Abstract
Progressive cerebral accumulation of amyloid β-protein (Aβ) is an early and invariant feature of Alzheimer's disease. Little is known about how Aβ, after being secreted, is degraded and cleared from the extracellular space of the brain. Defective Aβ degradation could be a risk factor for the development of Alzheimer's disease in some subjects. We reported previously that microglial cells release substantial amounts of an Aβ-degrading protease that, after purification, is indistinguishable from insulin-degrading enzyme (IDE). Here we searched for and characterized a role for IDE in Aβ degradation by neurons, the principal cell type that produces Aβ. Whole cultures of differentiated pheochromocytoma (PC12) cells and primary rat cortical neurons actively degraded endogenously secreted Aβ via IDE. However, unlike that in microglia, IDE in differentiated neurons was not released but localized to the cell surface, as demonstrated by biotinylation. Undifferentiated PC12 cells released IDE into their medium, whereas after differentiation, IDE was cell associated but still degraded Aβ in the medium. Overexpression of IDE in mammalian cells markedly reduced the steady-state levels of extracellular Aβ40 and Aβ42, and the catalytic site mutation (E111Q) abolished this effect. We observed a novel membrane-associated form of IDE that is ∼5 kDa larger than the known cytosolic form in a variety of cells, including differentiated PC12 cells. Our results support a principal role for membrane-associated and secreted IDE isoforms in the degradation and clearance of naturally secreted Aβ by neurons and microglia.
Keywords: neurons, Alzheimer's disease, amyloid β-protein degradation, insulin-degrading enzyme, oligomerization, membrane proteins
Alzheimer's disease (AD) is characterized by the progressive and severe accumulation in the brain of the amyloid β-protein (Aβ). Although much attention has been focused on the cellular production of Aβ, including the role of presenilin proteins therein (Selkoe, 1999), little is known about how Aβ, after being secreted, is degraded and cleared from tissues. Defective degradation of Aβ would be expected to be a risk factor for the development of AD. If brain-derived Aβ-degrading proteases can be identified, genetic changes in such proteases could be sought in as yet unidentified forms of familial AD, and their pharmacological upregulation could represent a therapeutic approach to AD in general.
Recent evidence suggests that insulin-degrading enzyme (IDE), a thiol metalloendopeptidase known to cleave insulin, glucagon, and other peptide hormones (for review, see Authier et al., 1996a), may be involved in the degradation of endogenous brain-derived Aβ peptides (Kurochkin and Goto, 1994; McDermott and Gibson, 1996; Qiu et al., 1998). The protease is expressed in a variety of tissues including brain and has a conformational rather than a sequence specificity for its substrates (Shii et al., 1985; Akiyama et al., 1990; Kuo et al., 1994). In mammalian cells, IDE has been principally localized to the cytosol and peroxisomes (McKenzie and Burghen, 1984; Authier et al., 1995, 1996b; Chesneau et al., 1997), raising the question of how the enzyme could degrade Aβ, because the peptide is not found in either of these locations. However, we have shown recently that intact IDE, like Aβ, can be released into the extracellular fluid by healthy cultured microglial (BV-2) cells and is also present in normal CSF (Qiu et al., 1998). Whether IDE is present in and released from neurons, widely considered to be the principal source of secreted Aβ in brain, is not known.
Here we used mixed primary rat brain cultures and differentiated pheochromocytoma (PC12) cells to investigate whether neuronal cells also possess Aβ-degrading activity. We demonstrate that neuronal-type cells indeed exhibit significant extracellular Aβ-degrading activity that is inhibited by competitive IDE substrates and other IDE inhibitors. Unlike the IDE in microglia, which is released into the medium, the IDE we detected in differentiated neurons was found in part on the cell surface, consistent with recent evidence that IDE can be localized to the surface of some non-neural cells expressing the enzyme (Seta and Roth, 1997). Surprisingly, we found that undifferentiated PC12 cells release some IDE into their medium, whereas differentiated PC12 cells retain it on the plasma membrane, where it can be biotinylated. We confirmed that IDE has a major role in Aβ degradation by showing that cellular overexpression of wild-type but not active site-mutated IDE markedly decreases the steady-state levels of naturally secreted Aβ40 and Aβ42 in the medium of APP-expressing cells. Interestingly, intracellular levels of Aβ were not affected by overexpressing IDE. Finally, detergent extraction of neuronal cells reveals the presence of a novel ∼115 kDa IDE isoform that is the major species in neuronal membranes but is absent in cytosol, which contains the known 110 kDa isoform.
MATERIALS AND METHODS
Cell culture. All growth media were supplemented with glutamine (2 mm), penicillin (500 units/ml), and streptomycin (500 μg/ml) except where otherwise indicated. Rat PC12 cells were routinely cultured in DMEM with 10% horse serum and 5% fetal bovine serum (FBS). For differentiation, the cells were plated at a density of 1 × 106 cells per collagen-coated 10 cm dish and cultured for 5 d in plain DMEM supplemented with 100 ng/ml nerve growth factor (NGF) and 0.5% FBS. The medium was changed every 2 d. Mouse BV-2 microglial cells were cultured in RPMI and 10% FBS (Qiu et al., 1998). Chinese hamster ovary (CHO) cells stably transfected with βAPP770cDNA containing the V717F AD-causing missense mutation (7PA2 cells) were cultured in DMEM and 10% FBS with G418 (200 μg/ml).
Primary brain cultures were prepared as described (Hartley et al., 1993), with slight modifications. Briefly, brain cells were isolated from the neocortex of embryonic day 16 rat embryos and plated at high density (200,000 cells/ml) in DMEM and Ham's F-12 (1:10 dilution of 1×) containing 10% FBS and 20 mm HEPES onto glial feeder layers in 3.5 cm dishes. Cultures were fed twice a week with the plating medium. At approximately day 9, glial growth was inhibited by the addition of 10−5mcytosine arabinoside, and the cultures were changed to reduced serum medium (5% bovine calf serum + MEM + 20 mm HEPES). Cultures were used after 3 weeks in vitro.
Cell-surface biotinylation. For biotinylation experiments, cells were placed on ice and washed three times with ice-cold Dulbecco's PBS (DPBS; BioWhittaker, Walkersville, MD). The cells were then incubated with 0.5 mg/ml biotinamidocaproic acid 3-sulfo-N-hydroxysuccinamide ester (Pierce, Rockford, IL) in DPBS for 10 min at 4°C. Free biotin groups were blocked by washing the cells twice with cold DMEM. Cells were lysed in STEN buffer (50 mm Tris, pH 7.6; 150 mmNaCl; 2 mm EDTA; 1% NP-40; 20 mm PMSF; 0.5 mg/ml each of leupeptin, aprotinin, and pepstatin A; 10 mmN-ethylmethylamide (NEM); and 10 mm1,10-phenanthroline) and then centrifuged for 10 min at 16,000 ×g at 4°C. Surface IDE was immunoprecipitated from the lysates using the IDE-specific monoclonal antibody 9B12 (kind gift of Dr. Richard Roth, Stanford University). Immunoprecipitates were electrophoresed on 10% Tris/Glycine gels, blotted onto nitrocellulose, and probed for biotin-labeled proteins with neutravidin-HRP (Jackson ImmunoResearch, West Grove, PA). Surface labeling in this protocol was verified by immunoprecipitating lysates with a polyclonal antibody (C7) specific to APP (positive control) and a monoclonal antibody specific to the cytoplasmic protein phosphatidylinositol 3-kinase (PI3-kinase; negative control).
Transient and stable transfections. 7PA2 cells were transfected using Lipofectamine (Life Technologies, Gaithersburg, MD) according to the manufacturer's instructions. Briefly, the cells were grown until 80% confluent and then transfected overnight with 5 μg of DNA and 15 μg of Lipofectamine reagent. Two constructs carrying the cDNA of N-terminal hemagglutinin (HA)-tagged human wild-type IDE or inactive mutant E111Q (glutamic acid 111 to glutamine) were used (Perlman et al., 1993). Empty vector was used as the control. After transfection, the cells were refed with normal culture medium for 24 hr, and aliquots of the medium were assayed for levels of total Aβ or Aβ1–42 peptides by highly specific sandwich ELISAs, using distinct capture antibodies as described previously (Johnsonwood et al., 1997; Qiu et al., 1997). For stable transfections, full-length wild-type (wt) human IDE was inserted into the vector pCDNA3.1 (Invitrogen, San Diego, CA), which contains a zeocin-resistant gene. CHO cell lines stably coexpressing the APP V717F AD mutation and IDE were generated from cell line 7PA2 (above) using zeocin and G418 for selection. Single clones were isolated as described previously (Citron et al., 1997), and cell extracts were analyzed for IDE and APP expression by Western blotting using the IDE-1 and C7 antibodies, respectively. To assay for levels of total Aβ or Aβ1–42 peptides in the medium of the APP and IDE double-transfected cells, we grew the cells to 90% confluency and then refed them with culture medium for 4 hr before sandwich ELISAs were performed. 7PA2 medium was used as the control.
Quantifying degradation of endogenous Aβ by immunoprecipitation. Confluent monolayers of 7PA2 cells in 10 cm dishes were preincubated for 2 hr in methionine- and serum-free medium and then labeled for 16 hr with 100 μCi of [35S]methionine. The labeled media were collected and centrifuged at 3000 × g for 30 min at 4°C. To characterize Aβ-degrading activity in the medium of neuronal cells, 2 ml of the labeled 7PA2 medium (containing abundant Aβ and p3) was mixed with an equal amount of neuronal conditioned medium alone or with unconditioned medium as a control, and the mixtures were incubated at 37°C for 24 hr. For quantifying Aβ degradation in whole cultures, 2 ml of labeled 7PA2 medium was mixed with 2 ml of fresh neuronal culture medium and incubated on the whole cultures for 24 hr at 37°C in the presence or absence of 10 μm insulin. The amount of labeled Aβ remaining in each condition was assessed by immunoprecipitation with the Aβ-specific antiserum R1282, followed by 10–20% Tris/Tricine SDS-PAGE and gel fluorography. For experiments in which purified IDE was examined, 7PA2 cells were incubated overnight with [35S]Met, and the conditioned medium was further incubated for 12 hr with 100 ng of the recombinant active or inactive (E111Q) enzymes before being precipitated with the Aβ-specific antiserum R1280.
Assaying degradation of 125I-Aβ by gel fluorography and trichloroacetic acid precipitation. These assays were performed as described previously (Garcia et al., 1989; Qiu et al., 1998). Briefly, 10,000 cpm of 125I-Aβ (IAβ; specific activity, 2000 μCi/mmol) were added per milliliter of medium to whole cultures or their collected conditioned media (CM) and incubated at 37°C for up to 24 hr. Aliquots were removed at specific times and examined by 10–20% Tris/Tricine gel fluorography. In addition, aliquots of the CM were also treated with 15% trichloroacetic acid (TCA) to precipitate undegraded Aβ. The precipitated samples were centrifuged, and the amounts of label in the supernatant (degradative products) and pellet (intact peptide) were counted.
Preparation of cell cytosol and cell extracts. Cells were washed twice in cold PBS buffer, suspended in cold homogenization buffer (PBS, supplemented with a protease inhibitor mixture), and disrupted using 10 strokes in a Dounce homogenizer followed by several passages through a 25G needle. Nuclei and cell debris were pelleted by centrifugation at 4000 × g at 4°C, and the resulting supernatant was centrifuged at 90,000 × g for 1 hr to separate the cytosol and membrane fractions. The membrane-containing pellets were homogenized in cold homogenization buffer supplemented with 1% NP-40. Undissolved material was pelleted by centrifugation at 16,000 × g and then resuspended in cold PBS containing 0.2% SDS. For total cell extracts (cell lysates), cells were homogenized in STEN/lysis buffer and incubated on ice for 20 min. The cell homogenate was then centrifuged at 16,000 × g for 20 min; the supernatant was kept and stored at −80°C.
Western blotting of IDE and Aβ. To detect IDE in the CM of different cells, cultures were washed and incubated with serum-free N2 medium for 16 hr at 37°C. CM were pooled (to ∼400 ml) and filtered to remove floating cells before being sequentially precipitated with 40 and 60% NH4SO4 (Qiu et al., 1998). The final 60% precipitate was dissolved in ∼5 ml of PBS and dialyzed in the same buffer overnight. Protein concentration was estimated using the Bio-Rad assay. Twenty micrograms of total protein were electrophoresed on 10% Tris and glycine gels. IDE was detected by immunoblotting with a new polyclonal antibody, IDE-1, we raised to a peptide comprising amino acids 62–73 of human IDE. To detect endogenous Aβ in lysates, samples were precleared twice with the APP-specific antibodies B5 and C7 and then immunoprecipitated with the high-titer Aβ antiserum R1280. Precipitates were electrophoresed on 16% Tris/Tricine gels and transferred to 0.2 μm nitrocellulose membranes. Membranes were treated for 10 min with boiling PBS and then immunoblotted with the Aβ monoclonal antibody 6E10 (Senetek).
Purification of recombinant IDE. Polyhistidine- and hemagglutinin- tagged wild-type and catalytically inactive (E111Q) IDEs were expressed in bacteria and purified by metal affinity chromatography as described by V. Chesneau and M. R. Rosner (unpublished observations).
RESULTS
Neuronal cells degrade extracellular Aβ via a protease that has the properties of insulin-degrading enzyme but is cell associated
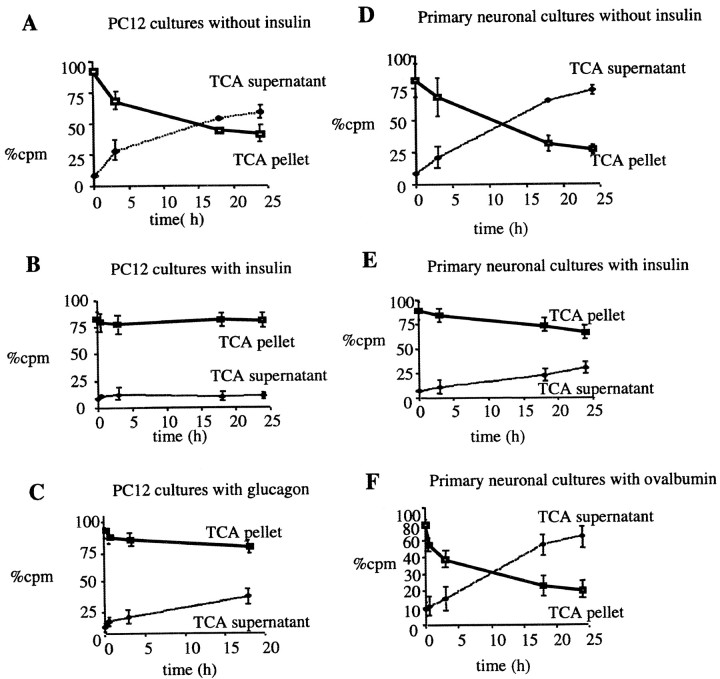
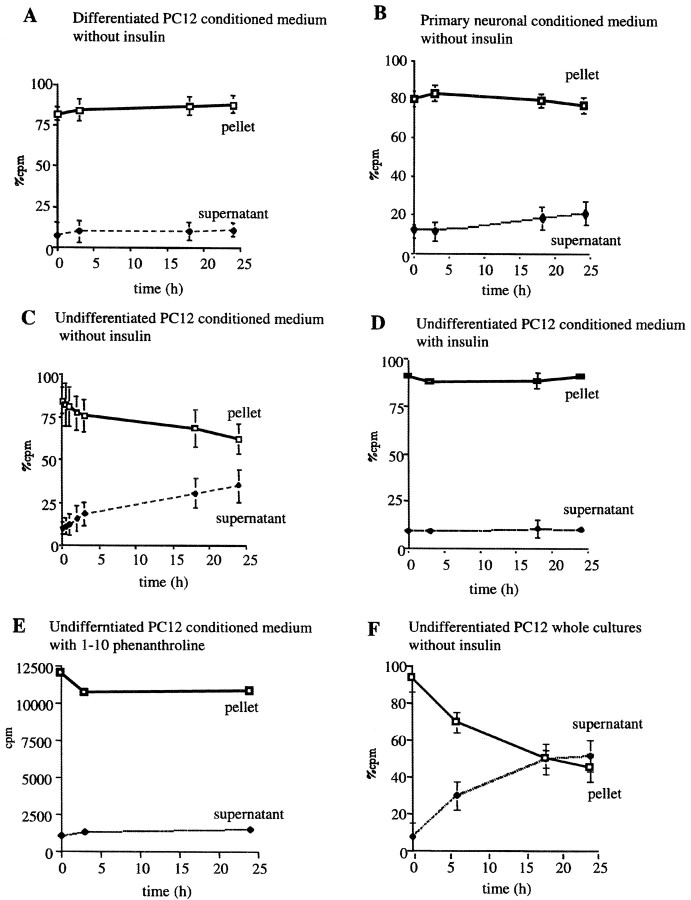
We recently purified and characterized an Aβ-degrading metalloprotease released into the medium of a microglial cell line (BV-2) and showed that it was indistinguishable from insulin-degrading enzyme. To extend this finding and determine whether neuronal cells, which appear to be the major producers of Aβ in the brain, degrade extracellular Aβ in a manner similar to that of microglia, we examined both differentiated PC12 cells and primary rat brain cultures. Differentiated PC12 cells (5 d in vitro) and embryonic rat mixed brain cultures (21 d in vitro) were washed and changed to fresh medium containing 300 pm125I-labeled Aβ1–40 (IAβ) in the presence or absence of 10 μm insulin. The loss of intact IAβ over the course of a 24 hr incubation was quantified by a TCA precipitation assay. Radioactivity in both the TCA-insoluble pellets and the supernatant was measured. Whole PC12 and mixed brain cultures degraded ∼50% of the extracellular IAβ during an 18 hr incubation, and insulin (10 μm) completely inhibited this in the PC12 cultures (Fig.1A,B) and partially inhibited it in the primary cultures (Fig. 1D,E). To confirm that the insulin was specifically inhibiting an IDE-like metalloprotease, we examined the effects of glucagon, a known IDE substrate with a higher Km than insulin, and ovalbumin, which is not a substrate of IDE. In agreement with the lower affinity of IDE for glucagon, degradation of the labeled Aβ by PC12 cells was only partially inhibited (∼40%) by the addition of 10 μm glucagon during the incubation (Fig. 1C). Ovalbumin (10 μm) had no effect on the degradation of IAβ (Fig. 1F). Although NEM was found to be toxic to these cultures even at low concentrations and very brief incubation times, treatment of both differentiated PC12 and rat brain cell cultures with 500 μm 1,10-phenanthroline for up to 3 hr inhibited >80% of the IAβ degradation observed otherwise in these neuronal cultures (data not shown). These results indicate that an IDE-like protease is also responsible for the degradation of Aβ in neuronal cultures. To confirm that this IDE-mediated Aβ-degrading activity was neuronal in origin, we stained rat brain cultures with IDE antibodies and double-labeled them with antibodies to the glial or neuronal markers GFAP or MAP-2, respectively. We found that unlike neurons, astrocytes are not reactive for IDE (data not shown). This is in agreement with a recent report that IDE does not localize to glia in AD brain sections but only to neurons (Bernstein et al., 1999). We then investigated whether the CM of our neuronal cultures were sufficient to mediate the degradation of extracellular Aβ seen in the above experiments. IAβ was incubated for 24 hr at 37°C solely in the CM of either differentiated PC12 cells or rat mixed brain cultures. Unlike that of microglia, the CM of differentiated PC12 cells showed no IAβ-degrading activity (Fig.2A). Very little IAβ-degrading activity was observed in the CM of the primary cultures (Fig. 2B), and this activity might be caused by IDE released from microglia present in our mixed brain cell cultures. Unlike the differentiated PC12 cells, their proliferating undifferentiated precursors exhibited Aβ-degrading activity in their medium (Fig. 2C) that could be inhibited either by insulin or 1,10-phenanthroline (Fig. 2D,E). Whole cultures of undifferentiated PC12 cells were also examined, and these exhibited substantial IAβ degradation, as expected (Fig. 2F). Our results suggest that in mature neuronal-type cells, the IDE-like extracellular Aβ-degrading activity is cell associated rather than released.
Fig. 1.
Degradation of Aβ in differentiated PC12 cell cultures and primary neurons is mediated by IDE. A–C, PC12 cells were differentiated in DMEM and 0.5% FBS supplemented with 100 ng/ml NGF for 5–7 d. Whole cultures were then incubated with 300 pm125I-Aβ in the absence (A) or presence of the known IDE substrates insulin (10 μm;B) or glucagon (10 μm;C). D–F, Primary mixed rat cortical cultures were maintained in DMEM supplemented with FBS and glucose for 3 weeks. For Aβ degradation assays, cultures were conditioned overnight and then incubated for up to 24 hr with 300 pm IAβ in the presence (D) or absence (E) of insulin or the presence of a control protein, ovalbumin (10 μm;F). Aliquots of CM were removed at the indicated times, and loss of intact peptide was assayed by TCA precipitation; the percents of total cpm recovered in supernatants and pellets are displayed. Curves represent means (± SEM) ofn = 4 experiments.
Fig. 2.
Differentiated neuronal cells do not release IDE into their medium. A, B, Differentiated PC12 cells (A) or 21 d primary mixed cortical cultures (B) were conditioned for 12 hr before the CM were collected and incubated for up to 24 hr with 300 pm IAβ.C–E, Undifferentiated PC12 culture medium that had been conditioned for 12 hr was similarly incubated with 300 pmIAβ in the absence (C) or presence (D) of 10 μm insulin or with 1 mm 1,10-phenanthroline (E).F, Whole cultures of undifferentiated PC12 cells were incubated with 300 pm IAβ in the absence of insulin. Aliquots of the CM were removed at the times indicated, and loss of intact IAβ was assayed by TCA precipitation. Curvesrepresent means (± SEM) of n = 4 experiments
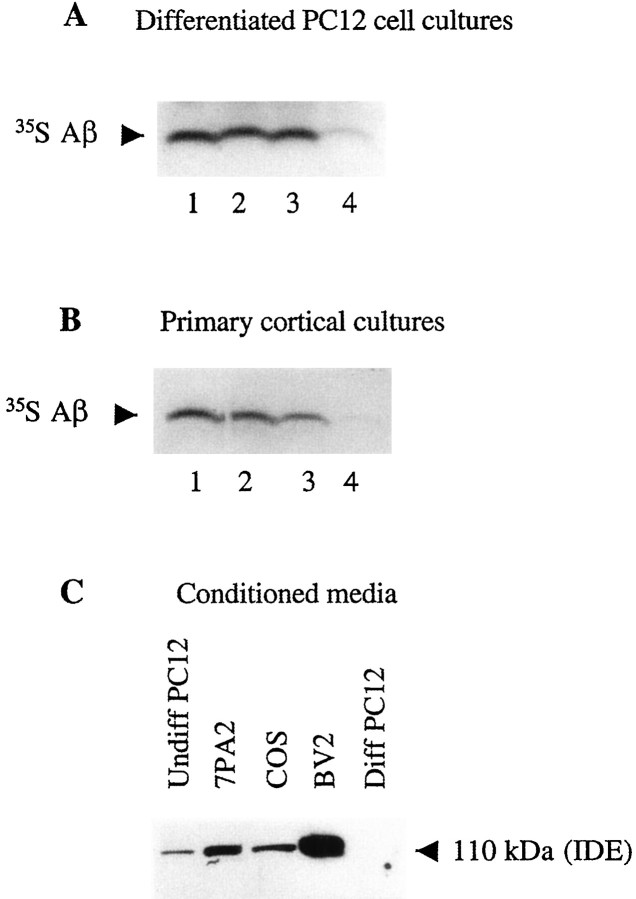
We next examined the effect of neuronal cell cultures or their media alone on the degradation of naturally secreted Aβ. CHO cells stably transfected with APP770 cDNA containing the V717F AD mutation (7PA2 cells) were metabolically labeled with [35S]methionine, and the resultant medium, containing abundant labeled Aβ and p3 peptides, was incubated on whole PC12 or primary cortical cultures or with just their CM for 24 hr at 37°C in the presence or absence of 10 μm insulin (Fig.3A,B). Immunoprecipitation of the media with an Aβ antibody (R1282) and gel fluorography revealed the expected marked decrease of Aβ in the whole neuronal cultures, and this was inhibited by 10 μm insulin. Similar to the TCA precipitation results obtained using IAβ, the CM alone had no effect on the degradation of secreted Aβ (Fig.3A,B). To confirm that IDE was present in the PC12 CM, we used a newly produced, affinity-purified IDE-specific polyclonal antibody, IDE-1, to immunoblot various media. Samples of microglial BV-2 and CHO 7PA2 conditioned media were used as positive controls (Qiu et al., 1998). As expected, the characteristic 110 kDa IDE band was detected in the CM of the BV-2 and CHO cells. The same band was also present in the CM of undifferentiated PC12 cells but was undetectable in that of differentiated PC12 cells (Fig. 3C), consistent with the Aβ degradation assays.
Fig. 3.
Naturally secreted Aβ is degraded by a cell-associated form of IDE present in whole neuronal cultures but not in their conditioned media. A, Conditioned media of 7PA2 CHO cells labeled with [35S]Met (as a source of [35S]Aβ) were incubated at 37°C for 24 hr with either unconditioned 7PA2 medium (lane 1), unconditioned PC12 cell medium (lane 2), conditioned PC12 medium (lane 3), or differentiated PC12 whole cultures (lane 4). By the use of R1282 immunoprecipitation, labeled secreted Aβ could only be degraded by differentiated PC12 cell whole cultures and not by their conditioned medium alone. B, [35S]Met-labeled 7PA2 cell medium was incubated for 24 hr either with unconditioned 7PA2 medium (lane 1), mixed cortical cultures plus insulin (lane 2), conditioned medium of the cortical cultures (lane 3), or the whole cortical cultures (lane 4). Note that the starting amount of labeled Aβ just before the 24 hr incubation (time 0) is actually what remains after degradation in the 7PA2 cultures by IDE during the initial 16 hr conditioning period. We observed no further Aβ degradation after 16 hr of conditioning in 7PA2 cultures (data not shown). C,Thirty micrograms of NH4SO4-concentrated conditioned medium from each of the indicated cell types were separated by 10% Tris/Glycine SDS-PAGE and immunoblotted with the IDE-specific antibody IDE-1. Note the absence of IDE in the medium of differentiated PC12 cells. Diff, Differentiated.
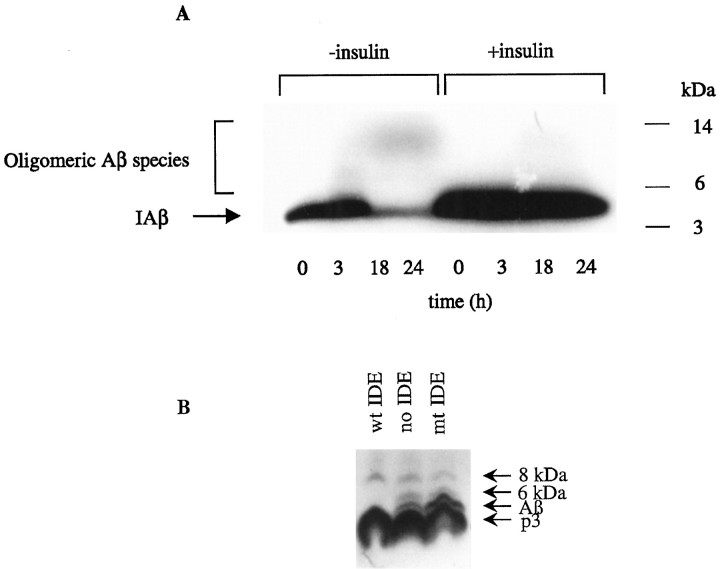
To visualize directly the fate of the Aβ species during the incubation of IAβ in whole neuronal cultures, samples of CM were collected at different times up to 24 hr and analyzed by fluorography of 10–20% Tris/Tricine SDS-PAGE gels (Fig.4A). As the incubation interval at 37°C increased, the amount of the monomeric (4 kDa) Aβ band decreased, consistent with the results of the TCA assay (above). As described previously in BV-2 cultures (Qiu et al., 1998), we also observed the time-dependent formation of small amounts of SDS-stable IAβ species migrating at 6–12 kDa. After a 3 hr incubation, SDS-stable IAβ oligomeric species appeared between 6 and 10 kDa. As the incubation interval increased to 18 and 24 hr, we observed the loss of the 6 kDa band and the formation of additional IAβ oligomers migrating at 12 kDa. Both the loss of Aβ and the formation of these apparent oligomers were abolished when 10 μminsulin was present in the incubation, implicating IDE in both the degradation and the apparent oligomerization of the iodinated peptide. When IAβ was incubated in differentiated neuronal CM alone and then assayed by SDS-PAGE and gel fluorography, Aβ degradation and oligomer formation were not observed (data not shown), consistent with the lack of secreted IDE in differentiated neuronal cultures documented above. Similar results were obtained using PC12 cultures and their CM (data not shown).
Fig. 4.
IDE stimulates the oligomerization of iodinated synthetic Aβ but not naturally secreted Aβ in neuronal cultures.A, Primary mixed cortical cultures were conditioned in serum-free medium overnight and incubated with IAβ (300 pm) for up to 24 hr in the presence or absence of insulin (10 μm). Aliquots of the reaction mixtures were removed at the times indicated and characterized by 10–20% Tris/Tricine gel fluorography. Note that the loss of Aβ monomer and the formation of small amounts of SDS-stable higher molecular weight species are almost abolished by insulin. B, 7PA2 CHO cells were labeled with [35S]Met for 12 hr in the absence (no IDE) or presence of 100 ng of purified recombinant active IDE (wt IDE) or mutant (E111Q) IDE (mt IDE). CM were collected, immunoprecipitated with the Aβ antibody R1282, and assayed by SDS-PAGE and fluorography. Note that the 6 kDa species has been shown to be a modified form of Aβ monomer that migrates anomalously in SDS-PAGE gels; it always follows the behavior of the 4 kDa conventional Aβ monomer, in contrast to that of the 8 kDa Aβ dimer (Podlisny et al., 1998) (D. Walsh, R. Wong, and D. J. Selkoe, unpublished observations).
To investigate the effect that purified IDE has on the oligomerization of endogenously secreted full-length Aβ, we metabolically labeled 7PA2 cultures and incubated the resulting labeled medium for 12 hr at 37°C in the absence or presence of 100 ng of purified active or inactive recombinant enzyme. The media were then immunoprecipitated with R1280, which has been shown to precipitate reproducibly oligomeric forms of secreted Aβ (Podlisny et al., 1995, 1998). As expected, IDE caused a significant reduction of the monomeric 4 kDa Aβ band (Fig.4B). As reported previously, p3 levels were not significantly affected by IDE (Qiu et al., 1998). This is consistent with the IDE cleavage sites being outside or near the N terminal of p3 (A. Savafi and L. Hersh, personal communication; V. Chesneau and M. R. Rosner, unpublished observations). However, we observed neither new formation of endogenous oligomeric species nor augmentation or reduction of the preexisting oligomers accompanying degradation of endogenous monomer by IDE. These data suggest that any IDE-mediated fragments of endogenous Aβ are unlikely to act as seeds for the aggregation of monomeric endogenous Aβ and that preformed endogenous Aβ oligomers are resistant to degradation by IDE.
A portion of IDE in neuronal cells is localized to the cell surface
The data described so far suggest that in differentiated neuronal cells, unlike microglia, Aβ-degrading activity remains cell associated rather than being released into the medium. To investigate whether this finding can be explained by the neurons internalizing Aβ and then degrading it intracellularly, we incubated synthetic IAβ (300 pm) with differentiated PC12 cultures for up to 24 hr at 37°C. At 0 and 24 hr, the CM were collected, and the cells were washed twice in 2 ml of cold PBS. The washes were kept, and the cells were then scraped and pelleted by centrifugation at 1000 ×g for 5 min at 4°C. Total125I counts in the medium, washes, and cell pellets showed that only ∼2% of the total counts were associated with the cell lysate at both 0 and 24 hr. Trypsin pretreatment of the cells for 15 min at 4°C removed approximately one-half of these cell-associated counts. The large majority of the total counts remained in the medium at 24 hr (data not shown). In a number of experiments, we searched for intracellular IAβ at earlier time points, specifically at 1, 5, and 18 hr. Again, the cell lysate-associated IAβ counts were consistently found to be <2% of the total (data not shown). These results indicate that the degradation of Aβ in our neuronal cultures and under our culture conditions takes place almost exclusively in the extracellular space.
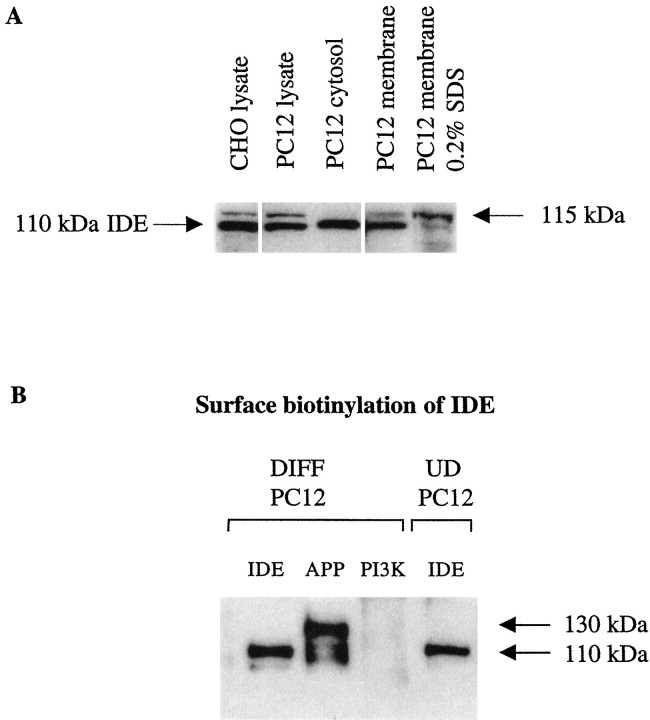
Although IDE has been classically localized to the cytosol and peroxisomes, recent data suggest that it can also be biotinylated on the plasma membrane of at least some non-neuronal cell types naturally expressing the enzyme (Seta and Roth, 1997). Indeed, IDE has been demonstrated previously on the cell surface, where it has been shown to degrade insulin actively (Duckworth, 1979; Yokono et al., 1982;Goldfine et al., 1984). It has also been reported that ∼10% of the total cellular IDE is associated with cell membranes (Duckworth, 1988). To examine the localization of IDE in neuronal cells, the cytosolic and membrane fractions of differentiated PC12 cells were isolated as described in Materials and Methods, electrophoresed on 10% SDS-PAGE gels, and immunoblotted with our polyclonal antibody IDE-1. As expected, an immunoreactive band of the correct molecular weight for IDE (110 kDa) was present in the cytosolic fraction of differentiated PC12 cells (Fig. 5A). Surprisingly an immunoreactive band of ∼115 kDa was also detected in the 1% NP-40-solubilized membrane extracts and in total cell lysates, although at lower levels. Further extraction of the membrane fraction with 0.2% SDS resulted in enrichment of this upper band and the loss of the 110 kDa IDE band (Fig. 5A). Whether this 115 kDa band represents a novel, membrane-specific isoform of IDE is under investigation. To confirm the surface localization of IDE, differentiated and undifferentiated PC12 cells were incubated with a membrane-impermeable biotinylating reagent, washed, and lysed, and the IDE was immunoprecipitated using 9B12 antibody. Biotinylated IDE was detected by blotting the immunoprecipitated IDE with neutravidin-HRP. A biotinylated band of 110 kDa was clearly observed in the anti-IDE immunoprecipitates. Simultaneous control experiments verified the expected biotinylation of cell-surface APP (Haass et al., 1992) but not that of the 85 kDa cytoplasmic protein PI3-kinase (Fig. 5B). Surprisingly, the higher 115 kDa membrane-associated species showed no detectable biotinylation. It may be that either this isoform is specifically localized on intracellular membranes and does not reach the cell surface or its plasma membrane levels are very low.
Fig. 5.
IDE can be detected on the surface of neuronal cells. A, Equal amounts of protein (30 μg) of CHO lysate as well as differentiated PC12 cytosol, lysate, membranes, and membranes extracted with 0.2% SDS were electrophoresed on 10% Tris/Glycine gels and Western blotted with the antibody IDE-1. Unlike cytosol in which only the 110 kDa IDE is detected, the whole lysate and membranes of PC12 cells also contain a higher (∼115 kDa) IDE-reactive band. Note that this higher band is also present in the CHO lysate.B, Differentiated and proliferating PC12 cells were biotinylated and lysed in STEN buffer, and equal amounts of protein were immunoprecipitated with the monoclonal antibody 9B12 against IDE (IDE), a monoclonal antibody specific to the cytosolic protein PI3-kinase (PI3K), or a polyclonal antibody (C7) to APP (APP). The immunoprecipitates were separated by SDS-PAGE and probed with neutravidin-HRP. Note that the large majority of biotinylated APP is the mature, N-plus O-glycosylated form (130 kDa), as expected. DIFF, Differentiated;UD, undifferentiated.
Cellular overexpression of IDE markedly reduces the levels of secreted Aβ
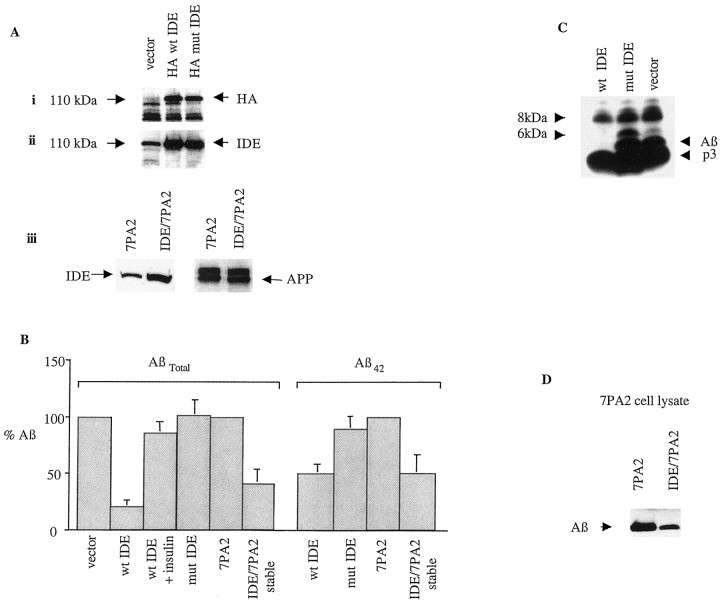
To confirm directly the effect of cellular expression of IDE on the steady-state levels of naturally secreted Aβ, we transfected 7PA2 cells with a pCMV expression vector encoding HA-tagged wild-type or mutant (inactive) IDE or with vector alone. The levels of endogenous Aβ (Aβtotal and Aβ42) in CM were quantified 24 hr after transfection by sandwich ELISA. Transfection efficiency was established by SDS-PAGE and autoradiography of the cell extracts using a specific monoclonal antibody to the HA epitope as well as our polyclonal IDE-1 antibody. IDE-transfected cells had a more than threefold increase in total IDE levels compared with those transfected with vector alone (Fig.6A). Cells overexpressing wild-type HA–IDE consistently exhibited a >70% decrease in the levels of total Aβ (principally Aβ40) in their media when compared with cells expressing either the mutant IDE or vector alone (Fig.6B; means of n = 7 experiments). In a manner similar to that of Aβtotal, Aβ42 levels in the CM of the wild-type IDE transfectants were ∼50% of those of vector- or mutant IDE-transfected cells (Fig. 6B). When we established stable APP and IDE double-transfected 7PA2 clones, these showed a more than twofold increase in their total IDE levels but equal APP levels when compared with plain 7PA2 cells (Fig. 6A, iii). In a manner similar to that of the transient transfectants, the stable IDE-transfected 7PA2 cells showed a reduction in Aβtotal and Aβ42 levels in their medium of 60 and 40%, respectively, within 4 hr compared with untransfected 7PA2 cells (Fig. 6B; means ofn = 3 experiments). These data clearly demonstrate that IDE is responsible for Aβ degradation in intact cells and that it can efficiently degrade naturally secreted Aβ40 and Aβ42.
Fig. 6.
Expression of IDE markedly reduces steady-state levels of endogenous Aβ in the media. 7PA2 CHO cells were grown to 80% confluency before being transfected overnight with a pCMV expression vector encoding HA-tagged wild-type or mutant IDE or with vector alone. After transfection, the cells were refed with growth medium. In addition, 7PA2 CHO cells that were stably transfected with IDE were grown to 90% confluency and then refed with growth medium for 4 hr. A, Transient and stable transfection efficiency was quantitated by SDS-PAGE and autoradiography of the cell extracts using a specific monoclonal antibody to the HA epitope (i) as well as our polyclonal antibody IDE-1 (ii). APP background expression was quantitated using the specific polyclonal antibody C7 (iii). Note that endogenous IDE in the vector lane is solely detected by IDE-1, not anti-HA, as expected. B, Levels of endogenous Aβtotal (primarily Aβ40) and Aβ42 in the conditioned media of transiently and stably transfected cultures as indicated were established by ELISA.Verticalbars indicate means (± SEM). Values were normalized to vector alone or 7PA2 alone as 100%.C, 7PA2 cells were transiently transfected as described above and then starved in methionine-free medium for 2 hr before labeling with 100 μCi of [35S]me-thionine overnight. Conditioned media were immunoprecipitated with R1282 and assayed by SDS-PAGE autoradiography. In agreement with the ELISA results, wt IDE expression markedly reduced the levels of secreted monomeric Aβ. Note that the 6 kDa anomalously migrating monomeric Aβ isoform behaves like the 4 kDa monomer, in contrast to the 8 kDa dimer, which is primarily resistant to IDE. p3 is also resistant to IDE as reported previously (Qiu et al., 1998). D, Lysates from five 10 cm dishes of CHO cells stably transfected with APP or with both IDE and APP were precleared with APP antibodies C7 and B5, immunoprecipitated with R1280, and blotted with the monoclonal Aβ antibody 6E10. mut, Mutant.
To confirm these findings, 7PA2 cells were [35S]Met labeled after transfection, and the conditioned media were immunoprecipitated with the Aβ antibody R1282. In agreement with the ELISA results, SDS-PAGE and gel fluorography showed that endogenous Aβ levels in the medium of wt IDE-transfected cells were markedly reduced, compared with that of the cells transfected with mutant IDE or with vector alone (Fig.6C). As expected, the smaller p3 peptide (residues 18–40/42 of Aβ) was degraded much less. To examine any effect that IDE overexpression might have on the levels of intracellular Aβ, we immunoprecipitated IDE/APP cell lysates with R1282 after transfection and electrophoresed the respective precipitates on 16% SDS-PAGE Tris/Tricine gels. After transfer to nitrocellulose, Aβ was visualized by immunoblotting with the monoclonal antibody 6E10. IDE expression reduced intracellular Aβ monomer levels by ∼58% (Fig.6D). Results were quantitated by computer densitometry (data not shown). This result suggests that degradation of Aβ by IDE occurs intracellularly as well as extracellularly.
DISCUSSION
Converging lines of evidence from many laboratories support the hypothesis that cerebral deposition of Aβ peptides is an early, invariant, and necessary step in the pathogenesis of AD. As a result, there is growing interest in decreasing the cerebral Aβ burden as a therapeutic or preventative approach to the disease. Quantitative biochemical studies suggest that IDE, a conserved neutral thiol metalloendopeptidase involved in the degradation of certain peptide hormones, is a major Aβ-cleaving protease in brain tissue (Kurochkin and Goto, 1994; McDermott and Gibson, 1997). We conducted previously an unbiased search for endogenous Aβ-degrading proteolytic activities in the media of several non-neuronal cell types and found that the principal such activity represented IDE, with particularly robust levels observed in microglial cell media (Qiu et al., 1997, 1998). More important, immunodepletion of IDE from the microglial medium markedly decreased the degradation of Aβ. Evidence that a metalloprotease with the properties of IDE is released into the medium of primary rat microglial cultures has also been reported (Mentlein et al., 1998).
In the present study, we examined differentiated PC12 cells and 21-d primary brain cultures and showed that these neuronal cells degrade secreted and synthetic Aβ via a 110 kDa protease that has the same inhibitor and substrate properties and immunoreactivity as IDE. Interestingly, this IDE-mediated Aβ degradation was confined to whole neuronal cultures and was absent from the conditioned media of the differentiated cells (Figs. 1, 2). In agreement with this finding, surface biotinylation revealed the presence of intact 110 kDa IDE on the plasma membrane of neuronal cells (Fig. 5). Western blotting further demonstrated that IDE was almost absent from the CM of differentiated PC12 cells, whereas it was abundant in the CM of BV-2 microglia and CHO cells. It appears, therefore, that the locus in which IDE effects degradation of extracellular Aβ can vary by cell type. One explanation might be that different cell types use different isoforms of IDE molecules, including a form that can be released and one that is cell associated. To address this possibility we used proliferating and differentiated PC12 cells and examined the ability of their CM alone to degrade Aβ. Only the proliferating (undifferentiated) PC12 cells exhibited Aβ-degrading activity in their media (Fig. 2). Accordingly, immunoblotting with the IDE antibody IDE-1 demonstrated the presence of intact IDE in the CM of just the proliferating PC12 cells (Fig. 3). It therefore appears that mature neuronal cells lose their ability to release IDE. The downregulation of an as yet unidentified IDE-releasing protease from the surface of differentiated neurons is one possible explanation for this difference. Alternatively, IDE might become membrane anchored via its association with other membrane proteins or lipids. The upregulation of such proteins or lipids during differentiation might further explain the lack of IDE release in differentiated PC12 cells.
It has been reported that synthetic Aβ can be internalized into PC12 cells and degraded to some extent intracellularly (Burdick et al., 1997). However, when we assayed for internalized Aβ in our PC12 cultures, we did not observe any significant internalization and degradation of IAβ under our culture conditions. The difference between these results may have several explanations. Serum is present, although at low concentrations, in our differentiated PC12 cultures. Regarding Aβ peptide length, we only examined iodinated Aβ1–40, which has been shown not to accumulate nearly as much as Aβ1–42 (Burdick et al., 1997; Morelli et al., 1999). Finally, we used picomolar concentrations of Aβ, which may have reduced its absorption rate, because this has been shown to be concentration dependent. However, although Aβ degradation in our neuronal cultures appears to occur extracellularly, we cannot exclude the possibility that a portion of Aβ is internalized and degraded intracellularly.
It has been reported that the first putative start codon of IDE is not a functional one and that initiation of translation occurs at the second ATG codon located 123 nucleotides downstream (Baumeister et al., 1993; Perlman et al., 1993; Perlman and Rosner, 1994). Here we report the detection of a slightly larger (∼115 kDa) IDE-immunoreactive species in cell lysates that seems to reside predominantly on membranes. Whether this is a product of translation at the first putative start codon is under study and will take some time to determine. Although we do not yet know the degradative function of this form, we speculate that its slightly larger size and its marked enrichment in membranes versus cytosol indicate that it may be a principal membrane-anchored isoform that degrades substrates on the plasma membrane. In any case, it is not yet clear how IDE could be routed to the cell surface and extracellular space, because it appears to lack a classical hydrophobic signal peptide (Baumeister et al., 1993). However, a number of prokaryotic and eukaryotic proteins have been shown to be trafficked via nonclassical pathways that involve the ATP-binding cassette transporters {for review, see Kuchler and Thorner, 1992). Examples include the yeast a-factor, the cytokines IL1α and IL1β, and certain growth factors like FGF.
Our cell culture data suggest that IDE is the major Aβ-degrading activity in cultured microglia and neuronal cells. To confirm that IDE can actively degrade naturally secreted Aβ and to quantify this effect, we transfected either wild-type IDE or an inactive form bearing a mutation (E111Q) of the active site into CHO cells stably expressing APP. Both transient and stable expression of wild-type IDE markedly reduced the steady-state levels of Aβ40 and Aβ42 in the media of our 7PA2 cells, whereas expression of the mutant enzyme had no effect. Immunoprecipitation and SDS-PAGE autoradiography of naturally secreted Aβ from IDE- and control-transfected cells confirmed the ELISA results. Interestingly, the SDS-stable oligomers of Aβ that we have previously described in these cells were not decreased. This finding suggests that IDE could play an important role in regulating the levels of Aβ monomers, but once aggregation begins, the protease would be unlikely to effectively control the progressive accumulation of insoluble Aβ species observed in the brains of AD subjects. Taken together, these data suggest a quantitatively important role for the degradation of secreted Aβ by IDE. However, recent reports have suggested that other Aβ-degrading metalloproteases might operate independently of IDE in some cell types and contribute to the overall regulation of extracellular Aβ levels (Yamin et al., 1999). An endogenous 14 kDa inhibitor of IDE has been identified (Ogawa et al., 1992). It is possible that like IDE, its release is also cell type specific, potentially explaining the lack of Aβ degradation in the CM of some cells in the presence of IDE.
The role of IDE, if any, in the pathogenesis of the excessive cerebral accumulation of Aβ that occurs in AD remains to be elucidated. In this regard, the CSF-to-plasma insulin ratio has been shown to be reduced in AD patients compared with nondemented controls (Stolk et al., 1997; Craft et al., 1998). Brain insulin levels have also been reported to decrease with age. However, no significant difference in brain insulin levels between AD and aged-matched control patients has been reported to date (Frolich et al., 1998). Interestingly, insulin receptors are upregulated in AD brain, suggesting some impairment of the signal transduction pathway between ligand and receptor. Identifying how the activity of IDE is regulated in brain and how this changes with age and in AD could provide new insights into the mechanisms of Aβ accrual in the brains of sporadic AD cases. In this respect, it was reported recently that IDE immunoreactivity is more prominent in AD than in normal human brain and can be localized to certain cortical neurons and to senile plaques (Bernstein et al., 1999).
Many studies have shown that soluble Aβ can be physiologically secreted from a variety of neural and non-neural cells (Selkoe, 1998). How this monomer accumulates sufficiently in the extracellular space to form first diffuse and then fibrillar deposits is not well understood. In this regard, we showed in a previous study that IDE appeared to potentiate the oligomerization of synthetic iodinated Aβ (Qiu et al., 1998). In the present study, we show that IDE readily cleaves naturally secreted Aβ without noticeably promoting its oligomerization. Transfection of CHO cultures with wt IDE or their incubation with the purified recombinant enzyme led to substantial degradation of secreted Aβ but did not appear to affect the levels of the preexisting endogenous Aβ oligomers we have characterized previously in these cultures (Podlisny et al., 1995, 1998) (Fig. 6C,D). These data, in combination with additional studies of the recombinant enzyme currently underway (Chesneau, Vekrellis, Rosner, and Selkoe, unpublished observations), suggest that the cleavage products of Aβ from natural IDE are unlikely to act as “seeds” for the aggregation of the endogenous secreted Aβ peptide and that the C-terminal fragments produced from the cleavage of endogenous Aβ by IDE do not themselves oligomerize under the conditions tested. It thus appears that synthetic and endogenous Aβ behave differently after cleavage by IDE. Interestingly, the intracellular pools of monomeric Aβ were also decreased by IDE overexpression, suggesting that the enzyme can degrade intracellular monomeric Aβ. We are currently carrying out cell-fractionation experiments to identify the intracellular compartment(s) in which Aβ degredation by IDE occurs. Of particular interest is our observation that whereas monomeric Aβ40and Aβ42 are both avidly degraded by IDE, stable oligomers in the same lysates and media are resistant to such degradation.
Footnotes
This work was supported by National Institutes of Health Grants AG 12749 (D.J.S.) and N 533858 (M.R.R.).
Correspondence should be addressed to Dr. Dennis J. Selkoe, Center for Neurological Diseases, Harvard Institutes of Medicine 730, 77 Avenue Louis Pasteur, Boston, MA 02115. E-mail: Selkoe@cnd.bwh.harvard.edu.
REFERENCES
- 1.Akiyama H, Yokono K, Shii K, Ogawa W, Taniguchi H, Baba S, Kasuga M. Natural regulatory mechanisms of insulin degradation by insulin degrading enzyme. Biochem Biophys Res Commun. 1990;170:1325–1330. doi: 10.1016/0006-291x(90)90539-y. [DOI] [PubMed] [Google Scholar]
- 2.Authier F, Bergeron JJ, Ou WJ, Rachubinski RA, Posner BI, Walton PA. Degradation of the cleaved leader peptide of thiolase by a peroxisomal proteinase. Proc Natl Acad Sci USA. 1995;92:3859–3863. doi: 10.1073/pnas.92.9.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Authier F, Cameron PH, Taupin V. Association of insulin-degrading enzyme with a 70 kDa cytosolic protein in hepatoma cells. Biochem J. 1996a;319:149–158. doi: 10.1042/bj3190149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Authier F, Posner BI, Bergeron JJM. Insulin-degrading enzyme. Clin Invest Med. 1996b;19:149–160. [PubMed] [Google Scholar]
- 5.Baumeister H, Muller D, Rehbein M, Richter D. The rat insulin-degrading enzyme. Molecular cloning and characterization of tissue-specific transcripts. FEBS Lett. 1993;317:250–254. doi: 10.1016/0014-5793(93)81286-9. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein HG, Ansorge S, Riederer P, Reiser M, Frolich L, Bogerts B. Insulin-degrading enzyme in the Alzheimer's disease brain: prominent localization in neurons and senile plaques. Neurosci Lett. 1999;263:161–164. doi: 10.1016/s0304-3940(99)00135-4. [DOI] [PubMed] [Google Scholar]
- 7.Burdick D, Kosmoski J, Knauer MF, Glabe CG. Preferential absorption, internalization and resistance to degradation of the major isoform of the Alzheimer's amyloid peptide, Aβ1–42, in differentiated PC12. Brain Res. 1997;740:275–284. doi: 10.1016/s0006-8993(96)01262-0. [DOI] [PubMed] [Google Scholar]
- 8.Chesneau V, Perlman RK, Li W, Keller G-A, Rosner MR. Insulin-degrading enzyme does not require peroxisomal localization for insulin degradation. Endocrinology. 1997;138:3444–3451. doi: 10.1210/endo.138.8.5344. [DOI] [PubMed] [Google Scholar]
- 9.Citron M, Westaway D, Xia W, Carlson G, Diehl T, Levesque G, Johnson-Wood K, Lee M, Seubert P, Davis A, Kholodenka D, Motter R, Sherrington R, Perry B, Yao H, Strome R, Lieberburg I, Rommens J, Kim S, Schenk D, Fraser P, St George-Hyslop P, Selkoe DJ. Mutant presenilins of Alzheimer's disease increase production of 42-residue amyloid β-protein in both transfected cells and transgenic mice. Nat Med. 1997;3:67–72. doi: 10.1038/nm0197-67. [DOI] [PubMed] [Google Scholar]
- 10.Craft S, Peskind E, Schwartz MW, Schellenberg GD, Raskind M, Porte D., Jr Cerebrospinal fluid and plasma insulin levels in Alzheimer's disease: relationship to severity of dementia and apolipoprotein E genotype. Neurology. 1998;50:164–168. doi: 10.1212/wnl.50.1.164. [DOI] [PubMed] [Google Scholar]
- 11.Duckworth WC. Insulin degradation by liver cell membranes. Endocrinology. 1979;104:1758–1763. doi: 10.1210/endo-104-6-1758. [DOI] [PubMed] [Google Scholar]
- 12.Duckworth WC. Insulin degradation: mechanisms, products and significance. Endocr Rev. 1988;9:319–345. doi: 10.1210/edrv-9-3-319. [DOI] [PubMed] [Google Scholar]
- 13.Frolich L, Blum-Degen D, Bernstein HG, Engelsberger S, Humrich J, Laufer S, Muschner D, Thalheimer A, Turk A, Hoyer S, Zochling R, Boissl KW, Jellinger K, Riederer P. Brain insulin and insulin receptors in aging and sporadic Alzheimer's disease. J Neural Transm. 1998;105:423–438. doi: 10.1007/s007020050068. [DOI] [PubMed] [Google Scholar]
- 14.Garcia JV, Stoppelli MP, Decker SJ, Rosner MR. An insulin epidermal growth factor-binding protein from Drosophila has insulin-degrading activity. J Cell Biol. 1989;108:177–182. doi: 10.1083/jcb.108.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldfine ID, Williams JA, Bailey AC, Wong KY, Iwamoto Y, Yokono K, Baba S, Roth RA. Degradation of insulin by isolated mouse pancreatic acini. Evidence for cell surface protease activity. Diabetes. 1984;33:64–72. doi: 10.2337/diab.33.1.64. [DOI] [PubMed] [Google Scholar]
- 16.Haass C, Koo EH, Mellon A, Hung AY, Selkoe DJ. Targeting of cell-surface β-amyloid precursor protein to lysosomes: alternative processing into amyloid-bearing fragments. Nature. 1992;357:500–503. doi: 10.1038/357500a0. [DOI] [PubMed] [Google Scholar]
- 17.Hartley DM, Kurth MC, Bjerkness L, Weiss JH, Choi DW. Glutamate receptor-induced 45Ca2+ accumulation in cortical cell culture correlates with subsequent neuronal degeneration. J Neurosci. 1993;13:1993–2000. doi: 10.1523/JNEUROSCI.13-05-01993.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnsonwood K, Lee M, Motter R, Hu K, Gordon G, Barbour R, Khan K, Gordon M, Tan H, Games D, Lieberburg I, Schenk D, Seubert P, McConlogue L. Amyloid precursor protein processing and a-beta(42) deposition in a transgenic mouse model of Alzheimer disease. Proc Natl Acad Sci USA. 1997;94:1550–1555. doi: 10.1073/pnas.94.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuchler K, Thorner J. Secretion of peptides and proteins lacking hydrophobic signal sequences: the role of adenosine triphosphate-driven membrane translocators. Endocr Rev. 1992;13:499–514. doi: 10.1210/edrv-13-3-499. [DOI] [PubMed] [Google Scholar]
- 20.Kuo W-L, Gehm BD, Rosner MR, Li W, Keller G. Inducible expression and cellular localization of insulin-degrading enzyme in a stably transfected cell line. J Biol Chem. 1994;269:22599–22606. [PubMed] [Google Scholar]
- 21.Kurochkin IV, Goto S. Alzheimer's β-amyloid peptide specifically interacts with and is degraded by insulin degrading enzyme. FEBS Lett. 1994;345:33–37. doi: 10.1016/0014-5793(94)00387-4. [DOI] [PubMed] [Google Scholar]
- 22.McDermott JR, Gibson AM. Degradation of Alzheimer's beta-amyloid protein by human cathepsin D. NeuroReport. 1996;7:2163–2166. doi: 10.1097/00001756-199609020-00021. [DOI] [PubMed] [Google Scholar]
- 23.McDermott JR, Gibson AM. Degradation of Alzheimer's beta-amyloid protein by serine protease activity in brain microvessels. Neurosci Res Commun. 1997;20:93–102. [Google Scholar]
- 24.McKenzie RA, Burghen GA. Partial purification and characterization of insulin protease and its intracellular inhibitor from rat liver. Arch Biochem Biophys. 1984;229:604–611. doi: 10.1016/0003-9861(84)90193-0. [DOI] [PubMed] [Google Scholar]
- 25.Mentlein R, Ludwig R, Martensen I. Proteolytic degradation of Alzheimer's disease amyloid β-peptide by a metalloproteinase from microglia cells. J Neurochem. 1998;70:721–726. doi: 10.1046/j.1471-4159.1998.70020721.x. [DOI] [PubMed] [Google Scholar]
- 26.Morelli L, Giambartolomei GH, Prat MI, Castaño EM. Internalization and resistance to degradation of Alzheimer's Aβ 1–42 at nanomolar concentrations in THP-1 human monocytic cell line. Neurosci Lett. 1999;262:5–8. doi: 10.1016/s0304-3940(99)00023-3. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa W, Shii K, Yonezawa K, Baba S, Yokono K. Affinity purification of insulin-degrading enzyme and its endogenous inhibitor from rat liver. J Biol Chem. 1992;267:1310–1316. [PubMed] [Google Scholar]
- 28.Perlman RK, Rosner MR. Identification of zinc ligands of the insulin-degrading enzyme. J Biol Chem. 1994;269:33140–33145. [PubMed] [Google Scholar]
- 29.Perlman RK, Gehm BD, Kuo WL, Rosner MR. Functional analysis of conserved residues in the active site of insulin-degrading enzyme. J Biol Chem. 1993;268:21538–21544. [PubMed] [Google Scholar]
- 30.Podlisny MB, Ostaszewski BL, Squazzo SL, Koo EH, Rydel RE, Teplow DB, Selkoe DJ. Aggregation of secreted amyloid β-protein into SDS-stable oligomers in cell culture. J Biol Chem. 1995;270:9564–9570. doi: 10.1074/jbc.270.16.9564. [DOI] [PubMed] [Google Scholar]
- 31.Podlisny MB, Walsh DM, Amarante P, Ostaszewski BL, Stimson ER, Maggio JE, Teplow DB, Selkoe DJ. Oligomerization of endogenous and synthetic amyloid β-protein at nanomolar levels in cell culture and stabilization of monomer by Congo red. Biochemistry. 1998;37:3602–3611. doi: 10.1021/bi972029u. [DOI] [PubMed] [Google Scholar]
- 32.Qiu WQ, Ye Z, Kholodenko D, Seubert P, Selkoe DJ. Degradation of amyloid β-protein by a metalloprotease secreted by microglia and other neural and non-neural cells. J Biol Chem. 1997;272:6641–6646. doi: 10.1074/jbc.272.10.6641. [DOI] [PubMed] [Google Scholar]
- 33.Qiu WQ, Walsh DM, Ye Z, Vekrellis K, Zhang J, Podlisny M, Rosner MR, Safavi A, Hersh LB, Selkoe DJ. Insulin-degrading enzyme regulates extracellular levels of amyloid β-protein by degradation. J Biol Chem. 1998;273:32730–32738. doi: 10.1074/jbc.273.49.32730. [DOI] [PubMed] [Google Scholar]
- 34.Selkoe DJ. The cell biology of beta-amyloid precursor protein and presenilin in Alzheimer's disease. Trends Cell Biol. 1998;8:447–453. doi: 10.1016/s0962-8924(98)01363-4. [DOI] [PubMed] [Google Scholar]
- 35.Selkoe DJ. Translating cell biology into therapeutic advances in Alzheimer's disease. Nature. 1999;399:A23–A31. doi: 10.1038/399a023. [DOI] [PubMed] [Google Scholar]
- 36.Seta KA, Roth RA. Overexpression of insulin degrading enzyme: cellular localization and effects on insulin signaling. Biochem Biophys Res Commun. 1997;231:167–171. doi: 10.1006/bbrc.1997.6066. [DOI] [PubMed] [Google Scholar]
- 37.Shii K, Baba S, Yokono K, Roth RA. Covalent linkage of 125I-insulin to a cytosolic insulin-degrading enzyme. J Biol Chem. 1985;260:6503–6506. [PubMed] [Google Scholar]
- 38.Stolk RP, Breteler MMB, Ott A, Pols HAP, Lamberts SWJ, Grobbee DE, Hofman A. Insulin and cognitive function in an elderly population. Diabetes Care. 1997;20:792–795. doi: 10.2337/diacare.20.5.792. [DOI] [PubMed] [Google Scholar]
- 39.Yamin R, Malgeri EG, Sloane JA, McGraw WT, Abraham CR. Metalloendopeptidase EC 3.4.24.15 is necessary for Alzheimer's amyloid-β peptide degradation. J Biol Chem. 1999;274:18777–18784. doi: 10.1074/jbc.274.26.18777. [DOI] [PubMed] [Google Scholar]
- 40.Yokono K, Roth RA, Baba S. Identification of insulin-degrading enzyme on the surface of cultured human lymphocytes, rat hepatoma cells, and primary cultures of rat hepatocytes. Endocrinology. 1982;111:1102–1108. doi: 10.1210/endo-111-4-1102. [DOI] [PubMed] [Google Scholar]