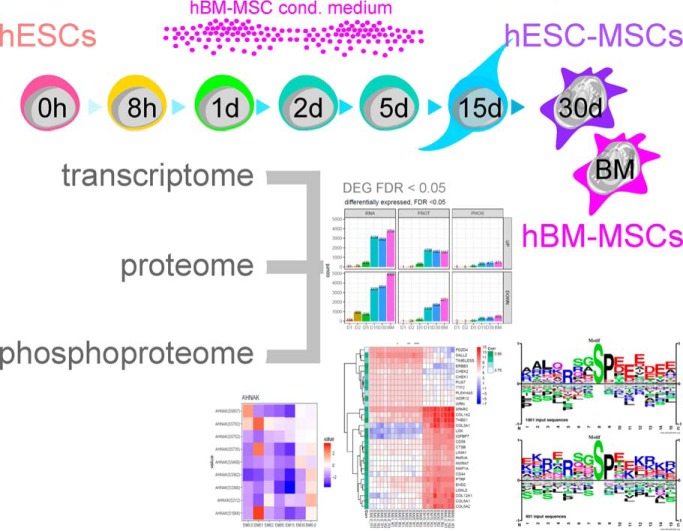
In this integrative, multi-omics study we characterized the differentiation of human embryonic stem cells into mesenchymal stem cells using transcriptomics, quantitative MS-based proteomics and phosphoproteomics. Based on RNA-to-protein correlation, we determined a set of high confidence genes that are important to differentiation with AHNAK hypothesized to be a defining factor in MSC biology. Additional central findings include two distinct expression waves of developmental HOX genes as well as an AGO2-to-AGO3 switch in gene silencing.
Keywords: RNA SEQ, cell differentiation, quantification, stem cells, systems biology, phosphoproteome, differentiation, developmental biology, gene expression, post-translational modifications, human embryonic stem cells, human mesenchymal stem cells, LC-MS/MS, quantitative proteomics
Graphical Abstract
Highlights
Integrative multi-omics study characterizing the differentiation from hESCs into hMSCs.
Set of high confidence genes important in hESC to hMSC differentiation defined.
Two distinct expression waves of HOX genes and a AGO2-to-AGO3 switch in gene silencing identified.
AHNAK hypothesized as a defining factor in MSC biology.
Abstract
Mesenchymal stem/stromal cells (MSCs) are self-renewing multipotent cells with regenerative, secretory and immunomodulatory capabilities that are beneficial for the treatment of various diseases. To avoid the issues that come with using tissue-derived MSCs in therapy, MSCs may be generated by the differentiation of human embryonic stems cells (hESCs) in culture. However, the changes that occur during the differentiation process have not been comprehensively characterized. Here, we combined transcriptome, proteome and phosphoproteome profiling to perform an in-depth, multi-omics study of the hESCs-to-MSCs differentiation process. Based on RNA-to-protein correlation, we determined a set of high confidence genes that are important to differentiation. Among the earliest and strongest induced proteins with extensive differential phosphorylation was AHNAK, which we hypothesized to be a defining factor in MSC biology. We observed two distinct expression waves of developmental HOX genes and an AGO2-to-AGO3 switch in gene silencing. Exploring the kinetic of noncoding ORFs during differentiation, we mapped new functions to well annotated long noncoding RNAs (CARMN, MALAT, NEAT1, LINC00152) as well as new candidates which we identified to be important to the differentiation process. Phosphoproteome analysis revealed ESC and MSC-specific phosphorylation motifs with PAK2 and RAF1 as top predicted upstream kinases in MSCs. Our data represent a rich systems-level resource on ESC-to-MSC differentiation that will be useful for the study of stem cell biology.
Mesenchymal stem/stromal cells (MSCs)1 are self-renewing multipotent cells with regenerative, secretory and immunomodulatory capabilities. They thus have great potential in cell-based therapy and have proven beneficial for the treatment of various diseases (1). MSCs may be isolated from multiple adult tissues with bone marrow as the most common source. Use of adult tissue-derived MSCs in therapy, however, is burdened with complications including inhomogeneity and senescence (2). Highly proliferative embryonic stem cell-derived MSCs (ESC-MSCs) have been proposed as an alternative source with benefits to availability, biosafety and standardized therapy (3). MSCs may be generated from ESCs or induced pluripotent stem cells (iPSCs) by protocols ranging from co-culture with OP9 cells, selection of mesenchymal-like cells after undirected differentiation, to directed differentiation using small molecular compounds (4–7). Here we improve our earlier published protocol, providing robust and reproducible differentiation (8). At the core of the improved protocol is the efficient initiation of ESC differentiation by bone marrow-derived MSC (BM-MSC)-conditioned medium. Despite the strong interest in MSCs over the last decade, their developmental origin remains unclear. Mesoderm commitment and differentiation is considered the major source of MSCs, and of the three main mesodermal branches, hematoendothelial (CD34+/KDR+/PDGFRα dim/neg; blood, endothelium), cardiovascular (CD34neg/KDR+/PDGFRα+; endothelium, cardiomyocytes, smooth muscle) and mesenchymal (CD34neg/KDR dim/neg/PDGFRα+/CD73+; fibroblasts, bone, cartilage, fat) (9), the latter is thought to give rise to MSCs.
Several large-scale studies including transcriptome, histone and/or DNA methylation have explored mesoderm commitment and differentiation (10–12). Advances in quantitative mass spectrometry based proteomics and phosphoproteomics have also led to investigation of system-wide ESC pluripotency and differentiation (13–17), lineage commitment, as represented by the differentiation of ESCs to MSCs, has, however, proteomicly been studied only for the neuronal lineage (16).
Using an integrative multi-omics approach combining transcriptome, proteome and phosphoproteome profiling of MSC differentiation from hESCs we here present a comprehensive view of MSC development yielding insights into molecular mechanisms, reveal players in MSC biology and include a phosphoproteomic angle, which lays a foundation to decipher the evolution of molecular events during differentiation. We extracted affected biological functions, signaling pathways, and differentiation potential in developing MSCs and identified important transcription factors, kinases and phosphatases, as well as noncoding transcripts associated with the process.
EXPERIMENTAL PROCEDURES
Cell Culture and Differentiation
Permission to use the human ESC line (hESC) ES04 (WiCell institute, Madison, WI) was obtained from the Cornell/Rockefeller/Sloan Kettering tri-institutional ESC research oversight committee. Funding was secured from nonfederal, US-external funding sources. ES04 were expanded feeder-free in mTeSR1 medium (Stem Cell Technologies, Vancouver, Canada) supplemented with penicillin/streptomycin on growth factor-reduced matrigel (Stem Cell Technologies). Cells were passaged 1:6 at 80% confluence using 1 mg/ml dispase. Medium was changed daily. Differentiation into MSCs was initiated at 10% confluence using conditioned MesenCult medium (Stem Cell Technologies) mixed 1:1 with unconditioned MesenCult medium. On day 6 of differentiation cells were passaged to noncoated flasks 1:3 using accutase, with an additional passage on day 12. All following passages were performed with 0.05% EDTA-trypsin. Differentiation medium was replaced every 3 days until day 18. Starting from differentiation day 19 normal MesenCult medium was used. Fully differentiated ESC-derived MSCs were obtained on day 30. Based on microscopic observation and FACS data of MSC markers the purity of ESC-derived MSCs from day 30 was around 90–95% and comparable to BM-MSCs. Bone marrow-derived MSCs (BM-MSCs) were expanded in MesenCult medium supplemented with penicillin/streptomycin. BM-MSCs were passaged 1:3 at 80% confluence using 0.05% trypsin with EDTA-trypsin. Medium was collected after 3 days as conditioned medium for ESC to MSC differentiation. Conditioned medium was cleared from cellular debris by centrifugation (300 × g, 10 min). BM-MSCs were purchased (Lonza, Basel, Switzerland, StemCell technologies) with the following donor details: 40y/m (MSC-001F, lot#BM2893), 39/m (PT2505, lot#1F3422), 27y/m (PT2505, lot#318006). To study the differentiation samples were harvested at 8h, after 1d, 2d, 5d, 15d and 30d. Undifferentiated ESCs and ESC-MSCs were harvested with dispase until day 2, with accutase until day 15 and at day 30 with trypsin according to the sampling schedule (Fig. 1). One limitation of the study is the use of a single hESC line for the ESC to MSC differentiation experiments.
Fig. 1.
ESC to MSC differentiation. A, Schematic of the differentiation protocol with sampling schedule for the different techniques. B, Summary of omics technologies and their analytical depth to profile the differentiation from ESCs to MSCs. C, Expression profiles of selected markers for ESCs, MSCs, and (D) mesoderm, as well as (E) RNA transcripts upregulated during differentiation from 8 h onwards.
Experimental Design and Statistical Rationale
All experiments were performed independently in biological triplicates. Differentiation was replicated using ESCs from different passages. For each differentiation experiment conditioned medium was obtained from independent BM-MSC donors (purchased from Lonza, Stem Cell technologies), also included per experiment as end point controls. Samples for transcriptome, proteome and phosphoproteome analysis were obtained according to the harvesting schedule in Fig. 1. Differential expression analyses for all three omics approaches were performed with the limma R package using the empirical Bayes moderated t test after ensuring normal distribution. For each time point all biological triplicates (n = 3) were included in the statistical analysis. Features with a FDR-corrected (Benjamini-Hochberg) p value < 0.05 were regarded as differentially expressed. Features with less than 50% completeness were excluded from the analysis.
Flow Cytometry
Cells were labeled with fluorescence-conjugated antibodies against CD73, CD105, and CD45 (BD Biosciences, San Jose, CA). Matching isotype controls were used as staining controls. Samples were measured on FACSAria II and Fortessa (BD Biosciences) with 10,000 events per acquisition. Data was analyzed with FACSDiva v6.3 (BD Biosciences).
Functional Differentiation for MSC Phenotyping
ESC-MSCs were differentiated into adipocytes, osteocytes and chondrocytes as described elsewhere (18) to prove a MSC phenotype. BM-MSCs were included as positive control. Cells were seeded in 6-well plates at a density of 50,000/cm2. For chondrogenic differentiation cells were seeded as pellets (0.5 × 106 cells), forming spheroids within 24 h of differentiation. Differentiation medium was exchanged every 3 days. Adipogenic differentiation: 1 μm dexamethasone, 500 μm IBMX, 100 μm indomethacin, 10 μg/ml insulin. Osteogenic differentiation: 100 nm dexamethasone, 50 μm l-ascorbic acid 2-phosphate and 10 mm glycerol 2-phosphate. Chondrogenic differentiation: 50 μm l-ascorbic acid 2-phosphate, 6.25 μg/ml insulin and 10 ng/ml TGFβ1. After 2–3 weeks of differentiation cells were stained with Oil Red O, Alizarin Red or Alcian Blue for evaluation of an adipogeneic, osteogenic or chondrogenic phenotype, respectively. Negative controls were either undifferentiated cells (adipocytes, osteocytes) or spheroids at day 7 (chondrocytes).
Transcriptome: Next Generation RNA Sequencing
Sample Preparation for RNA-seq
Following RNA isolation with TRIZOL, 10 ng of total RNA was used to generate RNA-seq libraries using the Ovation Single Cell RNA-Seq System (Nugen Technologies, Inc., San Carlos, CA) following the manufacturer's protocol. The final libraries were quantified using the Agilent Bioanalyzer High Sensitivity DNA (Agilent, Santa Clara, CA) assay then pooled using 8 libraries per pool. Paired-end 100 bp sequencing was carried out on the Illumina HiSeq 2500 (Illumina, Inc., San Diego, CA).
Data Analysis for RNA-seq
Quality check was performed for the raw reads with FastQC (version 0.10.1). The 100 bp paired reads were mapped to the human reference genome from GENCODE built GRCh38 patch release 25 with STAR (v 2.5.1a) using Ensembl 87 gene annotation. Aligned reads were quantified with featureCounts function from Rsubread (v 1.24.2), Bioconductor Package in R (v 3.3.3). All the read counts from conditions were combined into a data matrix based on gene identifiers. Differential expression was based on RNA count data after voom (20) and inverse normalization using empirical Bayes moderated t test (21) within the autonomics package (manuscript in preparation; https://github.com/bhagwataditya/autonomics).
Proteome and Phosphoproteome
Protein Preparation, Reduction, Alkylation, Digestion, Labeling
Cells were lysed in 2% SDS buffer containing 30 mm Tris, pH 8.5, protease inhibitors (Roche, Basel, Switzerland), phosphatase inhibitors (Roche), and benzonase (Sigma, 2 μl per 1 ml). After methanol/chloroform precipitation samples were resuspended in urea/thiourea buffer (6 m/2 m, 30 mm HEPES, pH 8). Protein concentration was determined by Bradford. After reduction (1 mm DTT, 30 min) and alkylation (5 mm IAA, 20 min, in the dark), proteins were digested at a protein to enzyme ratio of 100:1 first by endopeptidase Lys-C (Wako Chemicals, Richmond, VA) for 3 h and then by trypsin overnight after a 1:4 dilution with 10 mm tetraethylammonium bromide, pH 8. Peptides (200 μg) were labeled by reductive dimethylation. Time points were labeled as “medium” or “heavy,” a combined standard representing a mixture of all samples as “light.” Dimethyl labeled samples were combined as triplex and desalted on R3 columns. Labeled eluted peptides were split into equal aliquots for proteome and phosphoproteome analysis with 300 μg total peptides each.
Peptide Fractionation by In-solution Isoelectric Focusing
Peptides for proteome analysis were separated by in-solution isoelectric focusing (OFFGEL fractionator, Agilent) into 12 fractions within the pH range 3–10 according to manufacturer's instructions. The ampholyte and glycerol concentration were reduced to 0.1% and 0.3%, respectively (23). Peptides were harvested after 20 kVh focusing with an additional well wash of 50:49:1 methanol/dH2O/TFA. Peptides were desalted on reversed phase (RP) C18 STAGE Tips.
Phosphopeptide Enrichment
Phosphopeptides were enriched according to the TiSH protocol (24), a combination of TiO2 and sequential elution from IMAC enrichment combined with HILIC fractionation. Briefly, 300 μg dimethyl-labeled peptides were equilibrated in 1 ml loading buffer (80% acetonitrile, 5% TFA, 1 m glycolic acid) and phosphopeptides were enriched twice with 1.8 and 0.9 mg TiO2-beads. Phosphopeptides were eluted from the beads with ammonia solution (pH 11.3) and cleaned on R3 column. Phosphopeptides were evaporated to dryness, reconstituted in 50% acetonitrile, 2% TFA (pH ≤ 0.83) and added to washed IMAC beads. After 30 min low speed vortexing at RT unbound phosphopeptides were combined with eluates at 50 and 20% acetonitrile in 2% TFA. These monophosphorylated peptides were subjected to a second TiO2 enrichment using 1.8 mg and 0.9 mg beads. Multi-phosphorylated peptides were eluted from the IMAC beads with ammonia solution (pH 11.3). Mono- and multiphosphorylated peptides were cleaned on R3 columns before nanoLC-MS/MS analysis. Mono-phosphorylated peptides were further fractionated by HILIC (see below).
Hydrophilic Interaction Liquid Chromatography (HILIC) Fractionation
Mono-phosphorylated peptides were reconstituted in 0.45 μl 10% TFA and 3.5 μl H2O and 40 μl 100% acetonitrile were added. Samples were loaded onto an in-house packed TSKgel Amide 80 HILIC 320 μm × 170 mm capillary HPLC column using an Agilent 1260 HPLC system. Mono-phosphorylated peptides were eluted over a 48 min gradient starting from 90% acetonitrile, 0.1% TFA to 0% acetonitrile, 0.1% TFA. Fractions were collected automatically in one-minute intervals in a 96-well plate. According to the UV detection fractions were combined into 16 final fractions. Before nanoLC-MS/MS analysis, samples were dried by vacuum centrifugation and reconstituted in 0.5 μl of 100% formic acid followed by 4.5 μl H2O.
Mass Spectrometry
Peptide samples were analyzed by liquid chromatography (LC) using an EASY nLC-II coupled to a Q Exactive mass spectrometer (Thermo Scientific, Bremen, Germany) as previously described (25). Briefly, peptides were separated on 120 min gradients and mass spectra were acquired in data dependent mode (Top10), using higher energy collisional dissociation (HCD) (normalized collision energy 25) for fragmentation. Precursor scans (MS1 level) were acquired at a resolution of 70,000 (m/z 300) and an AGC target value of 3,000,000 charges (maximum ion injection time 20 ms). Fragmentation spectra were acquired at a resolution of 17,500 (m/z 300) and an AGC target value of 100,000 charges (maximum ion injection time 120 ms). All scan events were recorded in profile mode. A dynamic exclusion list of 25 s was employed and the exclude isotopes functionality was activated.
Data Analysis for Proteome and Phosphoproteome
Mass spectrometric raw data was processed and analyzed using MaxQuant v.1.5.0.0 combining proteome and phosphoproteome measurements. In total, 319 MS runs were included with 29 fractions per experiment (proteome: 12× IEF, phosphoproteome: 16× HILIC - monophosphorylated peptides, 1× multiple phosphorylated peptides). The following default search parameters were employed: enzyme specificity trypsin, maximum of 2 missed cleavages, first search mass accuracy tolerance 20 ppm, main search mass accuracy tolerance 4.5 ppm, FTMS MS/MS tolerance 20 ppm, minimum peptide length of 7 amino acids, peptide spectrum match FDR and protein FDR both set to 0.01 as calculated by the revert database approach. Protein quantification was based on a minimum of two ratio counts, originating from unique or razor peptides only. Additionally, unless explicitly stated otherwise, other parameters of the data analysis were not changed from their MaxQuant 1.5.0.0 default value. Search was performed with the match between run and re-quantify options set as TRUE. As fixed modifications cysteine carbamidomethylation was selected, as variable modifications: (STY) phosphorylation, asparagine deamidation and methionine oxidation. Samples were searched as triplex (multiplicity 3) with the corresponding dimethyl labels selected (light: DimethLys0, DimethNterm0, medium: DimethLys4, DimethNterm4, heavy: DimethLys8, DimethNterm8). Andromeda searches were performed against Uniprot Homo sapiens proteome database (canonical including isoforms) downloaded Aug 2014 (68,382 entries). Differential expression was performed on log-transformed normalized ratios extracted from the MaxQuant proteinGroups.txt and Phosho(STY)Sites.txt tables using empirical Bayes moderated t test from limma (21), implemented in the in-house built autonomics R package (manuscript in preparation; https://github.com/bhagwataditya/autonomics).
Bioinformatics
Linear Kinase Motif Analysis
Centered phosphomotif sequences were uploaded to PhosphoSitePlus to test for significantly over-represented motifs and to generate sequence logos. Upstream kinases were predicted by the NetworKIN algorithm implemented in the KSEAapp package (26). To generate a combined heatmap for all sample groups the NetworKIN cutoff was set to 5, with a minimum of 10 members per kinase and p value < 0.05.
Network Generation
The Phosphopath app (27) within Cytoscape v3.3 was used to visualize phosphoprofiling data in combination with the proteome for selected enriched pathways. Protein-protein interaction networks for clustered features were generated by STRING to determine hub proteins.
Enrichment Analysis
Enrichment analysis was performed with DAVID implemented in the in-house built autonomics R package (manuscript in preparation; https://github.com/bhagwataditya/autonomics).
Clustering
Unsupervised clustering analysis (fuzzy c-means) on differentially expressed features (early time points: p value < 0.01, otherwise: FDR < 0.05) was performed using the Mfuzz R package (28).
RESULTS
Phenotyping of ESC-derived MSCs
During the differentiation of ECSs into MSCs (supplemental Fig. S1A), two classical MSC markers (CD73, CD105) were longitudinally monitored by FACS analysis (supplemental Fig. S1B). At the final stage of differentiation, both show levels comparable to BM-MSCs. As expected, the hematopoietic marker CD45 was absent at all time points. Additionally, ESC-derived MSCs (day 30, D30) displayed tri-lineage differentiation potential (adipocytes, osteocytes, and chondrocytes) equivalent to BM-MSCs (supplemental Fig. S1C), thus satisfying the minimal criteria for defining multipotent MSCs (31).
Multi-omics Analysis: RNA-seq, Proteomics, Phosphoproteomics
Principal Component Analysis (PCA)
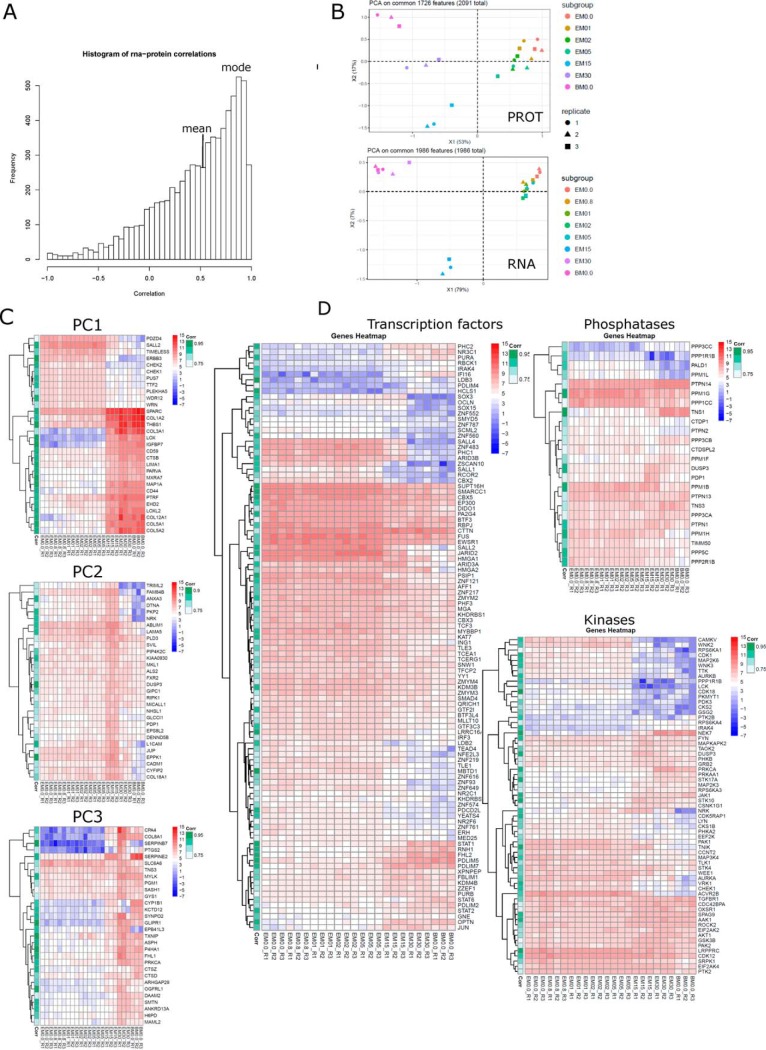
PCA analyzes of data from all approaches employed reflect the dynamics of ESC to MSC differentiation starting from closely grouping early stages, via the intermediate (D15) to the final ESC-MSC stage (D30) (supplemental Fig. S2A). For RNA-seq, fully differentiated ESC-MSCs and BM-MSCs are closely related. Day 15 ESC-MSCs represent a distinct state, equally distant to undifferentiated ESCs and fully-differentiated ESC-MSCs. This pattern was observed independent of RNA classes (protein-coding, lncRNA and antisense RNA) included in supplemental Fig. S2B. PCA for proteomics and phosphoproteomics is affected by the grouping inherent to the quantitation approach employed.
ESC, MSC, and Mesoderm Markers
The expression of classical ESC and MSC markers by RNA-seq and proteomics followed the expected pattern and further validated the differentiation protocol (Fig. 1C). Known mesoderm markers such as Brachyury (T) and EOMES peaked together with BMP2, HAND1 and TBX20 at the intermediate differentiation stage (D15) whereas other mesoderm markers such as FOXC2, GATA6, PDGFRA, PRRX1, SNAI1, and TWIST1 plateaued during differentiation. With the observed downregulation of KDR, the ESC-MSCs match the criteria for the mesenchymal branch of mesoderm commitment (CD34neg/KDR dim/neg/PDGFRα+/CD73+) (9) (Fig. 1D, supplemental Fig. S1D). Based on RNA-seq analysis, several markers showed persistent expression as early as 8h after induction of differentiation. Functionally, these are related to focal adhesion and differentiation. Among them are recognized MSC markers (PLAU, CD44), the serine/threonine kinase (PLK2) and a multifunctional scaffold protein with a role in early ESC fate determination (AHNAK) (75) (Fig. 1E).
Analytical Depth and Differential Expression Analysis
RNA-seq analysis detected an average of 95,038 transcripts, covering 17,665 genes (min: 11,800/max: 21,565). Of these 10,847 were present globally with read counts, 10,264 of which were annotated as protein-coding. Proteome profiling led to the identification of 9,470 proteins (supplemental Information SI 1), 7864 of which were quantified. Supplemental Fig. S3A summarizes their distribution across the dataset. A total of 4325 proteins were quantified in all samples and stages. We additionally identified 13,826 phosphosites (supplemental Information SI 2) mapping to 3621 phosphoproteins. Of the phosphosites 8390 had a localization probability of >0.75 (class I; 3,211 proteins) and yielded a STY distribution of 92:8:0. Conservatively, statistical analysis was performed on quantified class I sites following proteome normalization, which reduced the data set to 6204 sites (2398 proteins) (supplemental Information SI 3). The distribution of quantified class I phosphosites across the data set is shown in supplemental Fig. S3A with 1369 class I phosphosites quantified in all samples at all stages. Coverage and overlap between the three omics techniques is shown in supplemental Fig. S3B with 2269 features being quantified at transcriptome, proteome and phosphoproteome levels. Differentially expressed features (FDR <0.05) per technique and sample group are summarized in Fig. 2A based on comparisons to undifferentiated ESCs (D0). Examples of features continuously upregulated during differentiation are given in Fig. 1E, with AHNAK discussed in more detail below. Complete lists of differentially expressed features (FDR < 0.05) per technique are provided (supplemental Information SI 4–9).
Fig. 2.
Differential expression analysis. A, Differentially expressed features for each technique. B, Enrichment analysis on differentially expressed features (FDR < 0.05) of RNA, PROT and PHOS. Tile plots are shown for selected enriched GOBP terms. C, Expression profiles of selected lncRNA and antisense RNA transcripts.
Functional Enrichment, Differentiation and Signaling Pathways
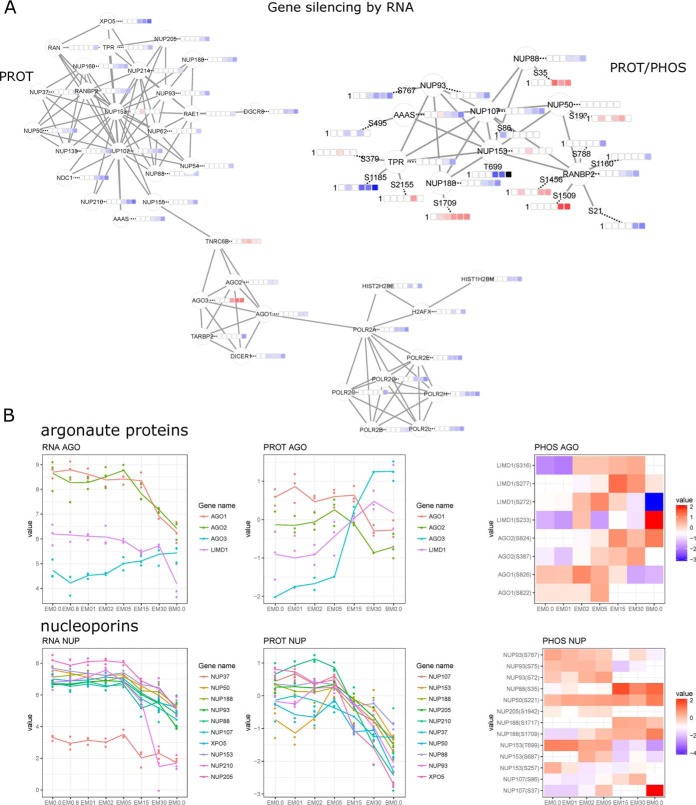
For downregulated genes, significant enrichment was observed for gene expression-related processes (Fig. 2B). Parallel to a general massive transcriptional downregulation, the observed protein interaction network for gene silencing by RNA (Fig. 3A) suggests a switch in argonaute isoforms, which as core components of the microRNA-induced silencing complex (miRISC) guide miRNAs to their targets. AGO3 appears to replace AGO1 and AGO2 during differentiation. Although originally AGO2 alone was believed to possess miRISC slicer activity, this has recently also been demonstrated for AGO3 in the context of specific miRNAs (32). AKT3-mediated phosphorylation on AGO2 S387 is necessary for miRISC assembly and interaction with the LIMD1 subunit (32). AGO3 can replace AGO2 independent of AKT3, recruiting LIMD1 proteins (LIMD1, WTIP, AJUBA). An AGO2 to AGO3 switch has been described in vitro following LIMD1 ablation (33). Our phosphorylation data supports this shift in the machinery (Fig. 3B) for both argonaute and LIM proteins, exemplified by increased phosphorylation from day 2 onwards on several positions on LIMD1 (S233, S272, S277, S316) and from day 5 on AGO2 (S387, S824). Although the shift from AGO2 to AGO3 during ESC to MSC differentiation is clear based on protein expression, the phosphorylation data suggests a complex regulatory interplay, that warrants further investigation.
Fig. 3.
Gene silencing by RNA during ESC to MSC differentiation. A, Protein-protein interaction network for proteome and phosphoproteome data. B, Expression (RNA/PROT) and phosphorylation profiles (PHOS) for argonaute proteins and nucleoporins.
Features upregulated during differentiation, display an enrichment for the terms vesicle-mediated transport, cytoskeleton organization, focal adhesion, extracellular matrix organization, development, differentiation and cell signaling (Fig. 2B). Filtering for developmental and differentiation terms reproduces our reported observation that MSCs express a plethora of tissue developmental markers (25), most notably for cardiovascular system, epithelium and neuron development (supplemental Fig. S2C).
Cell signaling pathways affected by differentiation were examined in detail (supplemental Fig. S2D). The enrichment of MSC signaling pathways such as integrin, EGF, VEGF, and TGFβ signaling further confirms differentiation. Among the affected signaling-related pathways were tyrosine and serine/threonine kinase signaling, which together with enriched MAPK and PKB signaling justifies our approach studying the global phosphoproteome during ESC to MSC differentiation.
LncRNAs and Antisense
Long noncoding (lnc) RNAs are known to play a crucial role in both maintenance of pluripotency and differentiation of ESCs (34, 35). During ESC to MSC differentiation 433 lncRNAs and 411 antisense transcripts were differentially expressed (FDR < 0.05) (supplemental Information SI 9, 10). Functional annotation is available for a few, but of these, CARMN, MALAT, NEAT1 and LINC00152 were among the most prominent induced lncRNAs. CARMN is associated with cardiomyocyte differentiation (36), MALAT1 with osteoblast differentiation (37), NEAT1 with adipogenesis (38) and LINC00152 with epithelial-mesenchymal transition (39). Commonly upregulated from D1 onward was lncRNA MIR4435–2HG, sequence homolog to LINC00152 (40). HOTAIR and HOTAIRM1 were among the most induced antisense transcripts during differentiation, both located in the HOX gene cluster. HOTAIR has been described as an important player in Snail-mediated epithelial-mesenchymal transition (41) and HOTAIRM1 has been linked to neurogenesis (42) (Fig. 2C). The antisense RNA MIAT found to be downregulated during differentiation, has been described as a suppressor of osteogenic differentiation in stem cells (43). LINC-ROR, also downregulated during differentiation, is involved in ESC self-renewal by trapping miRNA145, which targets SOX2, NANOG and POU5F1 transcripts (44). Besides these annotated lncRNAs observed as changing during differentiation and reflecting the loss of pluripotency and the gain of MSC specific features, many other lncRNA candidates were found to be regulated and warrant future evaluation.
Clustering - Identification of MSC-defining Transcription Factors
Differentially expressed features were grouped using unsupervised fuzzy c-means clustering (Fig. 4). PCA of the clusters for all techniques separates up and downregulated features (Fig. 4A). Clusters of interest are those with intense expression changes (red - increase, blue - decrease) or displaying peak expression on D15 (green). (Fig. 4A, 4B). Enrichment analysis was performed on them and protein interaction networks were generated. Hub proteins as well as the most enriched terms are displayed together with the expression profiles of transcription factors per cluster in Fig. 4C. For RNA-seq, blood vessel development and skeletal system development are top induced terms with the transcription factors FOXC1, FOXC2, FOXD1, FOXP2, NR2F1, NR2F2, PITX1, PITX2, RUNX1, RUNX2, SIX1, SIX2, and TWIST1 most prominently increased during differentiation. Although all are related to cell development, only a subset (RUNX1, RUNX2, TWIST1) are reported to be important in MSC self-renewal, mesodermal tissue development and lineage commitment (45–47). Transcription factors with a prominent decrease over the time course include the pluripotency factors NANOG, SOX2 and ZSCAN10, as well as ARID3B, RCOR2, SALL4 and ZNF483, which were also identified by proteome profiling as strongly downregulated. Several transcription factors, many of which belong to the homeobox gene family, known to be master regulators of pattern specification and tissue development, display distinct peak expression at D15 and include HOXB2, HOXB4, HOXB5, HOXB6, HOXC8, HOXC10, and HOXD10. Others with a distinct expression peak are the mesoderm markers EOMES and Brachyury (T). Differentiation-associated HOX transcription factors are only found in three clusters, namely cluster 20, 7, and 31. Specifically clusters 7 and 31 contain transcripts with peak expression at intermediate differentiation stage D15, whereas cluster 20 contains transcripts with a prominent induction during differentiation.
Fig. 4.
Fuzzy c-means clustering on differentially expressed features for each technique. Data was partitioned in either 32 (RNA) or 16 clusters (PROT, PHOS). A, PCA of clusters for each technique. Clusters with exceptional high increase (red), decrease (blue) or peak expression at intermediate differentiation stage (D15) (green) are highlighted. B, Expression profiles of selected RNA and PROT clusters. C, Selected clusters were filtered for transcription factors. D, Quantified and differentially expressed (DEGs) features per technique were filtered for transcription factors (TF), kinases (KIN) and phosphatases (PPT).
Likely because of dynamic range limitations of the technology in combination with low protein abundance, HOX proteins were not covered by proteome profiling. Most prominent induced transcription factors found by proteomics include the PDZ and LIM domain containing proteins PDLIM2, PDLIM5, and PDLIM7, as well as RBCK1, FHL2, NR3C1 and PURA.
From this plethora of transcription factors involved in differentiation (RNA: 777, PROT: 185, PHOS: 46) (Fig. 4D) we selected sentinels with extreme expression based on clustering results as defining the MSC phenotype.
RNA to PROT Correlation
Because RNA expression is not always a proxy for protein expression especially during embryonic development (48), we were interested in gene-wise correlation profiles between both techniques across time points (supplemental Information SI 10). The distribution for 6903 intersecting features is depicted in Fig. 5A with average correlation of 0.5 and a mode at 0.9 demonstrating better correlation than previously published for somatic cells (0.4) (49)or differentiating cells (0.2) (48), but in line with our previous study (50). PCA analysis was performed on differentially expressed features, which display good correlation (R > 0.7 and FDR < 0.05) (Fig. 5B). Proteomics data now also shows the distinct intermediate differentiation stage at day 15. The top 30 genes for the first 3 components are shown as heatmaps with correlation indicated (Fig. 5C). Genes which best distinguish ESCs (D0–5) from MSCs (D15–30, BM-MSCs) are described by principal component 1 and include e.g. SALL2, TIMELESS, ERBB3, CHEK1, and CHEK2 for ESCs; COL1A1, COL3A1, COL5A1, COL5A2, COL12A1, CD44, CD59, LOX, IGFBP7, and MAP1A for MSCs. Genes with expression peaking at the intermediate or late stage of differentiation are represented by principal component 2 and 3, respectively. We regard the set of 1,986 differentially expressed transcripts (RNA-seq) which display good correlation to the proteome (R> 0.7 and FDR <0.05) as high confidence genes involved in ESC to MSC differentiation. This gene set includes 105 transcription factors, 65 kinases and 24 phosphatases (Fig. 5D). Among upregulated transcription factors during differentiation are PDLIM proteins (PDLIM2, 4, 5, 7), PHC2, NR3C1, and PURA. The PDLIM proteins, although not yet associated with MSC physiology, are implicated in MSC related differentiation processes such as osteogenesis and cardiomyogenesis (51, 52).
Fig. 5.
RNA-PROT correlation. A, Distribution of RNA-PROT correlation with mean and mode indicated. B, PCA on features with R > 0.7 and FDR < 0.05 for PROT and RNA. C, Hierarchical clustering of top30 features per principal components 1–3. D, Hierarchical clustering of transcription factors, kinases and phosphatases, which show good RNA-PROT correlation (R > 0.7, FDR < 0.05). RNA transcripts are shown with RNA-PROT correlation indicated on the left. Legends for expression intensity and correlation are on the right.
To summarize, around 2,000 differentially expressed features are well correlated between RNA to protein (R > 0.7 and FDR < 0.05), allowing to cross-validate findings and focus on MSC specific signature gene products.
Phosphoproteome Profiling: Differentiation/Pluripotency and Nuclear Pore
Comparing our data set to PhosphoSitePlus (as of 08.2017, 234,166 entries for H. sapiens), 301 phosphosites are novel when amino acid position is extracted from canonical protein forms. Novel phosphosites were e.g. found for the transcription factors DACH1 (S392), FOXH1 (S16), NLRP10 (S585), SALL2 (T467), SALL3 (S917), SALL4 (S126, S129, S512), and TCEAL2 (S114). Knowledge about the human MSC phosphoproteome is limited to just 700 sites reported in PhosphoSitePlus. When comparing our data set with reported sites for hESCs and hMSCs, 5,157 phosphosites are novel (Fig. 6A). Among the proteins most extensively phosphorylated (phosphosites per protein) in our data (Fig. 6B) were proteins with the strongest induction of expression during differentiation (AHNAK, VIM, MAP1A, MAP1B) (Fig. 6B, 6C) and extensive differential phosphorylation (AHNAK, VIM, SRRM2, MAP1A, MAP1B) (Fig. 6, supplemental Fig. S3C). When ESCs differentiate and loose pluripotency, massive changes are observed for cell cycle, gene expression and RNA processing (both coding and noncoding). At early differentiation (D1) JARID2, a member of the PRC2 complex essential for histone methyltransferase recruitment and initiating ESC differentiation (53) becomes phosphorylated (S124). This observation is paralleled by a decrease of JARID2 RNA expression over time, as well as of EZH2, a critical subunit of the PRC2 complex (supplemental Fig. S4A). EZH2 decrease has been shown to be specific to mesoderm commitment by reducing H3K27me3 (54). AHNAK, mentioned above as a transcript strongly and stably induced from as early as 8 h of differentiation, is the top phosphorylated protein at D1. It is one of the most phosphorylated proteins in homo (source: PhosphoSitePlus) as well as in our data with strong phosphorylation changes on multiple sites (Fig. 6D). The transcriptional co-repressor TRIM28 is similarly phosphorylated starting with an early differentiation stage (D2: S19, S473, D5: S473, T544), followed by decreased expression at intermediate differentiation stages, which supports recent findings that TRIM28 expression is indispensable to pluripotency and repression of differentiation-inducible genes (55). Also strongly affected by phosphorylation changes from early differentiation on are proteins involved in RNA and transcription factor transport (e.g. nucleoporins NUP188, NUP50, NUP88) (Fig. 3A, 3B), RNA splicing (e.g. SRFS1, SRFS6, SRFS7, SRFS9, SRFS10, SRFS11, SRRM1) and gene expression in general with a vast majority of proteins associated with RNA-binding activity. Nuclear pore complexes consist of >30 nucleoporins (NUPs) with a tissue-specific composition. NUPs have recently been discussed as scaffolds regulating developmental gene expression (56). Together with the massive downregulation of specific NUPs during ESC to MSC differentiation described above, we observed bidirectional phosphorylation changes (e.g. increase in NUP50 S192, NUP88 S35, NUP188 S1709, decrease in NUP153 T699) (Fig. 3A, 3B), hinting at a more complex regulatory picture.
Fig. 6.
Phosphoproteome profiling - phosphomotif analysis. A, Class I phosphosites in our data set (PHOS) compared with the whole human PhosphoSitePlus database (PPSP) or entries for hESCs (PPSP_ESC) and hMSCs (PPSP_MSC). B, Top10 phosphoproteins with the most phosphosites per protein. Half of them displayed intense phosphorylation during differentiation (*). C, Expression profiles for top phosphoproteins. D, Differential phosphorylation for AHNAK. E, Counts of enriched phosphomotifs with reduced (left) or induced (right) phosphorylation during ESC to MSC differentiation. ESC or MSC specific phosphomotifs common for D15, D30, BM. F, Combined sequence logo for phosphomotifs with reduced phosphorylation (ESCs) or increased phosphorylation (MSCs) during differentiation.
Phosphorylation Motif Analysis
Differentially phosphorylated peptide sequences were subjected to motif analysis. Phosphomotif counts per time point are shown in Fig. 6E, separately for increased and decreased phosphorylation. Among the 37 motifs thus defined as specific for MSCs were most prominently sXXXXXE, sXD, sXE, and RXXsXS, whereas ESC-specific motifs were sXXXXXK, sXXK, sPXK, and sPS. As compared with the PhosphoSitePlus compendium of experimentally identified phospohosites, the most prominently enriched motive in ESCs was sPXK recognized by CDKs (CDK1,2,5), reflecting the proliferative nature of the cell type. ESC-specific motifs tended to basic c-termini, enriched in K and R (Fig. 6E, 6F). MSC-assigned motifs where mostly of an acidic nature c-terminal to the phosphorylated amino acid with high abundance of D and E, possibly recognized by the acidophilic kinase PLK2 (57), found to be upregulated by RNA-seq from 8 h onward.
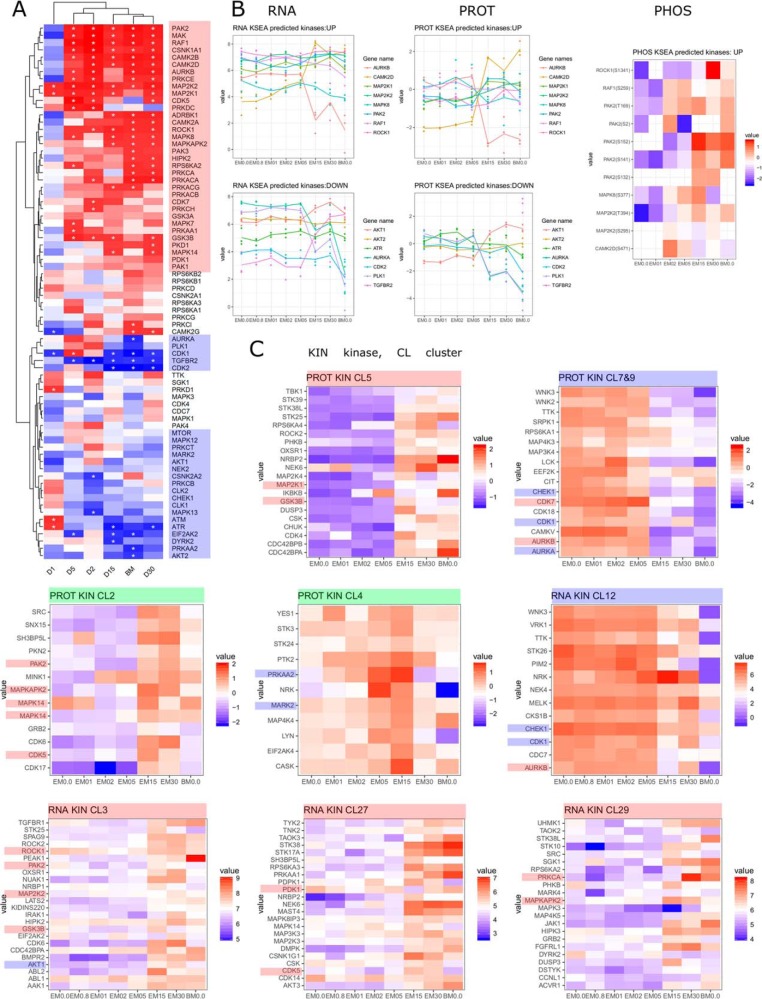
Predicted Upstream Kinases by KSEA
Based on differentially occupied phosphosites upstream kinases were predicted using KSEA (58); Fig. 7A). Hierarchical clustering of the predicted kinases reflects the time course of the differentiation. Expression profiles for these kinases (Fig. 7B) do not necessarily correlate to those changes, indicating abundance-independent activity modulation, exemplified by PAK2, the top predicted kinase. Although only moderately increased proteomically, phosphoproteome data revealed increased phosphorylation on several phosphosites (Fig. 7B), including S141, an autophosphorylation site known to be important for kinase activity (59). PAK2 is part of the TGFβ induced Raf/MEK/Erk pathway regulating SMAD2/SMAD3 signaling (60), known to act in MSC signaling and differentiation. RAF1, another top predicted kinase showed stable expression at RNA and protein level, but elevated phosphorylation of S259, important in signal transduction specificity, but decreasing kinase activity (61). Based on fuzzy c-means clustering, kinases with expression patterns changing during differentiation were identified (Fig. 7C). Several among them reflected the KSEA prediction (up/active/MSC specific: GSK3B, MAP2K1, MAP2K2, MAPK14, MAPKAP2K, PAK2, ROCK1; down/inactive/ESC specific: AUKA, CDK1, CHEK1). Kinases with prominent expression changes include e.g. upregulated NEK6, NEK7, STK17A, STK25, and STK38 (MSC specific), and downregulated CAMKV, CDK18, LCK, WNK2, and WNK3 (ESC specific).
Fig. 7.
Kinases during differentiation. A, Upstream kinase prediction by KSEA. B, Expression profiles of KSEA predicted kinases at RNA, protein and phosphoprotein level. C, Expression profiles of selected clusters filtered for kinases which show distinct induction (red), reduction (blue) or peak expression (green). Predicted kinases are highlighted according to their predicted activity.
DISCUSSION
In this integrative multi-omics study we present a detailed picture of the differentiation of hESCs into MSCs, covering differential expression of protein coding and noncoding ORFs as well as cell signaling events by examining the transcriptome, proteome and phosphoproteome over a 30-day time course. From a bird's eye view, focal adhesion, cytoskeleton organization and extracellular matrix formation are the earliest and most prominent biological terms induced during MSC differentiation, paralleled by a downregulation in DNA replication, RNA splicing, processing and chromosome organization.
Mesoderm markers such as Brachyury, EOMES, and BMP2 are peaking at an intermediate stage, verifying mesoderm lineage commitment. We also observed downregulation of epigenetic regulators including the PRC2 component EZH2, which has been found to promote mesoderm development specifically by reducing H3K27me3 (54). In a recent paper by the Weissman group, a roadmap of 12 mesodermal lineages was provided with detailed expression analysis from single cell RNA-seq (10). Based on mapping our data onto markers from that work, MSCs appear to share lineage characteristics with lateral mesendoderm (PRRX1, HAND1) derived cardiac mesoderm (GARP/LRRC32, NKX2–5). MSCs show high expression of GARP/LRRC32 and transient expression of NKX2–5 (D15 peak). In addition, they express markers of early somites (FOXC2, PDGFRAhigh), also defining paraxial mesoderm. This suggests that MSCs may represent the end point to another branch of the developmental roadmap between lateral and paraxial mesoderm (supplemental Fig. S5).
An early step in ESC to MSC differentiation includes the induction of AHNAK. Over 50 phosphorylation sites were identified for AHNAK, 9 of them differentially phosphorylated with early induction at D1 (S1068, S5735, S5782). AHNAK mediates TGFβ-induced downregulation of SMAD3 target genes such as pluripotency factor c-Myc (62), and accordingly iPSC generation is more efficient from AHNAK−/− MEFs. AHNAK is involved in adipogenesis by SMAD1 activation (63), plays a role in vascular healing and vesicle formation (64). Although not described as being functional important in MSCs, all those features link AHNAK to MSC physiology and lineage commitment. During embryonic development AHNAK is expressed in a tissue-specific manner in migratory mesenchyme and tissues which undergo epithelial mesenchymal transitions (65), placing it as an important protein in developing MSCs, most probably related to focal adhesion, one of the earliest enriched biological functions.
In general, many of the highest induced transcription factors belong to the homeodomain family, which are important in body plan pattern specification, organ development and cell fate (66). Developmental HOX transcription factors (HOXA2, HOXA5, HOXA13, HOXB3, HOXB4, HOXB5, HOXB6, HOXB9, HOXD9, HOXD10, HOXD11) and modulators of cell fate and body patterning cluster with pan-mesodermal markers characterized by transitional expression at an intermediate differentiation stage. Functionally the clusters are related to embryonic pattern and posterior/anterior specification. The concept of transient HOX expression is not new (67) and has been described e.g. for the 5′-end of the HOXA cluster during monocyte to macrophage differentiation (68).
Another group of HOX transcription factors (HOXA9, HOXA10, HOXA11, HOXB2, HOXC4, HOXC6, HOXC8, HOXC9, HOXC10) cluster together with the highest induced transcripts, are functionally related to skeletal system development, with TWIST1 as hub protein and the only transcription factor that to date has been functionally associated with MSC fate (45). Based on the assumption of a tissue specific HOX code (69) and their own HOX profiling, Klein and colleagues recently efficiently initiated the differentiation of iPSCs into vascular wall resident MSCs by a lentiviral induced combination of HOXB7, HOXC6 and HOXC8 (70), demonstrating the impact of HOX genes and partially supporting our findings on HOXC6 and HOXC8. HOX expression in MSCs from different origins has been investigated (63), linking our cells to BM-MSC-like cells. However, our data also indicates HOX expression to be a dynamic process, exemplified by a transient induction at the intermediate stage.
When ESCs loose pluripotency, massive gene repression and silencing follows. Interestingly, we observed a considerable increase in the argonaute protein AGO3, suggesting an AGO2 to AGO3 switch, likely regulated by phosphorylation-dependent recruitment of LIM domain-containing proteins (71).
Among MSC-defining factors are proteins of the PDZ and LIM domain-containing family (PDLIM2, PDLIM4, PDLIM5, PDLIM7). PDLIM proteins are associated with cytoskeleton organization, cell differentiation, organ development and migration (72). PDLIM2 is involved in EMT by regulating transcription factor activity through COP9 signalosome (73), whereas PDLIM5 and PDLIM7 are implicated in heart development (51, 74).
Phosphoproteome data with subsequent KSEA analysis suggested PAK2 and RAF1 as top active kinases during ESC to MSC differentiation supported by kinase autophosphorylation pattern. PAK2 is a potential new player in MSC differentiation and physiology, connecting TGFβ signaling and SMAD activation leading to cell proliferation. Motif analysis clearly showed ESC-specific phosphorylation motifs to be basic, whereas MSC-specific ones were acidic C-terminal to the phosphorylated amino acid. For ESCs this has been known because the cell cycle modulators CDK1 and CDK2, which recognize the sPXK motif, have been reported as the most active kinases in hESCs, being responsible for more than 25% of the ESC phosphoproteome (17). The characteristic found for MSC phosphomotifs is, however, a new finding and may be associated with the kinases found to be induced.
In summary, we present here a time-resolved analysis of the differentiation on hESC to hMSC using a systems-level approach combining transcriptome, proteome, and phosphoproteome profiling. Despite the strong interest in MSCs for stem cell-based therapies, the cell type is underrepresented in omics studies. With this data we provide a rich repository on ESC and MSC biology, specifically adding knowledge to the phosphoproteome of MSCs. We identify important players in the differentiation process, including transcription factors, kinases, phosphatases and noncoding transcripts.
Data Availability
The mass spectrometry proteomics data, including all raw files and data analysis tables produced by MaxQuant, have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (29) with the dataset identifier PXD004652. Individual spectra of the PRIDE-deposited data can be visually interrogated with MS viewer (30) (search key: xjvcljyla8). RNA-seq data have been deposited to the Sequence Read Archive (SRA) with the dataset identifier PRJNA507603. URLs for transcriptomics (https://www.ncbi.nlm.nih.gov/sra/PRJNA507603) and for proteomics/phosphoproteomics (https://www.ebi.ac.uk/pride/archive/projects/PXD004652).
Supplementary Material
Acknowledgments
J.G. and the Proteomics Core at WCM-Q are supported by “Biomedical Research Program” funds at Weill Cornell Medicine - Qatar, a program funded by Qatar Foundation.
Footnotes
* This work was partially supported by a grant from Qatar National Research Fund's National Priority Research Program (4-1267-1-194). The statements made herein are solely the responsibility of the authors. We thank the Flow Cytometry Facility within the Microscopy Core at Weill Cornell Medicine - Qatar for contributing. We also thank the Genomics Core at WCM-Q for carrying out the RNA-seq experiments. Both cores are supported by “Biomedical Research Program” funds at Weill Cornell Medicine - Qatar, a program funded by Qatar Foundation. K.E.-K. was supported by the Lundbeck Foundation (grant number R83-2011-8143). The authors declare no competing interests.
 This article contains supplemental Figures and Information.
This article contains supplemental Figures and Information.
1 The abbreviations used are:
- MSCs
- mesenchymal stem/stromal cells
- ESC
- embryonic stem cell-derived
- iPSCs
- induced pluripotent stem cells
- BM
- Bone marrow-derived
- HILIC
- hydrophilic interaction liquid chromatography.
REFERENCES
- 1. Wei X., Yang X., Han Z., Qu F., Shao L., and Shi Y. (2013) Mesenchymal stem cells: a new trend for cell therapy. Acta Pharmacol. Sin. 34, 747–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wagner W., Bork S., Horn P., Krunic D., Walenda T., Diehlmann A., Benes V., Blake J., Huber F.-X., Eckstein V., Boukamp P., and Ho A. D. (2009) Aging and replicative senescence have related effects on human stem and progenitor cells. PLoS ONE 4, e5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hematti P. (2011) Human embryonic stem cell–derived mesenchymal stromal cells. Transfusion 51, 138S–144S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arpornmaeklong P., Brown S. E., Wang Z., and Krebsbach P. H. (2009) Phenotypic characterization, osteoblastic differentiation, and bone regeneration capacity of human embryonic stem cell–derived mesenchymal stem cells. Stem Cells Dev. 18, 955–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barberi T., Willis L. M., Socci N. D., and Studer L. (2005) Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PLoS Med. 2, e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boyd N. L., Robbins K. R., Dhara S. K., West F. D., and Stice S. L. (2009) Human embryonic stem cell–derived mesoderm-like epithelium transitions to mesenchymal progenitor cells. Tissue Eng. 15, 1897–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Trivedi P., and Hematti P. (2008) Derivation and immunological characterization of mesenchymal stromal cells from human embryonic stem cells. Exp. Hematol. 36, 350–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raynaud C. M., Halabi N., Elliott D. A., Pasquier J., Elefanty A. G., Stanley E. G., and Rafii A. (2013) Human embryonic stem cell derived mesenchymal progenitors express cardiac markers but do not form contractile cardiomyocytes. PLoS ONE 6, e54524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Evseenko D., Zhu Y., Schenke-Layland K., Kuo J., Latour B., Ge S., Scholes J., Dravid G., Li X., MacLellan W. R., and Crooks G. M. (2010) Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc. Natl. Acad. Sci. 107, 13742–13747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loh K. M., Chen A., Koh P. W., Deng T. Z., Sinha R., Tsai J. M., Barkal A. A., Shen K. Y., Jain R., Morganti R. M., Shyh-Chang N., Fernhoff N. B., George B. M., Wernig G., Salomon R. E. A., Chen Z., Vogel H., Epstein J. A., Kundaje A., Talbot W. S., Beachy P. A., Ang L. T., and Weissman I. L. (2016) Mapping the pairwise choices leading from pluripotency to human bone, heart, and other mesoderm cell types. Cell 166, 451–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mendjan S., Mascetti V. L., Ortmann D., Ortiz M., Karjosukarso D. W., Ng Y., Moreau T., and Pedersen R. A. (2014) NANOG and CDX2 Pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell 15, 310–325 [DOI] [PubMed] [Google Scholar]
- 12. Ulfenborg B., Karlsson A., Riveiro M., Améen C., Åkesson, Andersson K. C. X., Sartipy P., and Synnergren J. (2017) A data analysis framework for biomedical big data: Application on mesoderm differentiation of human pluripotent stem cells. PLOS ONE 12, e0179613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Munoz J., Low T. Y., Kok Y. J., Chin A., Frese C. K., Ding V., Choo A., and Heck A. J. R. (2011) The quantitative proteomes of human-induced pluripotent stem cells and embryonic stem cells. Mol. Syst. Biol. 7, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Phanstiel D. H., Brumbaugh J., Wenger C. D., Tian S., Probasco M. D., Bailey D. J., Swaney D. L., Tervo M. A., Bolin J. M., Ruotti V., Stewart R., Thomson J. A., and Coon J. J. (2011) Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat. Methods 8, 821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rigbolt K. T. G., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V., and Blagoev B. (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4, rs3. [DOI] [PubMed] [Google Scholar]
- 16. Singec I., Crain A. M., Hou J., Tobe B. T. D., Talantova M., Winquist A. A., Doctor K. S., Choy J., Huang X., La Monaca E., Horn D. M., Wolf D. A., Lipton S. A., Gutierrez G. J., Brill L. M., and Snyder E. Y. (2016) Quantitative analysis of human pluripotency and neural specification by in-depth (phospho)proteomic profiling. Stem Cell Rep. 7, 527–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Van Hoof D., Muñoz J., Braam S. R., Pinkse M. W. H., Linding R., Heck A. J. R., Mummery C. L., and Krijgsveld J. (2009) Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5, 214–226 [DOI] [PubMed] [Google Scholar]
- 18. Jääger K., Islam S., Zajac P., Linnarsson S., and Neuman T. (2012) RNA-Seq analysis reveals different dynamics of differentiation of human dermis- and adipose-derived stromal stem cells. PLoS ONE 7, e38833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S. L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Law C. W., Chen Y., Shi W., and Smyth G. K. (2014) voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., and Smyth G. K. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., and Pachter L. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Billing A. M., Ben Hamidane H., and Graumann J. (2015) Quantitative proteomic approaches in mouse: stable isotope incorporation by metabolic (SILAC) or chemical labeling (reductive dimethylation) combined with high-resolution mass spectrometry. Curr. Protoc. Mouse Biol. 5, 1–20 [DOI] [PubMed] [Google Scholar]
- 24. Engholm-Keller K., Birck P., Størling J., Pociot F., Mandrup-Poulsen T., and Larsen M. R. (2012) TiSH–a robust and sensitive global phosphoproteomics strategy employing a combination of TiO2, SIMAC, and HILIC. J. Proteomics 75, 5749–5761 [DOI] [PubMed] [Google Scholar]
- 25. Billing A. M., Ben Hamidane H., Dib S. S., Cotton R. J., Bhagwat A. M., Kumar P., Hayat S., Yousri N. A., Goswami N., Suhre K., Rafii A., and Graumann J. (2016) Comprehensive transcriptomic and proteomic characterization of human mesenchymal stem cells reveals source specific cellular markers. Sci. Rep. 6, 21507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wiredja D. D., Koyutürk M., and Chance M. R. (2017) The KSEA App: a web-based tool for kinase activity inference from quantitative phosphoproteomics. Bioinformatics 33, 3489–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Raaijmakers L. M., Giansanti P., Possik P. A., Mueller J., Peeper D. S., Heck A. J. R., and Altelaar A. F. M. (2015) PhosphoPath: Visualization of phosphosite-centric dynamics in temporal molecular networks. J. Proteome Res. 14, 4332–4341 [DOI] [PubMed] [Google Scholar]
- 28. Kumar L., and Futschik E. M. (2007) Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2, 5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q.-W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baker P. R., and Chalkley R. J. (2014) MS-Viewer: A web-based spectral viewer for proteomics results. Mol. Cell. Proteomics 13, 1392–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop D., and Horwitz E. (2006) Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8, 315–317 [DOI] [PubMed] [Google Scholar]
- 32. Park M. S., Phan H.-D., Busch F., Hinckley S. H., Brackbill J. A., Wysocki V. H., and Nakanishi K. (2017) Human Argonaute3 has slicer activity. Nucleic Acids Res. 45, 11867–11877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bridge K. S., Shah K. M., Li Y., Foxler D. E., Wong S. C. K., Miller D. C., Davidson K. M., Foster J. G., Rose R., Hodgkinson M. R., Ribeiro P. S., Aboobaker A. A., Yashiro K., Wang X., Graves P. R., Plevin M. J., Lagos D., and Sharp T. V. (2017) Argonaute utilization for mirna silencing is determined by phosphorylation-dependent recruitment of LIM-domain-containing proteins. Cell Rep. 20, 173–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bergmann J. H., Li J., Eckersley-Maslin M. A., Rigo F., Freier S. M., and Spector D. L. (2015) Regulation of the ESC transcriptome by nuclear long noncoding RNAs. Genome Res. 25, 1336–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee S., Seo H.-H., Lee C. Y., Lee J., Shin S., Kim S. W., Lim S., and Hwang K.-C. (2017) Human long noncoding RNA regulation of stem cell potency and differentiation. Stem Cells Int. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ounzain S., Micheletti R., Arnan C., Plaisance I., Cecchi D., Schroen B., Reverter F., Alexanian M., Gonzales C., Ng S. Y., Bussotti G., Pezzuto I., Notredame C., Heymans S., Guigó R., Johnson R., and Pedrazzini T. (2015) CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol. 89, Part A, 98–112 [DOI] [PubMed] [Google Scholar]
- 37. Xiao X., Zhou T., Guo S., Guo C., Zhang Q., Dong N., and Wang Y. (2017) LncRNA MALAT1 sponges miR-204 to promote osteoblast differentiation of human aortic valve interstitial cells through up-regulating Smad4. Int. J. Cardiol. 243, 404–412 [DOI] [PubMed] [Google Scholar]
- 38. Gernapudi R., Wolfson B., Zhang Y., Yao Y., Yang P., Asahara H., and Zhou Q. (2016) MicroRNA 140 promotes expression of long noncoding RNA NEAT1 in adipogenesis. Mol. Cell. Biol. 36, 30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao J., Liu Y., Zhang W., Zhou Z., Wu J., Cui P., Zhang Y., and Huang G. (2015) Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle 14, 3112–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nötzold L., Frank L., Gandhi M., Polycarpou-Schwarz M., Groβ M., Gunkel M., Beil N., Erfle H., Harder N., Rohr K., Trendel J., Krijgsveld J., Longerich T., Schirmacher P., Boutros M., Erhardt S., and Diederichs S. (2017) The long non-coding RNA LINC00152 is essential for cell cycle progression through mitosis in HeLa cells. Sci. Rep. 7, 2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Battistelli C., Cicchini C., Santangelo L., Tramontano A., Grassi L., Gonzalez F. J., Nonno V., Grassi G., Amicone L., and Tripodi M. (2017) The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene 36, 942–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin M., Pedrosa E., Shah A., Hrabovsky A., Maqbool S., Zheng D., and Lachman H. M. (2011) RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLOS ONE 6, e23356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jin C., Zheng Y., Huang Y., Liu Y., Jia L., and Zhou Y. (2017) Long non-coding RNA MIAT knockdown promotes osteogenic differentiation of human adipose-derived stem cells. Cell Biol. Int. 41, 33–41 [DOI] [PubMed] [Google Scholar]
- 44. Wang Y., Xu Z., Jiang J., Xu C., Kang J., Xiao L., Wu M., Xiong J., Guo X., and Liu H. (2013) Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell 25, 69–80 [DOI] [PubMed] [Google Scholar]
- 45. Isenmann S., Arthur A., Zannettino A. C., Turner J. L., Shi S., Glackin C. A., and Gronthos S. (2009) TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. STEM CELLS 27, 2457–2468 [DOI] [PubMed] [Google Scholar]
- 46. Kim W., Barron D. A., Martin R. S., Chan K. S., Tran L. L., Yang F., Ressler S. J., and Rowley D. R. (2014) RUNX1 is essential for mesenchymal stem cell proliferation and myofibroblast differentiation. Proc. Natl. Acad. Sci. 111, 16389–16394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao Z., Zhao M., Xiao G., and Franceschi R. T. (2005) Gene transfer of the Runx2 transcription factor enhances osteogenic activity of bone marrow stromal cells in vitro and in vivo. Mol. Ther. 12, 247–253 [DOI] [PubMed] [Google Scholar]
- 48. Peshkin L., Wühr M., Pearl E., Haas W., Freeman R. M., Gerhart J. C., Klein A. M., Horb M., Gygi S. P., and Kirschner M. W. (2015) On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Dev. Cell 35, 383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337. [DOI] [PubMed] [Google Scholar]
- 50. Billing A. M., Ben Hamidane H., Bhagwat A. M., Cotton R. J., Dib S. S., Kumar P., Hayat S., Goswami N., Suhre K., Rafii A., and Graumann J. (2016) Complementarity of SOMAscan to LC-MS/MS and RNA-seq for quantitative profiling of human embryonic and mesenchymal stem cells. J. Proteomics 150, 86–97 [DOI] [PubMed] [Google Scholar]
- 51. Konze S. A., Werneburg S., Oberbeck A., Olmer R., Kempf H., Jara-Avaca M., Pich A., Zweigerdt R., and Buettner F. F. R. (2017) Proteomic analysis of human pluripotent stem cell cardiomyogenesis revealed altered expression of metabolic enzymes and PDLIM5 isoforms. J. Proteome Res. 16, 1133–1149 [DOI] [PubMed] [Google Scholar]
- 52. Pan H., Li X., Wang J., Zhang K., Yang H., Li Z., Zheng Z., and Liu H. (2015) LIM mineralization protein-1 enhances bone morphogenetic protein-2–mediated osteogenesis through activation of ERK1/2 MAPK pathway and upregulation of Runx2 transactivity. J. Bone Miner. Res. 30, 1523–1535 [DOI] [PubMed] [Google Scholar]
- 53. Landeira D., Bagci H., Malinowski A. R., Brown K. E., Soza-Ried J., Feytout A., Webster Z., Ndjetehe E., Cantone I., Asenjo H. G., Brockdorff N., Carroll T., Merkenschlager M., and Fisher A. G. (2015) Jarid2 coordinates nanog expression and PCP/Wnt signaling required for efficient ESC differentiation and early embryo development. Cell Rep. 12, 573–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu Y., Deng P., Yu B., Szymanski J. M., Aghaloo T., Hong C., and Wang C.-Y. (2017) Inhibition of EZH2 promotes human embryonic stem cell differentiation into mesoderm by reducing H3K27me3. Stem Cell Rep. 9, 752–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Klimczak M., Czerwińska P., Mazurek S., Sozańska B., Biecek P., Mackiewicz A., and Wiznerowicz M. (2017) TRIM28 epigenetic corepressor is indispensable for stable induced pluripotent stem cell formation. Stem Cell Res. 23, 163–172 [DOI] [PubMed] [Google Scholar]
- 56. Toda T., Hsu J. Y., Linker S. B., Hu L., Schafer S. T., Mertens J., Jacinto F. V., Hetzer M. W., and Gage F. H. (2017) Nup153 interacts with Sox2 to enable bimodal gene regulation and maintenance of neural progenitor cells. Cell Stem Cell 21, 618–634 [DOI] [PubMed] [Google Scholar]
- 57. Salvi M., Trashi E., Cozza G., Negro A., Hanson P. i., Pinna L. a. (2012) Tools to discriminate between targets of CK2 vs PLK2/PLK3 acidophilic kinases. BioTechniques 53, 1–5 [DOI] [PubMed] [Google Scholar]
- 58. Casado P., Rodriguez-Prados J.-C., Cosulich S. C., Guichard S., Vanhaesebroeck B., Joel S., and Cutillas P. R. (2013) Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci Signal 6, rs6–rs6 [DOI] [PubMed] [Google Scholar]
- 59. Jung J.-H., and Traugh J. A. (2005) Regulation of the interaction of Pak2 with Cdc42 via autophosphorylation of serine 141. J. Biol. Chem. 280, 40025–40031 [DOI] [PubMed] [Google Scholar]
- 60. Hough C., Radu M., and Doré J. J. E. (2012) TGF-Beta induced Erk phosphorylation of Smad linker region regulates Smad signaling. PLOS ONE 7, e42513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Romano D., Nguyen L. K., Matallanas D., Halasz M., Doherty C., Kholodenko B. N., and Kolch W. (2014) Protein interaction switches coordinate Raf-1 and MST2/Hippo signalling. Nat. Cell Biol. 16, 673–684 [DOI] [PubMed] [Google Scholar]
- 62. Lee I. H., Sohn M., Lim H. J., Yoon S., Oh H., Shin S., Shin J. H., Oh S.-H., Kim J., Lee D. K., Noh D. Y., Bae D. S., Seong J. K., and Bae Y. S. (2014) Ahnak functions as a tumor suppressor via modulation of TGFβ/Smad signaling pathway. Oncogene 33, 4675–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ackema K. B., and Charité J. (2008) Mesenchymal stem cells from different organs are characterized by distinct topographic Hox codes. Stem Cells Dev. 17, 979–992 [DOI] [PubMed] [Google Scholar]
- 64. Silva T. A., Smuczek B., Valadão I. C., Dzik L. M., Iglesia R. P., Cruz M. C., Zelanis A., Siqueira A. S., Serrano S. M. T., Goldberg G. S., Jaeger R. G., Freitas V. M., Silva T. A., Smuczek B., Valadão I. C., Dzik L. M., Iglesia R. P., Cruz M. C., Zelanis A., Siqueira A. S., Serrano S. M. T., Goldberg G. S., Jaeger R. G., and Freitas V. M. (2016) AHNAK enables mammary carcinoma cells to produce extracellular vesicles that increase neighboring fibroblast cell motility. Oncotarget 7, 49998–50016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kingsley P. D., McGrath K. E., Maltby K. M., Koniski A. D., Ramchandran R., and Palis J. (2001) Subtractive hybridization reveals tissue-specific expression of ahnak during embryonic development. Dev. Growth Differ. 43, 133–143 [DOI] [PubMed] [Google Scholar]
- 66. Banerjee-Basu S., and Baxevanis A. D. (2001) Molecular evolution of the homeodomain family of transcription factors. Nucleic Acids Res. 29, 3258–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zákány J., Kmita M., Alarcon P., de la Pompa J.-L., and Duboule D. (2001) Localized and transient transcription of Hox genes suggests a link between patterning and the segmentation clock. Cell 106, 207–217 [DOI] [PubMed] [Google Scholar]
- 68. Rousseau M., Crutchley J. L., Miura H., Suderman M., Blanchette M., and Dostie J. (2014) Hox in motion: tracking HoxA cluster conformation during differentiation. Nucleic Acids Res. 42, 1524–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kessel M., and Gruss P. (1991) Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell 67, 89–104 [DOI] [PubMed] [Google Scholar]
- 70. Steens J., Zuk M., Benchellal M., Bornemann L., Teichweyde N., Hess J., Unger K., Görgens A., Klump H., and Klein D. (2017) In vitro generation of vascular wall-resident multipotent stem cells of mesenchymal nature from murine induced pluripotent stem cells. Stem Cell Rep. 8, 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bridge K. S., Shah K. M., Li Y., Foxler D. E., Wong S. C. K., Miller D. C., Davidson K. M., Foster J. G., Rose R., Hodgkinson M. R., Ribeiro P. S., Aboobaker A. A., Yashiro K., Wang X., Graves P. R., Plevin M. J., Lagos D., and Sharp T. V. (2017) Argonaute utilization for miRNA silencing is determined by phosphorylation-dependent recruitment of LIM-domain-containing proteins. Cell Rep. 20, 173–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Krcmery J., Camarata T., Kulisz A., and Simon H.-G. (2010) Nucleocytoplasmic functions of the PDZ-LIM protein family: new insights into organ development. BioEssays 32, 100–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bowe R. A., Cox O. T., Ayllón V., Tresse E., Healy N. C., Edmunds S. J., Huigsloot M., and O'Connor R. (2014) PDLIM2 regulates transcription factor activity in epithelial-to-mesenchymal transition via the COP9 signalosome. Mol. Biol. Cell 25, 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Camarata T., Snyder D., Schwend T., Klosowiak J., Holtrup B., and Simon H.-G. (2010) Pdlim7 is required for maintenance of the mesenchymal/epidermal Fgf signaling feedback loop during zebrafish pectoral fin development. BMC Dev. Biol. 10, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lim H. J., Kim J., Park C.-H., Lee S. A., Lee M. R., Kim K.-S., Kim J., and Bae Y. S. (2016) Regulation of c-Myc Expression by Ahnak Promotes Induced Pluripotent Stem Cell Generation. J. Biol. Chem. 291, 752–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data, including all raw files and data analysis tables produced by MaxQuant, have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (29) with the dataset identifier PXD004652. Individual spectra of the PRIDE-deposited data can be visually interrogated with MS viewer (30) (search key: xjvcljyla8). RNA-seq data have been deposited to the Sequence Read Archive (SRA) with the dataset identifier PRJNA507603. URLs for transcriptomics (https://www.ncbi.nlm.nih.gov/sra/PRJNA507603) and for proteomics/phosphoproteomics (https://www.ebi.ac.uk/pride/archive/projects/PXD004652).