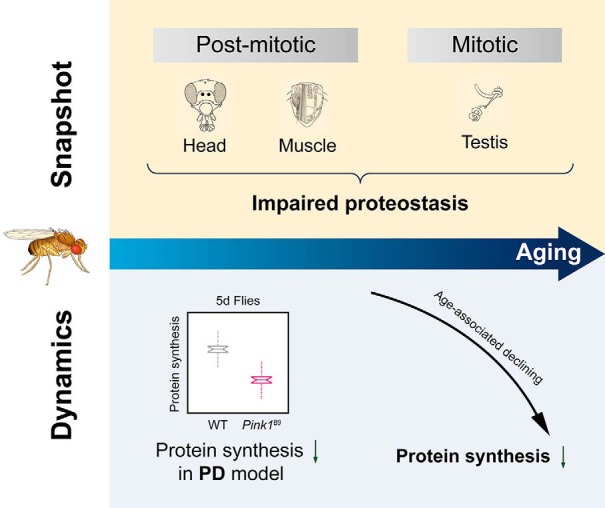
Yang et al. have profiled the snapshot and dynamics of the proteomes from Drosophila somatic and reproductive tissues during aging, revealing both tissue-specific and common pathways that are modulated with age. Analysis of proteome dynamics demonstrates that the global protein synthesis, particularly for those proteins functionally related to proteostasis and mitochondria, is down-regulated during normal aging, and that this decline becomes dramatically enhanced in the Drosophila model of human Parkinson's disease.
Keywords: aging, Drosophila melanogaster, quantification, protein synthesis, mitochondria function or biology, proteome homeostasis
Graphical Abstract
Highlights
Proteomic landscapes of Drosophila somatic and reproductive tissues during aging.
Pulsed metabolic labeling determines a decline in protein synthesis with age.
Drosophila model of human Parkinson's disease signifies an early-onset decline in protein synthesis.
Collapse of proteostasis and mitochondria are early signals for normal aging.
Abstract
Aging is characterized by a gradual deterioration in proteome. However, how protein dynamics that changes with normal aging and in disease is less well understood. Here, we profiled the snapshots of aging proteome in Drosophila, from head and muscle tissues of post-mitotic somatic cells, and the testis of mitotically-active cells. Our data demonstrated that dysregulation of proteome homeostasis, or proteostasis, might be a common feature associated with age. We further used pulsed metabolic stable isotope labeling analysis to characterize protein synthesis. Interestingly, this study determined an age-modulated decline in protein synthesis with age, particularly in the pathways related to mitochondria, neurotransmission, and proteostasis. Importantly, this decline became dramatically accelerated in Pink1 mutants, a Drosophila model of human age-related Parkinson's disease. Taken together, our multidimensional proteomic study revealed tissue-specific protein dynamics with age, highlighting mitochondrial and proteostasis-related proteins. We suggest that declines in proteostasis and mitochondria early in life are critical signals prior to the onset of aging and aging-associated diseases.
Aging and many human age-related diseases are linked to deterioration of proteome (1–5). The proteome, defined as all cellular proteins, is essential for biological processes. Proteome is not static, as evidenced by highly dynamic protein biosynthesis and degradation, which together constitute the proteome. Appropriate protein turnover ensures an optimized proteome by replenishing old dysfunctional proteins with functional new copies (6, 7). Mass spectrometry-based proteomic studies using stable isotope labeling by amino acids in cell culture (SILAC)1 have revealed widespread protein remodeling from the nematode, C.elegans and relative stable proteome from mouse muscle during the aging process (8, 9), likely reflecting the difference at the tissue type and the evolving complexity of model organisms. Drosophila melanogaster, the fruit fly, has been integral to the understanding of human neurodegenerative disease, revealing insight into numerous situations, including polyQ, Parkinson's and Alzheimer's diseases (10–12), as well as lifespan regulation (13, 14). However, the dynamic features of Drosophila proteomes from multiple tissue types during normal aging and in diseases remain poorly addressed. Importantly, protein synthesis has been closely linked with aging. Signaling pathways targeting protein translation machinery, such as the insulin growth factor 1 (IGF1) pathway (15), the target of rapamycin (TOR) pathway (16, 17), and the p38 mitogen-activated protein kinase (MAPK) pathway (18), have been linked with age-modulatory effects. However, quantitative insight of protein synthesis that occurs with normal aging and in age-related diseases remains to be explored.
To investigate the proteome in Drosophila, we first quantified the proteomes of head, muscle and testis tissues derived from flies of different ages, using 10-plex tandem mass tag (TMT) isotopic labeling reagents. Both common and tissue-specific changes were observed. The most significant pathway involved was the maintenance of proteostasis. To dissect age-associated protein synthesis, we used a pulsed isotope labeling strategy to study the change in protein synthesis during aging in fly heads. We further investigated the difference in protein synthesis using the Drosophila model of human Parkinson's disease (PD), an age-associated neurodegenerative disease. Flies deficient in the PTEN-induced putative kinase 1 (PINK1), a homolog of the familial PD gene, are short-lived. In this mutant background, we found an early-onset reduction in proteostasis and mitochondria in Pink1 deficient animals. Collectively, our data highlights that declines in proteostasis and mitochondria early in life are critical signals for aging and age-associated disease.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
Three biological replicates with ∼100 flies in each replicate were included for all experiments in this study. The Pearson correlation test, Student's t test and Fisher's exact test were performed and visualized using the R language (https://www.r-project.org/).
Fly Culture
Flies were cultured in standard media at 25 °C with 60% humidity in a 12-hour light/12-hour dark cycle. The WT (control) line used was 5905 (FlyBase ID FBst0005905, w1118). The mutant line was Pink1B9 (from Bloomington stock center, 34749). The Pclc421 Su(z)12c253 double mutants were generated as described (19).
Pulsed-SILAC Labeling
Flies were first grown on standard food until day 5 (5 d), day 15 (15 d), day 30 (30 d), day 45 (45 d), and day 60 (60 d) of age. Then, the flies were transferred to a medium consisting of 10% (w/v) 13C-lysine-labeled Saccharomyces cerevisiae (Silantes GmbH, Munich, Germany), 150 mm sucrose, 6 mm methylparaben, and 0.5‰ propionic acid. Head tissues were harvested after 5 d of 13C-lysine labeling.
Sample Preparation
Head, muscle and testis tissues from flies were homogenized in ice-cold urea lysis buffer containing 8 m urea, 100 mm Tris-HCl, pH 8.5, 150 mM NaCl, and protease inhibitor (Roche, Basel, Switzerland). The homogenate was centrifuged at 14,000 × g for 10 min, and the protein concentration of the supernatant was determined using the BCA protein assay (Thermo Fisher Scientific, MA). Proteins were digested with trypsin by a standard protocol of filter-aided sample preparation (20). For one given tissue, nine samples derived from three aging time points and three biological replicates per time point were labeled with TMT reagents (Thermo Fisher Scientific). Briefly, 100 μg of digested peptides was resuspended in 100 μl of 100 mm TEAB and labeled with different TMT reagents following the manufacturer's instructions. The TMT-labeled peptides were equally mixed for further processing.
Basic Reversed Phase (RP) Chromatography
Off-line basic RP HPLC fractionation was performed on a XBridge BEH C18 column (1 mm,150 mm,130 Å, 3.5 μm, Waters, Herts, UK) using an Agilent 1260 HPLC system. Peptides were fractionated by using a 72 min basic RP HPLC gradient. A flow rate of 70 μl/min was used for the entire LC elution. On sample injection, 72 fractions were collected into a 96-well plate (GE Healthcare, Chicago, IL). The resulting small fractions were then pooled noncontiguously into 6 or 7 large fractions, which were dried to completeness in a vacuum concentrator. Peptides were dissolved in water containing 0.1% FA, in preparation for LC-MS analysis.
LC-MS/MS
On-line LC-MS/MS analysis was performed on a Tribrid Orbitrap fusion mass spectrometer coupled with a NanoLC-1000 HPLC system (Thermo Fisher Scientific). The peptide mixture was separated by an in-house manufactured 15-cm fritless column packed with C18 resin (1.9 μm, Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) at a flow rate of 300 nl/min. For the TMT-labeled sample, mass spectra were acquired in a data-dependent mode with one full MS1 scan in the Orbitrap (m/z: 350–1500; resolution: 60,000; AGC target value: 400,000), followed by MS2 scan in the Orbitrap (37% normalized collision energy with 5% stepped collision energy; resolution: 30000; AGC target value: 50,000; maximal injection time: 60 ms). The isolation window was set as 1.5 and 1.6 m/z for TMT and SILAC samples, respectively. The dynamic exclusion for precursor selection was set as 45 s.
For pulsed-SILAC sample analysis, mass spectra were acquired in a data-dependent mode with one full scan in the Orbitrap (m/z: 350–1800; resolution: 120,000; AGC target value: 500,000), followed by MS2 scan in the linear trap (32% normalized collision energy with 5% stepped collision energy; AGC target value: 10,000; maximal injection time: 100 ms). The dynamic exclusion for precursor selection was set to 60 s.
TMT Data Analysis
The raw data from different fractions belonging to the same sample were searched together against the UniProt Drosophila database (download date: Feb 2016) using proteome discoverer 2.1 (PD 2.1), with the precursor and fragment mass tolerance set to 10 ppm and 0.1 Da, respectively. Trypsin was set as the enzyme, and the maximum allowed cleavage was 3. Static modifications included carbamidomethyl (C) (+57.02 Da) and 10-plex TMT (+229.16 Da) for lysine and the peptide N terminus, whereas dynamic modification could include the oxidation of methionine (+15.99 Da). The FDR was set to 0.01 for peptides. The FDR was defined as the ratio between false positive identifications and all identifications. Intensities for all channels were first normalized according to their median values. Protein fold changes were calculated as the ratios of the average intensities of the reporter ions, and the corresponding p values were calculated using Student's t test were used to assess the significance of differentially expressed proteins.
Pulsed-SILAC Data Analysis
The raw data from different fractions belonging to the same sample were searched together against the UniProt Drosophila database (download date: Feb 2016) using proteome discoverer 1.4 (PD 1.4), with the precursor and fragment mass tolerance set to 10 ppm and 0.6 Da, respectively. Trypsin was set as the enzyme, and the maximum allowed cleavage was 3. The static modification was carbamidomethyl (C) (+57.02 Da), whereas the dynamic modifications included the oxidation of methionine (+15.99 Da) and 13C labeling (+6.02 Da) on lysine. The FDR was set to 0.01 for peptides. The FDR was defined as the ratio between false positive identifications and all identifications. The percentage of 13C labeling was calculated as Lys(6)/(Lys(6)+ Lys(0)) to represent the synthesis rate for each time point. The newly synthesized protein fraction was calculated as the average of the ratios calculated from different replicates.
Pathway Analysis
Pathway analysis was performed by using ingenuity pathway analysis (IPA, Qiagen, Hilden, Germany) according to the vendor's guide. The ingenuity knowledge base (genes only) was used as reference.
Protein Complex Analysis
Proteins that clustered according to their translation rate change, were further analyzed to determine whether they were part of a complex defined in the published database (21). Proteins defined in the fly UniProt accession were first converted to the corresponding human Ensembl gene IDs using the biological DataBase network (22) then compared with complex database (21). Proteins that could be found in complex databases were components of a defined complex. Then, the percentage of proteins in defined complexes was calculated for each cluster.
Coexpression Analysis
For proteins that were part of complexes, the coexpression level with other components of the complexes were retrieved from the coexpression database COXPRESdb (23) (human: Hsa.v13–01.G20280-S73083). The coexpression values reflected the correlation of expression between components in each complex and were plotted for each component of a complex, as defined in complex analysis of each of the four clusters.
Lifespan Analysis
Adult male flies were collected on the day of eclosion, and 20 flies per vial were maintained at 25 °C with 60% humidity and a 12-hour light/12-hour dark cycle. Flies were transferred to new vials on alternate days and scored for survival.
RESULTS
Profiling the Static Abundance of Proteome with Age
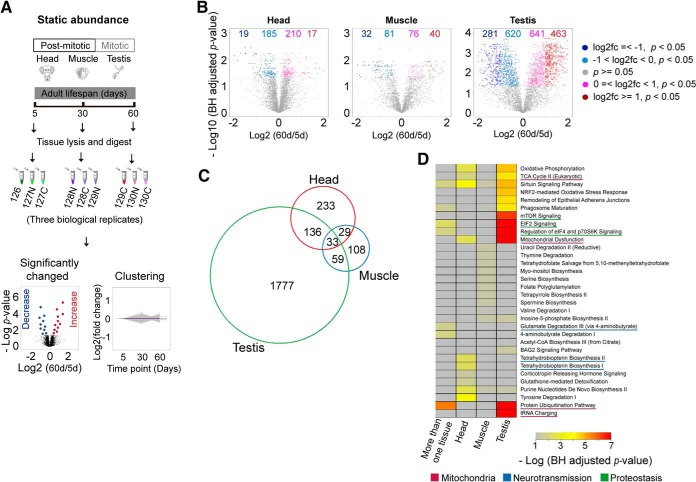
To study how protein abundance changes with age, we employed 10-plex TMT reagents to quantify the proteomes of wild-type (WT) Drosophila tissues from day 5 (5 d), day 30 (30 d), and day 60 (60 d) of age. We examined head and muscle tissues, which are primarily non-dividing somatic tissues, and testis tissue, which consists of mitotically-active germline cells (Fig. 1A). In total, we identified 4104, 2367, and 4251 proteins in the head, muscle and testis respectively, among which 3877, 2218, and 4008 proteins were quantitatively profiled at all age points and therefore used for the subsequent analysis (supplemental Table S1–S3). The assessment of quantification quality indicated a significant correlation between independent biological replicates (supplemental Fig. S1A–S1C). The median coefficients of variation (CVs) between replicates were 10.6%, 12.5%, and 10.4% for the head, muscle and testis, respectively (supplemental Fig. S1D–S1F).
Fig. 1.
Tissue-specific quantitative proteomic analysis of aging in WT Drosophila. A, Experimental scheme of quantitative aging proteome analysis. Three tissues including the post-mitotic head and muscle tissues, and the mitotic testis, were harvested at three different age time points. A tandem mass tag (TMT)-based quantitative method was employed. For each analysis, nine samples derived from three aging time points with three biological replicates for each tissue were labeled with different TMT reagents, mixed, and analyzed by the shotgun method. B, Volcano plots of tissue-specific quantitative aging proteomes between 5d and 60d. The numbers of significantly regulated proteins were indicated on the plots. C, Venn plot of proteins with significantly changed abundance (p value < 0.05) of three tissues. D, Pathway analysis for proteins with significantly changed abundance. The enriched pathways were shown for either tissue-specific proteins or proteins present in more than one tissues.
Comparison of the protein abundances between 5 d and 60 d revealed that 431,229 and 2005 proteins were differentially expressed (p value < 0.05) with age in the head, muscle, and testis, respectively (Fig. 1B, supplemental Fig. S2). Remarkably, more than half of the total proteins showed significant change in abundance during aging (p value < 0.05) in testis that contain mitotically-active cells, A further stringent filtering with fold-change no less than 2 (p value < 0.05, and fc ≥ 2) resulted in 36, 72, 744 proteins with altered levels in head, muscle, and testis, respectively. In addition to tissue-specific changes, we noted that 224 proteins exhibited altered abundance in more than one tissue (Fig. 1C). Interestingly, these commonly changed proteins were functionally linked to proteostasis such as the protein synthesis-related “EIF2 signaling,” protein degradation-related “protein ubiquitination pathway” (Fig. 1D), implicating common events that are associated with the progression of aging.
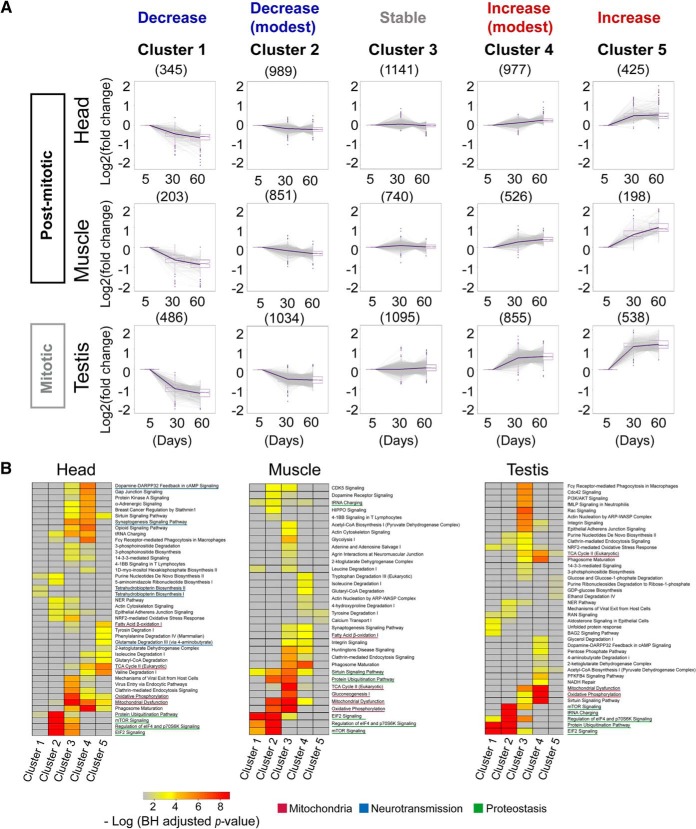
Pathway Analysis of Aging Proteomes
To characterize the aging proteome in detail, we first sorted proteins into five clusters according to their age-related patterns using the fuzzy c-means method (Fig. 2A). Notably, most proteins in head and muscle displayed minor changes or even unaltered levels with age. In contrast, up to ∼23% of testicular proteins showed substantial changes in protein levels (cluster 1 and cluster 5). We next used ingenuity pathway analysis (IPA) for functional annotation (Fig. 2B). In the head, proteins in “tetrahydrobiopterin biosynthesis” (p value = 1.24e-2), which plays important roles in the biosynthesis of dopamine and serotonin, two major monoamine neurotransmitters, were significantly decreased. Proteins that increased rapidly with age were linked to the metabolism of amino acids, such as the degradation of valine and isoleucine (Fig. 2B), two branched-chain amino acids (BCAAs) (24, 25). Additional enriched functional categories included the proteostasis-related “EIF2 signaling,” “protein ubiquitination pathway,” and “mTOR signaling” (Fig. 2B). On the other hand, proteins functionally related to “mitochondrial dysfunction,” and “oxidative phosphorylation” were mainly enriched in the clusters that had increased or unchanged abundance.
Fig. 2.
Tissue-specific functional analysis of aging in WT Drosophila. A, Clustering of proteins using the fuzzy c-means method according to their abundance changes during the aging process. The proteins localized in the middle cluster were generally stable. In contrast, the proteins in cluster 1 and cluster 5 changed dramatically with age, whereas proteins in cluster 2 and cluster 4 changed mildly. B, Pathway analysis for proteins in different clusters. Many tissue-specific and common pathways were identified for proteins in different clusters.
We then extended this analysis to the muscle and testis. Interestingly, these two tissues exhibited age-modulated proteins and biological pathways mainly in amino acid metabolism and proteostasis (Fig. 2B). Combined, this data highlights that proteins and pathways related to proteostasis were commonly decreased across all three tissues.
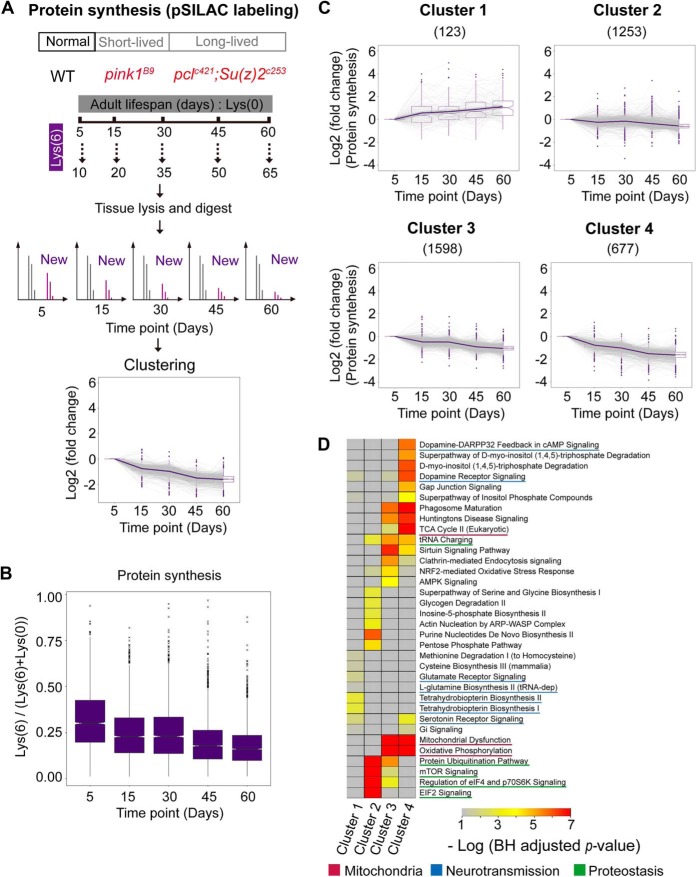
Pulsed Metabolic Labeling Analysis Determines a Progressive Reduction in Protein Synthesis with Age
Given the fact that proteostasis, the most significantly affected pathway in three tissues, is determined by protein synthesis and degradation, we subsequently interrogated how protein synthesis might be modulated with age by using the pulsed-SILAC (Lys6) strategy. In our study, we fed flies with Lys6 food for a fixed time of 5 days starting at 5 d, 15 d, 30 d, 45 d, and 60 d of age (Fig. 3A), such that the percentage of Lys6 signal relative to the total (Lys6 + Lys0) could quantitatively indicate the rate of protein synthesis. Three biological replicates per time point were conducted. Dissected heads were used for the analysis. A total of 9022 proteins were identified and 3651 proteins quantified from all aging time points (supplemental Table S4, supplemental Fig. S3). As noted, the median heavy labeling efficiencies (Lys6/(Lys6 + Lys0)) were 29.7%, 22.3%, 22.5%, 17.2%, and 15.9% for 5 d, 15 d, 30 d, 45 d, and 60 d flies, respectively (supplemental Table S4). The labeling efficiency differed greatly between different proteins. For examples, protein Histone 2B had only 2.3% labeled fraction, whereas 71.6% of Tyrosine-protein kinase (Q8INJ8) was labeled at 5 d time point.
Fig. 3.
Protein synthesis decreases during aging in Drosophila. A, Experimental scheme of quantitative proteomic synthesis. Wild-type, Pink1B9 mutant or Pclc421/+;Su(z)12c253 double mutant flies at different age time points were fed heavy stable isotope labeled lysine food for a fixed 5 d period. The newly synthesized proteins during this labeling period incorporated the heavy isotope lysine, thus, the percentage of each protein containing heavy lysine reflected its synthesis rate. B, Protein synthesis decreases during aging in Drosophila. The proportions of heavy isotope labeled proteins reflected protein synthesis rates at a given aging time point. A median turnover rate decreased from ∼30%/5 d at day 5 to ∼16%/5 d at day 60. (The median values were represented at the center) C, Clustering of proteins with different synthesis change trends. Only a small fraction of proteins in cluster 1 exhibited slightly increased synthesis with age. D, Pathway analysis for proteins in different clusters.
Interestingly, a large reduction in protein synthesis occurred between 5 d and 15 d at the early adult stage (Fig. 3B). We subsequently sorted proteins into four clusters using the fuzzy c-means method (Fig. 3C) according to the proportion of the Lys6 form. This analysis assigned only 123 proteins to cluster 1, which exhibited relatively increased synthesis with age. In contrast, proteins in other three clusters exhibited a reduction in synthesis. Notably, proteins in cluster 4, which had the most dramatic decrease in protein synthesis, accounted for more than 18% of total proteins.
We next performed pathway analysis for the clustered proteins. Proteins with upregulated synthesis were enriched in “tetrahydrobiopterin biosynthesis” (Fig. 3D, p value = 5.05e-4). On the other hand, proteins involved in “dopamine receptor signaling” were synthesized at a lower rate in aged compared with young animals (Fig. 3D, p value = 4.42e-6). Remarkably, many mitochondrial proteins were enriched in cluster 4, thus exhibiting a significant reduction in protein synthesis (Fig. 3D).
Negative Correlation Between Newly Synthesized Protein Fraction and Protein Abundance
To examine the relationship between newly synthesized protein fraction and protein abundance, we plotted the change in protein synthesis and protein abundance between 5 d and 60 d, revealing a weak but negative correlation (Fig. 4A). Further analysis found that proteins with decreased abundance tended to exhibit relatively increased synthesis, whereas proteins with increased abundance were more likely to have reduced synthesis during aging. For example, 48% of proteins from cluster 1, in which their synthesis relatively increased with age, showed a decline in abundance. In contrast, only 10% of the protein from cluster 4, in which their synthesis decreased dramatically with age, became less abundant during adult lifespan (Fig. 4B).
Fig. 4.
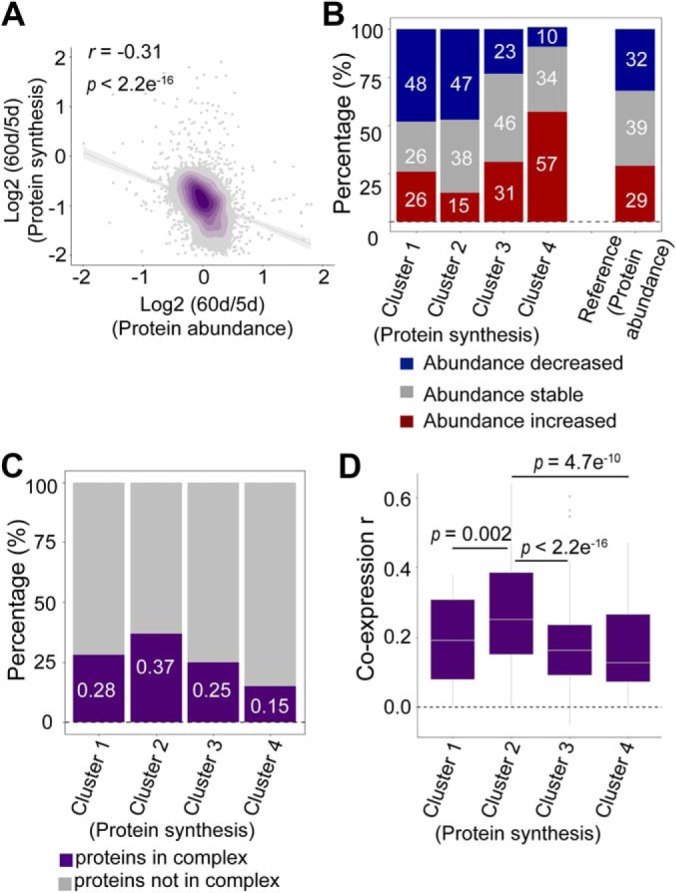
Factors that may impact the protein synthesis rate. A, Pearson correlation between protein abundance changes and protein synthesis changes. The respective ratios between day 5 and day 60 were compared. B, Protein synthesis affects protein abundance. Proteins in clusters 1 and 2 of Fig. 2A showed decreased abundance, and proteins belonging to clusters 4 and 5 in Fig. 2A showed increased abundance. Proteins in cluster 3 in Fig. 2A had stable abundance. C, Protein complexes stabilize protein synthesis. The proteins in cluster 2 exhibited the most stable synthesis during aging. A total of 37% of proteins in the cluster 2 were associated with a protein complex. D, Protein coexpression stabilizes protein synthesis. The proteins in cluster 2 were more likely to be coexpressed with other proteins. The median values were marked at the center.
Interestingly, proteins in cluster 2, whose synthesis was the most stable during aging, were more likely to interact with other proteins, as shown by the fact that 37% of proteins in this cluster normally exist as part of protein complexes. In contrast, 85% of proteins in cluster 4, which showed a dramatic reduction in protein synthesis, had no evidence to associate with protein complexes (Fig. 4C). We further examined the coexpression scores for proteins in different clusters. Consistently, the proteins in cluster 2 were more likely to be coexpressed with other proteins in the same complex. However, proteins with greater changes in synthesis tended to be present alone (Fig. 4D).
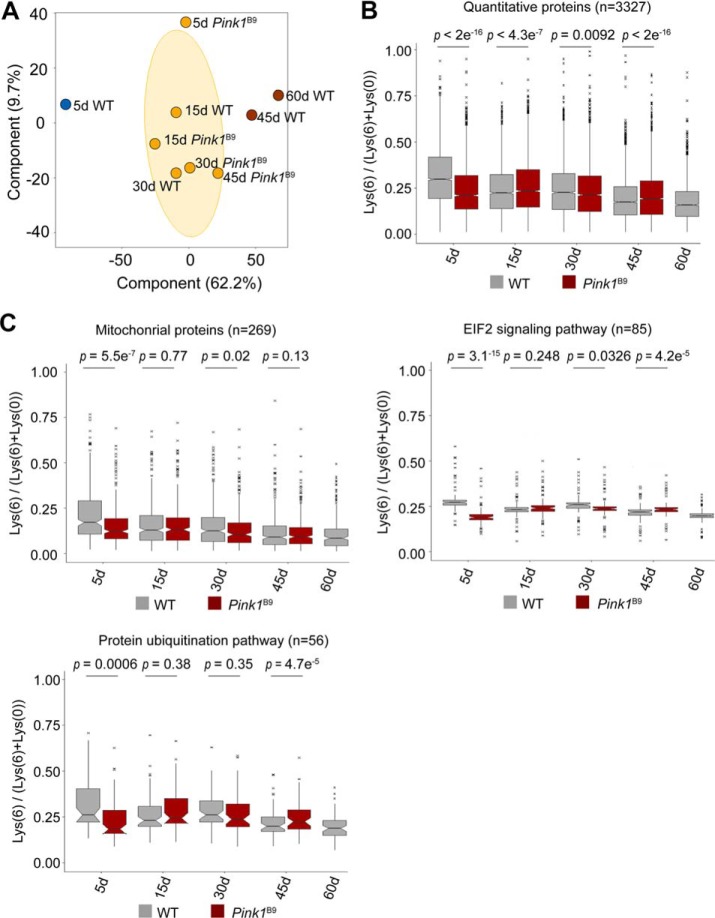
Early-onset Collapse of Protein Synthesis in Pink1B9 Mutant
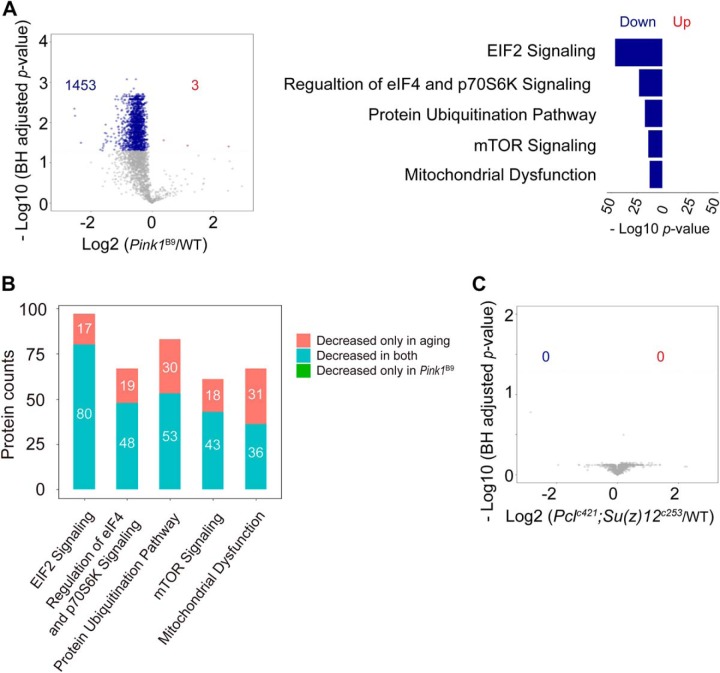
Because aging is the most significant risk factor for age-associated diseases, we asked how protein synthesis might be modulated in the context of late-onset disease. To address this, we examined the Pink1B9 mutant fly (supplemental Fig. S4), which is a well-established Drosophila model for human age-associated Parkinson's disease (PD). The Pink1B9 mutants were significantly short-lived compared with WT flies (supplemental Fig. S5). Because PD is a neurodegenerative disorder, we used fly head for the subsequent study. Based on the pulsed-SILAC strategy, we were able to quantify 3327 proteins at all time points in both WT and Pink1B9 flies (Fig. 3A, supplemental Table S5). Principal component analysis (PCA) was sufficient to separate the samples into three groups: 5 d WT flies represented a young and healthy state, whereas 45 d and 60 d WT flies exhibited age-modulated proteomes (Fig. 5A). It was important to note that Pink1B9 mutants at 5 d of age already displayed protein profiles reminiscent of WT animal at much older age (Fig. 5B). The most dramatic difference between Pink1B9 mutant and WT flies was observed on day 5, when the Pink1B9 mutants showed much lower rate in protein synthesis than that of age-matched WT (Fig. 5C). Quantitative analysis revealed that 1453 proteins in the Pink1B9 mutant showed significantly decreased protein synthesis, and that these proteins were enriched in proteostasis and mitochondrial function (Fig. 6A). Pathways related to protein synthesis, including “EIF2 signaling,” “protein ubiquitination pathway,” and “mitochondrial proteins,” was greatly affected at 5 d of age (Fig. 6A), but were only slightly changed or remained unchanged at later aging points. Interestingly, many common proteins decreased synthesis in both age and diseased conditions (Fig. 6B and supplemental Table S6). Collectively, these data suggest that defects in proteostasis and mitochondria at an early stage of life might represent critical indicators for late-onset deleterious phenotypes.
Fig. 5.
Early-onset collapse of protein synthesis in Pink1 mutant. A, PCA analysis of proteomic synthesis data of both WT and Pink1B9 mutant flies. B, Comparison of protein synthesis of WT and Pink1B9 mutant flies of different ages. The most dramatic difference was observed on day 5. C, Proteostasis and mitochondrial function are affected in the Pink1B9 mutant. The EIF2 pathway was a representative pathway in protein synthesis. The most significant difference was observed on day 5. The median values were marked at the center. Two-side student t-tests were used to compare WT and Pink1B9 mutant (B, C).
Fig. 6.
Protein translation shows different changes with shrinkage or extension of the lifespan. A, Volcano plot and functional analysis of protein synthesis between 5 d WT and Pink1B9 mutant flies. Many proteins in the Pink1B9 mutant had decreased protein synthesis compared with the WT levels, and these proteins functioned in proteostasis and mitochondria (Fisher's exact test). B, Protein synthesis is deceased in both aging and Pink1B9 mutant flies for some specific pathways. Proteins in clusters 2, 3 and 4 of Fig. 3C showed decreased protein synthesis with aging. C, Volcano plot of protein synthesis between 3 d WT and Pclc421/+;Su(z)12c253 mutant flies.
Protein Synthesis is Well-maintained in Long-lived Drosophila Mutants
Because protein synthesis is dramatically decreased in short-lived Pink1B9 mutant flies, we next asked how protein synthesis might change in long-lived animals. To address this, we applied the same pulsed-SILAC strategy to PRC2-deficient Pclc421 Su(z)12c253 double mutants that has been shown to have extended lifespan (19). Analysis using data from head tissues demonstrated that only mild differences were observed between WT flies and long-lived mutants (Fig. 6C and supplemental Table S7). No proteins showed altered synthesis in the PRC2-deficient flies. Combined, these data show that PRC2 long-lived mutants have relatively unchanged rate in protein synthesis as compared with age-matched WT animals.
DISCUSSION
Our proteomic analysis examined the change of protein with age in three tissue types including somatic tissues from head and muscle and the testis of germline tissues. Our data demonstrates both common and tissue-specific changes in protein abundance with age. Using Pink1B9 mutant of the Drosophila PD model, our finding further suggests that declines in proteostasis and mitochondria early in life are critical indicators prior to the onset of aging and perhaps age-associated diseases.
As revealed by this study, most proteins in head and muscle, as post-mitotic tissues, exhibit smaller than 2-fold change during lifespan. On the other hand, the mitotically-active testis contains comparatively more proteins that change in abundance with age. It's important to heighten the fact that the composition of proteome not just contributes but also reflects the state of cells. Post-mitotic cells, such as mature neurons and muscles, acquire their terminally differentiated states by expressing the proteome in a way that is required to maintain the cell identity and function. Mitotically active cells, such as testis, have mixed cell types as well as states of differentiation along the development of sperms, such that the proteomes in the testis would be expected to have more complicated and dynamic features than those of mature neurons and muscles. Notably, adult fruit flies are primarily made of postmitotic cells, except for the gut and germline. Because it has been reported that aging leads to a dramatic decline in germline (26), it is possible that cell types and differentiation would differ more dramatically in aged compared with young testis. However, the detailed mechanism by which aging markedly affects the protein expression in mitotic compared with post-mitotic tissues will need to be addressed by future studies. Apart from the tissue-specific changes, proteostasis is the most significantly affected biological function in all three tissues. Our data reveals that both protein synthesis and protein degradation are impaired in aged animals, implying that decreased ability in protein turnover may lead to functional decline of the proteome. Interestingly, proteins related to mitochondria and metabolic processes are significantly altered during aging. The metabolism of branched-chain amino acids (BCAAs), including leucine, isoleucine, and valine, are changed. This observation is consistent with previous studies in which BCAAs are shown to be involved in energy metabolism, mitochondrial biogenesis (25, 27), and pro-lifespan beneficial effects (24, 25).
As suggested by our data in Drosophila, although proteostasis is impaired, protein abundance may remain at a relatively steady-state level. Previous studies using stable isotope labeling suggest that low protein synthesis is a hallmark of aging in C. elegans (28, 29). Similarly, our study in Drosophila also reveals a global decline in protein synthesis that occurs in aged animals. Notably, the labeling efficiency (Lys6/(Lys6 + Lys0)) measured in this study can be affected by both protein synthesis and protein degradation. Given the fact that only a small fraction of proteins in fly heads have significantly changed abundance during aging, it is reasonable to predict that all proteins are under a steady-state level, in which the rate of protein synthesis is approximately equal to the rate of protein degradation. Further comparison between WT and Pink1B9-deficient flies shows that protein synthesis in short-lived mutant flies is dramatically reduced even at the young age. Our recent study revealed that stimulation of glycolytic pathway in PRC2-deficient flies promotes healthy longevity (19). In contrast, little changes are found between PRC2 long-lived flies and age-matched WT. This evidence highlights the possibility that the collapse of proteome is a critical signal for the onset of aging and age-associated disease, whereas proper maintenance of proteome would lead to normal aging or perhaps healthy longevity. Because protein synthesis is an efficient way to replenish damaged or misfolded proteins with new functional copies, animals with age or disease may have a decreased protein synthesis and consequently compromised ability to repair damaged proteins as well as cellular functions.
It is unclear how natural aging might lead to a decrease in protein translation. One possible reason is age-dependent reduction in energy production that reduces protein synthesis. It has been well-established that aging is associated with a dramatic decline in gene expression and bioenergetic process that yield less cellular energy, such as adenosine triphosphate (ATP) (19). Protein translation process, on the other hand, requires a high amount of energy (30). Thus, age-onset bioenergetic reduction may consequently lead to a global decrease in protein synthesis. Our quantitative assessment further indicates that, in Pink1B9 of a PD fly model, a decrease in protein synthesis rate becomes significantly accelerated, in a good congruence with the advanced aging status seen in Pink1B9 mutants. Notably, however, the pro-life benefits of mTOR pathways have been linked to their effects in down-regulation of protein translation (31). As a result of lowered protein synthesis, it has been suggested that long-lived mTOR mutants may actively reprogram cellular energy and metabolic process toward pro-life activities (32). Hence, the impact of protein homeostasis including protein synthesis on aging might be poised; the precise effect must be rather complicated. During natural aging or pathological conditions, the extent by which protein synthesis decreases appears to be a crucial indicator for aging and disease, whereas in long-lived mTOR mutants, reduction of protein synthesis would trigger a compensatory mechanism to promote lifespan.
Taken together, our proteomic analysis reveals both tissue-specific and common changes during aging. Interestingly, age-modulated proteins are generally related to proteostasis and mitochondria. Quantitative assessment of protein dynamics highlights early declines in proteostasis and mitochondria as critical indicators for both normal aging and PD-linked pathology in Drosophila.
Data Availability
The MS data have been deposited under the accession number IPX0001462000 at iProX (https://www.iprox.org/page/project.html?id=IPX0001462000).
Supplementary Material
Footnotes
* Funding support was provided by the National Natural Science Foundation of China to Y.Z. (31671428, 31500665, and 31530041) to N.L. (91849109), the 100 Talents Program of the Chinese Academy of Sciences to Y.Z., and the National Key R&D Program of China to Y.Z. and N.L. (2016YFA0501904 and 2016YFA0501900).
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- SILAC
- stable isotope labeling by amino acids in cell culture
- IGF1
- insulin growth factor 1
- TOR
- the target of rapamycin
- MAPK
- the p38 mitogen-activated protein kinase
- TMT
- tandem mass tag
- PINK1
- PTEN-induced putative kinase 1
- PD
- Parkinson's disease
- IPA
- ingenuity pathway analysis
- BCAAs
- branched-chain amino acids
- PCA
- Principal component analysis
- ATP
- adenosine triphosphate
- FDR
- false discovery rate
- PRC2
- polycomb repressive complex 2.
REFERENCES
- 1. Lopez-Otin C., Blasco M. A., Partridge L., Serrano M., and Kroemer G. (2013) The hallmarks of aging. Cell 153, 1194–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kirkwood T. B. L., and Austad S. N. (2000) Why do we age? Nature 408, 233–238 [DOI] [PubMed] [Google Scholar]
- 3. Sibille E. (2013) Molecular aging of the brain, neuroplasticity, and vulnerability to depression and other brain-related disorders. Dialogues Clin. Neurosci. 15, 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Forman D. E., Rich M. W., Alexander K. P., Zieman S., Maurer M. S., Najjar S. S., Cleveland J. C. Jr, Krumholz H. M., and Wenger N. K. (2011) Cardiac care for older adults. Time for a new paradigm. J. Am. Coll. Cardiol. 57, 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benz C. C. (2008) Impact of aging on the biology of breast cancer. Crit. Rev. Oncol. Hematol. 66, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldberg A. L. (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899 [DOI] [PubMed] [Google Scholar]
- 7. Hinkson I. V., and Elias J. E. (2011) The dynamic state of protein turnover: It's about time. Trends Cell Biol. 21, 293–303 [DOI] [PubMed] [Google Scholar]
- 8. Walther D. M., Kasturi P., Zheng M., Pinkert S., Vecchi G., Ciryam P., Morimoto R. I., Dobson C. M., Vendruscolo M., and Mann M. (2015) Widespread proteome remodeling and aggregation in aging C. elegans. Cell 161, 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walther D. M., and Mann M. (2011) Accurate quantification of more than 4000 mouse tissue proteins reveals minimal proteome changes during aging. Mol. Cell. Proteomics 10, M110.004523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bilen J., and Bonini N. M. (2005) Drosophila as a model for human neurodegenerative disease. Ann. Rev. Genetics 39, 153. [DOI] [PubMed] [Google Scholar]
- 11. Marsh J. L., and Thompson L. M. (2006) Drosophila in the study of neurodegenerative disease. Neuron 52, 169–178 [DOI] [PubMed] [Google Scholar]
- 12. Shulman J. M., Shulman L. M., Weiner W. J., and Feany M. B. (2003) From fruit fly to bedside: translating lessons from Drosophila models of neurodegenerative disease. Current Opinion Neurol. 16, 443–449 [DOI] [PubMed] [Google Scholar]
- 13. Tu M. P., and Tatar M. (2003) Juvenile diet restriction and the aging and reproduction of adult Drosophila melanogaster. Aging Cell 2, 327–333 [DOI] [PubMed] [Google Scholar]
- 14. Fontana L., Partridge L., and Longo V. D. (2010) Extending healthy life span—from yeast to humans. Science 328, 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holzenberger M., Dupont J., Ducos B., Leneuve P., Géloën A., Even P. C., Cervera P., and Le Bouc Y. (2003) IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 42, 182–187 [DOI] [PubMed] [Google Scholar]
- 16. Kapahi P., Chen D., Rogers A. N., Katewa S. D., Li P. W., Thomas E. L., and Kockel L. (2010) With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 11, 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harrison D. E., Strong R., Sharp Z. D., Nelson J. F., Astle C. M., Flurkey K., Nadon N. L., Wilkinson J. E., Frenkel K., Carter C. S., Pahor M., Javors M. A., Fernandez E., and Miller R. A. (2009) Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Troemel E. R., Chu S. W., Reinke V., Lee S. S., Ausubel F. M., and Kim D. H. (2006) p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2, e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma Z. W., Cai H., Wang Y., Niu H., Wu K., Ma X., Yang H., Tong Y., Liu W., Liu F., Zhang Z., Liu Y., Zhu R., and Liu Z. J. N. (2018) Epigenetic drift of H3K27me3 in aging links glycolysis to healthy longevity in Drosophila. Elife 7, e35368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wisniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 21. Ori A., Iskar M., Buczak K., Kastritis P., Parca L., Andres-Pons A., Singer S., Bork P., and Beck M. (2016) Spatiotemporal variation of mammalian protein complex stoichiometries. Genome Biol. 17, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mudunuri U., Che A., Yi M., and Stephens R. M. (2009) bioDBnet: the biological database network. Bioinformatics 25, 555–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Okamura Y., Aoki Y., Obayashi T., Tadaka S., Ito S., Narise T., and Kinoshita K. (2015) COXPRESdb in 2015: coexpression database for animal species by DNA-microarray and RNAseq-based expression data with multiple quality assessment systems. Nucleic Acids Res. 43, D82–D86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mansfeld J., Urban N., Priebe S., Groth M., Frahm C., Hartmann N., Gebauer J., Ravichandran M., Dommaschk A., Schmeisser S., Kuhlow D., Monajembashi S., Bremer-Streck S., Hemmerich P., Kiehntopf M., Zamboni N., Englert C., Guthke R., Kaleta C., Platzer M., Sühnel J., Witte O. W., Zarse K., and Ristow M. (2015) Branched-chain amino acid catabolism is a conserved regulator of physiological ageing. Nat. Communications 6, 10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. D'Antona G., Ragni M., Cardile A., Tedesco L., Dossena M., Bruttini F., Caliaro F., Corsetti G., Bottinelli R., Carruba M. O., Valerio A., and Nisoli E. (2010) Branched-chain amino acid supplementation promotes survival and supports cardiac and skeletal muscle mitochondrial biogenesis in middle-aged mice. Cell Metab. 12, 362–372 [DOI] [PubMed] [Google Scholar]
- 26. Wallenfang M. R., Nayak R., and DiNardo S. (2006) Dynamics of the male germline stem cell population during aging of Drosophila melanogaster. Aging Cell 5, 297–304 [DOI] [PubMed] [Google Scholar]
- 27. Valerio A., D'Antona G., and Nisoli E. (2011) Branched-chain amino acids, mitochondrial biogenesis, and healthspan: an evolutionary perspective. Aging 3, 464–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dhondt I., Petyuk V. A., Bauer S., Brewer H. M., Smith R. D., Depuydt G., and Braeckman B. P. (2017) Changes of protein turnover in aging Caenorhabditis elegans. Mol. Cell. Proteomics 16, 1621–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vukoti K., Yu X., Sheng Q., Saha S., Feng Z., Hsu A. L., and Miyagi M. (2015) Monitoring newly synthesized proteins over the adult life span of Caenorhabditis elegans. J. Proteome Res. 14, 1483–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aoyagi Y T. I., Okumura J., and Muramatsu T. (1988) Energy cost of whole-body protein synthesis measured in vivo in chicks. Comp. Biochem. Physiol. A Comp. Physiol. 91, 765–768 [DOI] [PubMed] [Google Scholar]
- 31. Bolster D. R., Crozier S. J., Kimball S. R., and Jefferson L. S. (2002) AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J. Biol. Chem. 277, 23977–23980 [DOI] [PubMed] [Google Scholar]
- 32. Wang X., and Proud C. G. (2006) The mTOR pathway in the control of protein synthesis. Physiology 21, 362–369 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS data have been deposited under the accession number IPX0001462000 at iProX (https://www.iprox.org/page/project.html?id=IPX0001462000).