Abstract
The ‘Immunogenetics of Aging’ project is a component introduced in the 14th International HLA and Immunogenetics Workshop (IHIW) and developed further within subsequent workshops. The aim was to determine the relevance of immunogenetic markers, focusing on HLA, cytokine genes, and some innate immunity genes, for successful aging and an increased capacity to reach the extreme limits of life-span. Within the 17th IHIW we applied Next Generation Sequencing methods to refine further HLA associations at allele level in longevity, and to extend our knowledge to additional loci such as HLA-DQA1, HLA-DPB1 and HLA-DPA1. Analysis of relatively small number of healthy elderly and young controls from four populations showed that some HLA class I and class II alleles were significantly positively associated with healthy aging. Additionally we observed statistically significant differences in HLA allele distribution when the analysis was performed separately in elderly females and males compared to sex-matched young controls. Haplotypes, probably associated with better control of viral and malignant diseases were increased in the elderly sample. These preliminary NGS data could confirm our hypotheses that survival and longevity might be associated with selection of HLA alleles and haplotypes conferring disease resistance or susceptibility. Therefore HLA alleles and haplotypes could be informative immunogenetic markers for successful ageing.
Keywords: HLA, Ageing, Longevity, NGS
1. Introduction
Longevity in the sense of a possibility of being long-lived, is limited by the ageing process. From a socioeconomic standpoint aging is becoming an important factor and it is expected that in the next 20 years people over 65 years will represent 20% of the world population. The increased frequency of chronic, age-related diseases leading to functional dependence will be expected to confer a significant burden to the public health care system. Therefore deeper knowledge on age-related disease and factors associated with ageing are required. Human longevity is at least partly heritable since it was shown that progeroid syndromes of accelerated aging certainly have a genetic basis [1,2]. The genetic component of longevity is under an intensive investigation. Different studies contribute to answering the question: “Does genetics really play a role in longevity?” Twin and family studies showed that between 15% and 30% of lifespan is controlled by our genes [3,4]. Gavrilova and Gavrilov [5] showed that millions of familial longevity records are available but so far are underused for research by carrying out a pilot search of data resources related to familial longevity using genealogical databases, published family history data, available scientific data resources, data sets on long-lived people and centenarians. McGue et al. studied 2872 pairs of Danish twins born between 1870 and 1900, and found that the hereditary component of the lifespan is moderate [6–8]. How long we live is determined also by environmental factors and epigenetic alterations during a lifetime. Studies on centenarians, on the other hand, suggest that they inherit a small number of powerfully acting longevity genes. Such genes might for example allow centenarians to control better the inflammation that is associated negatively with many of the diseases of old age. Furthermore the siblings of centenarians have a significantly higher likelihood of reaching old age, compared to the general population [9].
Many aspects of senescence in humans are characterized by increase inflammatory status, leading to extended tissue damage. Chronic inflammation, characterized by high circulating levels of a number of factors, including: C-reactive protein, tumor necrosis factor, and inter-leukin-6, have been associated with several age-related diseases such as atherosclerosis, obesity, type 2 diabetes and cancer [10–13] The inflammatory response may be a prime example of antagonistic pleiotropy, in which genes that are required to increase the inflammatory response to infectious organisms in early life are also associated with harmful effects, such as atherosclerosis in the post-reproductive period. Family segregation analysis has shown that the glycoproteins CD4, CD8 and CD4/CD8 ratio are under genetic control [14,15]. Longitudinal studies of the very elderly have documented an “immune risk profile”, a cluster of immune parameters predicting 2–8 years mortality, and characterized by inverted CD4:CD8 ratio [16]. All these data imply that aging process may be associated with alterations in the immune system and possibly with polymorphisms in the genes, regulating immune responses. Genes encoding molecules involved in the development of protective immunity are highly polymorphic, and this polymorphism is a result from an evolutionary adaptation of the organism driven by an ever-evolving pathogen environment. Such genes include: human leukocyte antigen (HLA) genes, those encoding “atypical” HLA-like molecules (CD1); killer cell immunoglobulin-like receptor genes (KIR); leukocyte Fcγ receptor genes; cytokine and cytokine receptor genes; Toll-like receptor gene family; TNF-receptor associated factors, MBL2.
The ‘Immunogenetics of Aging’ project was introduced as a component within the 14th International HLA and Immunogenetics Workshop (IHW) and further developed within the 15th, 16th and 17th. The overall aim of this component is to determine immunogenetic markers, such as HLA, cytokine genes, and some innate immunity genes, that might be relevant for successful aging and longevity. The following groups of samples were collected; families with long-lived members (octogenarians and nonagenarians), unrelated elderly (> 65 years old) individuals and ethnically matched young (< 35 years old) controls. As part of 14th and 15th IHIW, the effect of classical HLA class I and class II loci, analyzed by low-resolution methods, and cytokine polymorphisms in regulatory and/or coding regions with a possible impact on the level of gene expression of pro- and anti-inflammatory cytokines, was studied [17,18,29]. During the 16th IHIW innate immunity genes KIR and MBL2 and their possible correlation with CMV status were analyzed [19]. Our aim within the 17th IHIW was to apply next generation sequencing (NGS) to further clarify HLA associations at maximum allele resolution with longevity and to extend our knowledge to additional loci such as HLA-DQA1 and HLA-DPB1.
2. Material and methods
During the 17th IHIW additional samples were collected and one new population sample from Kuwait was included in the analysis. Two sample groups were analyzed: unrelated healthy elderly individuals and young controls. Elderly individuals were characterized according to the SENIEUR protocol [20]. The unrelated young controls were characterized according to the JUNIEUR protocol [20]. Written informed consent was obtained from all individuals included in the study. Data on four populations (Bulgarian, Romanian, Italian and Kuwaiti) were analyzed with a total number of 155 elderly (age 65–95 years; males n = 51; females n = 104) and 682 young controls (age 21–35 years; males n = 290; females n = 392). The number of individuals from each population is as follows; Italians – 292 young controls; Bulgarians – 367 young controls and 59 elderly individuals; Romanians – 29 elderly individuals; Kuwaiti – 21 young controls and 14 elderly individuals.
The following loci: HLA-A, HLA-C, HLA-B, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1 and HLA-DPB1 were typed by various NGS methods using commercially available kits: NGSgo and NGSengine software (GenDx, Utrecht, Netherlands); TruSight HLA v2 and Assign TruSight HLA software (Illumina, Inc., San Diego, CA); Holotype HLA and HLA Twin software (Omixon, Budapest, Hungary).
3. Statistical analysis
Python for Population Genomics (PyPop) software v.0.7.0 [21] was used to calculate allele frequencies, 2-locus haplotype frequencies, standardized linkage disequilibrium measure D′, D′ chi-square, and the Ewens-Watterson’s homozygosity test of neutrality was applied to each locus in separate populations. Comparisons across groups were performed using the R software version 3.5.1 by χ2-square or Fisher exact test when appropriate and OR was calculated. P-values lower than0.05 were considered to indicate a statistically significant difference between groups. Volcano plots were used for graphical data presentation (odds ratios according to P values). Statistical analyses were performed separately in each population: Bulgarians, Romanians and Italians, and Kuwaiti.
4. Results
In the present study we applied NGS approach for high resolution typing. Analysis of HLA allele and haplotype distribution in elderly and young controls was performed separately for the 4 populations included in the study: Bulgarians, Romanians, Italians and Kuwaiti.
Hardy-Weinberg equilibrium was tested in all populations and in all age-groups (Table 1). Single locus devitations were observed for Italian and Kuwait young samples. In Romanians and Bulgarians multiple loci showed daviations from Hardy – Weinberg equilibrium (Table 1).
Table 1.
Hardy-Weinberg equilibrium p-values.
| Population | n | A | C | B | DRB1 | DQA1 | DQB1 | DPA1 | DPB1 |
|---|---|---|---|---|---|---|---|---|---|
| Italian | 292 | 0.8447 | 0.0985 | 0.8679 | 0.2448 | – | 0.1224 | – | 0.0025 |
| Bulgarian | 479 | 0.0002 | 0.0043 | 0.0005 | 0.0000 | 0.0528 | 0.0000 | 0.0000 | 0.0000 |
| Bulgarian (Young) | 367 | 0.0022 | 0.0198 | 0.0072 | 0.0000 | 0.0005 | 0.0000 | 0.0000 | 0.0000 |
| Bulgarian (Young Female) | 179 | 0.0403 | 0.3459 | 0.0880 | 0.0014 | 0.0012 | 0.0000 | 0.0114 | 0.0002 |
| Bulgarian (Young Male) | 190 | 0.0037 | 0.1096 | 0.1394 | 0.0444 | 0.2082 | 0.0000 | 0.0000 | 0.0000 |
| Bulgarian (Old) | 59 | 0.3920 | 0.3380 | 0.0409 | 0.0000 | 0.2175 | 0.0001 | 0.3714 | 0.0023 |
| Bulgarian (Old Female) | 26 | 0.2393 | 0.1878 | 0.4317 | 0.0210 | 0.3946 | 0.0538 | 0.6322 | 0.0855 |
| Bulgarian (Old Male) | 17 | 0.2963 | 0.9685 | 0.7109 | 0.0203 | 0.9911 | 0.2182 | 0.0583 | 0.2639 |
| Romanian (Old) | 29 | 0.7206 | 0.2178 | 0.0391 | 0.1156 | 0.2267 | 0.0072 | 0.8505 | 0.0073 |
| Kuwait | 35 | 0.0703 | 0.7535 | 0.5891 | 0.7206 | 0.9865 | 0.2985 | 0.2106 | 0.1165 |
| Kuwait (Young) | 21 | 0.0149 | 0.8127 | 0.6451 | 0.8934 | 0.9920 | 0.6001 | 0.1563 | 0.3397 |
| Kuwait (Old) | 14 | 0.4433 | 0.7276 | 1.0000 | 0.5910 | 1.0000 | 0.5811 | 0.5760 | 0.4025 |
Probability values for Guo and Thompson Hardy-Weinberg equilibrium (HWE) tests. HLA loci that deviated significantly (P < 0.05) from HWE are shown in boldface type.
Ewens-Watterson test showed that with the exception of HLA-A locus all other HLA loci exhibit significantly lower homozygosity than expected under neutral evolution in different populations analyzed (Table 2). Negative Fnd values suggesting balancing selection were observed for HLA-C, -B, DRB1, -DQA1, -DQB1, -DPA1 and -DPB1 loci in different age groups studied. The only exception was HLA-A locus for which positive Fnd values were observed in all groups and subgroups.
Table 2.
Fnd values for HLA alleles. Slatkin’s implementation of Ewens –Watterson test.
| Group analyzed | HLA-A | HLA-C | HLA-B | HLA-DRB1 | HLA-DQA1 | HLA-DQB1 | HLA-DPA1 | HLA-DPB1 |
|---|---|---|---|---|---|---|---|---|
| Italian | 0.5874 | −0.8762 | −0.7186 | −0.9512 | – | −1.0724 | – | 0.9161 |
| Bulgarian | 2.0940 | − 0.6524 | 0.7113 | 2.6544 | −0.5590 | 1.5846 | −0.5899 | 2.2960 |
| Bulgarian (Young) | 1.2878 | − 0.5372 | 0.3941 | 1.3034 | −0.4844 | 2.0493 | −0.5233 | 2.0257 |
| Bulgarian (Young Female) | 0.9115 | − 0.5436 | 0.1138 | −0.0343 | −0.4137 | 0.4142 | −0.3315 | 2.0064 |
| Bulgarian (Young Male) | 1.5070 | −0.4033 | −0.1877 | 1.2614 | −0.4702 | 2.5982 | −0.6293 | 1.5517 |
| Bulgarian (Old) | 2.0747 | −0.0331 | 0.4870 | 0.7218 | −0.1492 | 1.1836 | −0.7169 | 1.0682 |
| Bulgarian (Old Female) | 0.2623 | 0.2907 | 0.4342 | −0.2258 | 0.8191 | 0.7991 | −0.7901 | 1.0527 |
| Bulgarian (Old Male) | 1.1210 | −0.5162 | −0.4632 | −0.6150 | 0.1956 | 0.9624 | −1.0790 | −0.5424 |
| Romanian (Old) | 1.0249 | − 0.6989 | 0.5502 | −0.3827 | −0.6756 | 0.0092 | −0.9923 | −0.0805 |
| Kuwait (Young) | 0.8900 | −1.2957 | −0.6639 | 0.2248 | −0.0500 | −0.9693 | −0.0869 | 1.7880 |
| Kuwait (Old) | 0.7249 | − 0.0658 | −0.3537 | −0.8843 | 0.1496 | −0.764 | −0.5212 | 0.1028 |
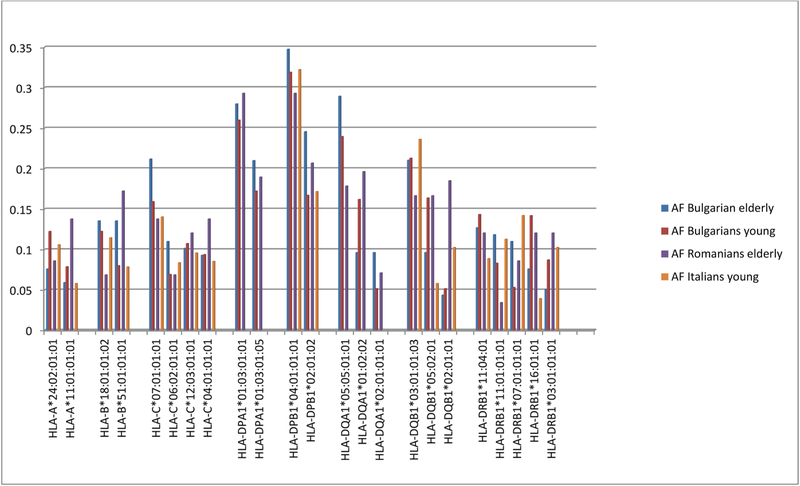
The most frequent HLA alleles observed in the 3 European populations included in the analysis were similar (Fig. 1).
Fig. 1.
The most frequent HLA alleles in the three European populations: Bulgarians (elderly, young), Romanian elderly and Italians young controls.
4.1. Allele level associations
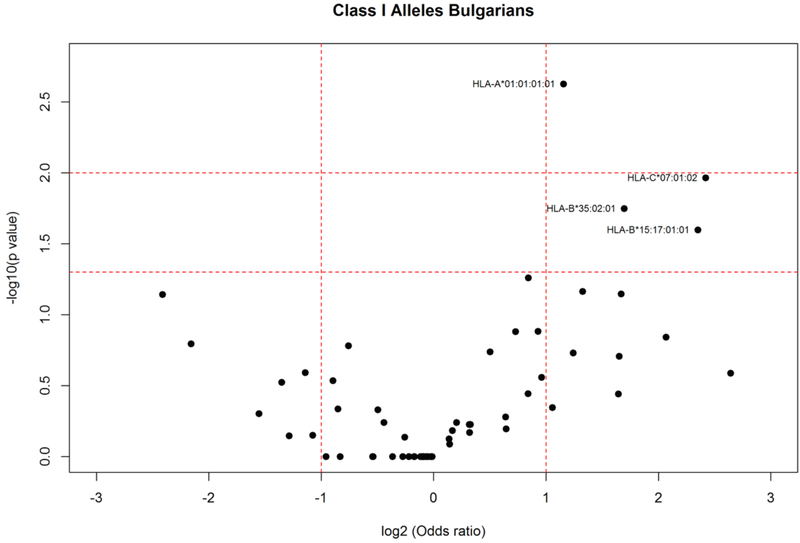
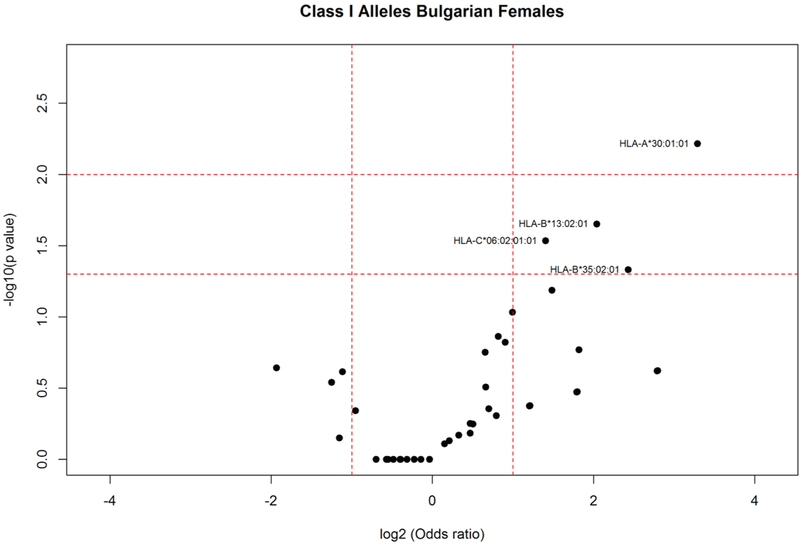
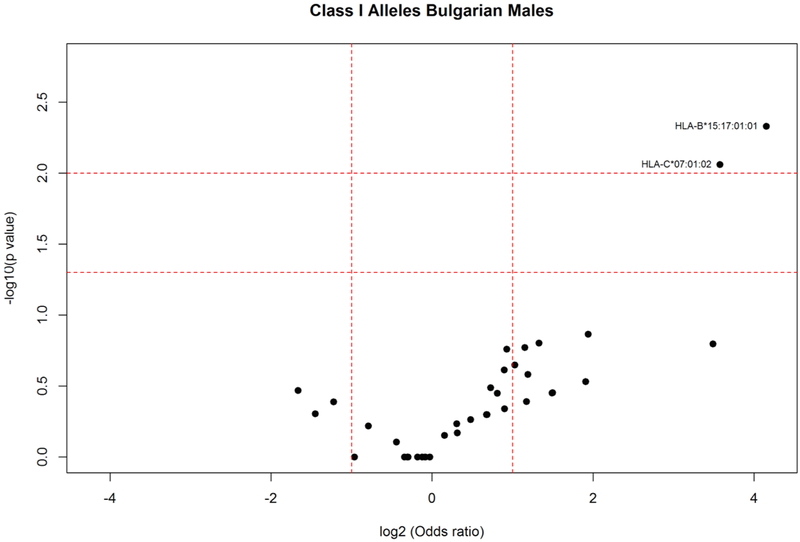
Analysis of HLA allele distribution in the Bulgarian population showed statistically significant increased frequencies in elderly (Fig. 2) of the following HLA class I alleles: HLA-A*01:01:01:01 (AF elderly: 0.212; AF young: 0.108; P = 0.002); HLA-C*07:01:02 (AF elderly: 0.042; AF young: 0.008; P = 0.011); HLA-B*35:02:01 (AF elderly:0.059; AF young: 0.019; P = 0.018), B*15:17:01:01 (AF elderly: 0.034; AF young: 0.007; P = 0.025). HLA-A*01:01:01:01 was among the most frequent HLA alleles in elderly Romanians (Fig. 1) and that was the only HLA-A*01:01:01 4th-field variant identified in the population. Stratification according to gender and age in the Bulgarians (Fig. 3) showed that HLA-B*35:02:01 (AF elderly females: 0.058; AF young females:0.011; P < 0.046) appeared as a specific marker for longevity in females since both alleles were increased only in elderly females compared to female young controls. On the other hand, as shown in Fig. 4, HLA-B*15:17:01:01 (AF elderly males: 0.088; AF young males: 0.005; P = 0.005) and HLA-C*07:01:02 were associated with longevity in men (AF elderly males: 0.088; AF young males: 0.008; P = 0.009). Additionally in elderly females alleles HLA-A*30:01:01 (AF elderly females: 0.077; AF young females: 0.008; P = 0.006); HLA-B*13:02:01 (AF elderly females: 0.096; AF young females: 0.025; P = 0.022); HLA-C*06:02:01:01 (AF elderly females: 0.173; AF young females: 0.073; P = 0.029) were observed with statistically significant increased frequencies (Fig. 3).
Fig. 2.
HLA class I allele distribution in elderly and young controls from the Bulgarian population. Volcano plot represents odds ratios according to P values.
Fig. 3.
HLA class I allele distribution in female elderly and young controls from the Bulgarian population. Volcano plot represents odds ratios according to P values.
Fig. 4.
HLA class I allele distribution in male elderly and young controls from the Bulgarian population. Volcano plot represents odds ratios according to P values.
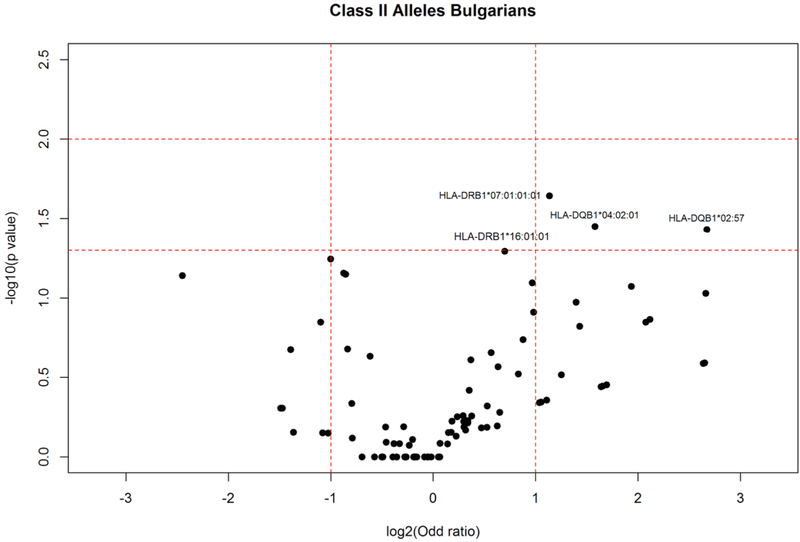
Comparison of HLA class II allele frequencies in elderly and young controls from Bulgarian population (Fig. 5) showed that: HLA-DRB1*07:01:01 (AF elderly: 0.110; AF young: 0.053; P = 0.023), HLA-DQB1*04:02:01 (AF elderly: 0.053; AF young: 0.018; P = 0.035), and HLA-DQB1*02:57 (AF elderly: 0.02; AF young: 0.004; P = 0.037) alleles were significantly increased in elderly. Alleles HLA-DRB1*04:10:01 (AF elderly: 0.018; AF young: 0.000; P = 0.019), HLA-DQB1*05:01:04 (AF elderly: 0.018; AF young: 0.000; P = 0.019), and HLA-DQA1*03:03:01:02 (AF elderly: 0.018; AF young: 0.000; P = 0.018) were observed only in elderly individuals and the differences were statistically significant. HLA-DRB1*07:01:01:01 was among the most frequent HLA alleles in elderly Romanians (Fig. 1)
Fig. 5.
HLA class II allele distribution in elderly and young controls from the Bulgarian population. Volcano plot represents odds ratios according to P values.
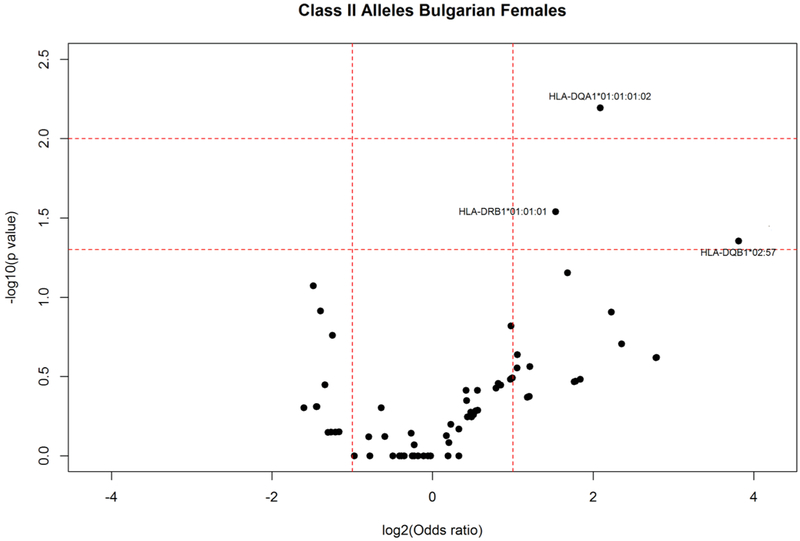
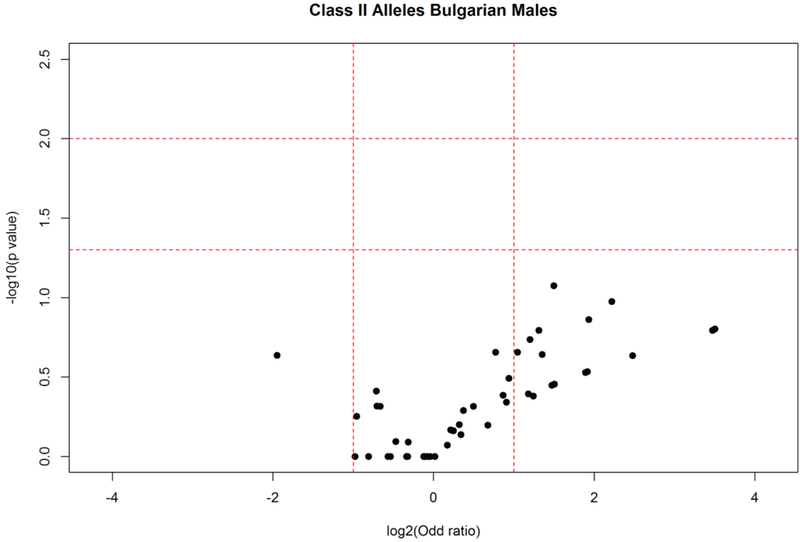
Among these alleles, stratification according to gender (Figs. 6 and 7) revealed that HLA-DQB1*02:57 (AF elderly females: 0.040; AF young females: 0.003; P = 0.044) and HLA-DQB1*05:01:04 (AF elderly: 0.040; AF young: 0.000; P = 0.016) were more frequent in the female elderly group. Additionally, alleles HLA-DRB1*01:01:01 (AF elderly females:0.135; AF young females: 0.051; P = 0.029); HLA-DQA1*01:01:01:02 (AF elderly females: 0.140; AF young females: 0.037; P = 0.006) were also increased in elderly females. No statistically significant differences in HLA class II allele frequencies were observed in males.
Fig. 6.
HLA class II allele distribution in female elderly and young controls from the Bulgarian population. Volcano plot represents odds ratios according to P values.
Fig. 7.
HLA class II allele distribution in male elderly and young controls from the Bulgarian population. Volcano plot represents odds ratios according to P values.
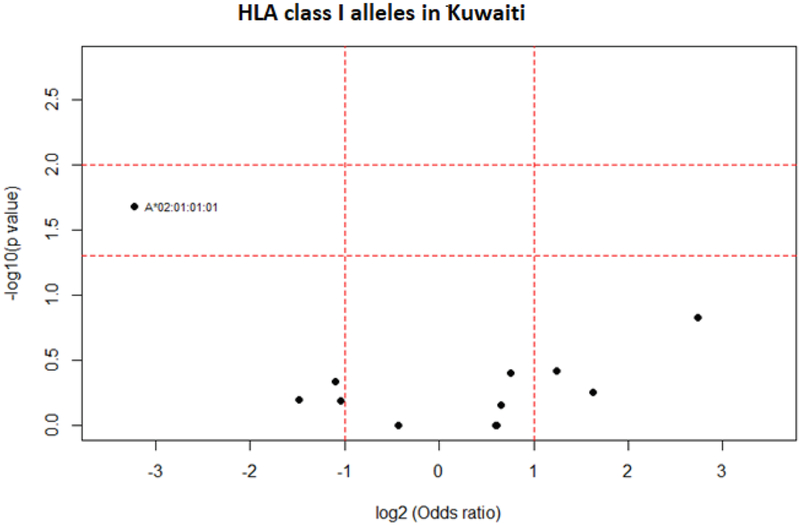
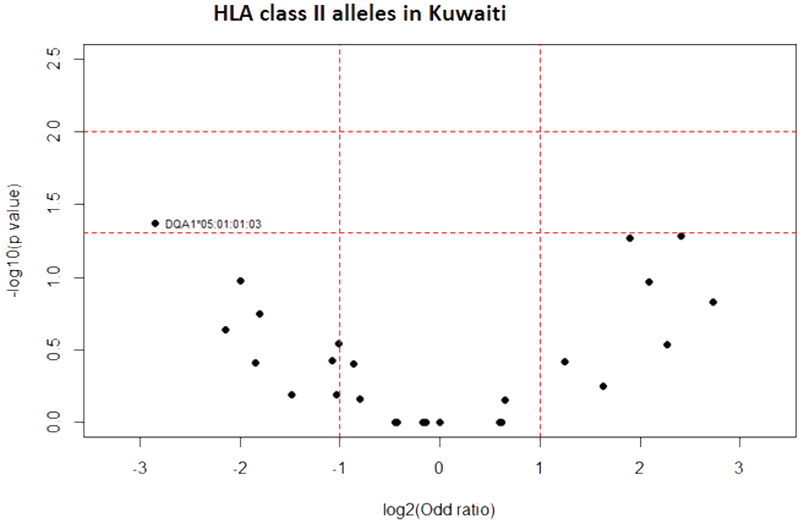
Analysis in a small cohort of elderly individuals compared to young controls from the Kuwaiti population showed statistically significant decreased frequency of HLA-A*02:01:01:01 (AF elderly: 0.035; AF young: 0.262; P = 0.035) and DQA1*05:01:01:03 (AF elderly: 0.036; AF young: 0.214; P = 0.042) in elderly (Figs. 8 and 9).
Fig. 8.
HLA class I allele distribution in elderly and young Kuwaiti. Volcano plot represents odds ratios according to P values.
Fig. 9.
HLA class II allele distribution in elderly and young Kuwaiti. Volcano plot represents odds ratios according to P values.
In the present study no significant associations between HLA-DPB1 alleles and longevity were observed.
4.2. Two-loci haplotype associations
Two-loci haplotypes that are observed with statistically significant frequencies in Bulgarian elderly and young controls are presented in Table 3. Four HLA-A*01:01:01 haplotypes were observed in significantly increased frequencies in elderly compared to the controls, from these HLA-A*01:01:01:03~HLA B*52:01:01:02; HLA-A*01:01:01:03~HLA B*35:02:01; HLA-A*01:01:01:03~HLA B*15:17:01:01 had significant high D′ raging from 0,634 to 1.000 and P < 0,01. Haplotypes HLA-A*26:01:01:01~HLA-DQA1*05:01:01:03; HLA-A*26:01:01:01~HLADQB1*03:01:01:01; HLA-B*35:01:01:01~HLA-DQB1*03:01:01:03; HLA-B*40:01:02~HLA-C*12:03:01:01; HLA-C*04:01:01:01~HLA-DQB1*06: 02:01; HLA-C*04:01:01:01~HLA-DRB1*07:01:01:01; HLA-C*16:02:01~ HLA-DRB1*11:03:01; HLA-DPB1*04:01:01:01~HLA-DQB1*05:01:01:01 were also significantly more frequent in elderly Bulgarians.
Table 3.
HLA haplotypes. observed with statistically significant differences (P < 0.01) in elderly Bulgarians and young controls.
| Haplotype | Elderly (n = 59) | Young controls (n = 367) | ||||
|---|---|---|---|---|---|---|
| count | HF | D′ | observed | HF | D′ | |
| HLA-A*01:01:01:03~HLA-B*52:01:01:02 | 1.000 | 0.009 | 1.000 | 1.000 | 0.001 | 0.253 |
| HLA-A*01:01:01:01~HLA-B*35:02:01 | 5.000 | 0.042 | 0.634 | 2.973 | 0.004 | 0.118 |
| HLA-A*01:01:01:01~HLA-B*13:02:01 | 3.000 | 0.025 | 0.274 | 0.000 | 0.000 | |
| HLA-A*01:01:01:01~HLA-B*15:17:01:01 | 3.000 | 0.025 | 0.682 | 0.000 | 0.000 | |
| HLA-A*68:01:01:02~HLA-C*04:01:01:01 | 0.000 | 0.000 | 1.000 | 0.001 | 0.448 | |
| HLA-A*26:01:01:01~HLA-DQA1*05:01:01:03 | 1.000 | 0.009 | 0.482 | 0.000 | 0.000 | |
| HLA-A*26:01:01:01~HLA-DQB1*03:01:01:01 | 1.000 | 0.009 | 0.216 | 0.000 | 0.000 | |
| HLA-B*35:01:01:01~HLA-DQBl*03:01:01:03 | 2.000 | 0.018 | 0.367 | 1.378 | 0.002 | 0.9 |
| HLA-B*40:01:02~HLA-C*12:03:01:01 | 1.000 | 0.008 | 1.000 | 0.000 | 0.000 | |
| HLA-B*35:02:01~HLA-C*04:01:01:06 | 5.000 | 0.042 | 0.696 | 13.000 | 0.018 | 0.924 |
| HLA-C*03:03:01:01~HLA-DQA1*01:02:02 | 0.000 | 0.000 | 2.587 | 0.003 | 0.103 | |
| HLA-C*07:01:01:01~HLA-DQA1*05:05:01:03 | 0.000 | 0.000 | 1.000 | 0.001 | 0.376 | |
| HLA-C*04:01:01:01~HLA-DQBl*03:01:02 | 1.000 | 0.009 | 0.000 | 0.000 | ||
| HLA-C*04:01:01:01~HLA-DQB1*06:02:01 | 1.000 | 0.009 | 1.000 | 3.016 | 0.004 | |
| HLA-C*04:01:01:01~HLA-DRB1*07:01:01:01 | 1.000 | 0.009 | 0.262 | 3.016 | 0.004 | 0.126 |
| HLA-C*16:02:01~HLA-DRBl*11:03:01 | 1.000 | 0.009 | 0.483 | 1.000 | 0.001 | 0.113 |
| HLA-DPB1*04:01:01:01~HLA-DQB1*05:01:01:01 | 1.000 | 0.009 | 1.000 | 1.781 | 0.003 | −0.065 |
The most frequent HLA two-loci haplotypes in elderly Romanians and young Italians are presented in Tables 4 and 5. These haplotypes are characteristic for South European Populations and of all these frequent haplotypes have positive D′ values.
Table 4.
The most Frequent HLA haplotypes in elderly Romanians (n = 29).
| Haplotype | Count | Haplotype Frequency | D′ | D′ Chi-square |
|---|---|---|---|---|
| HLA-A*02:01:01:01~HLA-B*51:01:01:01 | 5.000 | 0.086 | 0.356 | 5.288 |
| HLA-A*24:02:01:01~HLA-B*35:03:01 | 3.000 | 0.034 | 0.453 | 9.338 |
| HLA-A*01:01:01:01~HLA-B*18:01:01:02 | 2.000 | 0.034 | 0.420 | 4.737 |
| HLA-A*24:02:01:01~HLA-B*08:01:01:01 | 2.000 | 0.034 | 0.453 | 9.338 |
| HLA-A*11:01:01:01~HLA-B*35:01:01:01 | 2.000 | 0.034 | 1.000 | 12.946 |
| HLA-A*11:01:01:01~HLA-C*15:02:01:01 | 3.000 | 0.052 | 0.337 | 5.655 |
| HLA-A*01:01:01:01~HLA-C*07:01:01:01 | 3.000 | 0.052 | 0.275 | 4.386 |
| HLA-A*11:01:01:01~HLA-C*04:01:01:01 | 3.000 | 0.052 | 0.275 | 4.386 |
| HLA-A*02:01:01:01~HLA-C*01:02:01 | 3.000 | 0.052 | 0.678 | 6.832 |
| HLA-A*01:01:01:01~HLA-DPA1*01:03:01:05 | 4.000 | 0.069 | 0.383 | 5.816 |
| HLA-A*11:01:01:01~HLA-DPBl*04:01:01:01 | 5.000 | 0.086 | 0.470 | 4.934 |
| HLA-A*01:01:01:01~HLA-DPB1*02:01:02 | 4.000 | 0.069 | 0.370 | 4.859 |
| HLA-A*02:01:01:01~HLA-DQA1*01:02:02 | 6.000 | 0.107 | 0.408 | 7.539 |
| HLA-A*11:01:01:01~HLA-DQA1*01:01:01:02 | 3.000 | 0.054 | 0.708 | 12.968 |
| HLA-A*02:01:01:01~HLA-DQAl*03:01:01 | 3.000 | 0.054 | 0.479 | 4.168 |
| HLA-A*11:01:01:01~HLA-DQB1*05:03:O1:O1 | 3.000 | 0.056 | 0.707 | 12.399 |
| HLA-A*02:01:01:01~HLA-DRBl*16:01:01 | 4.000 | 0.069 | 0.448 | 5.521 |
| HLA-A*01:01:01:01~HLA-DRB1*11:04:01 | 3.000 | 0.052 | 0.337. | 5.655 |
| HLA-A*24:02:01:01~HLA-DRBl*03:01:01:01 | 3.000 | 0.052 | 0.545 | 11.845 |
| HLA-A*11:01:01:01~HLA-DRB1*01:01:01 | 4.000 | 0.052 | 0.710 | 13.536 |
| HLA-B*35:03:01~HLA-C*04:01:01:01 | 4.000 | 0.069 | 1.000 | 26.852 |
| HLA-B*51:01:01:01~HLA-C*15:02:01:01 | 4.000 | 0.069 | 0.482 | 8.883 |
| HLA-B*08:01:01:01~HLA-C*07:01:01:01 | 4.000 | 0.069 | 1.000 | 26.852 |
| HLA-B*51:01:01:01~HLA-C*01:02:01 | 4.000 | 0.069 | 1.000 | 20.622 |
| HLA-B*38:01:01~HLA-C*12:03:01:01 | 3.000 | 0.052 | 1.000 | 23.049 |
| HLA-B*27:05:02~HLA-C*02:02:02:01 | 3.000 | 0.052 | 1.000 | 42.709 |
| HLA-B*40:06:01:02~HLA-C*15:02:01:01 | 3.000 | 0.052 | 1.000 | 23.049 |
| HLA-B*18:01:01:02~HLA-DPAl*01:03:01:05 | 4.000 | 0.069 | 1.000 | 18.357 |
| HLA-B*40:06:01:02~HLA-DPA1*01:03:01:01 | 3.000 | 0.052 | 1.000 | 7.630 |
| HLA-B*08:01:01:01~HLA-DPA1*01:03:01:01 | 3.000 | 0.052 | 0.646 | 4.329 |
| HLA-B*38:01:01~HLA-DPBl*04:01:01:01 | 3.000 | 0.052 | 1.000 | 7.630 |
| HLA-B*18:01:01:O2~HLA-DPB1*04:02:01:01 | 3.000 | 0.052 | 0.726 | 24.031 |
| HLA-B*51:01:01:01~HLA-DQA1 “01:02:02 | 5.000 | 0.089 | 0.378 | 7.108 |
| HLA-B*08:01:01:01~HLA-DQAl*05:01:01:02 | 4.000 | 0.071 | 1.000 | 43.938 |
| HLA-B*40:06:01:02~HLA-DQAl*01:04:02 | 3.000 | 0.054 | 1.000 | 56.000 |
| HLA-B*18:01:01:02~HLA-DQA1*05:05:01:01 | 3.000 | 0.054 | 0.696 | 9.589 |
| HLA-B*51:01:01:01-HLA-DQBl*05:02:01 | 4.000 | 0.074 | 0.318 | 4.811 |
| HLA-B*18:01:01:02~HLA-DQB1*03:01:01:03 | 3.000 | 0.056 | 0.700 | 10.584 |
| HLA-A*01:01:01:03~HLA-B*52:01:01:02 | 4.000 | 0.069 | 0.482 | 8.883 |
| HLA-A*01:01:01:01~HLA-B*35:02:01 | 4.000 | 0.069 | 1.000 | 31.302 |
| HLA-A*01:01:01:01~HLA-B*13:02:01 | 3.000 | 0.052 | 1.000 | 58.000 |
| HLA-A*01:01:01:01~HLA-B*15:17:01:01 | 3.000 | 0.052 | 0.716 | 16.033 |
| HLA-C”15:02:01:01~HLA-DPAl*01:03:01:01 | 5.000 | 0.086 | 0.596 | 6.816 |
| HLA-C*07:01:01:01~HLA-DPA1*01:03:01:05 | 5.000 | 0.086 | 0.537 | 11.444 |
| HLA-C*06:02:01:01~HLA-DPAl*01:03:01:05 | 3.000 | 0.052 | 0.691 | 8.777 |
| HLA-C*07:01:01:01~HLA-DPBl*105:01 | 3.000 | 0.052 | 0.710 | 13.536 |
| HLA-C*02:02:02:01~HLA-DPBl*04:01:01:01 | 3.000 | 0.052 | 0.646 | 4.329 |
| HLA-C*07:01:01:01~HLA-DQAl*05:01:01:02 | 5.000 | 0.089 | 1.000 | 32.941 |
| HLA-C*12:03:01:01~HLA-DQAl*03:01:01 | 3.000 | 0.054 | 0.552 | 13.941 |
| HLA-C”15:02:01:01~HLA-DQA1*01:04:02 | 3.000 | 0.054 | 1.000 | 22.189 |
| HLA-C*01:02:01~HLA-DQA1*01:02:02 | 3.000 | 0.054 | 0.689 | 8.363 |
| HLA-C*01:02:01~HLA-DQBl*05:02:01 | 3.000 | 0.056 | 0.700 | 10.584 |
| HLA-C*07:01:01:01~HLA-DRBl*03:01:01:01 | 4.000 | 0.069 | 0.503 | 12.581 |
| HLA-C*15:02:01:01~HLA-DRBl*14:04:01 | 3.000 | 0.052 | 1.000 | 23.049 |
| HLA-C*01:02:01~HLA-DRB1*16:01:01 | 3.000 | 0.052 | 0.716 | 16.033 |
| HLA-DPA1 “01:03:01:01~HLA-DPBl*02:01:02 | 12.000 | 0.207 | 1.000 | 36.491 |
| HLA-DPA1*01:03:01:02~HLA-DPBl*04:01:01:01 | 10.000 | 0.172 | 1.000 | 29.142 |
| HLA-DPA1*01:03:01:04~HLA-DPB1*04:01:01:01 | 6.000 | 0.103 | 0.293 | 3.126 |
| HLA-DPA1*01:03:01:05~HLA-DPBl*04:02:01:01 | 5.000 | 0.086 | 1.000 | 23.379 |
| HLA-DPA1*01:03:01:04~HLA-DQA1*01:02:02 | 5.917 | 0.106 | 0.412 | 8.514 |
| HLA-DPA1*01:03:01:05~HLA-DQAl*05:05:01:01 | 4.196 | 0.075 | 0.278 | 3.842 |
| HLA-DPA1*01:03:01:05~HLA-DQAl*05:01:01:02 | 3.720 | 0.066 | 0.682 | 10.433 |
| HLA-DPA1*01:03:01:02~HLA-DQAl*01:01:01:02 | 3.000 | 0.054 | 0.702 | 11.090 |
| HLA-DPA1*01:03:01:04~HLA-DQAl*02:01:01:01 | 3.000 | 0.054 | 0.682 | 7.343 |
| HLA-DPA1*01:03:01:05~HLA-DQB1*03:01:01:03 | 4.775 | 0.088 | 0.410 | 7.111 |
| HLA-DPB1*04:01:01:01~HLA-DQA1*03:01:01 | 4.000 | 0.071 | 0.720 | 7.115 |
| HLA-DPB1*04:01:01:01~HLA-DQA1*01:01:01:02 | 4.000 | 0.071 | 1.000 | 10.769 |
| HLA-DPB1*04:02:01:01~HLA-DQA1*05:05:01:01 | 3.000 | 0.054 | 0.513 | 6.647 |
| HLA-DPB1*04:01:01:01~HLA-DQA1*02:01:01:01 | 3.000 | 0.054 | 0.650 | 4.550 |
| HLA-DPB1*04:01:01:01~HLA-DQB1*03:02:01 | 3.000 | 0.056 | 0.654 | 4.802 |
| HLA-DPB1*04:02:01:01~HLA-DQB1*03:01:01:03 | 3.000 | 0.056 | 0.520 | 7.450 |
| HLA-DPB1*04:01:01:01~HLA-DQB1*02:02:01:01 | 3.000 | 0.056 | 1.000 | 8.259 |
| HLA-DPB1*04:01:01:01~HLA-DRBl*01:01:01 | 4.000 | 0.069 | 1.000 | 10.362 |
| HLA-DPB1*04:02:01:01~HLA-DRB1*11:04:01 | 3.000 | 0.052 | 0.545 | 11.845 |
| HLA-DQA1*0l:02:02~HLA-DQBl*05:02:01 | 9.000 | 0.173 | 1.000 | 45.712 |
| HLA-DQA1*05:05:01:01~HLA-DQB1*03:01:01:03 | 8.000 | 0.154 | 0.862 | 34.000 |
| HLA-DQA1*05:05:01:01~HLA-DRB1*11:04:01 | 7.000 | 0.125 | 1.000 | 36.800 |
| HLA-DQA1*01:02:02~HLA-DRBl*16:01:01 | 7.000 | 0.125 | 1.000 | 32.727 |
| HLA-DQA1*01:01:01:02~HLA-DRBl*01:01:01 | 4.000 | 0.071 | 1.000 | 56.000 |
| HLA-DQA1*05:01:01:02~HLA-DRBl*03:01:01:01 | 4.000 | 0.071 | 0.771 | 22.871 |
| HLA-DQA1*02:01:01:01~HLA-DRB1*07:01:01:01 | 4.000 | 0.071 | 1.000 | 43.938 |
| HLA-DQB1*03:01:01:03~HLA-DRBl*11:04:01 | 7.000 | 0.130 | 1.000 | 40.213 |
| HLA-DQB1*05:02:01~HLA-DRB1*16:01:01 | 6.000 | 0.111 | 1.000 | 33.750 |
| HLA-DQB1*02:01:01~HLA-DRB1*03:01:01:01 | 5.000 | 0.093 | 0.795 | 18.793 |
Table 5.
The most frequent haplotypes in young Italians (n = 292).
| Haplotype | Count | Haplotype Frequency | D′ | D′ Chi-square |
|---|---|---|---|---|
| A*02:01:01:01~B*51:01:01:01 | 28.883 | 0.049 | 0.267 | 18.599 |
| A*01:01:01:01~B*08:01:01:01 | 22.118 | 0.038 | 0.670 | 98.410 |
| A*03:01:01:01~B*07:02:01 | 15.725 | 0.027 | 0.314 | 38.752 |
| A*01:01:01:01~C*07:01:01:01 | 29.085 | 0.050 | 0.282 | 42.492 |
| A*01:01:01:01~C*06:02:01:01 | 25.316 | 0.043 | 0.444 | 70.589 |
| A*02:01:01:01~C*04:01:01:06 | 18.643 | 0.032 | 0.233 | 9.399 |
| A*02:01:01:01~DPBl*04:01:01:01 | 49.989 | 0.096 | 0.134 | 5.951 |
| A*24:02:01:01~DPB1*04:01:01:01 | 24.304 | 0.047 | 0.188 | 4.498 |
| A*02:01:01:01~DQBl*03:01:01:03 | 40.776 | 0.070 | 0.098 | 5.259 |
| A*01:01:01:01~DQBl*02:01:01 | 22.989 | 0.039 | 0.291 | 37.816 |
| A*02:01:01:01~DRBl*11:01:01:01 | 22.106 | 0.038 | 0.143 | 5.233 |
| A*01:01:01:01~DRBl*03:01:01:01 | 21.649 | 0.037 | 0.265 | 31.434 |
| B*07:02:01~C*07:02:01:03 | 38.000 | 0.065 | 1.000 | 538.294 |
| B*08:01:01:01~C*07:01:01:01 | 30.000 | 0.051 | 0.962 | 185.659 |
| B*51:01:01:01~C*l 5:02:01:01 | 29.000 | 0.050 | 0.927 | 217.140 |
| B*18:01:01:02~C*07:01:01:01 | 26.96 | 0.046 | 0.518 | 82.171 |
| B*35:01:01:02~C*04:01:01:01 | 25.000 | 0.043 | 0.452 | 116.836 |
| B*18:01:01:02~DPBl*04:01:01:01 | 20.296 | 0.039 | 0.237 | 5.413 |
| B*51:01:01:01~DPBl*02:01:02 | 18.687 | 0.036 | 0.162 | 8.7949 |
| B*07:02:01~DPBl*04:01:01:01 | 17.998 | 0.035 | 0.242 | 4.908 |
| B*51:01:01:01~DQBl*03:01:01:03 | 26.126 | 0.045 | 0.201 | 9.900 |
| B*08:01:01:01~DQBl*02:01:01 | 23.635 | 0.041 | 0.735 | 154.548 |
| B*18:01:01:02~DQBl*03:01:01:03 | 23.531 | 0.040 | 0.360 | 20.962 |
| B*44:03:01:01~DRBl*07:01:01:01 | 23.971 | 0.041 | 0.832 | 122.961 |
| B*08:01:01:01~DRBl*03:01:01:01 | 23.95 | 0.041 | 0.747 | 159.349 |
| B*07:02:01~DRBl*l 5:01:01:01 | 23.649 | 0.040 | 0.544 | 168.409 |
| C*07:01:01:01~DQBl*02:01:01 | 28.753 | 0.049 | 0.394 | 63.595 |
| C*07:01:01:01~DQBl*03:01:01:03 | 27.547 | 0.047 | 0.130 | 5.248 |
| C*04:01:01:06~DQBl*03:01:01:03 | 21.148 | 0.036 | 0.293 | 13.815 |
| C*07:01:01:01~DRBl*03:01:01:01 | 28.645 | 0.049 | 0.392 | 62.923 |
| C*06:02:01:01~DRBl*07:01:01:01 | 24.795 | 0.042 | 0.424 | 58.091 |
| C*07:02:01:03~DRBl*15:01:01:01 | 21.529 | 0.037 | 0.533 | 148.9949 |
| DPB1*04:01:01:01~DQBl*03:01:01:03 | 50.218 | 0.097 | 0.122 | 5.091 |
| DPB1*04:01:01:01~DQBl*06:02:01 | 16.013 | 0.031 | 0.219 | 3.677 |
| DQB1*03:01:01:03~DRB1*11:01:01:01 | 64.000 | 0.110 | 0.960 | 221.776 |
| DQB1*02:01:01~DRBl*03:01:01:01 | 60.000 | 0.103 | 1.000 | 584.000 |
| DQB1*02:02:01:01~DRBl*07:01:01:01 | 56.000 | 0.096 | 0.922 | 343.342 |
| DQB1*03:01:01:03~DRBl*11:04:01 | 52.000 | 0.089 | 1.000 | 184.485 |
5. Discussion
Several studies during the last decades focused on the impact of HLA polymorphism in human longevity [22–27]. However data generated in different populations and ethnic groups are contradicting due to the major methodological problems such as limited sample size, typing approach and focusing on single loci different inclusion criteria and age limits, and neglecting of possible sex-related effects. Despite these limitations, HLA remains as one of the major genetics regions associated with human longevity as shown by GWAS [28], due to its central role in the development of adaptive immune response and modulation of individual’s response to life threatening diseases.
Here we present the data report from the collaborative study including four laboratories within the 17th IHIW component “Immunogenetics of Ageing” on unrelated elderly individuals and young controls. Some of the investigators involved in this study previously participated in this workshop component. Within the 17th IHIW data from additional populations were collected and the study was further extended including allele-level typing and examination of additional loci. However considering the relatively small number of elderly individuals, selected according to the SENIEUR protocol [20] and included in the analysis, the data presented here should be considered as preliminary and it is expected more individuals to be analyzed in the course of study within 18th IHIW. The SENIEUR protocol applied in the present study establishes a strict admission criteria based on clinical and laboratory data and restricts pharmacological interference. Therefore the protocol excludes elderly people with underlying disease and could be helpful to limit alterations caused by disease interference.
Analysis of data in four populations showed that some HLA class I and class II alleles had positive statistically significant association with healthy aging. Our previous studies within IHIW components “Ïmmunogenetics of Ageing” showed similar HLA allele and haplotype distributions in the Bulgarians, Romanians and Italians [17]. Since the number of individuals from the Kuwaiti populations was very small only HLA allele distribution was analyzed. Our data showing decreased frequencies of 2 alleles in this population have to be confirmed in a larger data set.
Our results on increased frequency of HLA-DRB1*07:01:01:01, confirmed and extended some of the previous reports showing positive association of HLA-DRB1*07 with longevity in the Greek and French populations [25]. Most of the studies on HLA association in ageing were focused on HLA class II region. However in our project we identified positive association of longevity with HLA-A*01:01:01:01, HLA-C*07:01:02, HLA-B*35:02:01, and HLA-B*15:17:01:01. HLA-A*01 and C*07 alleles as a part of the ancestral haplotype HLA-A1-C7-B8-DR3 [34] was shown to be associated with a shift for Th1 to Th2 immune response characteristic for ageing. Despite the role of HLA-B35 in HIV progression, HLA-B*35 has been confirmed as a protective allele for CML [30,31]. HLA-A*01:01 was described as a protective allele for CLL [32,33]. Stratification according to gender showed that some of the alleles appeared to be beneficial as longevity factors in males or females only, such as HLA-B*35:02:01 that was increased in elderly females as well as HLA-C*07:01:02 and HLA-B*15:17:01:01 increased only in the elderly male group. Also, we observed additional allelic associations such as increased frequency of HLA-A*30:01:01, HLA-B*13:02:01, HLA-C*06:02:01, HLA-DRB1*01:01:01, DQA1*01:01:01:02 in females. Alleles HLA-DQA1*01:01 and HLA-DRB1*01:01 were also increased in Japanese centenarians [26], although no sex-specific association was reported. The possible mechanisms responsible for the different HLA associations in elderly of both sexes are not clarified yet although it is likely to be related to the influence of on the immune reactivity [34].
Similarly to our previous study, haplotype analysis showed that haplotypes bearing HLA-DRB1*11 are prevalent among the elderly population [17]. Specifically, a statistically significant difference was observed for the HLA-C*16:02:01~HLA-DRB1*11:03:01 haplotype. HLA-DRB1*11 alleles and haplotypes were shown to be associated with protection to autoimmune diseases in Southern European populations and protection to HCV and HBV [35]. In the present study, we observed positive associations of longevity with haplotypes bearing HLA-C*04:01:01, HLA-DQB1*03:01 and HLA-DRB1*07:01:01 alleles, which are known to be associated with better control of hepatitis B and hepatitis C infection [35]. Additionally 4 haplotypes for the HLA-01:01:01 allele, described as a protective for hematological malignancies were statistically significantly increased in elderly form the Bulgarian population. Considering the number of subjects these results must be confirmed in a larger cohort of elderly individuals.
The present study to the best of our knowledge is one of the few using Next Generation Sequencing methods to clarify HLA associations at allele level in longevity. The major limitation of our study is a relatively small number of elderly subjects and deviations from Hardy Weinberg equilibrium. Therefore one of the tasks within the next IHIW is to collect additional samples of different age groups and different populations and to analyze HLA polymorphism at maximum resolution level. Studies of families with longevity members could be a useful tool in addition to cross sectional studies to characterize better accumulation of specific HLA alleles and haplotypes that are beneficial for reaching extreme limits of life span.
In conclusion, HLA allele level data generated under the 17th IHW component “Immunogenetics of Ageing” allowed further elucidation of HLA associations with longevity and suggested that survival and longevity might be related to natural selection of HLA alleles and haplotypes conferring disease resistance or susceptibility. Therefore, HLA alleles and haplotypes could be informative immunogenetic marker for successful ageing. Discovery of potential new markers that influence the longevity and susceptibility to age related diseases will contribute for development of strategies for rejuvenating the immune system and preventing replicative senescence.
Supplementary Material
Acknowledgements
We are thankful to Omixon, Budapest, Hungary and Illumina, Inc., San Diego, CA for supporting the project by providing some of the re-agents and technical support. LC (Stanford University) was supported by grant U19NS095774 from the U.S. National Institutes of Health (NIH). SAS and RA (Kuwait University) was supported by a grant 2012-130-204 from the Kuwait Foundation for Advancement of Science (KFAS), Kuwait.
Abbreviations:
- HLA
Human Leukocyte Antigen
- KIR
Killer cell Immunoglobulin-like Receptor
- TNF
Tumor Necrosis Factor
- MBL2
Mannose Binding Lectin 2
- IHW
International HLA and Immunogenetics Workshop
- TLR
Toll-Like Receptor
- TCR
T-cell Receptor
- BCR
B-cell Receptor
- GWAS
Genome-Wide Association Study
- NGS
Next Generation Sequencing
- CMV
Cytomegalovirus
- EBV
Epstein–Barr Virus
- HBV
Hepatitis B Virus
- HIV
Human Immunodeficiency Virus
- CLL
Chronic Lymphocytic Leukemia
- CML
Chronic Myelogenous Leukemia
- AML
Acute Myelogenous Leukemia
Footnotes
Declaration of Competing Interest
The authors declare no competing financial or other interests.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.humimm.2019.07.287.
References
- [1].Martin GM, Oshima J, Gray MD, Poot M, What geriatricians should know about the Werner syndrome, J. Am. Geriatr. Soc 47 (1999) 1136. [DOI] [PubMed] [Google Scholar]
- [2].Fossel M, Human aging and progeria, J. Pediatr. Endocrinol. Metab 13 (Suppl. 6) (2000) 1477. [DOI] [PubMed] [Google Scholar]
- [3].Herskind AM, McGue M, Holm NV, Sørensen TI, Harvald B, Vaupel JW, The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900, Hum. Genet 97 (1996) 319. [DOI] [PubMed] [Google Scholar]
- [4].Mitchell BD, Hsueh WC, King TM, et al. , Heritability of life span in the Old Order Amish, Am. J. Med. Genet 102 (2001) 346. [DOI] [PubMed] [Google Scholar]
- [5].Gavrilova N, Gavrilov L, Data resources for biodemographic studies in familial clustering of human longevity, Demogr. Res 1 (1999) 1. [Google Scholar]
- [6].McGue M, Vaupel JW, Holm N, Harvald B, Longevity is moderately heritable in a sample of Danish twins born 1870–1880, J. Gerontol 48 (1993) B237. [DOI] [PubMed] [Google Scholar]
- [7].Herskind AM, McGue M, Holm NV, Sorensen TI, Harvald B, Vaupel JW, The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900, Hum. Genet 97 (1996) 319. [DOI] [PubMed] [Google Scholar]
- [8].Yashin AI, Iachine IA, Genetic analysis of durations, Correlated frailty model applied to survival Danish Twins, Genet. Epidemiol 12 (1995) 529. [DOI] [PubMed] [Google Scholar]
- [9].Perls TT, Wilmoth J, Levenson R, et al. , Life-long sustained mortality advantage of siblings of centenarians, Proc. Natl. Acad. Sci. U.S.A 99 (2002) 8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chamorro A, Role of inflammation in stroke and atherothrombosis, Cerebrovasc. Dis 17 (Suppl. 3) (2004) 1–5. [DOI] [PubMed] [Google Scholar]
- [11].Dandona P, Aljada A, Bandyopadhyay A, Inflammation: the link between insulin resistance, obesity and diabetes, Trends Immunol. 25 (2004) 4. [DOI] [PubMed] [Google Scholar]
- [12].Franceschi C, Bonafe M, Valensin S, et al. , Inflamm-aging. An evolutionary perspective on immunosenescence, Ann. N. Y. Acad. Sci 908 (2000) 244. [DOI] [PubMed] [Google Scholar]
- [13].Cavallone L, Bonafe M, Olivieri F, et al. , The role of IL-1 gene cluster in longevity: a study in Italian population, Mech. Ageing Dev 124 (2003) 533. [DOI] [PubMed] [Google Scholar]
- [14].Amadori A, Zamarchi R, De Silvestro G, et al. , Genetic control of the CD4/CD8 T-cell ratio in humans, Nat. Med 1 (1995) 1279. [DOI] [PubMed] [Google Scholar]
- [15].Clementi M, Forabosco P, Amadori A, et al. , CD4 and CD8 T lymphocyte inheritance. Evidence for major autosomal recessive genes, Hum. Genet 105 (1999) 337. [DOI] [PubMed] [Google Scholar]
- [16].Ferguson FG, Immune parameters in a longitudinal study in a very old population of Swedish people: a comparison between survivors and non-survivors, J. Gerontol 50A (6) (1995) B378. [DOI] [PubMed] [Google Scholar]
- [17].Naumova E, Pawelec G, Ivanova M, Constantinescu I, Bogunia-Kubik K,Lange A, Qguz F, Carin M, 14th International HLA and Immunogenetics Workshop: report on the immunogenetics of aging, Tissue Antigens 69 (2007) 304. [DOI] [PubMed] [Google Scholar]
- [18].Naumova E, Ivanova M, Pawelec G, Constantinescu I, Bogunia-Kubik K, Lange A, Qguz F, Carin M, Franceschi C, Caruso C, Middleton D, ‘Immunogenetics of Aging’: report on the activities of the 15th International HLA and Immunogenetics Working Group and 15th International HLA and Immunogenetics Workshop, Tissue Antigens 77 (3) (2011) 187. [DOI] [PubMed] [Google Scholar]
- [19].Naumova E, Ivanova M, Pawelec G, Constantinescu I, Bogunia-Kubik K, Lange A, Oguz F, Ozdilli K, Franceschi C, Caruso C, Mishra M, Middleton D, 16(th) IHIW: immunogenetics of aging, Int. J. Immunogenet 40 (1) (2013) 77. [DOI] [PubMed] [Google Scholar]
- [20].Ligthart GJ, Corberand JX, Fournier C, Galanaud P, Hijmans W, Kennes B, Müller-Hermelink HK, Steinmann GG, Admission criteria for immunogerontological studies in man: the SENIEUR protocol, Mech. Ageing Dev 28(1) (1984) 47. [DOI] [PubMed] [Google Scholar]
- [21].Lancaster AK, Single RM, Solberg OD, Nelson MP, Thomson G, PyPop update—a software pipeline for large-scale multilocus population genomics, Tissue Antigens 69 (Suppl. 1) (2007) 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Soto-Vega E, Richaud-Patin Y, Llorente L, Human leukocyte antigen class I, class II, and tumor necrosis factor-alpha polymorphisms in a healthy elder Mexican Mestizo population, Immun. Ageing 32 (2005) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lagaay AM, D’Amaro J, Ligthart GJ, Schreuder GM, Van Rood JJ, Hijmans W, Longevity and heredity in humans. Association with the human leucocyte antigen phenotype, Ann. N. Y. Acad. Sci 621 (1991) 78–89. [DOI] [PubMed] [Google Scholar]
- [24].Henon N, Schachter F, Genetics of longevity CR Seances Soc. Biol. Fil 191 (1997) 553–562. [PubMed] [Google Scholar]
- [25].Ivanova R, Henon N, Lepage V, et al. , HLA-DR alleles display sex-dependent effects on survival and discriminate between individual and familial longevity, Hum. Mol. Genet 7 (1998) 187–194. [DOI] [PubMed] [Google Scholar]
- [26].Takata H, Suzuki M, Ishii T, et al. , Influence of major histocompatibility complex region genes on human longevity among Okinawan-Japanese centenarians and nonagenarians, Lancet 2 (1987) 824–826. [DOI] [PubMed] [Google Scholar]
- [27].Yong-Xing M, Yue Z, Zan-Shun W, Chuan-Fu W, Su-Ying C, Mao-Tong Z, et al. , HLA and longevity or aging among Shanghai Chinese, Mech. Ageing Dev. 94 (1–3) (1997) 191–198. [DOI] [PubMed] [Google Scholar]
- [28].Yang F, Sun L, Zhu X, Han J, Zeng Y, Nie C, Yuan H, Li X, Shi X, Yang Y, Hu C, Lv Z, Huang Z, Zheng C, Liang S, Huang J, Wan G, Qi K, Qin B, Cao S, Zhao X, Zhang Y, Yang Z, Correction: Identification of new genetic variants of HLA-DQB1 associated with human longevity and lipid homeostasis-a cross-sectional study in a Chinese population, Aging (Albany NY) 10 (3) (2018) 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Naumova E, Ivanova M, Pawelec G, Immunogenetics of ageing, Int. J. Immunogenet 38 (5) (2011) 373. [DOI] [PubMed] [Google Scholar]
- [30].Rea IM, Middleton D, Is the phenotypic combination A1B8Cw7DR3 a marker for male longevity? J. Am. Geriatr. Soc 42 (1994) 978–983. [DOI] [PubMed] [Google Scholar]
- [31].Naugler C, Liwski R, Human leukocyte antigen class I alleles and the risk of chronic myelogenous leukemia: a meta-analysis, Leuk. Lymphoma 51 (7) (2010) 1288. [DOI] [PubMed] [Google Scholar]
- [32].Fernández-Torres J, Flores-Jiménez D, Arroyo-Pérez A, Granados J, López-ReyesA. HLA-B*40 allele plays a role in the development of acute leukemia in Mexican population: a case-control study, Biomed. Res. Int (2013:) 705862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Urayama KY, Thompson PD, Taylor M, Trachtenberg EA, Chokkalingam AP, Genetic variation in the extended major histocompatibility complex and susceptibility to childhood acute lymphoblastic leukemia: a review of the evidence, Front. Oncol 3 (2013) 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Candore G, Balistreri CR, Listì F, Grimaldi MP, Vasto S, Colonna-Romano G, Franceschi C, Lio D, Caselli G, Caruso C, Immunogenetics, gender, and longevity, Ann. N. Y. Acad. Sci 1089 (2006) 516. [DOI] [PubMed] [Google Scholar]
- [35].Singh R, Kaul R, Kaul A, Khan K, A comparative review of HLA associations with hepatitis B and C viral infections across global populations, World J. Gastroenterol 13 (12) (2007) 1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.