Abstract
Introduction:
Cone-beam CT (CBCT) image quality is important for its quantitative analysis in adaptive radiation therapy. However, due to severe artifacts, the CBCTs are primarily used for verifying patient setup only so far. We have developed a learning-based image quality improvement method which could provide CBCTs with image quality comparable to planning CTs (pCTs). The accuracy of dose calculations based on these CBCTs is unknown. In this study, we aim to investigate the dosimetric accuracy of our corrected CBCT (CCBCT) in brain stereotactic radiosurgery (SRS) and pelvic radiotherapy.
Materials and Methods:
We retrospectively investigated a total of 32 treatment plans from 22 patients, each of whom with both original treatment pCTs and CBCTs acquired during treatment setup. The CCBCT and original CBCT (OCBCT) were registered to the pCT for generating CCBCT-based and OCBCT-based treatment plans. The original pCT-based plans served as ground truth. Clinically-relevant dose volume histogram (DVH) metrics were extracted from the ground truth, OCBCT-based and CCBCT-based plans for comparison. Gamma analysis was also performed to compare the absorbed dose distributions between the pCT-based and OCBCT/CCBCT-based plans of each patient.
Results:
CCBCTs demonstrated better image contrast and more accurate HU ranges when compared side-by-side with OCBCTs. For pelvic radiotherapy plans, the mean dose error in DVH metrics for PTV, bladder and rectum was significantly reduced, from 1% to 0.3%, after CBCT correction. The gamma analysis showed the average pass rate increased from 94.5% before correction to 99.0% after correction. For brain SRS treatment plans, both original and corrected CBCT images were accurate enough for dose calculation, though CCBCT featured higher image quality.
Conclusion:
CCBCTs can provide a level of dose accuracy comparable to traditional pCTs for brain and prostate radiotherapy planning and the correction method proposed here can be useful in CBCT-guided adaptive radiotherapy.
Keywords: Cone-beam CT, Adaptive radiation therapy
INTRODUCTION
Cone-beam computed tomography (CBCT) has been shown a promising imaging modality in radiation therapy, especially for adaptive radiation therapy. However, its routine use has been hindered by the artifacts in current clinical CBCT images, since those artifacts reduce its accuracy significantly. Our machine learning-based CBCT correction method has been able to generate CBCT images with image quality that is comparable to that of planning CTs (pCTs),1 but dose calculation accuracy using these corrected CBCTs has not been characterized. In this study, we aim to quantify the accuracy of dose calculation on corrected CBCT images generated by our machine learning-based method applied for brain stereotactic radiosurgery (SRS) and prostate radiotherapy cases.
CBCT has been increasingly utilized in image-guided radiation therapy to improve treatment performance. CBCTs are acquired at the time of treatment delivery and provide detailed anatomic information in the treatment position. In clinical practice, CBCT is primarily used to determine the degree of patient setup error and inter-fraction motion by comparing the displacement of anatomic landmarks from the treatment planning CT images.2 More demanding applications of CBCT have been proposed with the increasing use of adaptive radiation therapy, such as daily estimation of delivered dose based on CBCT images, and automatic contouring on CBCTs based on a deformable image registration (DIR) with the pCT.3,4
However, such potential uses of CBCT have been limited due in part to image quality that can be significantly degraded when compared to pCTs. One major source of degradation is the streaking and cupping artifacts caused mainly by scatter contamination, as well as by other physical non-idealities including beam-hardening effect, photon starvation, and detector lag, etc.5,6 These artifacts lead to significant CT number errors, which complicates the calibration process of CBCT Hounsfield Unit (HU) to electron density, an essential step in dose calculation.6 The degraded image contrast and the suppression of bone CT number can also cause large errors in DIR for contour propagation from pCT to CBCT.7 Thus, the severe distortion caused by scatter is considered one of the fundamental limitations of CBCT, and prevents CBCT from quantitative usage in radiation therapy.
Many correction methods for CBCT shading artifacts have been proposed in the literature, and in general these methods fall into two major categories. The first are hardware-based pre-processing methods, including airgap,8 bowtie filter9 and anti-scatter grid methods.10 By preventing part of the scattered photons from reaching the detector, these add-on devices successfully mitigate severe shading artifacts. To maintain the signal-to-noise ratio, the correction efficacy is limited by an associated increase in patient imaging dose because primary photons are attenuated by these devices as well as scattered photons. The second are post-processing techniques which correct image artifacts by estimating the scatter in the projection domain or the image domain. Typical methods of this type include analytical modeling,11 Monte Carlo simulation,12,13 measurement-based methods,14 and modulation methods.15 For example, high quality pCT images can be used as prior information to enhance the CBCT images of the same patient in either image domain6,16–18 or projection domain19,20. Instead of using such prior images as external helpers, other methods mitigate the shading artifacts by estimating the low-frequency shading field from the images, which is achieved by sophisticated image segmentation methods21–23 or ring-correction methods24. These methods enhance the scatter correction performance, while their implementations entail combined considerations of computational complexity, imaging dose, scan time, practicality, and efficacy.
In current commercial CBCT imaging systems, effective shading correction has been implemented by a combination of the above methods. For example, on the Varian TrueBeam On-Board Imager (OBI) CBCT system, a 10:1 anti-scatter grid is mounted on the flat-panel detector, and a model-based adaptive-scatter-kernel-superposition (ASKS) method is implemented in the image reconstruction process. However, residual artifacts are still commonly observed in clinical CBCT images. Moreover, most of the existing methods cannot restore the true Hounsfield Unit (HU) value in CBCT images; i.e., the pixel values in CBCT images are not calibrated identically to pCT images in the treatment planning system for dose calculation.
Recently, we developed a novel machine-learning based method to further improve CBCT image quality such that they are comparable to that of pCTs for potential application to CBCT-guided adaptive radiotherapy.1,25,26 By building a set of paired training images of pCT and CBCT and using the pCT as the learning target, the image quality of CBCT was improved significantly through a learning process. Compared with existing methods, our method not only mitigates non-uniform and streaking artifacts, but also restores true HU values on CBCT images such that the CBCT images after correction share the same calibration as pCTs for dose calculation. We evaluated our method with respect to image quality1. Our method has correction accuracy with mean absolute error of 12.81 ± 2.04 HU.
In this paper, we aim to study the dose calculation accuracy of the corrected CBCT images using our previously developed machine-learning-based method. We retrospectively investigated patient data of brain SRS and prostate radiotherapy treatments with both pCTs and CBCT images acquired during treatment. The dose maps of the same treatment plans were calculated on original pCT simulation images as ground truth, as well as on original (i.e., uncorrected) CBCT (OCBCT) and corrected CBCT (CBCCT) images. Clinically relevant DVH metrics and gamma analysis were extracted from both the ground truth and OCBCT/PCBCT results for comparison and evaluation on a total of 32 treatment plans from 22 patients.
METHODS AND MATERIALS
In this retrospective study, we analyzed the dataset of 12 patients with intracranial malignancies receiving frameless SRS and 10 patients with pelvic malignancies. These patients were randomly selected; each patient had at least one plan with both CT simulation and CBCT images acquired during the treatment. Some treatment plans included multiple target sites, while other some patients had multiple treatment plans each with its own isocenter. Our study was performed on individual plans, and a total of 16 brain SRS plans and 16 pelvic radiation therapy plans were analyzed. The specific location of the lesion varied from patient to patient.
The CT images were acquired using a Siemens SOMATOM Definition AS CT scanner at 120 kVp with the patient in treatment position. The image spacing was 0.586 mm × 0.586 mm × 0.6 mm and 0.98 mm × 0.98 mm × 2.0 mm for brain and pelvis patients, respectively. The CBCT images were acquired using the Varian On-Board Imager CBCT system. The image spacing was 1.17 mm × 1.17 mm × 2.5 mm for brain patients on Novalis Tx, and 0.908 mm × 0.908 mm × 2.0 mm for pelvis patients on TrueBeam. Physicians defined the target volume and organs-at-risk (OARs) on CT images, prescribed dose by protocols and registered the pCT and CBCT image datasets before treatment started. Treatment plans were created by dosimetrists based on pCT images using either Volumetric Arc Therapy (VMAT) or Dynamic Conformal Arc (DCA) technique for SRS patients, and VMAT plans for pelvis radiotherapy, using Eclipse 13.6 (Varian Medical Systems, Palo Alto, CA).
We have previously developed a learning-based method to improve the image quality of CBCTs.1 The OCBCT images of each patient were imported into a well-trained machine learning model to generate corresponding CCBCT images. Details in methods and implementation on this machine learning model developed by our group have been reported in our previous publication,1 and we would like to briefly introduce it here.
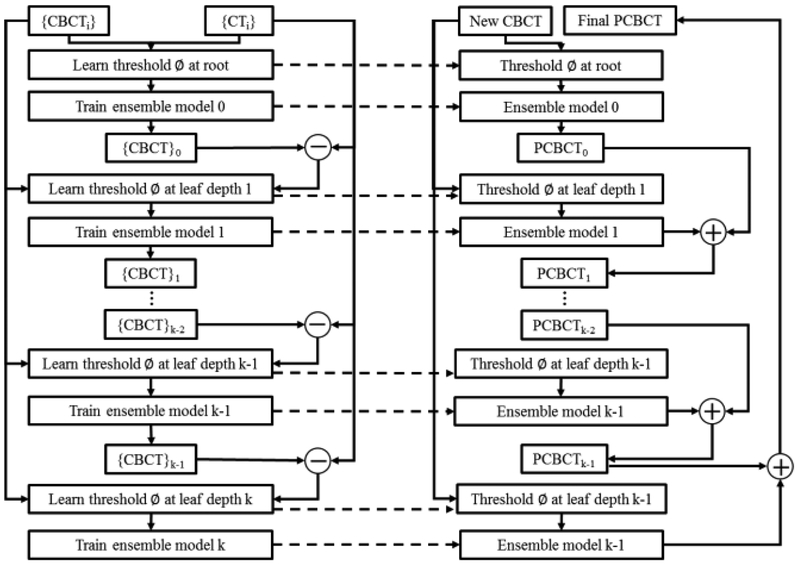
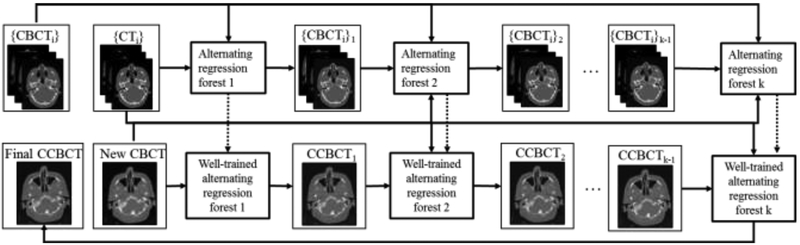
Suppose we have a set of pairs of CBCT and CT training images. For each pair, CT image is used as the regression target of CBCT image to improve the CBCT quality. CBCT and CT are preprocessed by removing noise and uninformative regions. The intra-subject registration is performed to align each pair of CT and CBCT images of the same patient, and a inter-patient registration is used to roughly map all patients onto a common space. We then register all the training data to the new CBCT image. In the training stage, patch-wise multi-level features, i.e., DCT27, LBP28, and pairwise voxel difference with multi-scale sensitivity29, i.e., original and down-sampled image with three down sampling factors (0.75, 0.5 and 0.25), are extracted from training CBCT. Secondly, fuzzy c-means labeling30 is utilized to generate the corresponding CT label automatically. Combining the extracted features with CT labels, we perform a feature selection using logistic LASSO algorithm31–34 to identify the most salient and informative features with patient-specific information. Thirdly, we utilize the selected features as well as the corresponding CT targets to train a random forest using alternating regression forest (ARF)35. During the training, we apply auto-context model (ACM)36 to incorporate the appearance information from original CBCT with the context information from the previously predicted CT for iterative refinement. In the test stage, anatomical features from the new CBCT are extracted and fed into the well-trained random forest for the prediction of an improved CBCT. Finally, we use all predicted patches together to reconstruct the final high-quality CBCT image prediction. Fig.1 shows the brief workflow of our prediction method.
Figure 1.
Schematic flow chart of the proposed algorithm for high-quality CBCT predication.
Alternating Regression Forest (ARF) is used in this study in training the regression model. Recent studies showed the efficacy of random forest in tackling medical image processing25,29,32. Classical random forest trains a bag of binary decision trees each of which is provided with a random subset of training data and trained independently from the others. The major drawback of classical random forest is its binary decision tree is only decided locally on the node level how the data is further split, without considering the state of the whole regressor. In this work, we propose ARF as a novel splitting procedure considering both the global and local optimization. The general learning objective can be written as a greedy stage-wise optimization:
| (1) |
where L(·) is a differentiable loss function, Rd−1(xi, ϕ) denotes the regressor trained up to depth d−1, rd(xi, ϕd) denotes the regressor for current depth, and ϕd is the split threshold optimized in the depth d. At root nodes, we start with an initial regressor R0 = r0(xi, ϕ) and add a new depth to the forest. At depth d, assume previous regressor Rd−1(xi, ϕ) predicts training samples xi as Rd−1(xi, ϕ) = Rd−2(xi, ϕ) +rd−1(xi, ϕd) by ensemble model according to stored samples in corresponding nodes. These predictions yield a stage-wise loss which should be minimized by (Eq. 1). After optimizing thresholds, the regressor rd(xi, ϕd) can be determined according to information gain criterion. Since the splitting function of each node and each depth is chosen by jointly reducing the uncertainty of training data and regularizing the whole regressors’ global loss, unlike the classical random forest whose path of splitting training data is independent beforehand for each node, our proposed ARF allows the splitting path to be always optimized globally and locally as we have a hierarchical splitting structure. Thus, the feedback of each depth reasonably reduces the uncertainty of binary decision tree and enhances the accuracy of the inference, as shown in Fig. 2.
Figure 2.
The flow chart of the alternating regression forest.
ACM is used to leverage the surrounding information with respect to the object of interest such as bone area for further improvement in the prediction performance36–39. We use the initial ARF to create context features for all training patients, which are then used in combination with the initial extracted features to train an improved ARF. The process is repeated to train a series of ARFs until the prediction criterion is met. It is proven that ACM can reduce the prediction error. In the testing stage, to predict the improved CBCT, the new CBCT image can follow the same concept of ACM to generate the final improved CBCT. The framework of ACM is showed in Fig. 3.
Figure 3.
The framework of the ACM.
To evaluate the dose calculation accuracy of the OCBCT/CCBCT images, we compared the differences between OCBCT/CCBCT and original pCT dose maps using the same plan parameters. To minimize the anatomy difference from pCT, we choose the CBCT from the first fraction of treatment. Both rigid and deformable registrations were performed between the pCT volume and OCBCT/CCBCT to align the patient anatomy. The aligned OCBCT/CBCCT images were interpolated into the same voxel size as the pCT images and then imported into the treatment planning system. The structures and treatment plan from the pCT volumes were duplicated onto the OCBCT/CBCCT volumes for dose calculation using the same algorithm (analytical anisotropic algorithm, AAA), grid size (0.1 cm for brain SRS and 0.25 cm for pelvis radiotherapy) and HU calibration. The original pCT-based plans served as ground truth.
For each patient, we first studied the image quality of OCBCT/CCBCT images to validate our method. We evaluated OCBCT/CCBCT image accuracy by two commonly used image quality matrices, i.e. mean absolute error (MAE) and peak signal-to-noise ratio (PSNR). MAE quantifies the absolute difference between OCBCT/CCBCT and pCT for each pixel. PSNR is an engineering term expressing the ratio between the maximum possible power of a signal to the power of corrupting noise that affects the fidelity of its representation, which is directly related to the relative difference between OCBCT/CCBCT and pCT. They are defined as follows:
| (2) |
| (3) |
where ICBCT and IpCT indicate image of OCBCT/CCBCT and pCT, i is the ith pixel in image volume, N is the total number of pixels in ICBCT or IpCT, Q is the maximum pixel intensity among ICBCT and IpCT. A lower MAE or a higher PSNR indicates higher quality of CBCT images.
For each plan, we visually checked the similarity of the dose distributions calculated on the OCBCT/CCBCT and the original pCT. Quantitatively, clinically relevant DVH metrics were extracted to compare dose to PTVs and relevant OARs. Specifically, Dmin, D10%, D50%, D95%, Dmean and Dmax are used as metrics for all PTVs and OARs, and D99% for brain SRS PTV additionally. For plans with multiple PTVs, each PTV is considered separately. Gamma analysis using the 1%/1mm criteria was performed on axial, coronal and sagittal planes at isocenter to compare the dose distributions computed on the OCBCT/CCBCT and the original pCT.
RESULTS
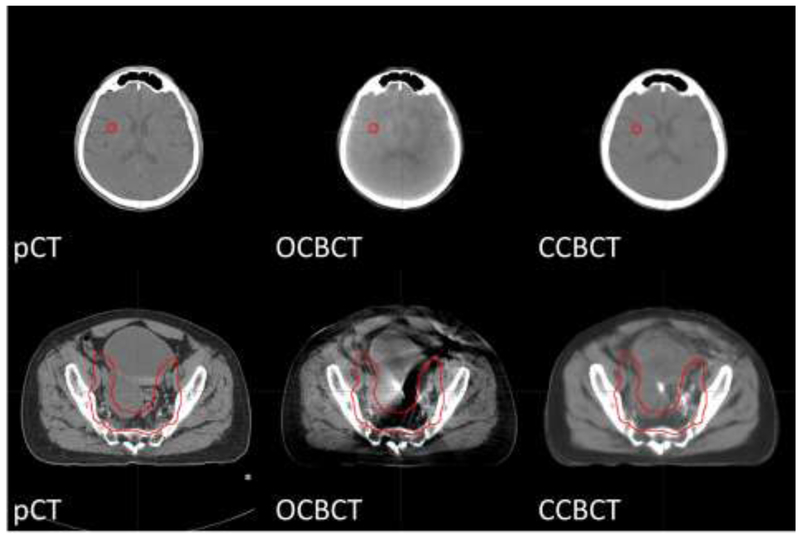
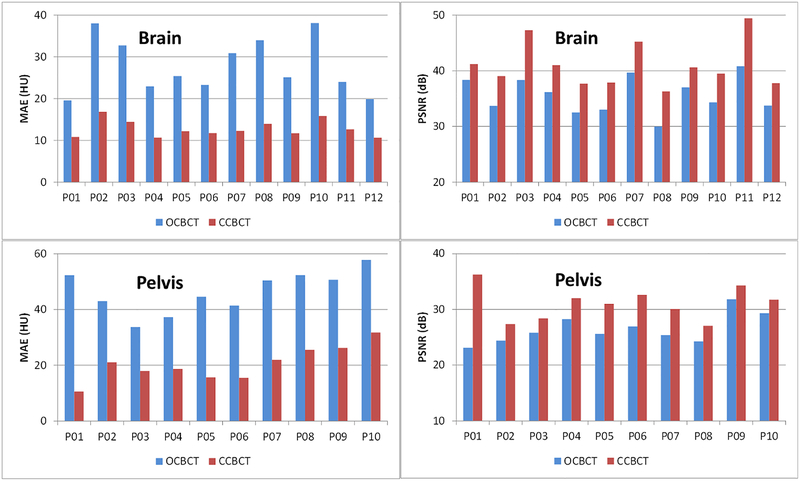
In Fig.4, the image quality improvement of CCBCT is shown using a side-by-side comparison with pCT and OCBCT; one example each is provided from a brain SRS case and a pelvic radiotherapy case with the same HU window level for each. Non-uniform artifacts and global HU shifts can be seen on OCBCT images in both cases, and are much more severe in the pelvic case. Our CCBCT images feature better image quality than OCBCT with comparable uniformity, contrast, and HU ranges to pCT, and its improvement in image quality in the pelvis case is more remarkable than that of the brain case. These findings are also consistent with the measured MAE and PSNR of OCBCT and CCBCT for each patient of brain or pelvis case plotted in Fig.5 and summarized in Table I. For all the patients, CCBCT has shown lower MAE and higher PSNR than OCBCT, and such improvement is larger for pelvis patients than brain SRS patients.
Figure 4.
The axial view of pCT/OCBCT/CCBCT images of brain patient (top) and pelvis patient (bottom). The red contours indicate PTVs. Display window: [−160 240] HU.
Figure 5.
Image quality matrices MAE (left) and PSNR (right) for each brain SRS (top) or pelvis (bottom) patient.
Table I.
Mean and standard deviation of MAE and PSNR among all patients of brain SRS and pelvis cases.
| Brain | Pelvis | |||||
|---|---|---|---|---|---|---|
| OCBCT | CCBCT | Improvement | OCBCT | CCBCT | Improvement | |
| MAE | 27.82 | 12.81 | 15.01 | 45.47 | 19.94 | 25.53 |
| PSNR | 35.64 | 41.07 | 15.24% | 26.71 | 31.31 | 17.22% |
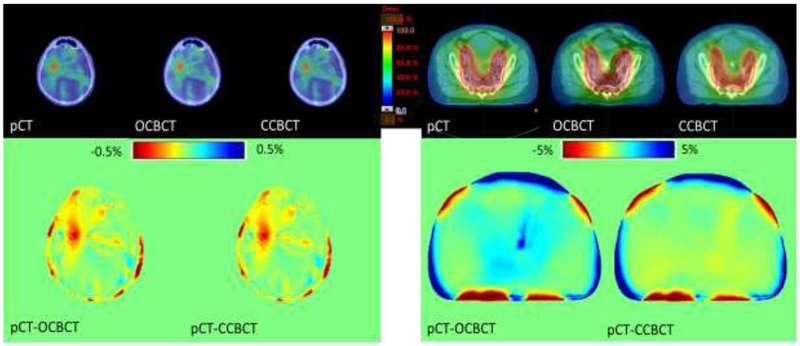
Fig. 6 compares the dose distribution in the axial plane of the patients shown in Fig. 4. For the radiosurgery case, the dose differences of OCBCT and CCBCT from pCT shown in Fig. 6 (bottom left) are both very small (~0.5%), and the dose calculation improvements of CCBCT can hardly be observed. However, for the pelvic case shown in the bottom right, the corresponding improvement is significant. Dose differences using the OCBCT occur mostly around streaking artifacts, which are largely mitigated for the majority of the volume in the CCBCT. The corresponding DVH curves of PTVs and relevant OARs shown in Fig. 7 indicate that the dose differences in target and avoidance structures are minimal. These results from the two patients are presented as examples, and similar results can be seen in other patients.
Figure 6.
Dose distribution calculated on pCT, OCBCT, and CCBCT. Top: dose maps in color wash on pCT, OCBCT, and CCBCT shown on the axial plane of isocenter. Bottom: dose difference map between pCT and OCBCT/CCBCT. Left and right are brain case and pelvis case, respectively.
Figure 7.
The DVH curves of PTVs and related OARs for brain case (top) and pelvis case (bottom) in Fig. 1. OARs for brain case are chiasm, brainstem, left optic nerve, and right optic nerve; OARs for pelvis are bladder, rectum, left femur and right femur.
The mean dose differences between OCBCT/CCBCT and pCT in relevant DVH metrics for both types of cases are shown by the chart in Fig. 8. All DVH metrics using the CCBCT are closer to pCT than those on OCBCT. For brain SRS cases, both OCBCT and CCBCT have very low dose difference for PTVs (~0.2%) and OARs (<0.05%). Compared with the 1% uncertainty in dose calculation from CT images in brain SRS cases,40 such dose difference between OCBCT/CCBCT and pCT is negligible, which indicates that OCBCT and CCBCT are both accurate enough for dose calculation for brain SRS cases. However, for pelvic cases, OCBCTs have relatively large dose difference in PTV, bladder and rectum (~1%), and the use of CCBCTs reduces that difference to ~0.3%. DVH differences among all plans are summarized in Table II. For brain SRS cases, there is no statistically significant improvement from the use of CCBCTs on dose calculation for all DVH metrics; however, such improvements are statistically significant for many pelvic DVH metrics.
Figure 8.
Mean differences in DVH metrics between OCBCT/CCBCT and pCT among all plans of brain SRS (top) and pelvis cases (bottom).
Table II.
Mean and standard deviation of differences in DVH metrics between OCBCT/CCBCT and pCT among all plans of brain SRS (top) and pelvis cases (bottom). The p-value of t-test of DVH differences between OCBCT and CCBCT are also listed.
| Dmin | D10% | D50% | D95% | D99% | Dmean | Dmax | ||
|---|---|---|---|---|---|---|---|---|
| PTV | OCBCT | 0.193 ± 0. 172 |
0.232 ± 0. 174 |
0.211 ± 0. 141 |
0.200 ± 0. 120 |
0.202 ± 0. 118 |
0.209 ± 0. 142 |
0.310 ± 0.321 |
| CCBCT | 0.136 ± 0.210 |
0.134 ± 0.158 |
0.105 ± 0. 124 |
0.096 ± 0. 115 |
0.122 ± 0. 117 |
0.104 ± 0. 124 |
0.210 ± 0.255 |
|
| P-value | 0.318 | 0.986 | 0.518 | 0.638 | 0.431 | 0.757 | 0.547 | |
| Brain Stem | OCBCT | 0.007 ± 0.014 |
0.031 ± 0.059 |
0.016 ± 0.037 |
0.008 ± 0.013 |
0.016 ± 0.031 |
0.051 ± 0.069 |
|
| CCBCT | 0.001 ± 0.002 |
0.006 ± 0.008 |
0.004 ± 0.005 |
0.002 ± 0.002 |
0.003 ± 0.003 |
0.013 ± 0.013 |
||
| P-value | 0.679 | 0.285 | 0.46 | 0.694 | 0.383 | 0.088 | ||
| Optic Nerve | OCBCT | 0.02 ±0.042 |
0.022 ±0.048 |
0.024 ±0.047 |
0.02 ±0.042 |
0.023 ±0.046 |
0.033 ±0.062 |
|
| CCBCT | 0.005 ±0.011 |
0.012 ±0.030 |
0.009 ±0.020 |
0.005 ±0.010 |
0.009 ±0.020 |
0.017 ±0.040 |
||
| P-value | 0.661 | 0.46 | 0.424 | 0.58 | 0.385 | 0.256 | ||
| Chiasm | OCBCT | 0.016 ±0.030 |
0.026 ±0.055 |
0.024 ±0.048 |
0.021 ±0.037 |
0.024 ±0.047 |
0.039 ±0.072 |
|
| CCBCT | 0.003 ±0.004 |
0.004 ±0.005 |
0.003 ±0.004 |
0.002 ±0.003 |
0.003 ±0.004 |
0.009 ±0.011 |
||
| P-value | 0.645 | 0.303 | 0.304 | 0.539 | 0.338 | 0.476 |
| Dmin | D10% | D50% | D95% | Dmean | Dmax | ||
|---|---|---|---|---|---|---|---|
| PTV | OCBCT | 0.924 ± 0.724 |
0.918 ± 0.548 |
0.774 ± 0.493 |
0.66 ± 0.434 |
0.791 ± 0.491 |
1.53 ± 1.235 |
| CCBCT | 0.452 ± 0.476 |
0.297 ± 0.176 |
0.284 ± 0.153 |
0.237 ± 0.168 |
0.281 ± 0.150 |
0.602 ± 0.898 |
|
| P-value | 0.559 | 0.001 | 0.001 | 0.002 | 0.001 | 0.002 | |
| Bladder | OCBCT | 0.068 ± 0.081 |
0.797 ± 0.434 |
0.443 ± 0.419 |
0.138 ± 0.185 |
0.468 ± 0.346 |
1.172 ± 0.868 |
| CCBCT | 0.042 ± 0.074 |
0.332 ± 0.143 |
0.143 ± 0.111 |
0.044 ± 0.037 |
0.161 ± 0.093 |
0.459 ± 0.434 |
|
| P-value | 0.021 | 0.001 | 0.01 | 0.073 | 0.002 | 0.001 | |
| Rectum | OCBCT | 0.037 ± 0.041 |
1.085 ± 1.456 |
1.054 ± 1.073 |
0.178 ± 0.324 |
0.485 ± 0.585 |
1.641 ± 1.870 |
| CCBCT | 0.025 ± 0.035 |
0.52 ± 0.839 |
0.545 ± 0.976 |
0.035 ± 0.053 |
0.21 ± 0.282 |
0.896 ± 1.343 |
|
| P-value | 0.021 | 0.013 | 0.007 | 0.311 | 0.003 | 0.02 | |
| Femur | OCBCT | 0.025 ± 0.050 |
0.295 ± 0.211 |
0.065 ± 0.049 |
0.013 ± 0.017 |
0.181 ± 0.280 |
0.447 ± 0.293 |
| CCBCT | 0.021 ± 0.057 |
0.111 ± 0.080 |
0.055 ± 0.047 |
0.016 ± 0.023 |
0.129 ± 0.294 |
0.209 ± 0.182 |
|
| P-value | 0.096 | 0.004 | 0.006 | 0.784 | 0.004 | 0.001 |
The dose accuracy of OCBCT/CCBCT is further evaluated by gamma analysis using 1%/1mm criteria on the three orthogonal dose planes cross isocenter between OCBCT/CCBCT and pCT for each plan. The passing rates for brain SRS are above 99% for both OCBCT and CCBCT for all cases. For pelvic cases, the average passing rates are 94.5% and 99.0% for OCBCT-based and CCBCT-based dose plans, respectively.
CONCLUSIONS AND DISCUSSION
In this study, we evaluated the dose calculation accuracy improvement of corrected CBCTs, made by our machine-learning-based method, in the context of brain SRS and pelvic radiotherapy. The side-by-side comparisons on image quality demonstrated better image contrast and more accurate HU ranges of corrected CBCTs compared to original CBCTs. Based on the statistical analysis of comparative DVH metrics between OCBCT, CCBCT and pCT for the same plans, we showed that the mean dose error in DVH metrics for PTV, bladder and rectum was reduced from 1% to 0.3% after CBCT correction in pelvic cases, and such improvements were statistically significant. The improvement in dose calculation accuracy is further supported by the gamma analysis study with an average pass rate increasing from 94.5% before correction to 99.0% after correction. For brain SRS treatments, both OCBCT and CCBCT images were accurate enough for dose calculation, while CCBCT features higher image quality. These quantitative results strongly indicate that the CCBCT could provide dose accuracy comparable to pCTs for brain SRS and prostate radiation therapy. It also demonstrates that the proposed correction method has great potential in CBCT-guided adaptive radiotherapy.
In the presented study, we found that the difference of dose distributions between CCBCT and pCT (Fig. 6) was very minimal for the majority of the volume, even in small areas with residual artifacts on the CCBCT images. It indicates that the artifacts are successfully mitigated for accurate dose calculation on CCBCT. Slightly larger errors were observed at the surface of body, especially for pelvic cases; this could be because the pCT images, which are chosen to be ground truth in this study, are not perfectly matched with CBCT images due to inter-fraction setup uncertainty. Such uncertainty includes misalignment in registration and deformation of body surface, both of which contribute to the image difference between CBCT and pCT, especially at the surface of body. Minimal image differences at the surface could lead to large dose differences since dose calculations were not performed outside the body contour. For pelvic cases, the soft tissue deformation of the body surface is more commonly seen than brain cases, which results in larger dose differences.
In this study, we evaluated our learning-based method on brain SRS and pelvic radiation therapy patients. Compared to other sites, brain SRS cases are special since CBCT scatter artifacts are minimized due to its smaller size, and photon plans are more forgiving due to its homogeneity. Thus, CBCT images are shown to be accurate enough for dose calculation without additional correction, even for SRS treatments that deliver large amount of dose to a very small volume. However, CBCT correction can still help provide better image quality for other tasks such as image registration and contour propagation in adaptive radiation therapy. For pelvic cases where CBCT contains complicated bone-tissue interface and severe artifacts, CBCT correction has been demonstrated to be necessary for both image quality and dose calculation.
In our clinic, the majority sites of CBCT taken on patients are pelvic and brain SRS treatment. Investigations in dose calculation using CBCT for these two cases would meet the most demands from clinical. Other cases involving CBCT in our clinic are lung. However, the CBCT images on thorax usually truncate patient body outside field of view. Thus, it is not an ideal case for this study.
ACKNOWLEDGMENT
This research is supported in part by the National Cancer Institute of the National Institutes of Health under Award Number R01CA215718 and Dunwoody Golf Club Prostate Cancer Research Award, a philanthropic award provided by the Winship Cancer Institute of Emory University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lei Y, Tang X, Higgins K, et al. Improving image quality of cone-beam CT using alternating regression forest. Proc of SPIE. 2018;10573:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barney BM, Lee RJ, Handrahan D, Welsh KT, Cook JT, Sause WT. Image-Guided Radiotherapy (IGRT) for Prostate Cancer Comparing kV Imaging of Fiducial Markers With Cone Beam Computed Tomography (CBCT). International Journal of Radiation Oncology • Biology • Physics. 2011;80(1):301–305. [DOI] [PubMed] [Google Scholar]
- 3.Zhu L, Xie Y, Wang J, Xing L. Scatter correction for cone-beam CT in radiation therapy. Medical physics. 2009;36(6):2258–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adam de la Z, Benjamin A, Lei X. Formulating adaptive radiation therapy (ART) treatment planning into a closed-loop control framework. Physics in Medicine and Biology. 2007;52(14):4137. [DOI] [PubMed] [Google Scholar]
- 5.Grimmer R, Kachelriess M. Empirical binary tomography calibration (EBTC) for the precorrection of beam hardening and scatter for flat panel CT [published online ahead of print 2011/06/02]. Medical physics. 2011;38(4):2233–2240. [DOI] [PubMed] [Google Scholar]
- 6.Marchant TE, Moore CJ, Rowbottom CG, MacKay RI, Williams PC. Shading correction algorithm for improvement of cone-beam CT images in radiotherapy. Phys Med Biol. 2008;53(20):5719. [DOI] [PubMed] [Google Scholar]
- 7.Hou J, Guerrero M, Chen W, D’Souza WD. Deformable planning CT to cone-beam CT image registration in head-and-neck cancer. Medical physics. 2011;38(4):2088–2094. [DOI] [PubMed] [Google Scholar]
- 8.Siewerdsen JH, Jaffray DA. Optimization of x-ray imaging geometry (with specific application to flat-panel cone-beam computed tomography). Medical physics. 2000;27(8):1903–1914. [DOI] [PubMed] [Google Scholar]
- 9.Mail N, Moseley DJ, Siewerdsen JH, Jaffray DA. The influence of bowtie filtration on cone-beam CT image quality. Medical physics. 2009;36(1):22–32. [DOI] [PubMed] [Google Scholar]
- 10.Siewerdsen JH, Moseley DJ, Bakhtiar B, Richard S, Jaffray DA. The influence of antiscatter grids on soft-tissue detectability in cone-beam computed tomography with flat-panel detectors. Medical physics. 2004;31(12):3506–3520. [DOI] [PubMed] [Google Scholar]
- 11.Boone JM, Seibert JA. An analytical model of the scattered radiation distribution in diagnostic radiology. Medical physics. 1988;15(5):721–725. [DOI] [PubMed] [Google Scholar]
- 12.Colijn AP, Beekman FJ. Accelerated simulation of cone beam X-ray scatter projections [published online ahead of print 2004/05/19]. IEEE Trans Med Imaging. 2004;23(5):584–590. [DOI] [PubMed] [Google Scholar]
- 13.Kyriakou Y, Riedel T, Kalender WA. Combining deterministic and Monte Carlo calculations for fast estimation of scatter intensities in CT [published online ahead of print 2006/09/06]. Phys Med Biol. 2006;51(18):4567–4586. [DOI] [PubMed] [Google Scholar]
- 14.Ning R, Tang X, Conover D. X-ray scatter correction algorithm for cone beam CT imaging. Medical physics. 2004;31(5):1195–1202. [DOI] [PubMed] [Google Scholar]
- 15.Zhu L, Bennett NR, Fahrig R. Scatter correction method for X-ray CT using primary modulation: theory and preliminary results [published online ahead of print 2006/12/16]. IEEE Trans Med Imaging. 2006;25(12):1573–1587. [DOI] [PubMed] [Google Scholar]
- 16.Brunner S, Nett BE, Tolakanahalli R, Chen GH. Prior image constrained scatter correction in cone-beam computed tomography image-guided radiation therapy [published online ahead of print 2011/01/25]. Phys Med Biol. 2011;56(4):1015–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu L, Bruesewitz MR, Thomas KB, Fletcher JG, Kofler JM, McCollough CH. Optimal tube potential for radiation dose reduction in pediatric CT: Principles, clinical implementations, and pitfalls. RadioGraphics. 2011;31(3):835–848. [DOI] [PubMed] [Google Scholar]
- 18.Shi L, Tsui T, Wei J, Zhu L. Fast shading correction for cone beam CT in radiation therapy via sparse sampling on planning CT. Medical physics. 2017. doi: 10.1002/mp.12190:In Press. [DOI] [PubMed] [Google Scholar]
- 19.Niu T, Sun M, Star-Lack J, Gao H, Fan Q, Zhu L. Shading correction for on-board cone-beam CT in radiation therapy using planning MDCT images [published online ahead of print 2010/11/26]. Medical physics. 2010;37(10):5395–5406. [DOI] [PubMed] [Google Scholar]
- 20.Niu T, Al-Basheer A, Zhu L. Quantitative cone-beam CT imaging in radiation therapy using planning CT as a prior: first patient studies [published online ahead of print 2012/04/10]. Medical physics. 2012;39(4):1991–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pengwei W, Xiaonan S, Hongjie H, et al. Iterative CT shading correction with no prior information. Physics in Medicine and Biology. 2015;60(21):8437. [DOI] [PubMed] [Google Scholar]
- 22.Zhao W, Vernekohl D, Zhu J, Wang L, Xing L. A model-based scatter artifacts correction for cone beam CT. Medical physics. 2016;43(4):1736–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang T, Zhu L. Image-domain non-uniformity correction for cone-beam CT. 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017). 2017. doi: 10.1109/ISBI.2017.7950611:680–683. [DOI] [Google Scholar]
- 24.Fan Q, Lu B, Park JC, et al. Image-domain shading correction for cone-beam CT without prior patient information [published online ahead of print 2015/12/25]. Journal of applied clinical medical physics / American College of Medical Physics. 2015;16(6):5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, Lei Y, Higgins KA, et al. A Leaning-Based Method to Improve Cone Beam CT Image Quality for Adaptive Radiation Therapy. International Journal of Radiation Oncology • Biology • Physics. 2017;99(2):S224. [Google Scholar]
- 26.Yang X, Liu T, Dong X, et al. A patch-based CBCT scatter artifact correction using prior CT. Proc of SPIE. 2017;10132:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sridhar D, Krishna IM. Brain Tumor Classification using Discrete Cosine Transform and Probabilistic Neural Network. Paper presented at: 2013 International Conference on Signal Processing , Image Processing & Pattern Recognition; 7–8 Feb. 2013, 2013. [Google Scholar]
- 28.Fehr J, Burkhardt H. 3D rotation invariant local binary patterns. Paper presented at: 2008 19th International Conference on Pattern Recognition; 8–11 Dec. 2008, 2008. [Google Scholar]
- 29.Huynh T, Gao YZ, Kang JY, et al. Estimating CT Image From MRI Data Using Structured Random Forest and Auto-Context Model. Ieee Transactions on Medical Imaging. 2016;35(1):174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bezdek JC, Ehrlich R, Full W. FCM: The fuzzy c-means clustering algorithm. Computers & Geosciences. 1984;10(2):191–203. [Google Scholar]
- 31.Liao S, Gao YZ, Lian J, Shen DG. Sparse Patch-Based Label Propagation for Accurate Prostate Localization in CT Images. Ieee Transactions on Medical Imaging. 2013;32(2):419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang X, Lei Y, Shu H-K, et al. Pseudo CT estimation from MRI using patch-based random forest. Proc SPIE. 2017;10133:101332Q–101337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X, Rossi P, Ogunleye T, Jani A, Curran W, Liu T. 3D transrectal ultrasound (TRUS) prostate segmentation based on optimal feature learning framework. Proc SPIE. 2016;9784:97842F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X, Wu S, Sechopoulos I, Fei B. Cupping artifact correction and automated classification for high-resolution dedicated breast CT images [published online ahead of print 2012/10/09]. Medical Physics. 2012;39(10):6397–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schulter S, Leistner C, Wohlhart P, Roth PM, Bischof H. Alternating Regression Forests for Object Detection and Pose Estimation. Paper presented at: 2013 IEEE International Conference on Computer Vision; 1–8 Dec. 2013, 2013. [Google Scholar]
- 36.Tu ZW, Bai XA. Auto-Context and Its Application to High-Level Vision Tasks and 3D Brain Image Segmentation. Ieee Transactions on Pattern Analysis and Machine Intelligence. 2010;32(10):1744–1757. [DOI] [PubMed] [Google Scholar]
- 37.Andreasen D, Edmund JM, Zografos V, Menze BH, Van Leemput K. Computed Tomography synthesis from Magnetic Resonance images in the pelvis using multiple Random Forests and Auto-Context features. Proc Spie. 2016;9784. [Google Scholar]
- 38.Wei L, Cao X, Wang Z, et al. Learning-based deformable registration for infant MRI by integrating random forest with auto-context model [published online ahead of print 2017/09/14]. Medical physics. 2017;44(12):6289–6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tu Z, Bai X. Auto-context and its application to high-level vision tasks and 3D brain image segmentation [published online ahead of print 2010/08/21]. IEEE Trans Pattern Anal Mach Intell. 2010;32(10):1744–1757. [DOI] [PubMed] [Google Scholar]
- 40.Verellen D, Linthout N, Bel A, et al. Assessment of the uncertainties in dose delivery of a commercial system for linac-based stereotactic radiosurgery [published online ahead of print 2000/04/13]. International journal of radiation oncology, biology, physics. 1999;44(2):421–433. [DOI] [PubMed] [Google Scholar]