Abstract
Primary management for head and neck cancers, including squamous cell carcinoma (SCC), involves surgical resection with negative cancer margins. Pathologists guide surgeons during these operations by detecting cancer in histology slides made from the excised tissue. In this study, 381 digitized, histological whole-slide images (WSI) from 156 patients with head and neck cancer were used to train, validate, and test an inception-v4 convolutional neural network. The proposed method is able to detect and localize primary head and neck SCC on WSI with an AUC of 0.916 for patients in the SCC testing group and 0.954 for patients in the thyroid carcinoma testing group. Moreover, the proposed method is able to diagnose WSI with cancer versus normal slides with an AUC of 0.944 and 0.995 for the SCC and thyroid carcinoma testing groups, respectively. For comparison, we tested the proposed, diagnostic method on an open-source dataset of WSI from sentinel lymph nodes with breast cancer metastases, CAMELYON 2016, to obtain patch-based cancer localization and slide-level cancer diagnoses. The experimental design yields a robust method with potential to help create a tool to increase efficiency and accuracy of pathologists detecting head and neck cancers in histological images.
Subject terms: Oral cancer detection, Cancer imaging
Introduction
Head and neck cancer is the sixth most common cancer world-wide, and majority of cancers of the upper aerodigestive tract are squamous cell carcinoma (SCC)1. Approximately two-thirds of SCC patients present with either stage III or IV advanced disease2. Surgical cancer resection remains the primary management SCC of the head and neck, with concurrent chemotherapy or radiation therapy depending on the extent of the disease3.
Commonly, the safe margin for surgical resection of oral squamous cell carcinoma (SCC) at sites including the surfaces of the lips, gums, mouth, plate, and anterior two-thirds of the tongue is considered 5 mm from the permanent edge of the tumor4. Alternative distances for resection margins have been proposed, as low as tumor clearance of 2.2 mm to be declared a “negative” margin5. However, closer margins, for example within 1 mm, are associated with significantly increased recurrence rates6. In head and neck surgical histology, there are two techniques to investigate surgical margin clearance: perpendicular sectioning and en-face technique. Perpendicular sectioning, also known as “bread loafing”, allows the margin clearance from the edge of the resected tissue to be easily quantified, but it is resource exhaustive and limited by the number of slices, which can create false negatives. The en-face technique evaluates the surface area in a longitudinal fashion to determine if there is any cancer on surface of the submitted specimen4. Both require examining a large quantity of histological slides and considerable diagnostic time.
Previous studies have implemented computer-assisted detection methods using histological images for machine learning7. Colorectal epithelial and stromal tissues have been classified on histological images using support vector machines with hand-crafted features, such as color and texture8,9. Additionally, convolutional neural networks (CNNs), which are a family of machine learning algorithms that learn to extract features from training images, have also been applied to classifying epithelium and stromal tissues from colorectal and breast cancers10. Non-small cell lung cancers, including metastatic SCC to the lungs, have been classified in histological images using CNNs that are trained to work regions of the image, called image-patches11. Another method for detecting lung cancers in histological images of needle core biopsies used morphological and color features for classification with an ensemble of artificial neural networks12. Head and neck SCC was investigated once before, but only in cell lines xenografted into mice, and a CNN was implemented with histological images to predict hypoxia of tumor-invaded microvessels7,13. Additionally, computerized methods have been developed for thyroid carcinomas to detect and classify malignant versus benign nuclei from thyroid nodules and carcinomas, including follicular and papillary thyroid carcinomas, in histological images on a cellular level with promising results14–16. However, most of the work involving thyroid carcinoma has been implemented on a cellular or nuclear level using hand-crafted features, such as texture or shape, and support-vector-machines are employed for nuclei classification, with many algorithms using an ensemble of classifiers15–19.
In the field of digital pathology, whole slide imaging (WSI) refers to the acquisition of high-resolution images of stained tissue slides, which retains the ability to magnify and navigate these digital slides just as standard microscopy20. After reviewing nearly 2,000 patient cases, it has been concluded that WSI is non-inferior to microscopy for primary diagnosis in surgical pathology across multiple staining types, specimen types, and organ systems21. Computer-assisted detection algorithms have recently been implemented using CNNs for diagnosis in WSI with considerable success for identifying metastasis in lymph nodes22,23. Several state-of-the-art methods using CNNs have been applied during a grand challenge hosted at the IEEE International Symposium for Biomedical Imaging in 2016 and 2017 to detect breast cancer metastasis in WSI of sentinel lymph nodes (CAMELYON) with AUCs up to 0.99, comparable to expert pathologists performing with an AUC of 0.81 to 0.97, with and without a time constraint22,24,25.
This study aims to investigate the ability of CNNs for detecting head and neck SCC and thyroid carcinomas in a novel dataset of digitized whole-slide histological images from surgical pathology. A recent literature review shows that this is the first work to investigate SCC and thyroid carcinoma detection on a WSI level in primary head and neck cancers7, and we implement state-of-the-art classification methods in an extensive dataset collected from our institution. The major contribution of this paper focuses on the first application of deep learning for the histological detection of H&N SCC and thyroid cancers in a sufficiently large head and neck cancer dataset that is best suited for a patch-based CNN approach. The anatomical variation of the head and neck is astonishingly complex. The inclusion of multiple, most common locations of SCC yields a successful and substantial generalization for this application. Additionally, three of the major forms of thyroid carcinoma are studied, and despite extensive morphological differences, the method allows successful performance. Altogether, the dataset and applied methodology of this work demonstrate the current potential to create a tool to increase the efficiency and accuracy of surgical pathologists performing real-time SCC cancer detection on WSI for intraoperative guidance during primary head and neck cancer resection operations.
Materials and Methods
In this section, the materials for this study, including the cancer histological datasets, are described. Additionally, the methods of image processing, convolutional neural networks, and performance evaluation are detailed.
Head and neck cancer dataset
Informed, written consent was obtained from all patients consented for our study. All experimental methods were approved by the Institutional Review Board (IRB) of Emory University under the Head and Neck Satellite Tissue Bank (HNSB, IRB00003208) protocol. In collaboration with the Otolaryngology Department and the Department of Pathology and Laboratory Medicine at Emory University Hospital Midtown, freshly excised, ex-vivo head and neck cancer tissue samples were obtained from previously consented patients undergoing surgical cancer resection26,27. Tissue specimens collected from patients were de-identified and coded by a clinical research coordinator before being released to our laboratory for research purposes only. Three tissue samples were collected from each patient: a sample of the tumor, a normal tissue sample, and a sample at the tumor-normal interface.
For this study, we present the first application of the histological component of this dataset of 381 WSI from 156 patients, which is detailed by dataset in Table 1. In the upper aerodigestive tract SCC group, there were 228 tissue samples collected from 97 patients. The number of patients and tissue specimens is enumerated per anatomical origin of the SCC in Table 2. The only tissues that were excluded in this study were from three patients that had SCC of Waldeyer’s ring. These tissues were excluded because they were comprised of entirely lymphoid tissue, and the samples from only 3 patients of this diverse tissue type was not sufficient for inclusion in this study. The normal specimens collected were non-dysplastic and non-cancerous, which may have inflammation, atypia, or reactive epithelium.
Table 1.
Summary of the number of patients and whole-slide images (WSI) included in this study for training, validation, and testing of the proposed method.
| Dataset | Training | Validation | Testing | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Patients | WSI | Patients | WSI | Patients | WSI | Patients | WSI | |
| Head & Neck SCC | 45 | 91 | 13 | 32 | 39 | 105 | 97 | 228 |
| Thyroid Carcinoma | 24 | 48 | 8 | 23 | 27 | 82 | 59 | 153 |
| Breast Cancer Mets. | 250 | 250 | 20 | 20 | 129 | 129 | 399 | 399 |
Table 2.
Summary of the number of patients in the SCC dataset and WSI obtained from tissue specimens per anatomical location of the head and neck. Tissue specimens refer to the ex-vivo samples used to construct the histological WSI (T: tumor, N: normal, TN: tumor-normal interface).
| Location | # Patients | # T | # N | # TN |
|---|---|---|---|---|
| Tongue | 18 | 9 | 17 | 17 |
| FOM | 12 | 7 | 10 | 13 |
| Larynx | 10 | 10 | 9 | 3 |
| Mucosal Gingiva | 9 | 7 | 9 | 4 |
| Mandible | 8 | 6 | 6 | 5 |
| Maxillary Sinus | 6 | 4 | 6 | 5 |
| Oral Cavity | 6 | 4 | 7 | 6 |
| Hypopharynx | 5 | 4 | 5 | 1 |
| RMT | 5 | 6 | 3 | 5 |
| Tonsil | 5 | 4 | 3 | 6 |
| Supraglottis | 4 | 2 | 4 | 3 |
| BOT | 4 | 0 | 4 | 4 |
| Nasal Cavity | 2 | 2 | 1 | 0 |
| Other | 3 | 2 | 2 | 3 |
| Total | 97 Patients | 228 WSI | ||
The thyroid carcinoma group was comprised of primary papillary, medullary, and follicular thyroid carcinomas. There were 153 tissue specimens collected from 59 patients, which included 47 patients with papillary thyroid carcinoma, 5 patients medullary thyroid carcinoma, and 7 patients with follicular carcinoma. Each dataset was subdivided into separate groups for training, validation, and testing of the proposed computer-assisted cancer detection algorithm.
Fresh ex-vivo tissues were collected from the surgical pathology department and fixed, paraffin embedded, sectioned, stained with haemotoxylin and eosin (H&E), and digitized using whole-slide scanning at an equivalent magnification to 40x objective using a NanoZoomer (Hamamatsu Photonics), which produces a final digital slide with pixel-level resolution of 0.23 μm × 0.23 μm. A board-certified pathologist with expertise in H&N pathology outlined the cancer margins on the digital slides using Aperio ImageScope (Leica Biosystems Inc, Buffalo Grove, IL, USA).
Breast cancer lymph node metastases dataset
For external validation, we implemented the proposed cancer detection algorithm on the open-source CAMELYON 2016 dataset23,28, in order to compare the results of our proprietary head and neck cancer dataset since currently no similar independent dataset exists. The CAMELYON 2016 dataset consists of 399 whole-slide digital images from sentinel lymph nodes (SLN) obtained from 399 patients, one SLN from each patient that underwent breast cancer surgical resection. The dataset is collected at two institutions: Radboud University Medical Center (RUMC) Netherlands and University Medical Center Utrecht (UMCU) Netherlands23,28. One slide was constructed from one SLN from each patient. Table 1 shows the numbers of patients and slides in each group.
The whole-slide images were digitized at each institution separately, so the different hospitals each use a different scanner. The slides that were digitally scanned at RUMC were produced at 20x objective magnification using a Pannoramic 250 Flash II digital slide scanner (3DHISTECH), which corresponds to the pixel-level resolution of 0.24 μm × 0.24 μm. The slides that were digitized at UMCU were acquired with a NanoZoomer-XR digital slide scanner at 40x objective magnification (Hamamatsu Photonics) with a pixel-level resolution of specimens of 0.23 μm × 0.23 μm23,28.
Histological image processing
The histological dataset presented consists of primary tumor specimens acquired from surgical resections. Our SCC and thyroid cancer datasets do not have fine cellular-level annotations. Instead, regions were broadly marked as cancer if there were any cancer cells present, even if surrounded by normal structures, to establish which areas would require surgical removal. For this task, cell-by-cell annotations are not necessary. Clinicians require accurate regional diagnosis of cancer invaded tissues with an estimate of border clearance distance to the edge of the resected tissue. Therefore, the nature of the ground-truth for this work necessitates a patch-based deep learning approach. Moreover, a fully-convolutional network (FCN), as is widely used in the literature, would be problematic for this approach. Firstly, the tissue specimens of primary cancers collected tend to have large regions of each class. Therefore, the large majority of patches tend to be one class (all normal or all tumor), with few border patches that contain both classes. This would create problems with loss calculation and gradient optimization for training an FCN. Lastly, as stated the ground truth is coarse, so if a FCN could be adequately trained to produce fine-level segmentations, not only are they not needed for this task, but the ground truth would call potentially correct areas as misclassifications.
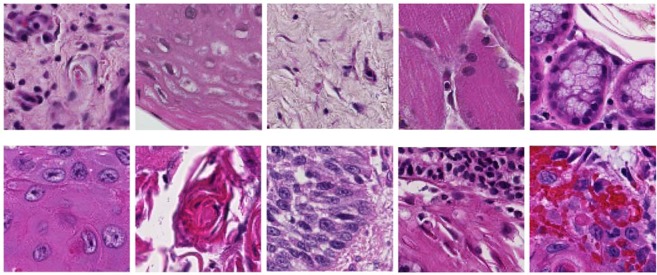
A ground-truth binary mask of the cancer area is produced from each outlined histology slide. The WSIs and corresponding ground-truths were down-sampled by a factor of four using nearest neighbor interpolation. The proposed method classifies the WSI in a patch-based method using a window that slides over the entire image. Due to the unique challenges of working with digital pathology images, which can create datasets of hundreds of images that are each tens of gigabytes, it is the current state-of-the-art to perform both down-sampling and patch-based image reconstruction approaches to computationally handle this type of data22,29–35. Image patches (I) are produced from each down-sampled H&E slide using 101 × 101 pixels and are labeled corresponding to the center pixel, where . Representative patches from H&N SCC are shown in Fig. 1 showing the histological variation of normal anatomical structures and various appearances of SCC of various identifiable difficulty. The SCC and thyroid carcinoma training groups were comprised of patches only from the tumor and normal tissue WSI, and the validation and testing groups were comprised of patches from all slides. Since the lymph node dataset contained more WSI but with smaller cancer areas, the training dataset was constructed by taking up to 5000 image patches from the cancerous area of each of the 101 cancer WSI in the training dataset, and using up to 1000 image patches from each slide of the 149 normal WSI. The training group was approximately balanced between cancer and normal patches for better performance.
Figure 1.
Histological images (101 × 101 pixel image-patches) showing anatomical diversity. Top: Patches of various normal structures, including chronic inflammation, stratified squamous epithelium, stroma, skeletal muscle, and salivary glands (from left to right). Bottom: Patches of SCC with varying histologic features: keratinizing SCC, keratinizing SCC with keratin pearls, basaloid SCC, SCC with chronic inflammation, SCC with hemorrhage (from left to right).
Histology slides have no canonical orientation, meaning the tissue will have the same diagnosis from all vantage points. Therefore, the number of image patches were augmented by 8x by applying 90-degree rotations and reflections to develop a more robust diagnostic method. Additionally, to establish a level of color-feature invariance and tolerance to differences in H&E staining between slides, the hue, saturation, brightness, and contrast of each patch were randomly manipulated to make a more rigorous training paradigm.
Convolutional neural network implementation
The three distinct cancer datasets in this study were employed to separately train, validate, and test a 2D-CNN classifier based on the Inception V4 architecture, implemented in TensorFlow on 8 Titan-XP NVIDIA GPUs36–39. The Inception V4 CNN architecture was modified slightly in the early layers, which is detailed in Table 3, in order to accommodate the patch-size selected for this study. The CNN architecture consisted of 3 convolutional layers and 1 max-pooling layer to accommodate the patch-size used, and in total the CNN contained 141 convolutional layers and 18 pooling layers37,39. Gradient optimization was performed using the Adadelta optimizer with an initial learning rate of 1.0 that was exponentially decayed by 0.95 every 3 epochs of training data40. The softmax cross entropy was used as the loss function. If the kth training patch is denoted as , , where K is the number of training patches in a batch, the CNN training process is to find a function that minimize the following cost function :
where
and and are the ground truth labels for cancer-negative and cancer-positive tissue classes, respectively, corresponding to the kth patch.
Table 3.
Schematic of the proposed modified Inception V4 CNN. The input size is given in each row, and the output size is the input size of the next row. All convolutions were performed with sigmoid activation and 40% dropout.
| Layer | Kernel size/Remarks | Input Size |
|---|---|---|
| Conv | 3 × 3/‘valid’ | 101 × 101 × 3 |
| Conv | 3 × 3/‘valid’ | 98 × 98 × 32 |
| Max Pool | 2 × 2/stride=2 ‘valid’ | 96 × 96 × 64 |
| Conv | 3 × 3/stride=2 ‘valid’ | 48 × 48 × 64 |
| 4 x Inception-A Block | 1 × 1 and 3 × 3/‘same’ | 23 × 23 × 80 |
| Reduction-A Block | 1 × 1 and 3 × 3/‘same’ | 23 × 23 × 384 |
| 7 × Inception Block | 1 × 1, 1 × 7, 7 × 1, and 3 × 3/‘same’ | 11 × 11 × 1024 |
| Reduction-B Block | 1 × 1, 1 × 7, 7 × 1, and 3 × 3/‘same’ | 11 × 11×1024 |
| 3 × Inception-C Block | 1 × 1, 1 × 3, 3 × 1, and 3 × 3/‘same’ | 5 × 5 × 1024 |
| Avg. Pool | 5 × 5/‘valid’ | 5 × 5 × 1536 |
| Linear | Logits | 1 × 1536 |
| Softmax | Classifier | 1 × 2 |
The validation groups were used to determine the optimal number of training epochs used for each of the three datasets. Each CNN was trained with a batch size of 128 image patches, and batches were converted from RGB to HSV before being passed into the CNN. Both RGB and HSV were tested in early validation experiments, and HSV without any other modification out-performed RGB results. One reason could be the separation of the image intensity from the color information in HSV color model. Additionally, one major challenge of H&E stained images is inconsistency of the stain quality. To demonstrate that color feature augmentation can solve this problem, working in HSV directly, the hue, saturation, and brightness were perturbed randomly in each channel independently. The SCC CNN was trained for 30 epochs of training data, equivalent to 295,000 steps using a batch-size of 128 patches. The random color augmentation was using the native color feature variance in the training group: hue 4%, saturation 15%, brightness 8%, and contrast 2%. The thyroid carcinoma CNN was trained for 70 epochs of training data (equivalent to 433,000 steps). HSV and contrast perturbation was 5%, 5%, 8%, and 5% respectively. The breast cancer SLN metastasis CNN was trained for 20 epochs (equivalent to 203,400 steps). HSV and contrast were each randomly perturbed in range of −10% to 10%.
Image reconstruction and post processing
Each of the N testing slides () were tiled into image patches of size 101 × 101 pixels (), produced with a stride of 50 pixels, which makes an overlap of 51 pixels.
where , , and Mt × Nt is the size of the tth testing slide. Therefore, the final classified whole-slide image is a cancer probability heat-map with a level of detail equal to 50 × 50 pixel blocks. Each image patch, , was classified in all 8 orientations with randomized HSV and contrast features and averaged to obtain a single cancer prediction value, according to the following equation.
where is the cancer prediction for the sth orientation of and is the average cancer prediction of the patch. Additionally, the results of overlapping image patches were averaged in the overlapping area, as follows.
where is the final probability of the final resolution block size () of the heat-map. The benefit of this post-processing method was to increase the resolution of the heat-map from 101-pixel image patches to 50-pixel image patches. Moreover, the image patches that constituted the free edge of the tissue were averaged less than four times because they did not have the complete number of neighboring patches. This image reconstruction and post-processing method was determined to increase accuracy by about 2% in early validation experiments.
To investigate the ability of the CNN to detect cancer on histological images, we implemented the gradient class-activated map (grad-CAM) method to visualize gradients activated by each class for the example input image patches41. We traced the gradients from the last convolutional layer before the inception modules to the logits layer to separately visualize cancer and normal components. This technique produces a weighted combination of the convolutional filters and gradients as the CNN is activated by a specific input image for each class.
Performance evaluation
The reference standard cancer margin was annotated by hand for all digital slides employed in this study. For the head and neck cancer database, a board-certified pathologist with expertise in H&N pathology outlined the cancer margins on the digital slides. For the breast cancer metastasis database, an experienced lab technician and a clinical Ph.D. student outlined the cancer margins, which were then confirmed by one of two board-certified pathologists with expertise in breast cancer28.
During training, the performance of the validation group was calculated and monitored. The optimal operating threshold was calculated from the validation group for generalizable results, and it was used for generating performance evaluation metrics for the testing group. To reduce bias in the experiment, the fully-independent testing group was only classified a single time at the end of the experiment, after all the network optimizing had been determined using the validation set. To test the ability to diagnose and localize cancer on WSI, we used AUC, F1 score, accuracy, sensitivity, and specificity to evaluate cancer detection on a patch-based level. Confidence intervals were calculated using a boot-strapping algorithm. Additionally, the ability of the proposed algorithm to diagnose slides with cancer from normal slides was investigated. This slide-level AUC was calculated by assigning the value of the image patch with the maximum cancer probability to the entire WSI.
Informed consent
Informed written consent was obtained from all patients prior to participation in this study. All methods were carried out in accordance with the approved Institutional Review Board protocols and the relevant guidelines and regulations of Emory University.
Results
Head and neck primary SCC was detected on digitized WSI with an AUC of 0.916 and 85% accuracy for patients in the testing group. The ideal threshold for distinguishing SCC from normal tissue was SCC probability of greater than 62%. Thyroid carcinoma was detected on digitized WSI with an AUC of 0.954 and 89% accuracy for patients in the testing group. The ideal threshold for distinguishing thyroid carcinoma from normal thyroid tissue was cancer probability of greater than 50%. Breast cancer lymph node metastasis was detected on digitized WSI with an AUC of 0.967 and 93% accuracy for patients in the testing group. The ideal threshold for identifying metastasis in SLNs was cancer probability of greater than 28%. Reported in Table 4 are the AUC for the validation groups and the AUC, accuracy, sensitivity, and specificity of the testing groups.
Table 4.
Cancer detection results, obtained from ROC curves using all histological images’ patch-level statistics. Reported are the AUC for the validation group and the AUC, F1 score, accuracy, sensitivity, and specificity of the testing group with 95% confidence intervals for all values. Also shown in the right-most column is the ability of the proposed method to distinguish slides that contain cancer from slides that are all normal.
| Group | Validation AUC | Test AUC | F1 Score | Accuracy | Sensitivity | Specificity | Slide Level AUC |
|---|---|---|---|---|---|---|---|
| SCC | 0.913 (0.90,0.93) | 0.916 (0.90, 0.93) | 84.8 ± 1.5% | 84.8 ± 1.6% | 84.7 ± 2.2% | 85.0 ± 2.2% | 0.944 (0.91, 0.97) |
| Thyroid | 0.927 (0.92, 0.94) | 0.954 (0.94, 0.97) | 89.4 ± 1.3% | 89.4 ± 1.3% | 89.6 ± 1.8% | 89.1 ± 1.9% | 0.995 (0.99, 1.00) |
| Lymph Node | 0.986 (0.96, 0.99) | 0.967 (0.96, 0.98) | 91.8 ± 1.3% | 93.4 ± 1.2% | 90.1 ± 1.8% | 93.6 ± 1.6% | 0.901 (0.86, 0.94) |
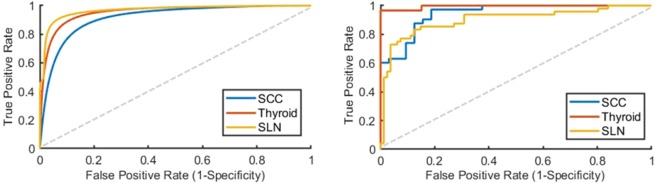
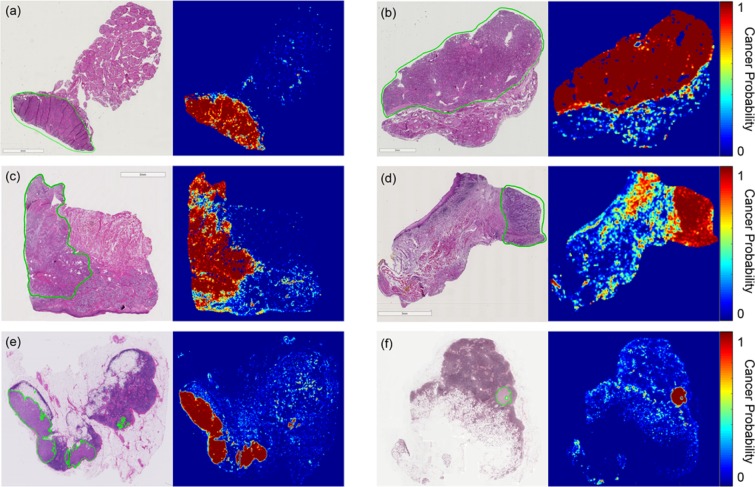
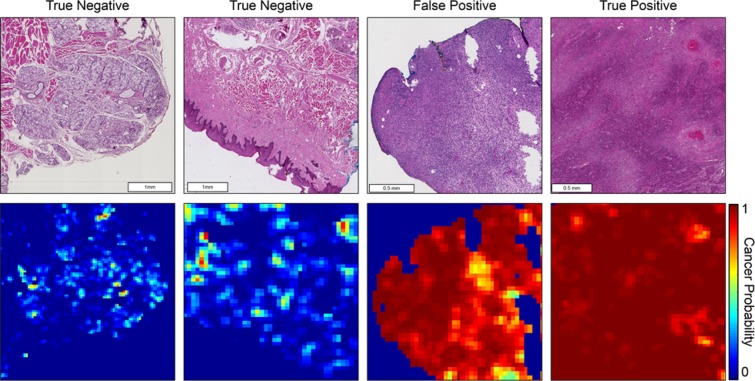
Receiver operator characteristic (ROC) curves for slide-level and patch-level cancer detection in the testing groups from all three datasets are shown in Fig. 2. Patch-level ROC curves are generated using all histological images’ patch-level data for cancer localization, and slide-level ROC curves demonstrate WSI diagnosis. Additionally, two representative WSI from each of the three testing groups and their corresponding predicted heat-maps are shown in Fig. 3. Several regions of interest (ROI) are detailed in Fig. 4 to identify the strengths and weaknesses of the proposed method in the detection of SCC. The ideal threshold for whole-slide level detection of SCC was above 95% probability, so the regions detailed as true negatives in Fig. 4 fall below this threshold. Additionally, the grad-CAM technique was used to visualize the contributing normal and cancerous components of a few example input images that were corrected classified with high probability (Fig. 5). This approach reveals that a contribution of the cancer prediction is made by nuclear features.
Figure 2.
The ROC curves are shown for patch-level cancer detection (left) and slide-level cancer diagnosis (right) in the testing groups from all three datasets. The dotted gray line corresponds to random guess.
Figure 3.
Representative whole-slide classification results. (a,b) Papillary thyroid carcinoma WSI from two patients. (c,d) SCC WSI from patients with tongue and retromolar trigone SCC. (e,f) Breast cancer metastasis to lymph node WSI from two patients. The cancer area is outlined in green on the H&E images, and the heat maps are shown of the cancer probability.
Figure 4.
Heat maps representing cancer probability of several regions of interest. From left to right, the CNN correctly identifies salivary gland and muscular components as having a low probability of SCC; stratified squamous epithelium correctly shown as a true negative; a false positive area representing inflammatory infiltration near the SCC border (not shown); correctly classified true positive SCC classified with a high probability of SCC.
Figure 5.
Representative, correctly-classified cancer image-patches from each dataset, visualized with cancer and normal components using the grad-CAM technique41. The gradients were traced from the last convolutional layer before the inception modules to the cancer and normal logits separately to visualize contributions from cancer and normal features leading to a correct cancer prediction. (a) Breast cancer SLN metastasis image patch correctly classified as cancer with 58% probability. (b) Papillary thyroid carcinoma image patch correctly classified as cancer with 89% probability. (c) SCC image patch correctly classified as cancer with 99% probability.
The ability of the proposed method to diagnose the entire WSI that contain any cancer was also investigated. WSIs with SCC were diagnosed with an AUC of 0.944. Thyroid carcinoma WSIs were diagnosed with an AUC of 0.995. WSI of SLN with breast cancer metastases were diagnosed with an AUC of 0.901.
Discussion
In this work, we present a new and extensive histological dataset of primary head and neck cancer and implement a state of the art Inception V4 CNN architecture for cancer detection and WSI diagnosis. The results are generalizable because of the division of patients across training, validation, and testing. To the best of our knowledge, this is the first work to investigate SCC detection in digitized whole-slide histological images from primary head and neck cancers.
The digitized, whole-slide histological images were saved as TIF files with resolution equivalent to 40x microscopic objective. After 4x down-sampling, the image patches correspond to 10x objective equivalence. Different down-sampling factors and patch-sizes were explored, but this method yielded the best validation group results, so it was used for testing. Similarly, pathologists detecting SCC in histology slides use a variety of objectives, not exclusively 40x, which may be too zoomed-in to determine if the region is cancerous or benign. We see this issue in our dataset as well. It is not only possible, but likely that in some slides labeled as ‘tumor only’, there may be some areas inside the tissue, or in between tumor nests, that is entirely normal. Therefore, it is understandable that classification using 4x down-sampled images obtains the highest accuracy. Other CNN architectures were explored in early experiments using the validation set only, and various patch-sizes were experimented with, but ultimately the Inception V4 CNN architecture with a patch-size of 101 × 101 × 3 in HSV color space, yielded the most promising validation results.
Additionally, the regions of interest that are presented show true negative, false positive, and true positive regions that vary from 1 to 3 mm in size. These results demonstrate the proposed method is able to distinguish normal anatomical structures like epithelium and salivary gland from SCC with high probability. Also, the most common false positive observed in the classified result is tissue areas that contain dense inflammation. This result is most likely a by-product of the training paradigm. As SCC develops, there is an accompanying immune response that leads to massive inflammatory infiltration into the tissue42. Therefore, the proposed algorithm learned the association between SCC and inflammation.
To our knowledge, there are no other studies that attempt to detect or diagnose H&N SCC or thyroid carcinoma on WSI, and we used a proprietary dataset collected from patients at our institution. Therefore, we wanted to test the proposed, diagnostic algorithm on a similar, open-source dataset for comparison. Our slide-level results would have placed 3rd in the original CAMELYON 201623,28.
The grad-CAM technique was used to visualize what components of the input image are determined as useful features with a significant contribution to the cancer prediction from the CNN, as shown in Fig. 5. This reveals that the decision is made by looking at the nuclei, just like a pathologist detects cancer. The proposed method does not segment all cancerous nuclei in the image patch, but it identifies a few cancerous nuclei with a high probability of being cancer and uses this information for making the decision. We did not train the proposed algorithm specifically with this in mind. Rather, this phenomenon was learned naturally by the training paradigm. The trained CNN model also has a level of stain invariance.
Limitations
One limitation of the presented approach results from the application of the down-sampled resolution and the patch-based Inception V4 CNN implementation. After down-sampling, each pixel represents approximately 0.91 microns, which produces a patch size that spans about 92 microns in each x-y dimension for the patch size of 101 × 101 that was implemented in this approach. The typical diameter of an SCC single-cell nucleus in our dataset was about 12 microns, which agrees with values reported in the literature of about 13 +/−2 microns43. Therefore, the theoretical limit of the smallest carcinoma that could be detected would be a nest of SCC cells with an approximate diameter of 92 microns. This value corresponds with an SCC nest on the order of tens of cells of SCC, depending on the cytoplasmic overlap in the arrangement of the SCC nest.
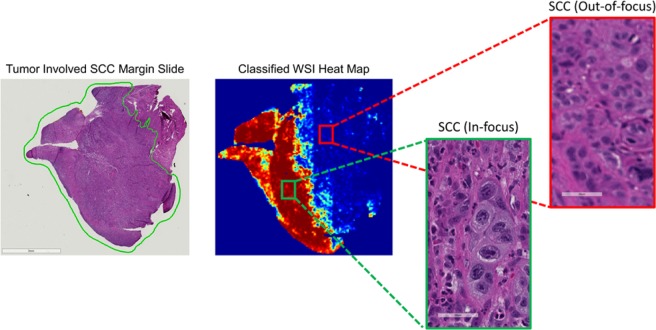
Another limitation of this approach was that the algorithm suffered from whole-slide scanning artifacts, such as out-of-focus regions and including errors from slide processing, such as tissue folding and tearing. This was discovered after the completion of the experiment, and the effect was substantial, accounting for the reason in misclassification of the lowest performing WSI in the SCC testing dataset, which is shown in Fig. 6. As can been in Fig. 6, the left side of the WSI is classified correctly as a true positive SCC region, but the out-of-focus regions result in misclassification a similar ROI (shown in the cut-out boxes on the right) to be classified as false negative incorrect result. These misclassifications were retained in the testing dataset to not manipulate or bias the results, but in future work, slide scanning artifact detection should be additionally performed to determine which slides cannot be classified because of limited quality.
Figure 6.
Representative false negative resulting from slide scanning artifacts. This accounts for the misclassification result of the lowest performing WSI in the SCC test group. The left side of the WSI was a correct true positive classification (green outlined box), but the out of focus artifact results in false negative misclassification (red outlined box).
Digital pathology with WSI allows pathologists to view high-resolution histological images, just as standard microscopy, and it was concluded that digital pathology is non-inferior to microscopy for primary diagnosis in surgical pathology cases across multiple institutions, staining types, and organ systems20,21. Therefore, we believe the robust experimental procedure of the proposed method, designed to eliminate bias, has demonstrated potential benefit in a modern, digitized clinical setting. However, primary diagnosis of surgical specimens for intraoperative guidance is performed on frozen-sections rather than formalin-fixed, paraffin embedded tissues, as were investigated in this study. Additionally, frozen-sections are typically lower quality than those created from fixed, embedded specimens because they suffer from many different artifacts and depend heavily on the skill of the operator. Therefore, we believe the presented work demonstrates potential for clinical benefit, but more investigation needs to be performed. Moreover, the generalization of the results beyond head and neck cancers to breast cancer metastasis in sentinel lymph nodes suggests this method is not limited to any organ system and could be adapted to serve multiple purposes if implemented in a more clinical setting.
Conclusion
In summary, this work focuses on the first application of deep learning for the histological detection of H&N SCC and thyroid cancers. The proposed method is able to detect and localize primary head and neck SCC on whole-slide, digitized histological images with an AUC of 0.916 for patients in the SCC testing group and 0.954 for patients in the thyroid carcinoma testing group. Moreover, the proposed method is able to discriminate WSI with cancer versus normal slides with an AUC of 0.944 and 0.995 for the SCC and thyroid carcinoma testing groups, respectively. The SCC detection method is performed across all anatomical locations, which indicates the algorithm is not limited to one location of the head and neck anatomy. For thyroid cancers, three major thyroid carcinoma are studied together which additionally demonstrates the generalizability of the method. For external validation, we tested the proposed method on an open-source dataset, CAMELYON 2016, and obtained good results. The agreement between validation and testing demonstrate that the technique is generalizable due to the robustness of the training paradigm and the careful experimental design to reduce bias. Together, the novel application to our dataset and promising results of this work demonstrate potential that such methods as the one proposed could help create a tool to increase efficiency and accuracy of pathologists performing head and neck cancer detection on histological slides for intraoperative guidance during head and neck cancer resection operations.
Acknowledgements
The authors would like to thank the surgical pathology team at Emory University Hospital Midtown for their help in collecting fresh tissue specimens. This research was supported in part by the U.S. National Institutes of Health (NIH) grants (R21CA231911, R01CA156775, R01CA204254, and R01HL140325). The project was also supported in part by an Early Translational Research Award (RP190588) from the Cancer Prevention and Research Institute of Texas (CPRIT). The authors thank the members of the Quantitative Bioimaging Laboratory (QBIL, www.fei-lab.org) for their helps.
Author Contributions
M.H. designed and performed the experiments, analyzed the results, and wrote the manuscript. M.S. provided software for image pre-processing and helped design the experiments. J.V.L. performed annotation of histological data. J.V.L., A.Y.C., B.S. and L.M. were involved with clinical input and study design. B.F. is the PI of the project and supervised the work.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Joseph LJ, et al. Racial disparities in squamous cell carcinoma of the oral tongue among women: a SEER data analysis. Oral Oncol. 2015;51(6):586–92. doi: 10.1016/j.oraloncology.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 2.Yao M, et al. Current surgical treatment of squamous cell carcinoma of the head and neck. Oral Oncol. 2007;43(3):213–23. doi: 10.1016/j.oraloncology.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 3.Kim, B. Y. et al. Prognostic factors for recurrence of locally advanced differentiated thyroid cancer. J Surg Oncol, 116(7) (2017). [DOI] [PubMed]
- 4.Baddour HM, Jr., Magliocca KR, Chen AY. The importance of margins in head and neck cancer. J Surg Oncol. 2016;113(3):248–55. doi: 10.1002/jso.24134. [DOI] [PubMed] [Google Scholar]
- 5.Zanoni D, et al. A proposal to redefine close surgical margins in squamous cell carcinoma of the oral tongue. JAMA Otolaryngol Head Neck Surg. 2017;143(6):555–560. doi: 10.1001/jamaoto.2016.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tasche, K. K. et al. Definition of “close margin” in oral cancer surgery and association of margin distance with local recurrence rate. JAMA Otolaryngol Head Neck Surg (2017). [DOI] [PMC free article] [PubMed]
- 7.Fuyong X, et al. Deep Learning in Microscopy Image Analysis: A Survey. IEEE Trans Neural Netw Learn Syst. 2018;29(10):4550–4568. doi: 10.1109/TNNLS.2017.2766168. [DOI] [PubMed] [Google Scholar]
- 8.Linder Nina, Konsti Juho, Turkki Riku, Rahtu Esa, Lundin Mikael, Nordling Stig, Haglund Caj, Ahonen Timo, Pietikäinen Matti, Lundin Johan. Identification of tumor epithelium and stroma in tissue microarrays using texture analysis. Diagnostic Pathology. 2012;7(1):22. doi: 10.1186/1746-1596-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianconi F, Álvarez-Larrán A, Fernández A. Discrimination between tumour epithelium and stroma via perception-based features. Neurocomputing. 2015;154:119–126. doi: 10.1016/j.neucom.2014.12.012. [DOI] [Google Scholar]
- 10.Xu J, et al. A Deep Convolutional Neural Network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing. 2016;191:214–223. doi: 10.1016/j.neucom.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou, L. A. S. et al. Patch-based Convolutional Neural Network for Whole Slide Tissue Image Classification, Proc IEEE Conf Comp Vis Pattern Recogn (2016). [DOI] [PMC free article] [PubMed]
- 12.Zhou Z-H, et al. Lung cancer cell identification based on artificial neural network ensembles,”. Artif Intell Med. 2002;24(1):25–36. doi: 10.1016/S0933-3657(01)00094-X. [DOI] [PubMed] [Google Scholar]
- 13.Carneiro, G. et al. Weakly-Supervised Structured Output Learning with Flexible and Latent Graphs Using High-Order Loss Functions. IEEE International Conference on Computer Vision (ICCV), 648–656, 10.1109/ICCV.2015.81, (2015).
- 14.Kim, E., Baloch, Z. & Kim, C. Computer assisted detection and analysis of tall cell variant papillary thyroid carcinoma in histological images. Medical Imaging: Digital Pathology,9420(8), 94200A, 10.1117/12.2082156, (2015).
- 15.Angel J, Rajam MA. Automatic classification of thyroid histopathology images using multi-classifier system. Multimed Tools Appl. 2017;76(18):18711–18730. doi: 10.1007/s11042-017-4363-0. [DOI] [Google Scholar]
- 16.Wang W, Ozolek JA, Rohde GK. Detection and classification of thyroid follicular lesions based on nuclear structure from histopathology images. Cytometry A. 2010;77A(5):485–494. doi: 10.1002/cyto.a.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gopinath B, Shanthi N. Computer-aided diagnosis system for classifying benign and malignant thyroid nodules in multi-stained FNAB cytological images. Australas Phys Eng Sci Med. 2013;36(2):219–230. doi: 10.1007/s13246-013-0199-8. [DOI] [PubMed] [Google Scholar]
- 18.Daskalakis A, et al. Design of a multi-classifier system for discriminating benign from malignant thyroid nodules using routinely H&E-stained cytological images. Comput Biol Med. 2008;38(2):196–203. doi: 10.1016/j.compbiomed.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Ozolek JA, et al. Accurate diagnosis of thyroid follicular lesions from nuclear morphology using supervised learning. Med Image Anal. 2014;18(5):772–780. doi: 10.1016/j.media.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantanowitz L, et al. Validating Whole Slide Imaging for Diagnostic Purposes in Pathology: Guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med. 2013;137(12):1710–1722. doi: 10.5858/arpa.2013-0093-CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukhopadhyay S, et al. Whole Slide Imaging Versus Microscopy for Primary Diagnosis in Surgical Pathology: A Multicenter Blinded Randomized Noninferiority Study of 1992 Cases (Pivotal Study) Am J Surg Path. 2018;42(1):39–52. doi: 10.1097/PAS.0000000000000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yun Liu, K. G. et al. Detecting Cancer Metastases on Gigapixel Pathology Images, arXiv: Computational Research Repository, abs/1703.02442 (2017).
- 23.Bejnordi BE, et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA. 2017;318(22):2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang AKD, Gargeya R, Irshad H, Beck AH. Deep Learning for Identifying Metastatic Breast Cancer. arXiv: Computational Research Repository. 2016;1606:05718. [Google Scholar]
- 25.de With PHN, Zinger S, Zanjani FG. Cancer detection in histopathology whole-slide images using conditional random fields on deep embedded spaces. Proc SPIE Int Soc Opt Eng. 2018;2015810I:17. [Google Scholar]
- 26.Fei B, et al. Label-free reflectance hyperspectral imaging for tumor margin assessment: a pilot study on surgical specimens of cancer patients. J Biomed Opt. 2017;22(8):7. doi: 10.1117/1.JBO.22.8.086009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halicek M, et al. Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging. J Biomed Opt. 2017;22(6):60503. doi: 10.1117/1.JBO.22.6.060503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litjens, G. et al. 1399 H&E-stained sentinel lymph node sections of breast cancer patients: the CAMELYON dataset. Gigascience7(6) (2018). [DOI] [PMC free article] [PubMed]
- 29.Bulten W, et al. Epithelium segmentation using deep learning in H&E-stained prostate specimens with immunohistochemistry as reference standard. Sci Rep. 2019;9(1):864. doi: 10.1038/s41598-018-37257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruz-Roa A, et al. Accurate and reproducible invasive breast cancer detection in whole-slide images: A Deep Learning approach for quantifying tumor extent. Sci Rep. 2017;7:46450. doi: 10.1038/srep46450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gertych A, et al. Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci Rep. 2019;9(1):1483. doi: 10.1038/s41598-018-37638-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen L, et al. Spatial Statistics for Segmenting Histological Structures in H&E Stained Tissue Images. IEEE Trans Med Imaging. 2017;36(7):1522–1532. doi: 10.1109/TMI.2017.2681519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sirinukunwattana K, et al. Locality Sensitive Deep Learning for Detection and Classification of Nuclei in Routine Colon Cancer Histology Images. IEEE Trans Med Imaging. 2016;35(5):1196–1206. doi: 10.1109/TMI.2016.2525803. [DOI] [PubMed] [Google Scholar]
- 34.Tellez, D. et al. Whole-Slide Mitosis Detection in H&E Breast Histology Using PHH3 as a Reference to Train Distilled Stain-Invariant Convolutional Networks. IEEE Trans Med Imaging (2018). [DOI] [PubMed]
- 35.Wei JW, et al. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci Rep. 2019;9(1):3358. doi: 10.1038/s41598-019-40041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abadi, M. et al. TensorFlow: Large-scale machine learning on heterogeneous systems, https://www.tensorflow.org (2015).
- 37.Szegedy SIC, Vanhoucke V. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. arXiv: Computational Research Repository. 2016;1602:07261. [Google Scholar]
- 38.Szegedy SIC, Vanhoucke V, Shlens J, Wojna Z. Rethinking the Inception Architecture for Computer Vision. arXiv: Computational Research Repository. 2015;1512:00567. [Google Scholar]
- 39.Szegedy C, et al. Going deeper with convolutions. arXiv: Computational Research Repository. 2014;1409:4842. [Google Scholar]
- 40.Zeiler MD. ADADELTA: An adaptive learning rate method. arXiv: Computational Research Repository. 2012;121:5701. [Google Scholar]
- 41.Selvaraju, R. R. et al. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization, Proc IEEE Int Conf Comput Vis, 618–626 (2017).
- 42.Gasparoto TH, et al. Inflammatory events during murine squamous cell carcinoma development. J Inflamm. 2012;9(1):46–46. doi: 10.1186/1476-9255-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee TK, et al. Overlap of nuclear diameters in lung cancer cells. Anal Quant Cytol Histol. 1990;12(4):275–8. [PubMed] [Google Scholar]