Abstract
Recently discovered mammalian clock genes are believed to compose the core oscillator, which generates the circadian rhythm. BMAL1/CLOCK heterodimer is the essential positive element that drives clock-related transcription and self-sustaining oscillation by a negative feedback mechanism. We examined BMAL1 protein expression in the rat suprachiasmatic nuclei (SCN) by immunoblot analysis. Anti-BMAL1 antiserum raised against rBMAL1 recognized 70 kDa mBMAL1b and detected a similar immunoreactivity (IR) as a major band in rat brains. Robust circadian BMAL1-IR oscillations with nocturnal peaks were detected in the SCN during a light/dark cycle and under constant darkness. A short duration light exposure at night acutely reduced BMAL1-IR in the SCN during photoentrainment. This might be attributable to the degradation of BMAL1 protein. Application of glutamate and NMDA to the SCN slices at projected night, a procedure mimicking photic phase delay shift, also acutely reduced BMAL1-IR in a similar manner. A rapid decrease of BMAL1 protein suggests that BMAL1 protein might be implicated in the light-transducing pathway within the SCN.
Keywords: circadian clock, light, phase shift, glutamate, NMDA, suprachiasmatic nucleus
Circadian clocks controlling various biological timings are widely observed in eukaryotes and some prokaryotes. Recent progress has revealed an important part of the mechanism, a set of mammalian circadian clock genes localized within the site of the master circadian pacemaker in the hypothalamic SCN (Dunlap, 1999).
The molecular core of the circadian clock is thought to be the feedback loop with positive and negative limbs. The mouseClock gene was first demonstrated to be an essential component of the mammalian circadian clock (Vitaterna et al., 1994;King et al., 1997). BMAL1 protein is a heterodimeric partner of CLOCK protein (Ikeda and Nomura, 1997; Gekakis et al., 1998). CLOCK/BMAL1 heterodimer is believed to bind E-box elements and drives and maintains circadian oscillations of mammalian orthologs ofDrosophila period genes, i.e., mPer1 transcript (Sun et al., 1997; Tei et al., 1997). BMAL1 transcript exhibits evident circadian oscillation, whereas Clocktranscripts do not (King et al., 1997; Honma et al., 1998). CYCLE, aDrosophila ortholog of BMAL1, is essential for the circadian rhythmicity (Rutila et al., 1998).
The negative limb in the circadian loop is believed to be composed of PER1, PER2, PER3, TIM, CRY1, and CRY2. These molecules, except TIM, show stronger circadian oscillation than that of Clock/BMAL1(Albrecht et al., 1997; Shigeyoshi et al., 1997; Sangoram et al., 1998;Zylka et al., 1998a,b; Kume et al., 1999; Okamura et al., 1999).mCry1, mCry2, and mPer2 genes encode a functional component of the circadian clock (van der Horst et al., 1999; Zheng et al., 1999).
Light is the most powerful external stimulus for connecting and entraining the circadian clock to the environment. In rodents, even a single brief exposure to light in the early (subjective) night causes a phase delay shift, whereas a light pulse during late (subjective) night induces a phase advance shift. The c-Fos gene was first to be identified as one of the immediate responsive genes to light in the SCN (Rea, 1989; Rusak et al., 1990). Rodent Per1 andPer2 transcripts are also immediately induced (Albrecht et al., 1997; Shigeyoshi et al., 1997; Yan et al., 1999).Drosophila dCRY protein is an essential transducer in photic phase shift (Emery et al., 1998). Light-induced degradation of dTimeless protein correlates with behavioral entrainment (Myers et al., 1996; Zeng et al., 1996; Naidoo et al., 1999). However, no mammalian gene has been proved essential in photoentrainment, nor have hypothetical light-responsive elements (LREs) upstream of the light-responsive genes been identified.
The function of BMAL1 in the photoentrainment and maintaining of the circadian clock is not clear. To understand further how the putative BMAL1 functions in the circadian clock cells, we have generated a specific antiserum against rBMAL1 and used it for the immunoblot analysis of the temporal regulation related to the clock mechanism. In this report, we discuss photic downregulation of BMAL1 protein during the resetting of the circadian clock.
MATERIALS AND METHODS
Animals. Male Wistar rats (Nippon Bio-Supply Center, Tokyo, Japan) aged 5–7 weeks were maintained at 25°C on a 12 hr light/dark (LD) cycle [light: zeitgeber time (ZT) 0–12; dark: ZT12–24] for at least 10 d before use. The animals were then transferred to a dim light (<1 lux) condition, and their circadian locomotor activities were monitored using the far-infrared monitor system (Supermex System, Muromachi-Kikai, Tokyo, Japan). The experiments under constant darkness (DD) conditions were performed 2–3 d after the transition. For the light pulse experiment, rats were exposed to white light (1000 lux) for 30 min, then they were killed at the experimental time point. As controls, we analyzed the locomotor activities of a number of rats under the same sampling conditions and confirmed that the phase shifts occurred only by light exposure at subjective night.
Materials. A glutathione-Sepharose, HiTrap column and pGEX-5X vector DNA were purchased from Amersham Pharmacia Biotech (Tokyo, Japan). Escherichia coli strain JM109 and pBluescript SK+ were from Clontech (Tokyo, Japan). Affi-Gel10 was from Bio-Rad (Hercules, CA). Glutathione and complete protease inhibitor cocktail tablets were from Boehringer Mannheim (Tokyo, Japan). pGEM-T Easy vector DNA and TNT T7 Quick Coupled Transcription/Translation System were from Promega (Tokyo, Japan). Bovine serum albumin, glutamate, NMDA, and tetrodotoxin (TTX) were from Sigma-Aldrich (Tokyo, Japan). TiterMax Gold was from CytRx (Atlanta, GA). The BCA protein assay system was from Pierce (Rockford, IL). Immobilon P membranes were from Millipore (Bedford, MA). HISTOFINE Simple Stain PO (MULTI) system was Nichirei (Tokyo, Japan). Anti-actin antibody was from Chemicon (Temecula, CA). Anti-c-fos antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). HIBERNATE A medium and B27 supplement were from Life Technologies (Tokyo, Japan). (R,S)-3,5-dihydroxyphenylglycine (DHPG) was from Tocris Cookson (Bristol, UK).
Preparation of recombinant BMAL1 proteins. A full-length of cDNA for mBMAL1b was amplified by PCR (94°C for 30 sec, 50°C for 1 min, 72°C for 2 min, 30 cycles), using mouse brain cDNA as a template. The PCR product was ligated into the plasmid pGEM-T Easy and analyzed. The construct was transformed into E. coli strain JM109. Recombinant mBMAL1b protein was produced from the construct byin vitro transcription and translation using the TNT T7 Quick Coupled Transcription/Translation System.
A fragment of cDNA for rBMAL1 (encoding amino acid 154–182; chosen as a region nonhomologous to rTIC, assumed to be a BMAL1 homolog or splice variant) was amplified by PCR (94°C for 30 sec, 52°C for 30 sec, 72°C for 1 min, 30 cycles), using rat brain cDNA as a template. The PCR product was ligated into the plasmid pBluescript SK+ and analyzed, and ligated into pGEX-5X1. The construct was transformed into E. coli strain JM109. Expression of glutathione S-transferase (GST)-rBMAL1 fragment fusion protein (GST-ΔBMAL1) was induced by a final concentration of 0.5 mm isopropyl-thiogalactoside for 12 hr at 25°C. The cells were harvested by centrifugation and disrupted by sonication in buffer A (in mm: 50Tris-HCl, pH 8.0, 50 NaCl, 1 dithiothreitol, and 1 EGTA). After centrifugation at 10,000 × g for 20 min at 4°C, the supernatant was incubated with glutathione-Sepharose for 1 hr at 4°C. After extensive washing with buffer B (0.14 mNaCl, 3 mm KCl, 8 mmNa2HPO4, and 1.5 mmKH2PO4, pH 7.4, and 0.1% Tween 20), the bound proteins were eluted with buffer A containing 20 mm glutathione. The eluate was dialyzed against 1000-fold volume of 20 mm HEPES, pH 7.5, containing 25% glycerol, 0.01% NP-40, and 1 mmdithiothreitol for 6 hr at 4°C.
Preparation of the antiserum against rBMAL1. Primary immunizations were performed with purified GST-ΔBMAL1 using TiterMax Gold, and boosts (at 2 week intervals) used Freund's incomplete adjuvant. Obtained antiserum was used for immunoblot analyses.
Preparation of the tissue extracts. Animals were decapitated at each experimental time point. They were killed under dim light (<1 lux) during the DD cycle or the dark phase during the LD cycle. The brain was removed from the skull and quickly frozen in dry ice. As described previously, the SCN region was punched out with a 2-mm-diameter needle that was inserted to a 1 mm depth into the surface of the coronal plane, an area that included the SCN (Tamaru et al., 1999). Statistical significance was confirmed by using sufficient numbers of SCN. The sample was homogenized in 30 μl of buffer C: 20 mm Tris-HCl, pH 7.5, 0.1% Triton X-100, 0.4 mNaCl, 1 mm phenylmethylsulfonyl fluoride, 5% glycerol, 2 mm EGTA, 20 mm NaF, 0.5 mmNa3VO4, 0.1 mmNa2MoO4, 1 mmdithiothreitol, and 20 mm sn-glycerol 2-phosphate and complete protease inhibitor cocktail. Immunoblot analysis was performed using 10 μl of the extract. For nuclear fractionation, the SCN sample was homogenized in 20 μl of buffer D: 20 mm Tris-HCl, pH 7.5, 0.1% Triton X-100, 0.32 m sucrose, 1.5 mmCaCl2, 10 mm KCl, 3 mmMgCl2, 1 mm phenylmethylsulfonyl fluoride, 25 mm NaF, 0.5 mmNa3VO4, 0.1 mmNa2MoO4, 1 mmdithiothreitol, and 20 mm sn-glycerol 2-phosphate and complete protease inhibitor cocktail. The homogenate was centrifuged at 1000 × g for 10 min. The pellet was solubilized with 20 μl of buffer C. The nuclear fraction was recovered in the supernatant after centrifugation at 10,000 × g for 20 min.
Brains and tissues were removed from rats during the day phase and homogenized using 5 volume buffer C, as described above. Protein concentration was determined using a BCA protein assay system, with bovine serum albumin as a standard. Twenty micrograms of the extracts were subjected to immunoblot analysis.
Immunoblot analysis. The samples were separated on SDS-10% polyacrylamide gels and transferred onto Immobilon P membranes. After blocking with 5% skim milk in Tris-buffered saline with 0.1% Tween 20 (TTBS), membranes were incubated at 4°C for 16 hr with a 1:200 anti-BMAL1 antiserum dilution in TTBS containing 3% bovine serum albumin. After washing with TTBS, membranes were developed with a HISTOFINE Simple Stain PO (MULTI) system according to the manufacturer's protocol. Quantification of bands of interest was performed by densitometry in a computerized image analyzer system (AE-6920; ATTO, Tokyo, Japan). As shown in figure legends, the relative density of BMAL1-IR was normalized against actin-IR (1 = the bottom level in each experiment). Student's t test was used for the statistic analysis of the relative values.
Analysis using the rat SCN slices. Rats aged 4–5 weeks were killed at 9–10 hr after light onset (ZT9–10), and their brains were removed. A region containing SCN located immediately dorsal to optic chiasm was processed into two sections cut at 500 μm. SCN-containing sections were transferred to HIBERNATE A medium supplemented with B27 and placed in an incubator and slowly brought to a final temperature of 35°C. Experiments were performed in the continued presence of 1 μm TTX, which reportedly reversibly blocks electrophysiological output from the circadian clock, but does not affect the timing of the circadian clock itself (Shibata and Moore 1993) to discount intrinsically released glutamate. To examine dose-responsive effects on BMAL1-IR reduction, the slices were treated with glutamate (0, 3, 10, 30, 100, 300 μm) or NMDA (0, 3, 10, 30, 100, 300 μm) for 40 min at projected ZT14. Slices were also treated with glutamate (100 μm), NMDA (100 μm), DHPG (100 μm) or without for the indicated times, which are shown in the figures and figure legends (see Fig. 6).
Fig. 6.
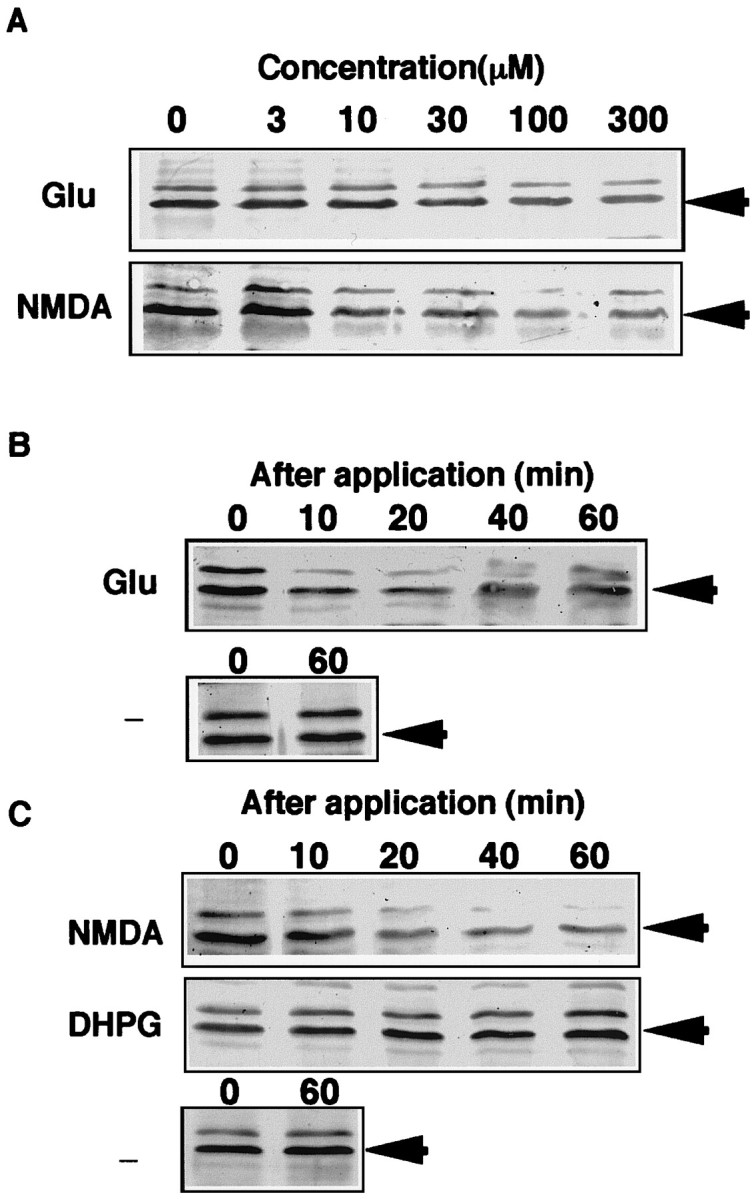
Effects of glutamate receptor agonists on BMAL1-IR in the rat SCN slices. Rats (n = 4 at each treatment) aged 4 weeks were killed at ZT9 to ZT10. At projected ZT14, slices were treated with glutamate (A; 0–300 μm) or NMDA (A; 0–300 μm) for 40 min. Slices were also treated with glutamate (B; 100 μm), NMDA (C; 100 μm) or without (B, C; −) for the indicated time. SCN extracts (A, B) and nuclear (C) fractions containing 10 μg of protein were subjected to immunoblot analysis probing with anti-BMAL1.
RESULTS
Characterization of anti-BMAL1-specific antibody
Polyclonal antibody was raised against recombinant GST-ΔBMAL1 protein by immunizing a rabbit. Specificity of anti-BMAL1 antiserum was then characterized. By immunoblot analysis, anti-BMAL1 antiserum cross-reacted with 70 kDa mouse BMAL1b protein, which was produced byin vitro translation from a cDNA of a full-length type of BMAL1 variant (Fig.1A). In mouse and rat brains, similar BMAL1-like immunoreactivities of ∼70 kDa were detected as a major band. BMAL1-IR in rat brain was specifically absorbed with GST-ΔBMAL1 antigen, but not with GST. Next, we examined the tissue distribution of BMAL1-IR. BMAL1-IR was detected in all regions of rat brain (Fig. 1Ba). BMAL1-IRs were highly detected in skeletal muscle, testis, liver, and spleen, as well as in brain. Brain and testis mainly contained ∼70 kDa BMAL1-IR, whereas the spleen mainly contained one of ∼71 kDa (Fig.1Bb).
Fig. 1.
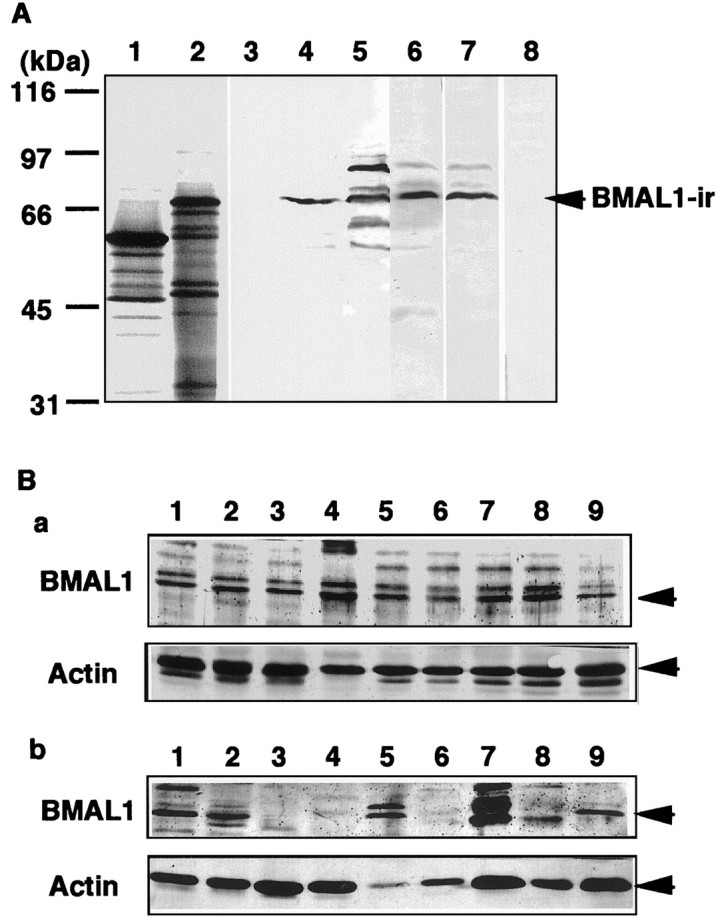
Characterization of anti-BMAL1 antiserum.A, Immunizing a rabbit with GST-ΔBMAL1 (amino acids 154–182) produced anti-BMAL1 antiserum. Recombinant mBMAL1b protein (lanes 2 and 4) and Luciferase (lanes 1 and 3) were produced byin vitro transcription and translation using methionine labeled with 35S (lanes 1 and2) and without (lanes 3 and4). The lysates containing mBMAL1b protein and luciferase, mouse brain extract (20 μg of protein, lane 5), and rat brain extract (20 μg of protein, lanes 6–8) were subjected to immunoblot analysis using anti-BMAL1 antiserum. BMAL1-IR was specifically absorbed by addition of GST-ΔBMAL1 (10 μ g/ml; lane 8) or not of GST (10 μg/ml; lane 7). Ba,Distributions of BMAL1-IR in rat brain. Lysates (20 μg of protein) of hypothalamus (lane 1), olfactory bulb (lane 2), hippocampus (lane 3), pineal body (lane 4), cerebral cortex (lane 5), basal ganglia (lane 6), thalamus (lane 7), superior colliculus (lane 8), and cerebellum (lane 9) were subjected to immunoblot analysis probing with anti-BMAL1 antiserum and anti-actin antibody. Bb, Distributions of BMAL1-IR in rat tissues. Lysates (20 μg of protein) of whole brain (lane 1), testis (lane 2), heart (lane 3), lung (lane 4), liver (lane 5), kidney (lane 6), skeletal muscle (lane 7), intestine (lane 8), and spleen (lane 9) were subjected to immunoblot analysis and probed with anti-BMAL1 antiserum and anti-actin antibody.
Circadian oscillation of BMAL1-IR content in the SCN
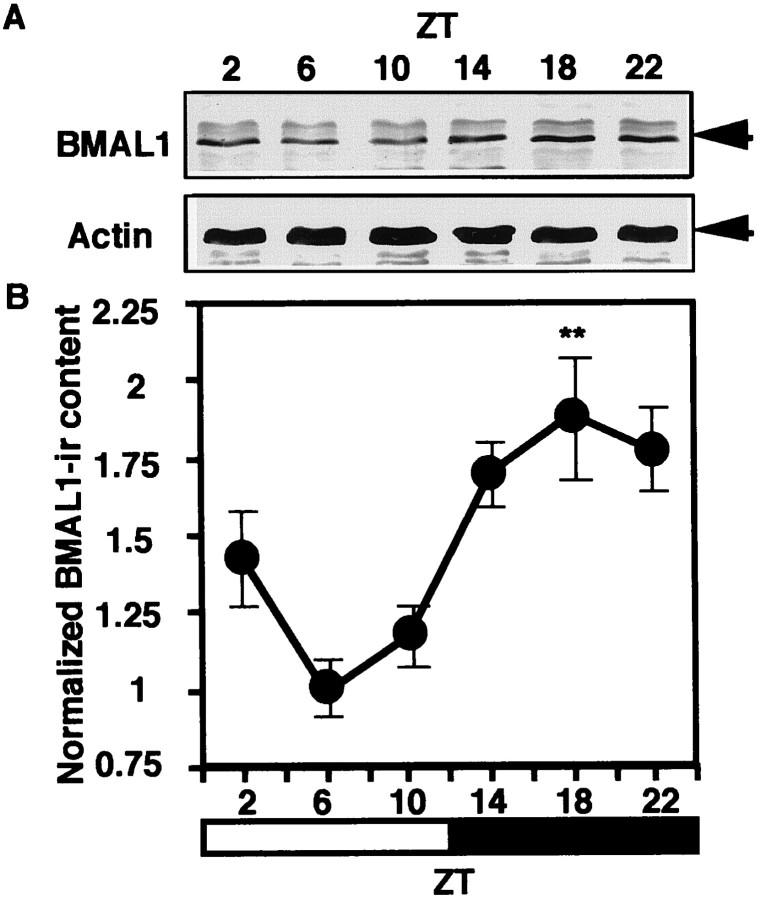
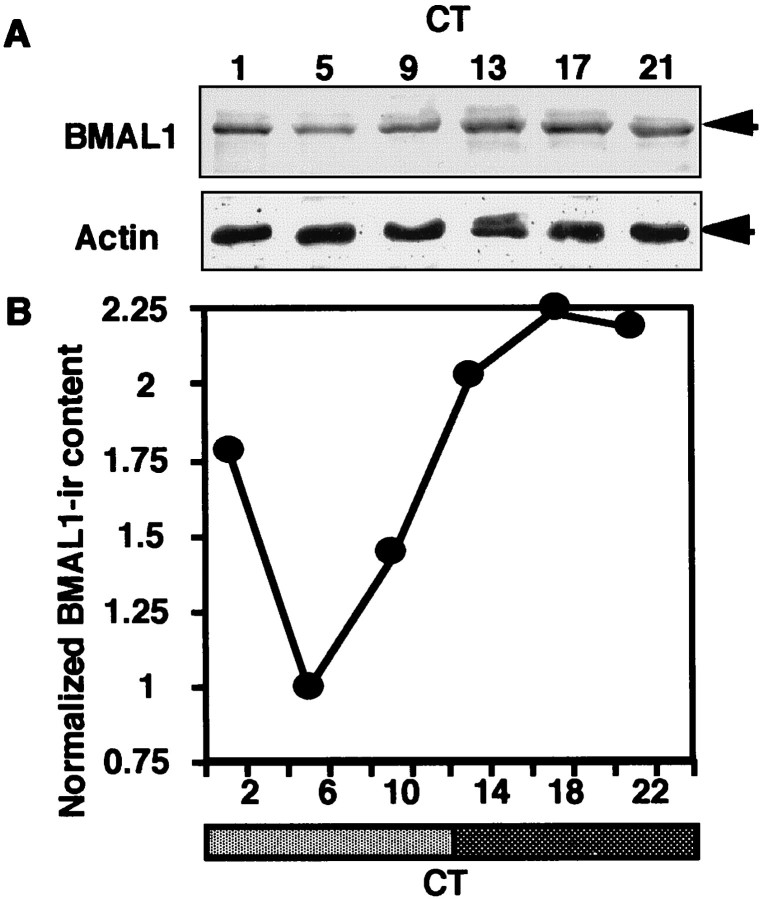
To investigate the roles of BMAL1 in the circadian clock, we examined the circadian profile of BMAL1-IR content in the rat SCN. By immunoblot analysis, robust circadian oscillation of BMAL1-IR content was observed in the SCN under both LD (Fig.2A) and DD conditions (Fig. 3A). A 70 kDa major band and a slightly larger minor band were detected. Normalizing BMAL1-IR content against actin-IR content as a constitutively expressed internal control was performed as the exact quantitative analysis. Circadian BMAL1-IR profiles during LD and DD cycles exhibited evident oscillation with a nocturnal peak (Figs. 2B, 3B). The peak/trough ratios for both cycles were ∼2:1.
Fig. 2.
Circadian profile of BMAL1-IR in the rat SCN during the LD cycle. A, Male Wistar rats aged 5–8 weeks were maintained under a 12 hr light/dark regimen. Rats were killed at ZT2, 6, 10, 14, 18, or 22. The SCN extracts from five animals at each time point were combined and subjected to immunoblot analysis.B, The relative density of BMAL1-IR from the analysis for individual SCN was normalized against actin-IR and plotted (1 = the bottom level at ZT6). Each point represents the mean ± SEM. One-way ANOVA was used for statistical analysis. Statistical significance value: **p < 0.001.
Fig. 3.
Circadian profile of BMAL1-IR in the rat SCN during the DD cycle. A, Male Wistar rats aged 5–8 weeks were maintained under a 12 hr light/dark regimen (light: ZT0–12; dark: ZT12–24), then transferred to dim light (<1 lux). Rats were killed at CT1, 5, 9, 13, 17, or 21. The SCN extracts from six animals at each time point were combined and subjected to immunoblot analysis.B, The relative density of BMAL1-IR was normalized against actin-IR and plotted (1 = the bottom level at CT5).
Light pulse-induced rapid reduction of BMAL1-IR in the SCN during the resetting of the clock
To investigate the roles of BMAL1 in the phase-shifting mechanism of the circadian clock by photic stimulus, we examined the profiles of BMAL1-IR in the rat SCN containing retinorecipient cells after light exposure of rats. Interestingly, a significant decrease in BMAL1-IR content in the SCN was observed within 30 min after 1000 lux light exposure delivered 2 hr after the onset of early night in the LD cycle (ZT14), a result known to cause behavioral phase delay shift (Fig.4A). BMAL1-IR in the SCN also decreased after a 30 min 1000 lux light pulse delivered from ZT14, a stimulus sufficient to cause the phase shift (Fig.4B). Without light illumination at ZT14, decrease of the basal level of BMAL1-IR was not observed from ZT14 to ZT18 (Fig.2). A significant trough in BMAL1-IR content, 48–49% lower than basal level, was observed 1–2 hr after the exposure. BMAL1-IR content returned to near basal level 3 hr after the pulse began. A similar pattern of reduction in BMAL1-IR content was observed after a light pulse during subjective early night [circadian time 15 (CT15)] (data not shown). As positive controls, we detected an acute increase of c-fos protein after the stimulus (data not shown).
Fig. 4.
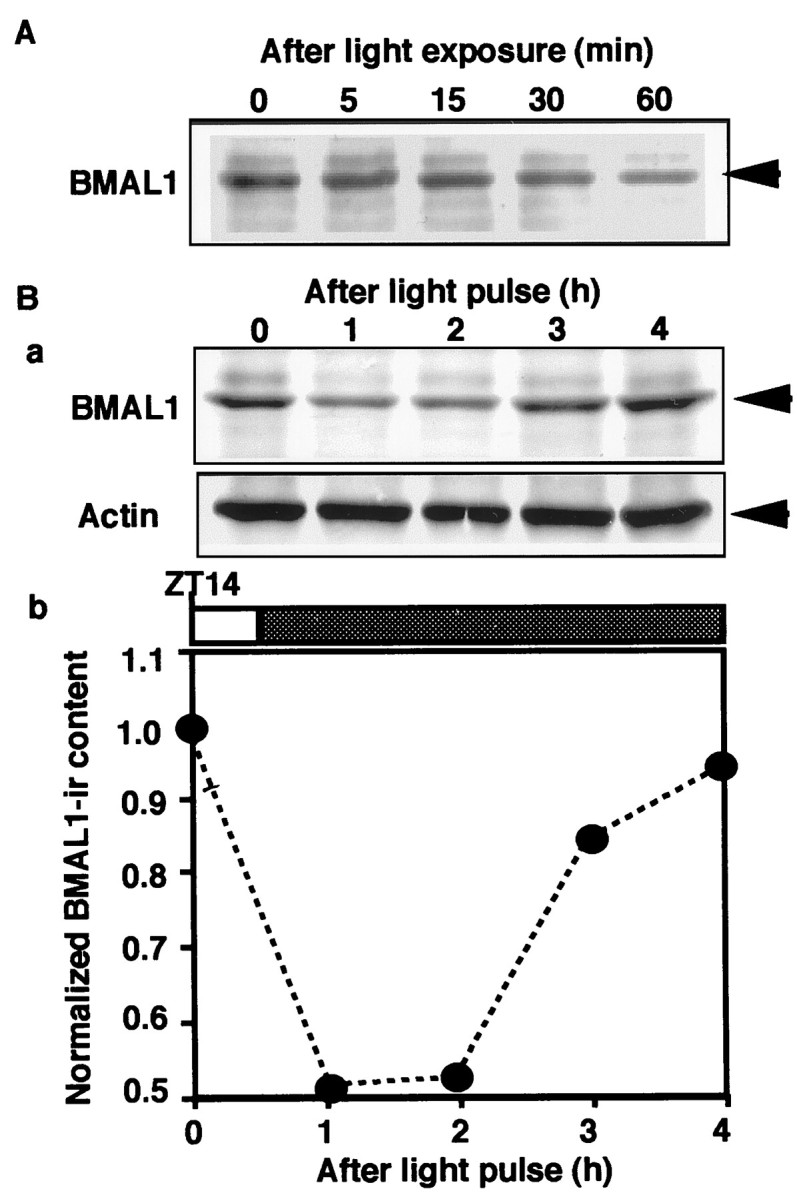
Temporal profile of BMAL1-IR in the rat SCN after light exposure at early night. A, Rats (n = 4 for each lane) were exposed to 1000 lux light delivered from ZT14, then killed at the indicated time. The SCN extract (10 μg of protein) from the rat was subjected to immunoblot analysis probing with anti-BMAL1 antiserum. B, Rats (n = 4 for each lane) were exposed to a 30 min 1000 lux light pulse delivered from ZT14 to ZT14.5, then killed at the indicated time. The SCN extract from the rat was subjected to immunoblot analysis probing with anti-BMAL1 antiserum and anti-Actin antibody (a). The relative densities of BMAL1-IR in the immunoblot membranes were normalized against actin-IR and plotted (1 = the basal level at ZT14) (b).
Nocturnal-specific reduction of BMAL1-IR
Because the circadian clock is entrained by photic stimulus only during subjective night, we examined the phase dependency of the photic reduction of BMAL1-IR. A number of rats (n = 5 at each treatment) were exposed to 30 min, 1000 lux light pulses delivered at early subjective night (CT15), late subjective night (CT21), and mid subjective day (CT6) (Fig.5A). We examined the samples at 2 hr after light exposure, because the level of BMAL1-IR became trough at that time (Fig. 4). SCN from individual rats killed 2 hr after exposure were all subjected to immunoblot analysis. A clear phase dependency was revealed in the photic reduction of BMAL1-IR. A light pulse at CT6 induced no significant change in BMAL1-IR. BMAL1-IR was significantly lower after exposure at CT15 (p < 0.001; 56% of basal level) and modestly lower after exposure at CT21 (p < 0.01; 74% of basal level).
Fig. 5.
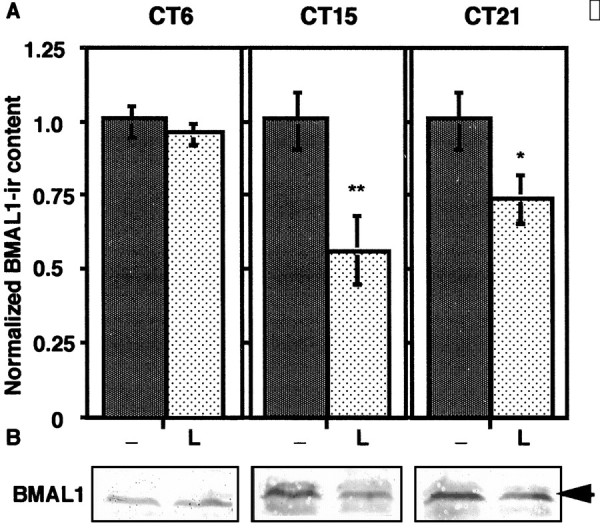
Phase-dependent effect of light exposure on BMAL1-IR in the rat SCN. A, Rats (n= 5 at each treatment) were exposed to 30 min 1000 lux light pulses delivered from CT15, CT21, and CT6 (L), then killed at the same time schedule (CT17). Control (−) rats were kept in darkness, then killed at 2 hr after the exposure. The SCN extracts from the rats were subjected to immunoblot analyses. The relative density of BMAL1-IR was normalized against actin-IR and plotted (1 = the basal level of control). Each column represents the mean ± SEM. Two-tailed Student's t test was used for statistical analysis. Statistical significance value: **p < 0.001; *p < 0.01. B, The SCN extracts from five animals at each time point were combined and subjected to immunoblot analysis. The figure represents the typical pattern for the phase-dependent effect of light exposure on BMAL1-IR in the SCN.
Glutamate and NMDA-induced rapid BMAL1-IR reduction in the SCN slices
To explore the mechanism of photic reduction of BMAL1-IR in the SCN, we performed in vitro pharmacological experiments on the rat SCN slices. Glutamate is believed to be a major neurotransmitter which transduces retinal photic information to the SCN via retinohypothalamic projection. Application of glutamate to the SCN can mimic the photic phase shift (Ding et al., 1994). NMDA alone substantially mimics photic phase shifts (Mintz et al., 1999). At projected ZT14, slices were treated with glutamate (0–300 μm) or NMDA (0–300 μm) for 40 min, and the dose-dependent effects of glutamate and NMDA on BMAL1-IR content in the SCN were examined. Glutamate and NMDA exhibited dose-responsive effects on BMAL1-IR reduction (Fig.6A). The effective concentration of NMDA (>10 μm) revealed slightly more sensitive than glutamate (>30 μm). The maximum reducing effects were observed by using ∼100 μm of glutamate and NMDA.
The precise temporal patterns of BMAL1-IR were analyzed after the application of glutamate receptor agonists. Similar to the photic reduction, rapid BMAL1-IR reduction in the SCN slices began within 10 min after the application of glutamate and continued to decrease for at least 1 hr (Fig. 6B). Evident reduction of BMAL1-IR also was observed even 2 hr after the application of glutamate (data not shown). Acute c-fos protein induction was clearly detected after the application of glutamate, confirming the effect of glutamate on the SCN slices under our experimental conditions (data not shown). Treatment with NMDA also promptly reduced BMAL1-IR in the SCN nuclear fraction, in a pattern similar to the glutamate application (Fig.6C). In addition to ionotropic NMDA receptors, metabotropic glutamate receptor subtypes are also clearly detected in the SCN (Gannon and Rea, 1994). In contrast, DHPG, an agonist for mGluR1 and mGluR5, did not significantly change BMAL1-IR content in the SCN nuclear fraction (Fig. 6C).
DISCUSSION
Using anti-BMAL1 antiserum raised against GST-ΔBMAL1, robust circadian oscillation in BMAL1-IR content in the SCN was detected under LD and DD conditions. BMAL1-IR content peaked during the night phase (but did not significantly fluctuate during the night phase) and reached a trough in the mid-period of the day phase. The peak/trough ratio was ∼2:1. As reported, rBMAL1 transcripts also exhibited evident circadian fluctuation in LD and DD conditions (Honma et al., 1998), with quite similar phase-angle and amplitude to our data for BMAL1 protein. These findings indicate that the change in BMAL1 protein content is a direct manifestation of the change in BMAL1 transcript, without significant time lag and post-translational modulation of the protein content throughout the circadian cycle. CLOCK, the partner of BMAL1, is essential for maintaining circadian rhythm, but the Clock transcript shows no circadian change in the SCN (King et al., 1997). Per gene expression peaks during the day phase (CT4–8) when BMAL1 content reaches a trough (Albrecht et al., 1997; Shigeyoshi et al., 1997; Zylka et al., 1998a).Cry gene expression peaks during the day/night boundary [mCry1 at CT (ZT) 12; mCry2 at CT (ZT) 8–16] (Okamura et al., 1999), when BMAL1 content is still rising to a peak. During this period, all PER and CRY protein levels are elevated (Hastings et al., 1999; Kume et al., 1999; Field et al., 2000). The current evidence does not permit a direct explanation of the generating mechanism of circadian oscillatory BMAL1, Per,and Cry expression. Additional mechanisms may be required for generating the circadian oscillation of the components in the core feedback loop. Nevertheless, our findings suggest that BMAL1 oscillation contributes to circadian oscillatory expression of some additional circadian clock-controlled genes in the SCN.
The specificity of anti-BMAL1 antiserum is discussed from several aspects. As reported, in mice, three splice variants, mBMAL1b (mArnt3), mBMAL1b′, and mBMAL1g′ encode 626, 636, and 222 amino acids, whose predicted molecular weights are 68.6, 69.4, and 25 kDa, respectively (Yu et al., 1999). The BMAL1b variant is assumed to be a full-length type and is used as a “standard” form because it contains all the functional regions, PAS A, PAS B, and basic helix-loop-helix, which are required for interaction with PAS-containing transcription factors like CLOCK and transcriptional activation via E-box. The rBMAL1 antigen was designed as a 28 residue peptide containing amino acids 154–182, corresponding to the C-terminal half of the PAS A domain in rBMAL1b. The antigenic peptide is completely included in mBMAL1b and mBMAL1b′, but not in mBMAL1g′. These variants are expressed widely and differentially in the tissues. A [35S]-labeled band of ∼70 kDa was determined to be a major band in the lysate containing the reaction mixture for in vitro translation using mBMAL1b cDNA as a template and might therefore represent mBMAL1b protein. In the lysate, anti-BMAL1 antiserum almost uniquely recognized the 70 kDa band, demonstrating that anti-BMAL1 antiserum cross-reacts with mBMAL1b protein. In rats, two variants, rBMAL1b and rTIC encoding 626 and 590 amino acids (Honma et al., 1998; Wolting and McGlade, 1998), have predicted molecular weights of 68.5 and 64.7 kDa. rTIC does not include the antigenic peptide, and thus does not contain the functional PAS A domain. In the rat brain extract, a similar 70 kDa band was determined to be a major band. A band (71 kDa) slightly larger than the 70 kDa was clearly detected in certain brain regions such as the SCN. These bands selectively disappeared by the addition of GST-ΔBMAL1, but not of GST, indicating that the bands represent immunoreactivity for ΔBMAL1 but not for GST. These two evidently detected bands of ∼70 kDa were also found in the mouse brain extract and were widespread in a number of the rat brain regions. We suspect that the 70 kDa protein represents BMAL1b, and 71 kDa represents BMAL1b′, but no evidence excludes the possibility that post-translational modifications caused such mobility shifts. In the SCN and retina, the major bands were detected at ∼70 kDa. These results demonstrate that intensities of the immunoreactive bands of ∼70 kDa detected with anti-BMAL1 antiserum might represent the expression levels of functional, typical BMAL1 protein. This has been given recent support by a report that describes the daily accumulation of a 70 kDa antigen in mouse liver detected with the anti-BMAL1 antibody raised against full-length hBMAL1 (Ripperger et al., 2000).
It is surprising that BMAL1-IR remarkably decreased promptly after a nocturnal light pulse. It has been reported that BMAL1transcript does not change for up to ∼1 hr after light pulse (Abe et al., 1998). This evidence strongly suggests that acute BMAL1 protein reduction might be caused by degradation. This implies a certain evolutionary conserved similarity to the fly photoentrainment process accompanying dTimeless degradation (Myers et al., 1996; Zeng et al., 1996; Naidoo et al., 1999). Degradation of dTimeless appears to be one of the pivotal events for the resetting of the fly clock. Thus, BMAL1 reduction (presumably degradation) is also a potential light-sensitive target of the mammalian clock. The effect of light on BMAL1 also provides a potential explanation for entrainment to a 24 hr light/dark cycle. It can account for phase-shifting light pulses having three different time-dependent effects. (1) Light delays the BMAL1 protein cycle at CT15 or ZT14 by causing a reduction of BMAL1. In the early night, reduction of BMAL1 after a light pulse was observed more clearly than in late night. Although BMAL1 protein increases steeply at the boundary from day to night, reduction of BMAL1 at the boundary from night to day seems to be relatively moderate (Figs. 2, 3). Because the difference in increasing and decreasing pattern of BMAL1 protein, we can assume that phase delay shift needs a larger level of BMAL1 protein reduction than phase advance shift. This assumption can explain the reason why light exposure at CT15 cause a larger BMAL1 protein reduction than at CT21. Recovery of BMAL1 content is probably attributable to the existence of substantial transcript levels at these times, so the reduced BMAL1 can be replaced. (2) In the late night, reduction of BMAL1 after a light pulse was observed modestly, probably because the slight circadian change in BMAL1 content during night/day boundary does not require much change to shift into a newly advanced phase. This notion is supported by the fact that BMAL1transcript is also decreasing at these times. (3) During subjective day, no significant reduction of BMAL1 protein is consistent with no evident effect of light on phase shifting at these times.
We cannot exclude the possibility that other components of the system influence light response. Acute induction of Per1 andPer2 transcripts in the SCN by photic stimulation is assumed to be mediated by transactivation of BMAL1/CLOCK heterodimer on their E-box elements or another unknown activating pathway downstream of hypothetical LRE. Induced Per1 and Per2transcripts peak after 1–1.5 hr of light onset, then rapidly decrease (Albrecht et al., 1997; Shigeyoshi et al., 1997; Yan et al., 1999). Our data reveal that the BMAL1 content in the SCN substantially decreases within ∼2 hr after a light pulse. Experiments using hamsters and mice demonstrate that resetting of the circadian oscillator occurs within ∼2 hr, indicating that the molecular photoentrainment process in rodents may be completed at ∼2 hr after photic stimulation (Best et al., 1999). This circumstantial evidence suggests the following hypothesis: during the photoentrainment, acute BMAL1 reduction temporally releases the major part of the E-box elements upstream ofPer genes from the control of the CLOCK/BMAL1 heterodimer for circadian expression. Simultaneously, another unknown LRE-binding factor replaces the major part of the controller upstream ofPer genes, enabling the LRE to induce Per genes acutely. Our findings may shed light on unknown modulating pathways potentially important in the phase resetting of the mammalian circadian clock.
The biochemical mechanism responsible for light-induced BMAL1 reduction (presumably degradation) is largely elusive. Glutamate is considered a major neurotransmitter mediating photoentrainment into the SCN (Ding et al., 1994). Glutamate causes phase shifts on the SCN slice and induces LRE-controlled immediate early genes, i.e., c-Fos andmPer1 (Akiyama et al., 1999), probably via intracellular signaling mainly downstream of an NMDA receptor (Mintz et al., 1999). Our results showed that application of glutamate and NMDA, but not DHPG, on SCN slices directly reduced BMAL1 protein content in a manner similar to the photic reduction in vivo. The reduction in BMAL1 protein appears to occur via certain unknown signal transduction pathways probably including activation of some protein kinases and proteases in the nucleus downstream of the ionotropic NMDA-gated calcium channel. Our findings strongly suggest the existence of novel modulating mechanisms, potentially the primary light-responsive targets before the elevation of LRE-evoked transcription during the resetting of the mammalian biological clock.
Footnotes
This work was supported by a Grant-in-Aid for Scientific Research from Uehara Memorial Foundation (T.T.), a Grant-in-Aid for Scientific Research from Nissan Science Foundation (T.T.), the Project Research Grant 10-20 of Toho University School of Medicine (T.T.), and the Scientific Research Promotion Fund from the Japan Private School Promotion Foundation (T.K.).
Correspondence should be addressed to Teruya Tamaru, Department of Physiology, Toho University School of Medicine, 5-21-16 Ohmori-nishi Ohta-ku, Tokyo 143–8540, Japan. E-mail: tetamaru@med.toho-u.ac.jp.
REFERENCES
- 1.Abe H, Honma S, Namihira M, Tanahashi Y, Ikeda M, Honma K. Circadian rhythm and light responsiveness of BMAL1 expression, a partner of mammalian clock gene Clock, in the suprachiasmatic nucleus of rats. Neurosci Lett. 1998;258:93–96. doi: 10.1016/s0304-3940(98)00877-5. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci. 1999;19:1115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 4.Best JD, Maywood ES, Smith KL, Hastings MH. Rapid resetting of the mammalian circadian clock. J Neurosci. 1999;19:828–835. doi: 10.1523/JNEUROSCI.19-02-00828.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding JM, Chen D, Weber ET, Failman LE, Rea MA, Gillette MU. Resetting the biological clock: nocturnal circadian phase shifts by glutamate and NO. Science. 1994;266:1713–1717. doi: 10.1126/science.7527589. [DOI] [PubMed] [Google Scholar]
- 6.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 7.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated Cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 8.Field MD, Maywood ES, O'Brien JA, Weaver DR, Reppert SM, Hastings MH. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- 9.Gannon RL, Rea MA. In situ hybridization of antisense mRNA oligonucleotides for AMPA, NMDA and metabotropic glutamate receptor subtypes in the rat suprachiasmatic nucleus at different phases of the circadian cycle. Mol Brain Res. 1994;23:338–344. doi: 10.1016/0169-328x(94)90244-5. [DOI] [PubMed] [Google Scholar]
- 10.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 11.Hastings MH, Field MD, Maywood ES, Weaver DR, Reppert SM. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock. J Neurosci. 1999;19(RC11):1–7. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honma S, Ikeda M, Abe H, Tanahashi Y, Namihira M, Honma K, Nomura M. Circadian oscillation of BMAL1, a partner of a mammalian clock gene CLOCK, in rat suprachiasmatic nucleus. Biochem Biophys Res Commun. 1998;250:83–87. doi: 10.1006/bbrc.1998.9275. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda M, Nomura M. cDNA cloning and tissue-specific expression of a novel basic Helix-Loop-Helix/PAS protein (BMAL1) and identification of alternatively spliced variants with alternative translation initiation site usage. Biochem Biophys Res Commun. 1997;233:258–264. doi: 10.1006/bbrc.1997.6371. [DOI] [PubMed] [Google Scholar]
- 14.King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TDL, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian Clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 16.Mintz EM, Marvel CL, Gillespie CF, Price KM, Albers HE. Activation of NMDA receptors in the suprachiasmatic nucleus produces light-like phase shifts of the circadian clock in vivo. J Neurosci. 1999;19:5124–5130. doi: 10.1523/JNEUROSCI.19-12-05124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 18.Naidoo N, Song W, Hunter-Ensor M, Sehgal A. A role for the proteasome in the light response of the Timeless clock protein. Science. 1999;285:1737–1741. doi: 10.1126/science.285.5434.1737. [DOI] [PubMed] [Google Scholar]
- 19.Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, Hoeijmakers JHJ, van der Horst GTJ. Photic induction of mPer1 and mPer2 in Cry-deficient mice lacking a biological clock. Science. 1999;286:2531–2534. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- 20.Rea MA. Light increases Fos-related protein immunoreactivity in the rat suprachiasmatic nuclei. Brain Res Bull. 1989;23:577–581. doi: 10.1016/0361-9230(89)90204-9. [DOI] [PubMed] [Google Scholar]
- 21.Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- 22.Rusak B, Robertson HA, Wisden W, Hunt SP. Light pulses that shift rhythms induce gene expression in the suprachiasmatic nucleus. Science. 1990;248:1237–1240. doi: 10.1126/science.2112267. [DOI] [PubMed] [Google Scholar]
- 23.Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 24.Sangoram AM, Saez L, Antoch MP, Gekakis N, Staknis D, Whiteley A, Fruechte EM, Vitaterna MH, Shimomura K, King DP, Young MW, Weitz CJ, Takahashi JS. Mammalian circadian autoregulatory loop: a Timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron. 1998;21:1101–1113. doi: 10.1016/s0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 25.Shibata S, Moore RY. Tetrodotoxin does not affect circadian rhythms in neuronal activity and metabolism in rodent suprachiasmatic nucleus in vitro. Brain Res. 1993;606:259–266. doi: 10.1016/0006-8993(93)90993-w. [DOI] [PubMed] [Google Scholar]
- 26. Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Lily Y, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 91 1997. 1043 1053. [DOI] [PubMed] [Google Scholar]
- 27.Sun ZS, Albrecht U, Zhunchenko O, Bailey J, Eichele G, Lee CC. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 28.Tamaru T, Okada M, Nagai K, Nakagawa H, Takamatsu K. Periodically fluctuating protein kinases phosphorylate CLOCK, the putative target in the suprachiasmatic nucleus. J Neurochem. 1999;72:2191–2197. doi: 10.1046/j.1471-4159.1999.0722191.x. [DOI] [PubMed] [Google Scholar]
- 29.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 30.van der Horst GTJ, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker APM, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JHJ, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 31.Vitaterna MH, King DP, Chang A-M, Kornhauser JM, Lowrey PL, McDonald D, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, CLOCK, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolting CD, McGlade CJ. Cloning and chromosomal localization of a new member of the bHLH/PAS transcription factor family. Mamm Genome. 1998;9:463–468. doi: 10.1007/s003359900797. [DOI] [PubMed] [Google Scholar]
- 33.Yan L, Takekida S, Shigeyoshi Y, Okamura H. PER1 and PER2 gene expression in the rat suprachiasmatic nucleus: circadian profile and the compartment-specific response to light. Neuroscience. 1999;94:141–150. doi: 10.1016/s0306-4522(99)00223-7. [DOI] [PubMed] [Google Scholar]
- 34.Yu W, Ikeda M, Abe H, Honma S, Ebisawa T, Yamauchi T, Honma K, Nomura M. Characterization of three splice variants and genomic organization of the mouse BMAL1 gene. Biochem Biophys Res Commun. 1999;260:760–767. doi: 10.1006/bbrc.1999.0970. [DOI] [PubMed] [Google Scholar]
- 35.Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 36.Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 37.Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998a;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 38.Zylka MJ, Shearman LP, Levine JD, Jin X, Weaver DR, Reppert SM. Molecular analysis of mammalian Timeless. Neuron. 1998b;21:1115–1122. doi: 10.1016/s0896-6273(00)80628-5. [DOI] [PubMed] [Google Scholar]