Abstract
Cell fate determination is influenced by interactions between master transcription factors (TFs) and cis-regulatory elements. Hepatocyte nuclear factor 4 alpha (HNF4A), a liver-enriched TF, acts as a master controller in specification of hepatic progenitor cells by regulating a network of TFs to control the onset of hepatocyte cell fate. Using analysis of genome-wide histone modifications, DNA methylation and hydroxymethylation in mouse hepatocytes, we show that HNF4A occupies active enhancers in hepatocytes and is essential for active histone and DNA signatures, especially H3K27ac and 5-hydroxymethylcytosine (5hmC). In mice lacking HNF4A protein in hepatocytes, we observed a decrease in both H3K27ac and hydroxymethylation at regions bound by HNF4A. Mechanistically, HNF4A-associated hydroxymethylation (5hmC) requires its interaction with TET3, a protein responsible for oxidation from 5mC to 5hmC. Furthermore, HNF4A regulates TET3 expression in liver by directly binding to an enhancer region. In conclusion, we identified that HNF4A is required for the active epigenetic state at enhancers that amplifies transcription of genes in hepatocytes.
Keywords: Histone modification, hydroxymethylation, hepatoblasts, hepatocytes, TET3
INTRODUCTION
Hepatocyte nuclear factor 4A (HNF4A, also known as NR2A1) is a member of the nuclear receptor superfamily of transcription factors (TFs) and is expressed in the liver, pancreas, gastrointestinal tract and kidneys (1). In liver, HNF4A is required for normal development (2), including morphogenesis (3) and differentiation (4), and controls other TFs critical in liver cell fate as well as genes necessary for hepatocyte functions. HNF4A deficiency in mouse embryonic liver causes failure in hepatocyte differentiation (2), while in adult mice loss of HNF4A affects lipid homeostasis leading to liver steatosis (5). In humans, HNF4A mutations have been linked to maturity-onset diabetes of young type1 (MODY-1), a monogenic form of type 2 diabetes (6). In addition to maintaining expression of various genes associated with metabolic processes in liver, several studies have connected reduced HNF4A expression with liver cancer progression (7). HNF4A disruption in mature hepatocytes has been shown to result in epithelial-to-mesenchymal transition (EMT), a process associated with cancer progression (8). Conversely, its overexpression in hepatocellular carcinoma rodent models shows its role in tumour suppression (4, 7) by repressing the proliferative capacity of cancer cells(9). Taken together, these studies strongly support a role for HNF4A as a central regulator of hepatocyte identity by playing a crucial function in regulating liver gene expression (5, 10). However, the mechanisms through which HNF4A exerts its functions are not well understood.
TFs, such as HNF4A, primarily function through directly binding to DNA at sequence-specific motifs; however, the number of potential binding sites for a given TF across the genome far exceeds bound regions observed in vivo. We have shown that HNF4A binding is highly dynamic and that bound regions can change based on tissue and differentiation state (11). Genome-wide epigenetic analysis has proven that active cis-regulatory regions are located in open chromatin regions (12), while condensed chromatin is inaccessible and thus inactive. How chromatin structure is regulated in tissue- and differentiation stage-specific patterns and how TFs play a role in the adaptation of chromatin structure is only beginning to be explored.
Modifications of histone and DNA are the two main mechanisms of epigenetic control influencing chromatin condensation and DNA accessibility. Histone modifications have been associated with repressed, poised and active enhancers and promoters, and ultimately control gene activity (13, 14). In particular, lysine 4 of histone 3 monomethylation (H3K4me1) and trimethylation (H3K4me3) mark enhancers and promoters respectively (15). In addition, acetylation of lysine 27 of histone 3 (H3K27ac) is associated with the active state of both elements. DNA methylation (5-methylcytosine or 5mC) is associated with gene repression (16) and can disrupt the binding of many TFs (17, 18). DNA hydroxymethylation or 5-hydroxymethylcytosine (5hmC) is a recently discovered DNA modification that is produced by the oxidation of 5mC due to activity of ten-eleven translocation proteins (TET1, TET2 and TET3). 5hmC plays an important role during mammalian development and cellular differentiation: Tet½/3 deficiency in ES cells (triple knockout cells) reduces their differentiation capabilities and also limits their developmental potential during embryogenesis (19). Genome-wide enrichment studies in human and mouse embryonic stem cells (ESCs) confirms the presence of 5hmC at promoters and enhancers in addition to gene bodies (20).
A mechanistic understanding of the interrelationship between TF binding, histone modification and DNA methylation states is only beginning to emerge. The establishment and maintenance of patterns of histone and DNA modifications in a tissue-specific manner may be controlled by TFs directly or indirectly interacting with epigenetic modifiers (21), such as the histone acetylase CBP/p300 (22). Recent reports have identified TFs that interact with TET proteins to maintain low 5mC and high 5hmC at enhancers (23, 24).
Studies transducing a combination of TFs into human fibroblasts to produce highly functional hepatocytes support HNF4A’s involvement in chromatin remodelling. These studies revealed that HNF4A is crucial for generating induced hepatocyte-like cells (iHeps) (25, 26) as HNF4A overexpression alone may be sufficient to generate these cells (27). These experiments suggest that HNF4A may help establish an appropriate epigenetic state to allow binding of additional TFs. Here, we show that HNF4A maintains an active epigenetic state at cis-regulatory elements in hepatocytes and during liver differentiation. Loss of HNF4A during liver differentiation confirmed that HNF4A is required for establishment and maintenance of H3K27ac. Furthermore, HNF4A-mediated oxidation of 5mC to 5hmC during differentiation of hepatoblasts to hepatocytes is achieved by its interaction with TET3, a newly identified HNF4A target gene in hepatoblasts. Overall, we confirm that HNF4A-driven epigenetic changes are required for expression of target genes during hepatocyte differentiation. These findings provide a novel mechanism through which HNF4A works as a master regulator to shape and maintain the epigenetic landscape in liver.
MATERIALS AND METHODS
Mice and cell preparation
Mouse protocols were approved by the University of British Columbia Animal Care committee. Livers were collected from 8 week old female C57BL6/J mice, perfused with 0.005% collagenase (STEMCELL Technologies) in PBS, minced with a razor blade and passed through a 40μm cell strainer. Hepatocytes were separated by low-speed centrifugation (50g x 30sec) (28). Hepatoblast isolation from E14.5 livers was previously described (11). AlbERT2cre- and Hnf4aFl/Fl mouse lines were described in an earlier study (4). These two mouse strains were crossed to produce Tamoxifen inducible, hepatocyte-specific cre recombinase expressing mouse. Mice used in this study were 6–8 weeks old and were treated with Tamoxifen (2mg/ml, intraperitoneal, subcutaneously) every other day. Mice were euthanized by cervical dislocations under anesthesia and livers were collected on the 7th day. Floxed, cre negative mice, without Tamoxifen were used as control. The HNF4A conditional knockout animals were housed at NIH and procedures were carried out in accordance with the Institute of Laboratory Animal Resources guidelines and approved by the National Cancer Institute Animal Care and Use Committee. They were fed NIH-31 chow diet (at NIH). C57BL6/J were house at BC Cancer, with procedures approved at UBC as stated above and fed Envigo-Teklad 2920X. All mice were housed in a pathogen-free animal facility under a standard 12-hour light/dark cycle.
5mC and 5hmC Analysis
Low input MeDIP-seq and hMeDIP-seq was previously described (29). DNA was sonicated to 200–400bp using E-220 sonicator (Covaris) followed by end repair adapter ligation, and immunoprecipitation using anti-5mC or anti-5hmC antibodies. Two PCR reactions of 12 cycles using paired-end, indexed, Illumina PCR primers amplified the DNA for library preparation. Size selection of fragments <700bp was achieved on 8% polyacrylamide gels. Paired-end, 125 nucleotide reads were generated on Illumina MiSeq or Illumina Hiseq 2500 platform (v3 chemistry). Bioinformatic analysis of MeDIP and hMeDIP-seq data was performed as described in supplementary material and methods section. MeDIP and hMeDIP for locus-specific qPCR analysis were performed as described in Supplementary materials and methods.
Histone modification and transcription factor analysis
ChIP was performed as previously described (30) with the following modifications. Cells or minced tissues were fixed with 1% formaldehyde (Sigma) for 10 minutes then quenched using 125 mM glycine for 5 minutes at room temperature. DLK1+ MACs purified hepatoblasts (31) were fixed prior to cell sorting. Cells were lysed for 15 min on ice in lysis buffer (10mM Tris-HCl pH8, 10mM NaCl, 3mM MgCl2, 0.5% Nonidet P-40) with proteinase inhibitor cocktail (1x PIC, Roche). Nuclei were pelleted and lysed for 30–60 min on ice with nuclear lysis buffer (50 mM Tris-HCl pH 8, 5 mM EDTA, 1% SDS, 1X PIC). Samples were sonicated with Q Sonica Sonicator Q700 for 10 min total in 30 second cycles to generate fragments of 200–500bp. Chromatin was diluted in ChIP dilution buffer (16.7 mM Tris-HCl pH 8, 167 mM NaCl, 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 1X PIC) and Protein A/G beads (ThermoFisher) was added to clear the samples prior to addition of antibody. Antibody (3μg for TF and 2ug for histone modifications) was added to samples and incubated overnight at 4°C. Protein A/G beads (ThermoFisher) were added and samples were placed on a rotator at 4°C for 4 hours. Samples were washed with low salt (20 mM Tris-HCl pH 8, 0.15 M NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), high salt (20 mM Tris-HCl pH 8, 0.5 M NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), lithium chloride (10 mM Tris-HCl pH 8, 0.25 M LiCl, 1 mM EDTA, 1% Nonidet P-40, 1% sodium deoxycholate) and TE buffer for 10 minutes each followed by elution with 0.1M sodium hydroxycholate in 1% SDS. Proteinase K and RNAse A (Invitrogen) and 0.192 M NaCl were added and incubated overnight at 65 °C. DNA purification and qPCR analysis were the same as above. ChIP-seq library construction and Illumina Genome Analyzer sequencing was performed as described (15). Bioinformatic analysis of ChIP-seq data was performed as described in Supplementary material and methods section.
Co-immunoprecipitation (co-IP)
HEK293T cells were transfected with an HNF4A expression vector (32) and HA-tagged TET3 vector (Addgene #49446) alone and in combination using Polyethylenimine (PEI). Cells were lysed using lysis buffer (10mM Tris-HCl pH8, 10mM NaCl, 3mM MgCl2, 0.5% Nonidet P-40) and nuclei were collected on ice. CHAPS buffer (FIVEphoton BiochemicalsTm) was used for nuclear extraction and the lysate was incubated with 2ug of HNF4A antibody overnight at 4°C on a rocker. Protein A/G beads (ThermoFisher) were added (20ul) and samples were rotated for 30 minutes at RT. Beads were collected by centrifugation and extensively washed with lysis buffer. Proteins were eluted into SDS gel loading buffer by heating at 65°C. Proteins were separated using NuPAGE 4–12% gels (ThermoFisher), transferred to PVDF membranes, and analysed using antibodies specified in Table S2.
In situ proximity ligation assay (PLA)
Embryonic (E14.5) and adult (8 weeks old) mouse livers were fixed with 4% paraformaldehyde (Sigma) at 4°C for 16–48 hrs, put through a sucrose gradient at 4°C, and embedded in OCT (TissueTek). Sections (4 μm) were permeabilized with Triton-X (0.1% v/v in PBS). PLA was performed according to manufacturer’s instructions using the Duolink In situ PLA Kit (Sigma Aldrich). Fluorescent foci were imaged using the Zeiss Axio Imager M2. Cells were seeded on coverslips and washed twice with PBS before fixation and permeabilization.
Statistical analysis
Results are calculated as mean ±SEM derived from at least three independent experiments. Student’s t-test was used for qPCR data with GraphPad software. A p-value <0.05 was considered statistically significant.
RESULTS
Regions bound by HNF4A are associated with active histone and DNA modifications in hepatocytes
HNF4A is a well-studied master regulator of hepatocyte identity; however, how it contributes to epigenetic plasticity in liver is not well understood. To address this, we investigated epigenetic modifications associated with HNF4A binding in adult mouse hepatocytes. We used our previously published HNF4A (11), H3K4me1 and H3K4me3 (15), and newly generated H3K27ac ChIP-seq datasets and examined the enrichment of histone modifications around HNF4A bound regions (HBRs) (Fig. 1A). HNF4A antibody (Table. S2) used for ChIP-seq identifies both embryonic and adult isoforms. Analysis of HNF4A ChIP-seq data revealed its preferential binding to intronic (47.35%), and distal intergenic regions (33.33%) with a small proportion at gene promoters (9.81%) (Fig. S1A). Examination of HBRs demonstrated a high enrichment of H3K4me1 and H3K27ac flanking these regions (Fig. 1A), suggestive of active enhancers. The active status of these enhancers was further confirmed by examination of mouse liver DNase-seq data (ENCODE), which shows very high signals, demonstrative of open chromatin at HBRs (Fig. S1B). It is well established that active enhancers are associated with high 5hmC and low 5mC DNA modifications (14, 33). To examine the level of 5mC and 5hmC at HBRs, we isolated hepatocytes from mouse liver and performed MeDIP (Methylated DNA Immunoprecipitation) and hMeDIP (5-hydroxymethylated DNA Immunoprecipitation) followed by high-throughput sequencing. We observed that HBRs are surrounded by high regions of 5hmC but not 5mC (Fig. 1A) as illustrated by the combined profile plot (Fig. 1B). The epigenetic status of HBRs, such as those near the genes Cdc42bpb, Rfx4 (Fig. S1C and D), Mgst3 and Ido2 (Fig. S1C & D), demonstrate features of active enhancers. To validate our sequencing data, we confirmed 5mC depletion and 5hmC enrichment by MeDIP- and hMeDIP-qPCR at Cdc42bpb, Rfx4, Mgst3, Ido2, and Tet3 HBRs (Fig. 1E). Dazl, Tbx15 and IGd were included as positive, negative and no CpG controls, respectively. As expected, the level of 5mC at CpG islands associated with Dazl and Tbx15 was very similar to what we observed in our sequencing data (Fig. S1E and F). Overall, our data show that HBRs are enriched for H3K4me1 and H3K27ac, DNaseI hyper-sensitivity and 5hmC, which together indicate that HNF4A is primarily bound at active enhancers in hepatocytes.
Fig.1. Active enhancer signatures are associated with HNF4A bound regions (HBRs).
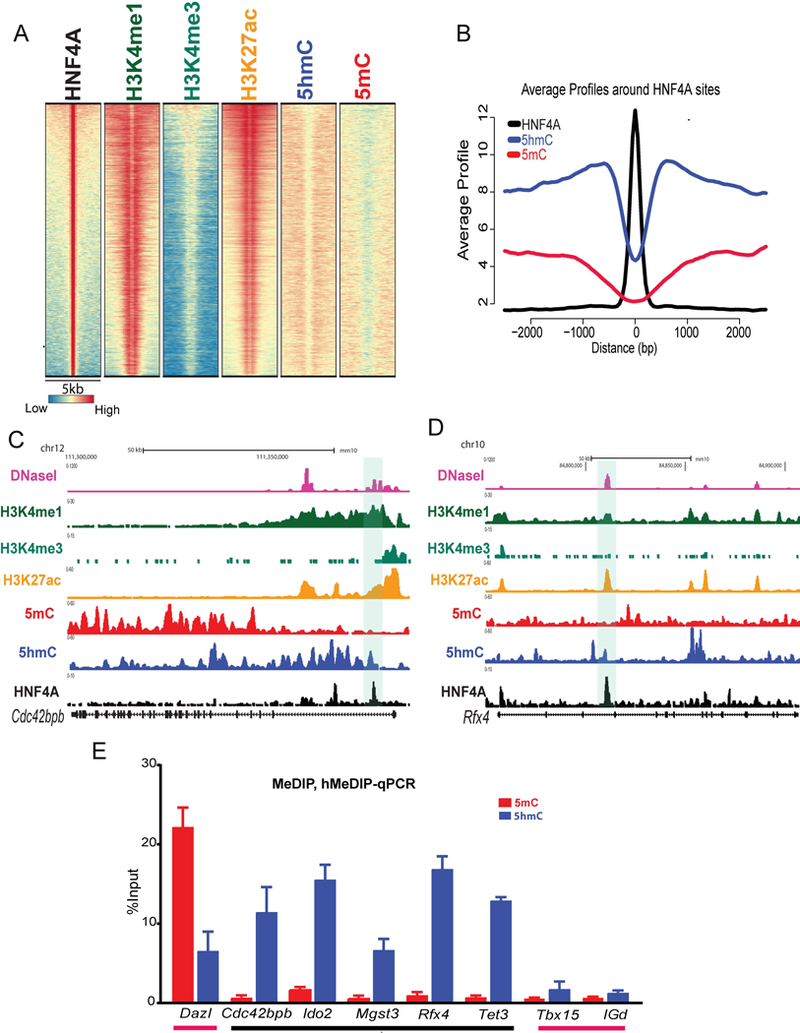
(A) Heatmap displaying the genomic representation of histone (H3K4me1, H3K4me3, and H3K27ac) and DNA modification (5mC and 5hmC) patterns at HBRs in mouse hepatocytes. Each line represents a 5kb window around the summit of a HNF4A bound region and the pattern of modifications around it. The colours, blue, yellow and red, represent low, moderate and high enrichment respectively. (B) Aggregation plots of MeDIP-seq (5mC,red) and hMeDIP-seq (5hmC, blue) signals at HBRs (black line). (C and D) Genome browser view of Cdc42bpb and Rfx4 loci showing enrichment of HNF4A (black), H3K27ac (orange), H3K4me1 (dark green), H3K4me3 (light green), 5mC (red), 5hmC (blue) and DNase-seq signals (pink) at HBR (highlighted). (E) Bar graph of qPCR enrichment following IP of 5mC (MeDIP, Red) and 5hmC (hMeDIP, blue) at HBR associated genes Cdc42bpb, Ido2, Mgst3, Rfx4 and Tet3 (black line). Dazl, Tbx15 and IGd genomic regions were used as 5mC positive, negative and non-CpG controls (pink line), respectively. (n=3). Values represent mean ±SEM.
Regions bound by HNF4A do not require FOXA2 to have a transcriptionally permissive epigenetic state in hepatocytes
We previously reported a significant overlap between HNF4A and FOXA2 bound regions (FBRs) in hepatocytes (11). As a pioneer factor, FOXA family members are capable of altering epigenetic modifications to regulate gene expression (24, 34). We, therefore, speculated that a high level of H3K4me1 and H3K27ac at HBRs could be due to FOXA2 occupancy at those sites. To address this, we performed K-means clustering (K=5) on HNF4A and FOXA2 bound regions in hepatocytes using ChAsE (35) and identified 5 clusters. As expected, a large overlap was observed between HBRs and FBRs (Cl2, Cl3 and Cl4), however, two clusters showed exclusive binding of either HNF4A or FOXA2 (Cl1 and Cl5, respectively) (Fig. 2A). Further, these clusters revealed distinctive histone and DNA signature patterns with Cl1 being enriched for H3K4me1, H3K27ac and 5hmC but depleted for 5mC compared to Cl5 (Fig. 2A). Given the importance of CpG density and its impact on 5mC, we performed CpG density analysis and found no significant difference between clusters, indicating 5mC enrichment in Cl5 is independent of CpG density (data not shown). We generated profile plots to highlight differences in clusters. These show decreased levels of H3K4me1 (Fig. 2B), H3K27ac (Fig. 2C) and 5hmC (Fig. 2D) but an increase in 5mC (Fig. 2E) at regions in Cl5, which do not have HNF4A bound. Furthermore, Cl5 regions show the least chromatin accessibility of all clusters based on DNaseI-seq data from mouse liver (Fig. S2A). Cluster 1, 2, 3 and 4 displayed almost identical patterns of histone and DNA modifications indicating open and active chromatin states. We looked at expression of genes with transcriptional start sites (TSSs) within 20kb of a bound region using RNA-seq data from hepatocytes (11) and not surprisingly found that genes associated with these active clusters showed higher expression than Cl5, which lacks evidence of active, open chromatin (Fig. 2F). Analysis of regions in Cl1 confirmed no enrichment of motifs for FOXA, suggesting no contribution of FOXA family members in maintaining a transcriptional permissive state at those regions (Fig. S2B). In contrast, motif analysis of cluster 5 revealed high enrichment for the FOXA2 motif and low enrichment for NFIC and CTCF (Fig. S2C). Interestingly, we also observed low, but significant, enrichment of HNF4A motifs in cluster 5, where our HNF4A ChIP-seq data confirmed no binding in hepatocytes. Gene classification analysis to highlight the functions of genes associated with these clusters showed that cluster 1 genes are frequently associated with metabolic functions (Fig. S2D) and cluster 5 genes are associated with various signaling pathways (Fig. S2E). Overall these findings, confirm that regions bound by HNF4A, irrespective of presence (Cl 2, 3, 4) or absence (Cl1) of FOXA2, are accessible and actively enhancing transcription.
Fig. 2. HNF4A bound regions show high H3K27ac and 5hmC in the absence of FOXA2.
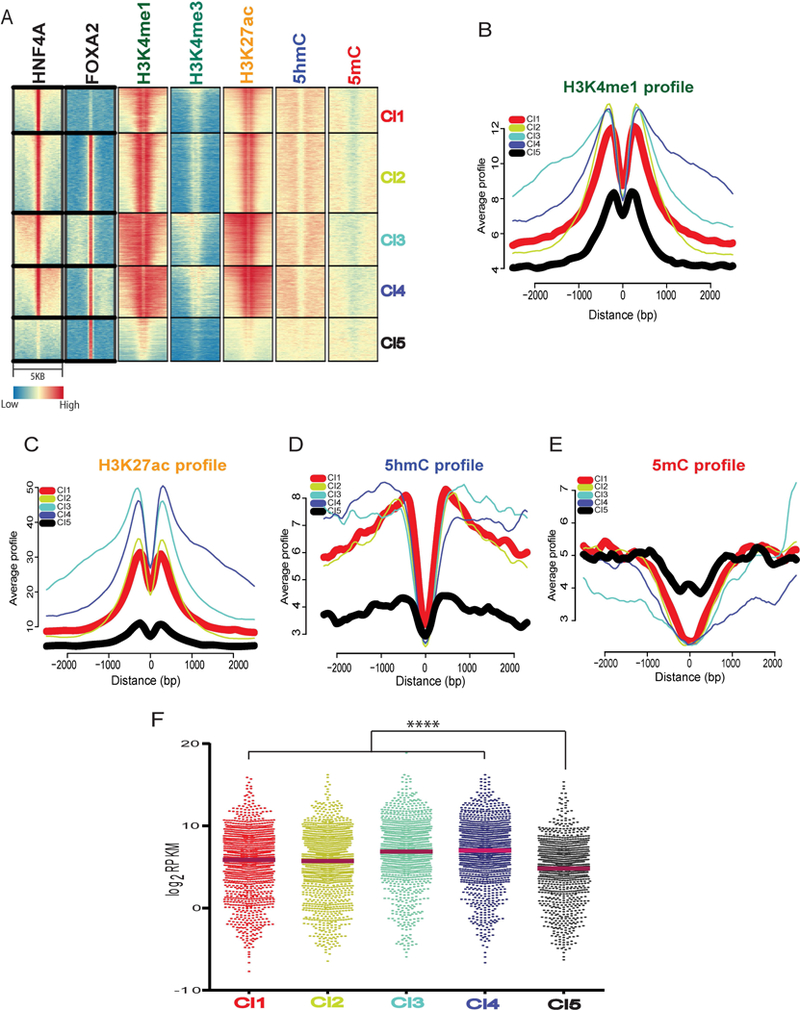
(A) Heatmap of K-means clustering (K=5) of genome wide patterns of TF (HNF4A and FOXA2) bound regions and H3K4me1, H3K4me3, H3K27ac, 5hmC and 5mC. Clusters Cl1 and Cl5 represent regions with only HNF4A or FOXA2 , respectively. Cl2/¾ represents HNF4A/FOXA2 co-bound regions. A 5kb window is shown with transcription factor binding in the middle of each panel. (B) Aggregation plots of H3K4me1 (C) H3K27ac (D) 5hmC (E) 5mC for Cl1, Cl2, Cl3, Cl4 and Cl5 represented by red, yellow, cyan, blue and black lines, respectively. (F) Scatter dot plot representing RNA-seq Log2 RPKM values of all genes associated with clusters. A significant difference was tested using Kruskal-Wallis one-way ANOVA with Dunn’s post-test correction; ****p < 0.001.
Without HNF4A, H3K27ac and 5hmC are significantly decreased at HBRs in hepatocytes
With the observation that H3K27ac and 5hmC are enriched at HNF4A bound regions, we hypothesized that HNF4A is required for appropriate enhancer modifications that ensure precise expression requirements independent of FOXA2. To test this, we took advantage of a Tamoxifen-inducible, hepatocyte-specific, HNF4A conditional knockout (cKO) mouse (4) and performed ChIP-, MeDIP- and hMeDIP-qPCRs on WT and cKO livers. For our analysis, we investigated sites bound by both HNF4A and FOXA2 (Cl2, Cl3 and Cl4, see Fig. 3A for example) as well as sites bound by HNF4A alone (Cl1, see Fig. 3B for example) for changes in H3K27ac patterns. H3K27ac ChIP-qPCR in WT and cKO livers showed that loss of HNF4A resulted in decreased H3K27ac at all sites, indicating a requirement for HNF4A in ensuring H3K27ac is maintained (Fig. 3C). Positive and negative controls regions (Gapdh and Six1) had no significant changes in H3K27ac levels. Furthermore, in cKO livers, the same genomic locations exhibited a reduction in 5hmC to the same extent (almost two fold change) for both HNF4A only and HNF4A/FOXA2 shared regions (Fig. 3D). We also observed an increase in 5mC levels in cKO livers at most loci examined (Fig. S3A), especially at those sites that are bound only by HNF4A. This investigation was extended in vitro using siRNA-mediated knockdown in HepG2 cells. We confirmed a reduction of HNF4A mRNA and protein expression (Fig. S3B) and validated that HNF4A bound to regions in HepG2 cells as identified in ENCODE data (e.g., RBKS, MGST2, SLCO4C1, APOA4, PRLR and AGXT) (Fig. S3C). After siRNA depletion of HNF4A, these sites had significantly decreased levels of 5hmC (Fig. S3D) and increased levels of 5mC (Fig. S3E). Together, these findings suggest that HNF4A is required to maintain high levels of H3K27ac and 5hmC in in vivo and cell lines.
Fig. 3. HNF4A is required for maintenance of H3K27ac and 5hmC at enhancers.
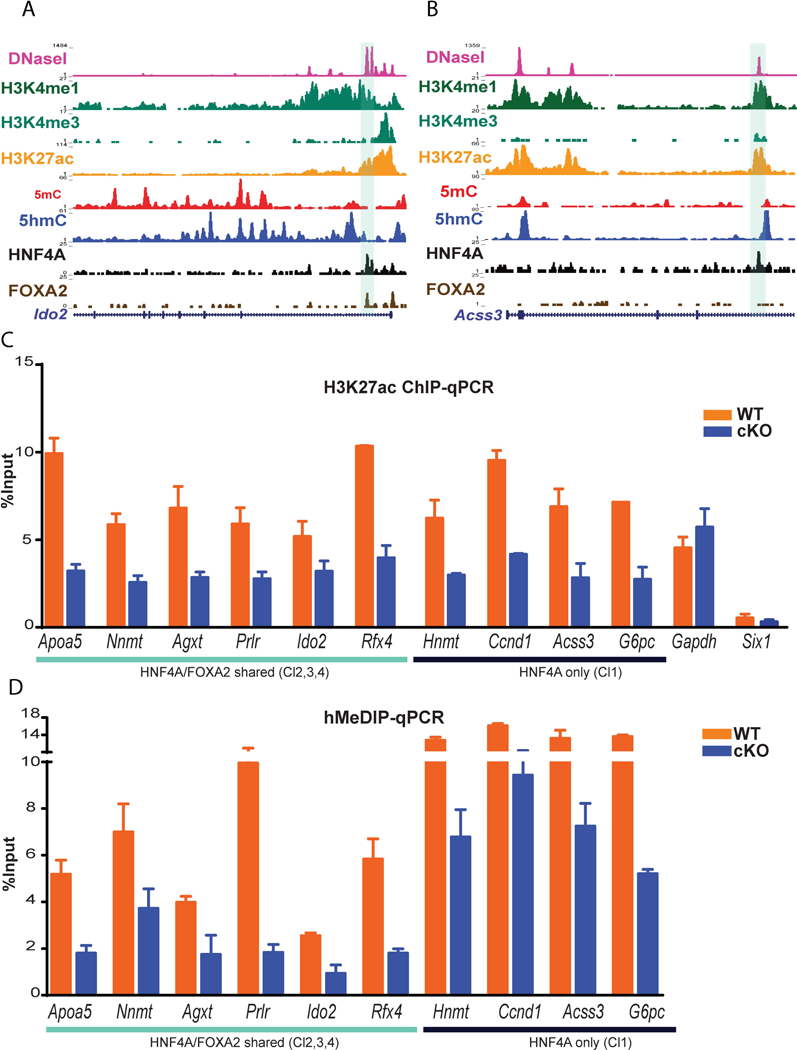
(A) Genome browser view displaying ChIP-seq (TF and histone modifications), MeDIP-seq, hMeDIP-seq and DNase-seq signals at HNF4A and FOXA2 shared sites at Ido2 locus (highlighted region) and (B) at Acss3 locus, HNF4A only bound region. (C) ChIP-qPCR displaying H3K27ac enrichment in WT and HNF4A conditional knockout (cKO, HNF4A F/F; AlbERT2cre) livers at HNF4A/FOXA2 co-occupied regions (Apoa5, Nnmt, Agxt, Prlr and Rfx4) and HNF4A only bound regions (Hnmt, Ccnd1, Acss3 and G6pc). Gapdh and Sixl1 regions are not bound by HNF4A or FOXA2 and represent H3K27ac positive and negative controls. (D) Bar plot showing the 5hmC level at HNF4A/FOXA2 co-occupied regions (Apoa5, Nnmt, Agxt, Prlr and Rfx4) and HNF4A only bound regions (Hnmt, Ccnd1, Acss3 and G6pc)
The gain of HNF4A during differentiation results in increased H3K27ac and 5hmC at HBRs
We have previously reported the phenomenon of enhancer switching during liver differentiation where TFs, including HNF4A, show distinct bound regions in embryonic hepatoblasts and adult hepatocytes (11). To determine if H3K27ac is altered in response to differential binding of HNF4A in hepatoblasts and hepatocytes, we compared ChIP-seq data for HNF4A and H3K27ac in hepatoblasts purified from E14.5 embryonic livers and in adult hepatocytes. We identified 2765 hepatocyte-specific HBRs that are evident in differentiated hepatocytes but not bound in hepatoblasts (examples are shown in Fig. 4A). Of note, these hepatocyte-specific HNF4A bound regions also gain H3K27ac (Fig. 4B) in hepatocytes compared to hepatoblasts. In contrast, regions that lose HNF4A-binding after differentiation (hepatoblast-specific) showed decreased H3K27ac in adult hepatocytes (Fig. S4A). Moreover, hepatocyte-specific HBRs show decreased levels of 5mC (Fig. 4C) and increased levels of 5hmC (Fig. 4D) in hepatocytes compared to hepatoblasts, suggesting that addition of HNF4A in those regions results in oxidation of 5mC to 5hmC. Notably, genes associated with HNF4A bound regions gained in hepatocytes were significantly more highly expressed in hepatocytes compared to hepatoblasts (individual and global examples are shown in Fig. 4E and 4F), whereas genes associated with regions no longer bound by HNF4A in hepatocytes showed the opposite trend (Fig. S4B). Overall these findings support an important role of HNF4A in cell type-specific gene regulation by ensuring high levels of H3K27ac and 5hmC at gene regulatory regions.
Fig. 4. Gain of HNF4A binding during hepatoblast to hepatocyte differentiation accompanies histone and DNA modifications.
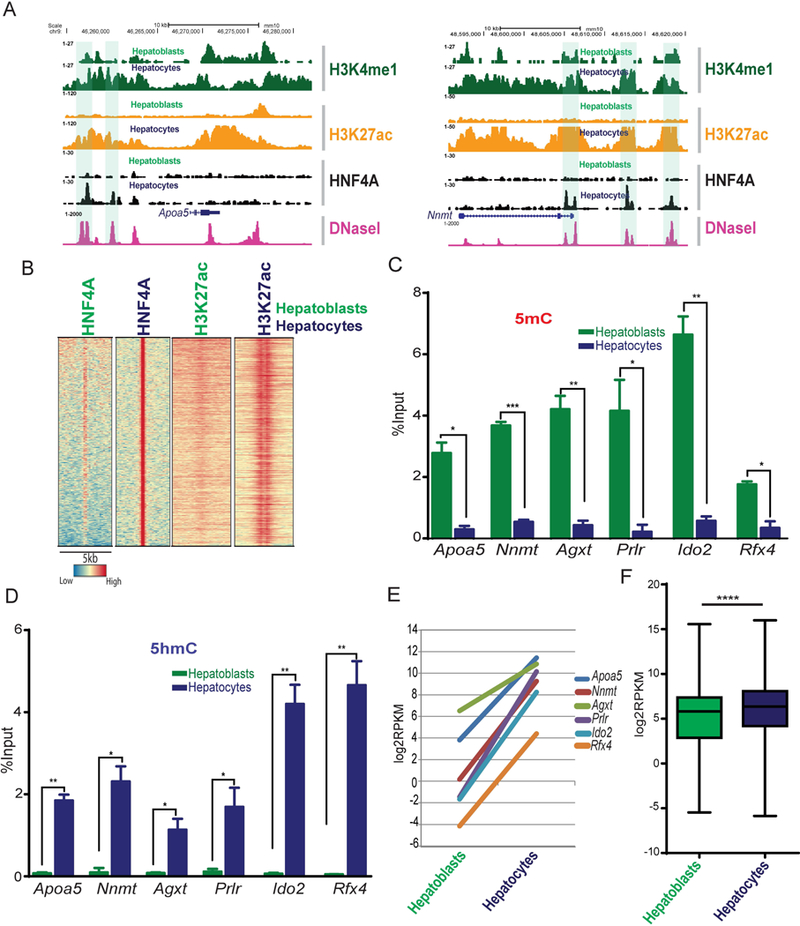
(A) Genome browser view for HNF4A target genes Apoa5 and Nnmt, with ChIP-seq signals for H3K4me1 (dark green), H3K27ac (orange) and HNF4A (black) from hepatoblasts and hepatocytes showing acquisition of HNF4A and increased H3K27ac at hepatocyte-specific HBRs (highlighted). DNase-seq data from the adult liver is shown in pink. (B) Heatmap of HNF4A bound regions that are only significant in hepatocytes, not hepatoblasts. H3K27ac accompanied this differentiation. Colours represent enrichment level. (C) MeDIP-qPCR and (D) hMeDIP-qpcr showing 5mC and 5hmC (respectively) levels at loci that gain HNF4A binding during differentiation from hepatoblasts and hepatocytes (n=3). Values represent mean ±SEM *P<0.05, **P<0.01 by t-test. (E) RNA-seq in hepatoblasts and hepatocytes (log2RPKM values) of genes associated with hepatocyte-specific HNF4A bound regions (Apoa5, Nnmt, Agxt, Prlr, Ido2 and Rfx4). (F) Box plot showing RNA-seq values (Log2RPKM) in hepatoblasts and hepatocytes for all genes associated with hepatocyte-specific HNF4A bound regions.
HNF4A and TET3 proteins physically interact
We speculated that HNF4A interacts with TET proteins to maintain low-levels of 5mC at bound regions through oxidation to 5hmC. We focused on TET3 due to its high expression in hepatocytes (Fig. 5A and 5B) and its known interactions with another nuclear receptor family member, TRα1 (36). Of note, TET3 co-immunoprecipitation with HNF4A in HepG2 cells, which express high levels of both HNF4A and TET3, confirmed that the two proteins interact (Fig. 5C). This interaction was also confirmed using expression of tagged proteins in HEK293T cells (Fig.S5B). Importantly, the TET3-HNF4A interaction was observed in nuclear lysates isolated from mouse hepatocytes (Fig. 5D) as well as hepatoblasts (Fig. 5E). Proximity ligation assay (PLA) on adult and embryonic mouse liver cryosections validated the close association between HNF4A and TET3 (Fig. 5F) in nuclei of hepatoblasts and hepatocytes in vivo. These PLA findings were further validated in HEK293T cells using the expression of Myc-HNF4A and Flag-TET3 (Fig. S5D). Together, our findings suggest that HNF4A physically interacts with TET3, in vitro and in vivo, to mediate the active 5hmC status and that presence of both are required for expression of genes associated with HBRs.
Fig. 5. HNF4A interacts with TET3 in vivo and in vitro.

(A) RNA-Seq RPKM values of Tet1, Tet2 andTet3 in hepatocytes. (B) Western blot showing expression of TET3 in hepatoblast and hepatocytes with β-Actin control. (C) Immunoprecipitation in HepG2 lysate using anti-HNF4A and negative control (IgG) followed by Western blots for TET3 and HNF4A. (D) Immunoprecipitation on hepatocytes isolated from 3 female livers using anti-HNF4A and negative control (IgG) followed by Western blots for HNF4A and TET3. (E) Immunoprecipitation and Western blot analysis of hepatoblast lysates using anti-HNF4A and IgG antibody to IP and TET3 and HNF4A antibodies to validate interaction. (F) Depiction of proximity ligation assay (PLA) technique (top). PLA on adult (middle) and embryonic liver (bottom) sections using an antibody against HNF4A and TET3 confirm endogenous interaction, shown as yellow dots inside DAPI stained nuclei (Blue).
TET3 is a direct transcriptional target of HNF4A
In hepatoblasts, we observed a strong intragenic HNF4A binding site approximately 19kb downstream from the Tet3 transcriptional start site. This HNF4A site co-localizes with histone modifications H3K4me1 and H3K27ac, suggesting an active enhancer (Fig. 6A). Knockdown of HNF4A in mouse HPPL cells (hepatic progenitor cells proliferating on laminin), which are capable of generating hepatocyte and cholangiocytes similar to mouse hepatoblasts, caused a reduction of Tet3 mRNA expression (p<0.0001) and protein levels (Fig. 6B), suggesting that HNF4A regulates Tet3 expression in hepatic cells. To determine if the HNF4A bound enhancer in Tet3 (mTET3E, a 405bp region) is functional; we used luciferase reporter assays in HEK293T cells. HNF4A overexpression significantly increased (p>0.0001) enhancer activity of mTET3E (Fig. 6C). We confirmed these results using HepG2 cells endogenously expressing HNF4A (Fig. 6D). Furthermore, ENCODE ChIP-seq data in HepG2 cells demonstrated a similar HNF4A bound region with active histone modifications between exon 2 and 3 of TET3 in the human genome (Fig. S6B). Luciferase assays with the human region (332bp) in HEK293T cells with HNF4A overexpression (Fig. S6C), and HepG2 cells with endogenous HNF4A (Fig. S6D) confirmed HNF4A-mediated regulation of TET3.
Fig. 6. Tet3 transcription is directly regulated by HNF4A.
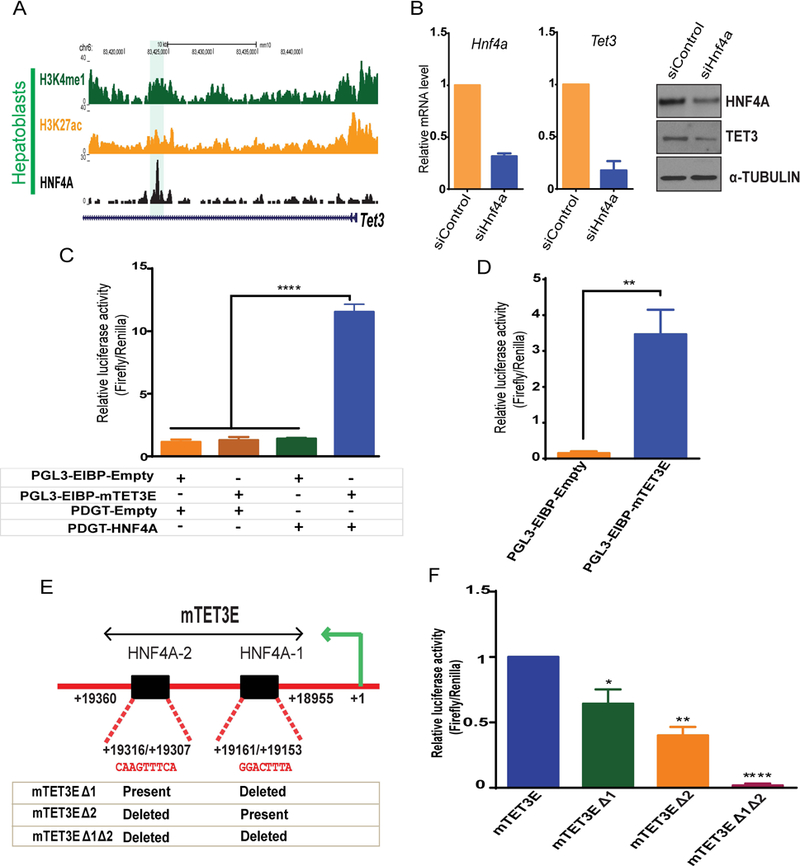
(A) Genomic browser view showing ChIP-seq of HNF4A, H3K4me1 and H3K27ac enrichment at Tet3 locus in hepatoblasts (highlighted). (B) Hnf4a and Tet3 mRNA and protein level in siControl and siHnf4a knockdown in HPPL cells (96 hrs). (C) Luciferase assays using control (PGL3-EIBP-Empty and PDGT-Empty), HNF4A overexpression (PDGT-HNF4A) and the mouse Tet3 enhancer (PGL3EIBP-mTet3E) constructs. Constructs were overexpressed in HEK293T cells in combinations shown below the graph (+ transfected and – not transfected). (D) Luciferase assays in HepG2 cells transfected with control (PGL3-EIBP-empty) or Tet3 enhancer luciferase constructs (PGL3EIBP-mTET3E). (E) Schematic representation of the Tet3 enhancer region (mTet3E) cloned for luciferase assays. Sequence alignments show two HNF4A motifs (HNFA-1 and HNF4A-2) within the Tet3 enhancer. The table represents the constructs generated by deleting HNF4A motifs. (F) Luciferase assays in HepG2 cells transfected with mTET3E or mTET3E carrying one deleted HNF4A motif (mTET3EΔ1 or mTET3EΔ2) or mTET3E with both HNF4A motifs deleted (mTET3Δ1Δ2). Values represent the mean ± SEM. p*<0.05, **p<0.01, ****p<0.0001 from t-test.
The mouse TET3 enhancer contains two HNF4A motifs (HNF4A-1 and HNF4A-2) (Fig. 6E). The motifs were deleted sequentially in the reporter construct containing the mouse enhancer and activity was measured in both HepG2 (Fig. 6F) and HEK293T cells (Fig. S6A). As expected, combinatorial deletion of these motifs diminished luciferase activity in HepG2 cells (Fig. 6F). Of note, in HEK293T cells, deletion of the two motifs did not have any additional effect over the single deletions, suggesting that the hepatic environment of HepG2 cells may contain additional factors required for this regulation. Overall, our results suggest that HNF4A directly regulates Tet3 in hepatic cells and TET3 interacts with HNF4A to control the epigenetic environment by ensuring 5hmC and H3K27ac are maintained.
DISCUSSION
HNF4A is a key TF in liver as its loss is associated with transcriptional repression of genes (5, 10). Although various TFs were shown to regulate gene expression by influencing histone (24, 37) and DNA modifications at gene regulatory regions (24, 37, 38), the role of HNF4A in epigenetic-mediated gene regulation in liver is not well defined.
Here, we show that HNF4A, a master regulator of hepatocyte differentiation, controls gene expression through its influence on epigenetic modifications in hepatoblasts and hepatocytes. In particular, we show that HNF4A binding in hepatocytes is almost exclusively associated with active enhancers marked by histone modifications (H3K4me1 and H3K27ac) and DNA hydroxymethylation (5hmC), supporting a primary role of HNF4A in transcriptional activation. Moreover, our data suggest that HNF4A is required for the presence of H3K27ac and 5hmC since loss of HNF4A in adult liver, through an inducible knockout model, leads to reduced levels of these marks at HNF4A bound regions. In addition, regions bound by HNF4A specifically in hepatoblasts but not hepatocytes have lower H3K27ac and 5hmC in hepatocytes. This correlates with less transcription from associated genes (Fig. 7). Finally, HNF4A bound regions acquired during differentiation in hepatocytes are enriched for H3K27ac and 5hmC in hepatocytes. Together this data establishes HNF4A as a facilitator of the active enhancer state.
Fig. 7. Role of HNF4A in establishing and maintaining epigenetic modifications at enhancers.
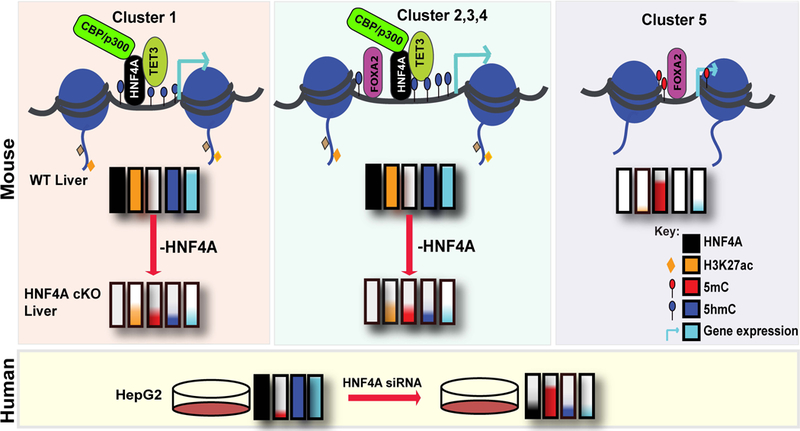
HNF4A is required for the maintenance of H3K27ac and 5hmC in mouse (top) and human (bottom). Coloured bars represent the TF (HNF4A- black) occupancy and level of H3K27ac (orange), 5mC (red) and 5hmC (blue) in WT and HNF4A cKO liver. Loss of H3K27ac and 5hmC at gene regulatory regions result in decreased expression (sky-blue) of genes with associated HNF4A bound regions. Cluster 1 (left panel) represents regions bound by HNF4A, but not FOXA2, where the interaction with CBP/p300 and TET3 maintains H3K27ac and 5hmC levels at those sites. Cluster 2, 3 and 4 (middle panel) represents HNF4A and FOXA2 co-occupied regions where 5hmC and H3K27ac levels were similarly maintained by HNF4A’s interaction with TET3 and CBP/p300. Without HNF4A, as in the cKO liver, regions in clusters 1–4 have less active marks (H3K27ac and 5hmC) and more 5mC resulting in reduced transcription from associated genes. Cluster 5 (right panel) represents regions occupied by FOXA2, not HNF4A, in hepatocytes, with very low H3K27ac and 5hmC and high 5mC. The genes associated with these regions show lower expression (sky blue bars) as compared to the other clusters. In human liver carcinoma cells (HepG2 cells), HNF4A KD also produced similar changes in the level of 5hmC and 5mC (shown below the three mouse panels).
Previous studies have suggested that HNF4A plays multiple roles in chromatin reorganization: ectopic HNF4A expression in fibroblasts was shown to induce DNase1 hypersensitivity of the Serpin gene cluster at 14q32.1 to transcriptionally regulate α1-antitrypsin and corticosteroid binding globulin (39). This supports a role of HNF4A in the establishment of active enhancers(39). In addition, HNF4A may regulate chromatin indirectly by controlling the expression of chromatin remodelling genes, such as Smarcd3 and Cdt1, as their expression is altered in HNF4A KO livers (40).
The mechanisms through which HNF4A alters the epigenome are likely to be complex. HNF4A was shown to directly interact with CBP/p300, which may account for its ability to regulate H3K27ac (2, 41, 42). Our data suggest that in the absence of HNF4A, histone acetylases either are not recruited or not able to function at HNF4A enhancers, despite the potential presence of other TF binding. For example, HNF4A frequently co-binds to regions with FOXA2; however, at regions that are bound by FOXA2 alone very little H3K27ac is evident. In addition, in HNF4A KO livers, H3K27ac was reduced at regions bound by either HNF4A alone or FOXA2/HNF4A, indicating HNF4A is primarily responsible for maintaining active epigenetic signatures at these locations. In addition to FOXA family members, HNF4A was previously shown to recruit several coactivators such as glucocorticoid receptor interacting protein 1, steroid receptor coactivator1 and cAMP response element binding proteins to regulate gene expression (41, 43). The role of these co-factors is not clear. Moreover, we observed that RXRA, ESRB and HINFP motifs were enriched in HNF4A bound regions which lack FOXA2. Thus, H3K27ac at HNF4A binding sites could be affected by interactions with other TFs. Alternatively, HNF4A, with or without other factors, could have a protective effect that disallows histone deacetylases to act.
In addition to H3K27ac, we observed depletion of 5mC and enrichment of 5hmC around HNF4 bound regions. In HNF4A cKO livers, 5hmC was reduced around HBRs. A recent study showed that HNF4A loss can impact global 5mC and 5hmC to a small extent (44). By first identifying HNF4A-bound regions, we were able to observe significant changes in 5hmC upon HNF4A depletion both in vivo and in vitro, demonstrating its role in the maintenance of this active mark at enhancers (Fig. 7). Interestingly, we observed only a small increase in 5mC at HBRs in HNF4A cKO livers, suggesting that either recovery of 5mC is slower than 5hmC loss or it does not occur. A previous study observed a similar effect at FOXA1 bound enhancers in LNCaP prostate cancer cells in which knockdown of FOXA1 reduced 5hmC without gain of 5mC (24). Furthermore, it has previously been shown that 5hmC abundance represents only ~10% of the 5mC level in embryonic stem cells,(45), thus changes in 5mC upon HNF4A depletion in the whole liver may be difficult to observe. However, we did observe a clear gain in 5mC in HepG2 cells upon HNF4A knockdown (Fig. S3E).
In the present study, we show that HNF4A functions to regulate DNA methylation in the surrounding genomic regions through interactions with TET3. Of interest, the HNF4A binding motif does not contain a CG which can be methylated, thus making it a methylation-insensitive TF (11, 17). Thus, HNF4A may bind to its motif and then recruit TET proteins to oxidize 5mC to 5hmC in surrounding regions. TET2 and TET3 are known to be required for this 5mC oxidation process in hepatocytes (46). Here, we focused on TET3 because it is more highly expressed compared to other TETs in liver and is known to interact with other TFs (36). Since TET2 is also expressed in liver, HNF4A may also interact with TET2 in vivo. It should be mentioned that while we focused on 5hmC at HBRs, other DNA modifications, such as 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) may also be present and important. A recent study showed that TET3 can identify and facilitate removal of 5caC by base excision repair to achieve demethylation of gene regulatory regions (47). Thus, future work is needed to examine the entire 5mC oxidation pathway in liver.
In summary, we show that HBRs are associated with a high level of H3K27ac and 5hmC, likely due to HNF4A interactions with CBP/p300 and TET3 respectively (Fig. 7). After examining the data for patterns (Fig. 2A) and how H3K27Ac and 5hmC levels vary, we identified that at these regions, HNF4A, but not FOXA2, is necessary for the active epigenetic state (Fig. 7). We also confirmed that loss of HNF4A in a human hepatoma cell line, HepG2, produced similar changes in DNA modifications as observed in Hnf4a cKO mouse livers, indicating similar mechanisms of gene regulation (Fig. S3 and Fig. 7).Thus, we have identified a mechanism through which HNF4A regulates epigenetic plasticity in hepatic cells. We have also confirmed HNF4A function in establishment and maintenance of H3K27ac and 5hmC at enhancers and that this function can be independent of FOXA2. Whether the two epigenetic alterations established and maintained by HNF4A occur concomitantly or are unrelated remains to be determined.
Supplementary Material
(A) Genomic distribution of HNF4A bound regions in hepatocytes (B) DNase-seq signals at all HBRs in mouse liver (C and D) UCSC genome browser view showing enrichment of HNF4A, H3K27ac H3K4me1, H3K4me3, 5mC, 5hmC and DNase-seq signals at HBR (highlighted). (E and F) UCSC maps of Dazl and Tbx15 displaying the level of 5mC at respective CpG Islands.
(A) DNase-seq signals for each cluster in hepatocytes (B) Motif analysis of Cluster 1 regions showing enrichment for potential transcription factor binding sites. (C) Motif analysis of Cluster 5 regions showing enrichment for potential transcription factor binding sites. (D) Pathway analysis of cluster 1 associated genes in liver. (E) Pathway analysis of cluster 5 genes in liver.
(A) The 5mC level at shared and HNF4A only gene loci in WT and HNF4A KO livers. n=3.Values represent mean ±SEM. (B) Left: qRT-PCR analysis of HNF4A in HepG2 cells 72 hours following HNF4A knockdown (siHNF4a) to control (siCntrl) data is normalised to GAPDH. (n=3). Values represent the mean ± SEM. **p<0.01 from t-test. Right: WB analysis of HNF4A in HepG2 cells 72 hours following HNF4A knockdown (siHNF4a). (C) ChIP-qPCR analysis displaying enrichment of HNF4A at RBKS, MGST2, SLCO4C1, APOA4, PRLR and AGXT loci. (n=3) Values represent the mean ± SEM. DAZL promoter and region on Chr17 serve as negative control. (D) hMeDIP-qPCR of HNF4A occupied regions at RBKS, MGST2, SLCO4C1, PRLR and AGXT in siCntrl and siHNF4A HepG2 samples. (E) MeDIP qPCR of HNF4A target genes in HepG2, DAZL serves as a 5mC positive control.
(A) heat map displaying HNF4A and H3K27ac ChIP signals in hepatoblasts and hepatocytes. Colour bar below indicates the strength of signals. (B) Box plot showing RNA-seq values (Log2RPKM) in hepatoblasts and hepatocytes for genes that lost HNF4A binding during differentiation. Significance was tested using a t-test, ****p<0.0001.
(A) Western showing protein expression of TET2 in hepatoblasts and hepatocytes (top) and β-Actin as loading control (bottom). (B) Co-immunoprecipitation in which Myc-HNF4A and flag-TET3 were ectopically expressed in 293T cells. IP was performed using anti-Myc and a negative control antibody (IgG), followed by western analysis with Flag (top) and Myc (bottom) antibody. (C) mRNA levels of TETs in HepG2 cells. (D) Proximity Ligation Assay for ectopically expressed Myc-HNF4A and Flag-TET3 confirming the interaction in HEK293T cells (shown by Red dots). (E) Secondary only probes for PLA analysis for embryonic (left) and adult liver (right) sections. Nuclei are stained with DAPI (blue).
Cells were transfected with mouse Tet3 enhancer construct (PGL3EIBP-mTET3E) or enhancer carrying one deleted HNF4A motif (mTET3EΔ1 or mTET3EΔ2) and both deleted HNF4A motifs (mTET3Δ1Δ2) with overexpression of HNF4A (PDGT-HNF4A). (B) UCSC maps using ENCODE ChIP-seq data displaying HNF4A, H3K4me1 and H3K27ac enrichment at TET3 locus in HepG2 cells. The highlighted region was cloned in luciferase vector. (C) Luciferase assay using control (PGL3-EIBPEmpty and PDGT-empty), HNF4A overexpression (PDGT-HNF4A) and cloned human TET3 (PGL3EIBP-hTET3) enhancer constructs. Constructs were overexpressed in HEK293T cells in combinations shown below the graph (+ transfected and – not transfected). (D) Luciferase assays in HepG2 cells transfected with control (PGL3-EIBP-empty) or human TET3 (PGL3EIBP-hTET3E) enhancer luciferase constructs.
Acknowledgments
Financial support: This work was funded by Genome BC and the Canadian Institutes of Health Research (FRN128092) to P.A.H and Terry Fox New Investigator award (1021) and Terry Fox New Frontiers Program Project Grant (1074) to M.H. and F.J.G is supported by National Cancer Institute Intramural Research Program.
List of Abbreviations:
- HNF4A
Hepatocyte nuclear factor 4-alpha
- TF
Transcription Factor
- FOXA2
Forkhead Box A2
- TET3
Ten-eleven translocation methylcytosine dioxygenase 3
- HBR
HNF4A bound region
- FBR
FOXA2 bound region
- 5mC
5-methylcytosine
- 5hmC
5-hydroxymethylcytosine
- PLA
Proximity ligation assay
REFERENCES
- 1.Sladek FM, Zhong WM, Lai E, Darnell JE. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–65. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- 3.DeLaForest A, Nagaoka M, Si-Tayeb K, Noto FK, Konopka G, Battle MA, et al. HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Development. 2011;138:4143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonzo JA, Ferry CH, Matsubara T, Kim J-H, Gonzalez FJ. Suppression of Hepatocyte Proliferation by Hepatocyte Nuclear Factor 4α in Adult Mice. J. Biol. Chem. 2012;287:7345–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 2001;21:1393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, et al. Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1). Nature. 1996;384:458–460. [DOI] [PubMed] [Google Scholar]
- 7.Ning B-F, Ding J, Yin C, Zhong W, Wu K, Zeng X, et al. Hepatocyte Nuclear Factor 4 Suppresses the Development of Hepatocellular Carcinoma. Cancer Res. 2010;70:7640–7651. [DOI] [PubMed] [Google Scholar]
- 8.Santangelo L, Marchetti A, Cicchini C, Conigliaro A, Conti B, Mancone C, et al. The stable repression of mesenchymal program is required for hepatocyte identity: A novel role for hepatocyte nuclear factor 4α. Hepatology. 2011;53:2063–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walesky C, Gunewardena S, Terwilliger EF, Edwards G, Borude P, Apte U. Hepatocyte-specific deletion of hepatocyte nuclear factor-4α in adult mice results in increased hepatocyte proliferation. Am. J. Physiol. Liver Physiol. 2013;304:G26–G37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watt A, Garrison WD, Duncan SA. HNF4: A central regulator of hepatocyte differentiation and function. Hepatology. 2003;37:1249–1253. [DOI] [PubMed] [Google Scholar]
- 11.Alder O, Cullum R, Lee S, Kan AC, Wei W, Yi Y, et al. Hippo signaling influences HNF4A and FOXA2 enhancer switching during hepatocyte differentiation. Cell Rep. 2014;9:261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller B, Mieczkowski J, Kundu S, Wang P, Sadreyev R, Tolstorukov MY, et al. Widespread changes in nucleosome accessibility without changes in nucleosome occupancy during a rapid transcriptional induction. Genes Dev. 2017;31:451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu H, Wang G, Qian J. Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet. 2016;17:551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calo E, Wysocka J. Modification of enhancer chromatin: what, how and why? 2013;Mol Cell. 2013;49:825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson AG, Bilenky M, Tam A, Zhao Y, Zeng T, Thiessen N, et al. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 2008;18:1906–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science (80-. ). 2017;356:eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schübeler D, Lorincz MC, Cimbora DM, Telling A, Feng YQ, Bouhassira EE, et al. Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol. Cell. Biol. 2000;20:9103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev. Cell. 2014;29:102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perera A, Eisen D, Wagner M, Laube SK, Künzel AF, Koch S, et al. TET3 Is Recruited by REST for Context-Specific Hydroxymethylation and Induction of Gene Expression. Cell Rep. 2015;11:283–294. [DOI] [PubMed] [Google Scholar]
- 22.Choi SH, Gearhart MD, Cui Z, Bosnakovski D, Kim M, Schennum N, et al. DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res. 2016;44:5161–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujiki K, Shinoda A, Kano F, Sato R, Shirahige K, Murata M. PPARγ-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nat. Commun. 2013;4:2262. [DOI] [PubMed] [Google Scholar]
- 24.Yang YA, Zhao JC, Fong K-W, Kim J, Li S, Song C, et al. FOXA1 potentiates lineage-specific enhancer activation through modulating TET1 expression and function. Nucleic Acids Res. 2016;44:8153–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. [DOI] [PubMed] [Google Scholar]
- 26.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. [DOI] [PubMed] [Google Scholar]
- 27.Nakamori D, Akamine H, Takayama K, Sakurai F, Mizuguchi H. Direct conversion of human fibroblasts into hepatocyte-like cells by ATF5, PROX1, FOXA2, FOXA3, and HNF4A transduction. Sci. Rep. 2017;7:16675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreamer BL, Staecker JL, Sawada N, Sattler GL, Hsia MT, Pitot HC. Use of a low-speed, iso-density percoll centrifugation method to increase the viability of isolated rat hepatocyte preparations. In Vitro Cell. Dev. Biol. 1986;22:201–11. [DOI] [PubMed] [Google Scholar]
- 29.Taiwo O, Wilson GA, Morris T, Seisenberger S, Reik W, Pearce D, et al. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat. Protoc. 2012;7:617–636. [DOI] [PubMed] [Google Scholar]
- 30.Wederell ED, Bilenky M, Cullum R, Thiessen N, Dagpinar M, Delaney A, et al. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 2008;36:4549–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanimizu N, Saito H, Mostov K, Miyajima A. Long-term culture of hepatic progenitors derived from mouse Dlk+ hepatoblasts. J. Cell Sci. 2004;117:6425–6434. [DOI] [PubMed] [Google Scholar]
- 32.Hong YH, Varanasi US, Yang W, Leff T. AMP-activated Protein Kinase Regulates HNF4α Transcriptional Activity by Inhibiting Dimer Formation and Decreasing Protein Stability. J. Biol. Chem. 2003;278:27495–27501. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Wu X, Zhou Y, Lee M, Guo L, Han W, et al. Decoding the dynamic DNA methylation and hydroxymethylation landscapes in endodermal lineage intermediates during pancreatic differentiation of hESC. Nucleic Acids Res. 2018;46:2883–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, et al. The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Mol. Cell. 2016;62:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Younesy H, Nielsen CB, Lorincz MC, Jones SJM, Karimi MM, Möller T. ChAsE: chromatin analysis and exploration tool. Bioinformatics. 2016;32:3324–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan W, Guyot R, Samarut J, Flamant F, Wong J, Gauthier KC. Methylcytosine dioxygenase TET3 interacts with thyroid hormone nuclear receptors and stabilizes their association to chromatin. Proc. Natl. Acad. Sci. U. S. A. 2017;114:8229–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perera A, Eisen D, Wagner M, Laube SK, Künzel AF, Koch S, et al. TET3 Is Recruited by REST for Context-Specific Hydroxymethylation and Induction of Gene Expression. Cell Rep. 2015;11:283–294. [DOI] [PubMed] [Google Scholar]
- 38.Ancey P-B, Ecsedi S, Lambert M-P, Talukdar FR, Cros M-P, Glaise D, et al. TET-Catalyzed 5-Hydroxymethylation Precedes HNF4A Promoter Choice during Differentiation of Bipotent Liver Progenitors. Stem cell reports. 2017;9:264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rollini P, Fournier RE. The HNF-4/HNF-1alpha transactivation cascade regulates gene activity and chromatin structure of the human serine protease inhibitor gene cluster at 14q32.1. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holloway MG, Miles GD, Dombkowski AA, Waxman DJ. Liver-specific hepatocyte nuclear factor-4alpha deficiency: greater impact on gene expression in male than in female mouse liver. Mol. Endocrinol. 2008;22:1274–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida E, Aratani S, Itou H, Miyagishi M, Takiguchi M, Osumu T, et al. Functional Association between CBP and HNF4 inTrans-activation. Biochem. Biophys. Res. Commun. 1997;241:664–669. [DOI] [PubMed] [Google Scholar]
- 42.Eeckhoute J, Formstecher P, Laine B. Hepatocyte nuclear factor 4alpha enhances the hepatocyte nuclear factor 1alpha-mediated activation of transcription. Nucleic Acids Res. 2004;32:2586–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J-C, Stafford JM, Granner DK. SRC-1 and GRIP1 coactivate transcription with hepatocyte nuclear factor 4. J. Biol. Chem. 1998;273:30847–30850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Q, Lei X, Lu H. Alterations of Epigenetic Signatures in Hepatocyte Nuclear Factor 4a Deficient Mouse Liver Determined by Improved ChIP-qPCR and (h)MeDIP-qPCR Assays. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reizel Y, Sabag O, Skversky Y, Spiro A, Steinberg B, Bernstein D, et al. Postnatal DNA demethylation and its role in tissue maturation. Nat. Commun. 2018;9:2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin S-G, Zhang Z-M, Dunwell TL, Harter MR, Wu X, Johnson J, et al. Tet3 Reads 5-Carboxylcytosine through Its CXXC Domain and Is a Potential Guardian against Neurodegeneration. Cell Rep. 2016;14:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Genomic distribution of HNF4A bound regions in hepatocytes (B) DNase-seq signals at all HBRs in mouse liver (C and D) UCSC genome browser view showing enrichment of HNF4A, H3K27ac H3K4me1, H3K4me3, 5mC, 5hmC and DNase-seq signals at HBR (highlighted). (E and F) UCSC maps of Dazl and Tbx15 displaying the level of 5mC at respective CpG Islands.
(A) DNase-seq signals for each cluster in hepatocytes (B) Motif analysis of Cluster 1 regions showing enrichment for potential transcription factor binding sites. (C) Motif analysis of Cluster 5 regions showing enrichment for potential transcription factor binding sites. (D) Pathway analysis of cluster 1 associated genes in liver. (E) Pathway analysis of cluster 5 genes in liver.
(A) The 5mC level at shared and HNF4A only gene loci in WT and HNF4A KO livers. n=3.Values represent mean ±SEM. (B) Left: qRT-PCR analysis of HNF4A in HepG2 cells 72 hours following HNF4A knockdown (siHNF4a) to control (siCntrl) data is normalised to GAPDH. (n=3). Values represent the mean ± SEM. **p<0.01 from t-test. Right: WB analysis of HNF4A in HepG2 cells 72 hours following HNF4A knockdown (siHNF4a). (C) ChIP-qPCR analysis displaying enrichment of HNF4A at RBKS, MGST2, SLCO4C1, APOA4, PRLR and AGXT loci. (n=3) Values represent the mean ± SEM. DAZL promoter and region on Chr17 serve as negative control. (D) hMeDIP-qPCR of HNF4A occupied regions at RBKS, MGST2, SLCO4C1, PRLR and AGXT in siCntrl and siHNF4A HepG2 samples. (E) MeDIP qPCR of HNF4A target genes in HepG2, DAZL serves as a 5mC positive control.
(A) heat map displaying HNF4A and H3K27ac ChIP signals in hepatoblasts and hepatocytes. Colour bar below indicates the strength of signals. (B) Box plot showing RNA-seq values (Log2RPKM) in hepatoblasts and hepatocytes for genes that lost HNF4A binding during differentiation. Significance was tested using a t-test, ****p<0.0001.
(A) Western showing protein expression of TET2 in hepatoblasts and hepatocytes (top) and β-Actin as loading control (bottom). (B) Co-immunoprecipitation in which Myc-HNF4A and flag-TET3 were ectopically expressed in 293T cells. IP was performed using anti-Myc and a negative control antibody (IgG), followed by western analysis with Flag (top) and Myc (bottom) antibody. (C) mRNA levels of TETs in HepG2 cells. (D) Proximity Ligation Assay for ectopically expressed Myc-HNF4A and Flag-TET3 confirming the interaction in HEK293T cells (shown by Red dots). (E) Secondary only probes for PLA analysis for embryonic (left) and adult liver (right) sections. Nuclei are stained with DAPI (blue).
Cells were transfected with mouse Tet3 enhancer construct (PGL3EIBP-mTET3E) or enhancer carrying one deleted HNF4A motif (mTET3EΔ1 or mTET3EΔ2) and both deleted HNF4A motifs (mTET3Δ1Δ2) with overexpression of HNF4A (PDGT-HNF4A). (B) UCSC maps using ENCODE ChIP-seq data displaying HNF4A, H3K4me1 and H3K27ac enrichment at TET3 locus in HepG2 cells. The highlighted region was cloned in luciferase vector. (C) Luciferase assay using control (PGL3-EIBPEmpty and PDGT-empty), HNF4A overexpression (PDGT-HNF4A) and cloned human TET3 (PGL3EIBP-hTET3) enhancer constructs. Constructs were overexpressed in HEK293T cells in combinations shown below the graph (+ transfected and – not transfected). (D) Luciferase assays in HepG2 cells transfected with control (PGL3-EIBP-empty) or human TET3 (PGL3EIBP-hTET3E) enhancer luciferase constructs.


