Abstract
DNA methylation is important for controlling the profile of gene expression and is catalyzed by DNA methyltransferase (MTase), an enzyme that is abundant in brain. Because significant DNA damage and alterations in gene expression develop as a consequence of cerebral ischemia, we measured MTase activity in vitro and DNA methylation in vivo after mild focal brain ischemia. After 30 min middle cerebral artery occlusion (MCAo) and reperfusion, MTase catalytic activity and the 190 kDa band on immunoblot did not change over time. However, [3H]methyl-group incorporation into DNA increased significantly in wild-type mice after reperfusion, but not in mutant mice heterozygous for a DNA methyltransferase gene deletion (DnmtS/+).DnmtS/+ mice were resistant to mild ischemic damage, suggesting that increased DNA methylation is associated with augmented brain injury after MCA occlusion. Consistent with this formulation, treatment with the MTase inhibitor 5-aza-2′-deoxycytidine and the deacetylation inhibitor trichostatin A conferred stroke protection in wild-type mice. In contrast to mild stroke, however, DNA methylation was not enhanced, and reduceddnmt gene expression was not protective in an ischemia model of excitotoxic/necrotic cell death. In conclusion, our results demonstrate that MTase activity contributes to poor tissue outcome after mild ischemic brain injury.
Keywords: cerebral ischemia, delayed cell death, DNA damage, DNA methylation, DNA methyltransferase, gene expression
Mammalian DNA methylation is a covalent, postreplicative modification of genomic DNA and has been implicated in development and differentiation (Li et al., 1992; Panning and Jaenisch, 1998), X-chromosome inactivation (Cedar and Razin, 1990), imprinting (Li et al., 1993; Razin and Cedar, 1994; Tucker et al., 1996a,b; Surani, 1998), and cancer (Baylin et al., 1991; Counts and Goodman, 1995; Laird et al., 1995; Jones, 1996). DNA methylation is catalyzed by the maintenance DNA (cytosine-5) methyltransferase enzyme (MTase, EC 2.1.1.37) and S-adenosyl methionine (SAM) as methyl donor (Jones, 1996; Lengauer et al., 1997; Okano et al., 1998). The primary substrates for MTase are CpG dinucleotides, which represent only 1–2% of the total genome, and ∼70–80% of all CpG sites are methylated in adult somatic mammalian cells (Cross and Bird, 1995). Generally, DNA methylation represses gene transcription (“gene silencing”), whereas hypomethylation is correlated with active transcription (Ehrlich and Wang, 1981; Adams and Burdon, 1982; Doerfler, 1983; Cedar, 1988; Bestor, 1990; Bird, 1992; Leonhard and Bestor, 1993; Laird and Jaenisch, 1996).
It is not known whether DNA methylation is important in ischemic brain damage, but it may be of particular interest in this context because (1) MTase activity is unexpectedly high in neurons, implying a neuron-specific function in these nonreplicating cells (Goto et al., 1993; Brooks et al., 1996). (2) Brain ischemia causes DNA damage including G-T mismatches generated by deamination of 5-methyl cytosine (5-MeC), a powerful endogenous mutagen; MTase may therefore remethylate newly incorporated cytosines after DNA repair in brain (Brooks et al., 1996; Liu et al., 1996; MacManus and Linnik, 1997; Cui et al., 1999). In addition, the formation of 8-hydroxyguanine by oxygen radical injury during ischemia/reperfusion may alter methylation of adjacent cytosines (Cerda and Weitzman, 1997). (3) DNA methylation could alter gene expression, although it has not been determined whether changes in DNA methylation develop after cerebral ischemia.
In this study we examined whether there are significant differences in MTase protein, enzyme activity, and DNA methylation after experimental brain ischemia in mice and determined whether reduced levels of MTase are associated with differences in stroke outcome in transgenic mice heterozygous for a dnmt1 gene deletion (DnmtS/+ mice).
MATERIALS AND METHODS
Transgenic mice. Mice heterozygous for the maintenance DNA methyltransferase gene (DnmtS/+) were generated as described (Li et al., 1992, 1993).DnmtS/+ mice are healthy, fertile, and phenotypically normal; dnmt1 null mice (DnmtS/S) die during embyronic development (Li et al., 1992, 1993). All experiments were performed using littermates (DnmtS/+ andDnmt+/+) in a pure 129/SVJae background.
Mouse model for focal cerebral ischemia. Animal experiments were performed according to National Institutes of Health and institutional guidelines and the policy on the use of animals in neuroscience research of the Society for Neuroscience. Mice (18–22 gm) were anesthetized with 1.5% halothane (induction) and maintained on 1.0% halothane in 70% N2O and 30% O2 by face mask. Focal cerebral ischemia was induced as described (Endres et al., 1998b). In brief, MCA occlusion was produced by a silicone-coated 8–0 monofilament into the internal carotid artery. Thirty minutes or 2 hr later, the filament was withdrawn to reperfuse the brain. To insure equivalent levels of ischemia between groups, regional cerebral blood flow (rCBF) was measured by laser Doppler flowmetry using a flexible skull probe (Huang et al., 1994; Endres et al., 1998a). In randomly selected animals the left femoral artery was cannulated for arterial blood pressure and blood gas determination (Huang et al., 1994; Endres et al., 1998a). Arterial blood samples (50 μl) were analyzed for pH, partial pressure of oxygen (Pao2) and partial pressure of carbon dioxide (Paco2) using a blood gas/pH analyzer (Corning 178, Ciba-Corning Diagnostics, Medford, MA). Core temperature was maintained at ∼37 ± 0.5°C with a thermostat (FHC, Brunswick, ME) and a heating lamp during the monitoring period until 1 hr after reperfusion.
Western blotting. MTase protein levels were determined by immunoblots of extracts of olfactory bulb, striatum, and hippocampus inDnmtS/+ and Dnmt+/+ mice. To examine for time-dependent changes in MTase protein, 129/SVEvTacBr wild-type mice (Taconic Farms, Germantown, NY) underwent 30 min MCAo as above followed by reperfusion (1–72 hr) (Endres et al., 1998b). After mice were killed, brains were immediately removed. Ischemic tissue was identified using a dissecting microscope, and ischemic tissue only (mostly striatum) was isolated along with corresponding tissue from the contralateral side, snap-frozen in liquid nitrogen, and stored at −80°C until further use. Proteins were isolated according to standard techniques, separated by a 7% SDS/PAGE gel, and transferred onto a nitrocellulose membrane. Subsequently, blots were incubated with rabbit polyclonal antiserum HM334 (1:2000 dilution) that recognizes the following polypeptide sequence within mouse maintenance DNA methyltransferase: CRSPRSRPKPRGPRRSK (Tucker et al., 1996a). Bound protein was detected by chemiluminescence (ECL, Amersham Pharmacia Biotech, Piscataway, NJ). Coomassie blue staining was performed to insure equivalent levels of protein loading.
MTase catalytic activity assay. Animals underwent 30 min MCAo/reperfusion as above. Cortical and striatal tissue from each hemisphere and cerebellum were separated, snap-frozen in liquid nitrogen, and stored at −80°C. Nuclear extracts were analyzed for MTase activity as described (Li et al., 1992). Nuclear extract (5 μg) was incubated with 20 mm Tris-HCl, pH 7.4, 10 mm EDTA, 10% glycerol, 2 mm DTT, 200 μm PMSF, 0.5 μm[3H]-SAM, 15 μg/ml RNase A, and 50 pmol hemimethylated oligo-probe dnMT1m2 (see below) in a total volume of 50 μl. The reaction was initiated by addition of nuclear extract and incubated for 1 hr at 37°C. Fifty microliters of 16 μm SAM were then added, and the [3H]dnMT1m2-probe was extracted with phenol/chloroform. Probes were applied to a positively charged Porablot plus membrane (pore size 0.45 μm; Macherey-Nagel, Düren, Germany) using a dot blot apparatus and washed three times with 300 μl sterile H2O. Membranes were air-dried, and the respective areas were counted in 2 ml nonliquid scintillation solution (OptiPhase “Hi-Safe” II) using a scintillation counter (LKB, EG&G Wallac, Gaithersburg, MD). MTase activity was linear over time and enzyme dependent (data not shown). The oligodeoxynucleotide duplex probe dnMT1m2 was as follows: 5′-GAXGXGATCXGGTTAAXGAGTCXGATGXGTAG-3′, 3′-CTGCGCTAGGCCAATTGCTCAGGCTACGCATC-5′; X = 5-MeC.
In vivo DNA methylation assay. Two hundred microliters of PBS, pH 7.4, and 200 μl of [methyl-3H]methionine (200 μCi, specific activity 513 mCi/mg; Amersham Life Science, Freiburg, Germany) were continuously infused over 2 hr via PE-10 interarterial catheter after 30 min MCA occlusion. Anesthesia was then stopped, and animals were placed in a cage for an additional 10 hr before they were killed. Cortex, striatum, and cerebellum were removed and prepared as described above and snap-frozen. Total DNA was isolated, phenol-chloroform-extracted, and dissolved in TE. Three micrograms DNA were blotted on positively charged Porablot plus membranes and measured as above. For controls, samples were treated with proteinase K, with RNase A (Life Technologies, Karlsruhe, Germany), or with DNase I (Invitrogen, NV Leek, Netherlands). Before blotting on membrane, DNase I treatment abolished the [3H]-signal, whereas RNase A and Proteinase K treatment were without effect.
Drug administration. Stock solutions were prepared in PBS/100% ethanol/bovine serum albumin (100:10:1) as vehicle. 5-Aza-′2-deoxycytidine (5-aza-dC) (Sigma, St. Louis, MO), an inhibitor of DNA methylation (Laird et al., 1995), was dissolved at a concentration of 10 μg/μl. Trichostatin A (TSA; Sigma), a specific inhibitor of histone deactylation (Yoshida et al., 1995), was dissolved at a concentration of 0.1 and 1 μg/μl, respectively. Two microliters of the solution or vehicle were administered intracerebroventricularly (bregma −0.9 mm lateral; −0.1 mm posterior; −3.1 mm deep) 10 min before ischemia using a Hamilton syringe (Fisher Scientific, Pittsburgh, PA) as described (Endres et al., 1998b). All inhibitor experiments were performed in 129/SV wild-type mice.
Determination of lesion size. Animals were killed at 72 hr reperfusion (30 min MCAo) or 24 hr reperfusion (2 hr MCAo), and brains were snap-frozen in isopentane on dry ice for cryostat sectioning. Infarction areas were quantitated with an image analysis system (M4, Imaging Research, St. Catherines, Ontario, Canada) on 20 μm hematoxylin and eosin (H&E)-stained cryostat sections. Infarction volume was calculated by summing the volumes of each section directly (Huang et al., 1994) or indirectly (only for 2 hr MCAo experiments) using the following formula: contralateral hemisphere (mm3) − undamaged ipsilateral hemisphere (mm3). The difference between direct and indirect infarct volumes reflects brain swelling.
Determination of neuronal survival. The density of viable cells with neuronal appearance was counted within ischemic striatum on coronal H&E-stained sections (20 μm) at the level of the anterior commissure using an established protocol (Fink et al., 1998).
Data analysis. Experiments (cerebral ischemia, evaluation of infarct size, cell counts) were performed in a blinded fashion. Data are presented as mean ± SE. Differences between groups were evaluated by paired or unpaired two-tailed Student's t test or by ANOVA followed by Scheffe's test (physiology). pvalues of <0.05 were considered statistically significant.
RESULTS
Effects of ischemia on MTase catalytic activity
We determined MTase catalytic activity ex vivo using an established model of 30 min MCAo followed by reperfusion (Endres et al., 1998b). As displayed in Figure1A, cortical and striatal levels in ischemic tissue did not differ from baseline (sham) and contralateral tissues, nor did they change with time (1, 3, 6, 18 hr) during reperfusion. Levels inDnmtS/+ mice were ∼50% compared with Dnmt+/+ mice and also did not change over time (1, 3, 6, 18 hr) (Fig.1B). Activity levels in cerebellum were considerably higher than in other brain regions, which agrees with the literature (Brooks et al., 1996). Hence, MTase, an enzyme with a short protein half life (Szyf, 1994), remains functionally intact after ischemia/reperfusion.
Fig. 1.
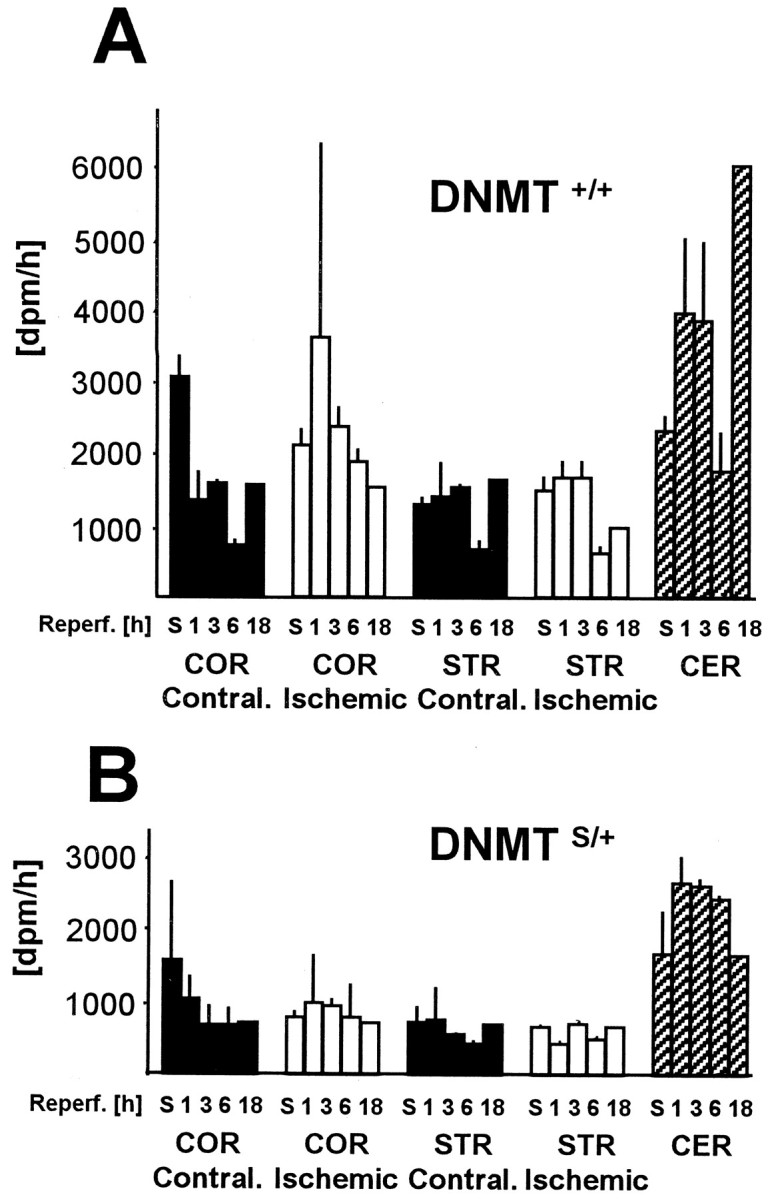
Time-dependent changes of MTase activity inDnmt+/+ (A) andDnmtS/+ mice (B) after 30 min middle cerebral artery occlusion and reperfusion. MTase activity was measured in ischemic cortex (COR) and striatum (STR) along with the respective contralateral tissue and sham (S)-operated animals. MTase activity was measured in nuclear extracts in vitro using a hemimethylated oligo-probe. Levels in cerebellum (CER) are shown for comparison. n = 2 animals per time point. Mean ± SE. p > 0.05.
Effects of ischemia on MTase protein levels
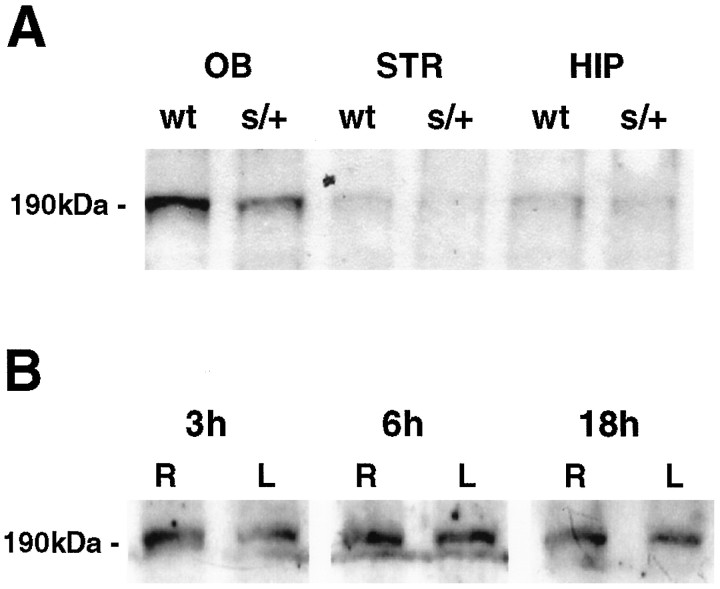
MTase protein was measured semiquantitatively using immunoblot analysis of brain lysates probed with rabbit antiserum (Tucker et al., 1996a). A band corresponding to molecular weight 190 kDa was detected on immunoblot. MTase protein levels were ∼50% inDnmtS/+ mice compared with the wild-type strain in olfactory bulb (OB), striatum (STR), and hippocampus (HIP) (Fig.2A). To characterize the fate of MTase after brain ischemia, we measured MTase protein after 30 min MCAo/reperfusion. No differences were found over time in ischemic striatum after recirculation of 0, 1, 3, 6, 12, 18, 24, 48, and 72 hr (Fig. 2B). Hence, MTase levels did not change over time after ischemia/reperfusion.
Fig. 2.
A, MTase protein levels in olfactory bulb (OB), striatum (STR), and hippocampus (HIP) in brain lysates from normal (wt) and DnmtS/+ mice (s/+). Brain lysates were subjected to SDS-PAGE and immunoblot analysis performed using a polyclonal MTase antibody. The experiment was repeated three times; a representative experiment is shown. B, Time-dependent changes in MTase protein expression in striatum during reperfusion after 30 min middle cerebral artery occlusion. MTase protein was present in normal brain (sham) and did not change over time (0, 1, 3, 6, 12, 18, 24, 48, and 72 hr) between left (L = ischemic) and right (R = non-ischemic) hemispheres. The experiment was repeated three to four times per time point; a representative experiment for the time points 3, 6, and 18 hr is shown.
Effects of cerebral ischemia on DNA methylationin vivo
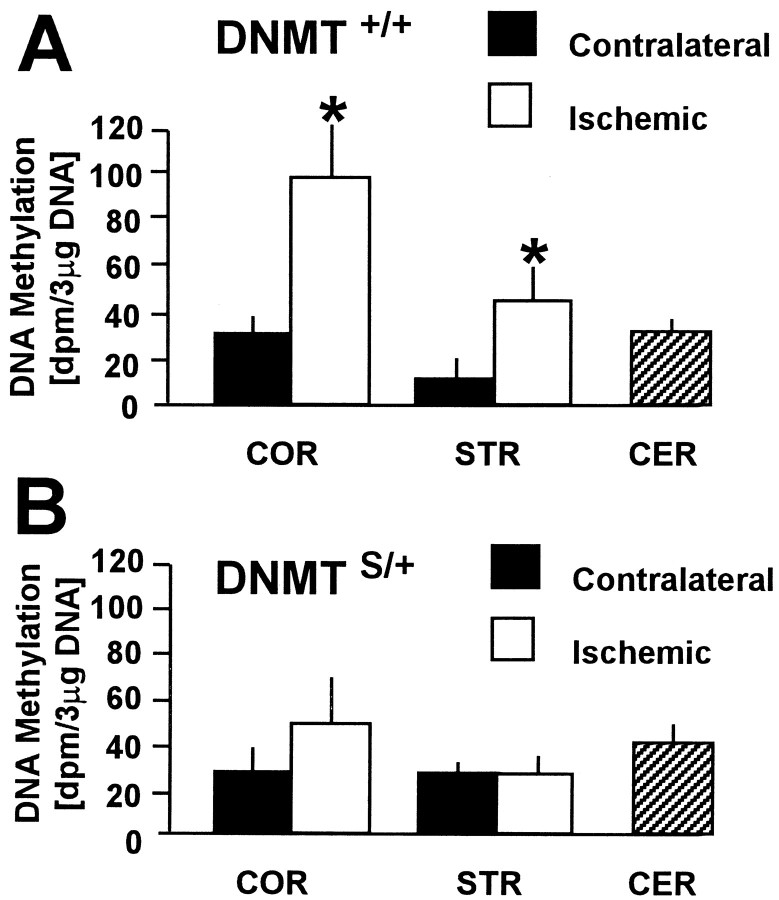
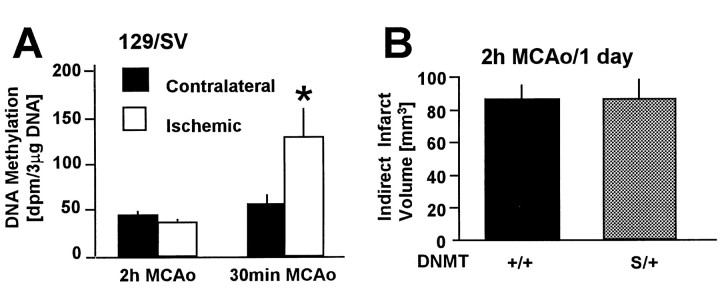
To measure newly incorporated methyl groups into DNA during brain ischemia/reperfusion in vivo, animals were first subjected to 30 min MCAo, and on reperfusion a labeled methyl-group precursor of SAM (l-[methyl-3H]methionine) was administered. Twelve hours after reperfusion, DNA methylation was significantly higher in ischemic striatum (4.1-fold) and cortex (3.2-fold) compared with the respective contralateral tissues in wild-type mice (Dnmt+/+littermates) (Fig. 3A). DNA methylation, however, did not increase in ischemic tissue ofDnmtS/+ mice (Fig.3B). Together, these results demonstrate that methyl group incorporation increases after ischemia/reperfusion in wild-type but not in DnmtS/+ mice.
Fig. 3.
Methyl group incorporation into brain DNAin vivo in Dnmt+/+(A) and DnmtS/+mice (B) after 30 min middle cerebral artery occlusion. On reperfusion,l-[methyl-3H]methionine was administered and [3H]methyl group incorporation was measured after 12 hr. DNA methylation was significantly increased in ischemic cortex (COR) and striatum (STR) compared with the contralateral side inDnmt+/+ (A) but not DnmtS/+ mice (B). Levels in cerebellum (CER) are shown for comparison. n = 4 and 5 animals. Mean ± SE. *p < 0.05. Paired Student'st test.
Smaller cerebral lesions and improved neuronal survival in DnmtS/+ mice after mild stroke
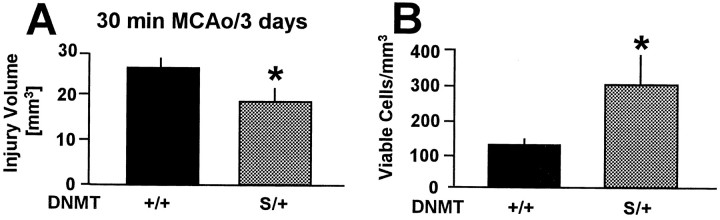
To determine whether the failure to increase DNA methylation altered the susceptibility ofDnmtS/+ mice to tissue injury, we quantitated lesion volume after 30 min MCAo/72 hr reperfusion. In fact, lesion size was significantly (29%) reduced compared with controls (Fig. 4A). Moreover, when neurons were counted within ischemic striatum, the density of viable cells was 2.4-fold higher in theDnmtS/+ mice compared with controls (Fig. 4B). These results demonstrate that reduced MTase activity is beneficial during mild cerebral ischemia.
Fig. 4.
A, Infarct volume was 29% smaller in DnmtS/+ mice compared withDnmt+/+ mice after 30 min filamentous middle cerebral artery occlusion (MCAo) and 72 hr reperfusion. Brain lesion volume was determined on serial coronal hematoxylin and eosin-stained cryostat sections (20 μm).B, DnmtS/+ mice had significantly higher numbers of viable cells in ischemic striatum after 30 min MCAo/72 hr reperfusion compared with controls. Viable cells with neuronal appearance were counted on coronal sections (20 μm) through the anterior commissure. Mean ± SE. n = 14 and 15 animals per group. *p < 0.05 compared withDnmt+/+ mice. Student'st test.
Systemic physiological parameters are unaltered inDnmtS/+ mice
Because alterations in systemic and cerebrovascular parameters can modify outcome after stroke, we excluded possible effects of such variables by careful physiological monitoring. Rectal temperature, arterial blood pressure, pH, partial pressure of oxygen and carbon dioxide before, during, and after ischemia did not differ between groups (Table 1). rCBF, measured with a laser Doppler flow probe, decreased to <20% of baseline ischemia in all animals and returned to ∼100% within 5 min after reperfusion (Table 1). Blood pressure after reperfusion was somewhat lower inDnmtS/+ mice compared with controls (82 ± 6 vs 101 ± 5 mmHg) (Table 1). Differences in blood pressure after ischemia between groups could possibly be explained by specific gene expression changes (e.g., of endothelial NO synthase); however, they are unlikely to explain improved outcome inDnmtS/+ mice because systemic hypotension normally aggravates ischemic damage (Harms et al., 2000).
Table 1.
Physiological variables at baseline and during ischemia inDnmt+/+ and DnmtS/+mice
| Parameter | Dnmt+/+mice | DnmtS/+ mice |
|---|---|---|
| MABP (mmHg) | ||
| Baseline | 89 ± 3 | 87 ± 4 |
| During | 91 ± 5 | 88 ± 6 |
| After | 101 ± 5 | 82 ± 6* |
| pH | ||
| Baseline | 7.32 ± 0.01 | 7.29 ± 0.02 |
| After | 7.35 ± 0.01 | 7.33 ± 0.02 |
| PaO2(mmHg) | ||
| Baseline | 164 ± 10 | 132 ± 21 |
| After | 130 ± 13 | 123 ± 20 |
| PaCO2(mmHg) | ||
| Baseline | 43 ± 2 | 46 ± 3 |
| After | 38 ± 1 | 43 ± 3 |
| rCBF (%) | ||
| Before | 100 ± 0 | 100 ± 0 |
| During | 14 ± 5 | 11 ± 3 |
| After | 103 ± 8 | 100 ± 7 |
| Weight (gm) | 21.1 ± 1.0 | 19.7 ± 0.9 |
| CT (°C) | ||
| During | 37.1 ± 0.1 | 37.1 ± 0.1 |
Animals were subjected to 30 min filamentous middle cerebral artery occlusion followed by reperfusion (Endres et al., 1998b). MABP (mean arterial blood pressure) and rCBF (regional cerebral blood flow) were measured at baseline, during ischemia, and after 30 min reperfusion (Huang et al., 1994). Fifty microliters of blood were withdrawn twice, before ischemia and directly after reperfusion for blood gas determination (pH, PaO2, PaCO2). Animals were weighed before the onset of the experiment (body weight in grams). Core temperature (CT) was controlled and recorded by means of a feedback temperature control unit. *p < 0.05 vs control; otherwise there were no statistically significant differences between groups. n = 5 per group. Mean and SE, ANOVA, and Scheffé's test.
Pharmacological inhibition of DNA methylation and histone deacetylation confers stroke protection in wild-type mice
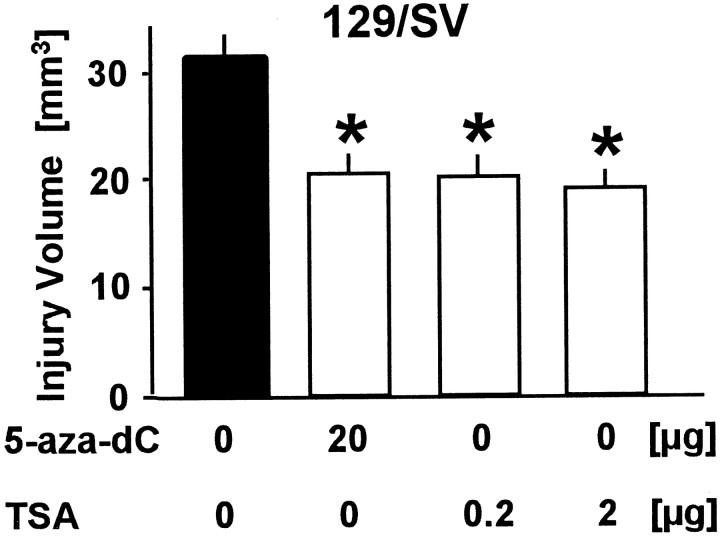
To test the theory that a pharmacological inhibitor of MTase confers stroke protection in wild-type mice, we treated 129/SV mice with the MTase inhibitor 5-aza-dC. Twenty micrograms given intracerebroventricularly 10 min before 30 min MCAo reduced lesion volume by 34% at 72 hr (Fig. 5). After a slightly different ischemia protocol (45 min MCAo followed by 48 hr reperfusion), lesion size was 54% smaller in the 5-aza-dC-treated animals (43.2 ± 12.6 vs 19.9 ± 0.8 mm3 in control vs treated mice, respectively; p < 0.001; n = 4 per group). We also determined methyl group incorporation into DNA (12 hr reperfusion) in 5-aza-dC-treated aniimals versus vehicle-injected animals. For this experiment, 400 μl [methyl-3H]methionine without additional PBS was used. A 2.72-fold increase compared with the contralateral side was found in the ischemic territory of vehicle-injected animals (90 ± 14 vs 244 ± 75 dpm in contralateral vs ischemic tissue, respectively; 3 μg DNA; n = 4). However, in 5-aza-dC-treated animals this increase was only 1.38-fold (101 ± 13 vs 140 ± 31 dpm in contralateral vs ischemic tissue, respectively; 3 μg DNA; n = 3).
Fig. 5.
Treatment with the MTase inhibitor 5-aza-2′-deoxycitide (5-aza-dC; 20 μg) or the specific deacetylation inhibitor trichostatin A (TSA; 1 or 10 μg) reduced lesion size after 30 min of filamentous middle cerebral artery occlusion and 72 hr of reperfusion compared with vehicle in 129/V mice. Drugs or vehicle was administered intracerebroventricularly 10 min before ischemia onset. Lesion volume was determined quantitatively. Data are presented as mean ± SE.n = 5–9 animals per group. *p< 0.01 versus vehicle. Student's t test.
Because deletion/inhibition of MTase may augment the level of gene transcription, we tested whether increasing gene transcription by a related yet distinct mechanism (i.e., inhibition of histone deacetylation) (Yoshida et al., 1995; Eden et al., 1998; Jones et al., 1998; Nan et al., 1998) would also protect from ischemic brain injury. Accordingly, when animals were pretreated with TSA, a highly specific inhibitor of histone deacetylase (Yoshida et al., 1995) (0.2 or 2 μg, i.c.v., given 10 min before ischemia), lesion size was significantly reduced after 30 min MCAo/72 hr reperfusion (Fig. 5). After 45 min MCAo/48 hr reperfusion, lesion size was 48% smaller in TSA-treated animals (43.2 ± 12.6 vs 22.4 ± 1.2 mm3 in control vs 2 μg TSA, respectively; p < 0.001; n = 4 per group).
For both the 5-aza-dC and the TSA experiments, we observed no significant differences in physiological parameters between groups (rCBF, blood pressure, blood gases, rectal temperatures) that could influence ischemia outcome (n = 5 animals per group; ANOVA plus Scheffe's test; data not shown).
Damage after severe stroke is not blunted inDnmtS/+ mice
Excitotoxic and free radical-mediated mechanisms predominate after prolonged periods of ischemia presumably because of necrotic mechanisms of cell death, whereas apoptotic mechanisms are unmasked in milder forms of ischemia (Choi, 1988; Endres et al., 1997, 1998b). We therefore tested the effects of dnmt1 heterozygosity in a model of severe stroke (2 hr MCAo and reperfusion).
First, we determined whether DNA methylation increases after more prolonged ischemia as it does after mild ischemia (30 min MCAo). Methyl group incorporation was not increased after 2 hr MCAo in wild-type mice (Fig. 6A). Moreover, there was no difference in infarct size betweenDnmtS/+ mice and wild-type littermates after 2 hr MCAo and 22 hr reperfusion (121.7 ± 11.5 vs 137.9 ± 19.6 mm3 in controls vsDnmtS/+, respectively;p > 0.05; n = 4 and 5). There was also no difference between groups when infarct size was corrected for brain swelling (Fig. 6B). Regional CBF during ischemia was <20% of baseline and did not differ between groups (13.8 ± 3.1 vs 18.6 ± 3.0% in controls vsDnmtS/+, respectively;p > 0.05; n = 4 and 5). Hence, DNA methylation does not increase after prolonged ischemia, and deletion of one gene copy of dnmt1 did not confer resistance to severe injury as it did after brief periods of MCAo.
Fig. 6.
A, Methyl group incorporation into brain DNA in vivo after 2 hr versus 30 min MCAo in 129/SV mice. On reperfusionl-[methyl-3H]methionine was administered, and [3H]methyl group incorporation into brain DNA was measured after 12 hr reperfusion. Unlike after 30 min MCAo (also see Fig. 3), DNA methylation did not increase in ischemic cortical tissue from the MCA territory compared with the contralateral side after 2 hr MCA occlusion. n = 5 (2 hr MCAo) and 4 (30 min MCAo) animals. Mean ± SE. *p < 0.05. Paired Student's t test. B, WhenDnmt+/+ andDnmtS/+ mice were subjected to 2 hr MCAo/24 hr reperfusion and infarct volume was quantitated, no difference in indirect infarct volume was noted between groups. This result was confirmed by a direct method to measure infarct volume (137 ± 19.6 vs 121.7 ± 11.5 mm3 forDnmtS/+ vsDnmt+/+ mice, respectively). Mean ± SE. n = 4 and 5. p> 0.05 compared with wild type. Student's ttest.
DISCUSSION
This is the first report to implicate DNA methylation in the evolution of ischemic brain injury. Ischemia/reperfusion generated a three- to fourfold increase in methyl group incorporation in brain, whereas this increase was not observed in transgenic animals expressing reduced MTase levels; furthermore, the mutant brain was resistant to injury as evidenced by decreased striatal damage and increased numbers of surviving striatal neurons. There were no group differences in genetic background or differences in physiological or cerebrovascular parameters to explain these results. Together, this may mean that DNA methylation may render the tissue more vulnerable to ischemic injury and may increase as a consequence of such injury. The former postulation is supported by the finding that ischemic brain injury can be reduced by treatment with an inhibitor of DNA methylation, 5-aza-dC.
DNA methylation after cerebral ischemia
An in vivo methylation assay was developed to measure newly incorporated methyl groups into DNA usingl-[methyl-3H]methionine as methyl donor. [3H]methyl groups became incorporated into DNA, and we expect that most of the incorporated methyl groups was added to cytosine residues, although thymine methylation is a theoretical possibility (Razin et al., 1970). Although there were significant differences in DNA methylation in vivo between Dnmt+/+ andDnmtS/+ mice after ischemia, baseline methylation (contralateral hemisphere and cerebellum) did not differ, which agrees with previous findings inDnmtS/+ cells (Li et al., 1992,1993; Laird and Jaenisch, 1996). Absolute numbers for methylation frequency could not be obtained with our assay [assuming that the endogenous SAM pool was completely replaced by [methyl-3H]-SAM and that [3H]methyl groups were incorporated only into CpG dinucleotides, we estimate the striatal methylation frequency as 4.5 × 106 (control) vs 12.2 × 106 (ischemia) per CpG dinucleotide]. Despite significant increases in methyl group incorporation into DNA, protein amount and enzymatic activity of MTase were unchanged after cerebral ischemia and reperfusion, although small differences may have been missed because of assay sensitivity. Possible explanations include induction of other MTases, which seems unlikely, however, in view of the results in DnmtS/+ mice. A more reasonable explanation is an increase in MTase substrate, i.e., hemimethylated DNA, the mechanisms of which are discussed below.
Ischemia-induced methylation may reflect MTase activity in neurons after DNA damage and repair. Ischemic stress facilitates deamination of 5′MeC leading to G-T mismatches. For example, after forebrain ischemia, the mutation spectrum indicates that 58% of mutants with base substitutions (or one-third of all mutations) involved G-T mismatches (Liu et al., 1996). MTase may remethylate cytosine residues after base-excision repair of mismatches (Brooks et al., 1996; Brooks, 1998). Cui et al. (1999) reported specific DNA damage 15 min after focal ischemia (i.e., in the c-fos gene), most of which was effectively repaired 60 min after ischemia by base-excision repair. Another important mechanism in this respect is the observation that the formation of oxygen radicals [i.e., 8-hydroxyguanine (oh8dG)] profoundly alters methylation of adjacent cytosines, suggesting a role for oxidative injury in the formation of aberrant DNA methylation patterns (Cerda and Weitzman, 1997). Interestingly, oh8dG levels increased fourfold after forebrain ischemia (Liu et al., 1996), and we observed a 2.7- to 4.1-fold increase of methylation in a focal model. However, it is unlikely that this mechanism can solely explain methylation changes in our model. Moreover, the recent discovery of a DNA demethylase (Bhattacharya et al., 1999) and the notion that DNA methylation may be a reversible biological phenomenon (Rachmandani et al., 1999) shed new light on DNA methylation in postmitotic cells such as neurons. Notably, our preliminary results indicate that ischemic resistance was also enhanced in genetically engineered mice in which the dnmt1 gene was selectively deleted in neurons (M. Endres, G. Fan, and R. Jaenisch, unpublished observation). Alternatively, increased DNA methylation after brain ischemia could relate to mitotic activity and DNA replication in non-neuronal cells such as glia or progenitor cells (Lee et al., 1996).
In addition, administration of 5-aza-dC, an inhibitor of MTase, was able to significantly inhibit DNA methylation when administered intracerebroventricularly before ischemia. Notably, 5-aza-dC, which is known to easily penetrate cells (Chabor et al., 1983), requires incorporation into DNA to inhibit MTase (Laird et al., 1995). In accordance with the above discussed mechanisms for DNA methylation in the adult ischemic brain, 5-aza-dC could be incorporated into DNA of replicating non-neuronal cells or newly repaired DNA of neurons (Brooks et al., 1996; Liu et al., 1996; Brooks, 1998; Cui et al., 1999).
Possible neuroprotective mechanisms
In our study we showed that suppression of DNA methylation conferred resistance to ischemia as well as specific inhibition of histone deacetylation. An inhibitor of DNA methylation, 5-aza-dC, caused significant reduction in ischemic injury as did a deficiency indnmt1 gene expression. In addition to methylation, histone deacetylation has also been linked to gene silencing and provides a second global mechanism by which genes are regulated (Yoshida et al., 1995; Eden et al., 1998; Jones et al., 1998;Nan et al., 1998). In fact, the two mechanisms have themselves been linked recently by data showing that transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex (Eden et al., 1998; Jones et al., 1998; Nan et al., 1998). We infer from our data that DNA methylation and histone deacetylation alter gene expression after ischemia in such a way as to render tissue susceptible to injury. According to a “good versus bad gene” theory, this could relate to overexpression of protective/antiapoptotic genes (e.g.,c-fos, bcl-2, sod) or repression of deleterious/proapoptotic genes (e.g., bad,bax). Cui et al. (1999) recently reported specific alterations in gene expression of c-fos after focal ischemia/reperfusion. Hence, neuroprotection may relate not only to the specific characteristics of delayed neuronal cell death after mild stroke but to ischemia-induced changes in gene expression mediated by DNA methylation and histone deacetylation. To further differentiate between possible necrotic and apoptotic mechanisms, we used two different models of cerebral ischemia (30 min vs 2 hr MCAo) (Endres et al., 1998b). The fact that methyl group incorporation did not increase after 2 hr MCAo and mutants partially deficient in MTase were not protected from severe stroke points to fundamental differences in the pathways of mild versus severe ischemic injury.
Experiments using DnmtS/+ mice cannot distinguish between effects of dnmt1 heterozygosity during development versus effects in the adult brain. However, the fact that methylation patterns are similar betweenDnmtS/+ andDnmt+/+ cells (Li et al., 1992,1993; Laird and Jaenisch, 1996) and that inhibiting MTase activity with 5-aza-dC was neuroprotective during ischemia suggests that effects in adult brain are responsible for protection. Experiments using conditional knockouts will help to resolve this issue.
In addition to the above epigenetic mechanisms, the enzyme MTase could enhance DNA mutagenesis. Under certain conditions, especially during SAM deficiency, C-T and C-U transitions can be facilitated by MTase directly (Wyszynski et al., 1994; Gonzalgo and Jones, 1997). Possibly, the mutagenic potency of MTase is compensated for by an effective DNA repair mechanism. This would imply a protective role for mismatch repair versus deleterious effects of MTase and DNA methylation. Therefore, although this mechanism has yet to be convincingly demonstrated in mammals, a direct genetic mechanism by which MTase contributes to ischemic damage is possible. Notably, SAM administration is neuroprotective in models of cerebral ischemia in rats and gerbils (Sato et al., 1988; Rao et al., 1997). It will be interesting to measure SAM levels after cerebral ischemia and conditions of MTase deficiency in our model.
In conclusion, we demonstrate that DNA methylation increases in vivo after ischemia/reperfusion and that reduced levels of MTase in brain protect from ischemic injury. These observations underscore the potential importance of DNA methylation to mechanisms of injury and repair in the postischemic brain.
Footnotes
This research was supported by Grant NS10828 from National Institutes of Health (M.A.M.), the Deutsche Forschungsgemeinschaft (En343/1-1 to M.E.; En343/4-1 to M.E. and A.M.; Me1562/1-1 to A.M. and U.D.; Di454/8-2 to U.D), the Humboldt-University of Berlin (M.E.), and the Hermann and Lilly Schilling Stiftung (U.D.). We thank Guoping Fan for assistance in the immunoblot assays.
M.E. and A.M. contributed equally to this work.
Correspondence should be addressed to Dr. Matthias Endres, Department of Neurology, Charité Hospital, Humboldt-University, D-10098 Berlin, Germany. E-mail:matthias.endres@charite.de.
REFERENCES
- 1.Adams RL, Burdon RH. DNA methylation in eukaryotes. CRC Crit Rev Biochem. 1982;13:349–384. doi: 10.3109/10409238209108714. [DOI] [PubMed] [Google Scholar]
- 2.Baylin SB, Makos M, Wu JJ, Yen RW, de Bustros A, Vertino P, Nelkin BD. Abnormal patterns of DNA methylation in human neoplasia: potential consequences for tumor progression. Cancer Cells. 1991;3:383–390. [PubMed] [Google Scholar]
- 3.Bestor TH. DNA methylation: evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes. Philos Trans R Soc Lond B Biol Sci. 1990;326:179–187. doi: 10.1098/rstb.1990.0002. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya SK, Rachmandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 5.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 6.Brooks PJ. Detection of excision nuclease in cell-free extracts from the adult mammalian brain. Mutat Res. 1998;408:37–46. doi: 10.1016/s0921-8777(98)00018-4. [DOI] [PubMed] [Google Scholar]
- 7.Brooks PJ, Marietta C, Goldman D. DNA mismatch repair and DNA methylation in adult brain neurons. J. Neurosci. 1996;16:939–945. doi: 10.1523/JNEUROSCI.16-03-00939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cedar H. DNA methylation and gene activity. Cell. 1988;34:5503–5513. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- 9.Cedar H, Razin A. DNA methylation and development. Biochim Biophys Acta. 1990;1094:1–8. doi: 10.1016/0167-4781(90)90076-e. [DOI] [PubMed] [Google Scholar]
- 10.Cerda S, Weitzman SA. Influence of oxygen radical injury on DNA methylation. Mutat Res. 1997;386:141–152. doi: 10.1016/s1383-5742(96)00050-6. [DOI] [PubMed] [Google Scholar]
- 11.Chabor GG, Rivard GE, Momparler RL. Plasma and cerebrospinal fluid pharmacokinetics of 5-aza-2′-deoxycytidine in rabbits and dogs. Cancer Res. 1983;43:592–597. [PubMed] [Google Scholar]
- 12.Choi DW. Glutamate neurotoxicity and diseases of the nervous system. Neuron. 1988;1:623–634. doi: 10.1016/0896-6273(88)90162-6. [DOI] [PubMed] [Google Scholar]
- 13.Counts JL, Goodman KI. Alteration in DNA methylation may play a variety of roles in carcinogenesis. Cell. 1995;83:13–15. doi: 10.1016/0092-8674(95)90228-7. [DOI] [PubMed] [Google Scholar]
- 14.Cross SH, Bird AP. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 15.Cui J, Holmes EH, Liu PK. Oxidative damage to the c-fos gene and reduction of its transcription after focal cerebral ischemia. J Neurochem. 1999;73:1164–1174. doi: 10.1046/j.1471-4159.1999.0731164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- 17.Eden S, Hashimshony T, Keshet I, Cedar H, Thorne AW. DNA methylation models histone acetylation. Nature. 1998;394:842. doi: 10.1038/29680. [DOI] [PubMed] [Google Scholar]
- 18.Ehrlich M, Wang R-Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981;212:1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- 19.Endres M, Wang Z-Q, Namura S, Waeber C, Moskowitz MA. Ischemic brain injury is mediated by activation of poly(ADP-ribose)polymerase. J Cereb Blood Flow Metab. 1997;17:1143–1151. doi: 10.1097/00004647-199711000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Endres M, Laufs U, Huang Z, Nakamura T, Huang PL, Moskowitz MA, Liao JK. Stroke protective effects of HMG-CoA reductase inhibitors mediated by upregulation of endothelial nitric oxide synthase. Proc Natl Acad Sci USA. 1998a;95:1880–1885. doi: 10.1073/pnas.95.15.8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endres M, Namura S, Shimizu-Sasamata M, Waeber C, Zhang L, Gomez-Isla T, Hyman BT, Moskowitz MA. Attenuation of delayed neuronal death after mild focal cerebral ischemia by inhibitors of the caspase family. J Cereb Blood Flow Metab. 1998b;18:238–247. doi: 10.1097/00004647-199803000-00002. [DOI] [PubMed] [Google Scholar]
- 22.Fink K, Zhu J, Namura S, Shimizu-Sasamata M, Endres M, Ma J, Yuan J, Moskowitz MA. Prolonged therapeutic window for ischemic brain damage due to delayed caspase-3 activation. J Cereb Blood Flow Metab. 1998;18:1071–1076. doi: 10.1097/00004647-199810000-00003. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalgo ML, Jones PA. Mutagenic and epigenetic effects of DNA methylation. Mutat Res. 1997;386:107–118. doi: 10.1016/s1383-5742(96)00047-6. [DOI] [PubMed] [Google Scholar]
- 24.Goto K, Numatas M, Momuram J-I, Ono T, Bestor T, Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1993;56:39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- 25.Harms H, Wiegand F, Megow D, Prass K, Einhäupl KM, Dirnagl U. Acute treatment of hypertension increases infarct sizes in spontaneously hypertensive rats. NeuroReport. 2000;11:355–359. doi: 10.1097/00001756-200002070-00027. [DOI] [PubMed] [Google Scholar]
- 26.Huang Z, Huang PL, Panahian N, Dalkara T, Fishman MC, Moskowitz MA. Effects of cerebral ischemia in mice deficient in neuronal nitric oxide synthase. Science. 1994;265:1883–1885. doi: 10.1126/science.7522345. [DOI] [PubMed] [Google Scholar]
- 27.Jones PA. DNA methylation errors and cancer. Cancer Res. 1996;56:2463–2467. [PubMed] [Google Scholar]
- 28.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 29.Laird PW, Jaenisch R. The role of DNA methylation in cancer genetics and epigenetics. Annu Rev Genet. 1996;30:441–464. doi: 10.1146/annurev.genet.30.1.441. [DOI] [PubMed] [Google Scholar]
- 30.Laird PW, Jackson-Grusby L, Fazeli A, Dickinson SL, Jung WE, Weinberg RA, Jaenisch R. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 31.Lee PJ, Wahser LL, Law DJ, Boland CR, Horon IL, Feinberg AP. Limited up-regulation of DNA methyltransferase in human colon cancer reflecting increased cell proliferation. Proc Natl Acad Sci USA. 1996;93:10366–10370. doi: 10.1073/pnas.93.19.10366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lengauer C, Kinzler KW, Vogelstein B. DNA methylation and genetic instability in colorectal cancer cells. Proc Natl Acad Sci USA. 1997;94:2545–2550. doi: 10.1073/pnas.94.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leonhard H, Bestor TH. Structure, function, and regulation of mammalian DNA methyltransferase. In: Jost JP, Saluz HP, editors. DNA methylation: molecular biology and biological significance. Birkhauser Verlag; Basel: 1993. pp. 109–119. [DOI] [PubMed] [Google Scholar]
- 34.Li E, Bestor T, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 35.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 36.Liu PK, Hsu CY, Dizdaroglu M, Floyd RA, Kow YW, Karakaya A, Rabow LE, Cui J-K. Damage, repair, and mutagenesis in nuclear genes after mouse forebrain ischemia-reperfusion. J Neurosci. 1996;16:6795–6806. doi: 10.1523/JNEUROSCI.16-21-06795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacManus JP, Linnik MD. Gene expression induced by cerebral ischemia: an apoptotic perspective. J Cereb Blood Flow Metab. 1997;17:815–832. doi: 10.1038/aj.jcbfm.9590266. [DOI] [PubMed] [Google Scholar]
- 38.Nan X, Ng H-H, Johnson CA, Laherty CD, Turner BT, Eisenmann RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 39.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferase. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 40.Panning B, Jaenisch R. RNA and the epigenetic regulation of X chromosome inactivation. Cell. 1998;93:305–308. doi: 10.1016/s0092-8674(00)81155-1. [DOI] [PubMed] [Google Scholar]
- 41.Rachmandani S, Bhattacharya SK, Cervoni N, Szyf M. DNA methylation is a reversible biological phenomenon. Proc Natl Acad Sci USA. 1999;96:6107–6112. doi: 10.1073/pnas.96.11.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao AM, Başkaya MK, Maley ME, Kindy MS, Dempsey RJ. Beneficial effects of S-adenosyl-l-methionine on blood-brain barrier breakdown and neuronal survival after transient cerebral ischemia in gerbils. Mol Brain Res. 1997;44:134–138. doi: 10.1016/s0169-328x(96)00245-8. [DOI] [PubMed] [Google Scholar]
- 43.Razin A, Cedar H. DNA methylation and genomic imprinting. Cell. 1994;77:473–476. doi: 10.1016/0092-8674(94)90208-9. [DOI] [PubMed] [Google Scholar]
- 44.Razin A, Sedat JW, Sinsheimer RL. Structure of the DNA of bacteriophage ΦX174. J Mol Biol. 1970;53:251–259. doi: 10.1016/0022-2836(70)90298-6. [DOI] [PubMed] [Google Scholar]
- 45.Sato H, Hariyama H, Moriguchi K. S-adenosyl-l-methionine protects the hippocampal CA1 neurons from the ischemic neuronal death in rat. Biochem Biophys Res Commun. 1988;150:491–496. doi: 10.1016/0006-291x(88)90547-5. [DOI] [PubMed] [Google Scholar]
- 46.Surani MA. Imprinting and the initiation of gene silencing in the germ line. Cell. 1998;93:309–312. doi: 10.1016/s0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- 47.Szyf M. DNA methylation properties: consequences for pharmacology. Trends Pharmacol Sci. 1994;15:233–238. doi: 10.1016/0165-6147(94)90317-4. [DOI] [PubMed] [Google Scholar]
- 48.Tucker KL, Talbot D, Lee MA, Leonhardt H, Jaenisch R. Complementation of methylation deficiency in embryonic stem cells by a DNA methyltransferase minigene. Proc Natl Acad Sci USA. 1996a;93:12920–12925. doi: 10.1073/pnas.93.23.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tucker KL, Beard C, Dausman J, Jackson-Grusby L, Laird PW, Lei H, Li E, Jaenisch R. Germ-line passage is required for establishment of methylation patterns of imprinted but not of nonimprinted genes. Genes Dev. 1996b;10:1008–1020. doi: 10.1101/gad.10.8.1008. [DOI] [PubMed] [Google Scholar]
- 50.Wyszynski M, Gabbara S, Bhagwat AS. Cytosine deaminations catalyzed by DNA cytosine methyltransferases are unlikely to be the major cause of mutational hot spots at sites of cytosine methylation in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:1574–1578. doi: 10.1073/pnas.91.4.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioassays. 1995;17:423–443. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]