Abstract
Terpene trilactones (TTLs) are the main secondary metabolites of Ginkgo biloba. As one of the rate-limiting enzymes in the mevalonic acid (MVA) pathway of TTL biosynthesis, 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) catalyzes the 3-hydroxy-3-methylglutaryl coenzyme A to form MVA. In this study, two cDNA sequences of HMGR genes, namely, GbHMGR2 and GbHMGR3, were cloned from G. biloba. The protein sequences of GbHMGR2 and GbHMGR3, which contain several functional domains, were analyzed. Regulatory elements related to light, hormone, and stress response were detected in the promoter regions of GbHMGR2 and GbHMGR3. The catalytic activity of these genes was verified by a functional complement experiment in yeast. Quantitative real-time PCR (qRT-PCR) showed the distinct expression patterns of the two genes in different organs. The TTL contents in the organs were detected by high-performance liquid chromatography– evaporative light scattering detector. GbHMGR2 and GbHMGR3 were responded to cold, dark, methyl jasmonate (MJ), abscisic acid (ABA), salicylic acid (SA), and ethephon (Eth) treatments. The TTL contents were also regulated by cold, dark, MJ, ABA, SA, and Eth treatment. In conclusion, GbHMGR2 and GbHMGR3 may participate in the MVA pathway of TTL biosynthesis.
Subject terms: Plant molecular biology, Plant biotechnology
Introduction
Ginkgo biloba is an important medicinal plant, and its leaf extracts are widely used in clinical medicine. The most popular G. biloba extract product is EGB761, which contains 24% ginkgo flavone glucosides and 6% terpene trilactones (TTLs) and exhibits protective properties against neuronal and vascular damages1. TTLs, including ginkgolide and bilobalide, are the main secondary metabolites of G. biloba and important active ingredients of EGB761. In addition, TTLs are natural antagonists of platelet-activating factor and are the preferred natural drug for the treatment of cardiovascular diseases2,3. However, the TTL content in G. biloba organs is extremely low4, and TTLs are difficult to synthesize chemically because of their complex chemical structures. Hence, methods that improve TTL content are the focal point in studies on G. biloba.
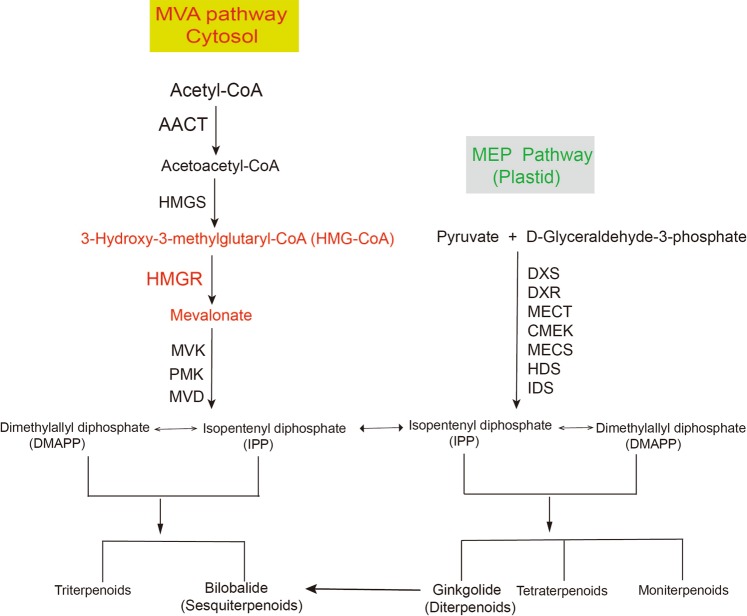
Terpenoids are generally synthesized through the mevalonic acid (MVA) and methylerythritol phosphate (MEP) pathways in plants. Monoterpenes, diterpenes, and tetraterpenes are generated through the MEP pathway, whereas sesquiterpenes and triterpene are produced through the MVA pathway5. Terpenoid content can be effectively increased by overexpressing a key gene in the terpenoid synthesis pathway or regulating the expression of multiple terpene synthase genes by transcription factors6,7. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) is one of the rate-limiting enzymes in the MVA pathway of terpenoid biosynthesis and catalyzes 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) to form MVA8 (Fig. 1). Overexpression of the HMGR gene can effectively increase the terpenoid content in plants. For example, the HMGR activity and sterol content of tobacco plants transgenic with rubber HMGR were increased by four to eight and six times, respectively9 (Schaller et al. 1995). Overexpressing Catharanthus roseus HMGR in Artemisia annua increased the artemisinin content by 38.9%10. Arabidopsis thaliana HMGR overexpression in Lavandula latifolia increased the essential oil and sterol contents11. Therefore, the overexpression of the HMGR gene shows potential in improving the TTL content of G. biloba.
Figure 1.
Metabolic pathways of terpenoids.
In 1989, Caelles et al.12 first isolated the HMGR gene from Arabidopsis. To date, the HMGR genes have been isolated from more than 60 plants, including angiosperms and gymnosperms4,13. The plant HMGR genes generally appear under HMGR gene family, and HMGR homologous genes of the same species are differentially expressed in various tissues under diverse external stimulations14. For example, methyl jasmonate (MJ) depresses the expression of Camptotheca acuminate HMGR1 (CaHMRG1) but not those of CaHMGR2 and CaHMGR315. Under normal conditions, the expression of CaHMRG1 is detected in the reproductive organs and leaves, but CaHMRG2 expression is found in all tissues16. Hence, a potential HMGR gene together with genetic engineering biotechnology can be used to increase terpenoid content by investigating the members of HMGR family and understanding the biological functions of each member and their relationship in G. biloba. In 2006, Shen et al.17isolated and characterized HMGR from G. biloba for the first time, they discussed that GbHMGR was lowly expressed in a tissue-specific manner, because this gene was found only in root. We have also previously cloned and characterized GbHMGR1 and demonstrated that this gene is significantly related to the TTL content in G. biloba4. However, whether other HMGR genes exist in G. biloba, and whether such genes are related to the synthesis of TTLs remains unclear. Thus, in this study, two novel cDNAs of HMGR (GbHMGR2 and GbHMGR3) were cloned and characterized from G. biloba. Yeast functional complementation of GbHMGR2 and GbHMGR3 was conducted to identify the functions of their proteins. Quantitative real-time PCR (qRT-PCR) was applied to determine the expression profiles of GbHMGR2 and GbHMGR3 in different tissues under various treatments, namely, cold, dark, MJ, salicylic acid (SA), ethephon (Eth), and abscisic acid (ABA) treatments. The TTL contents in the tissues and leaves under stress and hormone treatments were also determined. The results of this study will help in elucidating the functions of GbHMGR2 and GbHMGR3 and provide further insights into the biosynthesis of TTLs.
Results
Characterization and phylogenetic analysis of GbHMGR2 and GbHMGR3 proteins
GbHMGR2 measures a full length of 2260 bp and contained a 1746 bp open reading frame (ORF) encoding a 581-amino-acid protein, whereas GbHMGR3 features a full length of 2228 bp with a 1719 bp ORF encoding a 572-amino-acid protein. The sequences are similar to those of the unigenes filtered from the G. biloba transcriptome database (GeneBank accession number SRX1982952). BLASTn search revealed that these genes are highly homologous with the HMGR genes of other plants. Thus, these cloned genes are members of HMGR family and therefore were designated as GbHMGR2 (GeneBank accession number MH091702) and GbHMGR3 (GeneBank accession number MH091703).
The predicted GbHMGR2 and GbHMGR3 proteins comprise of 581 and 572 amino acids, respectively. The theoretical isoelectric point and molecular weight of the predicted GbHMGR2 protein were 8.18 and 61.7 KDa, respectively, whereas those of GbHMGR3 were 6.92 and 61.5 KDa, respectively. Both proteins belong to the HMGR I category according to protein conserved domain prediction analysis. This HMGR category is located in the cytoplasm and is the first rate-limiting enzyme in the MVA pathway6. BLASTp search revealed that GbHMGR2 and GbHMGR3 proteins feature 84%–71% identities with the HMGR proteins of G. biloba, Picea sitchensis, Taxus media, Trema orientalis, Ricinus communis, and Populus euphratica.
Transmembrane structural analysis of TMHMM Server 2.0 showed that the GbHMGR2 protein contains a transmembrane structure at positions 51–73 and 86–108, whereas those for GbHMGR3 protein are located at positions 26–48 and 61–83. Multiple alignment analysis revealed that GbHMGR2 and GbHMGR3 possess four conserved active motifs: two NADP(H) binding domains (TVGGGT and DAMGMNM) and two Hmg-CoA binding domains (TTEGCLVA and EMPVGYVQIP). These active motifs are highly conserved in the HMGRs from other plants and function as the catalytic active sites of the HMGR protein. Moreover, Glu of TTEGCLVA motif plays an important role in the HMGR catalytic action18 (Fig. 2). We speculate that GbHMGR2 and GbHMGR3 proteins are membrane-binding proteins with catalytic functions similar to those of other HMGR enzymes.
Figure 2.
Multiple sequence alignments of GbHMGR2 and GbHMGR3 with other plant HMGR proteins. Dark blue: identity = 100%; red: 75% ≤ identity < 100%; light blue: 50% ≤ identity < 75%; Four conserved active sites of HMGRs, including the two NADP(H) binding motifs (TVGGGT and DAMGMNM) and two HMG-CoA binding motifs (TTEGCLVA and EMPVGYVQIP), are highlighted in red square box.
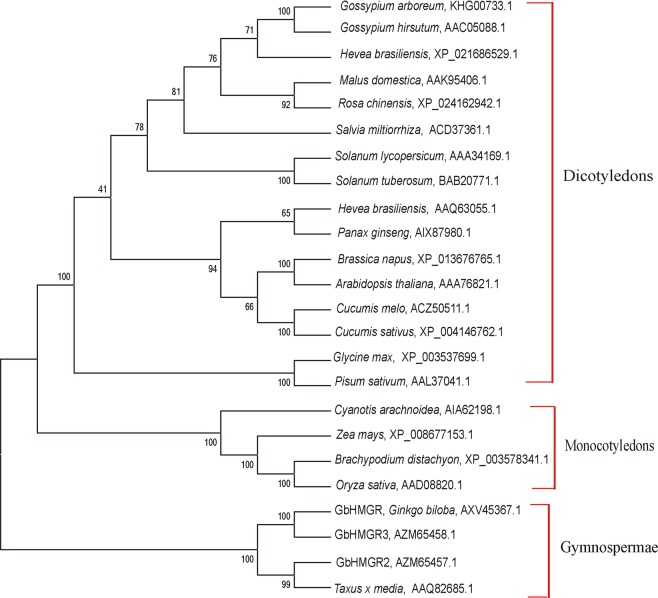
A phylogenetic tree from 24 HMGR proteins with a common ancestor was constructed to further understand the evolutionary relationship between GbHMGR2, GbHMGR3, and other HMGR proteins. The tree was divided into three branches, namely, dicotyledon, monocotyledon, and gymnosperm (Fig. 3). GbHMGR, GbHMGR2, GbHMGR3, and HMGR from T. media, are clustered as gymnosperms. This result indicates the evolutionary diversity and conservatism of HMGR proteins.
Figure 3.
Phylogenetic tree of HMGRs from different species using the neighbor-joining method. The reliability of the tree is measured by bootstrap analysis with 1000 replications. The bars represent evolutionary distance.
Promoter sequence analysis of GbHMGR2 and GbHMGR3
The promoter sequences of GbHMGR2 and GbHMGR3 span lengths of 825 and 686 bp, respectively (Supplementary Figs. S1 and S2). Table 1 displayed the cis-acting elements of GbHMGR2 and GbHMGR3. The TATA-box19 and CAAT-box20 were located at positions −28 bp and −173 bp of the GbHMGR2 promoter region, respectively. In addition, four light-responsive elements, [i.e., I-Box (position −87 bp), Sp1 (position −677 bp), GT1-motif (position −357 bp), and chs-CMA 1a (position −730 bp)];21–23 a low-temperature-responsive (LTR) element (position −163 bp)24, a gibberellin-responsive element GARE-motif (position −314 bp)25, and a development-related element HD-Zip1 (position −717 bp)26 were detected in the GbHMGR2 promoter region. This finding indicates that GbHMGR2 is regulated by various external signals, such as light, cold, and hormones.
Table 1.
Putative cis-elements in the promoters of GbHMGR2 and GbHMGR3.
| Element | Position | Signal Sequence | Expected function | Reference |
|---|---|---|---|---|
| GbHMGR2 | ||||
| TATA-box | −28 | TATAA | core promoter element around −30 of transcription start | 19 |
| I-box | −87 | CGAGAAGGCG | part of a light responsive element | 21 |
| LTR | −163 | CCGAAA | cis-acting element involved in low-temperature re | 24 |
| CAAT-box | −173 | CAAT | common cis-acting element in promoter and enhancer regions | 20 |
| GARE-motif | −313 | AAACAGA | gibberellin-responsive element | 25 |
| GT1-motif | −357 | GGTTAA | light responsive element | 22 |
| Sp1 | −678 | CCTCCCTCTT | light responsive element | 21 |
| HD-Zip 1 | −717 | CAATAATTGCTCC | element involved in differentiation of the palisade mesophyll cells | 26 |
| chs-CMA1a | −730 | TTACTTAA | part of a light responsive element | 23 |
| GbHMGR3 | ||||
| TATA-box | −25 | TATA | core promoter element around −30 of transcription start | 19 |
| AAGAA-motif | −59 | GAAAGAA | cis-acting element involved in light responsiveness | 31 |
| MNF1 | −152 | GTGCCCTCCTGTTCGA | light responsive element | 23 |
| TGA-element | −206 | AACGAC | auxin-responsive element | 30 |
| CAAT-box | −295 | CAAACAACTCC | common cis-acting element in promoter and enhancer regions | 20 |
| ACE | −346 | AAAACATTTA | cis-acting element involved in light responsiveness | 22 |
| Box III | −387 | ATCATTTACACT | protein binding site | 31 |
| Box I | −421 | TTTCAAA | light responsive element | 20 |
| ATCC-motif | −438 | CAATCCTC | part of a conserved DNA module involved in light responsiveness | 21 |
| TGACG-motif | −592 | TGACG | cis-acting regulatory element involved in the MeJA-responsiveness | 29 |
| TC-rich repeats | −656 | ATTCTCTCCA | cis-acting element involved in defense and stress responsiveness | 27 |
| ERE | −682 | ATTTCAAA | ethylene-responsive element | 28 |
Similar to GbHMGR2, the TATA-box19 and CAAT-box20 were detected at positions −25 bp and −295bp of the GbHMGR3 promoter region, respectively. Several other cis-acting elements, such as stress-responsive element TC-rich repeats (position −656 bp)27, light-responsive element ATCC-motif (position −438bp), Box I (position −421 bp), ACE (position −346 bp), MNF1 (position −152 bp)21–23, Eth-responsive element (ERE) (position −682 bp)28, MeJA-responsive element TGACG-motif (position −592 bp)29, auxin-induction-related element TGA-element (position −206 bp)30, conserved sequence AAGAA-motif (position −59 bp), and protein-binding site Box III (position −389 bp)31, were also predicted. Hence, the transcription of GbHMGR3 was speculated to be regulated by light, cold, Eth, MJ, and auxin stress.
Functional complementation of GbHMGR2 and GbHMGR3 in Saccharomyces cerevisiae
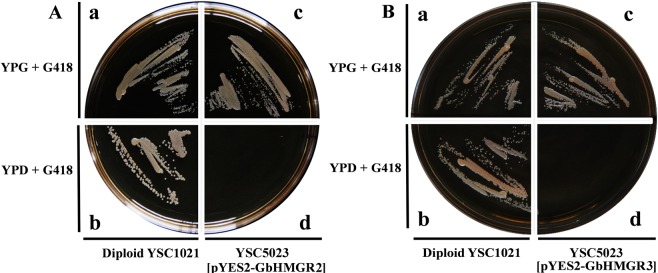
YSC5023 is an hmgr-deficient S. cerevisiae mutant strain that would die without mevalonate. We constructed yeast expression vectors, namely, GbHMGR2-pYES2 and GbHMGR3-pYES2 that could express GbHMGR2 and GbHMGR3, respectively, proteins through galactose induction to motivate MVA synthesis. Therefore, supplying MVA for YSC5023 can normalize the growth of the latter. As shown in Fig. 4, wild S. cerevisiae strain YSC1020 could grow on noninduced YPD (Fig. 4Ab,Bb) and induced YPG media (Fig. 4Aa,Ba), whereas the S. cerevisiae mutant YSC5023 containing GbHMGR2-pYES2 or GbHMGR3-pYES2 could only grow on the induced YPG + G418 galactose medium (Fig. 4Ac,Bc). These results revealed that the functional deficiency of YSC5023 could be remedied by the expressions of GbHMGR2 or GbHMGR3, indicating that GbHMGR2 and GbHMGR3 proteins can catalyze HMG-CoA to form MVA.
Figure 4.
Functional complementation of GbHMGR2 and GbHMGR3 for the hmgr-deficient yeast YSC5023. (A) Functional complementation of GbHMGR2; (B) Functional complementation of GbHMGR3; (a) Diploid YSC1021 strain was grown on YPG + G418 medium; (b) Diploid YSC1021 strain was grown on YPD + G418 medium; (c) Haploid YSC5023 strain harboring pYES2-GbHMGR1 or pYES2-GbHMGR2 was grown on YPG + G418 medium; (d) Haploid YSC5023 strain harboring pYES2-GbHMGR1 or pYES2-GbHMGR2 failed to grow on YPD + G418 medium. The strains were grown at 28 °C for 2 days.
TTL contents and expression profiles of GbHMGR2 and GbHMGR3 in the different organs of G. biloba
The contents of TTLs remarkably varied in the different organs of G. biloba. As shown in Fig. 5A, the content of TTLs in the roots, which was fivefold of that in the male strobili, was the highest, followed by those in the stems and leaves. The lowest content was found in the male and female strobili, indicating that TTLs are mainly biosynthesized in the roots.
Figure 5.
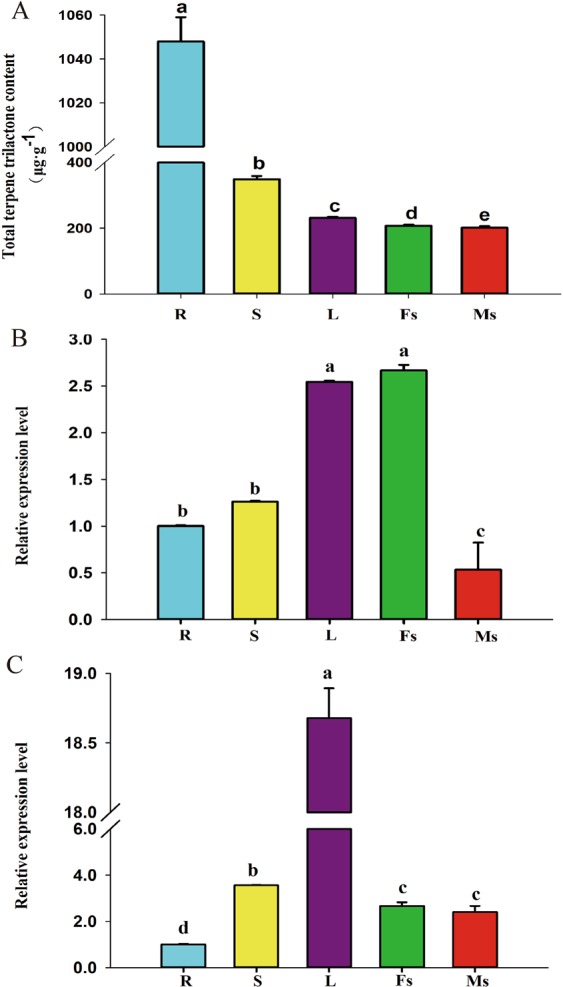
The accumulation pattern of TTLs and tissue expression patterns of GbHMGR2 and GbHMGR3 in different tissues of G.biloba. (A) Accumulation pattern of TTLs in different tissues; (B) Tissue expression patterns of GbHMGR2; (C) Tissue expression patterns of GbHMGR3. R: Root, S: Stem, L: Leaf, Fs: Female strobili, Ms: Male strobili; The gene expression level of GbHMGR2 and GbHMGR3 in the ginkgo root was set to 1. Data are shown as mean ± SE (n = 3); Means with different letters represent a Tukey’s honestly significant difference at p < 0.05.
qRT-PCR results indicated that both GbHMGR2 and GbHMGR3 genes are differentially expressed in various organs. GbHMGR2 was strongly expressed in the female strobili and leaves, followed by the stems and roots, and the lowest was observed in the male strobili (Fig. 5B). The expression pattern of GbHMGR3 in the different organs of G. biloba considerably differed from that of GbHMGR2. As shown in Fig. 5C, the expression level of GbHMGR3 in the leaves, which was 19-fold of that in the roots, was the highest, followed by those in stems, male and female strobili, and the lowest was noted in the roots.
Changes in the TTL contents and expression levels of GbHMGR2 and GbHMGR3 in G. biloba leaves under dark, cold, MJ, SA, ABA, and Eth treatments
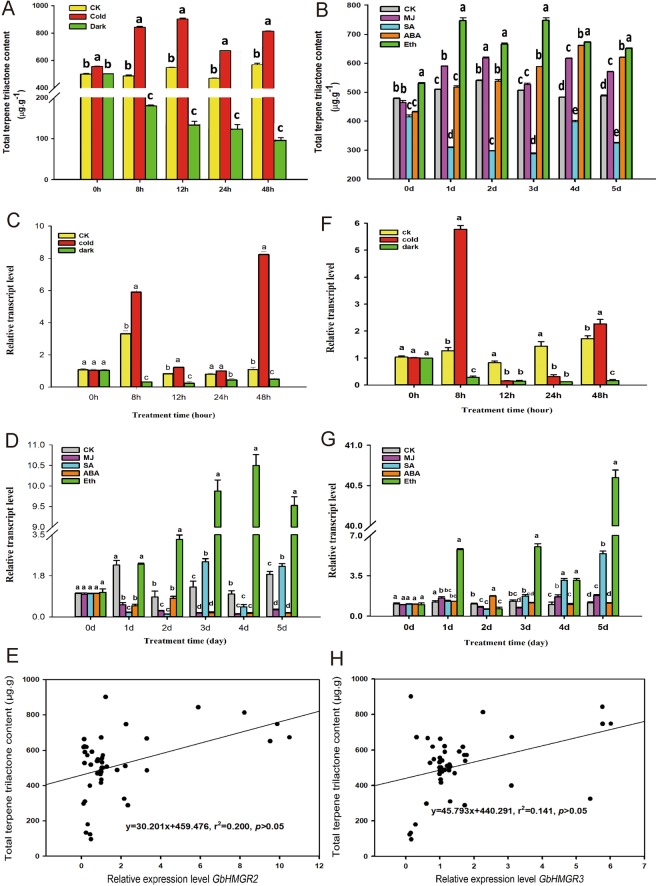
As shown in Fig. 6A, the TTL contents were inhibited by dark treatment. The most substantial inhibitory effect was observed at 48 h after treatment at 16% of the control. Low temperature considerably increased the TTL content by 65% compared with that of the control at 12 h. Except SA, plant hormones enhanced the accumulation of TTL (Fig. 6B). The TTL content peaked to a value 1.28-fold of the control 2 days after MJ treatment and increased constantly after being treated with ABA and reached the maximum value, which is 27.8% higher than that of the control after 4 days. Eth treatment increased the TTL content to the maximum value, which is 47.7% higher than that of the control at 3 days after treatment. The TTL content decreased by 76.35% after 3 days of SA treatment, and the value obtained was lower than that of the control.
Figure 6.
TTL content, expression changes of GbHMGR2 and GbHMGR3 under Dark, Cold, MJ, SA, ABA and Eth treatments, and correlation analysis of gene expression and TTL content. (A) TTL content changes by Dark and Cold; (B) TTL content changes by MJ, SA, ABA and Eth; (C) GbHMGR2 expression level changes by Dark and Cold; (D) GbHMGR2 expression level changes by MJ, SA, ABA and Eth; (E) correlation analysis of GbHMGR2 expression and TTL content; (F) GbHMGR3 expression level changes by Dark and Cold; (G) GbHMGR3 expression level changes by MJ, SA, ABA and Eth; H: correlation analysis of GbHMGR3 expression and TTL content. The expression levels were normalized to GbGAPDH gene. The transcript level of GbHMGR2 and GbHMGR3 at 0 h or 0 day post-treatment was set to 1. ALL the datas are shown as mean ± SE (n = 3); Means with different letters represent a Tukey’s honestly significant difference at p < 0.05.
As shown in Fig. 6C, the GbHMGR2 expression was substantially depressed under dark treatment and was 90% lower than that of the control at 8 h after treatment. Under cold treatment, its expression was enhanced by 6.6-fold compared with that of the control, and reached the highest level at 48 h. The expression level of GbHMGR2 was considerably influenced by plant hormones (Fig. 6D), particularly, the induction by SA and Eth and inhibition by MJ and ABA. MJ treatment caused the continuous downregulation of GbHMGR2 transcription level at only 5% that of the control at 4 days after treatment.Under SA treatment, the expression level of GbHMGR2 initially decreased first and increased, and the value was 80% higher than that of the control at 3 days after treatment. The expression level of GbHMGR2 was downregulated in response to ABA treatment, yielding a value 90% lower than that of the control at 5 days after treatment. Eth treatment induced the transcript of GbHMGR2, which continually increased and reached the highest expression value at 978% higher than that of the control at 4 days after treatment. Correlation analysis (Fig. 6E) showed that the changes in the expression of GbHMGR2 and TTL content feature a positive but nonsignificant correlation.
As shown in Fig. 6F, the expression level of GbHMGR3 was considerably downregulated under dark treatment, that is, 8% of the control at 24 h after treatment. However, this value was substantially enhanced under cold treatment. At 8 h after cold treatment, the transcription level of GbHMGR3 sharply increased to a peak value that is 4.5-fold that of the control. All hormone treatments, namely, MJ, SA, ABA, and Eth, remarkably induced the GbHMGR3 expression (Fig. 6G). GbHMGR3 expression reached the maximum value, which is 1.67-fold that of the control at 4 days after treatment with MJ. ABA treatment showed similarly increasing values until a a moderate peak at 2 days after treatment. SA and Eth treatments distinctly upregulated the expression of GbHMGR3 with the peak appering at 5 days after treatment. The former increased the expression by 3.74-fold and the latter by 34.6-fold compared with that of the control. Similar to GbHMGR2, a positive but nonsignificant correlation was observed between GbHMGR3 and TTL content (Fig. 6H).
Discussion
HMGR is one of the rate-limiting enzymes in MVA synthesis and catalyzes HMG-CoA to form MVA. This reaction can regulate terpene metabolism32. Plant HMGR genes typically contain multiple members, and the differences in the expression levels among various members can regulate the “carbon pool” in the MVA pathway6,33. In this study, two novel members of Ginkgo HMGR gene family, namely, GbHMGR2 and GbHMGR3, were cloned and identified. The promoter sequence, tissue, and stress expression patterns of these two genes were analyzed. The catalytic functions of GbHMGR2 and GbHMGR3 were further verified by yeast functional complementation experiment. Screening the genes relevant to TTL biosynthesis and the increase in TTL content bears importance in genetic engineering.
The highest TTL was observed in roots, followed by those in the stems and leaves. This finding agrees with the result indicating that TTL is synthesized in roots, translocated to the shoots through the phloem, and then stored in leaves27,28. Previous studies have shown that different members of the HMGR gene family exhibit various expression patterns33. For example, in Gentiana macrophylla, GmHMGR1 is expressed in the roots and flowers, whereas GmHMGR2 is expressed in the leaves34. Meanwhile, the PgHMGR1 and PgHMGR2 genes of Panax ginseng are highly expressed in the seedlings and rhizophore but lowly expressed in the other tissues35. In the present study, we observed that GbHMGR2 was highly expressed in the female strobili and leaves but lowly expressed in the male strobili. In addition, the expression level of GbHMGR3 in the leaves was higher than that in the other tissues. Therefore, GbHMGR2 and GbHMGR3 exhibited different expression patterns. These findings were inconsistent with the pattern of the TTL content in the different tissues. Correlation analysis showed a positive but nonsignificant relationship between the TTL content and GbHMGR2 or GbHMGR3. This results implied that GbHMGR2 and GbHMGR3 participated in the biosynthesis of TTLs in Ginkgo, but are not key genes. Diterpenes are the main components of TTLs in Ginkgo, and synthesized by the MEP pathway5.The biosynthesis of TTLs in Ginkgo is regulated by a complex network. The MVA and MEP pathways are the upstream pathways for the biosynthesis of TTLs, but genes in downstream pathway may play more important roles in this process. These may have caused the absence of significant correlations between GbHMGR2 or GbHMGR3 and TTL content. HMGRs have been reported to perform specific functions. For example, Song et al.36 proved that the overexpression of HMGR in Lactococcus lactis increases the production level of β-sesquiphellandrene by 1.25-fold to 1.60-fold. Kim et al.37 observed that the transformation of Platycodon grandiflorum with Panax ginseng HMGR (PgHMGR) induces a 1.2-fold to 1.5-fold increase in the levels of HMGR expression and total platycoside levels compared with those of the controls. Hence, we suggest that GbHMGR2 and GbHMGR3 might be involved in the biosynthesis of sesquiterpene and triterpene in G. biloba.
Gene expression is regulated to satisfy the requirements for the growth and development of plants and to resist external stress stimulated by the external environment stimulation. Darkness and low temperature are two usual abiotic stresses that affect the regulation of the plant HMGR genes. We previously showed that the GbHMGR1 expression level and TTL content in the Ginkgo callus are higher under light than those under dark after 24 h of treatment, and the GbHMGR1 expression and total TTL content were higher at 15 °C than at 24 °C4. These results are in accordance with that for GbHMGR1. Thus, GbHMGR2 and GbHMGR3 showed similar functions with GbHMGR1. Moreover, light- (i.e., I-box, Sp1, GT1-motif, and MNF1)21–23 and cold-responsive elements24 were found in the GbHMGR2 promoter region; and light- (i.e., ATCC-motif, I-box, ACE, and MNF1) and stress-responsive elements (i.e., TC-rich repeats)27 were similarly found in the GbHMGR3 promoter region. These findings explain the responses of GbHMGR2 and GbHMGR3 to cold and darkness.
Light corresponds with the synthesis of secondary metabolism in plants, but the effects vary among species. For example, in suspension-cultured Lithospermum erythrorhizon cells, white light strongly suppresses HMGR expression and shikonin formation38. Meanwhile, continuous dark exposure for 2–3 days increased the total ginsenoside content in a 3-year-old ginseng after the darkness-induced activity of PgHMGR17. In the present experiment, the expression levels of GbHMGR2 and GbHMGR3 and the TTL content substantially increased in the Ginkgo plant exposed to light compared with those under darkness. This finding reflects the diversity among species. In a dark environment, plants are under carbon starvation39, and the leaves are induced to undergo accelerated senescence because of the breakdown of chlorophyll, decline in carbohydrate and protein contents, and ROS accumulation in the plant cells40,41. Therefore, dark stress might starve the Ginkgo seedlings, and the synthesis of TTLs would be limited by the decline in carbonaceous matters. From these results, we deduce that GbHMGR2 and GbHMGR3 could regulate the synthesis of TTL by responding to light in the Ginkgo leaves.
Low temperature can stimulate Ginkgo plants to accumulate several secondary metabolites to resist stress. Low temperature induces the expression of various genes, such as GbPAL42, GbANS43, and GbFLS44, which are involved in the synthesis of G. biloba flavonoids. Other plants, such as Picrorhiza kurroa, also exhibit a similar trend of upregulating PkHMGR and picroside content under low temperature45. The expression levels of GbHMGR2 and GbHMGR3 and the TTL content were significantly increased under low temperature, and this phenomenon corresponded with the cold motif in their promoter region. We observed that low temperature could enhance GbHMGR2 and GbHMGR3 transcript and stimulate the biosynthesis of TTLs to a certain extent.
Chemical elicitors, such as MJ, SA, ABA, and Eth, play key roles in the defense responses and development of plants. MJ is a well-known elicitor of secondary metabolism in plants46 and can elicit the accumulation of total phenols, flavonoids, and acacetin, a flavonoid compound with multiple pharmaceutical values47. Moreover, applying MJ induces the expression of the genes involved in terpenoid synthesis, such as GbHMGS148, GbHMGS249, and GbIDS250. However, contrary to these genes, GbHMGR2 expression was suppressed after treatment with MJ. This discrepancy suggests the tissue specificity of GbHMGR2 expression, which presented a proportional expression level in the leaves and female strobili. Under MJ treatment, the TTL content in Ginkgo leaves substantially increased, and the expression of GbHMGR3 was stimulated and was higher than that in the other tissues. This result was consistent with the MJ-responsive element TGAGG-motif in the GbHMGR3 promoter region. This finding also implies that GbHMGR3 is involved in the formation of TTLs in Ginkgo leaves. SA is another critical signaling molecule that is closely related to local and systemic acquired resistance (SAR)51. Numerous plant resistance genes, such as the genes we previously characterized (i.e., GbANS43, GbFLS44, and GbHMGR4), are upregulated in response to SA. The transcription levels of GbHMGR2 and GbHMGR3 increased 3 days after SA treatment. This finding indicates that GbHMGR2 and GbHMGR3 are involved in the SAR. The TTL content decreased after SA treatment, although the GbHMGR2 and GbHMGR3 transcriptions were induced. This phenomenon suggests that SA might inhibit the formation of other substances in the MEP and MVA pathways. These findings require further investigation.
ABA controls stomatal movement, seed dormancy, germination, and plant development52 and is closely related to HMGR gene expression53. For example, ABA signals modulate the expression and/or activity of HMGR to control the fruit growth and final fruit size of “Hass” avocado54. ABA also induces the production of several secondary metabolites. In ABA-treated Artemisia annua plants, artemisinin content was considerably increased, and the expression of important genes, such as HMGR, in the artemisinin biosynthetic pathway, was remarkably induced55. The results showed the inhibited GbHMGR2 expression, slightly increased GbHMGR3 expression in response to exogenous ABA, and considerably increased TTL content compared with that of the control. These asynchronous phenomena might be attributed to the multiple roles of ABA in plants. Yang et al.56 discovered that tanshinone production and the mRNA level of HMGR in Salvia miltiorrhiza were considerably enhanced by ABA treatments. Moreover, exogenous applications of ABA trigger endogenous MJ accumulation, whereas exogenous MJ can directly induce tanshinone production mainly via the MEP pathway in the hairy roots of S. miltiorrhiza. Similar to tanshinone, Ginkgo TTLs are synthesized through the MEP or MVA pathway. Therefore, we suggest that the ABA treatment might trigger endogenous MJ accumulation in Ginkgo leaves, thereby enhancing the TTL content. However, GbHMGR2 and GbHMGR3 expression showed no increase similar to those under the exogenous MJ treatment.
Eth is also an important hormone in the activation of plant defenses and forms a signal network with jasmonic acid, SA, and ABA57. HMGR expression is also modulated by Eth. Lv et al.58 reported that MdHMGR2 from Malus domestica is significantly induced by Eth. Numerous studies indicated that Eth enhances the biosynthesis of terpenoids in plants. For example, Eth treatment increases ocimene, trans-caryophyllene, β-elemene, valencene, and α-panasinsene in citrus fruits59. In the present study, we noted that ERE was present in the promoter region of GbHMGR3. Eth treatment also drastically enhanced the expression levels of GbHMGR2 and GbHMGR3 and the TTL content. The results indicate that Eth induces the transcript of the novel genes and stimulates the biosynthesis of terpenoids in Ginkgo leaves.
Materials and Methods
Plant materials and treatments
Tissues from the roots, stems, leaves, and male and female strobili were collected from 30-year-old grafted Ginkgo trees (cv. Jiafoshou) from the Ginkgo Garden of Yangtze University, Jingzhou, China. The tissues were placed in a freezer at −80 °C after freezing in liquid nitrogen and used for RNA extraction.
Two-year-old grafted Ginkgo saplings were collected from the Ginkgo Garden of Yangtze University for the stress and hormone treatments, which included dark, cold (4 °C), MJ, SA, ABA, and Eth treatments. The saplings were placed in a completely dark plant growth chamber at 25 °C and 75% relative humidity (RH) for the dark treatment and in a luminous (600 μmol m−2·s−1) plant growth chamber at 4 °C and 75% RH with a 16 h/8 h light/dark photoperiod for the cold treatment. For the control, the Ginkgo saplings were exposed to light (600 μmol·m−2·s−1) with a 16 h/8 h light/dark photoperiod at 25 °C. The leaves of the treated Ginkgo saplings were sampled at 0, 8, 12, 24, and 48 h after treatment, frozen in liquid nitrogen, and placed in a freezer at −80 °C.
SA, ABA, MJ, and Eth were dissolved in 0.01% (v/v) Tween 20, and their final concentrations were 10, 100, 100, and 10 mM, respectively. The four hormone solutions were sprayed on both sides of the Ginkgo leaves for the hormone treatment group. For the control, 0.01% (v/v) Tween 20 was sprayed on the leaves. All the hormone-treated plants were placed in a growth chamber at 25 °C and 75% RH with a 16 h/8 h light/dark photoperiod. The leaves were harvested at 0–5 days after the hormone treatments, frozen with liquid nitrogen, and preserved in a freezer at −80 °C.
Cloning of the full-length cDNA of GbHMGR2 and GbHMGR3
The total RNA from each tissue was extracted using the TaKaRa MiniBEST Plant RNA Extraction Kit (TaKaRa, Dalian, China). The RNA extracted from the stems was used to synthesize the first-strand cDNA with the PrimeScript™ 1st Strand cDNA Synthesis Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. Two pairs of specific primers, namely, GbHMGR2-UP and GbHMGR2-DN and GbHMGR3-UP and GbHMGR3-DN (Supplementary Table S1), were designed based on the unigene sequences of HMGRs from the G. biloba transcriptome database (GenBank accession number SRX1982952). The full-length cDNAs of GbHMGR2 and GbHMGR3 were amplified using single-strand cDNA as a template. GbHMGR2 was amplified under the following conditions: 94 °C for 4 min, 32 cycles of 94 °C for 30 s, 54.7 °C for 30 s, and 72 °C for 90 s, and GbHMGR3 was amplified under the following conditions: 94 °C for 4 min, 32 cycles of 94 °C for 30 s, 57.2 °C for 30 s, and 72 °C for 90 s. The PCR products were purified and ligated into the pMD19-T vector (Takara Bio Inc., Dalian, China). The recombined plasmids were transformed into Escherichia coli DH5α competent cells and sequenced by Shanghai Sangon Biotech.
Promoter amplification of GbHMGR2 and GbHMGR3
According to the ORF sequence of the G. biloba GbHMGR2 and GbHMGR3 genes, two round-nested pairs of primers, GbHMGR2-qd1 and GbHMGR2-qd2 and GbHMGR3-qd1 and GbHMGR3-qd2 (Supplementary Table S1), were designed near the 5′ end. The promoter sequences of GbHMGR2 and GbHMGR3 were obtained using the Universal Genome Walker Kit (Clontech, CA, USA), which mainly included adapter construction, DNA digestion, promoter walking library construction, and nested PCR.
Bioinformatics analysis
A homologous search for the nucleic acid and predicted protein sequences of GbHMGR2 and GbHMGR3 was conducted with the online tools BLASTn and BLASTp in National Center for Biotechnology Information (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The ORFs of GbHMGR2 and GbHMGR3 were predicted using Vector NTI 11.5. The molecular weights and isoelectric points of GbHMGR2 and GbHMGR3 were analyzed using an online tool, ExPASy (https://web.expasy.org/protparam/). The Conserved Domains Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and TMHMM Server V.2.0 (https://www.cbs.dtu.dk/services/TMHMM-2.0/) were used to analyze the protein conserved domain and transmembrane domain of GbHMGR2 and GbHMGR3. DNAMAN 8.0 software was used for the the translation of GbHMGR2 and GbHMGR3 proteins and multiple alignment of these proteins with the HMGRs other plants. The phylogenetic tree of HMGR was constructed using MEGA7.0. The transcription start site (TSS) and cis-acting element of the GbHMGR2 and GbHMGR3 promoters were predicted by Berkeley Drosophila Genome Project (http://www.fruitfly.org/seq_tools/promoter.html) and Plant Cis-Acting Regulatory Element (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Functional complementation of GbHMGR2 and GbHMGR3 in S. cerevisiae
YSC5023 (Δ[hmg2 + hmg3]) is an hmgr-double-deficient mutant that requires MVA and uracil to grow; this mutant is obtained by mating two hmgr-single-deficient mutants, BY4741 (hmg2) and BY4742 (hmg3), and dissecting the tetrad60. The functional deficiency of YSC5023 could be remedied by transferring exogenous HMGR genes into YSC5023, because HMGR expression can be induced by galactose and promote MVA synthesis. The growth state of recombinant YSC5023 yeast was observed to verify whether or not the proteins of GbHMGR2 and GbHMGR3 can catalyze HMG-CoA to form MVA. Two pairs of primers were designed according to the full-length GbHMGR2 and GbHMGR3 cDNA and the restriction enzyme cutting sites of the pYES2 plasmid. These primers were GbHMGR2-gnup and GbHMGR2-gndn and GbHMGR3-gnup and GbHMGR3-gndn (Supplementary Table S1), which contained the ScaI and XbaI restriction sites. The oding region was amplification through PCR, and the target fragments were purified with Agarose Gel DNA Purification Kit Ver. 4.0 (Takara, Dalian, China). The purified PCR products, which were double-digested by ScaI and XbaI, were ligated into the pYES2 vector. The recombinant plasmid was transformed into DH5α competence cells, and the positive strains were confirmed by sequencing in Shanghai Sangon (China). The constructed GbHMGR2-pYES2 and GbHMGR3-pYES2 expression vectors were transformed into YSC5023 using Frozen-EZ Yeast Transformation II Kit (Zymo Research, Orange, CA, USA). The recombinant yeasts were spotted on a yeast amino-acid-deficient medium, and the positively transformed strains were further confirmed by PCR. The wild-type S. cerevisiae strain YSC1020 and recombinant YSC5023 strain, which contained the GbHMGR2-pYES2 or GbHMGR3-pYES2 vector, were spread on noninduced YPD + G418 and induced YPG + G418 media and then incubated for 2 days at 28 °C. The growth of the strains was observed.
Quantitative analysis of GbHMGR2 and GbHMGR3 gene expression
The expression levels of GbHMGR2 and GbHMGR3 were determined by qRT-PCR. RNA was extracted from the tissues and leaves of 2-year-old Ginkgo seedlings under different treatments by using TaKaRa MiniBEST Plant RNA Extraction Kit (TaKaRa, Dalian, China) and inverse transcribed using PrimeScriptTM RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China). The primers (i.e., GbHMGR2-dlF and GbHMGR2-dlR and GbHMGR3-dlF and GbHMGR3-dlR) were designed with Primer Premier 5.0, and reference primers were GAPDH-F and GAPDH-R (Supplementary Table S1). LineGene 9600 Plus Fluorescent Quantitative PCR System and BioEasy Master Mix (SYBR Green) Kit (Bioer, Hangzhou, China) were used for qRT-PCR analysis. Three biological replicates were set for each treatment, and each sample was analyzed thrice. The relative expression levels of GbHMGR2 and GbHMGR3 were presented as 2−ΔΔCt in line with the CT method61. Data for qRT-PCR are shown as mean ± SE (n = 3).
Determination of TTL content
The TTL content in the tissues and leaves of 2-year-old Ginkgo seedlings under different treatments were determined via high-performance liquid chromatography (UltiMate 3000, Thermo, USA) with evaporative light scattering detector (6000, Alltech, USA) according to the method of Ganzera et al.62.
Statistical analysis
Data analysis was performed with one-way analysis of variance with Tukey’s honestly significant difference test by using SPSS 11.0 (SPSS Inc., USA) for Windows. The level of significance was set to p < 0.05.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NO. 31901344 and No. 31370680) and the Special Projects for Technological Innovation in Hubei Province (NO. 2019ABA113).
Author Contributions
F.X. and S.C. designed the whole experiment and drafted the manuscript. X.M. performed the experiment and the yeast complementation. S.R. wrote the manuscript. Y. L. analyzed the data and drew the figures. T.Y. and J.C. contributed in cDNA cloning and qRT-PCR analysis. J.T. determined the TTL contents. All authors have reviewed and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shen Rao, Xiangxiang Meng and Yongling Liao contributed equally.
Contributor Information
Feng Xu, Email: xufeng198@126.com.
Shuiyuan Cheng, Email: s_y_cheng@sina.com.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50629-8.
References
- 1.McKeag K, Lyseng-Williamson KA. Ginkgo biloba extract egb 761® in the symptomatic treatment of mild-to-moderate dementia: a profile of its use. Drugs Ther. Persp. 2018;34:358–366. doi: 10.1007/s40267-018-0537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohanta TK, Tamboli Y, Zubaidha PK. Phytochemical and medicinal importance of Ginkgo biloba L. Nat. Prod. Res. 2014;28:746–752. doi: 10.1080/14786419.2013.879303. [DOI] [PubMed] [Google Scholar]
- 3.Diamond BJ, Bailey MR. Ginkgo biloba: indications, mechanisms, and safety. Psychiatr. Clin. N. Am. 2013;36:73–83. doi: 10.1016/j.psc.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Liao Y, et al. Promoter analysis and transcriptional profiling of Ginkgo biloba 3-hydroxy-3-methylglutaryl coenzyme a reductase (GbHMGR) gene in abiotic sress responses. Not. Bot. Horti. Agrobo. 2015;43:25–34. doi: 10.15835/nbha4319416. [DOI] [Google Scholar]
- 5.Lange BM, Rujan T, Martin W, Croteau R. Isoprenoid biosynthesis: The evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. USA. 2000;97:13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi D, Ward BL, Bostock RM. Differential induction and suppression of potato 3-hydroxy-3-methylglutaryl coenzyme a reductase genes in response to phytophthora infestans and to its elicitor arachidonic acid. Plant Cell. 1992;4:1333–1344. doi: 10.1105/tpc.4.10.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim YJ, Lee OR, Ji YO, Jang MG, Yang DC. Functional analysis of 3-hydroxy-3-methylglutaryl coenzyme a reductase encoding genes in triterpene saponin-producing Ginseng. Plant physiol. 2014;165:373–387. doi: 10.1104/pp.113.222596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darabi M, Izadi-Darbandi A, Masoudi-Nejad A, Naghavi MR, Nemat-Zadeh G. Bioinformatics study of the 3-hydroxy-3-methylglotaryl-coenzyme A reductase (HMGR) gene in Gramineae. Mol. Biol. Rep. 2015;39:8925–8935. doi: 10.1007/s11033-012-1761-2. [DOI] [PubMed] [Google Scholar]
- 9.Schaller H, et al. Expression of the Hevea brasiliensis (H.B.K) Mull. Arg. 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 in tobacco results in sterol overproduction. Plant physiol. 1995;109:761–770. doi: 10.1104/pp.109.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nafis T, et al. Enhancement of artemisinin content by constitutive expression of the HMG-CoA reductase gene in high-yielding strain of Artemisia annua L. Plant Biotechnol. Rep. 2011;5:53–60. doi: 10.1007/s11816-010-0156-x. [DOI] [Google Scholar]
- 11.Muñoz-Bertomeu J, Sales E, Ros R, Arrillaga I, Segura J. Up-regulation of an N-terminal truncated 3-hydroxy-3-methylglutaryl CoA reductase enhances production of essential oils and sterols in transgenic Lavandula latifolia. Plant Biotechnol. J. 2007;5:746–758. doi: 10.1111/j.1467-7652.2007.00286.x. [DOI] [PubMed] [Google Scholar]
- 12.Caelles C, Ferrer A, Balcells L, Hegardt FG, Boronat A. Isolation and structural characterization of a cDNA encoding Arabidopsis thaliana 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Mol. Biol. 1989;13:627–638. doi: 10.1007/BF00016018. [DOI] [PubMed] [Google Scholar]
- 13.Kalita R, Patar L, Shasany AK, Modi MK, Sen P. Molecular cloning, characterization and expression analysis of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Centella asiatica. Mol. Biol. Rep. 2015;42:1431–1439. doi: 10.1007/s11033-015-3922-6. [DOI] [PubMed] [Google Scholar]
- 14.Roberts SC. Production and engineering of terpenoids in plant cell culture. Nat. Chem. Biol. 2007;3:387–395. doi: 10.1038/nchembio.2007.8. [DOI] [PubMed] [Google Scholar]
- 15.Maldonadomendoza IE, Vincent RM, Nessler CL. Molecular characterization of three differentially expressed members of the Camptotheca acuminata 3-hydroxy-3-methylglutaryl CoA reductase (HMGR) gene family. Plant Mol. Biol. 1997;34:781–790. doi: 10.1023/A:1005866813347. [DOI] [PubMed] [Google Scholar]
- 16.Tiski I, Marraccini P, Pot D, Vieira LG, Pereira LF. Characterization and expression of two cDNA encoding 3-hydroxy-3-methylglutaryl coenzyme a reductase isoforms in coffee (coffea arabica L.) Omics: A Journal of Integrative Biology. 2011;15:719–727. doi: 10.1089/omi.2010.0140. [DOI] [PubMed] [Google Scholar]
- 17.Shen G, et al. Cloning and characterization of a root-specific expressing gene encoding 3-hydroxy-3-methylglutaryl coenzyme a reductase from Ginkgo biloba. Mol. Bio. Rep. 2006;33:117–127. doi: 10.1007/s11033-006-0014-7. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Darnay BG, Rodwell VW. Identification of the principal catalytically important acidic residue of 3-hydroxy-3-methylglutaryl coenzyme a reductase. J. Biol. Chem. 1990;265:21634–21641. [PubMed] [Google Scholar]
- 19.Joshi CP. An inspection of the domain between putative tata box and translation start site in 79 plant genes. Nucleic Acids Res. 1987;15:6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 21.Mongkolsiriwatana C, Pongtongkam P, Peyachoknagul S. In silico promoter analysis of photoperiod-responsive genes identified by DNA microarray in Rice (Oryza Sativa L.) Kasetsart J. (Nat Sci) 2009;43:164–177. [Google Scholar]
- 22.Zhou DX. Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci. 1999;4:210–214. doi: 10.1016/S1360-1385(99)01418-1. [DOI] [PubMed] [Google Scholar]
- 23.Yanagisawa S, Izui K. MNF1, a leaf tissue-specific DNA-binding protein of maize, interacts with the cauliflower mosaic virus 35S promoter as well as the C4 photosynthetic phosphoenolpyruvate carboxylase gene promoter. Plant Mol. Biol. 1992;19:545–553. doi: 10.1007/BF00026781. [DOI] [PubMed] [Google Scholar]
- 24.Dunn MA, White AJ, Vural S, Hughes MA. Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.) Plant Mol. Biol. 1998;38:551–564. doi: 10.1023/A:1006098132352. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Olofsson L, Lundgren A, Brodelius PE. Trichome-specific expression of amorpha-4,11-diene synthase, a key enzyme of artemisinin biosynthesis in Artemisia annua L., as reported by a promoter-GUS fusion. Am. J. Plant Sci. 2011;2:619–628. doi: 10.4236/ajps.2011.24073. [DOI] [Google Scholar]
- 26.Ariel F, et al. Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell. 2010;22:2171–2183. doi: 10.1105/tpc.110.074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cartayrade A, et al. Ginkgolide and bilobalide biosynthesis in Ginkgo biloba. I: Sites of synthesis, translocation and accumulation of ginkgolides and bilobalide. Plant Physiol. Bioch. 1997;35:859–868. [Google Scholar]
- 28.Lu X, et al. Combining metabolic profiling and gene expression analysis to reveal the biosynthesis site and transport of ginkgolides in Ginkgo biloba L. Front. Plant Sci. 2017;8:872. doi: 10.3389/fpls.2017.00872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruíz-Rivero OJ, Prat S. A-308 deletion of the tomato LAP promoters is able to direct flower-specific and MeJA-induced expression in transgenic plants. Plant Mol Biol. 1998;36:639–648. doi: 10.1023/A:1005980028203. [DOI] [PubMed] [Google Scholar]
- 30.Tao TT, et al. Molecular cloning, characterization, and functional analysis of a gene encoding 3-hydroxy-3-methylglutaryl-coenzyme a synthase from Matricaria chamomilla. Genes Genom. 2016;38:1–9. doi: 10.1007/s13258-016-0463-x. [DOI] [Google Scholar]
- 31.Parveen I, et al. Investigating sesquiterpene biosynthesis in Ginkgo biloba: molecular cloning and functional characterization of (E,E)-farnesol and α-bisabolene synthases. Plant Mol. Biol. 2015;89:451–462. doi: 10.1007/s11103-015-0381-3. [DOI] [PubMed] [Google Scholar]
- 32.Bach TJ. Hydroxymethylglutaryl-CoA reductase, a key enzyme in phytosterol synthesis? Lipids. 1986;21:82–88. doi: 10.1007/BF02534307. [DOI] [PubMed] [Google Scholar]
- 33.Ma CY, Liu CS, Wang WQ. Molecular cloning and characterization of GuHMGR, an HMG-CoA reductase gene from liquorice (Glycyrrhiza uralensis) Front. Agric. China. 2011;5:400–406. doi: 10.1007/s11703-011-1121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng P, Hua WP, Wang ZZ. Cloning and characterization of HMGR genes from Gentiana macrophylla. Journal of Shaanxi Normal University. (Nat. Sci. Edition) 2012;40:67–72. [Google Scholar]
- 35.Stermer BA, Bianchini GM, Korth KL. Regulation of HMG-CoA reductase activity in plants. J. Lipid Res. 1994;35:1133–1140. [PubMed] [Google Scholar]
- 36.Song AA, et al. Overexpressing 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) in the lactococcal mevalonate pathway for heterologous plant sesquiterpene production. PLoS One. 2012;7:e52444. doi: 10.1371/journal.pone.0052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim YK, et al. Enhanced accumulation of phytosterol and triterpene in hairy root cultures of Platycodon grandiflorum by overexpression of Panax ginseng 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Agric. Food Chem. 2013;61:1928–1934. doi: 10.1021/jf304911t. [DOI] [PubMed] [Google Scholar]
- 38.Lange BM, Severin K, Bechthold A, Heide L. Regulatory role of microsomal 3-hydroxy-3-methylglutaryl-coenzyme A reductase for shikonin biosynthesis in Lithospermum erythrorhizon cell suspension cultures. Planta. 1997;204:234–241. doi: 10.1007/s004250050252. [DOI] [PubMed] [Google Scholar]
- 39.Brouquisse R, Gaudillere JP, Raymond P. Induction of a carbon-starvation-related proteolysis in whole maize plants submitted to Light/Dark cycles and to extended darkness. Plant Physiol. 1998;117:1281–91. doi: 10.1104/pp.117.4.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Araujo WL, et al. Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell. 2010;22:1549–1563. doi: 10.1105/tpc.110.075630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez A, Shin J, Davis SJ. Abiotic stress and the plant circadian clock. Plant Signal Behav. 2011;6:223–231. doi: 10.4161/psb.6.2.14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu F, et al. Molecular cloning, characterization and expression of phenylalanine ammonia-lyase gene from Ginkgo biloba. Afr. J. Biotechnol. 2008;7:721–729. [Google Scholar]
- 43.Xu F, et al. Molecular cloning and function analysis of an anthocyanidin synthase gene from Ginkgo biloba, and its expression in abiotic stress responses. Mol. Cells. 2008;26:536–547. [PubMed] [Google Scholar]
- 44.Xu F, et al. Isolation, characterization, and function analysis of a flavonol synthase gene from Ginkgo biloba. Mol. Biol. Rep. 2012;39:2285–2296. doi: 10.1007/s11033-011-0978-9. [DOI] [PubMed] [Google Scholar]
- 45.Kawoosa T, et al. Light and temperature regulated terpene biosynthesis: hepatoprotective monoterpene picroside accumulation in Picrorhiza kurrooa. Funct. Integr. Genomics. 2010;10:393–404. doi: 10.1007/s10142-009-0152-9. [DOI] [PubMed] [Google Scholar]
- 46.Sivanandhan G, et al. Increased production of withanolide A, withanone, and withaferin A in hairy root cultures of Withania somnifera (L.) Dunal elicited with methyl jasmonate and salicylic acid. Plant Cell Tissue Organ Cult. 2013;114:121–129. doi: 10.1007/s11240-013-0297-z. [DOI] [Google Scholar]
- 47.Manivannan A, Soundararajan P, Park YG, Jeong BR. Chemical elicitor-induced modulation of antioxidant metabolism and enhancement of secondary metabolite accumulation in cell suspension cultures of Scrophularia kakudensis Franch. Int. J. Mol. Sci. 2016;17:399. doi: 10.3390/ijms17030399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meng X, Song Q, Ye J, Wang L, Xu F. Characterization, function, and transcriptional profiling analysis of 3-Hydroxy-3-methylglutaryl-CoA synthase gene (GbHMGS1) towards stresses and exogenous hormone treatments in Ginkgo biloba. Molecules. 2017;22:1706. doi: 10.3390/molecules22101706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng X, et al. Isolation, characterization and functional analysis of a novel 3-hydroxy-3-methylglutaryl-coenzyme a synthase gene (GbHMGS2) from Ginkgo biloba. Acta Physiol. Plant. 2018;40:72. doi: 10.1007/s11738-018-2650-7. [DOI] [Google Scholar]
- 50.Kang MK, Nargis S, Kim SM, Kim SU. Distinct expression patterns of two Ginkgo biloba 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase/isopentenyl diphospahte synthase (HDR/IDS) promoters in Arabidopsis model. Plant Physiol. Bioch. 2013;62:47–53. doi: 10.1016/j.plaphy.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 51.Delaney TP, et al. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 52.Hancock JT, Neill SJ, Wilson ID. Nitric oxide and ABA in the control of plant function. Plant Sci. 2011;181:555–559. doi: 10.1016/j.plantsci.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 53.Moore KB, Oishi KK. 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in the endosperm of Maize vivipary mutants. Plant Physiol. 1994;105:119–125. doi: 10.1104/pp.105.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richings EW, Cripps RF, Cowan AK. Factors affecting ‘Hass’ avocado fruit size: Carbohydrate, abscisic acid and isoprenoid metabolism in normal and phenotypically small fruit. Physiol. Plant. 2000;109:81–89. doi: 10.1034/j.1399-3054.2000.100112.x. [DOI] [Google Scholar]
- 55.Jing F, et al. Abscisic acid (ABA) treatment increases artemisinin content in Artemisia annua by enhancing the expression of genes in artemisinin biosynthetic pathway. Biologia. 2009;64:319–323. doi: 10.2478/s11756-009-0040-8. [DOI] [Google Scholar]
- 56.Yang D, et al. PEG and ABA trigger methyl jasmonate accumulation to induce the MEP pathway and increase tanshinone production in Salvia miltiorrhiza hairy roots. Physiol. Plant. 2012;146:173–183. doi: 10.1111/j.1399-3054.2012.01603.x. [DOI] [PubMed] [Google Scholar]
- 57.Adie B, Chico JM, Rubio-Somoza I, Solano R. Modulation of plant defenses by ethylene. J. Plant Growth Regul. 2007;26:160–177. doi: 10.1007/s00344-007-0012-6. [DOI] [Google Scholar]
- 58.Lv DM, Zhang TT, Deng S, Zhang YH. Functional analysis of the Malus domestica MdHMGR2 gene promoter in transgenic Arabidopsis thaliana. Biol. Plantarum. 2016;60:667–676. doi: 10.1007/s10535-016-0637-z. [DOI] [Google Scholar]
- 59.Herrera, A., Rodrigo, M. J., Gil, J. & Zacarías, L. Ethylene stimulates emission of terpenoids and aliphatic esters in citrus fruits. [Ramina, A., Chang, C., Giavannoni, J., Klee, H., Perata, P. & Woltering, E. (ed.)] Advances In Plant Ethylene Research: Proceedings of the 7th International Symposium on the Plant Hormone Ethylene. 257–259 (Springer, 2007).
- 60.Sando T, et al. Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant, Hevea brasiliensis. J. Agric. Chem. Soc. Japan. 2008;72:2049–2060. doi: 10.1271/bbb.80165. [DOI] [PubMed] [Google Scholar]
- 61.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 62.Ganzera M, Zhao J, Khan IA. Analysis of terpenelactones in Ginkgo biloba by high performance liquid chromatography and evaporative light scattering detection. Chem. Pharm. Bull. (Tokyo) 2001;49:1170–1173. doi: 10.1248/cpb.49.1170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.