Abstract
In Drosophila melanogaster four circadian clock proteins termed PERIOD (PER), TIMELESS (TIM), dCLOCK (dCLK), and CYCLE (CYC/dBMAL1) function in a transcriptional feedback loop that is a core element of the oscillator mechanism. dCLK and CYC are members of the basic helix-loop-helix (bHLH)/PAS (PER-ARNT-SIM) superfamily of transcription factors and are required for high-level expression ofper and tim and repression ofdClk, whereas PER and TIM inhibit dCLK–CYC-mediated transcription and lead to the activation of dClk. To understand further the dynamic regulation within the circadian oscillator mechanism, we biochemically characterized in vivo-produced CYC, determined the interactions of the four clock proteins, and calculated their absolute levels as a function of time. Our results indicate that throughout a daily cycle the majority of the dCLK present in adult heads stably interacts with CYC, indicating that CYC is the primary in vivo partner of dCLK. dCLK–CYC dimers are bound by PER and TIM during the late evening and early morning, suggesting the formation of a tetrameric complex with impaired transcriptional activity. Although dCLK is present in limiting amounts and CYC is by far the most abundant of the four clock proteins that have been examined, PER and TIM appear to interact preferentially with dCLK. Our results suggest that dCLK is the main component regulating the daily abundance of transcriptionally active dCLK–CYC complexes.
Keywords: circadian rhythms, clock proteins, Drosophila, transcription, PAS, protein–protein interactions
A transcriptional feedback loop composed of at least four proteins termed PERIOD (PER), TIMELESS (TIM), dCLOCK (dCLK), and CYCLE (CYC/dBMAL1) is a core element of the circadian clock in Drosophila melanogaster (for review, seeHardin, 1998; Reppert, 1998; Young, 1998; Dunlap, 1999; Edery, 1999). dCLK and CYC are members of the basic helix-loop-helix (bHLH)/PAS (PER-ARNT-SIM) superfamily of transcription factors (Allada et al., 1998; Bae et al., 1998; Darlington et al., 1998; Rutila et al., 1998) that heterodimerize to activate per and tim by binding E-box elements (Hao et al., 1997, 1999; Allada et al., 1998;Darlington et al., 1998; Rutila et al., 1998; Lee et al., 1999) and that act to repress dClk transcription (Glossop et al., 1999).
The biochemical activities of PER and TIM are less well understood than those of dCLK and CYC, but current evidence indicates that they repress their own transcription and activate dClk transcription (Bae et al., 1998; Dunlap, 1999). During the late day/early evening PER and TIM accumulate in the cytoplasm and eventually interact to form a complex (Lee et al., 1996; Zeng et al., 1996) that enters the nucleus in a temporally gated manner (Curtin et al., 1995). Nuclear entry of the PER–TIM complex is accompanied by decreases in the levels ofper and tim transcripts and increases indClk transcripts. In Drosophila tissue culture cells the ectopic coexpression of PER and TIM inhibit dCLK-mediated stimulation of a reporter gene driven by E-box elements found in 5′ regulatory regions of per or tim (Darlington et al., 1998). Furthermore, PER and TIM interact with dCLK or a dCLK-containing complex during times in the day (Lee et al., 1998) when the transcription rates of per and tim are decreasing (So and Rosbash, 1997) and dClk transcripts increase (Bae et al., 1998), consistent with the suggestion that dCLK-mediated transcriptional regulation (either positive or negative) is inhibited by the binding of PER and/or TIM. Recent results usingin vitro-synthesized clock proteins suggest that PER and TIM participate in the circadian feedback mechanism, at least partly, by abrogating the DNA binding activity of a dCLK–CYC heterodimer (Lee et al., 1999).
To gain further insight into the molecular circuitry underlying the circadian transcriptional feedback mechanism in Drosophila, we biochemically characterized CYC produced in vivo and defined its interactions with dCLK, PER, and TIM as a function of time. In addition, although previous studies have measured the relative levels of individual clock proteins, their absolute amounts are not known. To this end we calculated the molar concentrations of dCLK, CYC, PER, and TIM in adult fly heads as a function of time throughout a daily cycle. We show that in adult heads the majority of dCLK stably interacts with CYC throughout a daily cycle, strongly suggesting that the primary or perhaps only physiologically relevant partner of dCLK in this tissue is CYC. PER and TIM mainly interact with the dCLK–CYC transcription factor during the late night. dCLK is present in limiting amounts, and our findings suggest that PER and TIM preferentially interact with dCLK as compared with CYC. Similar to recent results obtained by using analogous proteins synthesized in vitro(Lee et al., 1999), the binding of PER and TIM do not affect the association of dCLK with CYC. Together, our findings strongly suggest that dCLK is the key “molecular bridge” regulating the dynamic interactions that activate or repress gene expression within theDrosophila circadian feedback mechanism.
MATERIALS AND METHODS
Fly strains and collections. The wild-type Canton-S (CS) flies and the mutant per01flies used in this study were descendants of stocks originally maintained in the laboratory of Dr. M. Rosbash (Brandeis University, Waltham, MA), and were described previously (Edery et al., 1994). The tim0 flies were descendants of stocks originally maintained in the laboratory of Dr. A. Sehgal (University of Pennsylvania Medical School, Philadelphia, PA) (Sehgal et al., 1994), and the cyc0flies were described previously (Rutila et al., 1998). All flies were grown and maintained in vials or bottles containing standard agar-cornmeal-sugar-yeast-tegosept media. Vials containing ∼100 young (2- to 6-d-old) adult flies were placed in incubators (Precision Scientific, Winchester, VA) at 25°C, exposed to at least two cycles of 12 hr light/dark [LD; in which Zeitgeber time 0 (ZT0) is lights-on and ZT12 is lights-off], and subsequently were maintained in the dark (DD). At selected times during LD and DD the flies were collected by rapid freezing in dry ice, and the heads were isolated.
In vitro transcription and translation. Recombinant plasmids used in this study for the in vitro synthesis of PER, TIM, dCLK, and CYC were described previously (Citri et al., 1987; Bae et al., 1998; Lee et al., 1999). To produce DOUBLE-TIME (DBT) in vitro (see Fig. 1A), we obtained a cDNA plasmid containing the entire open reading frame (ORF) ofdbt (GenBank accession number AF055583) from Genome Systems (St. Louis, MO). Subsequently, the plasmid was digested withSacI and SacII, and the entire dbt ORF was subcloned into pGEM-5Zf (+) (Promega, Madison, WI), which placed the expression of dbt under control of the SP6 promoter (yielding a plasmid termed pGEM-5Zf dbt). In vitro radiolabeled translation products were produced by using the appropriate circular plasmids (described above) to prime a coupled transcription/translation rabbit reticulocyte system (TNT; Promega) in the presence ofl-[35S]methionine (Amersham, Arlington Heights, IL) according to the manufacturer's protocol. The amount of protein produced in each translation was determined by subjecting an aliquot of the translation mixture to trichloroacetic acid precipitation and normalizing for methionine content. An incubation that did not contain exogenously added plasmid served as a background control for the translation reactions. In vitro-translated proteins were resolved by PAGE with 6 or 12% gel and visualized by either autoradiography (e.g., see Fig.1A) or immunoblotting (e.g., see Fig.1B).
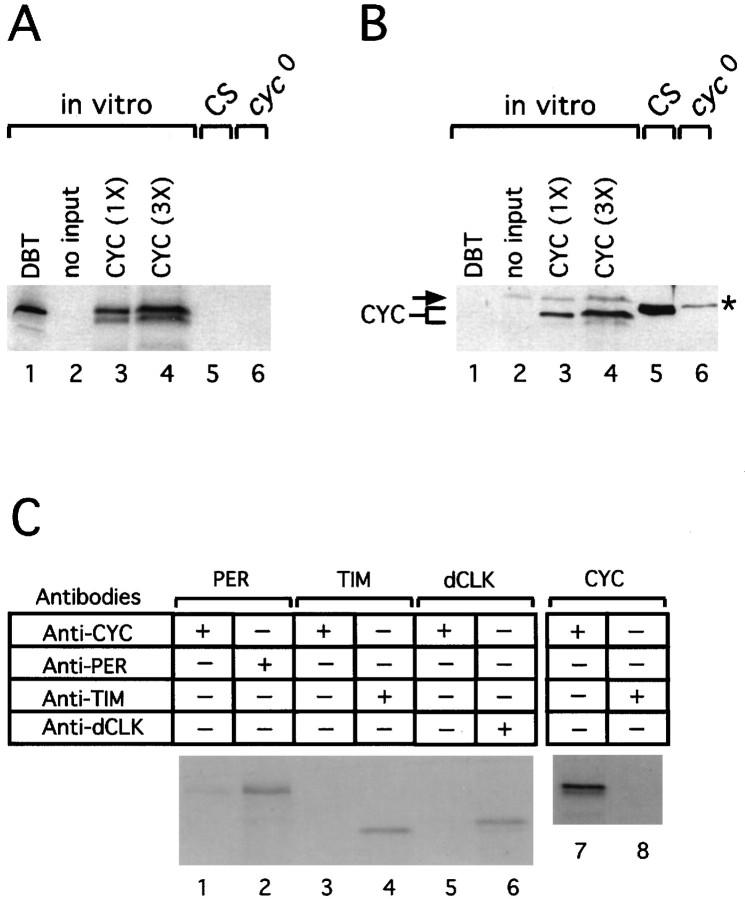
Fig. 1.
Biochemical detection of CYC in Drosophila melanogaster heads. Cell-free extracts were derived from eitherin vitro translation reactions performed in the presence of [35S]methionine (A,B,lanes1–4, andC) or adult fly heads (A,B,lanes5, 6). The radiolabeled target proteins produced in vitro (i.e., DBT, PER, TIM, dCLK, and CYC) and the genotype of flies that were used to prepare head extracts (i.e., wild-type CS flies orcyc0 mutants) are indicated ontop. Head extracts were prepared from flies collected at time 25 (in hours since the last dark/light transition at ZT0).C, In vitro translation products were incubated with the indicated antibodies, and immune complexes were recovered. Fly head extracts, in vitro translation reactions, and immune complexes were resolved by 6 or 12% (in the case of CYC) PAGE and either visualized by fluorography and autoradiography (A, C) or transferred to nitrocellulose; the immunoblots were probed with anti-CYC antibodies (B).A, B, Two different amounts of in vitro-translated CYC were resolved by PAGE (1× = 77 pg).B, The arrow (left) and asterisk (right) identify nonspecific bands that cross-react with the anti-CYC antibody in rabbit reticulocyte lysates (lanes1–4) and head extracts (lanes5, 6), respectively.
Antibodies and immunoblotting. To generate antibodies to CYC, we used PCR to amplify the entire open reading frame ofcyc by using a plasmid that contained a full-lengthcyc cDNA (EST plasmid obtained from Genome Systems; GenBank accession number AA695336). Then the PCR product was digested withNdeI and EcoRI and cloned upstream of sequences that encode a polyhistidine stretch (His) in the expression vector pET23b (Novagen, Madison, WI). The oligonucleotide primers used in the PCR were (cyc sequences are in italics) 5′-AAATCATATGGAAGTTCAGGAGTTCTGCG-3′ and 5′-GGATAAGAACACGGAATTCTTGGCG-3′. The CYC–His fusion protein was produced in bacteria according to the manufacturer's recommended procedure (Novagen) and purified under denaturing conditions (8 m urea), by using the Talon metal affinity resin from Clontech (Palo Alto, CA). The purified CYC–His fusion protein was used as an immunogen to produce antibodies in rats and guinea pigs (Cocalico Biologicals, Reamstown, PA). In this study we used a guinea pig anti-CYC antibody (GP-122) that strongly recognized in vitro-translated CYC (see Fig. 1).
Preparation of total fly head extract was essentially as described (Edery et al., 1994; Lee et al., 1998). For each time point ∼30 μl of heads isolated from frozen flies were placed in a Microfuge tube and homogenized at 4°C in 3 vol (relative to heads) of extraction buffer 1 [EB1; containing (in mm) 100 KCl, 20 HEPES, pH 7.5, 5 EDTA, 1 dithiothreitol (DTT), 0.25 PMSF, 5% glycerol, and 0.1% Triton X-100, plus 10 μg/ml aprotinin, 5 μg/ml leupeptin, and 1 μg/ml pepstatin A], in a battery-operated minihomogenizer (Kontes, Vineland, NJ). Subsequently, homogenates were centrifuged twice (12 min at 12,000 × g), and clarified supernatants were removed to new tubes. Protein concentration was determined by using a Coomassie protein assay according to the manufacturer's instructions (Pierce, Rockford, IL). An equal volume of 2× SDS-sample buffer was added to the supernatant fraction, and the mixture was boiled. Equal amounts of total protein (∼20 μg total at ∼4 μg/μl) from clarified supernatant fractions were resolved by PAGE and transferred to nitrocellulose paper; immunoblots were treated with chemiluminescence (ECL, Amersham) essentially as described (Lee et al., 1998; Sidote et al., 1998). To visualize dCLK, PER, and TIM, we used 6% SDS-polyacrylamide gels, whereas 12% SDS-polyacrylamide gels were used to detect CYC, as indicated in the figure legends. Immunoblots were incubated in the presence of anti-dCLK, anti-PER, anti-TIM, or anti-CYC antibodies at a final concentration of 1:2000. The antibodies to dCLK (dCGP90), PER (GP73), and TIM (TR1-E3 and GP72-2) used in this study were as described (Lee et al., 1998; Sidote et al., 1998). Bands on autoradiographs were quantified with a densitometer (Computing Densitometer Scan version 5.0) and ImageQuant software (Molecular Dynamics, Sunnyvale, CA). Scanned images of autoradiographs were manipulated with Adobe Photoshop 5.0 and Canvas 5.0.3 software.
To determine the concentrations of PER, TIM, dCLK, and CYC in extracts prepared from adult fly heads (see Fig. 4), we resolved (1) serial dilutions of different head extracts (4 μg of total protein/μl), each one corresponding to a time in LD when peak levels of either dCLK (i.e., ZT4), TIM (i.e., ZT16), or PER (i.e., ZT20) were attained, and (2) reticulocyte lysates containing serial dilutions of known amounts of target proteins by SDS-PAGE and immunoblotted them. The intensities of appropriate bands were measured with a densitometer, as described above. In each experiment the intensities of relevant immunoreactive bands in head extracts and reticulocyte lysates yielded linear doses–responses (data not shown). Absolute peak levels for each clock protein in head extracts were calculated by comparing their staining intensities with those from the relevant radiolabeled products, followed by correction for methionine content to obtain molar concentrations. For each protein the calculations of peak values derived from at least two independent experiments were pooled. Estimates of peak amounts for any given protein did not vary by >25% when comparing values obtained from the different independent experiments (data not shown). After the average peak levels for each of the target proteins were determined, the rest of the daily values were obtained by curve fitting, using the relative abundance profiles shown in Figure 3D. To ensure that our estimates of the absolute levels of individual clock proteins as a function of time were reliable, we also repeated the same procedure by using head extracts in which individual clock proteins attained ∼25 and 50% of peak levels (i.e., ZT12 and 16 for PER; ZT12 and 20 for TIM; ZT12, 16, 20, and 23.9 for dCLK). For each clock protein the results obtained were similar to those based on peak values (data not shown).
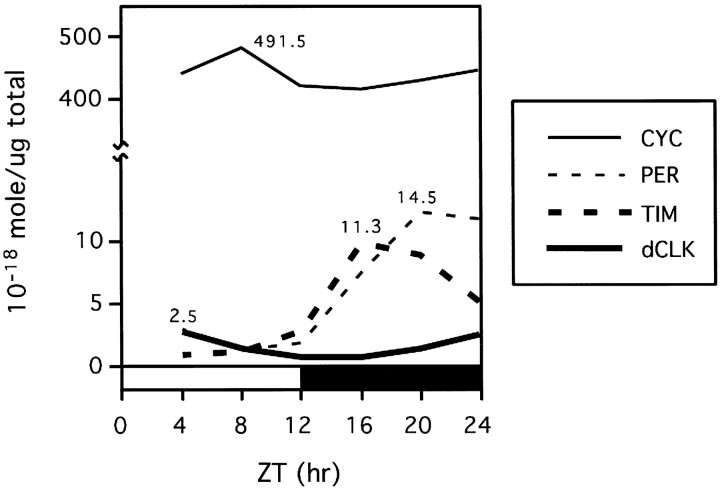
Fig. 4.
Average daily levels of dCLK are limiting. Shown are the molar concentrations (10−18 mol/μg total head protein) of dCLK, CYC, PER, and TIM during LD. For each protein the peak amounts were calculated by pooling results obtained from at least two independent experiments. The rest of the data points were generated by curve fitting, using the results shown in Figure3D (see Materials and Methods). Peak concentrations (10−18 mol/μg total head protein) for each protein also are indicated. Note that, because PER has a very broad electrophoretic mobility during the late night/early morning attributable to differential phosphorylation (Edery et al., 1994), we believe that PER levels during these times might be overestimated by up to 50% (data not shown).
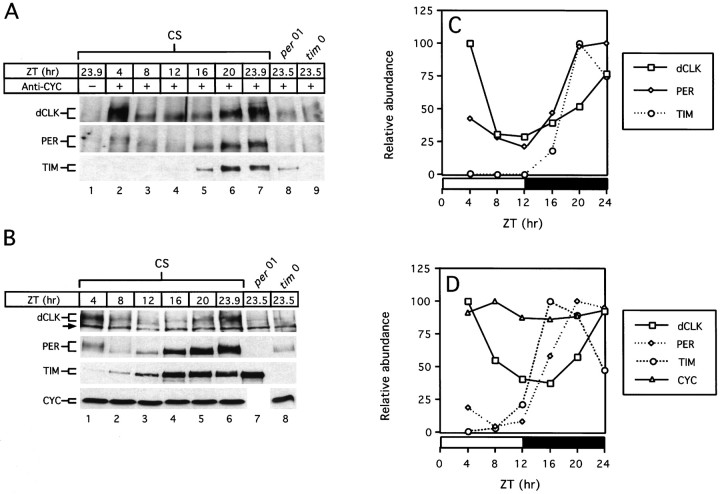
Fig. 3.
dCLK, PER, and TIM interact with CYC in a time-of-day specific manner. Wild-type (CS) flies were collected at the indicated times in LD (A, lanes1–7; B, lanes1–6). per01(A, lane 8; B, lane 7) and tim0(A, lane 9; B, lane 8) mutants were collected at time 23.5. Head extracts were prepared and either subjected to immunoprecipitation using antibodies against CYC (A) or analyzed directly (B). A, Immune pellets were divided into three equal aliquots; each fraction was probed for the presence of dCLK (top), PER (middle), or TIM (bottom). A,B, Twelve percent polyacrylamide gels were used to detect CYC, whereas 6% polyacrylamide gels were used to detect PER, TIM, and dCLK. The size ranges of the relevant proteins are indicated (left). The arrow (left, top panelin B) indicates a nonspecific band recognized by the anti-dCLK antibody that was used. C,D, Quantitation of results shown in A and B, respectively. Peak values for each protein were set to 100, and the rest of the values were normalized. Horizontal barsrepresent either 12 hr light (open bar) or 12 hr dark (filledbar).
RNase protection assay. For each time point, total RNA was extracted from ∼10 μl of fly heads by using TriReagent (Sigma, St. Louis, MO), as described previously (Majercak et al., 1997). The abundance of cyc and per transcripts was determined by RNase protection assays (Hardin et al., 1990) performed with the modification previously described (Zeng et al., 1994). To measure cyc RNA levels, we linearized the cyc EST plasmid (described above) with BglII and produced antisense radiolabeled probe in vitro by using T7 RNA polymerase. The radiolabeled antisense probe used to determine the levels ofper RNA was as previously described (Bae et al., 1998;Sidote et al., 1998). As a control for RNA loading in each lane, a ribosomal protein probe (RP49) was included in each protection assay (Hardin et al., 1990). Protected bands were quantified with a PhosphorImager from Molecular Dynamics, and values were normalized relative to those of RP49 (Hardin et al., 1990).
Immunoprecipitation. For each time point the fly heads were homogenized as described above, except that extraction buffer 2 (EB2) was used instead of EB1 [containing (in mm) 5 Tris-HCl, pH 7.5, 50 KCl, 10 HEPES, pH 7.5, 1 EDTA, 1 DTT, 0.25 PMSF, 10% glycerol, 0.05% Triton X-100, plus 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 μg/ml pepstatin A]. To remove nonspecific interactions, we first incubated head extracts (1.2 mg of total protein in a final volume of 0.4 ml) with 20 μl of Gammabind Plus beads (Pharmacia, Piscataway, NJ) for 25 min at 4°C; we centrifuged and then removed the clarified supernatant to a new tube. A slurry containing either 3 μl of anti-CYC (GP-122) or anti-dCLK (dCGP90) antibody and 20 μl of Gammabind Plus (Pharmacia) were added to the precleared head extracts and incubated with gentle rotation for 2 hr at 4°C. Subsequently, the beads were collected by centrifugation; in some cases (see Fig. 5) additional rounds of immunoprecipitation were performed on the clarified supernatants. Immune complexes were washed three times (0.5 ml of EB2 for 7 min each), mixed with 20 μl of 1× SDS-sample buffer, boiled, and centrifuged; the resulting supernatant was resolved by immunoblotting as described above. A similar procedure also was used to immunoprecipitate radiolabeled target proteins synthesized in vitro (see Fig. 1A).
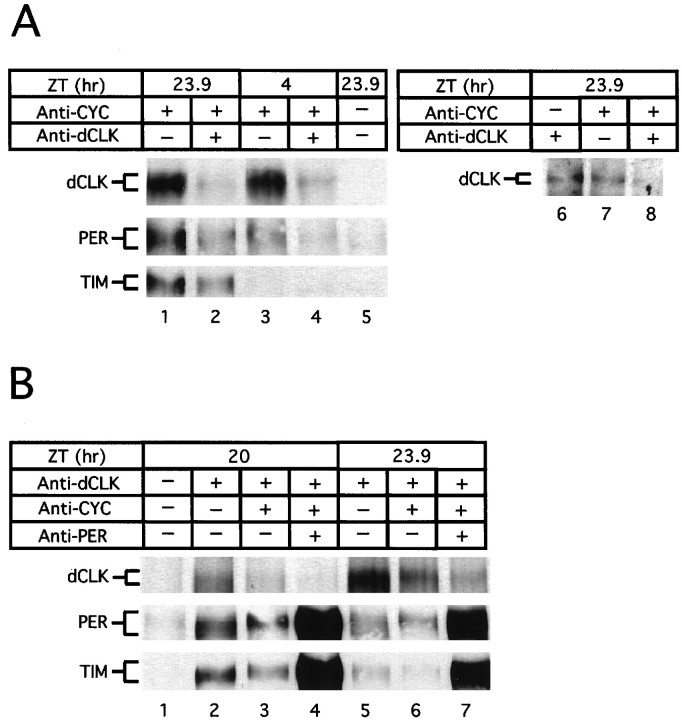
Fig. 5.
The majority of dCLK is bound to CYC during a daily cycle. A,B, Wild-type flies were collected at the indicated times during LD; head extracts were prepared and subjected to immunoprecipitation (IP), using antibodies against either CYC (A, lanes1–4, 7, 8) or dCLK (A, lane 6;B, lanes2–7). In some cases the remaining supernatant fraction subsequently was subjected to a second round of IP, using antibodies against either dCLK (A, lanes2, 4, 8) or CYC (B, lanes3, 4, 6, 7). Finally, in some cases the supernatant resulting from the second IP was subjected to a third round of IP, using antibodies against PER (B, lanes4, 7). Recovered immune complexes were probed for the presence of dCLK, PER, or TIM as indicated (left of panels). Control incubations using irrelevant antibodies were used to show specificity during IP (A, lane 5; B, lane 1) (data not shown).
RESULTS
The abundance of CYC is constant throughout a daily cycle
Previous work has demonstrated that in adult heads the levels of PER, TIM, and dCLK undergo daily oscillations (Edery et al., 1994;Hunter-Ensor et al., 1996; Myers et al., 1996; Zeng et al., 1996; Lee et al., 1998). Heads normally are used to investigate the temporal profiles of clock proteins in Drosophila because it is the anatomical location of the best-characterized circadian pacemaker in this species (Handler and Konopka, 1979; Ewer et al., 1992). To characterize CYC biochemically in vivo, we immunized guinea pigs and rats with bacterially produced CYC (see Materials and Methods). The immunoreactivity and specificity of the individual antiserum were tested initially by performing Western blots (immunoblots) of in vitro-translated CYC (Fig.1) (data not shown).
Figure 1B shows a typical immunoblotting result obtained with our strongest anti-CYC antibody; in vitro-translated CYC [Fig. 1A, lanes 3, 4; the appearance of a minor band that migrates slightly faster than the major in vitro-synthesized CYC product is attributable to the occasional use of an internal in-frame AUG (data not shown)], but not an equivalent molar amount of anotherDrosophila clock protein of similar predicted molecular weight, termed DOUBLE-TIME (DBT) (Kloss et al., 1998; Price et al., 1998) (Fig. 1A, lane 1), was recognized by the anti-CYC antibody (Fig. 1B, comparelanes 3, 4 with 1). In addition, the anti-CYC antibody used in this study immunoprecipitates little or no PER, TIM, or dCLOCK (Fig. 1C). When head extracts prepared from wild-type flies were probed by immunoblotting in the presence of this anti-CYC antibody, a strongly staining band of ∼45 kDa was detected (Fig. 1B, lane 5), in close agreement with the predicted molecular mass of CYC (Rutila et al., 1998). Importantly, this ∼45 kDa immunoreactive band is absent in head extracts prepared from either a presumptive null-mutant termedcyc0 (Fig. 1B,lane 6) or wild-type flies probed with preimmune sera (data not shown). Daily rhythms in locomotor activity and eclosion (emergence from pupal cases) as well as cycles in the protein and RNA products from per, tim, and dClk are abolished in cyc0 flies (Rutila et al., 1998; Glossop et al., 1999). Thecyc0 mutation introduces a premature stop codon that is predicted to remove the C-terminal 60% of this protein, including most of the PAS domain, consistent with its loss-of-function phenotype (Rutila et al., 1998). We did not observe the appearance of novel smaller molecular weight products in thecyc0 mutant when immunoblots were incubated with anti-CYC antibody (data not shown), suggesting that the putative truncated CYC protein produced in this mutant is highly unstable. The reason or reasons for the slight variation in electrophoretic mobility between CYC synthesized in vitro(Fig. 1B, lane 4) and that produced in the fly head (Fig. 1B, lane 5) are not known.
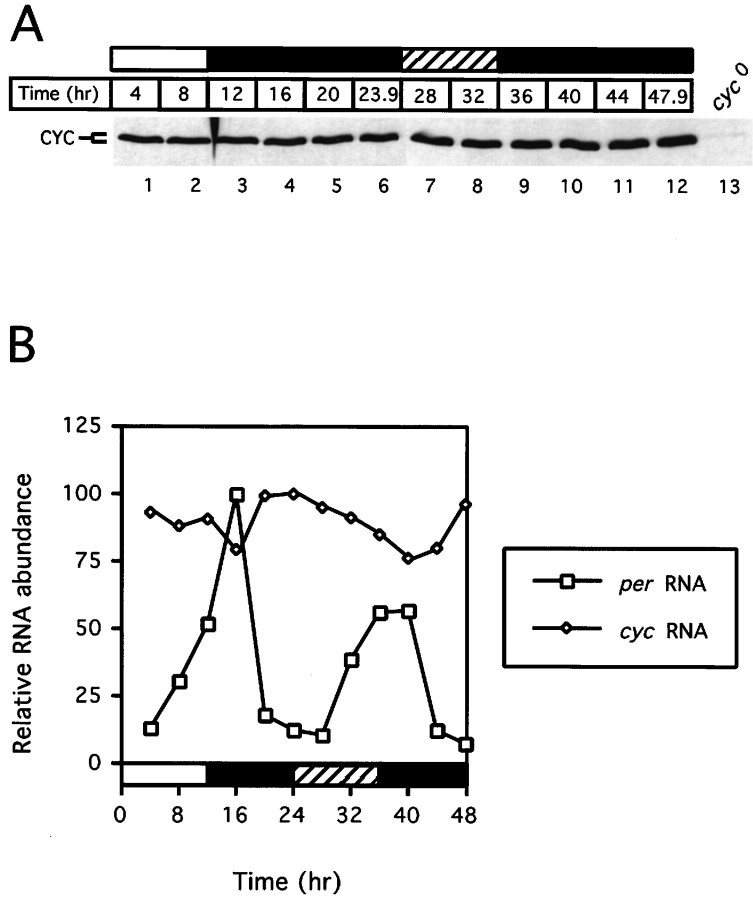
To test whether the abundance of CYC undergoes daily oscillations, we entrained wild-type flies for 3–4 d under standard conditions of 12 hr light/dark cycles [LD; in which Zeitgeber time 0 (ZT0) is lights-on and ZT12 is lights-off], followed by constant darkness (DD). Flies were collected at various times during LD and DD, and head extracts were analyzed for the presence of CYC by immunoblotting (Fig.2A). In sharp contrast to PER (Edery et al., 1994), TIM (Hunter-Ensor et al., 1996; Myers et al., 1996; Zeng et al., 1996), and dCLK (Lee et al., 1998), the abundance of CYC remains constant during LD and DD. This result is consistent with the observation that cyc mRNA levels are expressed constitutively (Fig. 2B), as previously reported (Rutila et al., 1998). We cannot rule out the possibility that in a limited number of cells CYC undergoes daily changes in abundance that are not detected by immunoblot analysis of total head extracts. Furthermore, we did not detect changes in the electrophoretic mobility of CYC as a function of time in a daily cycle (Fig.2A). In contrast, PER (Edery et al., 1994), TIM (Zeng et al., 1996), and dCLK (Lee et al., 1998) are modified by the addition of phosphate moieties in a time-of-day specific manner, which results in readily detectable changes in their electrophoretic mobilities. Nonetheless, the different apparent molecular weights of in vivo- and in vitro-produced CYC (see Fig.1B) could suggest that, in Drosophila, CYC undergoes post-translational modifications and/or that different size variants of CYC are generated by alternative splicing or differential usage of translation start sites.
Fig. 2.
Constitutive levels of CYC protein and RNA throughout a daily cycle. Wild-type flies were exposed to three 12 hr light/dark cycles (LD) and subsequently were kept in constant dark conditions (DD). Collections were done at the indicated times (time 0 is defined as the last dark-to-light transition), and head extracts either were analyzed by immunoblotting, using antibodies directed against CYC (A), or were subjected to RNase protection assays (B). A, As a control for specificity, head extracts prepared fromcyc0 flies were included (lane 13). B, Comparison of the relative amounts ofcyc (open diamond) and per(open square) RNA during the third day of LD and the first day of DD. Relative RNA levels refers to ratios ofcyc or per transcripts to the constitutively expressed RP49 RNA. Peak values for cycor per during a daily cycle were set to 100, and the rest of the values were normalized. Horizontal barsrepresent lights-on (open bar), lights-off (filled bar), or subjective day (hatched bar). Similar results were obtained in three independent experiments; representative examples are shown.
dCLOCK interacts with CYC throughout a daily cycle
To determine the interaction profile of CYC as a function of time in a daily cycle, we incubated head extracts with anti-CYC antibodies and probed the recovered immune complexes for the presence of PER, TIM, and dCLK (Fig. 3A). Unfortunately, we were not able to visualize immunoprecipitated CYC by Western blotting because of the presence of strongly staining nonspecific bands that comigrated with CYC (data not shown). To estimate the efficiency of CYC recovery from head extracts with our anti-CYC antibody, we added trace amounts of in vitro-generated radiolabeled CYC to head extracts. The results indicated that, under the conditions used in this study for immunoprecipitation, the majority of the radiolabeled CYC protein was recovered in the immune complex, with very little remaining in the supernatant (data not shown). This suggests that the bulk of thein vivo-produced CYC protein present in head extracts also is being recovered after incubation with anti-CYC antibodies.
The findings clearly indicate that, as expected, dCLK and CYC form a partnership in vivo, an interaction that is observed throughout a daily cycle (Fig. 3A, top panel). In agreement with previous findings that used anti-dCLK antibodies to perform immunoprecipitations (Lee et al., 1998), PER and TIM mainly associate with CYC (or a CYC-containing complex) during the night (Fig. 3A, bottom two panels) when the transcription rates of per andtim are low (So and Rosbash, 1997). Very little dCLK is present in immune complexes recovered from head extracts prepared fromper01 andtim0 mutant flies (Fig.3A, top panel, lanes 8, 9), which do not make functional PER and TIM, respectively (Konopka and Benzer, 1971; Sehgal et al., 1994). This is consistent with an earlier finding showing that the daily accumulation of dCLK requires PER and TIM (Lee et al., 1998) (Fig. 3B, top panel, lanes 7, 8). In sharp contrast, the abundance of the CYC protein does not appear to be regulated by PER or TIM (Fig. 3B, bottom panel, lane 8) (data not shown).
dCLOCK is present in limiting amounts in head extracts
A comparison of the curves for the daily changes in the levels of dCLK, PER, and TIM after recovery with anti-CYC antibodies with those obtained when total head extracts are analyzed directly revealed that the temporal abundance profiles of dCLK are very similar in both cases (compare Fig. 3C,D). Moreover, immune complexes recovered with antibodies that recognize either dCLK or CYC yield essentially identical profiles in the PER and TIM abundance rhythms (Fig.3A) [see Lee et al. (1998), their Fig. 4]. A possible explanation that could account for these observations is that the interaction of PER and TIM with CYC is mediated by dCLK and that dCLK is generally present in limiting amounts. However, molar concentrations have not been calculated for any Drosophila clock protein. To better understand the variables that might contribute to the dynamic changes underlying the interactions among CYC, dCLK, PER, and TIM, we determined the absolute concentrations of the different clock proteins in head extracts as a function of time in a daily cycle (Fig.4; see Materials and Methods).
In LD the peak molar concentration of dCLK is four- to sixfold lower than the highest values obtained for PER and TIM, and it is ∼200-fold lower than those of CYC (Fig. 4), making CYC by far the most abundant of the four clock proteins that have been examined. The steady-state molar concentrations of PER and TIM are similar in head extracts, supporting earlier calculations of the relative molar amounts of each protein (Zeng et al., 1996; Lee et al., 1998). On the basis of the large molar excess of CYC as compared with dCLK, we reasoned that throughout a daily cycle the majority of dCLK might be bound to CYC. This prediction was confirmed by the observation that even when dCLK is at peak levels little, if any, dCLK remains in the supernatant fraction after recovery of immune complexes with anti-CYC antibodies (Fig.5A, top panel, compare lanes 2 and 1, 4 and3). That the majority of dCLK in the adult head is stably bound to CYC also is indicated by the observation that approximately equal amounts of dCLK are immunoprecipitated from head extracts with antibodies against either dCLK or CYC (lanes 6, 7).
The results further indicate that, although significantly more PER and TIM interact with the dCLK–CYC complex at ZT23.9 as compared with ZT4 (Fig. 5A, bottom two panels, comparelanes 1, 3), there is no increase in the relative amount of dCLK that is free of CYC (Fig. 5A, top panel, compare lanes 1, 2 with 3, 4). The observation that the majority of dCLK stably associates with CYC despite the presence of significant amounts of PER and TIM provides biochemical evidence strongly suggesting that,in vivo, the binding of PER and/or TIM to dCLK and/or CYC does not disrupt the dCLK–CYC heterodimer. These in vivoresults are consistent with recent findings obtained by using the analogous proteins synthesized in vitro (Lee et al., 1999).
Because the molar concentrations of PER and TIM are substantially higher than those of dCLK during the night (see Fig. 4), we wondered whether PER and TIM interact with CYC that is not bound to dCLK. To address this possibility, we first recovered immune complexes with anti-dCLK antibodies (Fig. 5B, lanes 2, 5) and subjected the supernatant fraction to another round of immunoprecipitation by using anti-CYC antibodies (Fig. 5B,lanes 3, 6). Subsequently, the supernatant after incubation with anti-CYC antibodies was subjected to a further incubation with anti-PER (Fig. 5B, lanes 4, 7) or anti-TIM (data not shown) antibodies to measure the amounts of PER and TIM that are free of dCLK and CYC (Fig.5B). The results indicate that in the late night more PER and TIM associate with dCLK–CYC containing complexes as compared with CYC that is relatively free of dCLK (e.g., see results obtained at ZT20, Fig. 5B, lanes 2, 3). Importantly, despite the large excess of CYC the overwhelming majority of the PER and TIM proteins present in head extracts does not interact with CYC (Fig. 5B, bottom two panels, comparelanes 3, 4 and 6, 7). The most parsimonious explanation that can account for these observations is that PER and TIM preferentially interact with dCLK (or the dCLK–CYC heterodimer) as compared with “free” CYC. Because dCLK is present in limiting amounts (see Fig. 4) and most of it is bound to CYC (Fig.5A), only a small fraction of the PER and TIM proteins associates with the dCLK–CYC heterodimer (Fig. 5B). Our findings suggest that during the night dCLK acts as a molecular bridge or scaffold that simultaneously interacts with the PER–TIM complex and CYC. A caveat to this contention is that, although CYC is highly abundant in head extracts, it might be present in limiting amounts in PER/TIM-expressing cells. The apparent in vivo preference for dCLK is somewhat different from the situation observed in vitro whereby PER appears to interact equally well with both dCLK and CYC, but TIM only binds dCLK (Lee et al., 1999).
DISCUSSION
To understand further the molecular underpinnings governing the dCLK–CYC–PER–TIM four-component transcriptional feedback mechanism, we have undertaken direct biochemical studies of these proteins. We recently showed that in adult fly heads PER and TIM mainly interact with dCLK or a dCLK-containing complex during the night/early morning (Lee et al., 1998), in agreement with the observation that the transcription rates of per and tim are low and that dClk transcripts accumulate during these times in a daily cycle (So and Rosbash, 1997; Bae et al., 1998). PER, TIM, or both inhibit the DNA binding activity of dCLK–CYC in vitro (Lee et al., 1999), suggesting a mechanism for how PER and TIM participate in both transcriptional autoinhibition and dClk activation. This inhibition was not accompanied by the disruption of the dCLK–CYC heterodimer (Lee et al., 1999). In this report we biochemically characterized in vivo-produced CYC and determined its ability to interact with dCLK, PER, or TIM as a function of time in a daily cycle. In addition, we also determined the molar concentrations of the four clock proteins in adult head extracts. Throughout a daily cycle the majority of dCLK is stably bound to the constitutively expressed CYC protein, suggesting that in the adult fly head CYC is the major physiologically relevant bHLH/PAS partner of dCLK. Consistent with recent findings obtained in vitro (Lee et al., 1999), PER and TIM do not disrupt the dCLK–CYC interaction. In contrast to previous models based on the ability of the PER–PAS domain to mediate protein–protein dimerization (Huang et al., 1993), our results indicate that PER (and TIM) does not engage in autoinhibition by forming nonfunctional heterodimers with PAS-containing transcription factors. Furthermore, the results suggest that the PER–TIM complex mediates transcriptional autoinhibition and dClk activation by specifically targeting dCLK, raising the possibility that the highly abundant CYC protein interacts with other bHLH/PAS partners that function in pathways besides the circadian clock.
By comparing the staining intensities of known amounts of in vitro-synthesized clock proteins with their counterparts in head extracts, we were able to estimate the molar concentrations of PER, TIM, dCLK, and CYC present in adult heads throughout a daily cycle (see Fig. 4). The validity of our calculations is supported by the fact that our strategy for estimating the concentrations of the various clock proteins yielded approximately similar molar concentrations for PER and TIM during the mid-to-late night, consistent with earlier studies that used independent approaches (Zeng et al., 1996; Lee et al., 1998). dCLK is almost certainly the component limiting the maximum levels of dCLK–CYC complexes that can be assembled. Although calculations based on total head extracts do not take into account possible differences in spatial distributions, it is highly likely that, in all dCLK-containing cells in the adult head, CYC is also present and at levels higher than those of dCLK. In addition to our finding that CYC is present in at least a 200-fold molar excess relative to dCLK in head extracts (see Fig. 4), this contention is supported further by the observations that (1) at all times in a daily cycle essentially all of the dCLK is stably bound to CYC (see Fig. 5A), and (2) the temporal abundance profile of dCLK in immune complexes recovered with anti-CYC antibodies is very similar to that of dCLK present in head extracts (see Fig.3C,D).
PER and TIM mainly interact with dCLK/CYC during the night (see Fig.3A), consistent with previous findings showing that PER and TIM inhibit the activity of the dCLK–CYC heterodimer (Darlington et al., 1998; Lee et al., 1999) and that the transcription rates ofper and tim are low during the night (So and Rosbash, 1997). Nonetheless, during the night the majority of PER and TIM is not associated with dCLK or CYC (see Fig. 5B). Our findings suggest that this is because dCLK is present in limiting amounts (see Fig. 4) and that PER and TIM (or the PER–TIM complex) preferentially interact with dCLK (or the dCLK–CYC heterodimer) as compared with free CYC (see Fig. 5B). In addition to relative differences in levels, temporal changes in the subcellular distributions of PER and TIM also might regulate their interactions with the dCLK–CYC transcription factor. PER and TIM are detected first in the nucleus of key pacemaker neurons in the brain at approximately ZT17 (Curtin et al., 1995; Hunter-Ensor et al., 1996; Myers et al., 1996), coincident with times in a daily cycle when significant amounts of PER and TIM first interact with the dCLK–CYC complex (see Fig.3A). Immunohistochemical studies will be important in determining whether the subcellular distributions of dCLK and CYC are regulated and the extent to which their spatial distribution patterns overlap those of PER and TIM. Because most of the PER–TIM complex is not stably interacting with the dCLK–CYC heterodimer, our results raise the possibility that PER and/or TIM directly regulates other transcription factors.
Although our findings using head extracts suggest that PER and TIM preferentially interact with dCLK as compared with CYC, results obtained by using in vitro-generated products demonstrate that PER and CYC can interact stably (Lee et al., 1999). This difference in binding capabilities is especially curious because in head extracts CYC is present in at least a 40- to 200-fold molar excess relative to PER and dCLK, respectively. Although the reason underlying the different interaction profile obtained in vitro andin vivo is not clear, a speculative possibility is that,in vivo, CYC interacts with other bHLH/PAS proteins besides dCLK and that these heterodimers are not recognized by PER. At the very least, the high abundance of CYC as compared with dCLK, PER, and TIM in conjunction with the reasonable speculation that CYC interacts with bHLH/PAS proteins in a 1:1 molar ratio raises the strong possibility that CYC has other functions besides its established role in circadian clocks. In this context it is interesting to note that the putative ortholog of CYC in mammals, termed BMAL1, has a broad expression profile (highly expressed in skeletal muscle and brain), and circumstantial evidence suggests it is a general partner of a number of bHLH/PAS proteins (Hogenesch et al., 1997, 1998). This is consistent with the observation that the PAS domain of BMAL1 identifies this protein as a member of the ARNT-like group of bHLH/PAS factors that behave as general partners of other more specifically regulated bHLH/PAS proteins (Hogenesch et al., 1997). By analogy with the mammalian system, a similar scenario also might be occurring with CYC in flies. In agreement with this proposal, CYC is expressed constitutively in flies (Rutila et al., 1998) (see Fig.2A) and present in naïveDrosophila tissue culture cells (Schneider) in contrast todClk, per, or tim (Darlington et al., 1998; data not shown).
In addition to defining the dynamic interactions that occur between clock proteins involved in generating a circadian transcriptional feedback loop in Drosophila, our data suggest that stable structures can be assembled in vivo that comprise at least three PAS-containing proteins (i.e., PER, dCLK, and CYC), in agreement with recent findings obtained by using in vitro-synthesized proteins (Lee et al., 1999). These results are surprising in light of the well characterized ability of the canonical PAS domain to mediate the formation of heterodimers and in some cases homodimers (for review, see Crews, 1998). What the role of the PAS region might be in promoting the assembly of putative trimeric and tetrameric complexes comprising the dCLK–CYC heterodimer and PER and/or TIM is not clear. Nonetheless, findings based on determining interactions that can occur in vitro indicate that other regions on PER besides its PAS domain can interact stably with TIM (Saez and Young, 1996; Sangoram et al., 1998), suggesting that at least in the case of PER it might interact with PAS-containing proteins in a non-PAS-mediated manner.
The ability of PER and TIM to interact with a dCLK–CYC heterodimer can account for the inhibition of per and timtranscription and the activation of dClk transcription. Repression of per and tim transcription results from PER–TIM-mediated inhibition of dCLK–CYC activation (Darlington et al., 1998; Lee et al., 1999), whereas activation of dClkresults from the release of dCLK–CYC-dependent repression ofdClk transcription (Glossop et al., 1999). Thus, the same molecular interactions between PER–TIM and dCLK–CYC can account for the anti-phase cycling of dClk mRNA as compared withper and tim mRNAs (Bae et al., 1998; Lee et al., 1998). Based on the observations that dCLK is present in limiting amounts and that the PER-TIM complex likely interacts preferentially with dCLK as compared with CYC, our data indicate that the dynamic regulation of dCLK is a key variable governing both theper–tim and dClk feedback loops. In addition to our results suggesting the formation of a stable multimeric complex containing PER, TIM, dCLK, and CYC, other studies that used heterologous systems have shown that DBT can interact stably with PER (Kloss et al., 1998) and that the blue-light photoreceptor CRY can bind TIM in a light-dependent manner (Ceriani et al., 1999). Thus, there is the possibility that in Drosophila at least six clock proteins might interact simultaneously to form a light-responsive multimeric complex or “clockosome” that is the key biochemical entity in the timekeeping mechanism.
Footnotes
This work was supported by a grant from the National Institutes of Health (I.E.). We thank Dr. Jeffrey Hall (Brandeis University, Waltham, MA) for cyc0 flies.
Correspondence should be addressed to Dr. Isaac Edery, Department of Molecular Biology and Biochemistry, Center for Advanced Biotechnology and Medicine, 679 Hoes Lane, Piscataway, NJ 08854. E-mail:edery@cabm.rutgers.edu.
Dr. Lee's present address: Department of Biological Sciences, 600 Sherman Fairchild Center, Columbia University, New York, NY 10027.
REFERENCES
- 1.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 2.Bae K, Lee C, Sidote D, Chuang KY, Edery I. Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol Cell Biol. 1998;18:6142–6151. doi: 10.1128/mcb.18.10.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceriani MF, Darlington TK, Staknis D, Mas P, Petti AA, Weitz CJ, Steve AK. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- 4.Citri Y, Colot HV, Jacquier AC, Yu Q, Hall JC, Baltimore D, Rosbash M. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature. 1987;326:42–47. doi: 10.1038/326042a0. [DOI] [PubMed] [Google Scholar]
- 5.Crews ST. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 1998;12:607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- 6.Curtin KD, Huang ZJ, Rosbash M. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron. 1995;14:365–372. doi: 10.1016/0896-6273(95)90292-9. [DOI] [PubMed] [Google Scholar]
- 7.Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TDL, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 8.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 9.Edery I. Role of post-transcriptional regulation in circadian clocks: lessons from Drosophila. Chronobiol Int. 1999;16:377–414. doi: 10.3109/07420529908998716. [DOI] [PubMed] [Google Scholar]
- 10.Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells' influence on circadian behavioral rhythms. J Neurosci. 1992;12:3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glossop NRJ, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- 13.Handler AM, Konopka RJ. Transplantation of a circadian pacemaker in Drosophila. Nature. 1979;279:236–238. doi: 10.1038/279236a0. [DOI] [PubMed] [Google Scholar]
- 14.Hao H, Allen DL, Hardin PE. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol Cell Biol. 1997;17:3687–3693. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao H, Glossop NR, Lyons L, Qiu J, Morrish B, Cheng Y, Helfrich-Forster C, Hardin P. The 69 bp circadian regulatory sequence (CRS) mediates per-like developmental, spatial, and circadian expression and behavioral rescue in Drosophila. J Neurosci. 1999;19:987–994. doi: 10.1523/JNEUROSCI.19-03-00987.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardin PE. Activating inhibitors and inhibiting activators: a day in the life of a fly. Curr Opin Neurobiol. 1998;8:642–647. doi: 10.1016/s0959-4388(98)80093-7. [DOI] [PubMed] [Google Scholar]
- 17.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 18.Hogenesch JB, Chan WK, Jackiw VH, Brown RC, Gu YZ, Pray-Grant M, Perdew GH, Bradfield CA. Characterization of a subset of the basic helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J Biol Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 19.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang ZJ, Edery I, Rosbash M. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature. 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 21.Hunter-Ensor M, Ousley A, Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84:677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- 22.Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 23.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER–TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- 25.Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER–TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- 26.Lee C, Bae K, Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol Cell Biol. 1999;19:5316–5325. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majercak J, Kalderon D, Edery I. Drosophila melanogaster deficient in protein kinase A manifests behavior-specific arrhythmia but normal clock function. Mol Cell Biol. 1997;17:5915–5922. doi: 10.1128/mcb.17.10.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 29.Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 30.Reppert SM. A clockwork explosion! Neuron. 1998;21:1–4. doi: 10.1016/s0896-6273(00)80234-2. [DOI] [PubMed] [Google Scholar]
- 31.Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 32.Saez L, Young MW. Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 33.Sangoram AM, Saez L, Antoch MP, Gekakis N, Staknis D, Whiteley A, Fruechte EM, Vitaterna MH, Shimomura K, King DP, Young MW, Weitz CJ, Takahashi JS. Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK–BMAL1-induced transcription. Neuron. 1998;21:1101–1113. doi: 10.1016/s0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 34.Sehgal A, Price JL, Man B, Young MW. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- 35.Sidote D, Majercak J, Parikh V, Edery I. Differential effects of light and heat on the Drosophila circadian clock proteins PER and TIM. Mol Cell Biol. 1998;18:2004–2013. doi: 10.1128/mcb.18.4.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7145. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young MW. The molecular control of circadian behavioral rhythms and their entrainment in Drosophila. Annu Rev Biochem. 1998;67:135–152. doi: 10.1146/annurev.biochem.67.1.135. [DOI] [PubMed] [Google Scholar]
- 38.Zeng H, Hardin PE, Rosbash M. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J. 1994;13:3590–3598. doi: 10.1002/j.1460-2075.1994.tb06666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]