Herpesviruses are an important socioeconomic burden for both humans and livestock. Throughout their long evolutionary history, individual herpesvirus species have developed remarkable host specificity, while collectively the Herpesviridae family has evolved to infect a large variety of eukaryotic hosts. The development of approaches to fight herpesvirus infections has been hampered by the complexity of herpesviruses’ genomes, proteomes, and structural features. The data and insights generated by our study add to the understanding of the functional organization of herpesvirus-encoded proteins, specifically of family-wise conserved features defining essential components required for a productive infectious cycle across different hosts, which can contribute toward the conceptualization of antiherpetic infection strategies with an effect on a broader range of target species. All of the generated data have been made freely available through our HVint2.0 database, a dedicated resource of curated herpesvirus interactomics purposely created to promote and assist future studies in the field.
KEYWORDS: herpesviruses, intraviral network, protein-protein interactions, systems biology
ABSTRACT
Protein interactions are major driving forces behind the functional phenotypes of biological processes. As such, evolutionary footprints are reflected in system-level collections of protein-protein interactions (PPIs), i.e., protein interactomes. We conducted a comparative analysis of intraviral protein interactomes for representative species of each of the three subfamilies of herpesviruses (herpes simplex virus 1, human cytomegalovirus, and Epstein-Barr virus), which are highly prevalent etiologic agents of important human diseases. The intraviral interactomes were reconstructed by combining experimentally supported and computationally predicted protein-protein interactions. Using cross-species network comparison, we then identified family-wise conserved interactions and protein complexes, which we defined as a herpesviral “central” intraviral protein interactome. A large number of widely accepted conserved herpesviral protein complexes are present in this central intraviral interactome, encouragingly supporting the biological coherence of our results. Importantly, these protein complexes represent most, if not all, of the essential steps required during a productive life cycle. Hence the central intraviral protein interactome could plausibly represent a minimal infectious interactome of the herpesvirus family across a variety of hosts. Our data, which have been integrated into our herpesvirus interactomics database, HVint2.0, could assist in creating comprehensive system-level computational models of this viral lineage.
IMPORTANCE Herpesviruses are an important socioeconomic burden for both humans and livestock. Throughout their long evolutionary history, individual herpesvirus species have developed remarkable host specificity, while collectively the Herpesviridae family has evolved to infect a large variety of eukaryotic hosts. The development of approaches to fight herpesvirus infections has been hampered by the complexity of herpesviruses’ genomes, proteomes, and structural features. The data and insights generated by our study add to the understanding of the functional organization of herpesvirus-encoded proteins, specifically of family-wise conserved features defining essential components required for a productive infectious cycle across different hosts, which can contribute toward the conceptualization of antiherpetic infection strategies with an effect on a broader range of target species. All of the generated data have been made freely available through our HVint2.0 database, a dedicated resource of curated herpesvirus interactomics purposely created to promote and assist future studies in the field.
INTRODUCTION
Herpetic infections are ubiquitous and highly prevalent that affect a wide range of eukaryotic organisms. In humans and livestock, they are often associated with mild symptoms, yet they can develop into severe conditions through the lack of appropriate treatment or a weak host immune response. Serious conditions associated with these diseases include encephalitis, neurosensory birth defects, and several types of cancer (1). As a consequence, they represent an important socioeconomic burden (2–4). Despite the wide range of organisms that herpesviruses, as a family, infect, each species tends to show a high host specificity. This is the result of millions of years of subtly tuned coevolution with their respective hosts (1).
The human herpesviruses are a subgroup of currently nine known herpesvirus species that recurrently infect humans as their natural host and with which they are estimated to have been coevolving for over 400 million years (5). The nine species are found across the three subfamilies in which the Herpesviridae family is divided. Herpes simplex virus 1 (HSV-1, or human alphaherpesvirus 1 [HHV-1]), HSV-2 (or HHV-2), and varicella-zoster virus (VZV, or HHV-3) are members of the Alphaherpesvirinae subfamily. Human cytomegalovirus (HCMV, or human betaherpesvirus 5 [HHV-5]), human betaherpesviruses 6A and 6B (HHV-6A and HHV-6B, respectively), and human betaherpesvirus 7 (HHV-7) belong to the Betaherpesvirinae subfamily. Epstein-Barr virus (EBV, or human gammaherpesvirus 4 [HHV-4]) and Kaposi’s sarcoma-associated herpesvirus (KSHV, or human gammaherpesvirus 8 [HHV-8]) are part of the Gammaherpesvirinae subfamily. These nine species show different cell tropisms, and their infections result in distinct symptomatologies (1). As in other organisms, the great majority of these phenotypical features are not mediated by the product of one single gene but a number of them and, importantly, their physical and functional interactions. As a consequence, intraspecies interactions among virally encoded proteins have attracted interest over the years (6–9). However, comprehensive nonredundant compilations of these data are still largely missing.
Previously, we developed HVint (10), a centralized resource of curated protein-protein interaction (PPI) data for HSV-1, the prototypical species among human herpesviruses. The development of an improved bioinformatics protocol for PPI network inference led to the construction of our new database, HVint2.0, also dedicated to PPIs in HSV-1 (11). Here, we introduce the extension of HVint2.0 to contain curated PPI data across representative species of the Herpesviridae family. To this end, we applied our recently developed bioinformatics PPI network reconstruction pipeline (11) to generate PPIs for HCMV and EBV. Taking advantage of the family-wise representation of PPI networks, we inferred a conserved “central” interactome of the Herpesviridae family. This reconstructed central interactome portrays a large number of well-known conserved protein complexes within the family, which we deem indicative of its biological coherence. Coupling the information extracted from subfamily-specific intraviral networks and the central interactome could become a useful tool to assist experimental interactomics studies and reveal new insightful knowledge on the evolutionary dynamics of the family and identify system-level patterns that can explain some of the different phenotypical features observed across species.
RESULTS
HVint2.0 database.
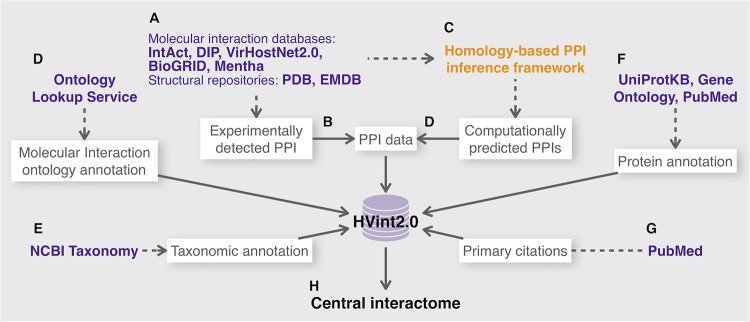
To populate the HVint2.0 database (Fig. 1; see Fig. S1 in the supplemental material), we generated curated compilations of PPI data for human herpesviruses HSV-1, HCMV, and EBV, representative members of the Alpha-, Beta-, and Gammaherpesvirinae subfamilies, respectively, using our recently described computational network reconstruction framework (11). In brief, our framework (see Materials and Methods and see Fig. S2 in the supplemental material) integrates, for each of these three (target) species, experimentally obtained evidence and computational PPI predictions (inferred using a conservative sequence similarity-based approach). The framework also integrates a refined scoring function that assesses the confidence of each interaction, taking into account the heterogeneity of its cumulative supporting evidence. The heterogeneity present among experimentally obtained supporting evidence is accounted for using the MIscore function (12). For those interactions that are computationally inferred using homology, we include an additional penalizing scaling factor, which is dependent on the family-wise conservation of the PPI: i.e., the wider the conservation of a PPI across the family, the higher the confidence score of the prediction. Below, for each reconstructed network, experimentally supported PPIs are referred to as “ePPIs,” computationally predicted ones as “pPPIs,” and those PPIs that had both computational and experimental support as ePPIs ∩ pPPIs.
FIG 1.
HVint2.0 data integration framework. Blue text indicates external resources (e.g., databases and Web servers), orange indicates data transformations (e.g., interaction prediction from raw input data), and text in white boxes refers to the data sets collated from these external resources. (A) Experimentally supported PPI evidence is collated from public repositories. (B to D) Experimentally supported PPIs are used as direct PPI input to HVint2.0 and to infer further PPIs based on sequence homology, which are then further introduced into HVint2.0. (F and G) From the resulting curated compilations, all corresponding unique identifiers for PSI-MI annotation codes, taxonomic ranks, protein species, and primary citations are collected, and their corresponding annotation data are included in HVint2.0. (H) The family-wise PPI data contained in HVint2.0 were used in this study to generate a candidate family-wise conserved interactome.
Physical schema of the HVint2.0 database. The data in HVint2.0 are organized into seven tables: i.e., “ppi,” “protein,” “evidence,” “evidence_to_psimi,” “psimi_ontology,” “taxon,” and “citation.” The content of each of table is detailed in the accompanying portion of Text S1. For each table, the figure displays the table name and the list of columns in the table, together with their data type. Primary keys are indicated with a key icon next to the corresponding column name; columns that can and cannot hold null values are indicated with empty and filled rhomboid icons, respectively. Download FIG S1, TIF file, 0.5 MB (505.3KB, tif) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Network reconstruction framework. (A) PPI data are collected from several public data resources and for the taxons of interest. These data include PPIs in the target taxon and in a number of orthologous species. (B) PPIs detected in orthologous species are the input for stage, where sequence-based homology assignments are used to predict new PPIs in the target taxon (green enlarged frame). (C) Predicted PPIs (pPPIs) and PPIs originally detected in the target taxon present in the input data set (tPPIs) are nonredundantly integrated in the final network. Each PPI in the final network is scored under the same confidence scoring scheme. (D) This framework was applied, separately, for each of the three representative target species of human herpesviruses (HSV-1, HCMV, and EBV) to reconstruct a corresponding network. Download FIG S2, TIF file, 0.7 MB (762.9KB, tif) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Database structure and content. Download Text S1, DOCX file, 0.02 MB (18.6KB, docx) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
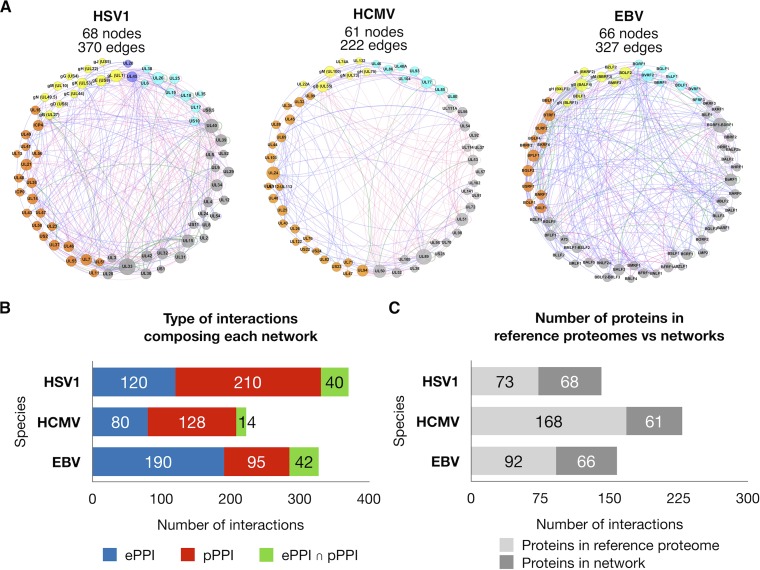
The data stored in HVint2.0 contain annotation for a total of 918 PPIs (369 for HSV-1, 222 for HCMV, and 327 for EBV), including protein annotation, confidence scores, supporting evidence, and cross references to external databases. Also present in the database, is annotation of all 704 herpesvirus protein identifiers gathered during interactome reconstructions. The latter include proteins from the reference proteomes (as defined in the UniProt Proteomes database [13]) to which the final interactomes were mapped, as well as identifiers of proteins involved in the interaction from homologous species used for the homology mapping and reference proteome mapping. Similarly, the database holds annotation on associated data, such as taxonomic groups, PSI-MI (Proteomics Standards Initiative Molecular Interactions) ontologies (14), and primary citations (Fig. S1).
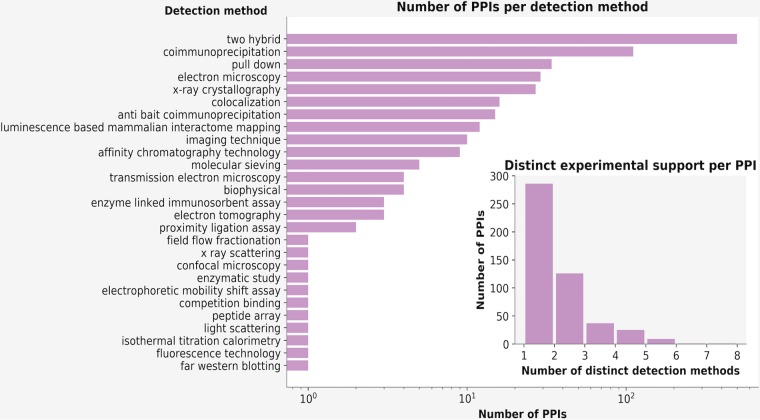
Examining the contributions of different type of supporting evidence in HVint2.0 (Fig. 2A), we observe, as expected, that for the reconstructed interactomes, experimentally obtained supporting evidence is strongly populated with data resulting from yeast two-hybrid studies (Fig. 2A; see Fig. S3 in the supplemental material). However, nearly half of the PPIs in HVint2.0 are supported by data obtained from at least two different types of experimental methods (Fig. 2B). To test the ability and reliability of our PPI prediction approach in highlighting potentially new PPIs (yet to be experimentally detected), we conducted experimental testing on a subset of our interactions. The encouraging results of this study are discussed in detail in a related article (11).
FIG 2.
HVint2.0 content statistics. (A) Number of PPIs in HVint2.0 detected by each detection method (according to PSI-MI annotation). The main plot shows the number of PPIs in logarithmic scale. The counts for detection method categories “two hybrid” and “two hybrid array” have been joined in the same bar, as done similarly in the case of categories “coimmunoprecipitation” and “anti tag coimmunoprecipitation.” (B) Number of PPIs as a function of the number of different types of experimental detection methods supporting an interaction. Note that the graph reflects different types of evidence per PPI: i.e., if an interaction was detected by the same experimental method (for instance Y2H) more than once, this would still be counted only once.
Number of interactions per primary citation. Here a large-scale study was defined as providing more than 50 PPIs. The graph shows that in the case of the reconstructed interactomes, only Y2H experiments provide that amount of interaction (corresponding to the four largest bars [6, 8, 68, 69]). Download FIG S3, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Herpesvirus species-specific interaction networks.
The PPI network for each of the target species generated from HVint2.0 (Table 1 and Fig. 3A) contains a similar total number of nodes (proteins), as well as a similar number of nodes representing core (family-wise conserved) and noncore (sublineage-specific) proteins (40, 38, and 39 core protein nodes and 28, 23, and 27 noncore protein nodes in the HSV-1, HCMV, and EBV networks, respectively). The best-represented proteome is that of HSV-1, with ∼93% coverage, followed by those of EBV and HCMV, with ∼72% and ∼36% coverage, respectively. Analogous rankings are obtained if one considers core and noncore protein node fractions separately (Table 1).
TABLE 1.
Reconstructed PPI networks in HSV-1, HCMV, and EBV
| Species | No. of unique: |
||
|---|---|---|---|
| Pieces of evidence | PPIs | Protein sequences | |
| HSV-1 | 644 | 370 | 68 (40 core, 28 noncore) |
| HCMV | 393 | 222 | 61 (38 core, 23 noncore) |
| EBV | 703 | 327 | 66 (39 core, 27 noncore) |
FIG 3.
Herpesvirus species-specific interaction networks. (A) Reconstructed PPI interaction networks of HSV-1, HCMV, and EBV. Nodes (circles) are color coded as follows: cyan for capsid and capsid-associated proteins, orange for tegument, blue for nonglycosylated envelope proteins, yellow for envelope glycoproteins, and gray for nodes representing proteins not present in the mature virions. Node size is scaled between minimum and maximum within each network. Node labels contain protein open reading frame identifiers and popular glycoproteins in addition to their frequently used alternative names. Edges (links) are color coded according to the type supporting evidence for each interaction: red for computationally predicted PPIs (pPPIs), blue for experimentally supported PPIs (ePPIs), and green for interactions that have been both experimentally supported and computationally predicted (ePPIs ∩ pPPIs). The thickness of the edges is proportional to the confidence score of the interaction. (B) Bar charts for each species illustrating the number of PPIs as a function of their type of supporting evidence: i.e., red for pPPIs, blue for ePPIs, and green for ePPIs ∩ pPPIs. (C) Bar charts illustrating the size of the reference proteome (light gray) for each species (i.e., 73, 168, and 92 proteins for HSV-1 strain 17, HCMV Merlin, and Epstein-Barr B95-8, respectively) and the number of proteins from the reference proteome present in the network (dark gray).
Inference of a herpesviral central interactome.
One of the features characterizing herpesvirus genomes across the Alpha-, Beta-, and Gammaherpesvirinae subfamilies, is the presence of a set of conserved proteins, referred to as “core” proteins. Following the consensus observed among available literature, we defined a list of 40 core proteins (see Table S1 and Table S2 in the supplemental material) (15, 16). Not surprisingly, these proteins are predominantly annotated with functional roles at early stages of the lytic life cycle, such as genome replication, packaging, capsid formation, and nuclear egress. However, they also include a few other components of the tegument and envelope, some of which are known to be responsible for processes that are imperative for an efficient viral production, such as glycoprotein B (gB) and the complex gH/gL, which are strictly required for entry into the host cell (17).
List of core herpesvirus proteins used in this study. Proteins in the same row are homologues. Download Table S1, XLSX file, 0.01 MB (8.3KB, xlsx) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Functional annotation for proteins in the central interactome. Data were retrieved from the HVint2.0 database. Download Table S2, XLSX file, 0.01 MB (7.8KB, xlsx) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
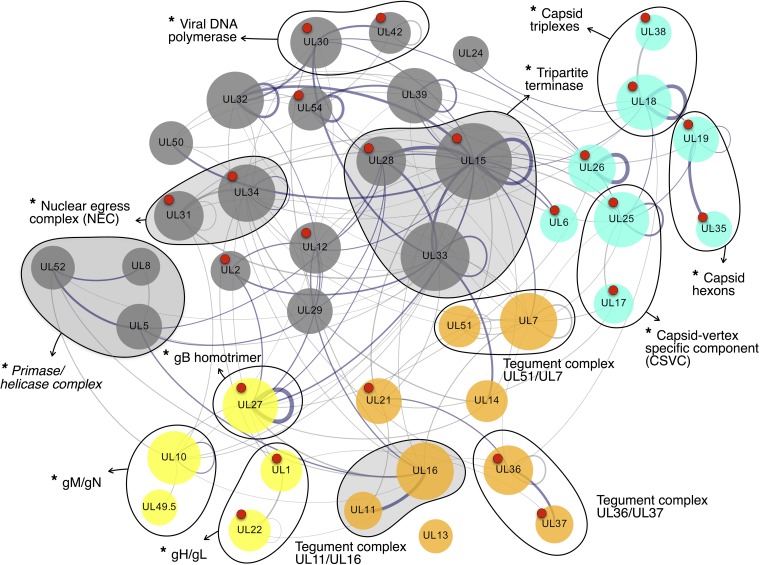
All three species-specific networks reconstructed above contain nearly the full subset of 40 core proteins defined in this study (Table 2). The intersection across all the three subsets is of 37 (out of 40) proteins (nodes) and 56 interactions. However, to infer a potential herpesviral central interactome, here we considered PPIs occurring in at least two out of the three subsets (i.e., instead of the intersection of all three). The resulting network contained the full set of 40 core proteins and a total of 134 nonredundant PPIs (Fig. 4). Out of these 134 PPIs, 56 (∼42%) had experimental support (ePPIs or ePPIs ∩ pPPIs) in all three species and 89 (∼66%) in at least one species. Additionally, 45 PPIs (∼34%) had only been computationally predicted in all three (pPPIs only). Importantly, by taking into account PPIs present in at least two species but not requiring them to be in all three, the central interactome was able to overcome the major limitation of our PPI prediction strategy (i.e., the dependence on sequence similarity between proteins to infer their evolutionary conservation). Consequently, our reconstructed central interactome is able to suggest interactions that could not be predicted using a sequence homology-based approach. One example of this is the interaction for the heterodimer gH/gL in HCMV, which was missing when we originally (in 2016) collated the input data set to HVint2.0. Because gL orthologues are positional and functional homologues but lack detectable sequence similarity (18), they could not be predicted using a sequence similarity-based strategy. However, in 2017, the crystal structure of gH/gL in HCMV was solved (PDB ID 5VOD) (19), confirming our prediction.
TABLE 2.
Species-specific core protein subnetworks
| Species | No. of: |
|
|---|---|---|
| Nodes | Edges | |
| HSV-1 | 40 | 192 |
| HCMV | 38 | 139 |
| EBV | 39 | 152 |
FIG 4.
Inferred herpesviral central interactome. The network was obtained by compiling PPIs taking place among core proteins (Table S2) in at least two of the reconstructed networks for HSV-1, HCMV, and EBV. Edge thickness indicates PPI confidence scores, which were calculated as the sum of the scores for the interaction in each species it was found over the maximum number of species where the interaction could exist (i.e., 3). Nodes are color coded as follows: cyan for capsid and capsid-associated proteins, orange for tegument, yellow for envelope glycoproteins, and gray for proteins not present in the mature virion. Node labels correspond to HSV-1 open reading frame nomenclature. Node size is proportional to the number of interactions of each protein in the central interactome network. Red dots on top of nodes indicate the availability of structural data (full or partial reconstructions [Table S3]) for the corresponding node. Nodes circled together by a solid black line indicate the 13 family-wise conserved complexes represented by the interactome. Asterisks at the beginning of complex labels indicate obligate complexes (i.e., complexes for which its stable assembly is required for the proteins involved be functional). Complexes also detected in at least two species in reference 8 are indicated with shaded circles. Correspondences between ORF nomenclature, UniProtKB identifiers, and functionally informative tags for each protein can be found in Table S1 and Table S2.
PDB entries associated with proteins and protein complexes appearing in the reconstructed Herpesviridae central interactome (Fig. 4). These entries can contain structural data for the full complex or only part of it (e.g., domains). Data obtained for homologous species are grouped in the same row using the HSV-1 nomenclature. Download Table S3, DOCX file, 0.02 MB (23.2KB, docx) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The generated central interactome contains 24 (60%) proteins that have some structural support (see Table S3 in the supplemental material). Furthermore, it is able to represent a total of 13 protein complexes widely accepted to be conserved across the Herpesviridae family, adding biological consistency to our results.
Functional annotation of the complexes in the herpesviral central interactome.
The 13 complexes composing the central interactome (Fig. 4 and 5) are predominantly involved in processes such as DNA replication and insertion of the genome into the viral capsid, formation of the latter, invasion of new host cells, as well as key aspects of virion structure and morphogenesis. Complexes involved in DNA replication and packages include the tripartite complexes helicase/primase (complex 1) and tripartite terminase (complex 2). These are responsible, respectively, for double-helix unwinding and genome packaging into new viral capsids, prior to and after genome replication by the DNA polymerase complex (complex 3), which is also present in the central interactome (20–22). Capsid complexes include interactions among hexon subunits (complex 4), heterotrimeric triplexes (complex 5) surrounding capsid pentons, and the capsid-vertex-specific component (CSVC) (complex 6) (23). Also present is the nuclear egress complex (NEC) (complex 7), which is required and sufficient to facilitate budding of the nuclear capsids at the inner nuclear membrane for primary envelopment (24). Tegument complexes known to participate at different stages of secondary envelopment include the UL36 and UL37 complex (UL36-UL37; complex 8), UL11-UL16 (complex 9), or UL51-UL7 (complex 10) (25–28). Finally, glycoprotein complexes such as gM/gN (complex 11) (29), and the abovementioned gH/gL (complex 12), suggested to act as the activity regulator to the trimeric fusogen gB (complex 13) (17), are also present. One complex not found in the central interactome, which surprised us initially, is the ribonucleotide reductase (RNR) complex, formed by homologues of HSV-1 proteins UL39 and UL40 (R1 and R2 subunits, respectively) (30). RNR is a holoenzyme that catalyzes the formation of deoxyribonucleotides required for DNA synthesis, and it is essential for viral growth in both Alpha- and Gammaherpesvirinae subfamilies. However, the reason this enzyme is missing from the central interactome is that an open reading frame (ORF) for the R2 subunit is missing in genomes of the Betaherpesvirinae subfamily (31). Consequently, the node corresponding to the R2 subunit does not appear in our network, and therefore the complex cannot be inferred.
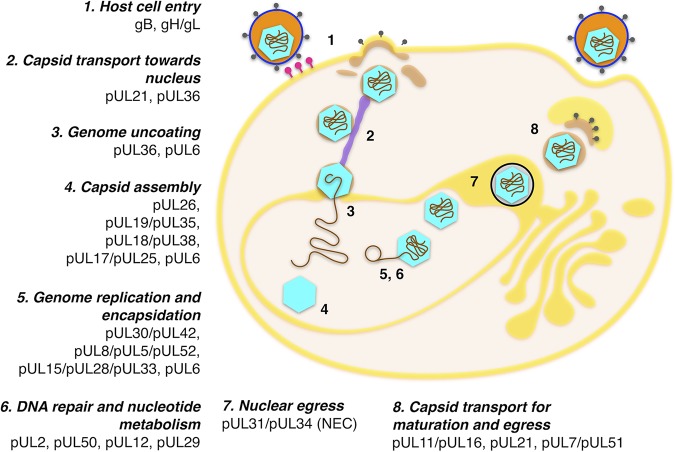
FIG 5.
Central interactome complexes in the herpesvirus life cycle. The sequential steps in a herpesvirus lytic life cycle are indicated with numbers. The complexes present in the reconstructed central interactome participating in each of the stages are also annotated. Upon viral entry mediated by virus envelope-cell membrane fusion (step 1), the tegument is delivered into the host cell and capsids are transported to the nucleus (step 2). Docking to the nucleopores triggers genome uncoating (i.e., delivery to the nucleus) (step 3). In a lytic replication, a procapsid is formed (step 4), and genome replication, capsid maturation, and genome encapsidation occur concomitantly (steps 5 and 6). Capsids egress from by budding at the nuclear membranes (step 7). The bulk of the tegument is acquired in the cytoplasm, primarily by fusion with trans-Golgi network-derived vesicles (step 8). Fusion of these vesicles with the plasma membrane upon virion maturation releases the virions to the extracellular space (step 8). Correspondences between ORF nomenclature, UniProtKB identifiers, and functionally informative tags for each protein can be found in Table S1 and Table S2.
Previously, published data on PPIs among core proteins in herpesviruses (8) defined a central interactome by including all PPIs taking place among core proteins in one or more of five analyzed herpesvirus species. To compare our data set to theirs, we first applied the same constraints as ours to their data set: i.e., we only considered interactions that were detected in at least two species among HSV-1, murine cytomegalovirus (MCMV), and EBV. (Note that the experiments in reference 8 were conducted 10 years ago, and there the authors used MCMV as a Betaherpesvirinae subfamily representative instead of HCMV.) We found that the coverage of the central interactome reconstructed in our study is in fact larger, both in terms of total number of PPIs (134 PPIs versus 17 PPIs in reference 8) and the number of conserved protein complexes.
HVint2.0 web interface.
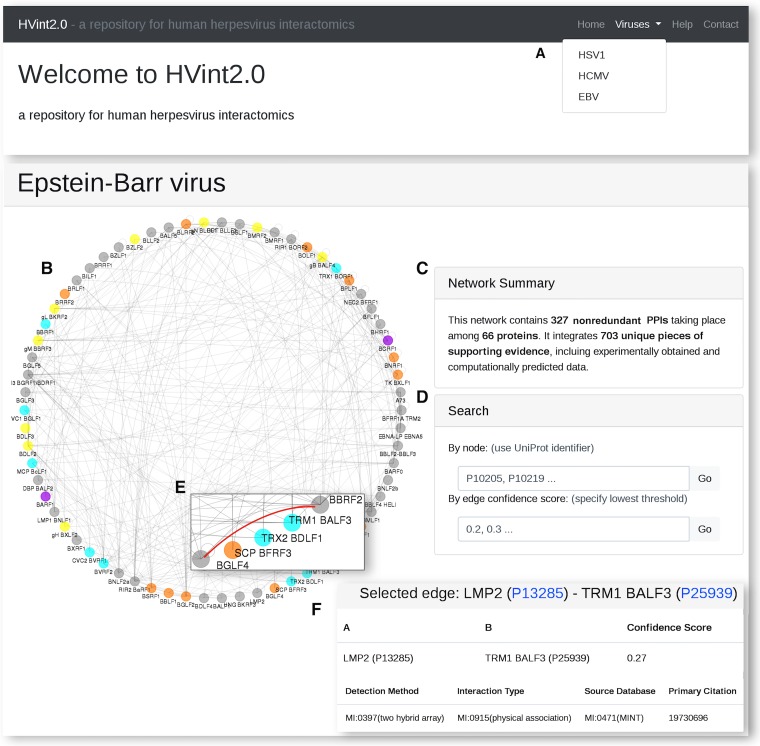
The HVint2.0 web interface (http://topf-group.ismb.lon.ac.uk/hvint2) provides an intuitive user-friendly access to the reconstructed interactomes (Fig. 6). From the main page, the user can access the reconstructed interactomes via a dropdown menu (Fig. 6A). Each interactome page is designed to provide the user with both a graphical representation of the network data (Fig. 6B) and a brief summary of the inspected interactome (Fig. 6C), as well as tools for further exploration, such as search boxes for both nodes and confidence score thresholds (Fig. 6D). However, one can also interactively explore the network by clicking nodes and edges in the graphical display window (Fig. 6E). Upon each query, the page is updated to provide both a graphical representation of the subnetwork associated with the query and further annotation data on these interactions in tabular format, including supporting evidence (Fig. 6F).
FIG 6.
HVint2.0 web interface. (A) Each interactome can be accessed from the top dropdown menu. (B and C) For each interactome, the website provides both a graphical representation of the network and a brief summary. (D) Search tools to subset the full network based on node identifiers or edge confidence threshold. (E) Alternatively, specific nodes and edges of interest can be selected from the graphical display. (F) For each selection, the corresponding subnetwork is rendered and its associated data provided in tabular format.
DISCUSSION
Study rationale.
The motivation behind our study was 2-fold. First, we wanted to introduce the extended HVint2.0 database, a centralized resource of curated PPI data across the Herpesviridae family. The database now includes data on archetypical species of all three phylogenetic herpesviral subfamilies: HSV-1 for the Alpha-, HCMV for the Beta-, and EBV for the Gammaherpesvirinae. To populate HVint2.0, we compiled new system-level PPI data sets for each of the three selected species using our recently updated network reconstruction framework (11).
Second, we wanted to use the compiled PPI data to extract new biological knowledge. Specifically, taking advantage of the fact that the collected PPI data span the Herpesviridae phylogeny, we sought to infer the family-wise conserved fraction of these networks, i.e., a “central” human herpesvirus intraviral interactome. PPIs are now consolidated as major driving forces behind observed phenotypes. Identification of the features that distinguish two or more biological networks can help explain species-specific pathogenic strategies and phenotypes. Equally important is to characterize the common network features across different species networks, as this fraction would presumably contain the scaffolding components that allow establishing an infection in different host systems.
Network coverage.
The total number of nodes, as well as core and noncore proteins represented in the three reconstructed networks, are comparable. However, they translate into different levels of proteome coverage for each network. The most comprehensively represented proteome is that of HSV-1, followed by EBV and HCMV. The same still holds for the fractions of the intraviral interactome formed by core and noncore proteins, for each species (which correspond, respectively, to the family-wise and sublineage-specific fractions of the proteomes). In the case of HSV-1 and EBV, a significant proportion of their sublineage-specific proteomes (∼85% and ∼52%, respectively) are present in the assembled networks. Instead, for HCMV, only around 18% of it is present. There are two main reasons for that. One is that the HCMV proteome is significantly larger (168 canonical protein sequences in the UniProt reference proteome) than those of other herpesviruses, including HSV-1 and EBV (with 73 and 92 proteins in their reference proteomes, respectively). The other reason is a remarkable underrepresentation of experimentally supported data for members of the Betaherpesvirinae subfamily in the input data set (Fig. 3B), which could derive, for instance, from experimental limitations in efficient culturing of species of the Betaherpesvirinae subfamily. Therefore, our results expose the areas in these herpesviral interactomes that are least represented by currently available data and would benefit most from further investigation.
Reconstruction of a central interactome and functional annotation.
To reconstruct our herpesviral central interactome, we used a cross-species network comparison and integrated all the interactions among core proteins. We have compared the central interactome with that of previously published data from reference 8. In the latter study, the authors conducted what constitutes, to the best of our knowledge, the largest experimentally based study on herpesviral networks to date and which has been a valuable input toward our network reconstructions. After applying the same constraints to their initial data set as the ones we used in our own reconstruction (i.e., PPIs among core proteins taking place in at least two of the three studied species), we found that the central interactome reconstructed in our study contains a larger number of PPIs, including a larger number of known conserved herpesviral protein complexes. These results are encouraging, indicating that our strategy of integrating experimentally and computationally generated data increases the coverage of our interactome in a biologically consistent and informative manner.
The choice of including PPIs present in only two of the three networks instead of all of them partially compensates for the limitations derived from a sequence-based PPI prediction method for orthologous proteins with very low or nonexistent sequence similarity, such as in the case of positional homologues like gL (18). Experimental data supporting the well-known interaction between gL and gH in HCMV specifically were missing in our input data set. Because of the lack of sequence similarity among gL homologues, the interaction between gL and gH could not be mapped from other species using a sequence-based approach. Yet, the reconstructed central interactome was able to highlight this interaction as family-wise conserved, which was later validated experimentally (19).
In total, the reconstructed central interactome contains representations of 13 protein complexes known to be conserved across the family. Unsurprisingly, these complexes are involved in essential functions for a productive infection, such as capsid formation, genome replication and encapsidation, virion egress, and viral entry. Together, it is easy to envision how these complexes could suffice as an essential toolkit for the creation of a minimal yet productive viral particle. This would consist of a basic self-assembling capsid shell containing a copy of the viral genome, which would then be able to egress the infected cell, circumvent the immune surveillance in the extracellular media, and finally deliver the viral genome into a new host cell. In contrast, the species-specific fractions from each of the reconstructed networks show well-defined functional differences from the central interactome (see Table S4 and Fig. S4 in the supplemental material). Comparatively, these species-specific fractions are enriched in proteins involved in pathways that are likely to have evolved, to a larger degree, to increase the efficiency of the infection in a host-specific manner. Examples of such processes include immune modulation, cell tropism, virulence, or gene expression regulation.
Inferred herpesviral central interactome and species-specific subnetwork. The inferred herpesviral central interactome is shown at the center, flanked by the subsets of species-specific interactions for each of the three species from the reconstructed interactomes. Gray edges and nodes indicate PPIs and proteins found in the central interactome. These are referred to using HSV-1 open reading frame nomenclature. Red, blue, and green edges and nodes indicate PPIs and proteins from the species-specific subnetworks in HSV-1, HCMV, and EBV, respectively. Edge thickness reflects confidence scores. For central interactome PPIs, this score was calculated as the sum of the scores for the interaction in each species it was found over the maximum number of species where the interaction could exist (i.e., 3). In species-specific subnetworks, this is calculated using the scoring function integrated in our network reconstruction framework. Node size is proportional to the number of interactions of each protein. Download FIG S4, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Functional annotation for proteins in all three species-specific network fractions. Data were retrieved from the HVint2.0 database. Download Table S4, DOCX file, 0.05 MB (50.1KB, docx) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The concept of a minimal biological system has been explored for a long time with the goal of unraveling the fundamental basis of life and the function of individual genes (32, 33). In this context, different strategies have been explored, including, but not limited to, minimal cells (32), in silico cell models (34), genomes (35), and minimal sets of genes (36). Within the virosphere, the initial interest largely focused on synthetically recreating minimal self-assembling capsid-like structures, mainly due to the potential as nanocarriers for drug delivery (37, 38). In 2016, Bale et al. (39) reported the creation of stable and engineerable icosahedral protein shells with enough internal volume to enclose biological macromolecules, such as their own encoding nucleic acids (39–41). Subsequent experiments also demonstrated the capacity of such RNA-protein complexes to evolve improving their fitness, providing new hypothetic evolutionary scenarios that could have given rise to ancestral viral particles (41). Contemporary viruses exhibit a much larger and complex array of functionalities that have allowed their adaptation to different environments and hosts, and at present, ample room remains to be explored to understand such evolutionary paths leading to such complexity. In a similar fashion to the case of cellular organisms, a first step to decode the currently observed viral complexity, of which herpesviruses are outstanding examples, is to identify those elements that are part of the essential scaffold (core genes and proteins), which viral evolution built upon over time.
In this study, we propose a scaffolding central interactome for the Herpesviridae family, one of the most structurally and genetically complex family of viruses (1). These data contribute to characterizing the evolutionary history of these human pathogens. Combining the central interactome with the species-specific networks, our data will assist in creating comprehensive system-level models of the functional architecture of this viral lineage. As computational models inspired on minimal cellular systems have already been demonstrated (42, 43), in this case, these models can bring new insights into pathogenic behaviors and phenotypical outcomes, guide experimental analysis, and potentially become important for designing and testing new molecules for industrial and clinical application.
Species-specific networks.
It is beyond the scope of this article to delve into the numerous species-specific features that characterize different herpesviruses. However, in the paragraphs below we elaborate on some of the examples that our species-specific network reconstruction highlights and encourage the reader to explore the vast and interesting literature available on the subject.
(i) Initiation of lytic replication.
We observe for instance, proteins involved in species-specific pathways that launch the gene expression cascades leading to lytic replication. In HSV-1, for instance, protein pUL48 (also called VP16) acts as a transcriptional activator, stimulating the expression of immediate-early (IE) genes (44), such as protein pUS1 or ICP0 (also present in the species-specific subnetwork).
In HCMV, proteins pUL122, pUL37, pUL38, and pUL112-113 constitute replication auxiliary factors only conserved in the Betaherpesvirinae subfamily. pUL122-123 is the precursor for the major immediate-early (MIE) genes, IE1 and IE2 (45). Proteins such as pUL37 and pUL38 have been suggested to contribute to viral replication by promoting the maintenance of essential cellular functions (46). The pUL112-113 genes encode four different phosphoproteins obtained through alternative splicing and can enhance DNA replication both by binding the DNA and promoting the transcriptional activation of IE genes, as well as by direct interaction with the latter products and the DNA polymerase (47).
The EBV species-specific subnetwork contains, in turn, the two IE transcriptional activators encoded by this virus, BRLF1 and BZLF1. Expression of either of these proteins has been shown to be sufficient to induce lytic replication from latency.
It is interesting to note that although the proteins referred above are encoded in a species- or sublineage-specific manner, some of the mechanisms they use to exert their functions are shared across species. For instance, the transactivators encoded in the three subfamily representatives (i.e., VP16 in HSV-1, IE2 in HCMV, and BRLF1/BZLF1 in EBV) all bind CREB-binding protein (CBP), to enhance transcription through histone acetylation (48).
(ii) Tropism.
In HSV-1, pUS6 (gD) is the HSV-1-specific receptor-binding protein that triggers the signal for membrane fusion to occur. pUL44 (gC) is an Alphaherpesvirinae-specific protein known for its role in host cell viral entry, where it promotes initial attachment of the virion to the cell surface by binding heparan sulfate proteoglycans, but it also exerts a function as a virulence factor by inhibiting classical and adaptive host immune responses (49). In EBV, we find BZLF2, also known as gp42, which controls cell tropism. The complex between gH/gL and gp42 enables infection of B cells; however, the gp42 N terminal inhibits entry in epithelial cells (50). Although the complexes driving HCMV tropism (i.e., the pentamer gH/gL/pUL128-131 [which includes UL128 through UL131] and the trimer gH/gL/gO) had been functionally characterized previously, conclusive evidence of the physical binary interactions taking place among the respective components was not available at the time of our data collection. The crystal structure of gH/gL/pUL128-131 was only solved in 2017 (PDB ID 5VOD) (19), and structural details for gH/gL/gO are, at this time, limited to low-resolution data (51).
(iii) Interference with host defense mechanisms.
Other proteins are involved in counteracting the host immune defense mechanisms. The pUS4 (gG) and pUS5 (gJ) genes in HSV-1 encode glycoproteins acting as immunomodulators. gG is a viral chemokine-binding protein (vCKBP), binding human chemokines expressed at the sides of viral replication and spread with high affinity (52), which translates into increased cell migration to these sites. The advantage that this mechanism confers to the virus is still unknown, but it has been suggested that it could help recruiting new infection targets, facilitating viral load and spread (52). Although the specific mechanisms by which gJ exerts its function are not fully understood, it is known to be sufficient to prevent apoptosis induced by several stimuli, including UV, fas, or cytotoxic T cells (53). The HCMV protein pUL25 antagonizes interferon responses, for instance by antagonizing the action of interferon-stimulated gene 15 protein (ISG15), which prevents the proteasomal degradation of other viral counterparts, such as pUL26 (54). In EBV, proteins BHRF1 and BALF1 are homologues of the mammalian cell death inhibitor BCL-2 proteins and participate in regulating apoptosis in infected cells (55).
(iv) Other.
We also find proteins like pUL23 in HSV-1 and BXLF1 in the EBV subnetworks, which encode the viral thymidine kinase. Thymidine kinase is commonly known for its role as an antiviral drug target. During infection, it participates in the metabolism of nucleic acids, catalyzing the conversion of thymidine (Thd) into dTDP (TDP) (56). To date, herpesvirus thymidine kinase ORFs have only been detected in the Alpha- and Gammaherpesvirinae subfamilies but could not be found in most Betaherpesvirinae genomes, including HCMV (57). In HCMV, the pUL32 gene encodes the basic phosphoprotein (BPP, or pp150), homologues of which are only found within the Betaherpesvirinae subfamily but not in the Alpha- or Gammaherpesvirinae lineages. pUL32 is closely associated with the capsid and constitutes a major constituent of the HCMV virion. Although not essential, this protein has been show to play a very important role in the correct morphogenesis of newly formed virions (58).
HVint2.0 usage.
This study has presented comprehensive compilations of curated system-level PPI data for three representative species across the Herpesviridae phylogeny. Full interactome coverage is, at present, still a distant target for individual techniques, and repositories and integrative pipelines like the one underlying the HVint2.0 data sets are needed to alleviate such shortcomings. The integration of computationally predicted PPIs does not only increase the coverage of the resulting networks. These data can also assist in more confidently identifying nonspecific interactions and false positives among experimentally generated data, such as those obtained from immunoprecipitation-mass spectrometry (IP-MS) studies (59). Our PPI network reconstruction framework presents limitations that are worth bearing in mind, such as the dependency on detectable sequence similarity between homologous proteins. However, the output of the current study already provides useful tools and data to help answering questions on the evolutionary dynamics of this important group of eukaryotic viruses and human pathogens. The data generated by our analysis could be the basis of several projects across a number of disciplines, including, but not limited to, PPI discovery, evolutionary dynamics, and structural modeling of the involved networks. Further, the availability of three-dimensional structural data for over half of the components in the central interactome encourages prospective endeavors to structurally model the full network. This will add another level of understanding of the dynamics and constraints that govern proteins’ connectivity.
Summary.
In the protocol described here, we strived to maximize not only the coverage of our reconstructed networks but also the degree of automation of the pipeline so that it is easily reproducible and generalizable to other species. This is the main reason why we minimized the use of manual literature curation in our protocol. Another reason is that, despite the advantages that such curation may bring (such as a larger degree of PPI coverage), this could also entail major drawbacks, especially regarding data quality validation and frequency of new database releases. Therefore, in our pipeline we used data repositories with dedicated curation teams that follow the curation rules established by the IMEx consortium (https://www.imexconsortium.org/) or that directly pull their data from such curated repositories. Taking advantage of the fact that our pipeline has been designed to be easily reproducible in other species (within the Herpesviridae and from other viral families), we plan to regularly release database updates, keeping up to date with new updates of the input PPI databases.
MATERIALS AND METHODS
Database design.
We shall first emphasize the distinction between the back-end HVint2.0 relational database (see Fig. S1 and Text S1 in the supplemental material) and the HVint2.0 web interface. The HVint2.0 relational database contains all the data collated during the study. These include a range of diverse annotation data, in addition to the finally reconstructed intraviral interactomes, such as for instance, functional annotations for proteins, primary citation details (like authors and journals), or protein sequence annotations, among others. While these additional data are certainly relevant, and we encourage the reader to explore them, navigating through these details can become cumbersome—especially at the early stage of network exploration. Our goal when creating the HVint2.0 web interface was precisely to ease the exploration of synthetic yet comprehensive representations of the interactomics data in our database, These representations hold all the key information to understand the evidence behind each interaction and trace its original source.
The HVint2.0 relational database was created using the MySQL server (version 5.1.73) as a relational database management system, within a Linux platform. The data are distributed across a total of seven tables. Six tables as named represent “protein,” “PPI,” “taxon,” “evidence,” “citation,” and “psimi_ontology” entities (Text S1 and Fig. S1). The remaining table allows referencing many-to-many relationships between “evidence” lines and the “psimi_ontology” table. Protein annotation, taxonomic data, and citation details were collated from the UniProtKB (13), NCBI Taxonomy (https://www.ncbi.nlm.nih.gov/taxonomy), and PubMed databases, respectively. Cross-references to the input databases are annotated following the PSI-MI format.
Although the PSI-MI format allows to allows annotation of each binary interaction with multiple pieces of annotation, for the implementation of our pipeline, the following are strictly necessary: identifiers for both of the proteins participating in the interaction, the PPI detection method (e.g., yeast two-hybrid [Y2H]), the type of PPI (e.g., colocalization), and the primary citation identifier (e.g., the PubMed identifier of the study reporting the interaction).
Data collection, PPI prediction, data integration, and scoring of PPIs.
To reconstruct the network for each of the three target species of human herpesviruses (i.e., HSV-1, HCMV, and EBV, respectively), we used our recently developed automated pipeline for collecting PPI data in HSV-1 in an unbiased fashion (11) (Fig. S2). In short, input data were retrieved from five molecular interaction repositories—BioGRID (60), the Database of Interacting Proteins (DIP) (61), IntAct (62), Mentha (63), and VirHostNet 2.0 (64)—and two structural databases—the Protein Data Bank (PDB) (65) and the Electron Microscopy Data Bank (EMDB) (66). To minimize bias, filters to ensure the quality of the PPI data were only applied by us to PPIs obtained from the structural repositories. These databases are not dedicated specifically to molecular interaction data, and therefore, we inspected each entry individually and selected binary PPI only where the following criteria were met:
Entries provided structural evidence of a physical interaction between protein chains and not other types of molecules: i.e., interactions involving nonpolypeptidic molecules, such as nucleic acids or small molecules, were disregarded.
Entries had an associated PubMed ID: i.e., deposited structures without associated publication at the time of data collation were disregarded.
The interacting chains belonged to one of the input species considered in our pipeline (i.e., human herpesviruses, as well as pseudorabies virus [PRV], MCMV, and murine gammaherpesvirus 68 [MHV68]).
In the case of PDB entries, we also looked for atomic contacts (as defined by default in UCSF Chimera [67] between protein chains belonging to the herpesvirus species considered in our study—i.e., all human herpesviruses, as well as pseudorabies virus, murine betaherpesvirus, and murine gammaherpesvirus). In the case of EMDB entries, we only considered those at near-atomic resolution at the time of data collation (i.e., a resolution of ≤5 Å).
For each of the target species, we used PPIs experimentally detected in orthologous species to predict new interactions in the target interactome. Mapping of binary interactions between two species relies on the existence of the corresponding homologous pairs of proteins in each of those species. Our homology mapping strategy combines information on the alignment coverage, sequence identity and similarity, and probabilistic scores to estimate the likelihood of a hit of being a true-positive homologue. The resulting predictions were then integrated (removing redundancy) with PPIs experimentally detected in the target species (11). It is worth noting that the input databases and the homology mapping sometimes involved protein isoforms, and in those cases, we mapped them to the canonical ORFs.
Each of the PPIs was scored using our recently implemented scoring function, which relies on MIscore (12), taking into account the heterogeneity in the evidence supporting an interaction, and an additional scaling factor to penalize for computational predictions (11). The latter includes information on the prediction method used (i.e., sequence-based orthology) and follows more closely the (nonlinear) mathematical description of the terms in the MIscore function.
The output data of all three networks are stored in HVint2.0 (http://topf-group.ismb.lon.ac.uk/hvint2). The entire pipeline, which is implemented as a series of python scripts using standard (default) libraries, is reproducible and can be generalized to include other species.
Inference of a herpesviral central intraviral interactome.
For each of the networks, we extracted the subnetwork composed of core proteins. Here we define the core protein data set as a total of 40 proteins based on previously published literature (Table S1 and Table S2) (15, 16). A central intraviral interactome was next assembled by gathering all PPIs present in at least two of the three subnetworks; both experimentally supported and computationally predicted were equally considered. To assess the completeness and biological coherence of the reconstructed central interactome, we assessed the extent to which widely accepted conserved protein complexes in herpesviruses found in the literature were represented in the central interactome network.
Construction of the HVint2.0 web interface.
The full pipeline is implemented as a series of python scripts using standard (default) libraries.
The HVint2.0 web interface was built using the open source library for web development Bootstrap (https://getbootstrap.com/), which provides an easy-to-implement framework for development of responsive databases. A static JSON-formatted representation of the network data is rendered using the JavaScript library vis.js (http://visjs.org/), designed to ensure correct rendering in several web browsers.
Data availability.
The data sets produced in this study are available by clicking on the “Viruses” dropdown menu at http://topf-group.ismb.lon.ac.uk/hvint2.
ACKNOWLEDGMENTS
We are grateful for funding from the Wellcome Trust (209250/Z/17/Z) to M.T. and K.G. and 107806/Z/15/Z and Wellcome Trust Core Award 203141/Z/16/Z to K.G., as well as funding from the MRC (MR/M019292/1) to M.T. and K.G.
A.H.D. collated and analyzed the data and wrote the initial manuscript. M.T. and K.G. supervised the conception of the analysis framework and the results of this work. All authors discussed the findings and contributed to the final manuscript.
The authors declare that no conflicts of interest exist.
REFERENCES
- 1.Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed). 2007. Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 2.Looker KJ, Magaret AS, May MT, Turner KME, Vickerman P, Gottlieb SL, Newman LM. 2015. Global and regional estimates of prevalent and incident herpes simplex virus type 1 infections in 2012. PLoS One 10:e0140765. doi: 10.1371/journal.pone.0140765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manicklal S, Emery VC, Lazzarotto T, Boppana SB, Gupta RK. 2013. The “silent” global burden of congenital cytomegalovirus. Clin Microbiol Rev 26:86–102. doi: 10.1128/CMR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ok CY, Li L, Young KH. 2015. EBV-driven B-cell lymphoproliferative disorders: from biology, classification and differential diagnosis to clinical management. Exp Mol Med 47:e132. doi: 10.1038/emm.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGeoch DJ, Gatherer D. 2005. Integrating reptilian herpesviruses into the family herpesviridae. J Virol 79:725–731. doi: 10.1128/JVI.79.2.725-731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uetz P, Dong Y-A, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E, Haas J. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311:239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- 7.Calderwood MA, Venkatesan K, Xing L, Chase MR, Vazquez A, Holthaus AM, Ewence AE, Li N, Hirozane-Kishikawa T, Hill DE, Vidal M, Kieff E, Johannsen E. 2007. Epstein-Barr virus and virus human protein interaction maps. Proc Natl Acad Sci U S A 104:7606–7611. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fossum E, Friedel CC, Rajagopala SV, Titz B, Baiker A, Schmidt T, Kraus T, Stellberger T, Rutenberg C, Suthram S, Bandyopadhyay S, Rose D, von Brunn A, Uhlmann M, Zeretzke C, Dong Y-A, Boulet H, Koegl M, Bailer SM, Koszinowski U, Ideker T, Uetz P, Zimmer R, Haas J. 2009. Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog 5:e1000570. doi: 10.1371/journal.ppat.1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Readhead B, Haure-Mirande J-V, Funk CC, Richards MA, Shannon P, Haroutunian V, Sano M, Liang WS, Beckmann ND, Price ND, Reiman EM, Schadt EE, Ehrlich ME, Gandy S, Dudley JT. 2018. Multiscale analysis of independent Alzheimer’s cohorts finds disruption of molecular, genetic, and clinical networks by human herpesvirus. Neuron 99:64–82.e7. doi: 10.1016/j.neuron.2018.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashford P, Hernandez A, Greco TM, Buch A, Sodeik B, Cristea IM, Grünewald K, Shepherd A, Topf M. 2016. HVint: a strategy for identifying novel protein-protein interactions in herpes simplex virus type 1. Mol Cell Proteomics 15:2939–2953. doi: 10.1074/mcp.M116.058552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernández Durán H, Greco TM, Vollmer B, Cristea IM, Grünewald K, Topf M. 2019. Protein interactions and consensus clustering analysis uncover insights into herpesvirus virion structure and function relationships. PLoS Biol 17:e3000316. doi: 10.1371/journal.pbio.3000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villaveces JM, Jiménez RC, Porras P, Del-Toro N, Duesbury M, Dumousseau M, Orchard S, Choi H, Ping P, Zong NC, Askenazi M, Habermann BH, Hermjakob H. 2015. Merging and scoring molecular interactions utilising existing community standards: tools, use-cases and a case study. Database (Oxford) 2015:bau131. doi: 10.1093/database/bau131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.UniProt Consortium. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, Tyers M, Salama JJ, Moore S, Ceol A, Chatr-Aryamontri A, Oesterheld M, Stümpflen V, Salwinski L, Nerothin J, Cerami E, Cusick ME, Vidal M, Gilson M, Armstrong J, Woollard P, Hogue C, Eisenberg D, Cesareni G, Apweiler R, Hermjakob H. 2007. Broadening the horizon—level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol 5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K, Davison AJ (ed). 2007. Human herpesviruses: biology, therapy, and immunoprophylaxis, chapter 2. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 16.Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K, Davison AJ (ed). 2007. Human herpesviruses: biology, therapy, and immunoprophylaxis, chapter 4. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 17.Spear PG, Longnecker R. 2003. Herpesvirus entry: an update. J Virol 77:10179–10185. doi: 10.1128/jvi.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaye JF, Gompels UA, Minson AC. 1992. Glycoprotein H of human cytomegalovirus (HCMV) forms a stable complex with the HCMV UL115 gene product. J Gen Virol 73:2693–2698. doi: 10.1099/0022-1317-73-10-2693. [DOI] [PubMed] [Google Scholar]
- 19.Chandramouli S, Malito E, Nguyen T, Luisi K, Donnarumma D, Xing Y, Norais N, Yu D, Carfi A. 2017. Structural basis for potent antibody-mediated neutralization of human cytomegalovirus. Sci Immunol 2:eaan1457. doi: 10.1126/sciimmunol.aan1457. [DOI] [PubMed] [Google Scholar]
- 20.Crute JJ, Tsurumi T, Zhu LA, Weller SK, Olivo PD, Challberg MD, Mocarski ES, Lehman IR. 1989. Herpes simplex virus 1 helicase-primase: a complex of three herpes-encoded gene products. Proc Natl Acad Sci U S A 86:2186–2189. doi: 10.1073/pnas.86.7.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heming JD, Conway JF, Homa FL. 2017. Herpesvirus capsid assembly and DNA packaging. Adv Anat Embryol Cell Biol 223:119–142. doi: 10.1007/978-3-319-53168-7_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarrouk K, Piret J, Boivin G. 2017. Herpesvirus DNA polymerases: structures, functions and inhibitors. Virus Res 234:177–192. doi: 10.1016/j.virusres.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 23.Dai X, Zhou ZH. 2018. Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science 360:eaao7298. doi: 10.1126/science.aao7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bigalke JM, Heldwein EE. 2015. Structural basis of membrane budding by the nuclear egress complex of herpesviruses. EMBO J 34:2921–2936. doi: 10.15252/embj.201592359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mettenleiter TC. 2004. Budding events in herpesvirus morphogenesis. Virus Res 106:167–180. doi: 10.1016/j.virusres.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Yeh P-C, Meckes DG, Wills JW. 2008. Analysis of the interaction between the UL11 and UL16 tegument proteins of herpes simplex virus. J Virol 82:10693–10700. doi: 10.1128/JVI.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newcomb WW, Brown JC. 2010. Structure and capsid association of the herpesvirus large tegument protein UL36. J Virol 84:9408–9414. doi: 10.1128/JVI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roller RJ, Fetters R. 2015. The herpes simplex virus 1 UL51 protein interacts with the UL7 protein and plays a role in its recruitment into the virion. J Virol 89:3112–3122. doi: 10.1128/JVI.02799-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Kasmi I, Lippé R. 2015. Herpes simplex virus 1 gN partners with gM to modulate the viral fusion machinery. J Virol 89:2313–2323. doi: 10.1128/JVI.03041-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frame MC, Marsden HS, Dutia BM. 1985. The ribonucleotide reductase induced by herpes simplex virus type 1 involves minimally a complex of two polypeptides (136K and 38K). J Gen Virol 66:1581–1587. doi: 10.1099/0022-1317-66-7-1581. [DOI] [PubMed] [Google Scholar]
- 31.Lembo D, Donalisio M, Hofer A, Cornaglia M, Brune W, Koszinowski U, Thelander L, Landolfo S. 2004. The ribonucleotide reductase R1 homolog of murine cytomegalovirus is not a functional enzyme subunit but is required for pathogenesis. J Virol 78:4278–4288. doi: 10.1128/JVI.78.8.4278-4288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morowitz HJ. 1984. The completeness of molecular biology. Isr J Med Sci 20:750–753. [PubMed] [Google Scholar]
- 33.Glass JI, Merryman C, Wise KS, Hutchison CA, Smith HO. 2017. Minimal cells—real and imagined. Cold Spring Harb Perspect Biol 9:a023861. doi: 10.1101/cshperspect.a023861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomita M, Hashimoto K, Takahashi K, Shimizu TS, Matsuzaki Y, Miyoshi F, Saito K, Tanida S, Yugi K, Venter JC, Hutchison CA. 1999. E-CELL: software environment for whole-cell simulation. Bioinformatics 15:72–84. doi: 10.1093/bioinformatics/15.1.72. [DOI] [PubMed] [Google Scholar]
- 35.Hutchison CA, Chuang R-Y, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, Pelletier JF, Qi Z-Q, Richter RA, Strychalski EA, Sun L, Suzuki Y, Tsvetanova B, Wise KS, Smith HO, Glass JI, Merryman C, Gibson DG, Venter JC. 2016. Design and synthesis of a minimal bacterial genome. Science 351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 36.Mushegian AR, Koonin EV. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci U S A 93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hernandez-Garcia A, Kraft DJ, Janssen AFJ, Bomans PHH, Sommerdijk N, Thies-Weesie DME, Favretto ME, Brock R, de Wolf FA, Werten MWT, van der Schoot P, Stuart MC, de Vries R. 2014. Design and self-assembly of simple coat proteins for artificial viruses. Nat Nanotech 9:698–702. doi: 10.1038/nnano.2014.169. [DOI] [PubMed] [Google Scholar]
- 38.Mateu MG. 2016. Assembly, engineering and applications of virus-based protein nanoparticles. Adv Exp Med Biol 940:83–120. doi: 10.1007/978-3-319-39196-0_5. [DOI] [PubMed] [Google Scholar]
- 39.Bale JB, Gonen S, Liu Y, Sheffler W, Ellis D, Thomas C, Cascio D, Yeates TO, Gonen T, King NP, Baker D. 2016. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 353:389–394. doi: 10.1126/science.aaf8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsia Y, Bale JB, Gonen S, Shi D, Sheffler W, Fong KK, Nattermann U, Xu C, Huang P-S, Ravichandran R, Yi S, Davis TN, Gonen T, King NP, Baker D. 2016. Corrigendum: design of a hyperstable 60-subunit protein icosahedron. Nature 540:150–150. doi: 10.1038/nature20108. [DOI] [PubMed] [Google Scholar]
- 41.Butterfield GL, Lajoie MJ, Gustafson HH, Sellers DL, Nattermann U, Ellis D, Bale JB, Ke S, Lenz GH, Yehdego A, Ravichandran R, Pun SH, King NP, Baker D. 2017. Evolution of a designed protein assembly encapsulating its own RNA genome. Nature 552:415–420. doi: 10.1038/nature25157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Ventura B, Lemerle C, Michalodimitrakis K, Serrano L. 2006. From in vivo to in silico biology and back. Nature 443:527–533. doi: 10.1038/nature05127. [DOI] [PubMed] [Google Scholar]
- 43.Karr JR, Sanghvi JC, Macklin DN, Gutschow MV, Jacobs JM, Bolival B, Assad-Garcia N, Glass JI, Covert MW. 2012. A whole-cell computational model predicts phenotype from genotype. Cell 150:389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wysocka J, Herr W. 2003. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem Sci 28:294–304. doi: 10.1016/S0968-0004(03)00088-4. [DOI] [PubMed] [Google Scholar]
- 45.Isomura H, Stinski MF. 2003. The human cytomegalovirus major immediate-early enhancer determines the efficiency of immediate-early gene transcription and viral replication in permissive cells at low multiplicity of infection. J Virol 77:3602–3614. doi: 10.1128/jvi.77.6.3602-3614.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith JA, Pari GS. 1995. Expression of human cytomegalovirus UL36 and UL37 genes is required for viral DNA replication. J Virol 69:1925–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schommartz T, Tang J, Brost R, Brune W. 2017. Differential requirement of human cytomegalovirus UL112-113 protein isoforms for viral replication. J Virol 91:e00254-17. doi: 10.1128/JVI.00254-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swenson JJ, Holley-Guthrie E, Kenney SC. 2001. Epstein-Barr virus immediate-early protein BRLF1 interacts with CBP, promoting enhanced BRLF1 transactivation. J Virol 75:6228–6234. doi: 10.1128/JVI.75.13.6228-6234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agelidis AM, Shukla D. 2015. Cell entry mechanisms of HSV: what we have learned in recent years. Future Virol 10:1145–1154. doi: 10.2217/fvl.15.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sathiyamoorthy K, Hu YX, Möhl BS, Chen J, Longnecker R, Jardetzky TS. 2016. Structural basis for Epstein-Barr virus host cell tropism mediated by gp42 and gHgL entry glycoproteins. Nat Commun 7:13557. doi: 10.1038/ncomms13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen CC, Kamil JP. 2018. Pathogen at the gates: human cytomegalovirus entry and cell tropism. Viruses 10:704. doi: 10.3390/v10120704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martínez-Martín N, Viejo-Borbolla A, Alcami A. 2016. Herpes simplex virus particles interact with chemokines and enhance cell migration. J Gen Virol 97:3007–3016. doi: 10.1099/jgv.0.000616. [DOI] [PubMed] [Google Scholar]
- 53.Aubert M, Chen Z, Lang R, Dang CH, Fowler C, Sloan DD, Jerome KR. 2008. The antiapoptotic herpes simplex virus glycoprotein J localizes to multiple cellular organelles and induces reactive oxygen species formation. J Virol 82:617–629. doi: 10.1128/JVI.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zimmermann C, Büscher N, Krauter S, Krämer N, Wolfrum U, Sehn E, Tenzer S, Plachter B. 2018. The abundant tegument protein pUL25 of human cytomegalovirus prevents proteasomal degradation of pUL26 and supports its suppression of ISGylation. J Virol 92:e01180-18. doi: 10.1128/JVI.01180-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bellows DS, Howell M, Pearson C, Hazlewood SA, Hardwick JM. 2002. Epstein-Barr virus BALF1 is a BCL-2-like antagonist of the herpesvirus antiapoptotic BCL-2 proteins. J Virol 76:2469–2479. doi: 10.1128/jvi.76.5.2469-2479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jamieson AT, Gentry GA, Subak-Sharpe JH. 1974. Induction of both thymidine and deoxycytidine kinase activity by herpes viruses. J Gen Virol 24:465–480. doi: 10.1099/0022-1317-24-3-465. [DOI] [PubMed] [Google Scholar]
- 57.Littler E, Stuart AD, Chee MS. 1992. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analogue ganciclovir. Nature 358:160–162. doi: 10.1038/358160a0. [DOI] [PubMed] [Google Scholar]
- 58.Baxter MK, Gibson W. 2001. Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J Virol 75:6865–6873. doi: 10.1128/JVI.75.15.6865-6873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jean Beltran PM, Federspiel JD, Sheng X, Cristea IM. 2017. Proteomics and integrative omic approaches for understanding host-pathogen interactions and infectious diseases. Mol Syst Biol 13:922. doi: 10.15252/msb.20167062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. 2006. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34:D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D. 2000. DIP: the database of interacting proteins. Nucleic Acids Res 28:289–291. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, Campbell NH, Chavali G, Chen C, del-Toro N, Duesbury M, Dumousseau M, Galeota E, Hinz U, Iannuccelli M, Jagannathan S, Jimenez R, Khadake J, Lagreid A, Licata L, Lovering RC, Meldal B, Melidoni AN, Milagros M, Peluso D, Perfetto L, Porras P, Raghunath A, Ricard-Blum S, Roechert B, Stutz A, Tognolli M, van Roey K, Cesareni G, Hermjakob H. 2014. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res 42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calderone A, Castagnoli L, Cesareni G. 2013. mentha: a resource for browsing integrated protein-interaction networks. Nat Methods 10:690–691. doi: 10.1038/nmeth.2561. [DOI] [PubMed] [Google Scholar]
- 64.Guirimand T, Delmotte S, Navratil V. 2015. VirHostNet 2.0: surfing on the web of virus/host molecular interactions data. Nucleic Acids Res 43:D583–D587. doi: 10.1093/nar/gku1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berman H, Henrick K, Nakamura H. 2003. Announcing the worldwide Protein Data Bank. Nat Struct Biol 10:980–980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 66.Lawson CL, Baker ML, Best C, Bi C, Dougherty M, Feng P, van Ginkel G, Devkota B, Lagerstedt I, Ludtke SJ, Newman RH, Oldfield TJ, Rees I, Sahni G, Sala R, Velankar S, Warren J, Westbrook JD, Henrick K, Kleywegt GJ, Berman HM, Chiu W. 2011. EMDataBank.org: unified data resource for CryoEM. Nucleic Acids Res 39:D456–D464. doi: 10.1093/nar/gkq880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 68.Stellberger T, Häuser R, Baiker A, Pothineni VR, Haas J, Uetz P. 2010. Improving the yeast two-hybrid system with permutated fusions proteins: the Varicella Zoster virus interactome. Proteome Sci 8:8. doi: 10.1186/1477-5956-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.To A, Bai Y, Shen A, Gong H, Umamoto S, Lu S, Liu F. 2011. Yeast two hybrid analyses reveal novel binary interactions between human cytomegalovirus-encoded virion proteins. PLoS One 6:e17796. doi: 10.1371/journal.pone.0017796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Physical schema of the HVint2.0 database. The data in HVint2.0 are organized into seven tables: i.e., “ppi,” “protein,” “evidence,” “evidence_to_psimi,” “psimi_ontology,” “taxon,” and “citation.” The content of each of table is detailed in the accompanying portion of Text S1. For each table, the figure displays the table name and the list of columns in the table, together with their data type. Primary keys are indicated with a key icon next to the corresponding column name; columns that can and cannot hold null values are indicated with empty and filled rhomboid icons, respectively. Download FIG S1, TIF file, 0.5 MB (505.3KB, tif) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Network reconstruction framework. (A) PPI data are collected from several public data resources and for the taxons of interest. These data include PPIs in the target taxon and in a number of orthologous species. (B) PPIs detected in orthologous species are the input for stage, where sequence-based homology assignments are used to predict new PPIs in the target taxon (green enlarged frame). (C) Predicted PPIs (pPPIs) and PPIs originally detected in the target taxon present in the input data set (tPPIs) are nonredundantly integrated in the final network. Each PPI in the final network is scored under the same confidence scoring scheme. (D) This framework was applied, separately, for each of the three representative target species of human herpesviruses (HSV-1, HCMV, and EBV) to reconstruct a corresponding network. Download FIG S2, TIF file, 0.7 MB (762.9KB, tif) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Database structure and content. Download Text S1, DOCX file, 0.02 MB (18.6KB, docx) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Number of interactions per primary citation. Here a large-scale study was defined as providing more than 50 PPIs. The graph shows that in the case of the reconstructed interactomes, only Y2H experiments provide that amount of interaction (corresponding to the four largest bars [6, 8, 68, 69]). Download FIG S3, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of core herpesvirus proteins used in this study. Proteins in the same row are homologues. Download Table S1, XLSX file, 0.01 MB (8.3KB, xlsx) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Functional annotation for proteins in the central interactome. Data were retrieved from the HVint2.0 database. Download Table S2, XLSX file, 0.01 MB (7.8KB, xlsx) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PDB entries associated with proteins and protein complexes appearing in the reconstructed Herpesviridae central interactome (Fig. 4). These entries can contain structural data for the full complex or only part of it (e.g., domains). Data obtained for homologous species are grouped in the same row using the HSV-1 nomenclature. Download Table S3, DOCX file, 0.02 MB (23.2KB, docx) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inferred herpesviral central interactome and species-specific subnetwork. The inferred herpesviral central interactome is shown at the center, flanked by the subsets of species-specific interactions for each of the three species from the reconstructed interactomes. Gray edges and nodes indicate PPIs and proteins found in the central interactome. These are referred to using HSV-1 open reading frame nomenclature. Red, blue, and green edges and nodes indicate PPIs and proteins from the species-specific subnetworks in HSV-1, HCMV, and EBV, respectively. Edge thickness reflects confidence scores. For central interactome PPIs, this score was calculated as the sum of the scores for the interaction in each species it was found over the maximum number of species where the interaction could exist (i.e., 3). In species-specific subnetworks, this is calculated using the scoring function integrated in our network reconstruction framework. Node size is proportional to the number of interactions of each protein. Download FIG S4, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Functional annotation for proteins in all three species-specific network fractions. Data were retrieved from the HVint2.0 database. Download Table S4, DOCX file, 0.05 MB (50.1KB, docx) .
Copyright © 2019 Hernández Durán et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The data sets produced in this study are available by clicking on the “Viruses” dropdown menu at http://topf-group.ismb.lon.ac.uk/hvint2.