Abstract
Estrogen receptor (ER) β plays a critical role in endometriosis progression because cytoplasmic ERβ stimulates proinflammatory signaling in ectopic lesions and prevents apoptosis to promote their survival. However, the role of “nuclear ERβ” in endometriosis progression is not known. This critical knowledge gap obscures our understanding of the full molecular etiology of ERβ-mediated endometriosis progression. To fill this void, we generated an ERβ-regulated transcriptome and ERβ cistrome in ectopic lesions and the eutopic endometrium of mice with endometriosis by using a new endometrium-specific FLAG-tagged human ERβ overexpression mouse model. The integration of these omics data sets revealed that ERβ stimulated the proliferation activities of ectopic lesions and the eutopic endometrium by directly upregulating MYC and E2 transcription factor target genes and genes associated with the G2/M transition. Additionally, ERβ stimulated gene expression associated with TNFα/nuclear factor κB (NF-κB) signaling, epithelial-mesenchymal transition, reactive oxygen species signaling, IL-6/Janus kinase (JAK)/signal transducer and activator of transcription (STAT)3 signaling, and hypoxia signaling and suppressed IFNα signaling in ectopic lesions to enhance endometriosis progression. ERβ also stimulated gene expression associated with the unfolded protein response and IL-6/JAK/STAT3 inhibitory signaling and suppressed TNFα/NF-κB signaling in the eutopic endometrium to cause endometriosis-associated endometrial dysfunction. Therefore, nuclear ERβ-regulated gene networks provide critical clues to understand the molecular etiology and complexity of endometriosis and endometriosis-associated endometrial dysfunction.
Endometriosis, an estrogen-dependent proinflammatory disease, is defined as the colonization and growth of endometrial tissues at anatomic sites outside of the uterine cavity, primarily on the pelvic peritoneum and ovaries (1). Up to 10% of reproductive-aged women worldwide chronically suffer from the symptoms of endometriosis (1, 2). For example, the prevalence of endometriosis increases dramatically to 50% in women with infertility (3), and up to 80% of women with this disease are infertile (4, 5). Owing to the severe chronic morbidity associated with this gynecological disorder, multiple studies have attempted to identify the distinguishing molecular features of endometriotic lesions and endometrial dysfunction with the aim of developing more effective prognostic, diagnostic, and treatment strategies for the clinical management of this debilitating disease and the amelioration of endometriosis-associated infertility (1).
Because endometriosis is an estrogen-dependent disease, estrogen receptors (ERs) play essential roles in the growth of ectopic lesions because endometriotic lesions are not well developed in either ERα- or ERβ-knockout mice (6, 7). Additionally, the inhibition of ER activity using ER-selective modulators, such as chloroindazole for ERβ and oxabicycloheptene sulfonate for ERα, can suppress the growth of endometriotic lesions in mice with endometriosis by inhibiting both estrogenic and inflammatory activities (8). Collectively, the estrogen/ERs axis plays essential roles in endometriosis progression. Despite their role in endometriosis, however, the molecular mechanisms of ER action in endometriosis progression and endometriosis-associated endometrial dysfunction are not fully known.
Among ERs, ERβ has interesting molecular properties associated with endometriosis progression. For example, the mRNA ratio of ERβ to ERα is significantly higher in ovarian endometriomas than in the normal uterine endometrium (9-13), suggesting that the overexpression of ERβ in conjunction with high levels of local estradiol in endometriotic lesions plays a critical role in the promotion of endometriosis. Immunohistochemistry analysis revealed that ERβ expression was detected in both the cytoplasmic and nuclear fractions in ectopic lesions (7). The cytoplasmic ERβ interacts with the apoptosis machinery, such as apoptosis signal-regulating kinase-1, caspase-9, and caspase-8, to prevent intrinsic and extrinsic apoptosis signaling in ectopic lesions and to evade the host immune surveillance system, improving their survival (7). Additionally, cytoplasmic ERβ interacts with the inflammasome complex, enhancing IL-1β-mediated proliferation and the adhesion activities of endometriotic lesions, improving their durability. ERβ also has a crucial role in the progress of progesterone resistance in endometriotic lesions by downregulating progesterone receptor in endometriotic lesions (11).
As a nuclear receptor, nuclear ERβ is likely to play an essential role in endometriosis progression along with its cytoplasmic function. However, a mechanistic role for nuclear ERβ in the pathogenesis of endometriosis is a long-standing enigma due to the lack of specific ERβ antibodies (14). In the absence of an understanding of nuclear ERβ function in endometriosis, targeted diagnostic, prognostic, and treatment strategies for endometriosis cannot be formulated logically. To overcome the ERβ antibody issue, we employed a new endometrium-specific FLAG-tagged ERβ overexpression (ERBOE) mouse model because the gain of FLAG-tagged ERβ gene function in the endometrium enhances endometriosis progression, and FLAG antibody can successfully immunoprecipitate FLAG-tagged ERβ from ectopic lesions (7). To define the molecular mechanism of nuclear ERβ in endometriosis, we generated ERβ-regulated transcriptome and cistrome that are specific for both ectopic lesions and the eutopic endometrium. In this way, we identified ERβ-regulated genomic networks that drive endometriosis progression and endometriosis-associated endometrial dysfunction in our mouse model.
Materials and Methods
Study design
ERβ is elevated in endometriotic tissues compared with the normal endometrium, and overexpression of ERβ in the endometrium enhances endometriosis progression and triggers endometriosis-associated endometrial dysfunction, causing infertility. Therefore, ERβ is a critical modulator of endometriosis. However, the molecular etiology of ERβ in endometriosis is not known. It is urgent to fill this gap in the understanding of the etiology of ERβ-mediated endometriosis and endometrial dysfunction. Therefore, we determined ERβ-regulated transcriptome and ERβ cistrome in ectopic lesions and the eutopic endometrium by using an endometrium-specific ERBOE mouse model and then identified directly ERβ-responsive cellular pathways for endometriosis progression and endometriosis-associated endometrial dysfunction.
To cause ectopic lesions and a eutopic endometrium, we used a mouse model with surgically induced endometriosis. To define ERβ-regulated gene networks for endometriosis progression, we used an endometrium-specific ERBOE mouse model and their control mice (7). We also employed immortalized human endometrial stromal and epithelial cells to validate the role of ERβ-responsive cellular pathways in endometriosis progression.
To obtain ERβ-overexpressing ectopic lesions, uterine fragments were generated from ERBOE female mice (C57BL/6J background, N = 9) and then implanted to syngeneic female mice (C57BL/6J, N = 9). To control ectopic lesions, uterine fragments were generated from progesterone receptor (PR)Cre/+ female mice (C57BL/6J background, N = 9) and then implanted to syngeneic female mice (C57BL/6J, N = 9). At the estrous cycle in the fourth week of endometriosis induction, ectopic lesions were harvested for further analyses.
To obtain an ERβ-overexpressing eutopic endometrium, uterine fragments were generated from C57BL/6J female mice (N = 9) and then implanted to ERBOE female mice (N = 9). For the control eutopic endometrium, uterine fragments were generated from C57BL/6J female mice (N = 9) and then implanted into PRCre/+ female mice (N = 9). At the estrous cycle in the fourth week of endometriosis induction, the eutopic endometrium was harvested for further analyses.
Animal size in each experiment was calculated by a power calculator (http://www.lasec.cuhk.edu.hk/sample-size-calculation.html). Based on this calculator, at least nine mice were required in each group to validate the gain of ERβ gene function in endometriosis progression with 95% probability of showing a statistically significant difference (P = 0.01).
All experiments in this project were blinded. For example, investigators who surgically induced endometriosis performed microarray and chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) and analyzed these omics data were blinded to information regarding the mouse and tissue samples.
Animals
ROSALSL:ERβ/+:PRCre/+ mice (ERBOE) (7), PRCre/+ mice (control) (15), and C57BL/6J mice were maintained in the designated animal care facility at Baylor College of Medicine according to the Institutional Animal Care and Use Committee guidelines for the care and use of laboratory animals. An Institutional Animal Care and Use Committee-approved protocol was followed for all animal experiments.
Surgical induction of endometriosis
Endometriosis in mice was surgically induced under aseptic conditions under anesthesia using a modified method as described previously (16). Briefly, one uterine horn from a donor female mouse (8 weeks old) was isolated under anesthesia. In a petri dish containing warmed DMEM/F-12 supplemented with 100 U/mL penicillin and 100 µg/mL streptomycin, the uterine horns were longitudinally cut with a pair of scissors. Using a 2-mm dermal biopsy punch, one endometrial fragment was generated from the isolated uterus and subsequently sutured to the mesenteric membrane attached to the intestine in the recipient mice through a midline incision (7-0 braided polypropylene suture). The abdominal incision was then closed with a 5-0 braided polypropylene suture continuously. Before harvesting ectopic lesions, the mouse estrous cycle was determined using vaginal cytology (17). At the estrous cycle in the fourth week after endometriosis induction, ectopic lesions or the eutopic endometrium were isolated from mice with endometriosis. The volume of ectopic lesions was determined using the modified ellipsoid equation ½(length × width2) (18).
RNA isolation from ERβ-overexpressing and control ectopic lesions
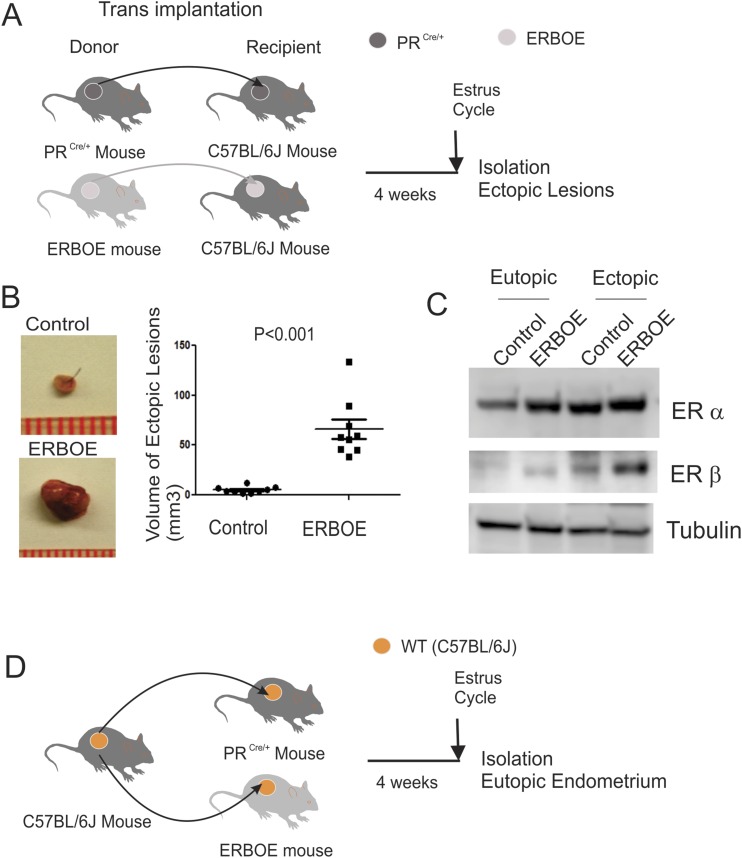
We isolated uteri from ERBOE female mice (8 weeks old, N = 9) and their female control mice (PRCre/+, 8 weeks old, N = 9) and then made endometrium fragments by 2-mm dermal biopsy punch. One uterus fragment isolated from an ERBOE female mouse was implanted in a wild-type intact C57BL/6J mouse (N = 9, 8 weeks old) by the surgically induced endometriosis method as described above (Fig. 1A). We also made endometrial pieces from uteri of female control mice (PRCre/+, N = 9) and then implanted one endometrial piece in a wild-type intact C57BL/6J mouse (N = 9, 8 weeks old) as the control by the surgically induced endometriosis method (Fig. 1A). At the estrous cycle in the fourth week of endometriosis induction, we harvested ERβ-overexpressing (N = 9) and ectopic control lesions (N = 9) and then isolated total RNA from them. After that, we combined total RNA isolated from three ectopic lesions to form a single group. Therefore, we made three RNA groups from nine ERβ-overexpressing ectopic lesions and nine ectopic control lesions.
Figure 1.
Endometrium-specific ERβ overexpression stimulates endometriosis progression. (A) Experimental scheme to isolate ERβ-overexpressing ectopic lesions (N = 9) and ectopic control lesions (N = 9). (B) Volume of ERβ-overexpressing ectopic lesions (N = 9) vs ectopic control lesions (N = 9) isolated from recipient C57BL/6J mice with endometriosis. (C) Experimental plan to separate the ERβ-overexpressing eutopic endometrium (N = 9) and eutopic control endometrium (N = 9). (D) Level of ERα and ERβ in eutopic control endometrium, ERBOE eutopic endometrium, ectopic control lesions, and ERBOE ectopic lesions was determined by Western blot analyses. Level of tubulin in this endometrium was determined as the loading control.
RNA isolation from ERβ-overexpressing and control eutopic endometrium
We isolated uteri from wild-type C57BL/6J female (8 weeks old, N = 18) and then made endometrium fragments by 2-mm dermal biopsy punch. The endometriosis was surgically induced by implanting one uterus fragment isolated from a wild-type C57BL/6J mouse into each ERBOE female mouse (8 weeks old, N = 9) and female control mouse (PRCre/+, N = 9 per group, 8 weeks old) by the surgically induced endometriosis method (Fig. 1C). At the estrous cycle in the fourth week of endometriosis induction, we harvested the eutopic endometrium and then isolated total RNA from them. After that, we combined total RNA from three eutopic endometria to form a single group. Therefore, we made three RNA groups from nine ERβ-overexpressing eutopic endometria and nine eutopic control endometria.
Microarray analyses
Total RNA was isolated from each type of endometrium by using TRIzol reagent, and then total RNA was purified using an RNeasy kit (Qiagen, Valencia, CA). Twenty-five nanograms of total RNA combined with RNA spike mix was reverse transcribed using a T7 primer mix to produce cDNA. The cDNA product was transcribed using T7 RNA polymerase, generating cyanine-3-labeled cRNA. The labeled cRNA was purified using a Qiagen RNeasy mini kit. Purified products were quantified using the NanoDrop spectrophotometer for yield and dye incorporation and tested for integrity on the Agilent Bioanalyzer. Six hundred nanograms of the labeled cRNA was fragmented. Approximately 480 ng of fragmented cRNA samples was loaded onto each of the mouse G3 v2 8×60K expression arrays. The arrays were hybridized in an Agilent hybridization chamber for 17 hours at 65°C with rotation at 10 rpm. The arrays were washed using the Agilent expression wash buffer sets 1 and 2, followed by acetonitrile, as per the Agilent protocol. The Gene Expression Omnibus accession no. for ERβ-regulated transcriptome is GSE114010 (19).
The Subio platform with quantiles for normalization and the log2 transformation was applied to generate signal values of all samples (https://www.subioplatform.com). The ANOVA model was used to compare the expression profiles from different groups. Differentially expressed genes were defined using the filters of ANOVA unadjusted P value <0.01 and absolute fold change >1.5.
Sampling for ERβ cistrome analysis in ectopic lesions
We isolated ERβ-overexpressing ectopic lesions (N = 9) as described above (Fig. 1A). Afterward, we pooled ERβ-overexpressing ectopic lesions (N = 9) and used them all for one FLAG-ERβ ChIP-seq analysis.
Sampling for ERβ cistrome analysis in eutopic endometrium
We isolated ERβ-overexpressing eutopic endometria (N = 9) as described above (Fig. 1C). Afterward, we pooled ERβ-overexpressing eutopic endometrium (N = 9) and used them all for one FLAG-ERβ ChIP-seq analysis.
FLAG-ERβ ChIP-seq
Tissue was submerged in PBS plus 1% formaldehyde, cut into small pieces, and incubated at room temperature for 15 minutes. The addition of 0.125 M glycine stopped fixation. The tissue pieces were then treated with a Tissue Tearor and spun down and washed twice in PBS. Chromatin was isolated by the addition of lysis buffer, followed by disruption with a Dounce homogenizer. Lysates were sonicated and the DNA was sheared to an average length of 300 to 500 bp. For each ChIP reaction, 50 µg of precleared chromatin was mixed with Flag M2 agarose (Sigma-Aldrich St. Louis, MO, A2220) and incubated for 3 hours. Immune complexes were washed, eluted from the beads with SDS buffer, and subjected to RNase and proteinase K treatment. Crosslinks were reversed by incubation overnight at 65°C, and ChIP DNA was purified by phenol-chloroform extraction and ethanol precipitation. Illumina sequencing libraries were prepared from the ChIP and input DNAs by the standard consecutive enzymatic steps of end-polishing, dA addition, and adaptor ligation. After a final PCR amplification step, the resulting DNA libraries were quantified and sequenced on a NextSeq 500. Standard Illumina software base-calling and quality-control filtering were applied. Sequences (75-nt reads, single end) were aligned to the mouse genome (mm10) using the BWA algorithm (v0.7.12, default parameters). Alignments were extended in silico at their 3′ ends to a length of 200 bp, which was the average genomic fragment length in the size-selected library, and assigned to 32-nt bins along the genome. The resulting histograms (genomic “signal maps”) were stored in bigWIG files. Peak locations were determined using the MACS algorithm (v2.1.0) with a cutoff of P = 1 × 10−7. Signal maps and peak locations were used as input data to the Active Motifs proprietary analysis program, which creates Excel tables containing detailed information on sample comparison, peak metrics, peak locations, and gene annotations. The FLAG-ERβ ChIP-seq was performed by Active Motif. The Gene Expression Omnibus accession no. for ERβ cistrome is GSE114047 (20).
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was used to analyze and interpret microarray and gene expression data integrated by microarray and ChIP-seq to propose biological properties. The data in question were analyzed regarding their differential enrichment in a predefined biological set of genes. GSEA was performed using GSEA version 2.2.2 from the Broad Institute at the Massachusetts Institute of Technology. Parameters used for the analysis were as follows. The hallmark gene set was used for running GSEA, 100 permutations were used to calculate P value, and permutation type was set to gene set. All basic and advanced fields were set to default. Enrichment score is the degree to which this gene set is overrepresented at the top or bottom of the ranked list of genes in the expression data set. Normalized enrichment score is the enrichment score for the gene set after it has been normalized across analyzed gene sets. False discovery rate is the estimated probability that the normalized enrichment score represents a false-positive finding.
Binding and expression target analysis
Binding and expression target analysis (BETA) was conducted to integrate the FLAG- ERβ ChIP-seq with gene expression data (ERβ transcriptome) that were differentially expressed in ERβ-overexpressing endometriotic tissues compared with the control endometriotic tissues to infer ERβ direct target genes that are statistically enriched near the ERβ binding sites (21).
Generation of ERβ-overexpressing immortalized human endometrial epithelial cells and immortalized human endometrial stromal cells
FLAG-tagged human ERβ cDNA gene was inserted into a lentiviral expression vector (pCDH-CMV-EF1-Neo, System Biosciences, Palo Alto, CA). A mixture of 2.5 µL of FuGENE, 0.75 µg of FLAG-tagged ERβ-expressing lentiviral vector, and 0.75 µg of VSVG lentiviral packaging mixture (composed of equal parts of pMDL, pRSV, and pVSVG at 0.25 µg/µL) in 100 µL of RPMI 1640 medium were incubated for 30 minutes at room temperature. After the 30-minute incubation, we added 100 µL of the plasmid/FuGENE complexes to 250,000 cells of 293T cells in each well of six-well plates. The culture medium containing the virus was harvested 48 hours after transduction. The multiplicity of infection of lentivirus in the culture medium was detected by using Lenti-X GoStix based on the manufacturer’s protocol (Takara Bio, Mountain View, CA). Immortalized human endometrial epithelial cells (IHEECs) (22) and immortalized human endometrial stromal cells (IHESCs) (23) were grown in 96-well plates with DMEM/F12 containing 10% FBS. At 70% confluence, the culture medium was replaced with DMEM/F12 containing 10% FBS, 8 µg/mL Polybrene, and lentivirus-expressing FLAG-tagged ERβ (multiplicity of infection of 2). At 24 hours after transduction, the culture medium was replaced with DMEM/F12 containing 10% FBS and 50 µg/mL G418 every 2 days until getting G418 resistance cells. Western blot analyses with FLAG antibody determined FLAG-tagged ERβ expression in these G418-resistant cells.
H2O2 assay
IHEECs, ERβ-overexpressing IHEECs (EBO-IHEECs), IHESCs, and ERβ-overexpressing IHESCs (EBO-IHESCs) were cultured in 96-well plates. At 90% confluence, the concentration of H2O2 in these cells was determined by a ROS-Glo™ H2O2 assay kit (Promega, Madison, WI). We also assessed the number of each type of recombinant human endometrial cell in 96-well plates by using a crystal violet assay (24). The H2O2 concentration per cell was determined by luciferase activity divided by cell number represented by optical density at 595 nm.
Western blot analysis
Primary antibodies against the following proteins were used: vimentin (RRID: AB_2178887) (25), N-cadherin (RRID: AB_2687616) (26), E-cadherin (RRID: AB_2291471) (27), Snail (RRID: AB_2255011) (28), Slug (RRID: AB_2239535) (29), β-catenin (RRID: AB_11127855) (30), ERα (RRID: AB_631469) (31), ERβ (RRID: AB_1593623) (32), tubulin (RRID: AB_2241191) (33), suppressor of cytokine signaling (SOCS)1 (RRID: AB_1861766) (34), SOCS2 (RRID: AB_304007) (35), SOCS3 (RRID: AB_304008) (36), vascular endothelial growth factor(RRID: AB_2212644) (37), hypoxia inducible factor (HIF)1A (RRID: AB_2116958) (38), angiopoietin (Ang)1 (RRID: AB_2772625) (39), and Ang2 (RRID: AB_2226215) (40). Membranes containing proteins were incubated with secondary horseradish peroxidase-tagged antibodies (Sigma-Aldrich), and the signals were visualized using ECL plus (Amersham Pharmacia Biotech, Piscataway, NJ)
Quantification of the expression level of ERβ target genes by quantitative PCR
Total RNA was reverse transcribed using the Moloney murine leukemia virus reverse transcription system (Invitrogen, Carlsbad, CA). The quantitative PCR (qPCR) reactions were conducted in the Applied Biosystems 7500 Fast real-time PCR system using RT2 SYBR Green Mastermix (Qiagen) and 0.5 μM of each primer, according to the manufacturer’s protocol. Cycle values from independently generated samples were normalized to the 18S rRNA levels. A Student t test was used to grade statistical significance in this section as follows: *P < 0.05, **P < 0.01. Primers for each gene are follows: fibrillin 1 (Fbn1; 5′-AATGAAGGCTATGAGGTGGC-3′ and 5′-TCTGTAGACTATACCCAGGCG-3′), catalase (Cat; 5′-CTCGTTCAGGATGTGGTTTTC-3′ and 5′-CTTTCCCTTGGAGTATCTGGTG-3′), ring finger protein 31 (Rnf31; 5′-AGCTTTCAGAGTTTCACCCC-3′ and 5′-GCACGAGACTTGGTTACAGG-3′), exosome component 1 (Exosc1; 5′-ATGAAGACCAGCGAGAATGG-3′ and 5′-CCGTGAGTTGATGCTAGAGAC-3′), protein tyrosine phosphatase nonreceptor type 1 (Ptpn1; 5′-AATGACTTCTGGCGGATGG-3′ and 5′-TGACCCCGTATTCTTTGAGC-3′), and musculin (Msc; 5′-CTACGAGGACAGCTATGTGC-3′ and 5′-ACCACAATCCATCTAACTGCC-3′).
Results
Stimulation of the endometriosis progression by ERβ overexpression in the endometrium
Our previous study revealed that endometrium-specific ERβ expression stimulates the growth of ectopic lesions in an ovariectomized mouse with surgically induced endometriosis (7). However, the ovariectomized mouse model does not precisely recapitulate the endometriosis progression of reproductive-aged women. To mimic human endometriosis progression in a mouse model, we surgically induced endometriosis with an intact mouse based on a previous study (41). To ensure the gain of ERβ gene function only in ectopic lesions without interference of the gain of ERβ gene function in other tissues during endometriosis progression, endometriosis was surgically induced by implanting uterine fragment isolated from an ERBOE female mouse (N = 9) or its female control mice (PRCre/+, N = 9) into a wild-type intact female C57BL/6J mouse (N = 9 per group). At the estrous cycle at 4 weeks of endometriosis induction, ectopic lesions were isolated from each group of mice with endometriosis (Fig. 1A). Consistent with our previous data, ERβ overexpression in the endometrium increased the volume of ectopic lesions in intact female mice with endometriosis compared with ectopic control lesions (Fig. 1B).
To evaluate ERβ function in the “eutopic” endometrium, endometriosis was surgically induced by implanting uterine fragments isolated from wild-type intact female C57BL/6J mice (N = 9 per group) into ERBOE female mice (N = 9) and female control mice (PRCre/+, N = 9). At the estrous cycle after 4 weeks of endometriosis induction, the eutopic endometrium was isolated from each group of mice with endometriosis (Fig. 1C).
Because ERα also has a role in endometriosis progression (6), we determined whether the ERβ overexpression alters ERα levels in endometriotic tissues. In both ectopic lesions and eutopic endometrium, however, ERβ overexpression did not change ERα levels (Fig. 1D). Therefore, the ERβ overexpression may not impact the ERα level in endometriotic tissues.
Activations of proliferation, epithelial-mesenchymal transition, hypoxia, inflammation, and lipid metabolism occur in ERβ-overexpressing ectopic lesions, stimulating their growth
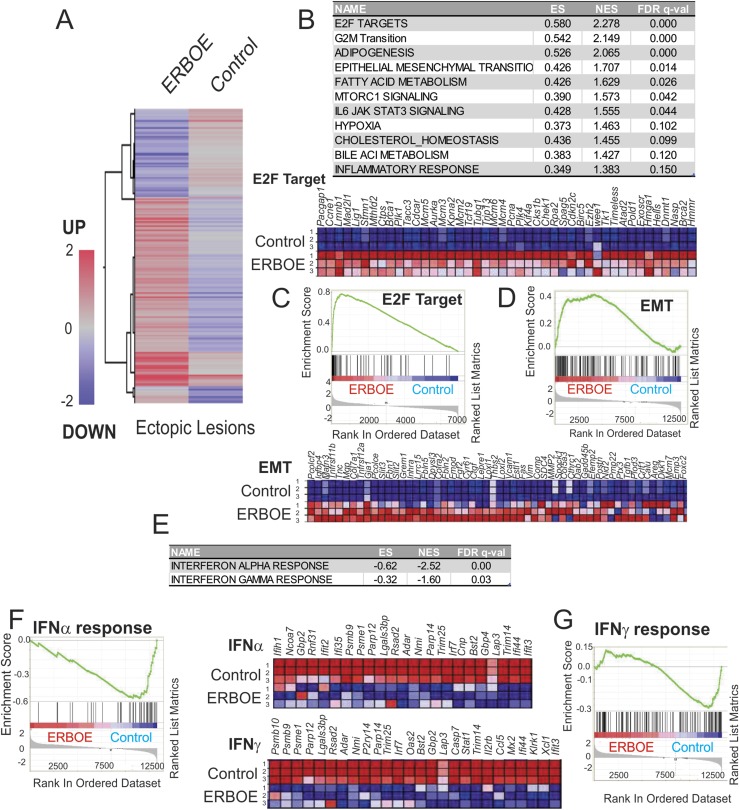
Using RNAs isolated from ERβ-overexpressing ectopic lesions and ectopic control lesions, we determined the gene expression profile in each group by using the Agilent mouse G3 v2 8×60K array (Fig. 2A) (19). To define the cellular pathways differentially regulated in ERβ-overexpressing ectopic lesions compared with ectopic control lesions, we conducted GSEA using the RNA expression profiles generated from ERβ-overexpressing ectopic lesions vs control ectopic lesions (Fig. 2B). The expression of genes regulated by E2 transcription factor (E2F) and mechanistic target of rapamycin (mTOR) complex 1 and genes associated with G2/M transition and IL-6/JAK/STAT3 signaling were significantly elevated in ERβ-overexpressing ectopic lesions compared with ectopic control lesions (Fig. 2B and 2C). Thus, the overexpression of ERβ in ectopic lesions should enhance their proliferation activity to stimulate endometriosis progression through activation of the above cellular pathways.
Figure 2.
Overexpression of ERβ changes cellular pathways in ectopic lesions to enhance endometriosis progression. (A) Heat map of differential gene expression profiles (>1.5-fold upregulation or downregulation) in ERβ-overexpressing ectopic lesions vs control ectopic lesions (average gene expression levels from nine samples per group). (B) Cellular pathways >1.5-fold upregulated in ERβ-overexpressing ectopic lesions compared with ectopic control lesions determined by GSEA. (C) GSEA analysis revealed that ERβ overexpression elevated genes signature associated with the E2F target genes in ectopic lesions. Relative expression of the E2F target gene set in control vs ERβ overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. (D) Genes involved in EMT signaling that is promoted in ERβ-overexpressing ectopic lesions compared with ectopic control lesions. Relative expressions of the EMT gene set in control vs ERβ overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. (E) Cellular pathways >1.5-fold downregulated in ERβ-overexpressing ectopic lesions compared with ectopic control lesions determined by GSEA. (F) GSEA analysis revealed that ERβ overexpression reduced gene signatures associated with the IFNα signaling in ectopic lesions. Relative expressions of the IFNα signaling gene set in control vs ERβ overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. (G) GSEA analysis revealed that ERβ overexpression reduced genes signature associated with the IFNγ signaling in ectopic lesions. Relative expressions of the IFNγ signaling gene set in control vs ERβ overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. EMT, epithelial-mesenchymal transition; ES, enrichment score; FDR, false discovery rate; NES, normalized enrichment score.
Additionally, gene expressions associated with the epithelial-mesenchymal transition (EMT), hypoxia, and the inflammatory response were significantly elevated in ERβ-overexpressing ectopic lesions compared with ectopic control lesions (Fig. 2B and 2D). Activation of these cellular pathways occurs in association with endometriosis progression (42, 43). Our observations imply that ERβ is a crucial regulator to coordinate for these cellular pathways to effectively enhance endometriosis progression.
We found that lipid metabolism was significantly elevated in ERβ-overexpressing ectopic lesions compared with ectopic control lesions through the activation of adipogenesis, fatty acid metabolism, cholesterol, and bile acid metabolism (Fig. 2B). Because altered lipid metabolism also has been associated with endometriosis progression (44), it appears that ERβ also has a critical role in this aspect of endometriosis progression.
IFN responses are downregulated in ERβ-overexpressing ectopic lesions
In addition to upregulated gene expression, a subset of genes was downregulated in ERβ-overexpressing ectopic lesions compared with ectopic control lesions (Fig. 2A). The IFNα and IFNγ response pathways were significantly downregulated in ERβ-overexpressing ectopic lesions compared with ectopic control lesions (Fig. 2E-2G). Activation of IFNα and IFNγ signaling pathways induces cell death in various cells (45, 46). Therefore, the overexpression of ERβ seemed able to reduce IFN-mediated cell death signaling in the endometrial fragments generated by retrograde menstruation in the peritoneal cavity to evade the immune surveillance system and promote their survival.
Proliferation, unfolded protein response, oxidative phosphorylation, and Wnt/β-catenin signaling are activated in the ERβ-overexpressing eutopic endometrium
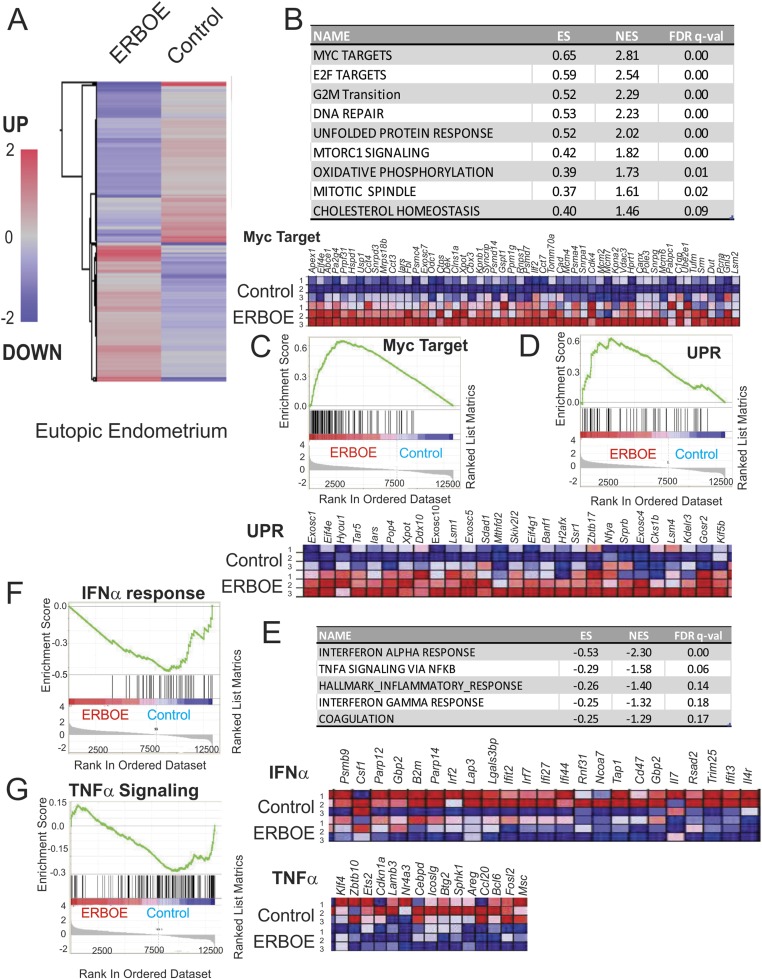
Using RNAs from the ERβ-overexpressing eutopic endometrium and eutopic control endometrium, we determined the gene expression profile in each group by using an Agilent mouse G3 v2 8×60K array (Fig. 3A) (19). The GSEA with these expression profiles revealed that Myc and E2F target genes and genes associated with mTOR complex 1 signaling and G2/M transition were elevated in the ERβ-overexpressing eutopic endometrium compared with the eutopic control endometrium (Fig. 3B and 3C). Compared with the normal endometrium, the eutopic endometrium revealed an enhanced proliferation genetic activity (47). Our results suggest that the overexpression of ERβ is the primary driver for the hyperproliferative activity of the endometrium of patients with endometriosis.
Figure 3.
Overexpression of ERβ changes cellular pathways in the eutopic endometrium to cause endometrial dysfunction. (A) Heat map of differential gene expression profiles (>1.5-fold upregulation or downregulation) in the ERβ-overexpressing eutopic endometrium vs control eutopic endometrium (average gene expression levels from nine samples per group). (B) Cellular pathways >1.5-fold upregulated in the ERβ-overexpressing eutopic endometrium compared with the eutopic control endometrium determined by GSEA. (C) List of Myc target genes elevated in the ERβ-overexpressing eutopic endometrium compared with the control eutopic endometrium. Relative expressions of the Myc target gene set in control vs ERβ overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. (D) Genes involved in UPR signaling that are upregulated in the ERβ-overexpressing eutopic endometrium compared with the eutopic control endometrium. Relative expressions of the UPR signaling gene set in control vs ERβ-overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. (E) Cellular pathways >1.5-fold downregulated in the ERβ-overexpressing eutopic endometrium compared with the eutopic control endometrium determined by GSEA. (F) GSEA analysis revealed that ERβ overexpression reduced gene signatures associated with the IFNα signaling in ectopic lesions. Relative expressions of the IFNα signaling gene set in control vs ERβ-overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. (G) GSEA analysis revealed that ERβ overexpression reduced gene signatures associated with TNFα/NF-κB signaling in ectopic lesions. Relative expressions of the TNFα/NF-κB signaling gene set in control vs ERβ overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. ES, enrichment score; FDR, false discovery rate; NES, normalized enrichment score; UPR, unfolded protein response.
Additionally, the overexpression of ERβ stimulated the unfolded protein response (UPR), oxidative phosphorylation, and WNT/β-catenin signaling in the eutopic endometrium (Fig. 3B and 3D). Alteration of the UPR, oxidative phosphorylation, and WNT/β-catenin signaling pathways causes endometrial dysfunction, reducing the fertility activity (48, 49). Therefore, overexpression of ERβ in the endometrium contributes to endometriosis-associated endometrial dysfunction by activating the UPR, oxidative phosphorylation, and WNT/β-catenin signaling in the eutopic endometrium.
IFN, nuclear factor κB, and inflammatory responses are downregulated in the ERβ-overexpressing eutopic endometrium
Inflammatory signaling was downregulated in the ERβ-overexpressing eutopic endometrium compared with the eutopic control endometrium by reducing gene expression linked to IFNα, IFNγ, and TNFα/nuclear factor κB (NF-κB) signaling (Fig. 3E-3G). Both IFNα and IFNγ signaling in the endometrium have essential roles in pregnancy, such as in embryo implantation (50, 51). During early pregnancy, NF-κB is activated via IκB kinase α-NF-κB-inducing kinase to regulate the gene expression profiles required for implantation and successful pregnancy (52). Overexpressing ERβ could further trigger endometrial dysfunction by downregulating inflammatory signaling, leading to infertility.
ERβ cistrome in ectopic lesions and the eutopic endometrium of mice with endometriosis
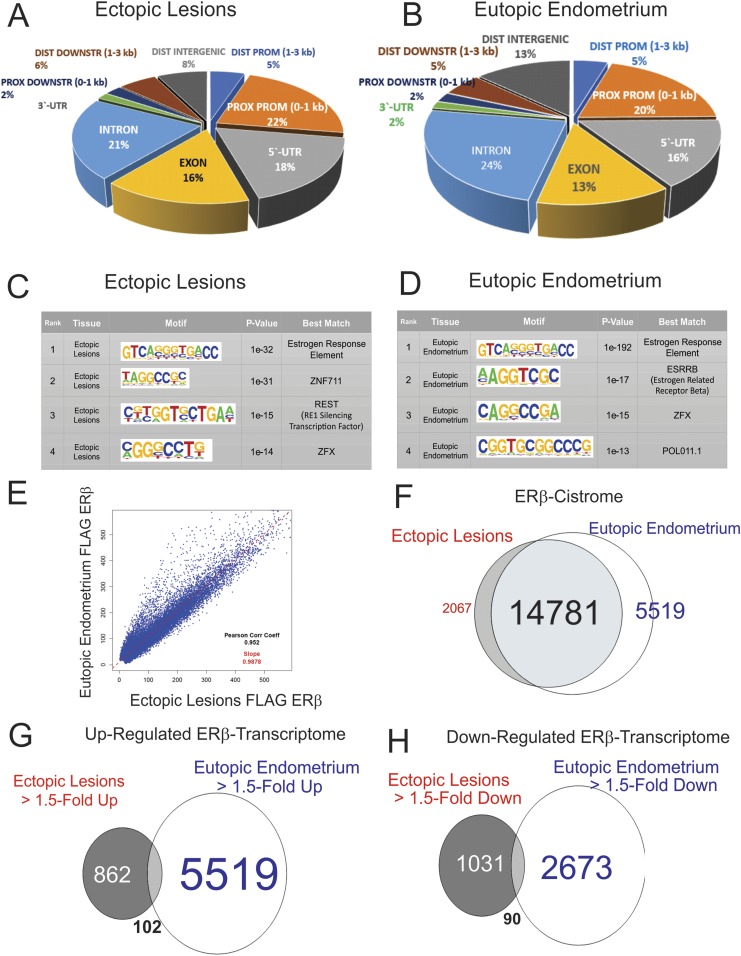
To define the ERβ cistrome in endometriotic tissues, we conducted FLAG ChIP-seq analysis with ectopic lesions and with eutopic endometrium isolated from ERBOE mice with endometriosis because FLAG-tagged ERβ was expressed in the ERBOE mouse (20). Compared with input data, ERβ binding sites were evenly distributed among intronic, exonic, 5′ untranslated region and promoter regions of target genes in both ectopic lesions and the eutopic endometrium of ERBOE mice with endometriosis (Fig. 4A and 4B). We next determined what transcription factor binding sites were enriched in DNA fragments coimmunoprecipitated with FLAG-ERβ from endometriotic tissues. The estrogen response element (ERE) was significantly enriched in DNA fragments coimmunoprecipitated by FLAG-ERβ from ectopic lesions (Fig. 4C). Therefore, the FLAG antibody successfully immunoprecipitated FLAG-ERβ bound to ERE in the genome from ectopic lesions. In addition to ERE, DNA fragments that had zinc finger protein (ZNF)711, RE1 silencing transcription factor (REST), and ZNF X-linked (ZFX) binding motifs were significantly coimmunoprecipitated by FLAG-tagged ERβ from ectopic lesions (Fig. 4C). Possibly, ERβ interacts with the ZNF, REST, and ZFX transcription factor family to corporately stimulate the endometriosis progression.
Figure 4.
ERβ cistrome in ectopic lesions and the eutopic endometrium of mice with endometriosis. (A and B) Distribution of ERβ-occupied sites in the genome of ectopic lesions (A) and the eutopic endometrium (B). (C and D) Determination of the sequence recognized by ERβ in ectopic lesions (C) and the eutopic endometrium (D). The MEME method (http://meme-suite.org) was used for identification of enriched sequences and is displayed with a correlation of the size of the character and the rate of enrichment. P value means the probability de novo-enriched sequences obtained from ChIP-seq– are matched to the displayed WebLogo and known consensus motifs by chance. (E) Peak correlation scatterplot for the tag of FLAG-tagged ERβ ectopic lesions against the eutopic endometrium. (F) Venn diagrams depicting the overlap of the tag of FLAG-ERβ within the upstream 5 kbp, transcription start site 1 kbp, 5′ untranslated region, and first intron between ectopic lesions and the eutopic endometrium. (G and H) Venn diagrams depicting the overlap of 1.50-fold-upregulated (G) or -downregulated (H) gene expression between ectopic lesions and the eutopic endometrium compared with their control. DIST DOWNSTR, distal downstream; DIST PROM, distal promoter; PROX DOWNSTR, proximal downstream; PROX PROM, proximal promoter; UTR, untranslated region; ZNF, zinc finger protein; ZFX, zinc finger protein x-linked.
In addition to ectopic lesions, DNA-binding motif analyses with DNA fragments coimmunoprecipitated with FLAG-ERβ from eutopic endometrium realized that DNA containing EREs also was significantly enriched in FLAG-ERβ immunoprecipitants from the ERβ-overexpressing eutopic endometrium (Fig. 4D). In addition to EREs, DNA fragments containing the binding motifs of estrogen-related receptor β, ZFX, and POL011.1 were significantly enriched in FLAG-ERβ immunoprecipitants from the eutopic endometrium (Fig. 4D). Therefore, ERβ functionally interacts with estrogen-related receptor, ZFX, and POL011.1 transcription factor family members to synergistically enhance endometriosis-associated endometrial dysregulation and infertility.
Based on these data, we compared ERβ binding sites between ectopic lesions and the eutopic endometrium; 87.7% (14,781 of 16,854) ERβ binding sites in ectopic lesions were detected in the eutopic endometrium, and 72.7% (14,782 of 20,300) ERβ binding sites in the eutopic endometrium also were identified in ectopic lesions (Fig. 4E and 4F). Thus, ectopic lesions and the eutopic endometrium of a mouse with endometriosis had substantial overlap in ERβ binding sites. Collectively, the ectopic lesion-specific ERβ cistrome had a similar pattern to the ERβ cistrome in the eutopic endometrium. In addition to the ERβ cistrome, we compared the upregulated and downregulated transcriptomes in ectopic lesions vs the eutopic endometrium (Fig. 4G and 4H). Interestingly, only a few upregulated or downregulated gene expression changes were shared in common by ectopic lesions and the eutopic endometrium. Although ERβ had similar binding sites in the genomes of ectopic lesions and the eutopic endometrium, ERβ differentially regulated the gene expression profiles in a context-specific manner.
Proliferation, EMT, reactive oxygen species, TNFα/NF-κB, and hypoxia signaling are upregulated by ERβ in ectopic lesions
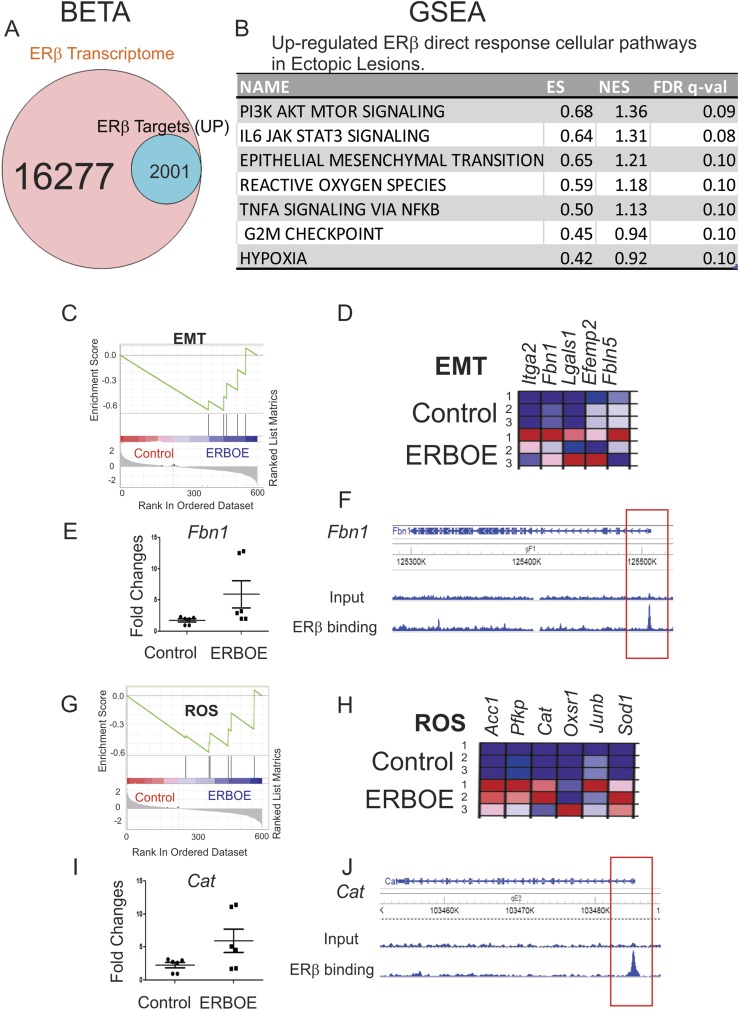
To define upregulated ERβ direct target genes that are statistically enriched near the ERβ binding sites in ectopic lesions, we conducted BETA with ERβ transcriptome that is differentially regulated in ERβ-overexpressing ectopic lesions compared with control ectopic lesions (P < 0.05) and the ERβ cistrome in ectopic lesions. BETA revealed that 12.3% (2001 of 16,277) of genes were upregulated by ERβ and statistically enriched near the ERβ binding sites (P < 0.05) in ectopic lesions (Fig. 5A). GSEA analyses with 2001 transcripts revealed that G2/M transition and IL-6/JAK/STAT3 signaling were ERβ direct responsive cellular pathways in ectopic lesions (Fig. 5B). Therefore, elevated proliferation activity in ectopic lesions was generated by ERβ. Additionally, the expression levels of genes involved in EMT signaling (Fig. 5C and 5D) were significantly elevated in ectopic lesions by ERβ overexpression. For example, the expression level of Fbn1 was elevated in ERβ-overexpressing ectopic lesions compared with wild-type ectopic lesions (Fig. 5E). FLAG ERβ ChIP-seq revealed that ERβ was recruited into the FbnI promoter region in ectopic lesions (Fig. 5F). Because Fbn1 is known as a critical regulator for the EMT (53), ERβ directly stimulates the EMT signaling in ectopic lesions. Also, the expression levels of genes involved in reactive oxygen species (ROS) were elevated in ERBOE ectopic lesions compared with ectopic control lesions (Fig. 5G and 5H). For example, ERβ overexpression promoted Cat level in ectopic lesions (Fig. 5I). ERβ was recruited to the promoter region of the Cat gene (Fig. 5J). Cat has an essential role in the modulation of ROS (54). In this manner, ERβ can directly regulate ROS signaling for the survival of ectopic lesions. The elevation of EMT, ROS, and hypoxia signaling can have a critical role in the initiation and progression of endometriosis (55-57). Our findings suggest that the estrogen/ERβ axis directly stimulates EMT, ROS, and hypoxia signaling in ectopic lesions to enhance their growth. Additionally, TNFα/NF-κB signaling directly responded to ERβ in ectopic lesions (Fig. 5B). TNFα signaling can induce apoptosis but also can enhance proliferation based on the type of stimulus (58). In endometriosis progression, cytoplasmic ERβ prevents TNFα-induced apoptosis in ectopic lesions to escape the immune surveillance system for their survival (7). In addition to cytoplasmic ERβ, nuclear ERβ in ectopic lesions also changes the properties of TNFα signaling from apoptosis to survival in ectopic lesions to enhance endometriosis progression.
Figure 5.
Cellular pathways directly upregulated to ERβ in ectopic lesions. (A) BETA with ERβ transcriptome and ERβ cistrome in ectopic lesions showed that 12.3% (2001 of 16,277) of genes were upregulated by ERβ and statistically enriched near the ERβ binding sites (P < 0.05) in ectopic lesions. (B) GSEA analysis with 2001 transcripts revealed specific cellular pathways upregulated by direct ERβ target genes in ERβ-overexpressing ectopic lesions. (C) GSEA analysis revealed that ERβ overexpression elevated genes signature associated with the EMT signaling in ectopic lesions. (D) Relative expressions of the EMT gene set in control vs ERβ-overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. (E) The expression of Fbn1 determined by RT-qPCR was elevated in ERBOE ectopic lesions compared with ectopic control lesions. The fold changes are shown relative to Fbn1 level in ectopic control lesions. (F) WashU Epigenome Browser image of ChIP-seq peak data revealed that ERβ bound to the promoter region of Fbn1 gene loci in ectopic lesions compared with the input signal. (G) GSEA analysis revealed that ERβ stimulated the gene signature associated with ROS in ectopic lesions. (H) Relative expressions of the ROS gene set in control vs ERβ overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. (I) Expression of Cat was elevated in ERBOE ectopic lesions compared with ectopic control lesions. The fold changes are shown relative to Cat level in ectopic control lesions. (J) WashU Epigenome Browser image of ChIP-seq peak data revealed that ERβ bound to the promoter region of Cat gene loci ectopic lesions compared with the input signal. ES, enrichment score; FDR, false discovery rate; NES, normalized enrichment score; ROS, reactive oxygen species.
IFNα signaling is downregulated by ERβ in ectopic lesions
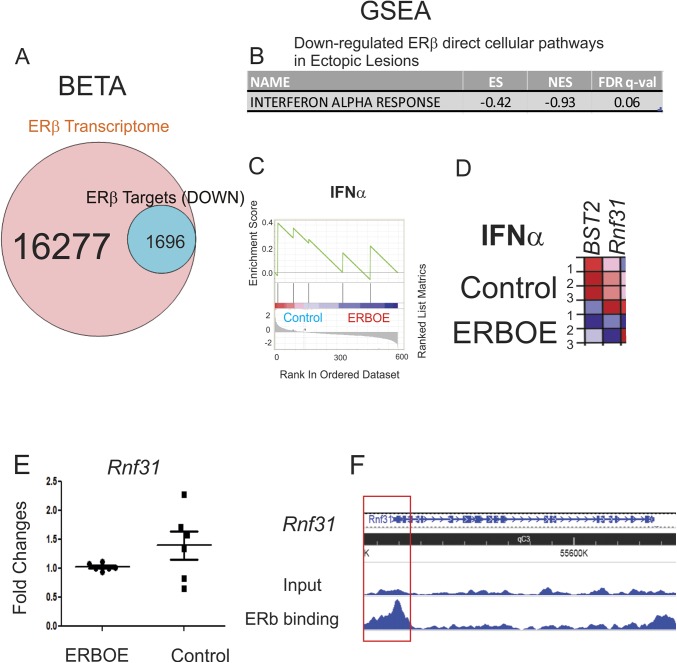
BETA analysis also revealed that 10.4% (1696 of 16,277) of genes were downregulated by ERβ and statistically enriched near the ERβ binding sites (P < 0.05) in ectopic lesions (Fig. 6A). The GSEA with 1696 transcripts showed that INFα signaling was downregulated in response to ERβ in ectopic lesions (Fig. 6B-6D). For example, the expression level of Rnf31 was significantly reduced in ERβ-overexpressing ectopic lesions compared with ectopic control lesions (Fig. 6E). FLAG-ERβ ChIP revealed that ERβ bound to the promoter region of the Rnf31 gene (Fig. 6F). Because RNF31 has a critical role in immune response (59), ERβ can directly regulate the INFα signaling in ectopic lesions. The downregulation of IFNα signaling in ectopic lesions is associated with endometriosis progression, and activation of IFNα signaling suppresses endometriosis progression (60). Therefore, ERβ directly suppressed IFNα signaling in ectopic lesions to promote their survival.
Figure 6.
Cellular pathways directly downregulated by ERβ in ectopic lesions. (A) BETA with ERβ transcriptome and ERβ cistrome in ectopic lesions showed that 10.4% (1696 of 16,277) of genes were downregulated by ERβ and statistically enriched near the ERβ binding sites (P < 0.05) in ectopic lesions. (B) GSEA analysis with 1696 transcripts revealed specific cellular pathways downregulated by direct ERβ target genes in ERβ-overexpressing ectopic lesions compared with ectopic control lesions. (C) GSEA analysis revealed that ERβ overexpression downregulated gene signatures associated with the IFNα response signaling in ectopic lesions. (D) Relative expressions of the IFNα response gene set in control vs ERβ-overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. (E) The expression Rnf31 determined by RT-qPCR was reduced in ERBOE ectopic lesions compared with ectopic control lesions. The fold changes are shown relative to Rnf31 level in ERBOE ectopic lesions. (F) WashU Epigenome Browser image of ChIP-seq peak data revealed that ERβ bound to the promoter region of Rnf31 gene loci in ectopic lesions compared with the input signal. ES, enrichment score; FDR, false discovery rate; NES, normalized enrichment score.
Proliferation, JAK/STAT3, and the UPR are upregulated by ERβ in eutopic endometrium
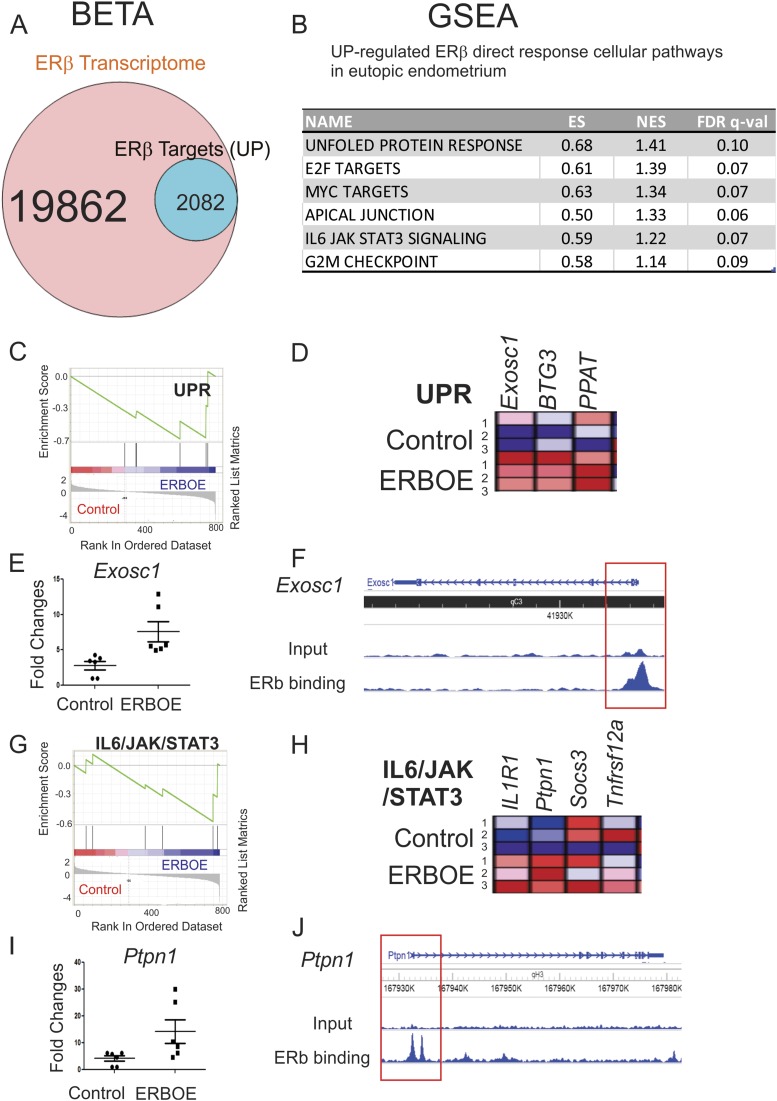
To define upregulated ERβ direct target genes that are statistically enriched near the ERβ binding sites in eutopic endometrium, we performed BETA with transcriptome that is differentially regulated in ERβ-overexpressing eutopic endometrium compared with control ectopic lesions (P < 0.05) and the ERβ cistrome in eutopic endometrium. BETA revealed that 10.5% (2082 of 19,862) of genes were upregulated by ERβ and statistically enriched near the ERβ binding sites (P < 0.05) in ectopic lesions (Fig. 7A). GSEA with 2018 transcripts revealed that genes associated with proliferation, such as G2/M transition and E2F and MYC target genes, were directly upregulated by ERβ in the eutopic endometrium (Fig. 7B). Therefore, ERβ stimulates the proliferation activity in both ectopic lesions and the eutopic endometrium during endometriosis progression.
Figure 7.
Cellular pathways directly upregulated by ERβ in the eutopic endometrium. (A) BETA with ERβ transcriptome and ERβ cistrome in eutopic endometrium showed that 10.5% (2082 of 19,862) of genes were upregulated by ERβ and statistically enriched near the ERβ binding sites (P < 0.05) in eutopic endometrium. (B) GSEA with 2082 transcripts predicted cellular pathways upregulated >1.5-fold by direct ERβ target genes in the eutopic endometrium. (C) GSEA analysis revealed that ERβ overexpression elevated genes signature associated with the UPR signaling in eutopic endometrium. (D) Relative expressions of the UPR gene set in control vs ERβ-overexpressing eutopic endometrium (N = 3) are shown in order of their signal/noise ratio rank. (E) The expression of Exosc1 determined by RT-qPCR was elevated in ERBOE eutopic endometrium compared with control eutopic endometrium. The fold changes are shown relative to Exosc1 level in control eutopic endometrium. (F) WashU Epigenome Browser image of ChIP-seq peak data revealed that ERβ bound to the promoter region of Exosc1 gene loci in eutopic endometrium compared with the input signal. (G) GSEA analysis revealed that ERβ overexpression elevated gene signatures associated with the IL-6/JAK/STAT3 signaling in eutopic endometrium. (H) Relative expressions of the IL-6/JAK/STAT3 gene set in control vs ERβ-overexpressing eutopic endometrium (N = 3) are shown in order of their signal/noise ratio rank. (I) The expression of Ptpn1 determined by RT-qPCR was elevated in ERBOE eutopic endometrium compared with control eutopic endometrium. The fold changes are shown relative to Ptpn1 level in control eutopic endometrium. (J) WashU Epigenome Browser image of ChIP-seq peak data revealed that ERβ bound to the promoter region of Ptpn1 gene loci in eutopic endometrium compared with the input signal. ES, enrichment score; FDR, false discovery rate; NES, normalized enrichment score.
Additionally, GSEA analysis revealed that ERβ instantly elevated the expression of genes involved in the UPR in eutopic endometrium (Fig. 7C and 7D). For example, the expression level of Exosc1 was significantly elevated in ERβ-overexpressing ectopic lesions compared with ectopic control lesions (Fig. 7E). FLAG-ERβ ChIP analysis revealed that ERβ bound to the promoter region of Exosc1 in eutopic endometrium (Fig. 7F). Therefore, ERβ directly can stimulate Exosc1 levels to enhance the UPR signaling in ectopic lesions for their survival. In addition to UPR-associated genes, expression of inhibitors for IL-6/JAK/STAT3 signaling were elevated in the eutopic endometrium by ERβ (Fig. 7G and 7H). For example, Ptpn1 level was elevated in eutopic endometrium (Fig. 7I). Additionally, ERβ was deposited onto the promoter region of Ptpn1 gene in eutopic endometrium (Fig. 7J). Ptpn1 has an inhibitory effect on JAK/STAT signaling (61). Therefore, ERβ can directly suppress IL-6/JAK/STAT3 signaling in eutopic endometrium.
Elevation of the UPR in the endometrium increases endoplasmic reticulum stress and prevents embryo implantation (48, 62). The inhibition of IL-6/JAK/STA3 signaling in the endometrium also causes embryo implantation defects (63, 64). Collectively, endometriosis-associated endometrial dysfunction is likely enhanced by ERβ through activation of UPR-mediated endoplasmic reticulum stress and IL-6/JAK/STA3 inhibitory signaling in the eutopic endometrium.
TNFα/NF-κB is downregulated by ERβ in eutopic endometrium
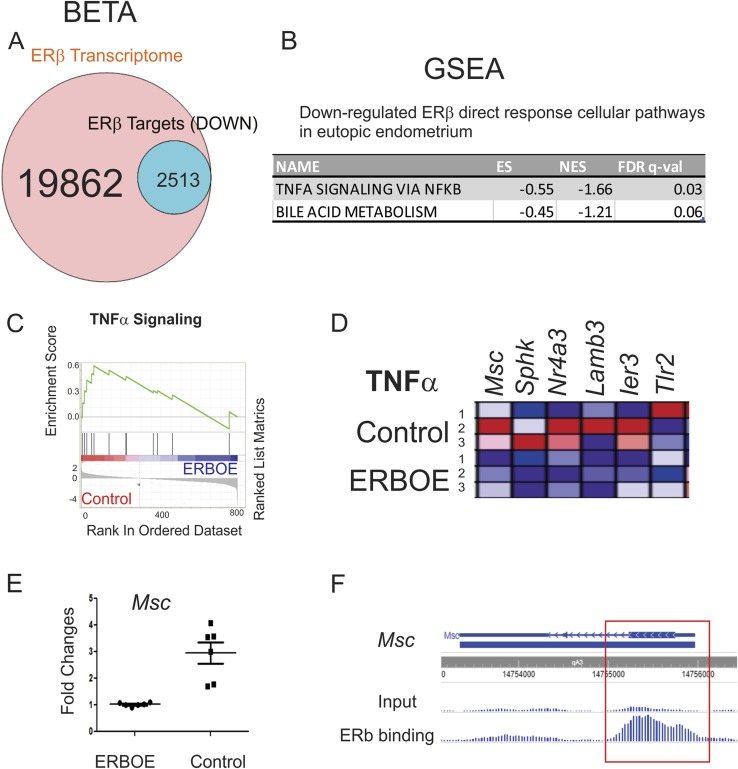
BETA analysis also revealed that 12.7% (2513 of 16,277) of genes were downregulated by ERβ and statistically enriched near the ERβ binding sites (P < 0.05) in eutopic endometrium (Fig. 8A). GSEA with 2513 transcripts revealed that the TNFα/NF-κB signaling and bile acid metabolism were directly downregulated by ERβ in the eutopic endometrium (Fig. 8B-8D). For example, the expression level of Msc was significantly reduced in ERβ-overexpressing eutopic endometrium compared with control eutopic endometrium (Fig. 8E). FLAG-ERβ ChIP revealed that ERβ bound to the Msc gene locus in eutopic endometrium (Fig. 8F). Therefore, the ERβ directly suppresses Msc level in eutopic endometrium to downregulate TNFα/NF-κB signaling in eutopic endometrium. As described above, activation of NF-κB in early pregnancy modulates the gene expression required for implantation and successful pregnancy (52). Therefore, ERβ-mediated downregulation of TNFα/NF-κB signaling in the eutopic endometrium seems to be a driver of endometriosis-associated infertility by impairing embryo implantation in early pregnancy.
Figure 8.
Cellular pathways directly downregulated by ERβ in eutopic endometrium. (A) BETA with ERβ transcriptome and ERβ cistrome in eutopic endometrium showed that 12.7% (2513 of 19,862) of genes were downregulated by ERβ and statistically enriched near the ERβ binding sites (P < 0.05) in eutopic endometrium. (B) GSEA with 2513 transcripts revealed specific cellular pathways downregulated in ERβ-overexpressing eutopic endometrium. (C) GSEA analysis revealed that ERβ overexpression downregulated gene signatures associated with the TNFα signaling in ectopic lesions. (D) Relative expressions of the TNFα signaling gene set in control vs ERβ overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. (E) The expression of Msc determined by RT-qPCR was reduced in ERBOE eutopic endometrium compared with control eutopic endometrium. The fold changes are shown relative to Msc level in ERBOE eutopic endometrium. (F) WashU Epigenome Browser image of ChIP-seq peak data revealed that ERβ bound to the Msc gene loci in eutopic endometrium compared with the input signal. ES, enrichment score; FDR, false discovery rate; NES, normalized enrichment score.
Stimulation of an EMT gene signature by overexpression of ERβ in IHEECs
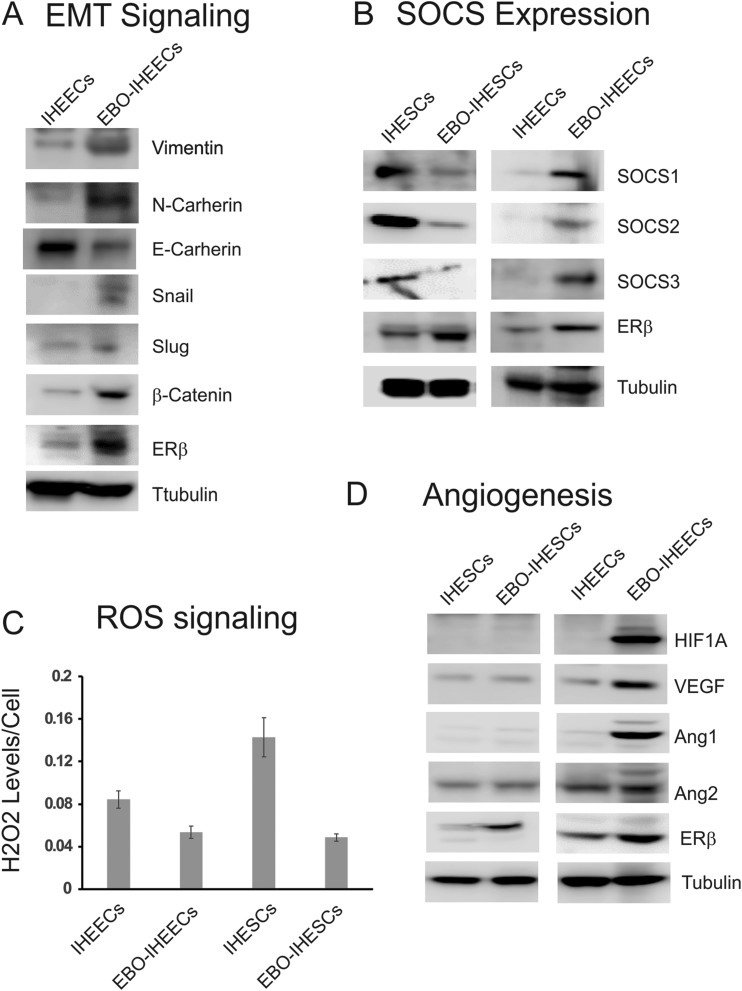
Our omics data revealed that ERβ directly enhances EMT signaling to increase the growth of ectopic lesions. To further validate this observation, we generated recombinant EBO-IHEECs by using a lentiviral expression vector containing FLAG-tagged human ERβ cDNA. ERβ was highly elevated in EBO-IHEECs compared with parental IHEECs (Fig. 9A). We then compared the expression of EMT marker genes between parental IHEECs and EBO-IHEECS by using Western blot analysis. The EBO-IHEECs had elevated levels of marker genes for mesenchymal cells, such as vimentin, N-cadherin, Snail, Slug, and β-catenin compared with parental IHEECs (Fig. 9A). However, EBO-IHEECs had reduced expression of epithelial marker gene expression, such as E-cadherin, compared with the control IHEECs (Fig. 9A). Therefore, ERβ overexpression enhanced the EMT signaling in IHEECs. Our study revealed that the ERβ overexpression stimulated cell adhesion and invasion activity of IHEECs compared with parental IHEECs (7). Therefore, the overexpression of ERβ enhances EMT signaling in endometrial epithelial cells to enhance the development of ectopic lesions in endometriosis.
Figure 9.
Overexpression of ERβ dysregulates EMT, JAK/STAT3, ROS, and angiogenesis signaling in human endometrial cells. (A) Expression profile of genes involved in EMT signaling in IHEECs vs EBO-IHEECS determined by Western blot. (B-D) Expression profiles of SOCSs (B), levels of H2O2 (C), and proangiogenic factors (D) in IHESCs, EBO-IHESCS, IHEECs, and EBO-IHEECs determined by Western blot or luciferase activity. VEGF, vascular endothelial growth factor.
Differential JAK-STAT inhibitor expression in human endometrial cells by overexpression of ERβ
Our omics data revealed that IL-6/JAK/STAT3 signaling is a target of ERβ in both ectopic lesions and the eutopic endometrium. However, the regulation pattern of IL-6/JAK/STAT3 signaling is different between them. For example, IL-6/JAK/STAT3 signaling is activated by ERβ in the ectopic lesions, but ERβ reduces IL-6/JAK/STAT3 signaling in the eutopic endometrium by enhancing IL-6/JAK/STAT3 inhibitor expression. To validate this observation, we generated recombinant EBO-IHESCs that stably expressed FLAG-tagged human ERβ. ERβ was highly elevated in EBO-IHESCs compared with parental IHESCs (Fig. 9B). Because ERβ directly elevated the expression of SOCS1 in the eutopic endometrium, we determined the expression of JAK/STAT inhibitors (such as SOCS1, SOCS2, and SOCS3) in IHESCs, EBO-IHESCs, IHEECs, and EBO-IHEECs by Western blot analysis. In human endometrial stromal cells, the ERβ overexpression significantly reduced all JAK/STAT inhibitors compared with the parental IHESCs (Fig. 9B). Therefore, this observation implies that the overexpression of ERβ in stromal cells of the uterus activates JAK/STAT signaling. In contrast with endometrial stromal cells, however, the overexpression of ERβ elevated JAK/STAT inhibitors in IHEECs compared with the control of IHEECs (Fig. 9B). Thus, the overexpression of ERβ might inhibit the JAK/STAT3 signaling in epithelial cells of the uterus. Collectively, these data show that IL-6/JAK/STAT3 signaling is regulated by ERβ in an endometrial cell type-dependent manner.
Reduction of H2O2 in human endometrial cells by overexpression of ERβ
Our omics data also revealed that ERβ directly upregulated ROS detoxification enzymes in ectopic lesions to promote survival under high levels of fatal ROS during endometriosis progression. To validate this observation, we determined the concentration of H2O2 in both human endometrial epithelial and stromal cells because the level of H2O2 negatively correlated with the levels of ROS detoxification enzymes (65). The overexpression of ERβ reduced H2O2 levels in both endometrial stromal and epithelial cells compared with control cells (Fig. 9C). Therefore, ERβ could have a critical role in the survival of shedding endometrial fragments in a high-ROS environment generated by iron overload during retrograde menstruation.
Activation of proangiogenic factors in human endometrial cells by overexpression of ERβ
Our omics data also revealed that ERβ directly upregulated hypoxia-induced angiogenesis signaling in ectopic lesions to promote survival. To validate this observation, we compared the expression of HIF1A between ERβ-overexpressing human endometrial cells and parental human endometrial cells by using Western blot analyses because HIF1A has a critical role in the regulation of angiogenesis by hypoxia (66). The ERβ overexpression elevated HIF1A levels in IHEECs, but not in IHESCs (Fig. 9D). In addition to HIF1A, the ERβ overexpression also raised levels of proangiogenic factors, such as vascular endothelial growth factor and ANG1, in IHEECs (67) (Fig. 9D). However, ANG2 expression was not elevated by ERβ overexpression in IHEECs. In contrast to epithelial cells, however, levels of proangiogenic factors were not raised in IHESCs by ERβ overexpression (Fig. 9D). Therefore, the overexpression of ERβ in human endometrial epithelial cells, but not in stromal cells, enhances angiogenesis signaling to improve the development of ectopic lesions in endometriosis.
Discussion
Genomic ERβ function in ectopic lesions
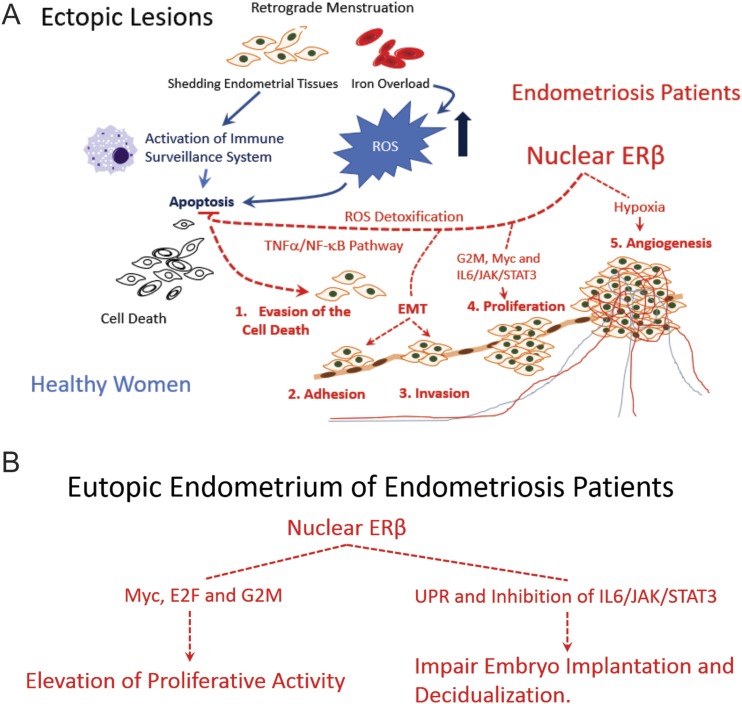
Retrograde menstruation is the primary hypothesis that can explain the etiology of endometriosis. The shedding of endometrial fragments triggers the host immune surveillance system in the peritoneal cavity. Because menstrual blood contains high levels of iron, the retrograde menstruation also overloads iron into peritoneal fluid and elevates ROS signaling. In healthy women, their host immune surveillance system and high levels of fatal ROS signaling clean the shedding endometrial tissues by activating apoptosis signaling (Fig. 10A, blue line). In patients with endometriosis, however, shedding of endometrial tissues effectively escapes this immune surveillance system in part because cytoplasmic ERβ prevents the apoptosis signaling in shedding endometrial tissues (7). In addition to cytoplasmic ERβ, we expected that nuclear ERβ also might play an essential role in the evasion of immune surveillance because TNFα/NF-κB signaling is a direct target of nuclear ERβ in ectopic lesions. NF-κB signaling is a crucial survival factor, and constitutive activation of NF-κB has been shown in endometriotic lesions (68, 69). Therefore, we found that cytoplasmic and nuclear ERβ synergistically enhance the cell survival signaling in shedding endometrial tissues to escape the immune surveillance system and sustain ectopic lesions (Fig. 10A, red line).
Figure 10.
Model for ERβ direct response cellular pathways in ectopic lesions and eutopic endometrium. (A) Model for direct ERβ-responsive pathways in ectopic lesions driving the progression of ectopic lesions. Nuclear ERβ directly activates TNFα/NF-κB and the ROS detoxification system in shedding endometrium fragments to evade the host immune surveillance system and then enhances EMT, proliferation, and hypoxia signaling to promote endometriosis progression. (B) Model for directly ERβ-responsive pathways in the eutopic endometrium. Nuclear ERβ directly activates proliferation, UPR, and IL-6/JAK/STAT3 inhibitory signaling in the eutopic endometrium, driving endometrial dysfunction.
Iron overload has been detected in the cells and peritoneal fluid of women with endometriosis compared with normal endometrial tissues, and excessive iron can induce deleterious ROS in the peritoneal environment (70, 71). In healthy women, high levels of deleterious ROS signaling enhances elimination of endometrial fragments from the pelvic area by activating apoptosis (Fig. 10A, blue line) (72). In patients with endometriosis, however, ERβ-overexpressing endometrial fragments can escape this fatal ROS signaling due to the activation of the ROS detoxification system (such as Abcc1, Pfkp, Jnb, Cat, Oxsr1, and Sod1) (Fig. 10A, red line). After surviving, however, a high level of ROS can lead to stimulation of the ERK and PI3K/AKT/mTOR signaling pathways in ectopic lesions, thus promoting adhesion, angiogenesis, and proliferation of endometriotic lesions and subsequent endometriosis progression (73). In the early stage of endometriosis progression, therefore, nuclear ERβ stimulates a gene network of shedding endometrial tissues to escape cell death signaling generated from the immune surveillance system and deleterious ROS signaling in peritoneal fluid.
The activation of EMT signaling has an essential role in endometriosis progression because activation of EMT has a critical role in the invasion of ectopic lesions into the mesothelial cell layer (74). The overexpression of thrombin-activated fibrinolytic inhibitor, enhancer of Zeste homolog 2, lipoxin A4, and hypoxia-inducible factor 1 are involved in activation of EMT in endometriosis progression (75-78). Our data reveal that overexpression of ERβ directly upregulates cytoskeleton components (such as Itga2, Fbn1, Efemp2, and Fbln5) in ectopic lesions. The cytoskeleton network has an essential role in the EMT process, and genes associated with endometriosis are direct regulators of the actin cytoskeleton, which coordinates mesothelial barrier integrity (79, 80). Thus, the dynamics of the cytoskeleton by ERβ is the new aspect to explain the estrogen-mediated activation of EMT in endometriotic lesions.
Hypoxia signaling is activated in patients with endometriosis and then induces the expression of many critical downstream genes to regulate the survival and maintenance of ectopic lesions (81). For example, the activated hypoxia signaling might have an essential role in the angiogenesis process for the establishment of ectopic lesions because hypoxia signaling is a driver for angiogenesis (82). Therefore, the hypoxia could be a critical concept delineating the etiology of endometriosis and designing new therapeutic strategies for endometriosis. In this study, we revealed that ERβ directly regulated HIF1A target genes (such as Ctsf, Wsb1, Sdc45, Pdk3, Klf4, and Pgm1) in ectopic lesions to induce hypoxia-induced angiogenesis signaling. The ERβ/HIF1A axis should have an essential role in merging hypoxia and estrogen signaling in ectopic lesions to promote the endometriosis progression via activation of angiogenesis.
Genomic ERβ function in eutopic endometrium
Infertility is one of the symptoms associated with endometriosis progression. The etiology of endometriotic survival and infertility are notably complex. Several hypotheses have been generated to explain how endometriosis leads to infertility (83). For example, dysregulation of the gene expression of Wnt7 and Hoxa10 and progesterone resistance in the eutopic endometrium can derive endometrial dysfunction, causing infertility (84-86). Our previous study revealed that ERBOE mice are infertile because of defective decidualization (7). Overexpressing ERβ stimulated the proliferation activity of the eutopic endometrium by activating G2/M transition and Myc and E2F target genes. The proliferative activity in the endometrium of women with endometriosis is higher than that in the endometrium of women without endometriosis (87) and likely could be caused by the overexpression of ERβ.
In addition to proliferation, Myc and E2F also induce the apoptosis signaling, and the Myc- and E2F-mediated apoptosis is dependent on the specific cell type and physiological status of the cells (88, 89). For the survival of endometriotic lesions, antiapoptosis signaling is elevated in endometriotic lesions compared with normal endometrium (90). In the endometriotic tissues, therefore, Myc and E2F stimulate the proliferation rather than apoptosis to stimulate endometriosis progression.
Other molecular properties associated with the eutopic endometrium of women with endometriosis are defects in embryo implantation and decidualization (64). We revealed that overexpressing ERβ significantly downregulates IL-6/JAK/STAT3 signaling by directly increasing the expression of JAK/STAT3 inhibitors (such as Socs3 and Ptpn1) in the eutopic endometrium of mice with endometriosis. Because IL-6/JAK/STAT3 signaling has an essential role in embryo implantation and decidualization of endometrial stromal cells (64), ERβ-mediated inhibition of IL-6/JAK/STAT3 signaling in the endometrium could induce embryo implantation defects and impair decidualization, causing endometriosis-associated infertility.
During endometriosis progression, the UPR is elevated in the endometrium of mice with endometriosis by activated hypoxia signaling (91). We revealed that the estrogen/ERβ axis has a critical role in the activation of UPR in the eutopic endometrium. Interestingly, activation of the UPR prevents blastocyst formation during preimplantation embryo development in vitro (92). Therefore, hyperactivation of the UPR by ERβ in the eutopic endometrium could promote endometriosis-associated infertility by antagonizing blastocyst formation through activation of the UPR.
In contrast to ectopic lesions, NF-κB signaling was significantly reduced in the eutopic endometrium by ERβ. The activation of NF-κB has an essential role in pregnancy because NF-κB signaling is activated in the implantation window of the mouse uterus (93). Downregulation of NF-kB signaling by ERβ also can contribute to endometriosis-associated infertility in women by impairing embryo implantation.
The goal of our study was to uncover comprehensively the direct ERβ response cellular pathways in ectopic lesions and the eutopic endometrium that accompany endometriosis progression and endometriosis-associated endometrial dysfunction. The ectopic lesions are generated from uterine endometrium. However, the ERβ-regulated cellular pathways in ectopic lesions are quite different from those in eutopic endometrium. Ectopic lesions consist of endometrial stromal cells, and glandular epithelial cells originated from the uterus (94). In normal uterine endometrium, endometrial stromal and glandular epithelial cells are not exposed to the host immune system. However, endometrial stromal and glandular epithelial cells in shedded endometrium generated by retrograde menstruation are exposed to the host immune system and then actively interact with immune cells in the peritoneal cavity for their survival. Therefore, ERβ-regulated cellular pathways in ectopic lesions are different from those in eutopic endometrium.
Our new findings should help us to understand better the complex molecular etiology of endometriosis induced by the estrogen/ERβ axis. ERβ and ERβ direct-response cellular pathways may serve as new molecular therapeutic targets to effectively suppress the growth of ectopic lesions and elevate the fecundity of women with endometriosis.
Acknowledgments
We thank the Active Motif company for conducting and analyzing FLAG ERβ ChIP-seq with ectopic lesions and eutopic endometrium of mice with endometriosis. We also thank Dr. Lisa D. White at Baylor College of Medicine for conducting microarray analyses.
Financial Support: This work was supported by National Institutes of Health/Eunice Kennedy Shriver National Institute of Child Health and Human Development Grants R01HD082786 and R01HD008188 (to B.W.O.) and a pilot grant (to S.J.H.); grants from the Brockman Medical Research Foundation and the Mike Hogg Foundation (to S.J.H.); the Genomic and RNA Profiling Core at Baylor College of Medicine with funding from the National Institutes of Health/National Cancer Institute Grant P30CA125123; and National Research Foundation of Korea Grant NRF-2018R1D1A1A02085827 (to Y.J.C.).
Author Contributions: S.J.H. led the entire project. S.J.H. and B.W.O. contributed to planning and evaluated all data. S.J.H., J.E.L, Y.J.C., and M.J.P. designed and performed the experiments. S.J.H. and B.W.O. wrote the manuscript.
Additional Information
Disclosure Summary: The authors have nothing to disclose.
Data Availability: The datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
Glossary
Abbreviations:
- Ang
angiopoietin
- BETA
binding and expression target analysis
- Cat
catalase
- ChIP
chromatin immunoprecipitation
- ChIP-seq
chromatin immunoprecipitation sequencing
- E2F
E2 transcription factor
- EBO-IHEEC
estrogen receptor β-overexpressing immortalized human endometrial epithelial cell
- EBO-IHESC
estrogen receptor β-overexpressing immortalized human endometrial stromal cell
- EMT
epithelial-mesenchymal transition
- ER
estrogen receptor
- ERBOE
estrogen receptor β overexpression
- ERE
estrogen response element
- Exosc1
exosome component 1
- Fbn1
fibrillin 1
- GSEA
gene set enrichment analysis
- HIF
hypoxia inducible factor
- IHESC
immortalized human endometrial stromal cell
- JAK
Janus kinase
- Msc
musculin
- mTOR
mechanistic target of rapamycin
- NF-κB
nuclear factor κB
- PR
progesterone receptor
- Ptpn1
protein tyrosine phosphatase nonreceptor type 1
- qPCR
quantitative PCR
- Rnf31
ring finger protein 31
- ROS
reactive oxygen species
- SOCS
suppressor of cytokine signaling
- STAT
signal transducer and activator of transcription
- UPR
unfolded protein response
- ZFX
zinc finger protein X-linked
- ZNF
zinc finger protein
References and Notes
- 1. Bulun SE. Endometriosis. N Engl J Med. 2009;360(3):268–279. [DOI] [PubMed] [Google Scholar]
- 2. Karaman K, Pala EE, Bayol U, Akman O, Olmez M, Unluoglu S, Ozturk S. Endometriosis of the terminal ileum: a diagnostic dilemma. Case Rep Pathol. 2012;2012:742035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Practice Committee of the American Society for Reproductive Medicine. Endometriosis and infertility: a committee opinion. Fertil Steril. 2012;98(3):591–598. [DOI] [PubMed] [Google Scholar]
- 4. Tsuji I, Ami K, Miyazaki A, Hujinami N, Hoshiai H. Benefit of diagnostic laparoscopy for patients with unexplained infertility and normal hysterosalpingography findings. Tohoku J Exp Med. 2009;219(1):39–42. [DOI] [PubMed] [Google Scholar]
- 5. Nakagawa K, Ohgi S, Horikawa T, Kojima R, Ito M, Saito H. Laparoscopy should be strongly considered for women with unexplained infertility. J Obstet Gynaecol Res. 2007;33(5):665–670. [DOI] [PubMed] [Google Scholar]
- 6. Burns KA, Rodriguez KF, Hewitt SC, Janardhan KS, Young SL, Korach KS. Role of estrogen receptor signaling required for endometriosis-like lesion establishment in a mouse model. Endocrinology. 2012;153(8):3960–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Han SJ, Jung SY, Wu SP, Hawkins SM, Park MJ, Kyo S, Qin J, Lydon JP, Tsai SY, Tsai MJ, DeMayo FJ, O’Malley BW. Estrogen receptor β modulates apoptosis complexes and the inflammasome to drive the pathogenesis of endometriosis. Cell. 2015;163(4):960–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao Y, Gong P, Chen Y, Nwachukwu JC, Srinivasan S, Ko C, Bagchi MK, Taylor RN, Korach KS, Nettles KW, Katzenellenbogen JA, Katzenellenbogen BS. Dual suppression of estrogenic and inflammatory activities for targeting of endometriosis. Sci Transl Med. 2015;7(271):271ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matsuzaki S, Uehara S, Murakami T, Fujiwara J, Funato T, Okamura K. Quantitative analysis of estrogen receptor alpha and beta messenger ribonucleic acid levels in normal endometrium and ovarian endometriotic cysts using a real-time reverse transcription-polymerase chain reaction assay. Fertil Steril. 2000;74(4):753–759. [DOI] [PubMed] [Google Scholar]
- 10. Juhasz-Böss I, Fischer C, Lattrich C, Skrzypczak M, Malik E, Ortmann O, Treeck O. Endometrial expression of estrogen receptor β and its splice variants in patients with and without endometriosis. Arch Gynecol Obstet. 2011;284(4):885–891. [DOI] [PubMed] [Google Scholar]
- 11. Trukhacheva E, Lin Z, Reierstad S, Cheng YH, Milad M, Bulun SE. Estrogen receptor (ER) β regulates ERα expression in stromal cells derived from ovarian endometriosis. J Clin Endocrinol Metab. 2009;94(2):615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fazleabas AT, Brudney A, Chai D, Langoi D, Bulun SE. Steroid receptor and aromatase expression in baboon endometriotic lesions. Fertil Steril. 2003;80(Suppl 2):820–827. [DOI] [PubMed] [Google Scholar]
- 13. Fujimoto J, Hirose R, Sakaguchi H, Tamaya T. Expression of oestrogen receptor-α and -β in ovarian endometriomata. Mol Hum Reprod. 1999;5(8):742–747. [DOI] [PubMed] [Google Scholar]
- 14. Nelson AW, Groen AJ, Miller JL, Warren AY, Holmes KA, Tarulli GA, Tilley WD, Katzenellenbogen BS, Hawse JR, Gnanapragasam VJ, Carroll JS. Comprehensive assessment of estrogen receptor beta antibodies in cancer cell line models and tissue reveals critical limitations in reagent specificity [published correction appears in Mol Cell Endocrinol.2017;443:175]. Mol Cell Endocrinol. 2017;440:138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Soyal SM, Mukherjee A, Lee KY, Li J, Li H, DeMayo FJ, Lydon JP. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis. 2005;41(2):58–66. [DOI] [PubMed] [Google Scholar]
- 16. Cummings AM, Metcalf JL. Induction of endometriosis in mice: a new model sensitive to estrogen. Reprod Toxicol. 1995;9(3):233–238. [DOI] [PubMed] [Google Scholar]
- 17. Byers SL, Wiles MV, Dunn SL, Taft RA. Mouse estrous cycle identification tool and images. PLoS One. 2012;7(4):e35538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Euhus DM, Hudd C, LaRegina MC, Johnson FE. Tumor measurement in the nude mouse. J Surg Oncol. 1986;31(4):229–234. [DOI] [PubMed] [Google Scholar]
- 19. Sang H. Data from: ER beta regulated transcriptome in endometriotic tissues. Gene Expression Omnibus 2019. Deposited 3 May 2018. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114010.
- 20. Sang H. Data from: Estrogen receptor beta cistrome in mouse endometriotic tissue. Gene Expression Omnibus 2019. Deposited 4 May 2018. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114047.
- 21. Wang S, Sun H, Ma J, Zang C, Wang C, Wang J, Tang Q, Meyer CA, Zhang Y, Liu XS. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc. 2013;8(12):2502–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bono Y, Kyo S, Takakura M, Maida Y, Mizumoto Y, Nakamura M, Nomura K, Kiyono T, Inoue M. Creation of immortalised epithelial cells from ovarian endometrioma. Br J Cancer. 2012;106(6):1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krikun G, Mor G, Alvero A, Guller S, Schatz F, Sapi E, Rahman M, Caze R, Qumsiyeh M, Lockwood CJ. A novel immortalized human endometrial stromal cell line with normal progestational response. Endocrinology. 2004;145(5):2291–2296. [DOI] [PubMed] [Google Scholar]
- 24. Feoktistova M, Geserick P, Leverkus M.. Crystal violet assay for determining viability of cultured cells. Cold Spring Harb Protoc. 2016(4):pdb.prot087379. [DOI] [PubMed] [Google Scholar]
- 25. RRID:AB_2178887, https://scicrunch.org/resolver/AB_2178887.
- 26. RRID:AB_2687616, https://scicrunch.org/resolver/AB_2687616.
- 27. RRID:AB_2291471, https://scicrunch.org/resolver/AB_2291471.
- 28. RRID:AB_2255011, https://scicrunch.org/resolver/AB_2255011.
- 29. RRID: AB_2239535, https://scicrunch.org/resolver/AB_2239535.
- 30. RRID:AB_11127855, https://scicrunch.org/resolver/AB_11127855.
- 31. RRID:AB_631469, https://scicrunch.org/resolver/AB_631469.
- 32. RRID:AB_1593623, https://scicrunch.org/resolver/AB_1593623.
- 33. RRID:AB_2241191, https://scicrunch.org/resolver/AB_2241191.
- 34. RRID:AB_1861766, https://scicrunch.org/resolver/AB_1861766.
- 35. RRID:AB_304007, https://scicrunch.org/resolver/AB_304007.
- 36. RRID:AB_304008, https://scicrunch.org/resolver/AB_304008.
- 37. RRID:AB_2212644, https://scicrunch.org/resolver/AB_2212644.
- 38. RRID:AB_2116958, https://scicrunch.org/resolver/AB_2116958.
- 39. RRID:AB_2772625, https://scicrunch.org/resolver/AB_2772625.
- 40. RRID:AB_2226215, https://scicrunch.org/resolver/AB_2226215.
- 41. Pelch KE, Sharpe-Timms KL, Nagel SC. Mouse model of surgically-induced endometriosis by auto-transplantation of uterine tissue. J Vis Exp. 2012; (59):e3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cho YJ, Lee SH, Park JW, Han M, Park MJ, Han SJ. Dysfunctional signaling underlying endometriosis: current state of knowledge. J Mol Endocrinol. 2018;60(3):R97–R113. [DOI] [PubMed] [Google Scholar]
- 43. Aznaurova YB, Zhumataev MB, Roberts TK, Aliper AM, Zhavoronkov AA. Molecular aspects of development and regulation of endometriosis. Reprod Biol Endocrinol. 2014;12:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dutta M, Anitha M, Smith PB, Chiaro CR, Maan M, Chaudhury K, Patterson AD. Metabolomics reveals altered lipid metabolism in a mouse model of endometriosis. J Proteome Res. 2016;15(8):2626–2633. [DOI] [PubMed] [Google Scholar]
- 45. Thyrell L, Arulampalam V, Hjortsberg L, Farnebo M, Grandér D, Pokrovskaja Tamm K. Interferon alpha induces cell death through interference with interleukin 6 signaling and inhibition of STAT3 activity. Exp Cell Res. 2007;313(19):4015–4024. [DOI] [PubMed] [Google Scholar]
- 46. Rakshit S, Chandrasekar BS, Saha B, Victor ES, Majumdar S, Nandi D. Interferon-gamma induced cell death: regulation and contributions of nitric oxide, cJun N-terminal kinase, reactive oxygen species and peroxynitrite. Biochim Biophys Acta. 2014;1843(11):2645–2661. [DOI] [PubMed] [Google Scholar]
- 47. Liu H, Lang JH. Is abnormal eutopic endometrium the cause of endometriosis? The role of eutopic endometrium in pathogenesis of endometriosis. Med Sci Monit. 2011;17(4):RA92–RA99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guzel E, Arlier S, Guzeloglu-Kayisli O, Tabak MS, Ekiz T, Semerci N, Larsen K, Schatz F, Lockwood CJ, Kayisli UA. Endoplasmic reticulum stress and homeostasis in reproductive physiology and pathology. Int J Mol Sci. 2017;18(4):E792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sonderegger S, Pollheimer J, Knöfler M. Wnt signalling in implantation, decidualisation and placental differentiation—review. Placenta. 2010;31(10):839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Murphy SP, Tayade C, Ashkar AA, Hatta K, Zhang J, Croy BA. Interferon gamma in successful pregnancies. Biol Reprod. 2009;80(5):848–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Seaward AV, Burke SD, Croy BA. Interferon gamma contributes to preimplantation embryonic development and to implantation site structure in NOD mice. Hum Reprod. 2010;25(11):2829–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. King AE, Critchley HO, Kelly RW. The NF-κB pathway in human endometrium and first trimester decidua. Mol Hum Reprod. 2001;7(2):175–183. [DOI] [PubMed] [Google Scholar]
- 53. Parsana P, Amend SR, Hernandez J, Pienta KJ, Battle A. Identifying global expression patterns and key regulators in epithelial to mesenchymal transition through multi-study integration. BMC Cancer. 2017;17(1):447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Birben E, Sahiner UM, Sackesen C, Erzurum S, Kalayci O. Oxidative stress and antioxidant defense. World Allergy Organ J. 2012;5(1):9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bartley J, Jülicher A, Hotz B, Mechsner S, Hotz H. Epithelial to mesenchymal transition (EMT) seems to be regulated differently in endometriosis and the endometrium. Arch Gynecol Obstet. 2014;289(4):871–881. [DOI] [PubMed] [Google Scholar]
- 56. Ngô C, Chéreau C, Nicco C, Weill B, Chapron C, Batteux F. Reactive oxygen species controls endometriosis progression. Am J Pathol. 2009;175(1):225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhan L, Wang W, Zhang Y, Song E, Fan Y, Wei B. Hypoxia-inducible factor-1alpha: a promising therapeutic target in endometriosis. Biochimie. 2016;123:130–137. [DOI] [PubMed] [Google Scholar]
- 58. Gaur U, Aggarwal BB. Regulation of proliferation, survival and apoptosis by members of the TNF superfamily. Biochem Pharmacol. 2003;66(8):1403–1408. [DOI] [PubMed] [Google Scholar]
- 59. Zhang Y, Li LF, Munir M, Qiu HJ. RING-domain E3 ligase-mediated host-virus interactions: orchestrating immune responses by the host and antagonizing immune defense by viruses. Front Immunol. 2018;9:1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Acién P, Quereda F, Campos A, Gomez-Torres MJ, Velasco I, Gutierrez M. Use of intraperitoneal interferon α-2b therapy after conservative surgery for endometriosis and postoperative medical treatment with depot gonadotropin-releasing hormone analog: a randomized clinical trial. Fertil Steril. 2002;78(4):705–711. [DOI] [PubMed] [Google Scholar]
- 61. Seif F, Khoshmirsafa M, Aazami H, Mohsenzadegan M, Sedighi G, Bahar M. The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun Signal. 2017;15(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Michalak M, Gye MC. Endoplasmic reticulum stress in periimplantation embryos. Clin Exp Reprod Med. 2015;42(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sakurai T, Takai R, Bürgin H, Ishihara K, Sakamoto Y, Amano J, Higuchi Y, Chiba S, Singer T, Kawamura A, Suzuki M, Müller L. The effects of interleukin-6 signal blockade on fertility, embryo-fetal development, and immunization in vivo. Birth Defects Res B Dev Reprod Toxicol. 2012;95(4):304–317. [DOI] [PubMed] [Google Scholar]
- 64. Suman P, Malhotra SS, Gupta SK. LIF-STAT signaling and trophoblast biology. JAK-STAT. 2013;2(4):e25155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Abdal Dayem A, Hossain MK, Lee SB, Kim K, Saha SK, Yang GM, Choi HY, Cho SG. The role of reactive oxygen species (ROS) in the biological activities of metallic nanoparticles. Int J Mol Sci. 2017;18(1):E120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003;9(6):677–684. [DOI] [PubMed] [Google Scholar]
- 67. Ucuzian AA, Gassman AA, East AT, Greisler HP. Molecular mediators of angiogenesis. J Burn Care Res. 2010;31(1):158–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kaponis A, Iwabe T, Taniguchi F, Ito M, Deura I, Decavalas G, Terakawa N, Harada T. The role of NF-kappaB in endometriosis. Front Biosci (Schol Ed). 2012;4:1213–1234. [DOI] [PubMed] [Google Scholar]
- 69. Piva R, Belardo G, Santoro MG. NF-κB: a stress-regulated switch for cell survival. Antioxid Redox Signal. 2006;8(3-4):478–486. [DOI] [PubMed] [Google Scholar]
- 70. Carvalho LF, Samadder AN, Agarwal A, Fernandes LF, Abrão MS. Oxidative stress biomarkers in patients with endometriosis: systematic review. Arch Gynecol Obstet. 2012;286(4):1033–1040. [DOI] [PubMed] [Google Scholar]
- 71. Van Langendonckt A, Casanas-Roux F, Donnez J. Iron overload in the peritoneal cavity of women with pelvic endometriosis. Fertil Steril. 2002;78(4):712–718. [DOI] [PubMed] [Google Scholar]
- 72. Fang S, Yu X, Ding H, Han J, Feng J. Effects of intracellular iron overload on cell death and identification of potent cell death inhibitors. Biochem Biophys Res Commun. 2018;503(1):297–303. [DOI] [PubMed] [Google Scholar]
- 73. McKinnon BD, Kocbek V, Nirgianakis K, Bersinger NA, Mueller MD. Kinase signalling pathways in endometriosis: potential targets for non-hormonal therapeutics. Hum Reprod Update. 2016;22(3):382–403. [DOI] [PubMed] [Google Scholar]
- 74. Yang YM, Yang WX. Epithelial-to-mesenchymal transition in the development of endometriosis. Oncotarget. 2017;8(25):41679–41689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cai Y, Jin H, Cao LQ, Gao Q, Tao J. Overexpression of TAFI promotes epithelial mesenchymal transition in endometriosis. Eur Rev Med Pharmacol Sci. 2017;21(24):5527–5533. [DOI] [PubMed] [Google Scholar]
- 76. Zhang Q, Dong P, Liu X, Sakuragi N, Guo SW. Enhancer of Zeste homolog 2 (EZH2) induces epithelial-mesenchymal transition in endometriosis. Sci Rep. 2017;7(1):6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wu RF, Huang ZX, Ran J, Dai SJ, Lin DC, Ng TW, Chen QX, Chen QH. Lipoxin A4 suppresses estrogen-induced epithelial–mesenchymal transition via ALXR-dependent manner in endometriosis. Reprod Sci. 2018;25(4):566–578. [DOI] [PubMed] [Google Scholar]
- 78. Xiong Y, Liu Y, Xiong W, Zhang L, Liu H, Du Y, Li N. Hypoxia-inducible factor 1α-induced epithelial-mesenchymal transition of endometrial epithelial cells may contribute to the development of endometriosis. Hum Reprod. 2016;31(6):1327–1338. [DOI] [PubMed] [Google Scholar]
- 79. Hsiao KY, Wu MH, Chang N, Yang SH, Wu CW, Sun HS, Tsai SJ. Coordination of AUF1 and miR-148a destabilizes DNA methyltransferase 1 mRNA under hypoxia in endometriosis. Mol Hum Reprod. 2015;21(12):894–904. [DOI] [PubMed] [Google Scholar]
- 80. Albertsen HM, Ward K. Genes linked to endometriosis by GWAS are integral to cytoskeleton regulation and suggests that mesothelial barrier homeostasis is a factor in the pathogenesis of endometriosis. Reprod Sci. 2017;24(6):803–811. [DOI] [PubMed] [Google Scholar]
- 81. Hsiao KY, Lin SC, Wu MH, Tsai SJ. Pathological functions of hypoxia in endometriosis. Front Biosci (Elite Ed). 2015;7:309–321. [DOI] [PubMed] [Google Scholar]
- 82. Krock BL, Skuli N, Simon MC. Hypoxia-induced angiogenesis: good and evil. Genes Cancer. 2011;2(12):1117–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Macer ML, Taylor HS. Endometriosis and infertility: a review of the pathogenesis and treatment of endometriosis-associated infertility. Obstet Gynecol Clin North Am. 2012;39(4):535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gaetje R, Holtrich U, Karn T, Cikrit E, Engels K, Rody A, Kaufmann M. Characterization of WNT7A expression in human endometrium and endometriotic lesions. Fertil Steril. 2007;88(6):1534–1540. [DOI] [PubMed] [Google Scholar]
- 85. Zanatta A, Rocha AM, Carvalho FM, Pereira RM, Taylor HS, Motta EL, Baracat EC, Serafini PC. The role of the Hoxa10/HOXA10 gene in the etiology of endometriosis and its related infertility: a review. J Assist Reprod Genet. 2010;27(12):701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Patel BG, Rudnicki M, Yu J, Shu Y, Taylor RN. Progesterone resistance in endometriosis: origins, consequences and interventions. Acta Obstet Gynecol Scand. 2017;96(6):623–632. [DOI] [PubMed] [Google Scholar]
- 87. Park JS, Lee JH, Kim M, Chang HJ, Hwang KJ, Chang KH. Endometrium from women with endometriosis shows increased proliferation activity. Fertil Steril. 2009;92(4):1246–1249. [DOI] [PubMed] [Google Scholar]
- 88. Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27(50):6462–6472. [DOI] [PubMed] [Google Scholar]
- 89. Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol. 2007;19(6):649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Taniguchi F, Kaponis A, Izawa M, Kiyama T, Deura I, Ito M, Iwabe T, Adonakis G, Terakawa N, Harada T. Apoptosis and endometriosis. Front Biosci (Elite Ed). 2011;3(2):648–662. [DOI] [PubMed] [Google Scholar]
- 91. Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8(11):851–864. [DOI] [PubMed] [Google Scholar]
- 92. Basar M, Bozkurt I, Guzeloglu-Kayisli O, Sozen B, Tekmen I, Schatz F, Arici A, Lockwood CJ, Kayisli UA. Unfolded protein response prevents blastocyst formation during preimplantation embryo development in vitro. Fertil Steril. 2014;102(6):1777–1784. [DOI] [PubMed] [Google Scholar]
- 93. Nakamura H, Kimura T, Ogita K, Nakamura T, Takemura M, Shimoya K, Koyama S, Tsujie T, Koyama M, Murata Y.. NF-κB activation at implantation window of the mouse uterus. Am J Reprod Immunol. 2004;51(1):16-21. [DOI] [PubMed] [Google Scholar]
- 94. Klemmt PAB, Starzinski-Powitz A. Molecular and cellular pathogenesis of endometriosis. Curr Womens Health Rev. 2018;14(2):106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]