Abstract
Background.
Enzyme-mediated biotransformation of pharmacological agents is a crucial step in xenobiotic detoxification and drug disposition. Herein, we investigated the metabolism and physicochemical properties of the top 200 most prescribed drugs (established) as well as drugs approved by the US Food and Drug Administration (FDA) between 2005 and 2016 (newly approved). Objective Our objective was to capture the changing trends in the routes of administration, physicochemical properties, and prodrug medications, as well as the contributions of drug-metabolizing enzymes and transporters to drug clearance.
Methods.
The University of Washington Drug Interaction Database (DIDB®) as well as other online resources (e.g., CenterWatch.com, Drugs.com, DrugBank.ca, and PubChem.ncbi.nlm.nih.gov) was used to collect and stratify the dataset required for exploring the above-mentioned trends.
Results.
Analyses revealed that ~ 90% of all drugs in the established and newly approved drug lists were administered systemically (oral and/or intravenous). Meanwhile, the portion of biologics (molecular weight > 1 kDa) was 15 times greater in the newly approved list than established drugs. Additionally, there was a 4.5-fold increase in the number of compounds with a high calculated partition coefficient (cLogP > 3) and a high total polar surface area (> 75 Å2) in the newly approved drug vs. the established category. Further, prodrugs in established or newly approved lists were found to be converted to active compounds via hydrolysis, demethylases, and kinases. The contribution of cytochrome P450 (CYP) 3A4, as the major biotransformation pathway, has increased from 40% in the established drug list to 64% in the newly approved drug list. Moreover, the role of CYP1A2, CYP2C19, and CYP2D6 was decreased as major metabolizing enzymes among the newly approved medications. Among non-CYP major metabolizers, the contribution of alcohol dehydrogenases/aldehyde dehydrogenases (ADH/ALDH) and sulfotransferases decreased in the newly approved drugs compared with the established list. Furthermore, the highest contribution among uptake and efflux transporters was found for Organic Anion Transporting Polypeptide 1B1 (OATP1B1) and P-glycoprotein (P-gp), respectively.
Conclusions
The higher portion of biologics in the newly approved drugs compared with the established list confirmed the growing demands for protein- and antibody-based therapies. Moreover, the larger number of hydrophilic drugs found in the newly approved list suggests that the probability of toxicity is likely to decrease. With regard to CYP-mediated major metabolism, CYP3A5 showed an increased involvement owing to the identification of unique probe substrates to differentiate CYP3As. Furthermore, the contribution of OATP1B1 and P-gp did not show a significant shift in the newly approved drugs as compared to the established list because of their broad substrate specificity.
1. Introduction
Metabolic biotransformation is the critical process that mediates the conversion of parent xenobiotic compounds into their metabolites. Metabolism is generally a detoxifying process that assists in either elimination of a drug compound or inactivation of a pharmaceutical ingredient. Drug metabolism is typically categorized as phase I and phase II reactions. Phase I involves the introduction of polar groups to the parent compound via oxidation, reduction, conjugation and/or hydrolysis. Whereas, phase II comprises the conjugation with hydrophilic moieties including the addition of glucuronide, sulfate, glutathione, or amino acids [1]. Metabolites generated by phase I and phase II enzymes are excreted into the bile, via the canalicular lumen in hepatocytes, or eliminated from the body by kidneys [2]. Through these processes, the biologically active agents are rendered less active, effluxed by transporters, and excreted from the body by renal or biliary routes. In 2004, Williams et al. [3] reported that cytochrome P450 (CYP) enzymes were responsible for the biotransformation of two-thirds of the most prescribed drugs in the USA. This is followed by UDP-glucuronosyltransferases (UGTs), esterases namely carboxylesterases (CES), arylacetamide deacetylase, butyrylcholinesterase, paraoxonases, and flavin-containing monooxygenases (FMOs) [3, 4]. The CYPs belong to a superfamily of heme-containing membrane proteins. In the human genome, CYPs are represented by 57 putatively functional genes including 18 families and 44 subfamilies (https://www.pharmvar.org/genes) [5, 6]. Despite the broad overlapping substrate specificity among the families, most of the drugs are metabolized by CYP families 1, 2, and 3, namely CYP 1A2, 2B6, 2C8, 2C9, 2C19, 2D6, 3A4, and 3A5 isoforms [7–9]. In recent years, the pharmaceutical industry has devoted a significant amount of resources to understand and subsequently improve the absorption, distribution, metabolism, and excretion properties of new chemical entities. In 1991, it was reported that the compounds’ failure rate from discovery to registration was as high as 40% because of an incomplete understanding of pharmacokinetic (PK) properties and sub-optimal bioavailability [10]. However, the failure of new chemical entities in the clinic has decreased to 10% in the following decade because of the emphasis on structural optimization resulting in enhanced PK properties [10, 11]. Therefore, an increasing number of new chemical entities that undergo non-CYP metabolism including oxidation, reduction, hydrolysis, and conjugation have recently emerged [12–15] with slightly higher lipophilicity [13, 16]. Despite these propositions, supporting data are sparse, and a systematic evaluation of the major and minor drug metabolism pathways is lacking. Along with drug-metabolizing enzymes, membrane transporters are additional major determinants of PK and safety profiles. Over 400 different transporters have been annotated in the human genome [17]. Transporters are responsible for the hepatocellular disposition of polar or charged molecules, without which these molecules could not pass through the lipid bilayer of the cell membrane [18]. Following the uptake of molecules into the intracellular space, a different class of efflux transporters aid the movement of molecules from the cell to the extracellular space [14]. Many clinically relevant xenobiotic transporters belong to two superfamilies: solute carrier (SLC) and ATP-binding cassette (ABC) transporters, which mediate the uptake and efflux of chemical entities, respectively [19, 20]. Therefore, coordinated operation of ABC and SLC transporters appears to be essential for drug absorption and elimination processes. The primary objective of this report is to conduct a systematic analysis of the established drugs in the US market [from the top 200 most prescribed drug list in 2014] (established drugs) and to compare this list with the US Food and Drug Administration (FDA)-approved drugs from 2005 to 2016 (newly approved drugs). We assumed that these lists represent a collection of drugs that have gained FDA approval in previous decades vs. more recent years. Our goal was to determine a possible shift, if any, in the routes of administration, lipophilicity, as well as changes in the contributions of major and minor metabolizing enzymes. Biological therapeutics were not considered in this review and excluded from both established and newly approved lists. The current review also aims to demonstrate the relative contribution of each transporter to drug disposition and elimination.
2. Methods
2.1. Sources of Established and Newly Approved Drug Lists
The sources of the information for this review are summarized in Table 1. Established drugs originated from the list of “Top 200 Drugs of 2014” from Symphony Health’s IDV® (Integrated Dataverse) [21]. This list ranks drugs based on the total prescription count in a given year. The newly FDA-approved drugs were obtained from the CenterWatch website [22] and included drugs that received FDA approval between 2005 and 2016.
Table 1.
List of the public and subscription based online resources from which data were extracted
| Database | Type of information | Domain Availability |
URL | Reference |
|---|---|---|---|---|
| DrugBank | Pharmacokinetics/metabolism | Public | https://www.drugbank.ca/ | [47] |
| Drugs.com | Pharmacokinetics/metabolism | Public | https://www.drugs.com/ | [48] |
| PubChem | Chemical data | Public | https://pubchem.ncbi.nlm.nih.gov/ | [49] |
| CenterWatch | FDA-approved list (2005-16) | Public | https://www.centerwatch.com/ | [22] |
| SymphonyHealth’s IDV® | Top 200 list | Public | http://symphonyhealth.com/wpcontent/uploads/2015/05/Top-200-Drugs-of-2014.pdf | [21] |
| The University of Washington drug interaction database (DIDB®) | Pharmacokinetics/metabolism and transporters | Subscription based | http://www.centerwatch.com/drug-information/fda-approved-drugs/year | [24] |
2.2. Allocation of Drugs to Each List
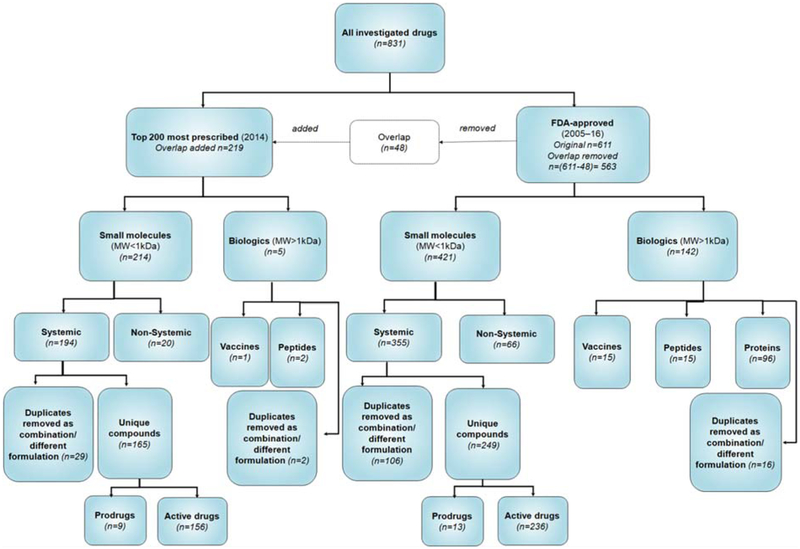
Figure 1 shows a flowchart describing the reasoning behind the allocation of compounds to a particular list. In total, the names of 831 drugs were retrieved from both lists. For combination therapies, each active ingredient was separated and considered individually. When the active ingredient was repeated in multiple combination therapies, the shared compound was considered as a duplicate and eliminated from the assessment. Considering this rationale, 219 drugs, including 214 small molecules with a molecular weight (Mw) < 1 kDa and five biologics (Mw > 1 kDa) were included in the established drug list. Moreover, information on a total of 563 drugs, including 421 small molecules (Mw < 1 kDa) and 142 biologics (Mw > 1 kDa) were extracted from the newly FDA-approved drug list. Considering that 48 drugs were shared between the two lists, the common drugs were only considered in the established drug list and excluded from the newly approved drug list. Among small molecules administered systemically (oral and/or intravenous routes), only compounds without duplication were analyzed. Using such filters, we finally obtained 165 (established) and 249 (newly approved) compounds possessing unique structures (active compounds and prodrugs) for further investigations.
Figure 1.
The workflow for inclusion of drugs in established and newly-approved drug lists. The metabolism pathways were analyzed using the unique compounds from “top 200” most prescribed medications in 2014 (established drugs; n=165) as well as FDA approved drugs from “2005–2016” (newly-approved drugs; n=249). n, denotes the number of compounds analyzed at each stage.
2.3. Combination therapy
We noticed 21 drugs in the established list and 57 drugs in the newly approved list that comprised more than one active compound. To address combinatory drugs, we separated active ingredients in each combination and assigned a unique identification for them in Microsoft Excel® to avoid any duplication for the same compound in entire lists. For example, Byvalson® is a combination of nebivolol and valsartan, while Entresto® is a combination of sacubitril and valsartan. According to our criteria, valsartan was accounted for only once to avoid repetition.
2.4. Physicochemical properties
Physiochemical parameters such as calculated partition coefficient (cLogP), Total Polar Surface Area (TPSA) in angstroms squared, Å2), and Mw were acquired for the drugs included in both lists. These parameters were retrieved from the PubChem website and analyzed based on the criteria proposed by Price et al. [23] using a threshold of 3.0 for cLogP and 75 Å2 for TPSA. Thus, we categorized and compared the drugs based on the suggested cut-off criteria and assessed the percentage change between the different groups.
2.5. Information on drug elimination pathways
The University of Washington Drug Interaction Database (DIDB®) was used to retrieve metabolic and other PK information for the established and newly approved drugs [24]. If a monograph was not available in DIDB® for a given drug, other online resources, including Drugs.com, DrugBank.ca, and PubChem.ncbi.nlm.nih.gov were used to support the evidence. Using this strategy, the major and minor contributions of enzymes to drug metabolism as well as potential drug–drug interactions (DDIs) and the contributions of uptake and efflux transporters were explored. Based on the FDA guideline for DDI studies, which is followed by DIDB®, “The contribution of a specific metabolizing enzyme to an investigational drug’s clearance is considered significant if the enzyme is responsible for ≥ 25% of the drug’s elimination based on the in vitro phenotyping studies and human PK data” [25]. Hence, all metabolic enzymes addressed as “major”, “predominant”, “main”, and “significant” in the DIDB® monograph were classified as the major metabolic pathway for a corresponding drug. Similarly, the minor pathway was recognized based on labels such as “minor” and “lesser extent”. Drugs were also ranked based on their potential risk of DDIs as a substrate and/or inhibitor/inducer. This risk comes from PK (metabolism and transporter) aspects but not pharmacodynamic interactions. The defined DDI risk levels for substrates (DDI risk/substrate) were based on a combination of the following features: (i) extent of metabolism/transport and sensitivity to induction/inhibition of the contributed enzyme and/or transporter; (ii) therapeutic range; and (iii) clinical interactions reflected in new drug application (NDA) reviews and publication [see data in the Electronic Supplementary Material (ESM)] [26–29].
2.6. Data analysis
The current evaluation is focused on the assessment of CYP and non-CYP enzyme contributions as major and/or minor metabolism pathways for drugs. Moreover, the routes of drug administration, lipophilicity, prodrug metabolism, and transporter-mediated uptake/efflux were also explored. The fractional clearance through a particular enzyme, if multiple enzymes were involved, was not defined for most of the drugs in DIDB®. Hence, fractional clearance was not included in the current analysis. Instead, the contribution of each enzyme was weighed equally. In this way, a score of 1.0 was assigned for each enzyme responsible for the major or minor metabolic pathways. For instance, assume that the database contains only two hypothetical drugs (i.e., A and B). Drug A undergoes major metabolism by two CYPs and one non-CYP, while drug B is metabolized by two CYPs. In our analysis, the number of involved CYPs and non-CYPs were summed up to estimate the overall contribution of each enzyme within the dataset. Consequently, 80% (4/5) and 20% (1/5) were assigned for the contributions of CYPs and non-CYPs, respectively. The contributions of enzymes in the metabolism of drugs were categorized namely by CYPs, non-CYPs, unknown metabolism, metabolized by both CYPs/non-CYPs (mixed), and no metabolism. These categories were analyzed under both major and minor metabolism, separately. All analyses were carried out using Microsoft Excel®. In the established and newly approved drug lists, 9 and 13 compounds were recognized as prodrugs, respectively. In all cases, prodrug bioactivation was determined independently from the metabolism of its active compound. For instance, isavuconazonium sulfate is converted to its active form, isavuconazonium, via plasma esterases. Subsequently, isavuconazonium is metabolized by hepatic CYP3A4, CYP3A5, and UGT enzymes. Thus, the active agent generated from the prodrug was included in the major and minor metabolism assessment, meanwhile, the role of esterases was accounted for prodrug bioactivation, separately.
3. Results
Figure 1 shows a flowchart demonstrating the criteria applied in this study to allocate pharmaceutical compounds to each subcategory. The total number of all investigated drugs from the established and newly approved lists was 831. From the total number of drugs, 219 compounds (including small molecules and biologics) belong to the established drug list, while 563 compounds account for the newly approved drug list.
3.1. Portion of Biologics
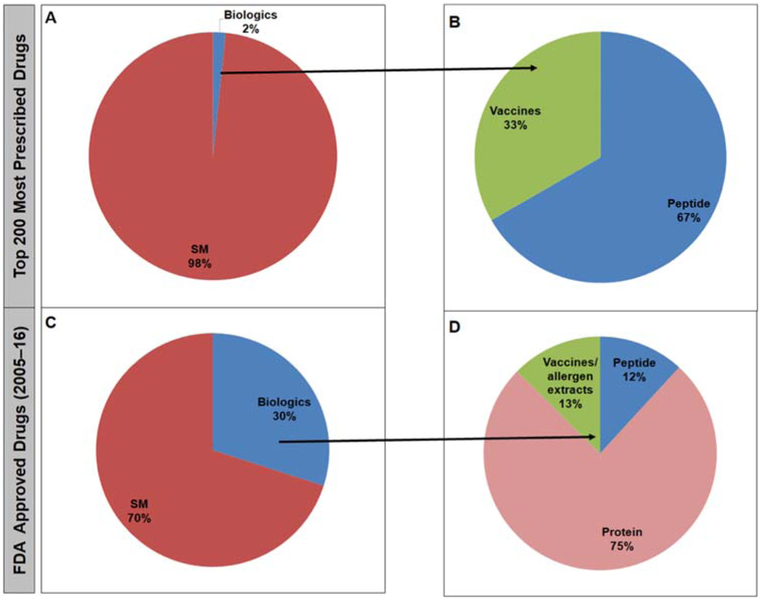
Among all active ingredients from the established and the newly approved lists, 5 and 142 drugs were considered as biologics. Figure 2 shows a substantial increase in the number of approved large molecules from 2% for the established list to 30% for the newly approved list. The biologics in the established list were predominantly indicated as hormone replacement therapy and vaccines. However, in the newly approved list, most of the biologics belong to the monoclonal antibodies.
Figure 2.
The contribution of small molecules versus biological therapeutics to the established (A) and newly-approved (B) drugs lists.
3.2. Route of drug administration
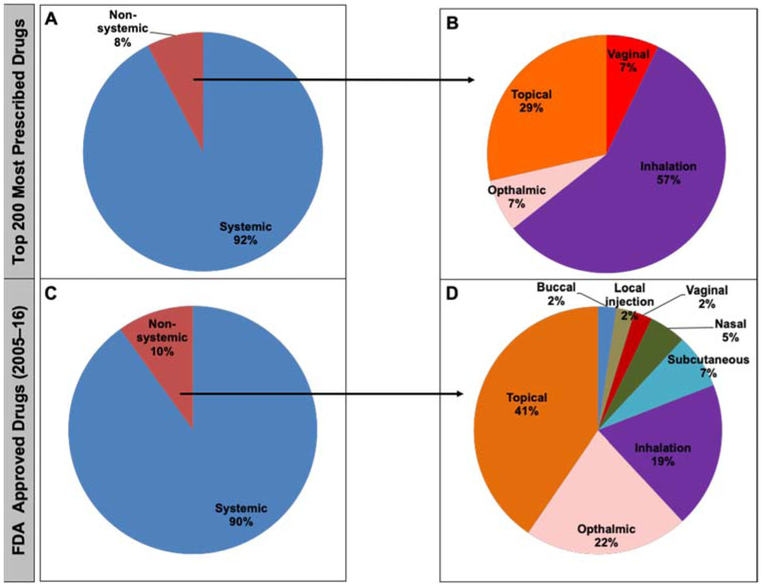
The route of administration was compared between 214 and 421 small molecules from the established and newly approved lists, respectively (Fig. 3). No major changes were found between the systemic and non-systemic routes of administration. Among the non-systemic administered drugs, an increase in the formulation of topically administered agents was observed from the established (29%) to the newly approved (41%) drug lists. In addition, there was an increase in the ophthalmic route from 7 to 22%. However, there was a reduction in the inhalation route from 57% in the established list to 19% in the newly approved list. Moreover, other routes of administration were found for the newly approved drugs, such as buccal (2%), local injection (2%), nasal (5%), and subcutaneous (7%) that were not present in the established list.
Figure 3.
The systemic and non-systemic routes of drug administration illustrated for the “top 200” (established) and “2005-16” (newly-approved) lists in panels A/B and C/D, respectively.
3.3. Physicochemical properties (cLog P, TPSA, and Mw)
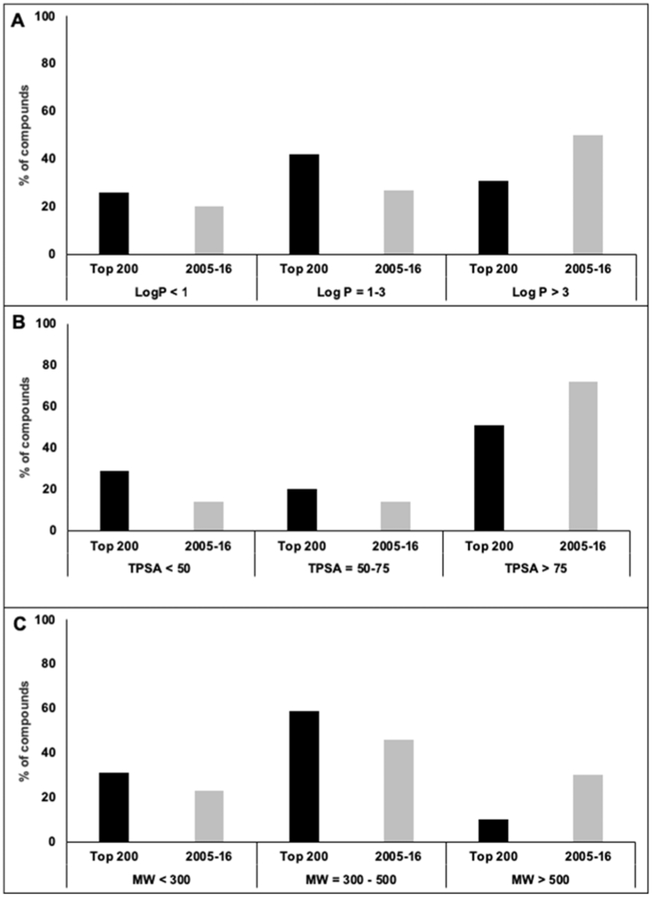
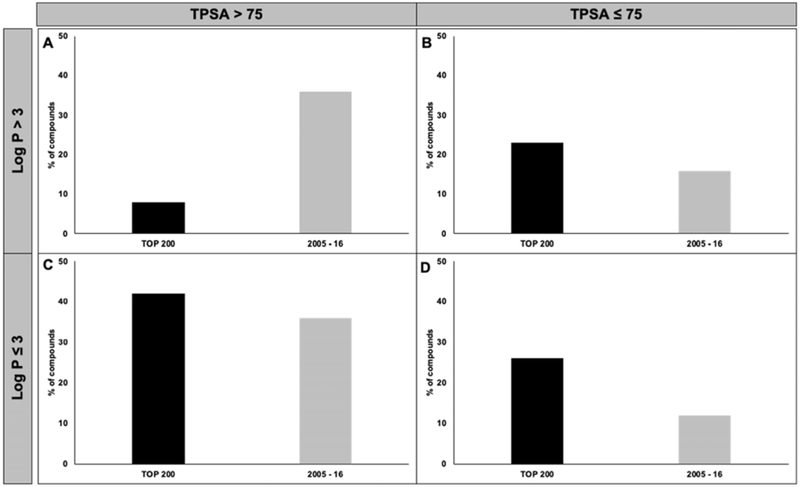
Figure 4a–c indicate the trends of cLogP, TPSA, and Mw values for the two drug lists, respectively. According to Fig. 4a, the percentage of drugs with cLogP > 3 in the newly approved drugs (2005–16) was 1.6-fold higher than that in the established list (Top 200). Moreover, a 21% increase in the drugs with TPSA > 75 Å2 was observed in the newly approved list compared with the established list (Fig. 4b). Figure 4c shows a three-fold increase in the percentage of compounds with Mw > 500 Da in the newly approved list vs. the established list. Meanwhile, the percentage of drugs with Mw < 300 Da and Mw ~ 300 to 500 Da decreased in the newly approved list compared with the established list (Fig. 4c). Figure 5 shows the classification of compounds into four groups based on the criteria proposed by Price et al. (cLogP > 3 or ≤ 3 as well as TPSA > 75 Å2 or ≤ 75 Å2). The most noticeable change in the “2005–16” list was a 4.5-fold increase in compounds with cLogP > 3 and TPSA > 75 Å2, while an ~ 2-fold decrease in the number of compounds in the opposite category (cLogP ≤ 3 and TPSA ≤ 75 Å2) was observed.
Figure 4.
The physicochemical properties of drugs from the “top 200” (established) and “2005-16” (newly-approved) lists, such as Log P, total polar surface area (TPSA), and molecular weight (Mw), were compared. Panel A (cLog P), B (TPSA), and C (Mw) illustrate the relative abundance of drugs in each list according to the criteria suggested by Price et al. [23].
Figure 5.
The lipophilicity was compared between the “top 200” most prescribed drugs (established) and newly-FDA approved (newly-approved) drugs “2005-16”. According to Price et al. [23], a cut-off criterion of 3 for cLog P and 75 Å2 for TPSA were established states that the compounds with high cLog P/low TPSA are ~ 2.5 times more likely to cause toxicity.
3.4. Prodrug metabolism
The metabolism of 9 and 13 prodrugs from the established and newly approved drugs were compared, and results are shown in Tables 1 and 2 of the ESM. Results show that prodrugs in the established list were activated via hydrolases (78%) and demethylase (22%); however, the newer drugs were converted to their active form through hydrolases (75%) and kinases (25%).
3.5. Major biotransformation enzymes
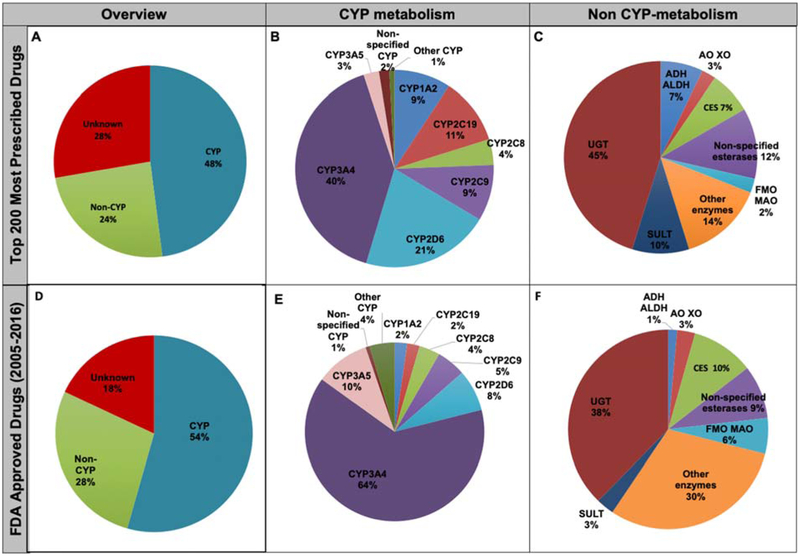
Figure 6 and Table 2 summarize the major contribution of enzymes in the biotransformation of small molecules from the two drug lists. A slight increase in CYP-mediated metabolism of xenobiotics from 48% to 54% was observed, while non-CYP metabolism shifted from 24% to 28% in established vs. newly approved categories. The sequence of CYP isoforms as the major metabolizing enzyme responsible for the biotransformation of established drugs is ranked as: CYP3 A4 > CYP2D6 > CYP2C19 > CYP1A2 = CYP2C9 > CYP2C 8. This sequence was changed in newly approved drugs as: CYP3A4 > CYP3A5 > CYP2D6 > CYP2C9 > CYP2C8 > C YP2C19 = CYP1A2. Consequently, the percentage of drugs that undergo major metabolism by CYP3A4 increased from 40 to 64%. In addition, the contribution of CYP3A5 increased from 3 to 10% from established vs. newly approved lists, respectively. With the aid of CYP3cide, the CYP3A isoenzyme specificity has been clearly evident and the relative contribution of CYP3A4 and CYP3A5 in drug metabolism could be easily distinguished [30]. The CYP3A5 polymorphism greatly affects the pharmacokinetics of tacrolimus in patients undergoing organ transplantation, which confirms the clinically significant role of CYP3A5 genotyping [31, 32]. If one considers the contribution of CYP3A as CYP3A4 and CYP3A5 combined, the increase in the CYP3A contribution becomes even larger from 43% in the established list vs. 74% in the newly approved list. Meanwhile, the contributions of CYP2C19 and CYP2D6 decreased. Figure 6c, f show the contribution of different nonCYP enzymes to the major metabolism. The contribution of non-CYPs as major enzymes contributing to biotransformation of established drugs was ranked as: UGTs > other enzymes > esterase > sulfotransferases (SULTs) > alcohol dehydrogenases (ADH)/aldehyde dehydrogenases (ALDH) = CES > aldehyde oxidase (AO)/xanthine oxidase (XO) > FMO/monoamine oxidase (MAO). The “other enzymes” category includes biotransformation reactions such as oxidative dealkylation, acetylation, deiodinase, de-ethylation, pyrimidine catabolism, dehydropeptidase, and thymidine phosphorylase. This rank was slightly changed in newly approved drugs as: UGT > other enzymes > CES > esterase > FMO/MAO > SULTs = AO/XO > ADH/ALDH.
Figure 6.
The contribution of major enzymes (A, D), major cytochrom P450s (CYPs) (B, E), and major non-CYP enzymes (C, F) contributed in the metabolism of “top 200 most prescribed” and "2005-2016" FDA approved drugs.
Table 2.
The contribution of cytochrom P450s (CYPs) and non-CYPs in the major metabolism of drugs
| Metabolizing enzymes | The number of drugs | |
|---|---|---|
| Established (n=156) | Newly-approved (n=237) | |
| Overall metabolism | ||
| CYP | 83 | 136 |
| Non-CYP | 42 | 69 |
| Unknown | 48 | 46 |
| CYP metabolism | ||
| CYP1A2 | 11 | 3 |
| CYP2C8 | 5 | 5 |
| CYP2C9 | 11 | 7 |
| CYP2C19 | 13 | 3 |
| CYP2D6 | 25 | 10 |
| CYP3A4 | 48 | 85 |
| CYP3A5 | 3 | 13 |
| Non-specified CYPs | 2 | 1 |
| Other CYPs | 1 | 6 |
| Non-CYP metabolism | ||
| ADH ALDH | 3 | 1 |
| AO XO | 1 | 2 |
| CESs | 3 | 7 |
| Esterase | 5 | 6 |
| FMO MAO | 1 | 4 |
| SULTs | 4 | 2 |
| UGTs | 19 | 26 |
| Unknown | 6 | 21 |
Top 200 most prescribed drugs (Established) and newly-FDA approved drugs from 2005 to 2016 (Newly-approved).
Drugs that are weighed once under CYP metabolism can be metabolized by multiple CYP isoforms like CYP3A4, CYP2C9 and CYP2C19. Hence, the CYP subtypes did not add up to the overall CYP and non-CYP mediated metabolism.
3.6. Minor biotransformation enzymes
The minor contribution of enzymes to the metabolism of small molecules is summarized in Fig. S1 of the ESM and Table 3. Minor contributing enzymes were not known for a large majority of drugs in both lists. However, the contribution of CYP was 4.5- and 2.8-fold higher than nonCYP enzymes as the minor enzymes contributing to drug metabolism in established and newly approved drug lists, respectively. The contribution of CYP enzymes to the minor metabolism of the established drugs is ranked as: CYP3A4 > CYP2D6 > CYP1A2 > CYP2C9 = CYP2C19 > CYP3A5 > CYP2B6 = CYP2C8. This contribution was changed for the newly approved drugs as: CYP2C19 > CYP2D6 > CYP 3A4 = CYP2C9 > CYP2C8 > CYP1A2 > CYP2E1 = CYP2 B6 > CYP3A5. According to Figs. S1B and 1E of the ESM, the percentage of drugs that undergo minor metabolism by CYP3A4 decreased from 32% to 14%, while the contribution of CYP2C19 and CYP2C8 increased from established to newly approved drugs. Furthermore, CYP2E1 is involved as a minor enzyme contributor in the newly approved list, which was absent in the metabolism of the established list.
Table 3.
The contribution of CYPs and non-CYP enzymes in the minor metabolism of drugs
| Metabolizing enzymes | The number of drugs | |
|---|---|---|
| Overview | Established* | Newly-approved* |
| CYP | 28 | 78 |
| Non-CYP | 6 | 27 |
| Unknown | 124 | 144 |
| CYP metabolism | ||
| CYP1A2 | 5 | 11 |
| CYP2B6 | 2 | 4 |
| CYP2C8 | 2 | 15 |
| CYP2C9 | 3 | 20 |
| CYP2C19 | 3 | 26 |
| CYP2D6 | 8 | 23 |
| CYP3A4 | 14 | 19 |
| CYP3A5 | 2 | 3 |
| Non-specified CYPs | - | 3 |
| Other CYPs | 3 | 11 |
| Non-CYP metabolism | ||
| ADH ALDH | - | 1 |
| CESs | 1 | 1 |
| FMO MAO | - | 6 |
| Unknown | - | 1 |
| SULTs | 1 | 2 |
| UGTs | 4 | 16 |
Top 200 most prescribed drugs (established) and newly FDA approved drugs from 2005 to 2016 (newly-approved)
Drugs, that are weighed once under P450 metabolism can be metabolized by multiple CYP isoforms like CYP3A4, CYP2C9 and CYP2C19. Hence the P450 subtypes will not add up in most cases to overall P450 and non-P450 mediated metabolism.
The contribution of non-CYP enzymes in the metabolism of therapeutic agents varied significantly from 4% to 11% in established vs. newly approved drug lists. Figures S1C and S1F of the ESM show the contribution of non-CYPs in the major metabolism of established drugs ranked as: UGTs > SULTs > CES. This sequence changed in newly approved drugs to: UGTs > FMO/MAO > SULTs > ADH/ ALDH = CES.
3.7. Contribution of Uptake and Efflux Transporters in Drug Disposition
Figure S2 of the ESM and Table 4 show the role of uptake and efflux transporters in the disposition of the small molecules. According to DIDB®, the data on the involvement of xenobiotic transporters were mainly adopted from in vitro studies using Madin-Darby canine kidney cell lines. As such, it is not clear that the result of in vitro studies directly translates to the same transporters in the human body. Moreover, no information was available on transporters for 69% of established drugs and 58% of newly approved drugs. Nevertheless, the available results indicate that organic anion transporting polypeptide 1B1 (OATP1B1) is the highest contributing uptake transporter with 44% and 30% for both the established and newly approved drugs, respectively. Figure S2 of the ESM shows the contribution of uptake transporters in the disposition of the established drugs ranked as: OATP1B1 > OATP1B3 = OATP2B1 = OAT1 > organic cation transporter (OCT) N1 = OCT2 = OAT3 > OATP. The contribution of uptake transporters to the disposition of newly approved drugs is ranked as: OATP1B1 > OAT3 > OAT1 = OA T > OATP1 B3 > O CT1 = OCT2 > OATP2B1 = OATP1A2 = LAT1 = OCTN1 = apical sodium dependent bile acid transporter. Figures S2C and SF of the ESM show the contribution of efflux transporters to the disposition of established drugs ranked as: P-glycoprotein (P-gp) > Breast Cancer Resistance Protein (BCRP)> Multidrug Resistance-associated Protein (MRP)-2. The contribution of efflux transporters to the disposition of newly approved drugs is ranked as: P-gp > BCRP > Bile Salt Export Pump (BSEP) = MRP2 > MRP4.
Table 4.
The contribution of uptake and efflux transporters in the disposition of drugs
| Transporters | The number of drugs | |
|---|---|---|
| Overview | Established* | Newly-approved* |
| Uptake | 18 | 30 |
| Efflux | 30 | 68 |
| Unknown/no transporter | 108 | 139 |
| Uptake | ||
| OCT1 | - | 1 |
| OCT2 | 1 | 1 |
| Non-specified OCT | - | 1 |
| ASBT | - | 1 |
| LAT1 | - | 1 |
| OATP1A2 | - | 1 |
| OATP1B1 | 8 | 9 |
| OATP1B3 | 2 | 2 |
| OATP2B1 | 2 | 1 |
| Non-specified OAT | - | 3 |
| OAT 1 | 2 | 3 |
| OAT3 | 1 | 5 |
| Non-specified OATP | 1 | - |
| OCTN1 | 1 | 1 |
| Efflux | ||
| P-gp | 23 | 49 |
| BCRP | 5 | 14 |
| BSEP | - | 2 |
| MRP2 | 2 | 2 |
| MRP4 | - | 1 |
Top 200 most prescribed drugs (established) and newly FDA approved drugs from 2005 to 2016 (newly-approved).
4. Discussion
The goal of this systematic review was to determine if there has been a shift in the routes of administration, lipophilicity, enzyme-mediated metabolism (major and minor), and the role of xenobiotic transporters in drug disposition between the most prescribed drugs in the US market vs. medications that received FDA approval between 2005 and 2016. To achieve this goal, the portions of drugs administered via oral or intravenous routes in both established and newly approved lists were compared. No significant change was found in the percentage of drugs administered via oral and intravenous routes between the established and newly approved lists. Meanwhile, drugs with the newly approved route of administration including buccal, nasal, local injection, and subcutaneous was not found in the established list.
Lipophilicity is one of the most important physicochemical characteristics of pharmaceuticals, which control their absorption, distribution, metabolism, and excretion properties in the body. Trends have been reported between an increase in lipophilicity and a higher propensity of adverse drug reaction [33–35]. Price et al. proposed thresholds for cLogP and TPSA at 3.0 and 75 Å2, respectively, as a unique predictor for drug-related toxicity in vivo [23]. He suggested that compounds with high-cLogP (cLog P > 3) and low TPSA (< 75 Å2) were ~ 2.5 times more likely to show an adverse reaction in vivo. However, some reports stated that the fundamental physiochemical properties of drugs have not changed over time [36]. To understand the conflicting information in the literature, we explored if any shift is present in the physiochemical properties of pharmaceuticals.
In our analysis, we have noted that a larger number of the newly approved drugs had Mw> 500 Da, suggesting an increased probability of hepatic/biliary vs. renal elimination [37, 38]. We also noticed a significant increase in newly approved drugs with cLogP > 3 and TPSA > 75 Å2, illustrating a trend towards an increase in newer compounds compared with older drugs under this category. A small decrease in compounds with high-cLogP and low TPSA (23% vs. 16% in established and newly approved lists, respectively) was found, which may be a testimony to the concerted effort by the pharmaceutical industry to market less toxic drugs. These observations suggest that newer drugs are more likely to exhibit an improved safety profile.
Cerny evaluated the contribution of CYP and non-CYP enzymes in the metabolism of newly FDA-approved drugs based on major metabolite formation [14]. However, this analysis focused on the FDA-approved drugs in the preceding 10 years. Furthermore, prodrugs were excluded from the overall contribution of metabolic enzymes. Similarly, Rendic and Guengerich discussed the role of UGTs and esterases in non-CYPs accounting for 25% of major metabolism [39]. Nevertheless, they focused mainly on the chemical carcinogens and their metabolic biotransformation. According to the review presented by Williams et al., the prominent role of UGT in non-CYPs was reported to contribute to oneseventh of the overall major metabolism of the top 200 most prescribed drugs from 2002 (1 in 13 compounds) [3]. However, again, the assessment had been made 15 years prior to this review, and it was critical to summarize and compare the trend with newer compounds. Furthermore, among the reported literature accounting for metabolism, data are driven by different approaches and criteria making it challenging to compare the analysis between different publications. This warranted a more systematic assessment, comparing the established vs. newly approved drugs using similar criteria, which we have addressed within this evaluation.
In this review, we have also included the metabolism of the active form of prodrugs in major and minor pathway assessments. Additionally, the contribution of CYP and non-CYP enzymes in major and minor metabolism were accounted for using DIDB® monographs. Regardless of the different approach employed, comparisons were made with other literature based on major metabolism. We observe the contribution of CYPs and non-CYPs (54% and 28%) in newly approved compounds is consistent with the reported literature [14]. As reported previously, the main CYPs are still 3A4, 2D6, 2C8, 2C9, 2C19, and 1A2 [3, 39]. However, a predominant role of CYP3A5 in major metabolism was observed in the newer drugs. Prominence of CYP3A5 could be the result of increased awareness and/or differentiating reagents not previously available [30–32, 39]. Within the non-CYP enzymes, glucuronidation plays a major role in metabolism. However, among the newly approved drugs, the role of UGT as major metabolizing enzymes was decreased by 7% while a significant increase (16%) in the role of “other enzymes” was noted.
With regard to minor metabolism, there is a significant shift in CYPs from 18% to 31% and non-CYPs from 4 to 11% in established vs. newly approved drug categories, respectively. All effective non-CYPs involved in the metabolism of established drugs correspond to phase II drug metabolizing enzymes such as UGTs and SULTs. However, among the newly approved drugs, metabolism by FMO/MAO has exceeded that of SULT enzymes with ~ 22% of minor metabolism by non-CYPs that can be attributed to FMO/MAO enzymes.
Among 156 and 237 compounds in established vs. newly approved drug lists, 21 and 74 drugs were substrates for xenobiotic transporters, respectively. OATP1B1 emerged as the most characterized uptake transporter. The number of active agents that were the substrate for OATP1B1 were 8 and 9 for the established and newly approved drugs, respectively. Among the efflux transporters, P-gp played a pivotal role in drug transport followed by BCRP and MRP. The OATP1B1 influx transporter is hepatocytes and consists of 691 amino acids, which is encoded by the solute carrier family 1B1 (SLCO1B1) gene [40]. Two common and clinically relevant single nucleotide polymorphisms have been reported for SLCO1B1 that may play a significant role in drug–drug interactions [41]. The c.521T>C (p.Val174Ala) and c.388A>G (p.Asn130Asp) variant alleles have been shown to alter the PK parameters of several statins [42]. A higher contribution of OATP1B1 in the metabolism of drugs may arise from the SLCO1B1 polymorphism, which can affect the pharmacokinetics of a wide range of drug substrates including statins, antidiabetic agents (e.g., repaglinide), and antihistamines (e.g., fexofenadine).
OATP and P-gp were considered as the highest uptake and efflux transporters in both drug lists not only owing to the availability of well-characterized assays in comparison to the other transporters, but also because of their broad range of substrate specificity [40, 43–45]. Because of limited available in vivo data on transporters, in vitro transport studies conducted on Madin–Darby canine kidney MDR1 multidrug resistance gene 1 (MDCK-MDR1) and Caco-2 cell lines were employed to show the contribution of different transporters. Notably, the MDCK-MDR1 cell line represents superior advantages in the expression of active P-gp and circumvents the complexities of multiple transporters that are present in Caco-2.
It is important to acknowledge the limitations of this analysis. For several established drugs, only a portion of the mass balance studies was reported, or the available metabolism data were incomplete owing to a lack of in-depth screening of the drugs in the earlier years. In the absence of mass-balance studies, in vitro data reported in the literature, as well as from DIDB® were used for the analysis of metabolic pathways. Glucuronide conjugates are not stable in feces, and therefore, their contribution to the total metabolism may be underappreciated. Moreover, some drugs, such as buprenorphine, are metabolized by multiple UGTs such as UGT1A1, UGT1A3, and UGT2B7 as well as CYP3A4. As a result, all enzymes are given an equal weight of 1 when assigning major and minor metabolic pathways. However, this could introduce a bias towards overestimating the role of UGTs and underestimating the role of CYP3A4. This issue could be mitigated if information on the fraction metabolized was available; however, the fraction metabolized value for each enzyme was not available for the majority of the drugs included in this review. In addition, special case scenarios such as compounds with low metabolism have not been addressed because of the sparse information available for most drugs. In the case of transporters, it is important to note that older drugs were evaluated in Caco-2, while MDCK may have been used for newer drugs. This shift in in vitro assays could introduce a bias in evaluation. However, this represents a preliminary evaluation, as the information obtained is not from in vivo systems and hence their tissue-specific role is yet to be explored.
5. Conclusion
Our systematic analysis demonstrates the growing significance of protein-based therapeutics in recent years with a clear majority of monoclonal antibodies approved in the last decade for the treatment of cancer, inflammatory disorders diseases, and rare diseases. Our analysis confirmed a 4.5-fold increase in drugs with cLog P > 3 and TPS A ≥ 75 Å2 along with a threefold increase in drugs with a Mw > 500 Da in established vs. newly approved drug lists. With this analysis, we anticipate that the newly approved drugs may show a decreased incidence of adverse effects in vivo. Our analysis also shows an increase in the CYP3A4/5-mediated major metabolism in recent years, along with the increased involvement of non-CYP-mediated metabolism in drugs approved from 2005 to 2016. Other results show the role of transporters from in vitro data with OATP1B1 being the main uptake transporter in both lists and P-gp as the most widely studied efflux transporter. With this analysis, we maintained the same set of criteria to compare the older drugs and more recently approved drugs in the market. However, this analysis is focussed on the approved drugs in the market, whose drug development and design are at least a decade behind that of molecules currently undergoing drug development. Therefore, further reviews on the investigational agents are warranted to establish the future direction of pharmaceutical products with respect to clearance pathways.
Supplementary Material
Key points.
Routes of administration, metabolism/physicochemical properties, and transporter-mediated disposition of the top 200 most prescribed drugs in 2014 (established drugs) were compared with the drugs that had received Food and Drug Administration (FDA) approval between 2005 and 2016 (newly approved drugs).
The higher rate of small molecules with a molecular weight >300 in the newly approved list confirmed that the newer drugs are more prone to undergo hepatic clearance than the established drugs.
The newly approved drugs showed a lower probability to be toxic than the established drugs based on their physicochemical properties (cLogP and total polar surface area).
The contribution of cytochromes P450 (CYPs), as major metabolizing enzymes, showed an increase in newly approved drugs in comparison to established drugs.
A greater contribution of CYPs, as minor metabolizing enzymes, was identified in the newly approved lists, with CYP3A4 and CYP2C19 being most prominent, for the established and newly approved drugs, respectively.
The emerging role of flavin-containing monooxygenase/monoamine oxidase (FMO/MAO), as the minor non-CYP enzymes, in the newly approved drug list, was notable.
The uptake transporter, Organic Anion Transporting Polypeptide 1B1 (OATP1B1) and the efflux transporter, P-glycoprotein were the most prevalent transporters in both lists.
Acknowledgements
The authors thank Mr. Timothy Lee, PharmD student for assistance in retrieving information for this study. The authors thank the University of Washington Drug Interaction Database, specifically Dr. Isabelle Ragueneau-Majlessi for providing feedback of the criteria for defining metabolism pathways. The authors also sincerely thank Dr. Kristina Ward for her contribution to the data extraction from the CenterWatch database.
Funding Support from Grant No. R15 GM101599 from the National Institutes of Health is gratefully acknowledged.
Footnotes
Conflict of interest Anitha Saravanakumar, Armin Sadighi, Rachel Ryu, and Fatemeh Akhlaghi have no conflicts of interest that are directly relevant to the content of this article.
References
- 1.Oda S, Fukami T, Yokoi T, Nakajima M. A comprehensive review of UDP-glucuronosyltransferase and esterases for drug development. Drug Metab Pharmacokinet. 2015;30(1):30–51. [DOI] [PubMed] [Google Scholar]
- 2.Iyanagi T Molecular mechanism of phase I and phase II drugmetabolizing enzymes: implications for detoxification. Int Rev Cytol. 2007;260:35–112. [DOI] [PubMed] [Google Scholar]
- 3.Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, et al. Drug-drug interactions for UDP-glucuronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab Dispos. 2004;32(11):1201–8. [DOI] [PubMed] [Google Scholar]
- 4.Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14(1):1–18. [DOI] [PubMed] [Google Scholar]
- 5.Nebert DW, Russell DW. Clinical importance of the cytochromes P450. Lancet. 2002;360(9340):1155–62. [DOI] [PubMed] [Google Scholar]
- 6.Sim SC, Ingelman-Sundberg M. Update on allele nomenclature for human cytochromes P450 and the human cytochrome P450 allele (CYP-allele) nomenclature database. Methods Mol Biol. 2013;987:251–9. [DOI] [PubMed] [Google Scholar]
- 7.Walsky RL, Obach RS. Validated assays for human cytochrome P450 activities. Drug Metab Dispos. 2004;32(6):647–60. [DOI] [PubMed] [Google Scholar]
- 8.Wienkers LC, Heath TG. Predicting in vivo drug interactions from in vitro drug discovery data. Nat Rev Drug Discov. 2005;4(10):825–33. [DOI] [PubMed] [Google Scholar]
- 9.Guengerich FP, Cheng Q. Orphans in the human cytochrome P450 superfamily: approaches to discovering functions and relevance in pharmacology. Pharmacol Rev. 2011;63(3):684–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3(8):711–5. [DOI] [PubMed] [Google Scholar]
- 11.Arrowsmith J, Miller P. Trial watch: phase II and phase III attrition rates 2011–2012. Nat Rev Drug Discov. 2013;12(8):569 10.1038/nrd4090. [DOI] [PubMed] [Google Scholar]
- 12.Gan J, Ma S, Zhang D. Non-cytochrome P450-mediated bioactivation and its toxicological relevance. Drug Metab Rev. 2016;48(4):473–501. [DOI] [PubMed] [Google Scholar]
- 13.Foti RS, Dalvie DK. Cytochrome P450 and non-cytochrome P450 oxidative metabolism: contributions to the pharmacokinetics, safety and efficacy of xenobiotics. Drug Metab Dispos. 2016;44(8):1229–45. [DOI] [PubMed] [Google Scholar]
- 14.Cerny MA. Prevalence of non-cytochrome P450-mediated metabolism in food and drug administration-approved oral and intravenous drugs: 2006–2015. Drug Metab Dispos. 2016;44(8):1246–52. [DOI] [PubMed] [Google Scholar]
- 15.Pryde DC, Dalvie D, Hu Q, Jones P, Obach RS, Tran TD. Aldehyde oxidase: an enzyme of emerging importance in drug discovery. J Med Chem. 2010;53(24):8441–60. [DOI] [PubMed] [Google Scholar]
- 16.Leeson PD. Molecular inflation, attrition and the rule of five. Adv Drug Deliv Rev. 2016;101:22–33. [DOI] [PubMed] [Google Scholar]
- 17.DeGorter MK, Xia CQ, Yang JJ, Kim RB. Drug transporters in drug efficacy and toxicity. Annu Rev Pharmacol Toxicol. 2012;52:249–73. [DOI] [PubMed] [Google Scholar]
- 18.International Transporter Consortium, Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9(3):215–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusuhara H, Sugiyama Y. In vitro-in vivo extrapolation of transporter-mediated clearance in the liver and kidney. Drug Metab Pharmacokinet. 2009;24(1):37–52. [DOI] [PubMed] [Google Scholar]
- 20.Shitara Y, Horie T, Sugiyama Y. Transporters as a determinant of drug clearance and tissue distribution. Eur J Pharm Sci. 2006;27(5):425–46. [DOI] [PubMed] [Google Scholar]
- 21.Symphony Health Solutions. 2015. http://symphonyhealth.com/wp-content/uploads/2015/05/Top-200-Drugs-of-2014.pdf. Accessed 18 Dec 2017.
- 22.CenterWatch. 2018. Available from: http://www.centerwatch.com/drug-information/fda-approved-drugs/year. Accessed Mar 2018.
- 23.Price DA, Blagg J, Jones L, Greene N, Wager T. Physicochemical drug properties associated with in vivo toxicological outcomes: a review. Expert Opin Drug Metab Toxicol. 2009;5(8):921–31. [DOI] [PubMed] [Google Scholar]
- 24.University of Washington Metabolism and Transport Drug Interaction Database. 2019. https://sop.washington.edu/department-ofpharmaceutics/research/metabolism-and-transport-drug-interaction-database. Accessed Feb 2019. [DOI] [PMC free article] [PubMed]
- 25.US Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER). 2017. Draft guidance for industry. In vitro metabolism and transporter mediated drug-drug interactions, https://www.fda.gov/downloads/Drugs/Guidances/UCM581965.pdf. Accessed Feb 2019. [Google Scholar]
- 26.Hachad H, Ragueneau-Majlessi I, Levy RH. A useful tool for drug interaction evaluation: the University of Washington Metabolism and Transport Drug Interaction Database. Hum Genomics. 2010;5(1):61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu J, Ritchie TK, Mulgaonkar A, Ragueneau-Majlessi I. Drug disposition and drug-drug interaction data in 2013 FDA new drug applications: a systematic review. Drug Metab Dispos. 2014;42(12):1991–2001. [DOI] [PubMed] [Google Scholar]
- 28.Yu J, Ritchie TK, Zhou Z, Ragueneau-Majlessi I. Key findings from preclinical and clinical drug interaction studies presented in new drug and biological license applications approved by the Food and Drug Administration in 2014. Drug Metab Dispos. 2016;44(1):83–101. [DOI] [PubMed] [Google Scholar]
- 29.Yu J, Zhou Z, Owens KH, Ritchie TK, Ragueneau-Majlessi I. What can be learned from recent new drug applications? A systematic review of drug interaction data for drugs approved by the US FDA in 2015. Drug Metab Dispos. 2017;45(1):86–108. [DOI] [PubMed] [Google Scholar]
- 30.Tseng E, Walsky RL, Luzietti RA, Harris JJ, Kosa RE, Goosen TC, et al. Relative contributions of cytochrome CYP3A4 versus CYP3A5 for CYP3A-cleared drugs assessed in vitro using a CYP3A4-selective inactivator (CYP3cide). Drug Metab Dispos. 2014;42(7):1163–73. [DOI] [PubMed] [Google Scholar]
- 31.Chen SY, Li JL, Meng FH, Wang XD, Liu T, Li J, et al. Individualization of tacrolimus dosage basing on cytochrome P450 3A5 polymorphism: a prospective, randomized, controlled study. Clin Transplant. 2013;27(3):E272–81. [DOI] [PubMed] [Google Scholar]
- 32.Thervet E, Anglicheau D, King B, Schlageter MH, Cassinat B, Beaune P, et al. Impact of cytochrome P450 3A5 genetic polymorphism on tacrolimus doses and concentration-to-dose ratio in renal transplant recipients. Transplantation. 2003;76(8):1233–5. [DOI] [PubMed] [Google Scholar]
- 33.McEuen K, Borlak J, Tong W, Chen M. Associations of drug lipophilicity and extent of metabolism with drug-induced liver injury. Int J Mol Sci. 2017;18(7):1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leeson PD, Davis AM. Time-related differences in the physical property profiles of oral drugs. J Med Chem. 2004;47(25):6338–48. [DOI] [PubMed] [Google Scholar]
- 35.Amidon GL, Lennernas H, Shah VP, Crison JR. A theoretical basis for a biopharmaceutic drug classification: the correlation of in vitro drug product dissolution and in vivo bioavailability. Pharm Res. 1995;12(3):413–20. [DOI] [PubMed] [Google Scholar]
- 36.Reichert JM. Trends in development and approval times for new therapeutics in the United States. Nat Rev Drug Discov. 2003;2(9):695–702. [DOI] [PubMed] [Google Scholar]
- 37.Yang X, Gandhi YA, Duignan DB, Morris ME. Prediction of biliary excretion in rats and humans using molecular weight and quantitative structure-pharmacokinetic relationships. AAPS J 2009;11(3):511–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hosey CM, Broccatelli F, Benet LZ. Predicting when biliary excretion of parent drug is a major route of elimination in humans. AAPS J. 2014;16(5):1085–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rendic S, Guengerich FP. Survey of human oxidoreductases and cytochrome P450 enzymes involved in the metabolism of xenobiotic and natural chemicals. Chem Res Toxicol. 2015;28(1):38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth M, Obaidat A, Hagenbuch B. OATPs, OATs and OCTs: the organic anion and cation transporters of the SLCO and SLC22A gene superfamilies. Br J Pharmacol. 2012;165(5):1260–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalliokoski A, Niemi M. Impact of OATP transporters on pharmacokinetics. Br J Pharmacol. 2009;158(3):693–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romaine S, Bailey K, Hall A, Balmforth A. The influence of SLCO1B1 (OATP1B1) gene polymorphisms on response to statin therapy. Pharmacogenomics J. 2010;10(1):1–11. [DOI] [PubMed] [Google Scholar]
- 43.Amin ML. P-glycoprotein inhibition for optimal drug delivery. Drug Target Insights. 2013;19(7):27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.König J, Seithel A, Gradhand U, Fromm MF. Pharmacogenomics of human OATP transporters. Naunyn Schmiedebergs Arch Pharmacol. 2006;372(6):432–43. [DOI] [PubMed] [Google Scholar]
- 45.Lin JH, Yamazaki M. Role of P-glycoprotein in pharmacokinetics: clinical implications. Clin Pharmacokinet. 2003;42(1):59–98. [DOI] [PubMed] [Google Scholar]
- 46.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drugs.com. 2015. Available from: https://www.drugs.com. Accessed 18 Dec 2017.
- 48.Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(D1):D1202–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.