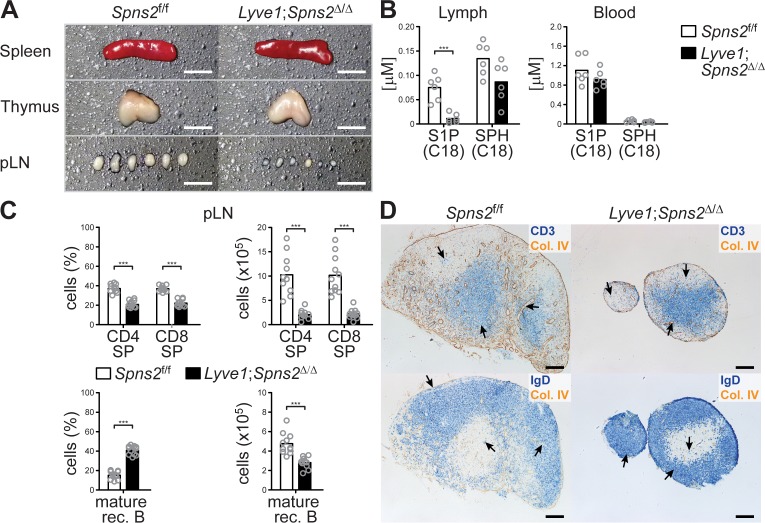
Figure 1. Lyve1CRE-mediated Spns2-deletion in endothelial cells causes hypotrophy in pLNs.
(A) Macroscopic view of spleen, thymus and pLNs of wildtype Spns2f/f (left) and Lyve1;Spns2Δ/Δ (right) mice. (B) Quantification of S1P and sphingosine (SPH) concentrations in lymph and blood. (C) FACS analysis of CD4+ and CD8+ SP T-cells (top) and mature rec. B-cells (bottom) of pLNs of Spns2f/f and Lyve1;Spns2Δ/Δ mice. (D) Light microscopy of pLNs of Spns2f/f (left) and Lyve1;Spns2Δ/Δ (right) mice for CD3+ T-cells (top, blue) and IgD+ mature rec. B-cells (bottom, blue) counterstained for collagen-IV+ (brown) tissue frameworks. Each circle (B, C) represents an individual mouse; bars indicate the mean. Scale bars, 0.2 cm (A) or 200 μm (D). ***p<0.0005 (two-tailed unpaired Student’s t-test (B, C)). Data are representative for six mice per group (A, B), for 2x inguinal, 2x brachial and 2x axial LNs of six mice per group (D) or are pooled from three independent experiments (C) with n = 3 or n = 4 mice per group.