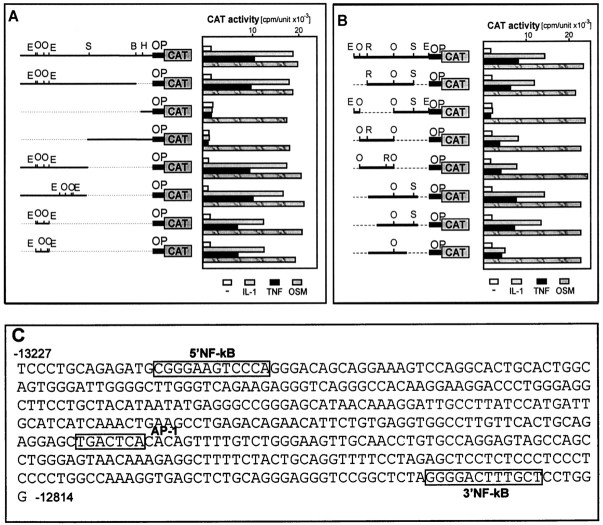
We have previously concluded that activation of the ACT gene by IL-1 is a transcriptional event (Kordula et al., 1998). However, the 3.6-kb-long 5′ flanking region of the ACT gene conferred responsiveness to neither IL-1 nor TNF, although it was fully responsive to OSM (Kordula et al., 1998) (Fig.2B). To identify regulatory elements that mediate the response to IL-1 and TNF, we cloned different fragments of the ACT gene (containing all introns and exons) in front of the tk promoter-driving transcription of the reporter CAT gene. Constructs were transfected into astrocytes, and their responsiveness to IL-1 and TNF was determined. However, neither construct was regulated by IL-1 or TNF (Fig.2B). We conclude that regulatory elements that mediate response to IL-1 and TNF are most likely located distal to the mRNA coding sequence. To obtain DNA fragments more 5′ to the ACT gene, we screened a human genomic library and purified a single phage harboring a DNA fragment containing 14719 bp of the 5′ flanking sequence of the ACT gene (Fig. 2A). Next, we cloned DNA fragments containing either the 5′ flanking region of the ACT gene or ACT coding sequences, in front of a short ACT promoter that is responsive to OSM. This approach enabled the analysis of cloned DNA fragments together with elements specific for the ACT promoter and thus would likely allow any specific interactions between IL-1/TNF-induced transcription factors and factors binding to the ACT promoter. The constructs obtained were analyzed in transfection experiments (Fig.2C) and, as expected, because of the presence of the ACT promoter, all were responsive to OSM. In addition, the construct containing the 7407-bp-long fragment from the 5′ flanking region of the ACT gene located at −14719 to −7312 was also responsive to IL-1 and TNF. Next, we analyzed shorter DNA fragments derived from the −14719 to −7312 fragment and found that a 413-bp-long fragment located at −13227 to −12814 still conferred responsiveness to IL-1 and TNF; however, further truncation led to a decrease or loss of responsiveness (Fig. 3B).
Fig. 3.
Detailed analysis of the putative IL-1/TNF response element of the ACT gene. Human astrocytes were transfected with p'a'StCAT, pΔBglIIACTCAT, pΔHindIIIACTCAT, pΔSphIACTCAT, pSSCAT, prSSCAT, p1EECAT, or p2EECAT (A); p1EECAT, pER-SCAT, pdelee1EECAT, pee1CAT, pee2CAT, pΔACTCAT, pΔ5ACTCAT, or pΔ3ACTCAT (B). An internal control plasmid pCH110 encoding β-galactosidase was included in all transfection experiments. Cells were treated with cytokines and harvested as described in the legend to Figure 2. OPmarks 244-bp-long ACT promoter. Restriction sites forBglII (B), EcoO109I (O), EcoRI (R), EcoRV (E), HindIII (H), and SacI (S) are indicated. Nucleotide sequence of the ACT IL-1/TNF response element is shown (C). The NF-kB and AP-1 elements are boxed.
Lack of effect of the ACT mRNA 5′ untranslated region (5′UTR) on responsiveness to IL-1 and TNF
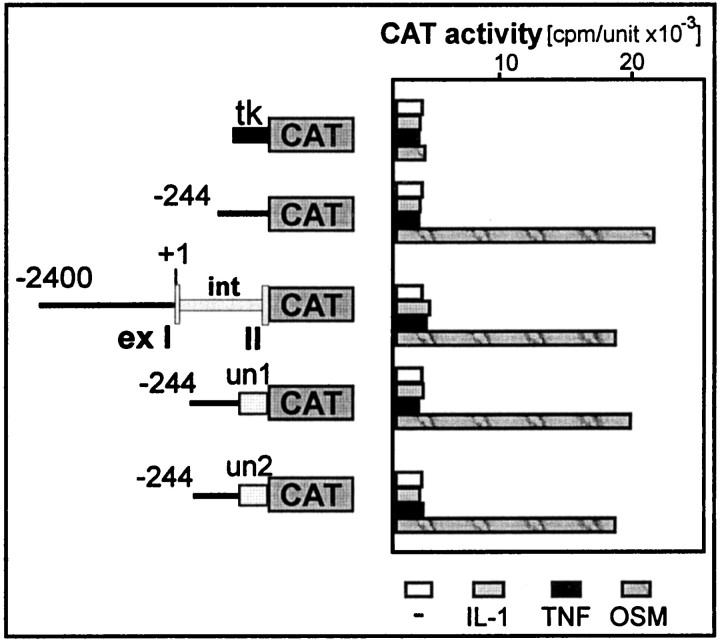
Recently, 5′UTRs of human β-amyloid precursor protein mRNA and ferritin mRNA have been shown to confer responsiveness to IL-1 (Rogers et al., 1994, 1999). Because multiple mechanisms could regulate expression of ACT by IL-1 and TNF, we analyzed the effect of its 5′UTR on the activation by both cytokines. First, we generated a construct containing a 2400-bp-long ACT 5′ flanking region, first exon (containing ACT 5′UTR), entire first intron, and the sequence coding for the first three amino acids of ACT followed by a CAT reporter gene. Transcription from this construct, followed by splicing, should result in production of a chimeric mRNA containing the ACT 5′UTR and the protein coding sequence that encodes the first three amino acids of ACT and the entire CAT protein. This construct, analyzed in transfection experiments (Fig. 4), proved to be responsive to OSM but not to IL-1 or TNF. The pattern of cytokine responsiveness indicated that chimeric mRNA was properly transcribed and spliced, whereas the protein synthesized was expressed in an active form. Moreover, the observed activation by OSM indicates the expected normal function of the ACT promoter in this “splicing” construct. However, the ACT 5′UTR proved to be incapable of mediating any response to IL-1 or TNF. Although by searching an EST database we found a second type of ACT 5′UTR, which differs from that analyzed by four nucleotides (additional GCAG at −12 to −9 upstream from first methionine). To investigate the role of this 5′UTR, we inserted both types of 5′UTR in front of the CAT gene downstream from the ACT promoter. However, these constructs were also not responsive to IL-1 and TNF in transfection experiments, although they did respond to OSM (Fig. 4). We conclude that neither of the ACT 5′UTRs can confer a response to IL-1 or TNF and that activation by these cytokines is most likely mediated in full by the distal regulatory element that we identified above.
Fig. 4.
The untranslated region of ACT mRNA does not confer any response to IL-1 and TNF. Human astrocytes were transfected with either ptkCATΔEH, pACT-244CAT, p-2431ACT(ex-in-ex)CAT, p-244un1CAT, or p-244un2CAT and an internal control plasmid pCH110 encoding β-galactosidase. Cells were treated with cytokines and harvested as described in the legend to Figure 2. CAT activities were normalized to β-galactosidase activities (cpm/unit × 10−3). The two variants of the untranslated region (un1, un2), first intron (int), and first and second exon (exI,II) of the ACT gene are marked. tkindicates thymidine kinase minimal promoter from ptkCATDEH.
Identification of regulatory elements binding IL-1- and TNF-induced factors
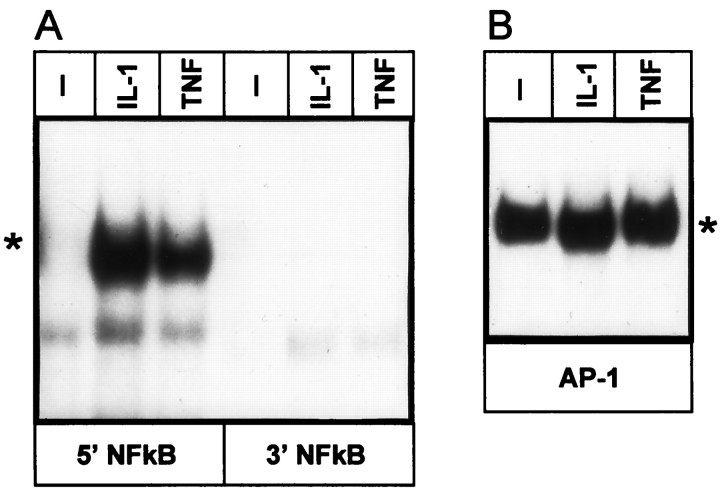
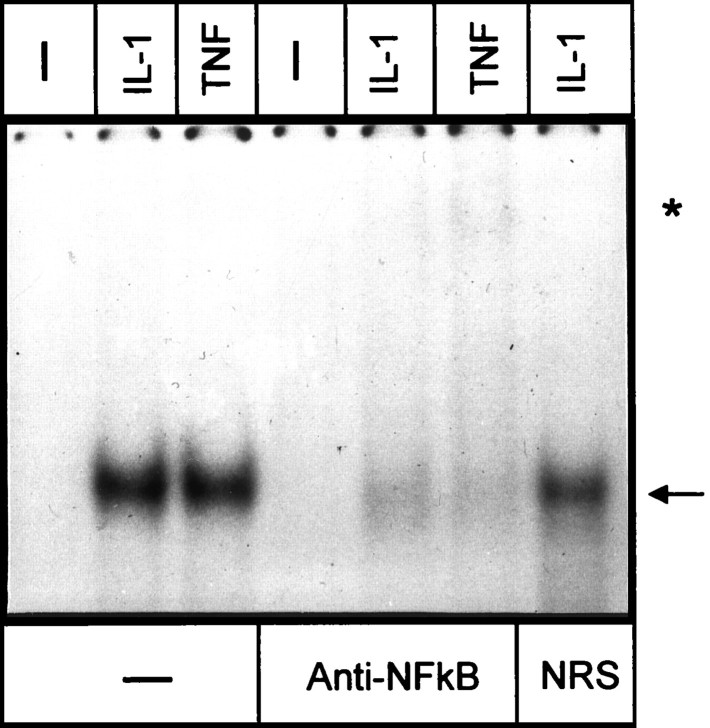
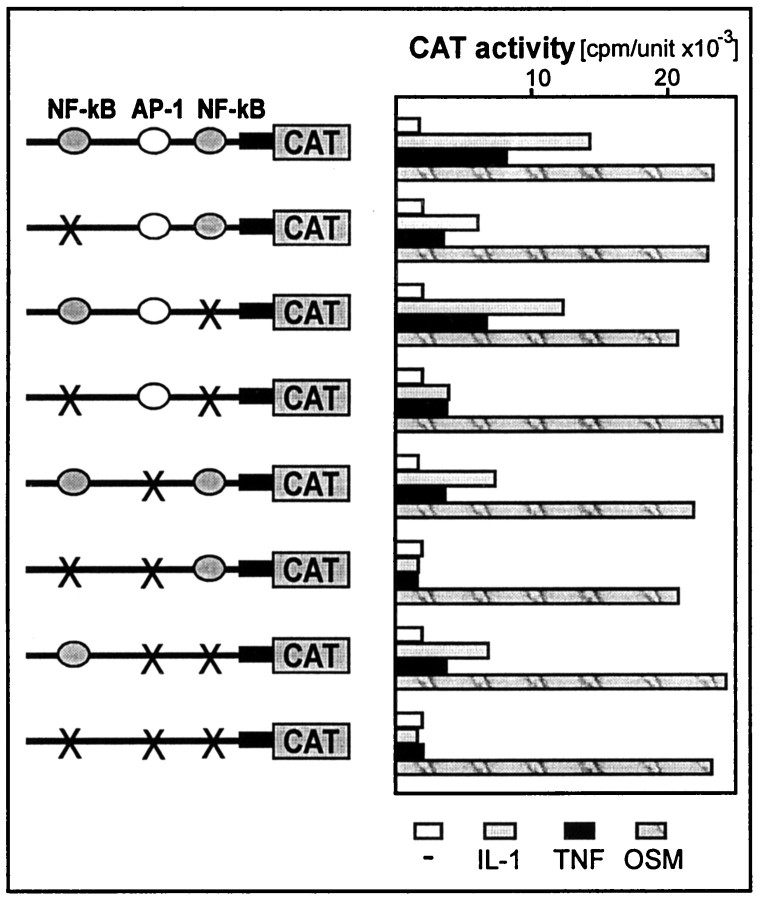
The 413-bp-long element that conferred a response to IL-1 and TNF was searched for the presence of putative binding sites for transcription factors. Two possible binding sites for NF-kB at −13213 to −13202 (5′NF-kB) and −12831 to −12820 (3′NF-kB) and a single putative AP-1 element at −12985 to −12979 were located (Fig. 3A). Binding of transcription factors to these elements was then analyzed by EMSA (Fig. 5). Treatment of astrocytes with IL-1 and TNF resulted in activation of a protein that bound to the 5′NF-kB binding site, and this protein was recognized by anti-NF-kB antibodies (Fig. 6). In contrast, we did not observe any proteins binding to the 3′NF-kB binding site. However, binding of a protein to the AP-1 site was also detected in control astrocytes, and treatment of these cells with IL-1 or TNF further enhanced this binding. To evaluate the contribution of each of these elements to the overall responsiveness to IL-1 and TNF, we constructed a series of mutants with mutations introduced into NF-kB and AP-1 elements (Fig. 7). Mutations introduced into 5′NF-kB resulted in a reduction of responsiveness to both IL-1 and TNF by 60%, whereas mutation of the 3′NF-kB site only slightly diminished this response (15%). These results correlated with both binding studies (Fig. 5) and analysis of deletion mutants (Fig. 3). The mutation of the AP-1 element reduced responsiveness to both cytokines by 50%. The mutant with an intact AP-1 site but that was mutated at both NF-kB sites had a reduced ability to respond to IL-1 and TNF (reduction by 70 and 50%, respectively). In addition, a mutant with all three sites changed was no longer responsive to IL-1 and TNF. We conclude that (1) all three elements contribute to the full activity of the 413-bp-long fragment, and (2) the 5′NF-kB and AP-1 elements mediate most of the response.
Fig. 5.
EMSA of nuclear extracts from human astrocytes using fragments from the IL-1/TNF response element. Human astrocytes were incubated with IL-1 or TNF for 40 min (A) or 4 hr (B). Nuclear extracts were prepared and analyzed using 32P-labeled double-stranded fragments derived from the IL-1/TNF response element;5′NF-kB, 3′NF-kB, andAP-1. Gels were exposed for 48 (A) or 24 (B) hr. Asterisks indicate IL-1-, TNF-induced bands.
Fig. 6.
The IL-1-, TNF-induced protein is recognized by anti-NF-kB antibodies. Human astrocytes were stimulated with IL-1 or TNF for 40 min. Nuclear extracts were prepared and analyzed using a 5′NF-kB double-stranded oligonucleotide. Extracts were incubated with anti-NF-kB antibodies or normal rabbit serum (NRS) for 10 min when indicated. Arrow indicates position of the IL-1-, TNF-induced band. Asterisk marks super-shifted complexes.
Fig. 7.
Effect of point mutations introduced into the NF-kB and AP-1 elements. Point mutations were introduced into putative binding sites of the ACT IL-1/TNF response element as described in Materials and Methods. Human astrocytes were transfected with either pΔ5ACTCAT, p(mut5′)ACTCAT, p(mut3′)ACTCAT, p(dm)ACTCAT, p(mutAP)ACTCAT, p(mut5′+AP) ACTCAT, p(mut3′+AP)ACTCAT, or p(muttriple)ACTCAT, and β-galactosidase expression vector as internal control for transfection efficiency. One day after transfection, cells were stimulated with indicated cytokines, cultured for another 24 hr, and harvested. CAT activities were normalized to β-galactosidase activities (cpm/unit × 10−3).
The 413-bp-long 5′ distal element of the ACT gene is an IL-1/TNF enhancer
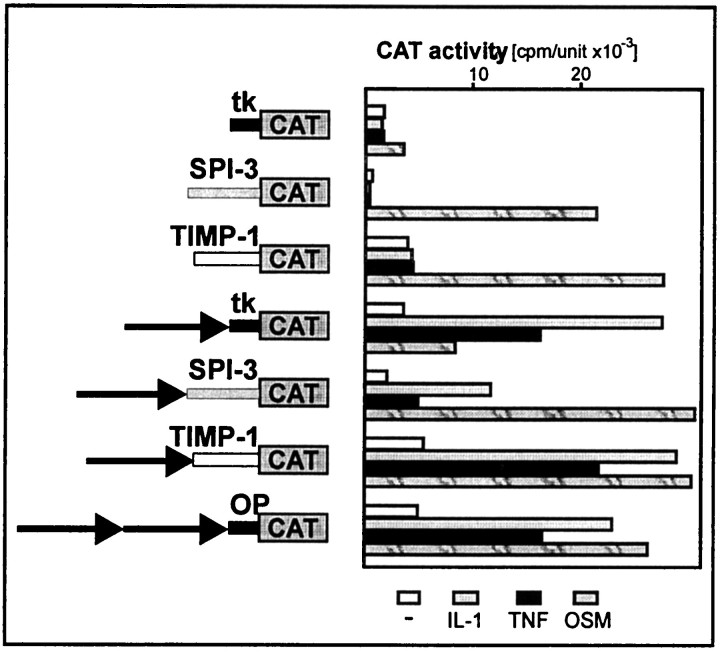
The 413-bp-long distal element of the ACT gene mediated a response to IL-1 and TNF when linked to the short ACT promoter. To determine whether the ACT promoter is indispensable for IL-1 and TNF response, we also linked the distal element to several promoters normally not responsive to IL-1 and TNF. These promoters were chosen to be diverse. The SPI-3 promoter contains a TATA box and is characterized by very low basal expression. In contrast, the TIMP-1 promoter does not contain a TATA box and has a relatively high basal activity. As shown in Figure8, the 413-bp-long element is an IL-1/TNF enhancer that conferred a responsiveness for both cytokines to all tested promoters. In addition, fusion of this enhancer to these promoters increased their basal activity.
Fig. 8.
The IL-1/TNF response element of the ACT gene confers responsiveness to IL-1 and TNF onto other promoters. The IL-1/TNF response element was cloned in front of the SPI-3, TIMP-1, andtk promoters. Human astrocytes were transfected with ptkCATΔEH, pSPI-3 (-148)CAT, prT-61, p(IL-1enh)CAT, pSPI-3(IL-1enh)TCAT, pTIMP-1(IL-1enh)CAT, or p2x(IL-1enh)CAT, and β-galactosidase expression vector as an internal control for transfection efficiency. One day after transfection, cells were stimulated with indicated cytokines, cultured for another 24 hr, and harvested. CAT activities were normalized to β-galactosidase activities (cpm/unit × 10−3).
Crucial role of NF-kB in regulating ACT gene expression
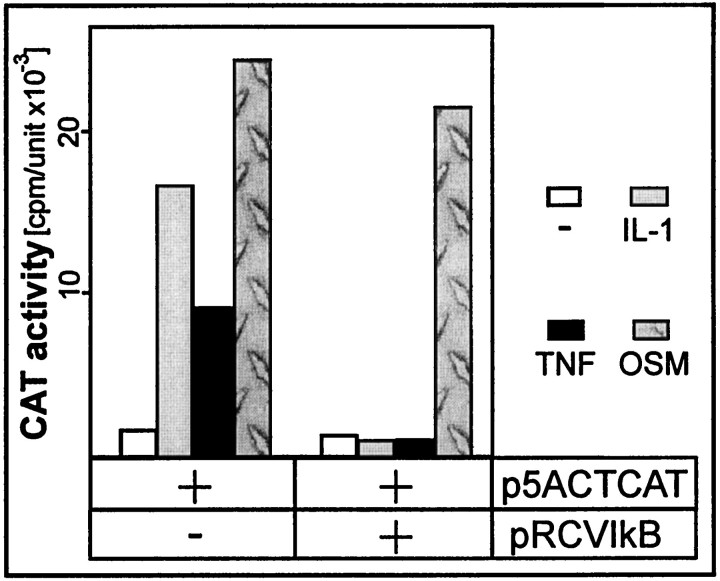
Strong binding of NF-kB to the 5′NF-kB site and substantially reduced activation of enhancer lacking this binding site suggested that NF-kB might be the most important factor regulating ACT gene expression. To prove this hypothesis we blocked activation of NF-kB by overexpressing inhibitor of NF-kB (IkB). We cotransfected into astrocytes a reporter plasmid containing the ACT IL-1/TNF enhancer and an expression plasmid encoding IkB (Fig.9). The activation of the reporter gene by IL-1 and TNF was totally blocked by expression of IkB. We conclude that NF-kB is a key regulatory transcription factor mediating activation of the ACT gene by IL-1 and TNF via a distal 5′ enhancer.
Fig. 9.
Expression of IkB inhibits activation mediated by the ACT IL-1/TNF response element. Human astrocytes were transfected with pΔ5ACTCATH, IkB expression vector, and β-galactosidase expression vector as an internal control for transfection efficiency. One day after transfection, cells were stimulated with the indicated cytokines, cultured for another 24 hr, and harvested. CAT activities were normalized to β-galactosidase activities (cpm/unit × 10−3).
DISCUSSION
Both IL-1 and TNF have been implicated as key regulatory molecules in a number of normal physiological and pathological processes within the CNS, including astrogliosis, inflammatory reactions, and induction of expression of “cerebral” acute phase genes (Giulian et al., 1988; Merrill, 1991; Vandenabeele and Fiers, 1991). Activation of the target genes by these cytokines can be either direct or mediated via activation of other signaling molecules such as IL-6 and IL-8, which in turn induce their target genes (Benveniste et al., 1990; Kasahara et al., 1991). The activation of the ACT gene by IL-1 and TNF in astrocytes seems, however, to be directly mediated by both cytokines, and this apparently occurs at the level of transcription (Kordula et al., 1998). Similarly, regulation of ACT expression in astrocytes by OSM or complexes of IL-6/sIL-6R is mediated on the level of transcription, as described previously (Kordula et al., 1998). However, low levels of OSM, IL-6, and sIL-6R in cerebrospinal fluids suggest that OSM and IL-6, although contributing to ACT regulation, might not be the major regulators of its expression in the brain (Frieling et al., 1994; Kordula et al., 1998). In contrast, IL-1 and TNF can readily be detected in the CNS, and thus, it is likely that the enhanced expression of ACT in astrocytes localized in affected areas of the brain is induced by these cytokines. Here, we demonstrate that the transcriptional mechanism of ACT activation is not accompanied, at least in astrocytes, by an additional translational regulation mediated by the 5′UTR of ACT mRNA (Fig. 4).
Recently, it has been shown by use of specific kinase inhibitors, that protein kinases A and C are not involved in the regulation of ACT gene expression by IL-1 or TNF in astrocytoma U373-MG cells (Lieb et al., 1996). It was also proposed that NF-kB might regulate ACT expression in response to these cytokines, because activation of ACT mRNA could be inhibited by pyrrolidine dithiocarbamate, a known inhibitor of NF-kB activation. However, neither of the regulatory elements mediating activation by IL-1 or TNF have been identified nor has any transcription factor binding to any elements near the ACT gene been shown. Moreover, IL-1 and TNF have been found to regulate target genes by activating a number of different transcription factors, including AP-1, NF-kB, CAAT enhancer binding protein, LPS, IL-1-induced STAT (LIL-STAT), and octamer binding factor 1 (Mukaida et al., 1990; Tsukada et al., 1996; Tseng and Schuler, 1998; Fukuoka et al., 1999). Thus, the mode of ACT regulation by IL-1 and TNF was unclear and needed identification of regulatory elements.
An increased expression of IL-1 has been reported in AD, and this finding correlates with an increased expression of ACT (Griffin et al., 1989). Moreover, polymorphism of the IL-1 gene that results in higher expression of this proinflammatory cytokine has been shown to correlate with a higher risk of developing AD (Grimaldi et al., 2000; Nicoll et al., 2000). These data suggest that inflammatory processes that involve enhanced expression of IL-1 and upregulation of IL-1-dependent target genes, including the ACT gene, can contribute to the development and progression of AD.
Here, we identify the 5′ distal enhancer located at −13227 to −12814 that mediates the response of the ACT gene to IL-1 and TNF. This element also confers responsiveness to both cytokines onto other promoters normally not responsive to IL-1 or TNF. Regulatory elements that control expression of most of the known genes are located within several kilobases upstream from the transcription start site. However, distant and very distant regulatory elements have also been described for several genes. The most distant regulatory elements include that for the human apolipoprotein B gene located at −60 kb (Nielsen et al., 1998), the bx enhancer of the Drosophila Ubx gene located at −30 kb (Qian et al., 1991), and the 3′ α E region located 70 kb downstream of the human IgH locus (Lieberson et al., 1995). It seems very probable that distant elements are common and regulate a great number of genes; however, their identification is much more difficult using currently available methodology. The ACT enhancer that we have identified seems to be one of these distant regulatory elements. This 413-bp-long enhancer contains at least three regulatory elements that contribute to its full activity (two NF-kB and one AP-1). The mutational analysis indicates that the 5′NF-kB site contributes the most to the full responsiveness, whereas the effect of the 3′NF-kB element is marginal. The AP-1 element contributes greatly to the response to both cytokines, and this is confirmed by the fact that the response of the mutant lacking the AP-1 binding site was greatly diminished (by 70 and 50% in response to IL-1 and TNF, respectively).
ACT produced within the CNS has been shown to be essentially identical to that secreted by hepatocytes (Hwang et al., 1999). However, its increased levels in the brain may have potentially drastic effects on both the degradation and polymerization of βA. We now know that IL-1 and TNF as well as OSM and IL-6/sIL-6R complexes control expression of ACT in astrocytes. Here, we have identified regulatory elements and transcription factors that mediate responses to IL-1 and TNF. It remains to be seen whether interruption of their regulatory function might also affect amyloid deposition associated with AD.
The enhancer that we identified is located ∼13 kb upstream of the ACT promoter and only 6 kb below the PCI gene (protein C inhibitor) (Rollini and Fournier, 1997). Both genes are positioned in the head to tail orientation, and the distance between the PCI promoter and the IL-1/TNF enhancer is only 18 kb. In astrocytes, PCI mRNA is not expressed (data not shown); however, both ACT and PCI mRNAs are expressed in liver, and production of PCI is constitutive. In hepatoma HepG2 cells, neither IL-1 nor TNF regulates expression of PCI, whereas synthesis of ACT is only barely upregulated by both cytokines (data not shown). Clearly, two questions remain to be answered in the future: (1) why is the enhancer not fully active in hepatoma cells, and (2) what are the mechanisms that allow activation of the ACT gene but not the PCI gene via the IL-1/TNF enhancer?
Footnotes
This work was supported by research Grants HL26148 and HL37090 from National Institutes of Health (J.T.) and Grant PB 0925/P04/97/12 (T.K.) from the Committee of Scientific Research (KBN, Warsaw, Poland).
Correspondence should be addressed to Dr. James Travis, Department of Biochemistry and Molecular Biology, The University of Georgia, Athens, GA 30602. E-mail: jtravis@arches.uga.edu.
REFERENCES
- 1.Abraham CR, Selkoe DJ, Potter H. Immunochemical identification of the serine protease inhibitor alpha 1-antichymotrypsin in the brain amyloid deposits of Alzheimer's disease. Cell. 1988;52:487–501. doi: 10.1016/0092-8674(88)90462-x. [DOI] [PubMed] [Google Scholar]
- 2.Baeuerle PA, Baltimore D. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- 3.Benveniste EN, Sparacio SM, Norris JG, Grenett HE, Fuller GM. Induction and regulation of interleukin-6 gene expression in rat astrocytes. J Neuroimmunol. 1990;30:201–212. doi: 10.1016/0165-5728(90)90104-u. [DOI] [PubMed] [Google Scholar]
- 4.Bugno M, Graeve L, Gatsios P, Koj A, Heinrich PC, Travis J, Kordula T. Identification of the interleukin-6/oncostatin M response element in the rat tissue inhibitor of metalloproteinases-1 (TIMP-1) promoter. Nucleic Acids Res. 1995;23:5041–5047. doi: 10.1093/nar/23.24.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das S, Potter H. Expression of the Alzheimer amyloid-promoting factor antichymotrypsin is induced in human astrocytes by IL-1. Neuron. 1995;14:447–456. doi: 10.1016/0896-6273(95)90300-3. [DOI] [PubMed] [Google Scholar]
- 6.Delegeane AM, Ferland LH, Mellon PL. Tissue-specific enhancer of the human glycoprotein hormone alpha- subunit gene: dependence on cyclic AMP-inducible elements. Mol Cell Biol. 1987;7:3994–4002. doi: 10.1128/mcb.7.11.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnelly RJ, Friedhoff AJ, Beer B, Blume AJ, Vitek MP. Interleukin-1 stimulates the beta-amyloid precursor protein promoter. Cell Mol Neurobiol. 1990;10:485–495. doi: 10.1007/BF00712843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eikelenboom P, Zhan SS, van Gool WA, Allsop D. Inflammatory mechanisms in Alzheimer's disease. Trends Pharmacol Sci. 1994;15:447–450. doi: 10.1016/0165-6147(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 10.Fraser PE, Nguyen JT, McLachlan DR, Abraham CR, Kirschner DA. Alpha 1-antichymotrypsin binding to Alzheimer A beta peptides is sequence specific and induces fibril disaggregation in vitro. J Neurochem. 1993;61:298–305. doi: 10.1111/j.1471-4159.1993.tb03568.x. [DOI] [PubMed] [Google Scholar]
- 11.Fried M, Crothers DM. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frieling JT, Sauerwein RW, Wijdenes J, Hendriks T, van der Linden CJ. Soluble interleukin 6 receptor in biological fluids from human origin. Cytokine. 1994;6:376–381. doi: 10.1016/1043-4666(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 13.Fukuoka T, Kitami Y, Okura T, Hiwada K. Transcriptional regulation of the platelet-derived growth factor alpha receptor gene via CCAAT/enhancer-binding protein-delta in vascular smooth muscle cells. J Biol Chem. 1999;274:25576–25582. doi: 10.1074/jbc.274.36.25576. [DOI] [PubMed] [Google Scholar]
- 14.Giulian D, Young DG, Woodward J, Brown DC, Lachman LB. Interleukin-1 is an astroglial growth factor in the developing brain. J Neurosci. 1988;8:709–714. doi: 10.1523/JNEUROSCI.08-02-00709.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorman C. DNA cloning: a practical approach (Glover D, ed), pp 143–190. IRL; Location: 1985. [Google Scholar]
- 16.Griffin WS, Stanley LC, Ling C, White L, MacLeod V, Perrot LJ, White C, Araoz C. Brain interleukin 1 and S-100 immunoreactivity are elevated in Down syndrome and Alzheimer disease. Proc Natl Acad Sci USA. 1989;86:7611–7615. doi: 10.1073/pnas.86.19.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimaldi LM, Casadei VM, Ferri C, Veglia F, Licastro F, Annoni G, Biunno I, De Bellis G, Sorbi S, Mariani C, Canal N, Griffin WS, Franceschi M. Association of early-onset Alzheimer's disease with an interleukin-1alpha gene polymorphism. Ann Neurol. 2000;47:361–365. [PubMed] [Google Scholar]
- 18.Hwang SR, Steineckert B, Kohn A, Palkovits M, Hook VY. Molecular studies define the primary structure of alpha1- antichymotrypsin (ACT) protease inhibitor in Alzheimer's disease brains. Comparison of act in hippocampus and liver. J Biol Chem. 1999;274:1821–1827. doi: 10.1074/jbc.274.3.1821. [DOI] [PubMed] [Google Scholar]
- 19.Janciauskiene S, Rubin H, Lukacs CM, Wright HT. Alzheimer's peptide Abeta1–42 binds to two beta-sheets of alpha1-antichymotrypsin and transforms it from inhibitor to substrate. J Biol Chem. 1998;273:28360–28364. doi: 10.1074/jbc.273.43.28360. [DOI] [PubMed] [Google Scholar]
- 20.Kasahara T, Mukaida N, Yamashita K, Yagisawa H, Akahoshi T, Matsushima K. IL-1 and TNF-alpha induction of IL-8 and monocyte chemotactic and activating factor (MCAF) mRNA expression in a human astrocytoma cell line. Immunology. 1991;74:60–67. [PMC free article] [PubMed] [Google Scholar]
- 21.Kordula T, Travis J. The role of Stat and C/EBP transcription factors in the synergistic activation of rat serine protease inhibitor-3 gene by interleukin-6 and dexamethasone. Biochem J. 1996;313:1019–1027. doi: 10.1042/bj3131019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kordula T, Rydel RE, Brigham EF, Horn F, Heinrich PC, Travis J. Oncostatin M and the interleukin-6 and soluble interleukin-6 receptor complex regulate alpha1-antichymotrypsin expression in human cortical astrocytes. J Biol Chem. 1998;273:4112–4118. doi: 10.1074/jbc.273.7.4112. [DOI] [PubMed] [Google Scholar]
- 23.Lieb K, Fiebich BL, Schaller H, Berger M, Bauer J. Interleukin-1 beta and tumor necrosis factor-alpha induce expression of alpha 1-antichymotrypsin in human astrocytoma cells by activation of nuclear factor-kappa B. J Neurochem. 1996;67:2039–2044. doi: 10.1046/j.1471-4159.1996.67052039.x. [DOI] [PubMed] [Google Scholar]
- 24.Lieberson R, Ong J, Shi X, Eckhardt LA. Immunoglobulin gene transcription ceases upon deletion of a distant enhancer. EMBO J. 1995;14:6229–6238. doi: 10.1002/j.1460-2075.1995.tb00313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma J, Yee A, Brewer HB, Jr, Das S, Potter H. Amyloid-associated proteins alpha 1-antichymotrypsin and apolipoprotein E promote assembly of Alzheimer beta-protein into filaments. Nature. 1994;372:92–94. doi: 10.1038/372092a0. [DOI] [PubMed] [Google Scholar]
- 26.Merrill JE. Effects of interleukin-1 and tumor necrosis factor-alpha on astrocytes, microglia, oligodendrocytes, and glial precursors in vitro. Dev Neurosci. 1991;13:130–137. doi: 10.1159/000112150. [DOI] [PubMed] [Google Scholar]
- 27.Mukaida N, Mahe Y, Matsushima K. Cooperative interaction of nuclear factor-kappa B- and cis-regulatory enhancer binding protein-like factor binding elements in activating the interleukin-8 gene by pro-inflammatory cytokines. J Biol Chem. 1990;265:21128–21133. [PubMed] [Google Scholar]
- 28.Namba Y, Tomonaga M, Kawasaki H, Otomo E, Ikeda K. Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer's disease and kuru plaque amyloid in Creutzfeldt-Jakob disease. Brain Res. 1991;541:163–166. doi: 10.1016/0006-8993(91)91092-f. [DOI] [PubMed] [Google Scholar]
- 29.Nicoll JA, Mrak RE, Graham DI, Stewart J, Wilcock G, MacGowan S, Esiri MM, Murray LS, Dewar D, Love S, Moss T, Griffin WS. Association of interleukin-1 gene polymorphisms with Alzheimer's disease. Ann Neurol. 2000;47:365–368. [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen LB, Kahn D, Duell T, Weier HU, Taylor S, Young SG. Apolipoprotein B gene expression in a series of human apolipoprotein B transgenic mice generated with recA-assisted restriction endonuclease cleavage-modified bacterial artificial chromosomes. An intestine-specific enhancer element is located between 54 and 62 kilobases 5′ to the structural gene. J Biol Chem. 1998;273:21800–21807. doi: 10.1074/jbc.273.34.21800. [DOI] [PubMed] [Google Scholar]
- 31.Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J Biol Chem. 1994;269:15957–15960. [PubMed] [Google Scholar]
- 32.Qian S, Capovilla M, Pirrotta V. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J. 1991;10:1415–1425. doi: 10.1002/j.1460-2075.1991.tb07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogers JT, Andriotakis JL, Lacroix L, Durmowicz GP, Kasschau KD, Bridges KR. Translational enhancement of H-ferritin mRNA by interleukin-1 beta acts through 5′ leader sequences distinct from the iron responsive element. Nucleic Acids Res. 1994;22:2678–2686. doi: 10.1093/nar/22.13.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers JT, Leiter LM, McPhee J, Cahill CM, Zhan SS, Potter H, Nilsson LN. Translation of the alzheimer amyloid precursor protein mRNA is up-regulated by interleukin-1 through 5′-untranslated region sequences. J Biol Chem. 1999;274:6421–6431. doi: 10.1074/jbc.274.10.6421. [DOI] [PubMed] [Google Scholar]
- 35.Rollini P, Fournier RE. A 370-kb cosmid contig of the serpin gene cluster on human chromosome 14q32.1: molecular linkage of the genes encoding alpha 1- antichymotrypsin, protein C inhibitor, kallistatin, alpha 1- antitrypsin, and corticosteroid-binding globulin. Genomics. 1997;46:409–415. doi: 10.1006/geno.1997.5077. [DOI] [PubMed] [Google Scholar]
- 36.Rose-John S, Dietrich A, Marks F. Molecular cloning of mouse protein kinase C (PKC) cDNA from Swiss 3T3 fibroblasts. Gene. 1988;74:465–471. doi: 10.1016/0378-1119(88)90179-5. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 38.Sawadogo M, Van Dyke MW, Gregor PD, Roeder RG. Multiple forms of the human gene-specific transcription factor USF. I. Complete purification and identification of USF from HeLa cell nuclei. J Biol Chem. 1988;263:11985–11993. [PubMed] [Google Scholar]
- 39.Seed B, Sheen JY. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene. 1988;67:271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- 40.Selkoe DJ. The molecular pathology of Alzheimer's disease. Neuron. 1991;6:487–498. doi: 10.1016/0896-6273(91)90052-2. [DOI] [PubMed] [Google Scholar]
- 41.Sinha S, Anderson JP, Barbour R, Basi GS, Caccavello R, Davis D, Doan M, Dovey HF, Frigon N, Hong J, Jacobson-Croak K, Jewett N, Keim P, Knops J, Lieberburg I, Power M, Tan H, Tatsuno G, Tung J, Schenk D, Seubert P, Suomensaari SM, Wang S, Walker D, John V. Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 42.Snow AD, Mar H, Nochlin D, Kimata K, Kato M, Suzuki S, Hassell J, Wight TN. The presence of heparan sulfate proteoglycans in the neuritic plaques and congophilic angiopathy in Alzheimer's disease. Am J Pathol. 1988;133:456–463. [PMC free article] [PubMed] [Google Scholar]
- 43.Tseng YH, Schuler LA. Transcriptional regulation of interleukin-1beta gene by interleukin-1beta itself is mediated in part by Oct-1 in thymic stromal cells. J Biol Chem. 1998;273:12633–12641. doi: 10.1074/jbc.273.20.12633. [DOI] [PubMed] [Google Scholar]
- 44. Tsukada J, Waterman WR, Koyama Y, Webb AC, Auron PE. A novel STAT-like factor mediates lipopolysaccharide, interleukin 1 (IL-1), and IL-6 signaling and recognizes a gamma interferon activation site-like element in the IL1B gene. Mol Cell Biol 16 1996. 2183 2194[Erratum (1996) 16:3233] . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vandenabeele P, Fiers W. Is amyloidogenesis during Alzheimer's disease due to an IL-1-/IL-6-mediated “acute phase response” in the brain? Immunol Today. 1991;12:217–219. doi: 10.1016/0167-5699(91)90032-O. [DOI] [PubMed] [Google Scholar]
- 46.Yamin R, Malgeri EG, Sloane JA, McGraw WT, Abraham CR. Metalloendopeptidase EC 3.4.24.15 is necessary for Alzheimer's amyloid-beta peptide degradation. J Biol Chem. 1999;274:18777–18784. doi: 10.1074/jbc.274.26.18777. [DOI] [PubMed] [Google Scholar]