Abstract
Background
Persistent cervical high-risk human papillomavirus (hrHPV) infection is a necessary cause of cervical cancer. However, the host genetic factors underlying its risk are not well understood. We hypothesized that immunogenetic variation plays a role in hrHPV infection and persistence. Therefore, we conducted a study of classical HLA alleles and their association with hrHPV infection and persistence among women.
Methods
We characterized HPV infection using SPF10/LiPA25 in Nigerian women at baseline and at 6 months follow-up visits in 2014. hrHPV infection was prevalent if at least one carcinogenic HPV genotype was detected at the baseline visit and persistent if at least one carcinogenic HPV genotype was detected at the baseline and follow-up visits. Classical HLA alleles were imputed from genotypes in the MHC region using the HLA genotype imputation with attribute bagging (HIBAG) algorithm. HLA association tests were conducted under additive genetic models.
Results
The mean (±SD) age of the 517 study participants was 38 (±8) years, 48% were HIV negative, 24% were hrHPV positive at baseline and 10% had persistent hrHPV infections. In multivariate regression models adjusted for age, HIV status and the first principal component, DQA1*01:02 and DQA1*02:01 were positively associated with prevalent but not persistent hrHPV infections, while DQA1*05:01 was negatively associated with prevalent hrHPV but positively associated with persistent cervical hrHPV infections. Four haplotypes (A*30:01-DQA1*05:01, B*07:02-C*07:02, B*07:02-DQA1*05:01 and C*07:02-DQA1*05:01) were significantly associated with prevalent cervical hrHPV infections and several haplotypes that included the DQA1*05:01 allelic variant were significantly associated with persistent cervical hrHPV infections. Six amino acid positions on DQα1 were associated with prevalent but not persistent cervical hrHPV infections.
Conclusions
In this first study to investigate the association between HLA alleles and persistent hrHPV in African women, we identified important risk alleles that merit further investigation. Our findings provide new insights into risk factors for hrHPV infection in African ancestry women.
Introduction
Persistent cervical infection with high-risk (hr) human papillomavirus (HPV) is necessary for the development of cervical cancer, the fourth leading cancer in women globally. In 2018, there were 569,847 incident cases and 311,365 deaths from cervical cancer globally [1]. While several behavioral and phenotypic cofactors for persistent cervical hrHPV infection have been identified, including cigarette smoking, multiple sexual partners and co-infection with HIV [2–4], the genetic factors associated with cervical hrHPV persistence are not well elucidated. It has been hypothesized that the human leucocyte antigen (HLA) complex, a set of genes involved in activating the cell-mediated immune response and identifying autologous and foreign proteins, is a cofactor for persistence of cervical hrHPV infections.
The HLA loci is the most polymorphic genetic loci in humans [5]. The highly polymorphic HLA molecules play a major role in immune response by binding and presenting foreign antigens, including HPV peptides, to T lymphocytes, thus promoting immune recognition and subsequent clearance of infected cells [6]. Individuals with HLA polymorphisms may produce proteins that have lower binding affinity to HPV antigens, are less likely to recognize cervical HPV infections, and thus have higher likelihood of associated persistent infections.
While most studies have focused on the associations between HLA and cervical cancer [7–16], few studies have examined the distribution of HLA alleles and their associated risk with hrHPV infections. One study among Canadian women showed that HLA-G alleles or genotypes were not significantly associated with persistence of alpha group 2 hrHPV or any HPV types [17]. On the other hand, Brazilian women who had cervical HPV infections were shown to be more likely to have DRB1*1503, DRB1-0405 and DQB1*0602 HLA alleles, compared to women without cervical HPV infections [18]. Till date, there are no previous studies on the association between HLA and cervical hrHPV infections among women in sub-Saharan Africa (SSA). In this study, we investigated the association between classical HLA alleles and cervical hrHPV infections, among African women.
Materials and Methods
Study Population
We examined 544 women from a cohort study of HPV infection and cervical cancer at National Hospital, Abuja and University of Abuja Teaching Hospital, Nigeria, who were enrolled between 2012 and 2014, as previously described [2, 19–21]. All the study participants were 18 years or older, had a history of vaginal sexual intercourse, were not currently pregnant and had no history of cervical disease or hysterectomy. We collected data on socio-demographic characteristics, sexual and reproductive history, and self-reported HIV status of the participants. We confirmed participants’ HIV status from hospital medical records. We collected venous blood samples and performed pelvic examinations on all study participants at each study visit. Elution swab system (Copan, Italy) was used to collect exfoliated cervical cells, which were inserted in 1 ml Amies’ transport media (Copan). Participants were asked to return for follow-up after six months, at which time, the history, physical examinations and sample collections were repeated.
HPV detection by SPF10/LiPA25
We extracted DNA from the cervical exfoliated cells as previously described [22]. Samples were tested for HPV DNA by hybridization of SPF10 amplimers to a mixture of general HPV probes recognizing a broad range of high-risk, low-risk, and possible hrHPV genotypes in a microtiter plate format, as described previously [23]. All samples determined to be HPV DNA positive by SPF10 DNA Enzyme Immunoassay (DEIA) were genotyped using the LiPA25 version 1. The LiPA25 assay provides type-specific information for 25 different HPV genotypes simultaneously and identifies infection by one or more of 13 hrHPV genotypes: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 [24, 25]. However, as this assay does not differentiate between HPV 68 and 73, we defined this HPV genotype (i.e. HPV68/73) as low-risk. We defined hrHPV infection as prevalent if at least one hrHPV genotype was detected in the baseline sample and persistent if at least one hrHPV genotype was detected in samples provided at both the baseline and follow-up visits. We defined persistently negative as absence of hrHPV genotype in the baseline or follow-up visit samples.
Statistical Analysis
From the original set of 544 women, we excluded 27 women from the baseline analyses because of incomplete data (5 missing HPV, 22 missing both HPV and HIV results), leaving only 517 women in the baseline analyses. Of the 517 women, we excluded those who did not return for the follow-up visit (n=9) and those with missing HPV results (n=35) and included the remaining 473 women in the analyses for persistent hrHPV infections. To describe the baseline characteristics of the study population, we used t-tests to assess differences in the distribution of continuous variables, and χ2 and Fisher’s exact tests for categorical variables. Using dense SNP genotype data available on the same women, we computed principal components based on the variance-standardized relationship matrix using PLINK 1.9 [26, 27]. We found that the first principal component was significant and included it in downstream association analyses.
HLA Genotype Imputation
Samples from the study participants were genotyped using the lllumina Multi-Ethnic Global Array (MEGA). After technical quality control, sample-level genotype call rate was at least 0.95 for all the study participants. The HLA genotype imputation with attribute bagging (HIBAG) algorithm was used to derive high-resolution HLA types [28]. Attribute bagging is a technique for improving the accuracy and stability of classifier ensembles deduced using bootstrap aggregating and random subsets of variables [29]. HIBAG makes predictions by averaging HLA-type posterior probabilities over an ensemble of classifiers for European, Asian, Hispanic and African ancestries [28]. The prediction accuracies for HLA-A, B, C, DRB1 and DQB1 ranged from 92.2% to 98.1%[28].
Association analysis
Association analysis was done using PyHLA [30], a software package designed for association analysis of the HLA types imputed from genome-wide genotyping and next-generation sequencing (NGS) data. We used additive logistic regression models adjusted for age, HIV status and the first principal component, to test for associations between four-digit HLA allele and both prevalent and persistent cervical hrHPV infections. Fisher’s exact tests and calculated odds ratio with Haldane’s correction of Woolf’s method were used to examine the association between HLA haplotype, amino acid sequence, allele and residual zygosity, and both prevalent and persistent cervical hrHPV infections. The zygosity tests evaluated the frequency difference of carrying the homozygous and heterozygous alleles and the absence of a particular allele or amino acid in women with hrHPV infections compared to those without hrHPV. When an allele or residual was associated (p < 0.05) with the disease, these tests were performed to identify whether a homozygote or heterozygote condition differentiates susceptibility to the disease, p values were adjusted for multiple comparisons with the false discovery rate (FDR) method.
Ethics
The study was conducted according to the Nigerian National Code for Health Research Ethics. Ethical approval to conduct this study was obtained from the Institute of Human Virology Nigeria research ethics committee. Written informed consent was obtained from all participants before enrollment in the study.
Results
The mean (±SD) age of the 517 participants was 38 (±8) years while their mean (±SD) body mass index (BMI) was 27 (±6) kg/m2. About half of the participants were HIV positive (52%, 270/517). The characteristics of the study participants at baseline are shown in Table 1. Participants returned for follow-up visits at a median (IQR) time of 6.0 (5.8 – 8.1) months.
Table 1:
Characteristics of the Study Participants at Baseline by Cervical hrHPV Status
| Characteristics | Total (n = 517) | hrHPV Negative (n = 392) | hrHPV Positive (n = 125) |
|---|---|---|---|
| Mean (± standard deviation) |
|||
| Age, years | 38 (± 8) | 38 (± 8) | 37 (± 7) |
| Body Mass Index, kg/m2 | 27 (± 6) | 27 (± 8) | 27 (± 8) |
| Age at sexual initiation, years | 20 (± 4) | 20 (± 4) | 19 (± 3) |
| Total sex partners | 3 (± 3) | 3 (± 3) | 4 (± 3) |
| n (%) |
|||
| Age, years | |||
| - 18 - 29 | 55 (11) | 40 (10) | 15 (12) |
| - 30 - < 45 | 357 (69) | 273 (70) | 84 (67) |
| - ≥ 45 | 105 (20) | 79 (20) | 26 (21) |
| Body Mass Index, kg/m2 | |||
| - Normal Weight, 18.5 - 24.9 | 173 (35) | 127 (34) | 46 (40) |
| - Overweight, 25.0 - 29.9 | 171 (35) | 130 (35) | 41 (35) |
| - Obese, ≥ 30.0 | 145 (30) | 116 (31) | 29 (25) |
| Age at sexual initiation, years | |||
| - <18 | 106 (21) | 80 (21) | 26 (21) |
| - 18 - 21 | 248 (49) | 188 (49) | 60 (49) |
| - ≥ 22 | 150 (30) | 114 (30) | 36 (29) |
| Total sex partners | |||
| - 1 | 122 (24) | 101 (26) | 21 (17) |
| - 2 - 4 | 202 (40) | 152 (39) | 50 (41) |
| - ≥ 5 | 186 (36) | 135 (35) | 51 (42) |
| HIV Status | |||
| - Positive | 270 (52) | 182 (46) | 88 (70) |
| - Negative | 247 (48) | 210 (54) | 37 (30) |
hrHPV= high-risk human papillomavirus
Prevalent hrHPV Infection
At baseline, about a quarter of the study participants (24%, 125/517) had hrHPV infections. The distribution of type-specific cervical hrHPV Infections by HIV Status at Baseline is shown in Table 2. For subsequent analyses, we compared 125 women with cervical hrHPV infections to 392 women without cervical hrHPV infections at baseline. DQA1*01:02 was the most prevalent HLA allele in the study population (frequency = 0.39). This allele was present in 60% of the women who were hrHPV negative (frequency = 0.36) and 71% of the women who were hrHPV positive (frequency = 0.46), Table 3. Supplemental table 1 shows the frequency distribution of all the HLA alleles identified among the study participants at baseline.
Table 2.
Distribution of Type-Specific Cervical High-Risk HPV Infections by HIV Status at Baseline
| HPV Type | Total (n = 517) | HIV Positive (n = 270) | HIV Negative (n = 247) |
|---|---|---|---|
| High-risk HPV Positive | 125 (24.2 %) | 88 (32.6 %) | 37 (14.9 %) |
| HPV 16 | 10 (1.9 %) | 8 (2.9 %) | 2 (0.8 %) |
| HPV 18 | 20 (3.9 %) | 15 (5.5 %) | 5 (2.0 %) |
| HPV 31 | 11 (2.1 %) | 10 (3.7 %) | 1 (0.4 %) |
| HPV 33 | 16 (3.1 %) | 13 (4.8 %) | 3 (1.2 %) |
| HPV 35 | 27 (5.2 %) | 22 (8.1 %) | 5 (2.0 %) |
| HPV 39 | 7 (1.3 %) | 5 (1.8 %) | 2 (0.8 %) |
| HPV 45 | 9 (1.7 %) | 5 (1.8 %) | 4 (1.6 %) |
| HPV 51 | 9 (1.7 %) | 8 (2.9 %) | 1 (0.4 %) |
| HPV 52 | 37 (7.1 %) | 25 (9.2 %) | 12 (4.8 %) |
| HPV 56 | 9 (1.7 %) | 5 (1.8 %) | 4 (1.6 %) |
| HPV 58 | 10 (1.9 %) | 8 (2.9 %) | 2 (0.8 %) |
| HPV 59 | 7 (1.3 %) | 7 (2.5 %) | 0 (0.0 %) |
Table 3.
Frequency Distribution of Top 10 HLA Alleles by Cervical hrHPV Status
| Total (n=517) | hrHPV Negative (n=392) | hrHPV Positive (n=125) | ||||||
|---|---|---|---|---|---|---|---|---|
| HLA Allele | Count | Frequency | Allele Count | Allele Frequency | Population Frequency | Allele Count | Allele Frequency | Population Frequency |
| Prevalent hrHPV | ||||||||
| DQA1*01:02 | 402 | 0.388 | 286 | 0.364 | 0.602 | 116 | 0.464 | 0.712 |
| DPB1*01:01 | 355 | 0.343 | 273 | 0.348 | 0.553 | 82 | 0.328 | 0.560 |
| DQB1*06:02 | 284 | 0.274 | 204 | 0.260 | 0.443 | 80 | 0.320 | 0.520 |
| C*04:01 | 282 | 0.272 | 210 | 0.267 | 0.451 | 72 | 0.288 | 0.472 |
| DRB1*15:03 | 187 | 0.180 | 134 | 0.170 | 0.298 | 53 | 0.212 | 0.360 |
| DPB1*02:01 | 181 | 0.175 | 133 | 0.169 | 0.301 | 48 | 0.192 | 0.360 |
| B*53:01 | 180 | 0.174 | 133 | 0.169 | 0.306 | 47 | 0.188 | 0.320 |
| DQB1*03:01 | 174 | 0.168 | 138 | 0.176 | 0.324 | 36 | 0.144 | 0.248 |
| A*23:01 | 168 | 0.162 | 125 | 0.159 | 0.290 | 43 | 0.172 | 0.336 |
| DQA1*05:05 | 146 | 0.141 | 113 | 0.144 | 0.262 | 33 | 0.132 | 0.208 |
| Persistent hrHPV | ||||||||
| Total (n=406) | Persistent hrHPV Negative (n=355) | Persistent hrHPV Positive (n=51) | ||||||
| HLA Allele | Count | Frequency | Allele Count | Allele Frequency | Population Frequency | Allele Count | Allele Frequency | Population Frequency |
| DQA1*01:02 | 298 | 0.367 | 257 | 0.362 | 0.594 | 41 | 0.402 | 0.666 |
| DPB1*01:01 | 276 | 0.339 | 234 | 0.3296 | 0.521 | 42 | 0.411 | 0.666 |
| C*04:01 | 216 | 0.266 | 192 | 0.2704 | 0.459 | 24 | 0.235 | 0.333 |
| DQB1*06:02 | 214 | 0.263 | 187 | 0.2634 | 0.445 | 27 | 0.264 | 0.431 |
| DRB1*15:03 | 147 | 0.181 | 131 | 0.1845 | 0.318 | 16 | 0.156 | 0.254 |
| DQB1*03:01 | 142 | 0.174 | 129 | 0.1817 | 0.329 | 13 | 0.127 | 0.254 |
| B*53:01 | 140 | 0.172 | 124 | 0.1746 | 0.312 | 16 | 0.156 | 0.294 |
| DPB1*02:01 | 137 | 0.168 | 122 | 0.1718 | 0.307 | 15 | 0.147 | 0.274 |
| A*23:01 | 134 | 0.165 | 112 | 0.1577 | 0.290 | 22 | 0.215 | 0.392 |
| DQA1*05:05 | 122 | 0.150 | 112 | 0.1577 | 0.281 | 10 | 0.098 | 0.196 |
hrHPV= high-risk human papillomavirus
Several HLA alleles were associated with prevalent hrHPV infections (Table 4, Figure 1). In unadjusted regression models (model 1), the odds ratio (OR), 95% confidence interval (CI) and p values were 0.35 (0.15 – 0.84), p 0.13 for C*07:02, 1.52 (1.13 – 2.03), p 0.03 for DQA1*01:02, 1.78 (1.08 – 2.92), p 0.05 for DQA1*02:01 and 0.49 (0.26 – 0.91), p 0.05 for DQA1*05:01. With adjustment for age and HIV status (model 2), most of these effect sizes became larger. With further adjustment for the first principal component in the fully adjusted models (model 3), DQA1*01:02 (OR: 1.46, 95% CI: 1.08 – 1.97, p 0.03), DQA1*02:01 (OR: 1.89, 95% CI: 1.12 – 3.15, p 0.03) and DQA1 *05:01 (OR: 0.45, 95% Cl: 0.24 – 0.85, p 0.03) were significantly associated, while C*07:02 (OR: 0.30, 95% Cl: 0.13 – 0.75, p 0.06) was borderline significantly associated with cervical hrHPV infection at baseline (Table 4). Results of the zygosity test for these HLA alleles showed that among women with one C*07:02 or DQA1*01:02 allele, heterozygosity for that allele was associated with higher odds of cervical hrHPV infections. While the presence of one homozygous and one heterozygous DQA1 *02:01 allele, was associated with higher odds of cervical hrHPV infections (Table 5). Additionally, four haplotypes, A*30:01 - DQA1*05:01, B*07:02 - C*07:02, B*07:02 - DQA1*05:01 and C*07:02 - DQA1*05:01, were significantly associated with prevalent cervical hrHPV infections at baseline (Table 6).
Table 4.
Association between HLA Alleles and Cervical hrHPV Infection
| Prevalent hrHPV | Persistent hrHPV | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Models | Allele | Frequency Controls | Frequency Cases | Odds Ratio (95% CI) | P value* (Adjusted) | Allele | Frequency Controls | Frequency Cases | Odds Ratio (95% CI) | P value* (Adjusted) |
| Model 1 | 1.00 (Ref) | |||||||||
| B*07:02 | 0.077 | 0.036 | 0.429 (0.207 – 0.887) | 0.157 | B*15:10 | 0.042 | 0.117 | 2.736 (1.381 – 5.421) | 0.027 | |
| C*07:02 | 0.063 | 0.024 | 0.353 (0.149 – 0.842) | 0.132 | DQA1*05:01 | 0.078 | 0.166 | 2.560 (1.359 – 4.819) | 0.021 | |
| DQA1*01:02 | 0.364 | 0.464 | 1.518 (1.132 – 2.035) | 0.031 | DQB1*02:01 | 0.080 | 0.156 | 2.304 (1.215 – 4.361) | 0.044 | |
| DQA1*02:01 | 0.059 | 0.104 | 1.779 (1.084 – 2.920) | 0.049 | DQB1*05:01 | 0.146 | 0.058 | 0.337 (0.141 – 0.808) | 0.044 | |
| DQA1*05:01 | 0.096 | 0.052 | 0.489 (0.262 – 0.912) | 0.049 | DRB1*03:01 | 0.071 | 0.156 | 2.604 (1.366 – 4.962) | 0.032 | |
| Model 2 | 1.00 (Ref) | 1.00 (Ref) | ||||||||
| B*07:02 | 0.077 | 0.036 | 0.400 (0.191 – 0.839) | 0.108 | B*15:10 | 0.042 | 0.117 | 2.585 (1.259 – 5.307) | 0.067 | |
| C*07:02 | 0.063 | 0.024 | 0.304 (0.126 – 0.737) | 0.058 | DQA1*05:01 | 0.078 | 0.166 | 2.470 (1.277 – 4.777) | 0.043 | |
| DQA1*01:02 | 0.364 | 0.464 | 1.454 (1.076 – 1.196) | 0.030 | DQB1*02:01 | 0.080 | 0.156 | 2.099 (1.078 – 4.083) | 0.087 | |
| DQA1*02:01 | 0.059 | 0.104 | 1.900 (1.136 – 3.177) | 0.030 | DQB1*05:01 | 0.146 | 0.058 | 0.336 (0.138 – 0.881) | 0.087 | |
| DQA1*05:01 | 0.096 | 0.052 | 0.456 (0.242 – 0.859) | 0.030 | DRB1*03:01 | 0.071 | 0.156 | 2.375 (1.212 – 4.657) | 0.105 | |
| Model 3 | 1.00 (Ref) | 1.00 (Ref) | ||||||||
| B*07:02 | 0.077 | 0.036 | 0.394 (0.188 – 0.828) | 0.099 | B*15:10 | 0.042 | 0.117 | 2.602 (1.264 – 5.356) | 0.065 | |
| C*07:02 | 0.063 | 0.024 | 0.309 (0.128 – 0.749) | 0.065 | DQA1*05:01 | 0.078 | 0.166 | 2.465 (1.274 – 4.768) | 0.043 | |
| DQA1*01:02 | 0.364 | 0.464 | 1.457 (1.076 – 1.972) | 0.033 | DQB1*02:01 | 0.080 | 0.156 | 2.103 (1.080 – 4.093) | 0.085 | |
| DQA1*02:01 | 0.059 | 0.104 | 1.880 (1.122 – 3.151) | 0.033 | DQB1*05:01 | 0.146 | 0.058 | 0.339 (0.140 – 0.823) | 0.085 | |
| DQA1*05:01 | 0.096 | 0.052 | 0.449 (0.238 – 0.847) | 0.033 | DRB1*03:01 | 0.071 | 0.156 | 2.376 (1.212 – 4.657) | 0.105 | |
hrHPV= high-risk human papillomavirus
Reference Category = hrHPV negative
Model 1 = Unadjusted model
Model 2 = Adjusted for age and HIV status
Model 3 = Adjusted for age, HIV status and the first principal component
= FDR-adjusted P value
Figure 1.
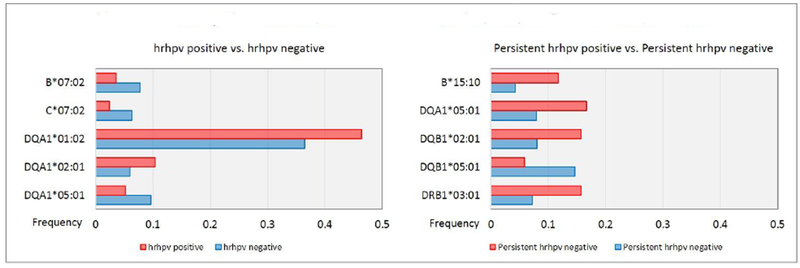
Frequency Distibution of Alleles Associated with Prevalent and Persistent Cervical hrHPV
Table 5.
Zygosity Tests for HLA Alleles Associated with Cervical hrHPV
| Homozygosity | Heterozygosity | Zygosity | ||||
|---|---|---|---|---|---|---|
| Allele | OR | P value | OR | P value | OR | P value |
| Prevalent hrHPV | ||||||
| B*07:02 | 0.492 | 0.999 | 1.876 | 0.079 | 0.262 | 0.336 |
| C*07:02 | 2.087 | 0.999 | 2.194 | 0.022 | 0.951 | 0.999 |
| DQA1*01:02 | 2.487 | 0.999 | 2.595 | 0.018 | 0.958 | 0.999 |
| DQA1*02:01 | 1.625 | 0.107 | 0.695 | 0.133 | 2.334 | 0.006 |
| DQA1*05:01 | 1.933 | 0.606 | 0.564 | 0.061 | 3.423 | 0.224 |
| Persistent hrHPV | ||||||
| B*15:10 | 5.588 | 0.155 | 0.426 | 0.062 | 13.11 | 0.036 |
| DQA1*05:01 | 10.93 | 0.222 | 0.432 | 0.017 | 25.30 | 0.107 |
| DQB1*02:01 | 11.89 | 0.208 | 0.484 | 0.046 | 24.53 | 0.110 |
| DQB1*05:01 | 2.164 | 0.999 | 2.709 | 0.015 | 0.799 | 0.999 |
| DRB1*03:01 | 10.65 | 0.227 | 0.425 | 0.022 | 25.02 | 0.108 |
hrHPV = high-risk human papillomavirus; ID = HLA Gene_Position_Amino acid residue; A = Alanine; D = Aspartic acid; E = Glutamic acid; H = Histidine; I = Isoleucine; K = Lysine; L = Leucine; M = Methionine; R = Arginine; S = Serine.
Table 6.
HLA Haplotypes Associated with Cervical hrHPV Infections
| Prevalent hrHPV | Persistent hrHPV | ||||
|---|---|---|---|---|---|
| Allele | OR | P value | Allele | OR | P value |
| A*30:01 - DQA1*05:01 | 0.99 | 0.025 | B*15:10 - DQA1*05:01 | 4.53 | 0.005 |
| B*07:02 - C*07:02 | 0.33 | 0.009 | B*15:10 - DQB1*02:01 | 4.38 | 0.006 |
| B*07:02 - DQA1*05:01 | 0.09 | 0.025 | B*15:10 - DRB1*03:01 | 4.47 | 0.005 |
| C*07:02 - DQA1*05:01 | 0.10 | 0.024 | DQA1*05:01 - DQB1*02:01 | 2.49 | 0.013 |
| DQA1*05:01 - DRB1*03:01 | 2.54 | 0.012 | |||
| DQB1*02:01 - DRB1*03:01 | 2.46 | 0.013 | |||
hrHPV= high-risk human papillomavirus
Next, we examined the association between amino acid positions and cervical hrHPV infections. We found that 6 amino acid positions on DQα1 were significantly associated with prevalent hrHPV infections at baseline. The position_amino acid residues were 230_Methionine, 70_Lysine, 75_Histidine, 77_Leucine, 75_Arginine, 98_Serine (Table 7).
Table 7.
Amino Acids Associated with Cervical hrHPV Infections
| Prevalent hrHPV | |||||||
|---|---|---|---|---|---|---|---|
| Number of Controls | Number of Cases | Odds Ratio (OR) | P value (Unadjusted) | P value Adjusted | |||
| ID | Carriers | Non-Carriers | Carriers | Non-Carriers | |||
| C_123_S | 49 | 343 | 6 | 119 | 0.38 | 0.012 | 0.012 |
| DQA1_230_M | 236 | 156 | 89 | 36 | 1.62 | 0.033 | 0.033 |
| DQA1_70_K | 45 | 347 | 24 | 101 | 1.84 | 0.034 | 0.034 |
| DQA1_75_H | 45 | 347 | 24 | 101 | 1.84 | 0.034 | 0.034 |
| DQA1_77_L | 45 | 347 | 24 | 101 | 1.84 | 0.034 | 0.034 |
| DQA1_75_R | 253 | 139 | 63 | 62 | 0.56 | 0.006 | 0.042 |
| DQA1_98_S | 170 | 222 | 37 | 88 | 0.55 | 0.006 | 0.012 |
| Persistent hrHPV | |||||||
| Number of Controls | Number of Cases | Odds Ratio (OR) | P value (Unadjusted) | P value Adjusted | |||
| ID | Carriers | Non-Carriers | ID | Carriers | |||
| DQB1_98_D | 130 | 225 | 31 | 20 | 2.66 | 0.001 | 0.003 |
| DQB1_99_I | 130 | 225 | 31 | 20 | 2.66 | 0.001 | 0.003 |
| DQB1_89_A | 107 | 248 | 26 | 25 | 2.4 | 0.004 | 0.012 |
| DQB1_103_K | 101 | 254 | 24 | 27 | 2.23 | 0.009 | 0.018 |
| DQB1_106_A | 101 | 254 | 24 | 27 | 2.23 | 0.009 | 0.018 |
| DQB1_60_S | 101 | 254 | 24 | 27 | 2.23 | 0.009 | 0.018 |
| DQB1_62_S | 101 | 254 | 24 | 27 | 2.23 | 0.009 | 0.018 |
| DQB1_69_I | 101 | 254 | 24 | 27 | 2.23 | 0.009 | 0.018 |
| DQB1_78_E | 101 | 254 | 24 | 27 | 2.23 | 0.009 | 0.018 |
| DQB1_84_L | 101 | 254 | 24 | 27 | 2.23 | 0.009 | 0.009 |
| DQB1_87_L | 101 | 254 | 24 | 27 | 2.23 | 0.009 | 0.009 |
| DQB1_62_H | 150 | 205 | 10 | 41 | 0.35 | 0.001 | 0.009 |
| DQB1_103_A | 125 | 230 | 8 | 43 | 0.36 | 0.006 | 0.012 |
| DQB1_46_L | 125 | 230 | 8 | 43 | 0.36 | 0.006 | 0.012 |
hrHPV = high-risk human papillomavirus; ID = HLA Gene_Position_Amino acid residue; A = Alanine; D = Aspartic acid; E = Glutamic acid; H = Histidine; I = Isoleucine; K = Lysine; L = Leucine; M = Methionine; R = Arginine; S = Serine
Persistent hrHPV Infection
We compared 51 women with hrHPV infection at both the baseline and follow-up visits to 355 women without hrHPV infections at either the baseline or follow-up visits. DQA1*01:02 was the most common HLA allele among the 406 women (frequency = 0.37). This allele was present in 59% of the women who were persistently cervical hrHPV negative (frequency = 0.36) and 67% of the women who were persistently cervical hrHPV positive (frequency = 0.40) – Figure 1. The frequency distribution of the top 10 alleles and all the alleles identified are shown in Table 3 and Supplemental Table 2, respectively.
In the unadjusted regression analyses, four alleles were significantly associated with higher odds of persistent cervical hrHPV infections (B*15:10, OR: 2.73, 95% Cl: 1.38 – 5.42, p 0.03; DQA1*05:01, OR: 2.56, 95% Cl: 1.36 – 4.82, p 0.02; DQB1*02:01, OR: 2.30, 95% Cl: 1.21 – 4.36, p 0.04; DRB1*03:01, OR: 2.60, 95% Cl: 1.37 – 4.96, p 0.03), while one allele (DQB1*05:01, OR: 0.34, 95% Cl: 0.14 – 0.81, p 0.04) was significantly associated with lower odds of persistent cervical hrHPV infections (Table 4). With further adjustments for age, HIV status and the first principal component, all the effect estimates were attenuated and the associations became borderline statistically significant (B*15:10, DQB1*02:01, DQB1*05:01) or not significant (DRB1*03:01), except for DQA1*05:01, which remained relatively the same in all the regression models. In the fully adjusted model, DQA1*05:01 (OR: 1.46, 95% Cl: 1.08 – 1.97, p 0.04) was still significantly associated with higher odds of persistent cervical hrHPV infection. The results of the zygosity tests showed that among women with one DQA1*05:01 allele, heterozygosity for that allele was associated with lower odds of persistent cervical hrHPV infections, OR: 0.45, p value 0.02 (Table 5). Several haplotypes that included the DQA1*05:01 allele, were also found to be significantly associated with persistent cervical hrHPV infections, including B*15:10 - DQA1*05:01, DQA1*05:01 - DQB1*02:01 and DQA1*05:01 - DRB1*03:01 (Table 6). While we found several amino acids associated with persistent cervical hrHPV infections, none of the significant amino acid residues were located on DQα1 (Table 7). The results of the zygosity tests for the amino acid residue are shown in Supplemental table 3.
In sensitivity analysis, we observed that DPB1*01*01 was the most prevalent allele (frequency=0.42) among women with type-specific cervical hrHPV infections (n=37) and DQA1*05*05 was significantly associated with lower odds of type-specific persistent cervical hrHPV infections (OR: 0.09, 95% Cl: 0.01 – 0.47, p 0.01).
Discussion
This is the first study to examine the association between HLA alleles, haplotypes and amino acids, and cervical hrHPV infections among African women. Notably, DQA1*01:02 and DQA1*02:01 were positively associated with prevalent cervical hrHPV infections. On the other hand, DQA1*05:01 was associated with 55% lower odds of prevalent cervical hrHPV infections. However, hrHPV infections were more likely to be persistent among women with DQA1*05:01. Several haplotypes that include DQA1*05:01 were associated with persistent hrHPV but none of the amino acid residues located on DQα1 were associated with persistent cervical hrHPV infections in this study population.
HLA-DQA1*01:02 was the most common allele in the study population (allele frequency 39%, population frequency 63%). Although DQA1*01:02 has not been previously reported in Nigeria, it’s frequency is high in other populations in SSA, including Gabon (allele frequency 50%), Congo (allele frequency 42%, population frequency 53%), Cameron (allele frequency 38%) and Kenya (allele frequency 32%, population frequency 57%), as well as in African Americans in the United States (allele frequency 31%, population frequency 48%) [31]. DQA1 *01:02 was associated with higher odds of prevalent cervical hrHPV infection in the present study. This HLA allele was also associated with cervical intraepithelial neoplasia (CIN) in American Indian women, OR 4.5, 95% Cl 1.3-5.3 [32]. It has also been linked to other viral diseases including JC polyomavirus and Merkel cell polyomavirus [33], immune-mediated disorders including type 1 diabetes [34], systematic lupus erythematous (SLE) [35], multiple sclerosis and celiac disease [36], as well as other disorders including tuberculosis [35] and narcolepsy [36, 37]. DQA1*02:01 was also associated with higher odds of prevalent cervical hrHPV infection in this study. This variant has been linked to other cancers including nasopharyngeal carcinoma [38] and chronic myelogenous leukemia [39]. It was also associated with seropositivity of HPV 77 [40], an α cutaneous HPV genotype linked to common warts and skin cancer [41–43]. Therefore, these two HLA alleles seem to be pleiotropic for several virus infections, virus-associated cancers and autoimmune disorders.
The frequency of DQA1*05:01 was lower in our population (allele frequency 8%), than in other populations, with frequencies of 60% reported from Argentina, 78% in Colombia, and 89% in the United States [31]. We were unable to compare our finding that DQA1*05:01 and several haplotypes that include this variant were associated with hrHPV persistence to results from other studies, given the dearth of data. Nonetheless, the association between DQA1*05:01 and cervical neoplasia has been examined in some studies and the results have been inconsistent. A study among Honduran women reported that the frequency of DQA1*05:01 was lower among women with CIN III or cervical cancer (n=5), compared to those with CIN I or II (n=12), RR: 0.30, p 0.03 [44]. This allele was more common among Norwegian women with CIN II-III compared to those without CIN (OR: 2.4, 95% CI: 1.0-5.4 p 0.02) [45]. While one study found DQA1*05:01-DQB1*0301 haplotype was negatively associated with cervical dysplasia and neoplasia in HPV16-positive women (OR: 0.19, 95% Cl: 0.06 – 0.50 p < 0.001) [46], another study showed that this haplotype was positively associated with cervical neoplasia among HPV16-seronegative women (OR: 3.00 p <0.01) [47]. These results may be due to non-16 hrHPV types that were not examined in the previous studies, such as HPV 31, 33, 35, 45, 52 and 58, which account for 20% of cervical cancers worldwide. These non-16 hrHPV are more common than HPV16 in our study population [2, 4, 20, 21, 48] and may explain the association between C and hrHPV persistence observed in this study. This variant and other haplotypes that include it have also be linked polycystic ovary syndrome [49], ovarian cancer [50] and autoimmune diseases including type 1 diabetes and celiac disease [36]. DQA1*05:05, which was associated with significantly lower odds of type-specific persistent hrHPV infections in our study, was also found to be associated with reduced odds in female patients with early onset breast cancer, compared to healthy controls [51].
Although we observed an association between DQB1*02:01 and persistent cervical hrHPV infections, the association was borderline significant after adjustment for age, HIV status and the first principal component (OR: 2.10, 95% Cl: 1.08 – 4.09, p 0.08). Alifu et al. reported that the frequency of DQB1 *02:01 was higher among Uyghur women with advanced squamous cell cervical cancer compared to otherwise healthy controls in China[8]. The small size of our study may be responsible for attenuation of the p values we observed. Conversely, we observed DQB1*05:01 was associated with 66% lower odds of persistent hrHPV infection and this association was also attenuated to become borderline significant in the fully adjusted models (OR: 0.34, 95% Cl: 0.14-0.82, p 0.08). In support of this finding, DQB1*05:01 allelic variant has been shown to be protective of cervical cancer in different populations. Brazilian [18] and British [52] women with DQB1*05:01 and Chinese women with DQB1*05:02:01 [53], were less likely to have cervical cancer compared to otherwise healthy women.
The results of our present study provide evidence for the role of specific HLA alleles in determining susceptibility to persistent cervical hrHPV infections, a precursor for cervical cancer. To promote immune recognition and subsequent clearance of infected cells, Class I HLA molecules present foreign antigens, such as HPV peptides to CD8 cytotoxic T lymphocytes, while class II HLA molecules present antigenic peptides to CD4 T helper cells [54]. HLA molecules that bind HPV-specific antigens facilitate viral clearance , while HLA molecules that fail to recognize and present HPV-specific antigens may promote viral persistence. Cytotoxic T lymphocytes responses to HPV infection have been well documented [55, 56]. Downregulation and loss of HLA class I antigen expression enable HPV-infected cells to escape detection by the immune system by becoming nonimmunogenic and this has been reported in cervical cancer [57, 58]. Our results suggest that DQA1*05*01 may be associated with immune evasion of host defense and promotion of hrHPV persistence, thereby facilitating development of cervical malignancy.
Our study has several important limitations. First, even though our sample size was bigger than that in some of the previous studies, we may have missed associations with smaller effect sizes, and we had inadequate power to examine the relationship between HLA alleles and type-specific hrHPV. However, examining the associations between HLA alleles and group hrHPV identified significant results and confirmed findings in other populations. Second, our study was limited by the short duration of follow up of 6 to 8 months. While a longer follow up of up to 12 months may have be useful, a meta-analysis of 86 previous studies which provided data on over 100,000 women globally, showed that a follow-up period of at least 6 months was sufficient to define persistent hrHPV infections [59]. Lastly, although we did not have HIV clinical information such as duration of HIV infection, use of anti-retroviral therapy, viral load or CD4 count that may be associated with clearance of HPV infection among HIV-positive women, we excluded HIV-positive women who were severely ill from our analyses. However, in another study where clinical features of HIV were examined in relation to HPV, no association was found [60]. The strengths of our study include studying a well-characterized longitudinal cohort, collecting detailed data on numerous exposures using validated procedures and conducting multiple hrHPV assessments in the participants.
In summary, we report the frequency distribution of HLA alleles and the association between the alleles and haplotypes and their association with cervical hrHPV infections in Nigerian women. We found some HLA allelic variants, haplotypes and amino acid positions were significantly associated with prevalent and persistent cervical hrHPV infections. Future large-scale studies that examine the association between HLA allelic variants and persistence of type-specific hrHPV infections and cervical cancer would be informative. These studies may contribute to a better understanding of the role of HLA allelic variants in individualized medicine and clinical decision making and the relationship between HPV infections and the host immune response that underpin the development of persistent infection and cervical carcinogenesis.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bray F, et al. , Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 2018. 68(6): p. 394–424. [DOI] [PubMed] [Google Scholar]
- 2.Adebamowo SN, et al. , Persistent Low-Risk and High-Risk Human Papillomavirus Infections of the Uterine Cervix in HIV-Negative and HIV-Positive Women. Front Public Health, 2017. 5: p. 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giuliano AR, et al. , Clearance of oncogenic human papillomavirus (HPV) infection: effect of smoking (United States). Cancer Causes Control, 2002. 13(9): p. 839–46. [DOI] [PubMed] [Google Scholar]
- 4.Adebamowo SN, et al. , Clearance of Type-Specific, Low-Risk, and High-Risk Cervical Human Papillomavirus Infections in HIV-Negative and HIV-Positive Women. J Glob Oncol, 2018(4): p. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams TM, Human leukocyte antigen gene polymorphism and the histocompatibility laboratory. J Mol Diagn, 2001. 3(3): p. 98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hildesheim A and Wang SS, Host and viral genetics and risk of cervical cancer: a review. Virus Res, 2002. 89(2): p. 229–40. [DOI] [PubMed] [Google Scholar]
- 7.Safaeian M, et al. , Human Leukocyte Antigen Class I and II Alleles and Cervical Adenocarcinoma. Front Oncol, 2014. 4: p. 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alifu M, et al. , Frequency distribution of HLA alleles and haplotypes in Uyghur women with advanced squamous cell cervical cancer and relation to HPV status and clinical outcome. Arch Gynecol Obstet, 2018. 297(3): p. 757–766. [DOI] [PubMed] [Google Scholar]
- 9.Wang SS, et al. , Comprehensive analysis of human leukocyte antigen class I alleles and cervical neoplasia in 3 epidemiologic studies. J Infect Dis, 2002. 186(5): p. 598–605. [DOI] [PubMed] [Google Scholar]
- 10.Leo PJ, et al. , Defining the genetic susceptibility to cervical neoplasia-A genome-wide association study. PLoS Genet, 2017. 13(8): p. e1006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen D, et al. , Genome-wide association study of susceptibility loci for cervical cancer. J Natl Cancer Inst, 2013. 105(9): p. 624–33. [DOI] [PubMed] [Google Scholar]
- 12.Hildesheim A, et al. , Human leukocyte antigen class I/II alleles and development of human papillomavirus-related cervical neoplasia: results from a case-control study conducted in the United States. Cancer Epidemiol Biomarkers Prev, 1998. 7(11): p. 1035–41. [PubMed] [Google Scholar]
- 13.Wang SS, et al. , Human leukocyte antigen class I and II alleles and risk of cervical neoplasia: results from a population-based study in Costa Rica. J Infect Dis, 2001. 184(10): p. 1310–4. [DOI] [PubMed] [Google Scholar]
- 14.Madeleine MM, et al. , Comprehensive analysis ofHLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci and squamous cell cervical cancer risk. Cancer Res, 2008. 68(9): p. 3532–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang YC, et al. , HLA-DRB1 alleles and cervical squamous cell carcinoma: experimental study and meta-analysis. Hum Immunol, 2006. 67(4–5): p. 331–40. [DOI] [PubMed] [Google Scholar]
- 16.Castro FA, et al. , Association of HLA-DRB1, interleukin-6 and cyclin D1 polymorphisms with cervical cancer in the Swedish population--a candidate gene approach. Int J Cancer, 2009. 125(8): p. 1851–8. [DOI] [PubMed] [Google Scholar]
- 17.Metcalfe S, et al. , The association between human leukocyte antigen (HLA)-G polymorphisms and human papillomavirus (HPV) infection in Inuit women of northern Quebec. Hum Immunol, 2013. 74(12): p. 1610–5. [DOI] [PubMed] [Google Scholar]
- 18.Maciag PC, et al. , Major histocompatibility complex class II polymorphisms and risk of cervical cancer and human papillomavirus infection in Brazilian women. Cancer Epidemiol Biomarkers Prev, 2000. 9(11): p. 1183–91. [PubMed] [Google Scholar]
- 19.Adebamowo SN, et al. , Mycoplasma hominis and Mycoplasma genitalium in the Vaginal Microbiota and Persistent High-Risk Human Papillomavirus Infection. Front Public Health, 2017. 5: p. 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akarolo-Anthony SN, et al. , HIV associated high-risk HPV infection among Nigerian women. BMC Infect Dis, 2013. 13: p. 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akarolo-Anthony SN, et al. , Age-specific prevalence of human papilloma virus infection among Nigerian women. BMC Public Health, 2014. 14: p. 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Famooto A, et al. , RPS19 and TYMS SNPs and Prevalent High Risk Human Papilloma Virus Infection in Nigerian Women. PLoS One, 2013. 8(6): p. e66930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Hamont D, et al. , Evaluation of the SPF10-INNO LiPA human papillomavirus (HPV) genotyping test and the roche linear array HPV genotyping test. J Clin Microbiol, 2006. 44(9): p. 3122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleter B, et al. , Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J Clin Microbiol, 1999. 37(8): p. 2508–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melchers WJ, et al. , Short fragment polymerase chain reaction reverse hybridization line probe assay to detect and genotype a broad spectrum of human papillomavirus types. Clinical evaluation and follow-up. Am J Pathol, 1999. 155(5): p. 1473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang CC, et al. , Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience, 2015. 4: p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S and Chang C, PLINK 1.9. [Google Scholar]
- 28.Zheng X, et al. , HIBAG--HLA genotype imputation with attribute bagging. Pharmacogenomics J, 2014. 14(2): p. 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryll R, Gutierrez-Osuna R, and Quek F, Attribute bagging: improving accuracy of classifier ensembles by using random feature subsets. Pattern Recognition, 2003. [Google Scholar]
- 30.Fan Y and Song YQ, PyHLA: tests for the association between HLA alleles and diseases. BMC Bioinformatics, 2017. 18(1): p. 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez-Galarza FF, et al. , Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res, 2015. 43(Database issue): p. D784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiff MA, et al. , HLA alleles and risk of cervical intraepithelial neoplasia among southwestern American Indian women. Hum Immunol, 2005. 66(10): p. 1050–6. [DOI] [PubMed] [Google Scholar]
- 33.Hammer C, et al. , Amino Acid Variation in HLA Class II Proteins Is a Major Determinant of Humoral Response to Common Viruses. Am J Hum Genet, 2015. 97(5): p. 738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pugliese A, et al. , HLA-DRB1*15:01-DQA1*01:02-DQB1*06:02 Haplotype Protects Autoantibody-Positive Relatives From Type 1 Diabetes Throughout the Stages of Disease Progression. Diabetes, 2016. 65(4): p. 1109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian C, et al. , Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat Commun, 2017. 8(1): p. 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones EY, et al. , MHC class II proteins and disease: a structural perspective. Nat Rev Immunol, 2006. 6(4): p. 271–82. [DOI] [PubMed] [Google Scholar]
- 37.Mignot E, et al. , DQB1*0602 and DQA1*0102 (DQ1) are better markers than DR2 for narcolepsy in Caucasian and black Americans. Sleep, 1994. 17(8 Suppl): p. S60–7. [DOI] [PubMed] [Google Scholar]
- 38.Karanikiotis C, et al. , HLA Class II alleles and the presence of circulating Epstein-Barr virus DNA in Greek patients with nasopharyngeal carcinoma. Strahlenther Onkol, 2008. 184(6): p. 325–31. [DOI] [PubMed] [Google Scholar]
- 39.Amirzargar AA, et al. , Association of HLA class II allele and haplotype frequencies with chronic myelogenous leukemia and age-at-onset of the disease. Pathol Oncol Res, 2007. 13(1): p. 47–51. [DOI] [PubMed] [Google Scholar]
- 40.Chen D, et al. , A systematic investigation of the contribution of genetic variation within the MHC region to HPVseropositivity. Hum Mol Genet, 2015. 24(9): p. 2681–8. [DOI] [PubMed] [Google Scholar]
- 41.Burd EM, Human papillomavirus and cervical cancer. Clin Microbiol Rev, 2003. 16(1): p. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iannacone MR, et al. , Case-control study of cutaneous human papillomavirus infection in Basal cell carcinoma of the skin. J Invest Dermatol, 2013. 133(6): p. 1512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purdie KJ, et al. , The promoter of a novel human papillomavirus (HPV77) associated with skin cancer displays UV responsiveness, which is mediated through a consensus p53 binding sequence. EMBO J, 1999. 18(19): p. 5359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferrera A, et al. , HLA DOA1 and DOB1 loci in Honduran women with cervical dysplasia and invasive cervical carcinoma and their relationship to human papillomavirus infection. Hum Biol, 1999. 71(3): p. 367–79. [PubMed] [Google Scholar]
- 45.Helland A, et al. , An increased risk of cervical intra-epithelial neoplasia grade II-III among human papillomavirus positive patients with the HLA-DQA1*0102-DQB1*0602 haplotype: a population-based case-control study of Norwegian women. Int J Cancer, 1998. 76(1): p. 19–24. [DOI] [PubMed] [Google Scholar]
- 46.Apple RJ, et al. , Comparison of human leukocyte antigen DR-DQ disease associations found with cervical dysplasia and invasive cervical carcinoma. J Natl Cancer Inst, 87(6): p. 427–36. [DOI] [PubMed] [Google Scholar]
- 47.Sanjeevi CB, et al. , Different HLA-DR-DQ haplotypes are associated with cervical intraepithelial neoplasia among human papillomavirus type-16 seropositive and seronegative Swedish women. Int J Cancer, 1996. 68(4): p. 409–14. [DOI] [PubMed] [Google Scholar]
- 48.Jedy-Agba EE, et al. , The burden of HPV associated cancers in two regions in Nigeria 2012–2014. Cancer Epidemiol, 2016. 45: p. 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ober C, et al. , Increased risk for polycystic ovary syndrome associated with human leukocyte antigen DQA1*0501. Am J Obstet Gynecol, 1992. 167(6): p. 1803–6. [DOI] [PubMed] [Google Scholar]
- 50.Kubler K, et al. , HLA-class II haplotype associations with ovarian cancer. Int J Cancer, 2006. 119(12): p. 2980–5. [DOI] [PubMed] [Google Scholar]
- 51.Mahmoodi M, et al. , HLA-DRB1,-DQA1 and -DQB1 allele and haplotype frequencies in female patients with early onset breast cancer. Pathol Oncol Res, 2012. 18(1): p. 49–55. [DOI] [PubMed] [Google Scholar]
- 52.Cuzick J, et al. , Association between high-risk HPV types, HLA DRB1* and DQB1* alleles and cervical cancer in British women. Br J Cancer, 2000. 82(7): p. 1348–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Y, et al. , Polymorphic amino acids at codons 9 and 37 of HLA-DQB1 alleles may confer susceptibility to cervical cancer among Chinese women. Int J Cancer, 2006. 118(12): p. 3006–11. [DOI] [PubMed] [Google Scholar]
- 54.Apple RJ, et al. , HLA DR-DQ associations with cervical carcinoma show papillomavirus-type specificity. Nat Genet, 1994. 6(2): p. 157–62. [DOI] [PubMed] [Google Scholar]
- 55.Nakagawa M, et al. , T-cell proliferative response to human papillomavirus type 16 peptides: relationship to cervical intraepithelial neoplasia. Clin Diagn Lab Immunol, 3(2): p. 205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakagawa M, et al. , Cytotoxic Tlymphocyte responses to E6 and E7proteins of human papillomavirus type 16: relationship to cervical intraepithelial neoplasia. J Infect Dis, 175(4): p. 927–31. [DOI] [PubMed] [Google Scholar]
- 57.Glew SS, et al. , HLA expression in pre-invasive cervical neoplasia in relation to human papilloma virus infection. Eur J Cancer, 1993. 29A(14): p. 1963–70. [DOI] [PubMed] [Google Scholar]
- 58.Cromme FV, et al. , Analysis of MHC class I and II expression in relation to presence of HPVgenotypes in premalignant and malignant cervical lesions. Br J Cancer, 1993. 67(6): p. 1372–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rositch AF, et al. , Patterns of persistent genital human papillomavirus infection among women worldwide: a literature review and meta-analysis. Int J Cancer, 2013. 133(6): p. 1271–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moscicki AB, et al. , Prevalence of and risks for cervical human papillomavirus infection and squamous intraepithelial lesions in adolescent girls: impact of infection with human immunodeficiency virus. Arch Pediatr Adolesc Med, 2000. 154(2): p. 127–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


