Abstract
Substrates for embryonic stem cell culture are typified by poorly defined xenogenic, whole proteins or cellular components that are difficult and expensive to generate, characterize, and recapitulate. Herein, the generation of well-defined scaffolds of Gly-Tyr-Ile-Gly-Ser-Arg (GYIGSR) peptide-functionalized poly(ԑ-caprolactone) (PCL) aligned nanofibers are used to accelerate the neural lineage commitment and differentiation of D3 mouse embryonic stem cells (mESCs). Gene expression trends and immunocytochemistry analysis were similar to laminin-coated glass, and indicated an earlier differentiation progression than D3 mESCs on laminin. Further, GYIGSR-functionalized nanofiber substrates yielded an increased gene expression of Sox1, a neural progenitor cell marker, and Tubb3, Cdh2, Syp, neuronal cell markers, at early time points. In addition, guidance of neurites was found to parallel the fiber direction. Herein, we demonstrate the fabrication of a well-defined, xeno-free functional nanofiber scaffold and demonstrates its use as a surrogate for xenogenic and complex matrixes currently used for the neural differentiation of stem cells ex vivo.
Graphical Abstract
Introduction
The differentiation of stem cells in vitro yields novel sources of cells for neural tissue replacement or repair. Complex protein solutions, such as Matrigel®, have been utilized extensively as substrates for differentiation.[1] However, Matrigel® is xeno-produced, differs from batch to batch, and contains a mixture of extracellular matrix (ECM) proteins, including laminin, collagen IV, entactin, heparin sulfate proteoglycans, and a multitude of growth factors.[2] While xenogenic factors and additives have proven useful in vitro for mechanistic evaluation and development, the translation potential of this approach into clinical environments is limited, as xeno-derived components can initiate potent immune responses.[3–4] In addition, several regulatory challenges exist to using these factors clinically. In order to push the translation of stem cells to clinical practice, synthetic xeno-free culture systems with defined concentration and spatial presentation of bioactive species for directed differentiation of ESCs and maintenance of cell maturity are required.[5]
In response to these challenges, polymeric substrates mimicking ECM elasticity, stiffness,[6–7] geometrical architecture,[8–9] chemical cues[8, 10–11] and a combination of these factors[12–14] have been explored to push stem cell differentiation into neural lineages with some success. However, the relative contributions of each these microenvironment parameters and how their combinations control cell behavior is still not completely understood. For neural tissue engineering, aligned fibers are of particular interest due to a highly polarized pattern of nerve cells. Aligned substrates have been shown to improve neural cell alignment and migration, guide neural progenitor differentiation, and direct neurite extension during development and regeneration.[8, 15–21]
Electrospinning affords the fabrication of polymeric fiber meshes with nano- to micrometer topologies that mimic the architecture of native ECM.[22–25] Electrospun fibers influence stem cell behavior by mimicking ECM properties including fiber diameter and alignment (via modification of voltage, tip-to-collector distance, solvent composition and solution concentration[26–30]) and controlling the concentration and spatial placement of bioactive species. Electrospinning of ECM adhesive proteins including collagen,[31] gelatin[32–33] or laminin[34] has been used widely to produce cellular substrates, but most of the bioactive molecules are hidden in the bulk and unavailable for cell-substrate interactions, and are expensive to manufacture. Furthermore, ECM proteins often lose their structural functionality during electrospinning due to the stretching of molecules and denaturation.[35–36] In contrast, most synthetic substrates lack biological signaling found in the natural ECM,[37–38] but can be modified with bioactive species including peptides, growth factors and carbohydrates to yield simple, scalable and cost-effective substrates with improved cell-matrix interactions.[39]
Laminin is the most abundant glycoprotein present in basement membranes, appears at the very early stage during embryogenesis,[40–41] and is a major component of Matrigel®.[1] It has various structural and biological activities including promotion of cell adhesion, migration, growth and differentiation.[41–42] Substituting short synthetic peptides corresponding to binding domains of long protein chains[43] for full proteins enables scalable, cost-effective substrate fabrication. For example, the six amino acid GYIGSR sequence, found in the B1 laminin chain, has been shown to exhibit cell adhesion, attachment, migration and binding to the 67 kDa laminin receptor.[44–46]
Recently, we investigated strain-promoted azide-alkyne cycloaddition (SPAAC),[47–50] for the post-electrospinning attachment of bioactive species to degradable polyesters.[26, 51–54] This approach affords facile, quantitative modification of 4-dibenzocyclooctynol (DIBO)-functionalized PCL with azide-derivatized compounds with no catalyst or chemical activation. Post-electrospinning surface modification method is the most efficient way to attach bioactive species to nanofibers. It affords control of concentration and spatial presentation in contrast to adsorbed bioactive species. Unlike conjugation methods that occur prior to electrospinning, where a significant fraction of bioactive species is hidden within the fiber and not available for interacting with target cells, post-electrospinning surface modification results in the bioavailability of the tethered groups.[54]
PLLA nanofiber scaffolds with tethered GYIGSR have previously been shown to enhance mESCs commitment to neural lineage within 3 days.[26] However, further characterization regarding the commitment and maturation of the mESC over longer times were not reported. Therefore, this study investigated mESC commitment, differentiation, and maturation on aligned PCL nanofiber substrates functionalized with GYIGSR peptide for up to 14 days. By changing the degradable polyester to PCL, this work will enable the introduction of multiple functionalities in the polymer chain for post-electrospinning modification with biomolecules in a controlled manner.[53–54]
Materials and methods
Materials
All materials were used as received unless otherwise stated. Tetrahydrofuran (anhydrous, ≥99.9%, inhibitor-free), chloroform (anhydrous, contains amylenes as stabilizer, ≥99%), and calcium hydride (reagent grade, 95%) were purchased from Sigma-Aldrich (St. Louis, MO). Phenylacetaldehyde (98%, stabilized), lithium diisopropylamide mono(tetrahydrofuran) (1.5 M solution in cyclohexane, AcroSeal™), iodotrimethylsilane (95–97%), n-butyllithium (2.5 M solution in hexanes, AcroSeal™), hexanes and methylene chloride were purchased from Fisher Scientific (Houston, TX). Sodium thiosulfate pentahydrate (Proteomics grade, 99%) was purchased from Amresco, LLC (Solon, OH). 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) was purchased from Oakwood Products, Inc. (Estill, SC). Sodium sulfate anhydrous (ACS grade) and methanol (ACS grade), hydrochloric acid (36.5–38%, ACS Grade) were purchased from VWR International (Radnor, PA). Dry toluene (HPLC Grade, 99.7%, Alfa Aesar) for polymerization was purified and dried on an Inert Pure Solv system (MD Solvent Purification system, model PS-MD-3) and degassed using three cycles of the freeze-vacuum-thaw. ε-Caprolactone (ε-CL, 99%, ACROS Organics™) was dried over calcium hydride under nitrogen overnight and distilled under reduced pressure. Magnesium 2,6-di-tert-butyl-4-methylphenoxide catalyst [Mg(BHT)2(THF)2] was synthesized using methods described previously.[55–56] 4-dibenzocyclooctynol (DIBO) initiator was synthesized using methods described previously.[48, 50, 52, 57–58] Resins for peptide synthesis (Novabiochem®) were purchased from EMD Millipore (Billerica, MA). Fmoc-amino acids were purchased from Aapptec (Louisville, KY). Flash chromatography was performed on silica gel (Sorbent Technologies Inc., 70–230 mesh).
Square (22 × 22 mm) and round (8 mm) Fisherbrand™ borosilicate cover glasses (#1.5) were washed with methanol/toluene/methanol, dried with nitrogen and cleaned with UV light (355 nm) for 3 min prior to use. After nanofibers were collected on the glass coverslips, the nanofiber mats were glued to the edges of a glass slide by a silicone sealant and dried under vacuum overnight.
Experimental Methods
Proton 1H nuclear magnetic resonance (NMR) (300 MHz and 500 MHz) spectra were recorded on Varian Mercury 300 and 500 spectrometers. The polymers were dissolved in CDCl3 solvent at 15 mg/mL, the relaxation time was 2 sec with 64 transients.
Size exclusion chromatography (SEC) was used to determine molecular mass and molecular mass distributions (ĐM). Eluograms were collected on a Tosoh EcoSEC HLC-8320GPC using N,N-dimethylformamide (DMF) containing 0.1 M lithium bromide as the eluent. The 2 columns were calibrated using narrow molecular mass polystyrene standards (20 standards from 0.5 kDa to 5,480 kDa).
Nanofiber scaffolds were sterilized by ethylene oxide using an Anprolene benchtop sterilizer (Anderson Products, Inc., Haw River, NC) according to the manufacturer’s protocol for 12 h at room temperature and 35% humidity (concentration of ethylene oxide is about 0.5 g/L), purged for at least 48 h and stored in vacuum desiccator until cell study.
Materials for cell study
Mouse embryonic stem cells (D3) were obtained from ATCC, and cultured without a cell feeder support layer. ES-fetal bovine serum (ES-FBS, ES009B), 0.1% gelatin (ES006b), β-mercaptoethanol (100X, ES007E), sodium bicarbonate (S6014), sodium pyruvate (S8636), retinoic acid (R2625), insulin (I1882), apo-transferrin (T1147), progesterone (P8783), putrescine (P5780), sodium selenite (S5261), trypsin-EDTA (T4174), and bisbenzimide H 33342 fluorochrome, trihydrochloride (H33342, 382065) were obtained from MilliporeSigma (St. Louis, MO). Dulbecco’s Modification of Eagle’s Medium (90–013-PB), DMEM/F-12 (MT 15090CM) was obtained from Corning (Corning, NY). L-glutamine (100X, 25030149), neurobasal medium (12349–015), paraformaldehyde (04042–500), bovine serum albumin (BP9706–100), donkey anti-rabbit IgG Alexa Fluor 647 (A10040), goat anti-mouse IgM Alexa Fluor 546 (A-21045), donkey anti-mouse Alexa Fluor 546 (A-10036), RNase-free glycogen (R0551), Trizol® (15596026), and molecular biology grade isopropanol (BP2618–500) were purchased from ThermoFisher Scientific (Waltham, MA). mESC-qualified recombinant human recombinant leukemia inhibitory factor (LIF) (GSR-7001) and PluriQ serum replacement (GSM-6102) were purchased from ThermoFisher Scientific. ES-qualified HEPES buffer (SH30851.01) was purchased from GE Healthcare (Chicago, IL). Triton X-100 (8698.5–16) was purchased from RICCA Chemical Company (Arlington, TX). Sodium borohydride (02102894) was purchased from MP Biomedicals (Santa Ana, CA). Perfecta SYBR Green SuperMix, Low ROX (95056–050) was obtained from Quanta Biosciences (Beverly, MA). The following antibodies were purchased from Abcam: anti-beta III tubulin (TUBB3, ab107216), goat anti-chicken IgY Alexa Fluor 488 (ab150169), anti-SOX1 (ab22572), anti-nestin (NES, ab134017), anti-OCT-4 (ab19857), anti-MAP2 (ab11267), anti-oligodendrocyte specific protein (ab53041), anti-CNPase (ab6319), antiGAP43 (ab16053). The following antibodies were purchased from Biolegend: PE antimouse/human CD15 (SSEA-1) antibody (125606) and PE mouse IgM isotype control (401611).
Synthesis of DIBO-end functionalized poly(ԑ-caprolactone)
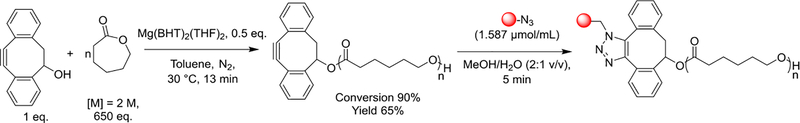
The synthesis of DIBO-end functionalized poly(ԑ-caprolactone) and post electrospinning modification of the nanofibers with peptides via strain-promoted azide-alkyne cycloaddition is shown in Scheme 1. Using standard drying techniques, a glass ampoule was filled with ԑ-caprolactone (22.16 mL, 0.200 mol), toluene (76.83 mL, 0.723 mol) DIBO (0.0678 g, 0.308 mmol) and Mg(BHT)2(THF)2 (0.0941 g, 0.155 mmol). The ampoule was sealed and heated at 30 °C for 13 min. The polymerization was quenched with the addition of acidified (5 % v/v HCl) methanol, dissolved into chloroform and precipitated into cold methanol. The crude polymer was re-dissolved in methylene chloride, precipitated into cold methanol and dried under high vacuum. The purified polymer was then stored in a desiccator. The monomer conversion (90%) and product (yield 65%) were determined by 1H NMR spectroscopy (Figure S2), UV-visible spectrophotometry (306 nm) and SEC (Figure S3, Mn = 60,600 Da, Mw = 83,700 Da, ĐM = 1.38). 1H NMR (500 MHz, CDCl3, 303 K): δ = 7.51 – 7.45 (m, aromatic), 5.56 (dd, 3JH-H = 3.2, 2.5 CHOH), 4.15 – 3.98 (m, CH2CH2OCH2), 3.10 (dd, 3JH-H = 15.2, 2.1 Hz, CH(H)CH), 2.93 (dd, 3JH-H = 15.1, 3.9 Hz, CH(H)CH), 2.38 – 2.22 (m, (C=O)CH2CH2), 1.72 – 1.54 (m, (C=O)CH2CH2CH2CH2CH2), 1.45 – 1.28 (m, (C=O)(CH2)2CH2((CH2)2) ppm.
Scheme 1.
DIBO-end-functionalized poly(ԑ-caprolactone) was synthesized via ring-opening polymerization of ԑ-caprolactone using DIBO as an initiator and was modified postelectrospinning with peptides via strain-promoted azide-alkyne cycloaddition.
Electrospinning conditions and nanofiber collection
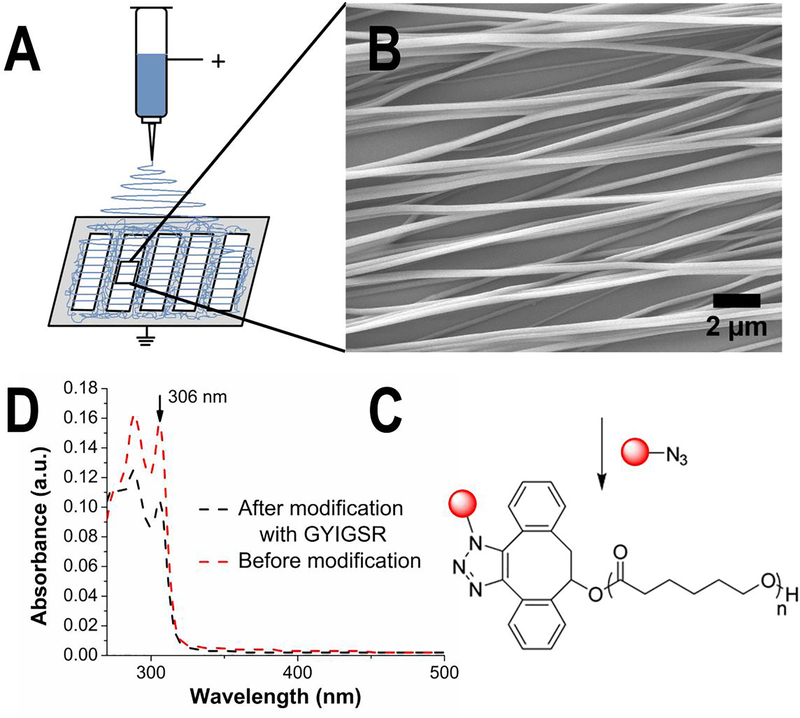
The electrospinning setup for aligned nanofiber scaffolds is shown in Figure 1 (A). For aligned fiber scaffolds, the DIBO-terminated PCL was dissolved in HFIP (17% (w/v)) to yield a clear, slightly viscous solution. The solution was placed in a 2 mL glass syringe with a 22 gauge needle (JG22–0.5X, Jensen Global Dispensing Solutions). A voltage of 15 kV was applied to the solution, and the tip-to-collector distance was set to 10 cm. The gap size for the metal collector plate was 24 × 110 mm. Aligned nanofibers were collected by placing cover glasses in between the gaps of the collector. The collected nanofiber mats were glued to the edges of a glass slide by a silicone sealant and dried under vacuum overnight.
Figure 1.
Nanofibers were fabricated by electrospinning from a solution of DIBO-terminated poly(ԑ-caprolactone) in HFIP (17% w/v) and a voltage of 15 kV. (A) Highly aligned nanofibers were collected on cover glasses in the gaps of metal collector plate. (B) Analysis of SEM images was performed to estimate topography of nanofibers. NIH ImageJ was used to estimate fiber diameter (ᴓ = 212 ± 63 nm) and alignment (Directionality™ plugin, average angle = 0 ± 6°). (C) Post-electrospinning modification with GYIGSR peptide via strain-promoted azide-alkyne cycloaddition. The concentration of peptide (17.3 ± 6.6 pmol/cm2) was measured using (D) UV-visible spectrophotometry by comparison of absorbance at 306 nm (peak corresponding to DIBO groups) before (red curve) and after (black curve) post-electrospinning modification based on calibration curve of DIBO in chloroform.
Characterization of diameter and orientation
Nanofiber dimensions and alignment were imaged by scanning electron microscope (SEM) with an applied voltage of 5 kV (JSM-7401F, JEOL, Peabody, MA). Samples were sputter coated for 30 seconds with silver under nitrogen atmosphere prior to imaging. A UVO Cleaner, Model #42A UV light unit was used to clean the glass coverslips for nanofiber collection. High voltage power supply (ES30P-5W, Gamma High Voltage, Ormond Beach, FL) was used for electrospinning. The variation in nanofiber diameters was measured on at least 3 independent samples (5 images of each sample with >150 fibers per sample) using NIH ImageJ[59] and reported as an average ± standard deviation. Distributions of fiber diameters are shown in Figure S4 (A). The Directionality™ plugin of ImageJ[60] was used to quantify the relative degree of alignment of the scaffolds by analyzing the angle distribution of fibers (Figure S4 (B)). The value is reported as an average ± standard deviation. Fityk 0.9.8 was used to fit a Gaussian function (red curve), and calculate average angle as the peak of the fit distribution.[61] Angles were normalized to 0. The highest peak was normalized to 1. Angle distribution of diameter directions was calculated using Gaussian fitting parameters. The quality (goodness) of fit to the Gaussian distribution curve calculated by Directionality™ plugin was reported as average ± standard deviation.
Solid phase peptide synthesis
N3-GYIGSR peptide was synthesized using standard FMOC conditions on a CEM Discovery microwave peptide synthesizer. The N-terminus was derivatized with 6-azidohexanoic acid.[62] The desired peptide product was confirmed by electrospray ionization mass spectrometry for N3-GYIGSR [M + Na]+ = 813.4 Da, yield = 71% (Figure S5).
Electrospray ionization (ESI) mass spectrometry experiments
Synthesized peptide was analyzed using mass spectrometry. The spectra were recorded using a Bruker HCT ultra II quadrupole ion trap mass spectrometer (Bruker Daltonics, Billerica, MA) by direct infusion with a syringe pump at a flow rate of 250 μL/h. The temperature and flow rate of the drying gas (N2) were 300 °C and 8 L/min, respectively; the pressure of the nebulizing gas (N2) was set at 10 psi. Stock solutions of the peptides were prepared in H2O at 10 mg/mL. The sprayed samples were prepared by adding 1 μL of the peptide solution to 500 μL H2O and 500 μL of MeOH to obtain a final peptide concentration of 0.01 mg/mL in 1:1 (v/v) H2O:MeOH.
Nanofiber functionalization
Nanofiber covered glass slides were dipped into a solution of the respective azide-functionalized peptide (1.587 μmol/mL) in 1:2 water/methanol (v/v) solution for 5 min. The cover slips with functionalized nanofibers were rinsed with 1:2 water/methanol (v/v) solution, blown with nitrogen and dried overnight in a desiccator. Scaffolds were sterilized using an ethylene oxide exposure cycle for 12 h, degassed for 2 days and stored in a vacuum desiccator until the cell studies.
The extent of functionalization with each peptide (reported as an average ± standard deviation) was confirmed using UV−visible spectrophotometry using chloroform as a solvent. The peak intensity at 306 nm (which corresponds to π-π* transition in alkyne bond in DIBO-functionalized polymer) decreases after reaction with azide-functionalized peptide in comparison with fibers before functionalization. The concentration of GYIGSR peptide was measured using UV-visible spectrophotometry (Synergy™ MX plate reader from BioTek, with spectral resolution 1 nm).
D3 mouse embryonic stem cell culture and seeding
D3 mESC were maintained feeder-free using 0.1% gelatin coated flasks in pluripotent media (DMEM with high glucose, 10% FBS, 0.1mM 2-mercaptoethanol, 4 mM L-glutamine, 4.7 mM HEPES, and 1000 U/mL LIF). Pluripotent cells were passaged every other day. All experiments utilized cells with less than 15 passages. The expression of SSEA-1 was utilized to confirm pluripotency at the time of seeding by flow cytometry. Cells were seeded onto scaffolds at 125,000 cells/cm2 in neural differentiation media (80% 1:1 DMEM:F-12, 20% neurobasal-A medium, 1X N2 (50 μg/mL insulin, 1 mg/mL apo-transferrin, 60 ng/mL progesterone, 160 μg/mL putrescine, 0.3 μM sodium selenite, 0.5 mg/mL BSA, 6 mg/mL D-Glucose, 5mM HEPES, diluted in DMEM/F12), 1X PluriQ, 1mM sodium pyruvate, 2 mM L-glutamine, and 2 μM retinoic acid). Differentiation status was determined at day 1, 3, 7, and 14 using both gene and protein expression.
Quantitative polymerase chain reaction (qPCR)
RNA isolation was performed using Trizol® according to the manufacturer’s protocol. Two samples were combined prior to RNA isolation for scaffolds, while one sample was utilized for RNA isolation from cells on laminin substrates. UV-visible spectrophotometry was utilized to quantify RNA and gel electrophoresis was performed on every RNA sample to confirm quality using 2% agarose gel with ethidium bromide. Quanta qScript DNase kit was used following the manufacturer’s protocol to digest any genomic DNA. Quanta qScript reverse transcriptase (RT) kit was used following manufacturer’s protocol to synthesize cDNA. The synthesized cDNA was stored at 4 °C until qPCR was performed. Real time PCR was performed using primers (Integrated DNA Technologies, Coralville, IA) in Table 1. No-template controls and no RT controls were tested on each sample at the same primer concentration for housekeeping genes. Reactions were prepared using Perfecta SYBR Green SuperMix, Low ROX and Applied Biosystems 7500 real time PCR system was used at a standard run.
Table 1.
Primers used for real time qPCR with accession numbers, forward and reverse sequences.
| Gene | Primer Accession Number | Forward Sequence | Reverse Sequence |
|---|---|---|---|
| Gapdh | NM_008084.3 | AAT GGT GAA GGT CGG TGT G | GTG GAG TCA TAC TGG AAC ATG TAG |
| Actb (β-actin) | NM_007393.5 | GCT GTA TTC CCC TCC ATC GTG | CAC GGT TGG CCT TAG GGT TCA G |
| Pou5f1 (Oct-4) | NM_013633.3 | GGC ACT TCA GAA ACA TGG TCT | GAA GCC GAC AAC AAT GAG AAC |
| Sox1 | NM_009233 | GGC AGT CAT ACA AAA GTT GGC | GTA CAG TAT TTA TCG TCC GCA GA |
| Pax6 | NM_001310144.1 | AAG GGC GGT GAG CAG ATG T | CAT GCT GGA GCT GGT TGG |
| Nes | NM_016701.3 | CAC CTC AAG ATG TCC CTT AGT C | GGA AAG CCA AGA GAA GCC T |
| Tubb3 | NM_023279.2 | CCT CCG TAT AGT GCC CTT TG | GTG GAC TTG GAA CCT GGA AC |
| Map2 | NM_001039934.1 | CCA CTA ATG CCA GTT TCT CTC T | GAC CCA GAG TGT GTG AGT TTA T |
| Gfap | NM_010277.3 | CCA CCA GTA ACA TGC AAG AGA | GCG ATA GTC GTT AGC TTC GTG |
| Th | NM_009377.1 | CCC TAC CAA GAT CAA ACC TAC C | CTG GAT ACG AGA GGC ATA GTT C |
| Foxo4 | NM_018789.2 | GCTCTTGGTGGATGCTGAAC | AACTGCTTCGTGGACGGAAA |
| Cdh2 (n-cadherin) | NM_007664 | GCCCGCTATTTGTTACCAGC | CACAGACGCCT GAAGCAAGG |
| Gap43 | NM_008083.2 | AGG AGG AGA AAG ACG CTG TA | TCA GGC ATG TTC TTG GTC AG |
| Syp | NM_009305.2 | TTT GGA GGG TGA GCG AAA TG | AGA GAA AGG GTG GAG AAG GTA G |
| Olig1 | NM_016968 | TCC AGA CTT CTC TCC CAG AC | AGC AAC TAC ATC GCT CCT TG |
Immunocytochemistry
Protein expression was evaluated using immunocytochemistry (ICC). Cells were fixed at the appropriate timepoint with freshly prepared 4% paraformaldehyde in 1X PBS for 10 minutes at ambient temperature. Cells were permeated with 0.5% Triton X-100 for 10 minutes, quenched with 1 mg/mL sodium borohydride for 8 minutes, and blocked with 2 mg/mL BSA for 40 minutes. Cells were incubated with primary antibodies at 4 °C for 10 hours, washed 3 times, and incubated with secondary antibodies for ~8 hours at 4 °C. All cells were labeled with H33342. Day 1 samples were labeled with the pluripotency markers POU5F1 and SSEA-1, early neural markers NES and SOX1, and the neural marker TUBB3. Day 3 samples were labeled with the pluripotency marker POU5F1, early neural markers NES and SOX1, neuronal marker TUBB3, and glial marker GFAP. Day 7 and day 14 samples were labeled with the neuronal markers TUBB3, MAP2 and GAP43, and the glial markers FOXO4, OLIG1, GFAP and CNPase.
Images were taken on Zeiss AxioObserver Z1 microscope (Zeiss, Thornwood, NY) or Olympus FV1000 (Tokyo, Japan) at exposures relative to controls with only secondary antibodies. Up to five images were taken per substrate. To provide semi-quantitative information from the images, we utilized several techniques within ImageJ including pixel quantification, percent labeled cells, and neurite alignment.
Pixel Quantification
The control images, taken on substrates processed with no primary antibodies, were thresholded to less than 0.1% area fraction of fluorescence, and the thresholded values were used on the sample images. The area fraction of fluorescent pixels after thresholding was recorded for each fluorophore and image and normalized to area and the number of cells in the ROI. This pixel quantification was performed for images taken on the same microscope at the same magnification.
Percent labeled cells
To determine percentages of labeled cells, the number of cells expressing various markers on day 1 and day 3 were manually counted and expressed as % cells ± standard error of the mean.
Neurite alignment
Finally, to quantify the alignment of the neurites, all TUBB3 labeled neurites in images were traced using NeuronJ plugin of ImageJ. The fiber orientation was measured in the phase channel, and all tracings were oriented to the fibers at 0°. The tracings were then processed through Directionality™ plugin of ImageJ. Goodness of fit to a Gaussian curve was used to measure alignment of the neurites and reported as average angle ± standard deviation and by the width of the Gaussian peak at half its maximum intensity. This process also provided a measure of total neurite length per area, but as starting and ending positions of neurites could not be identified in each case, only the total neurite length was reported.
Statistics
All experiments were conducted at least 3 times (n ≥ 3). Cellular experiments have a minimum of 3 biological replicates. PCR data are presented as the average ± standard deviation and image analysis is expressed as average ± standard error of the mean. Two-way ANOVA with Bonferroni’s post hoc analysis was used to express statistical differences with 95% confidence interval and a significance value of p>0.05. Two-sample t-test was performed to show statistical difference between neurite alignment cultured on different substrates with 95% confidence interval and a significance value of p>0.05.
Results
PCL nanofibers functionalized with GYIGSR
DIBO-end functionalized poly(ԑ-caprolactone) was synthesized by ring-opening polymerization of ԑ-caprolactone using Mg(BHT)2(THF)2 as a catalyst using standard techniques. This method yielded high molecular mass PCL with high end group fidelity. High (90%) monomer conversion was obtained within 13 min at 30 °C and yielded polymer with high molecular mass and narrow molecular mass distribution (Mn = 60,600 Da, Mw = 83,700 Da, ĐM = 1.38). This polymer was used to fabricate highly aligned nanofiber scaffolds with a narrow angular distribution of fibers (0 ± 6°, average ± standard deviation). The average diameter of fibers was ᴓ = 212 ± 63 nm. Aligned nanofibers were modified post-electrospinning with GYIGSR peptide via strain-promoted azide-alkyne cycloaddition and the surface concentration of GYIGSR peptide was determined to be 17.3 ± 6.6 pmol/cm2.
mESC response
To investigate whether PCL nanofiber substrates with a tethered synthetic laminin mimic (GYIGSR peptide) supported neural differentiation of mESC, pluripotent D3 mESC (>95% SSEA-1 positive via flow cytometry) were seeded on the YIGSR nanofiber substrates and laminin-coated cover slips as a positive control. Cells were then cultured for 14 days in presence of neural differentiation medium. The progress of neural differentiation was assessed using gene expression by qPCR and protein expression by ICC at day 1, day 3, day 7 and day 14. Cultured cells formed aggregates on both GYIGSR-functionalized PCL nanofibers and laminin-coated glass (Figure S11).
Initially, the adhesion was investigated to assure sufficient cell interactions with the scaffold. The number of cells at day 1 was determined by counting the nuclei on each substrate sample. Of the seeded cells, 1.1 ± 0.2% adhered to the YIGSR-aligned scaffolds and 53.6 ± 7.0% of the cells adhered to laminin-coated glass. In a preliminary study on aligned nanofibers made from the same material but without any peptide or non-bioactive peptide RGES, only a few cells adhered to the surface (≤0.2% after 24 h, with fewer cells at day 3, data not shown), which was an insufficient amount to perform qPCR or ICC.
Gene Expression
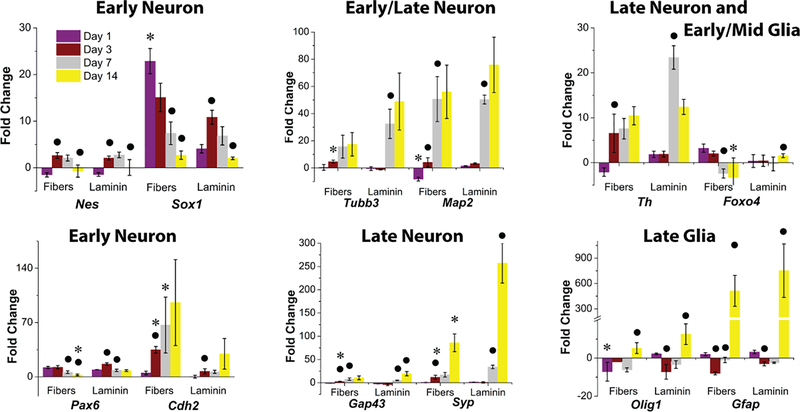
To characterize neural differentiation by gene expression, stage specific pluripotency markers, neural progenitors, early and late neural markers, and glial markers were analyzed by qPCR. Figure 2 and Figure S7 show fold change (normalized to average of two housekeeping genes – Gapdh and Actb and day 0) over time on YIGSR aligned fibers and laminin-coated glasses.
Figure 2.
Comparison of gene expression of D3 mESCs cultured on GYIGSR-functionalized aligned nanofibers and laminin-coated glass at day 1, 3, 7 and 14. Expression of neural progenitor (Nes, Sox1, Pax6) and neuronal (Tubb3, Map2, Cdh2, Gfap, Th, Gap43, Syp) as well as glial (Foxo4, Olig1, Gfap) gene markers demonstrated that aligned GYIGSR-functionalized nanofiber scaffolds have similar neuronal differentiation into neural lineage compared to laminin-coated glass. No significant differences were found between fibers and laminin glasses in most of the gene expression, with similar differentiation states at day 7 and day 14. Two exceptions of higher expression of neuronal genes on aligned GYIGSR fiber scaffolds at earlier time points were Sox1 at day1 and Cdh2 at day 3 and day 7. ● indicates statistically significant difference (p < 0.05) in comparison to the previous time point for the same substrate, * indicates that gene expression on fibers is statistically significantly different (p < 0.05) than on laminin for the same time point.
The pluripotent marker (Pou5f1) was downregulated in both groups (Figure S7) implying that cells were undergoing differentiation, with downregulation at day 14. Higher levels of Pou5f1 gene expression were found from cells on fibers compared to cells on laminin on day 1 and day 3, but the result was not significantly different on day 7 and 14.
Neural commitment was confirmed by expression of neural progenitor genes (Sox1, Pax6, Nes). Expression of these neural progenitor markers was upregulated at day 1 and day 3 and then down-regulated by day 14. Expression of Sox1 was significantly higher for cells on fibers than cells on laminin at day 1, but no significant differences were found between samples at each later time point (day 3, 7 and 14). Expression of Pax6 was not significantly different between samples for day 1, 3 and 7, but had a significantly lower level from cells on fibers than cells on laminin at day 14. No significant difference was noticed on Nes expression trends and on either substrate.
Cdh2, responsible for neural cell to cell interaction, had the same expression for both samples at day 1, but a significantly higher level of expression from cells on fibers at days 3 and 7. At day 14, the difference between the two samples was not significant.
After neural commitment, cells start to express neuronal markers (Tubb3, Map2, Gap43, Syp, Th), which indicated differentiation into neural lineage. Both Tubb3 and Map2 were up-regulated on day 7 in comparison to day 3. Expression of Tubb3 at day 7 and 14 by cells on laminin and cells on fibers had no significant differences from each other. Similarly, no differences were found at days 7 and 14 for Map2. However, early stage neuronal marker Tubb3 was expressed earlier by cells on fibers, with significant differences compared to cells on laminin at day 3. Gap43 was expressed at a higher level on fibers than on laminin at day 3. Late stage neuronal marker Syp, a protein that plays a role in synaptic plasticity[63], was upregulated during the differentiation, with the highest expression on day 14. The expression of Syp was increased for cells on laminin over cells on fiber substrates for day 3 and 14. Expression of tyrosine hydroxylase (Th), associated with dopaminergic neurons, demonstrated statistical increases at day 3 compared to day 1 on fibers (at day 3), but not until day 7 on laminin.
Glial cell markers expressed similarly on both substrates at similar time points, except Foxo4 at day 14 and Olig1 at day 1, where the gene expression was lower for cells on fibers. Gfap had significant increases (511-fold increase on fibers and 565-fold increase on laminin) by cells on both substrates by day 14.
Protein Expression
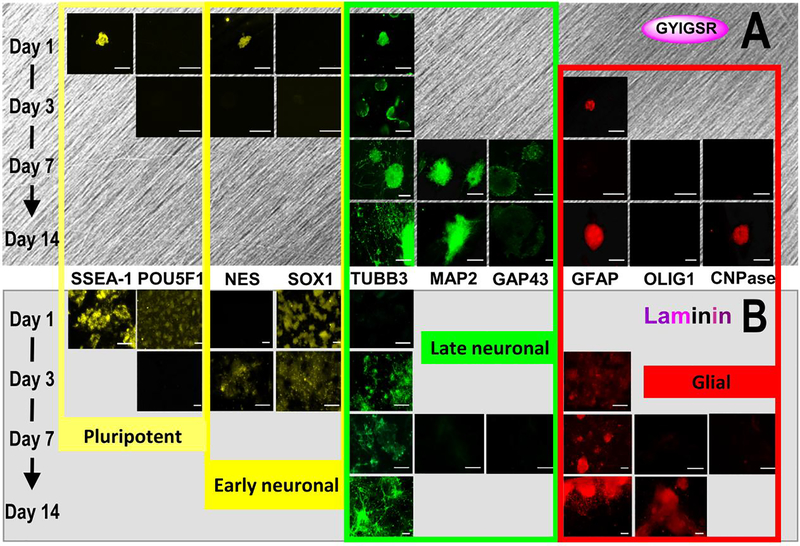
The expression of proteins typical for pluripotent state (SSEA-1, POU5F1), neural progenitors (NES, SOX1), neural (TUBB3 for early neuronal, MAP2 and GAP43 for late neuronal stage), and glial cells (GFAP for astrocytes, OLIG1 and CNPase for oligodendrocytes) was visualized and quantified. Third quartile images, based on pixel quantification, are shown in Figure 3.
Figure 3.
Protein expression during differentiation on YIGSR-aligned fibers and laminin over 14 days. Displayed images are from the 3rd quartile of fluorescent intensity for each protein. Expression is noted as typical for pluripotent state (SSEA-1, POU5F1), neural progenitors (NES, SOX1) neural (TUBB3, MAP2, GAP43) and glial cells (GFAP, OLIG1, CNPase), scale bars = 100 μm. Images indicate similar neural differentiation on (A) aligned GYIGSR-functionalized nanofiber scaffolds and (B) laminin glasses but with faster rates on fibers (earlier expression of NES, TUBB3, MAP2 and GAP43 on aligned fibers). ICC images were enhanced (+40% brightness, +20% contrast).
The pluripotent marker SSEA-1 was present on cells after day 1 of neural differentiation on both fiber and laminin substrates. While quantification showed a statistically increased number of cells expressing SSEA-1 on fibers (73 ± 4.7%) over laminin (38 ± 3.1%), the number of cells expressing pluripotent marker POU5F1 was not statistically different between fibers and laminin, 11 ± 4% and 0.59 ± 4%, respectively. Neural progenitor markers, such as NES, should be up- and then down-regulated during neural commitment. On fibers, NES appeared on day 1 (71 ± 6.6% of cells) and was minimally detectable by day 3 (0.8 ± 0.3%). On laminin, only 0.01% ± 0.01% of cells were expressing NES at day 1; this result was statistically lower than the expression on fibers. The percent of cells expressing NES on laminin statistically increased on day 3 to 5.6 ± 1.8%. A low number of cells expressed SOX1, resulting in no statistical differences between laminin and fibers at day 1.
TUBB3 appeared on day 1 on cells seeded on fiber substrates in comparison to cells cultured on laminin-substrates, where TUBB3 began to appear by day 3. On day 1, 29 ± 8.3% of cells expressed TUBB3 on fibers, which was statistically higher than 0.03 ± 5.3% of cells expressing TUBB3 on laminin. Compared to day 3, TUBB3 expression statistically increased by day 7 on laminin and fibers with more visible expression by cells on fibers than cells on laminin, although little visual differences were seen on day 14. For fiber scaffolds, the neurites primarily followed the path of the fibers, while neurites extended in all directions on laminin-coated substrates (Figure 4). The alignment relative to the fibers was demonstrated by the orientation angle of the neurites (−2.8 ± 21.5° on fibers in comparison to 21.5 ± 43.5° on laminin glass) and the goodness of the fit, which was statistically improved on fibers, 0.82 ± 0.14, compared to laminin 0.48 ± 0.21. The width of the Gaussian peak at half its maximum intensity was more narrow for neurites on fibers (50.6°) than on laminin glass (102.5°). By 7 and 14 days of differentiation, the neurites were extensively networked on glass substrates and elongated on fibers. Average neurite length and number of neurites could not be measured directly due to the inability to determine starting and ending positions of the actual neurites. In addition, many of the individual neurites could not be distinguished as they intertwined, particularly on fibers, making a measurement of total neurite length inaccurate. Total neurite length was statistically higher on fibers (55.4 ± 30.2 mm per 1 mm2) than on laminin glass (31.5 ± 21.6 mm per 1 mm2), with p = 0.016. Mature neuronal markers GAP43 and MAP2 were expressed earlier by cells on fibers than by cells on laminin, where day 7 protein expression is noticeably increased by cells on fibers in contrast to cells on laminin. Pixel quantification of MAP2 expression of the images at day 7 showed no statistical differences between laminin and fibers (p=0.11), although 2 of 5 images of cells on laminin had no fluorescence.
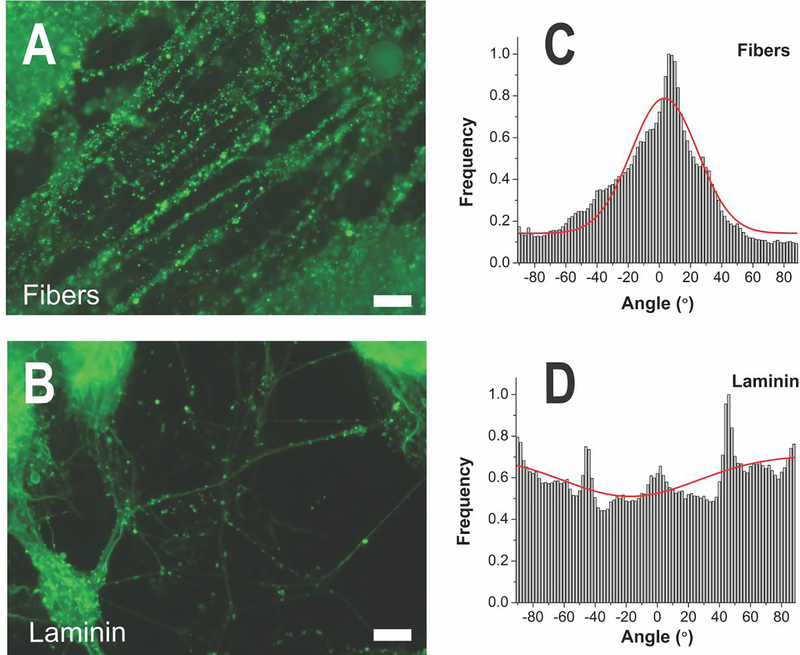
Figure 4.
(A) ICC of neural marker TUBB3 at day 14 showed the primary effect of the fibers to guide spreading of neurites along the fiber direction, while (B) neurites on laminin-coated glass spread in all directions. Scale bars = 50 μm. (C) Orientation distribution of neurites on aligned fibers and on (D) laminin coated glass with Gaussian fitting. Average angle of neurite orientation on fibers relative to the fiber direction was found to be −2.8 ± 21.5° and 21.5 ± 43.5° and on laminin glass. The goodness of fit (r2) to the Gaussian curve (0.82 ± 0.14) was statistically increased on aligned fibers in comparison to laminin glass (0.48 ± 0.21). The width of the Gaussian peak at half its maximum intensity was more narrow for neurites on fibers (50.6°) than on laminin glass (102.5°), also demonsrating the alignment.
GFAP was present on cells on both fiber and laminin substrates by day 3. While GFAP was present on cells on fiber substrates, the expression of GFAP by cells on laminin-coated surfaces showed more distinct glial morphology at day 14 (Figure 3). Pixel quantification for GFAP at day 7 was statistically higher on laminin substrates than YIGSR-aligned fibers. The expression of oligodendrocyte marker OLIG1 was slight for cells on laminin at day 7, and more prominent at day 14, but was not visibly expressed on cells on fibers (Figure 3). Another oligodendrocyte marker CNPase was also slightly expressed by day 7 on laminin, but was not noticeably expressed on cells on fibers until day 14. Pixel quantification of CNPase was not statistically different (p=0.07), with 12/12 images for cells on fibers had no labeling and 2/5 images for cells on laminin had no labeling.
Discussion
To translate the use of stem cells to clinical practice, xenogenic components of differentiation media need to be substituted by recombinant proteins or synthetic mimics.5 One method to reduce these components is to develop material-based systems that recapitulate the culture microenvironment currently utilized for stem cells. Cells produced via substrate-directed differentiation may improve access to neural cells for cell-based therapies by reducing exposure to potential immunogens. To address this concern, we designed a synthetic nanofibrous substrate with surface-tethered GYIGSR that increased the rate of mESC differentiation compared to laminin-coated surfaces, demonstrating an improved xeno-free substrate for neural differentiation of mESC. While current papers investigated neurite length and cell number as criteria for improved differentiation, in this work we provided a variety of gene and protein expression profiles in order to demonstrate a broader picture of differentiation capability of substrates and to have an insight in the mechanism of neural differentiation. To the best of our knowledge, no reports exist in literature that provide such a comprehensive comparison of cell differentiation on peptide-functionalized substrates to its whole protein as presented here.
To first characterize the D3 mESC neural differentiation culture, we seeded dissociated cells, directly from pluripotent culture, onto laminin-coated substrates to investigate the commitment and differentiation. Protein-coated substrates are commonplace in stem cell culture, particularly when investigating neural differentiation. In the literature, a variety of protein substrates have been studied as a means of simplifying culture away from embryoid body formation or use of feeder cells to induce differentiation, including laminin,[64] Matrigel®, heparan sulfate,[65] gelatin[66] and sulfated chitosan.[67] The time course of this differentiation protocol is demonstrated through down regulation of both gene expression (Figure 2, Figure S6) and protein expression (Figure 3). By culturing the mESCs out to 14 days, transient gene expression for early and late markers was noted. For example, by 3 days post-seeding, the gene expression of the pluripotent marker Pou5f1, was downregulated, while Nes, Sox1 and Pax6, neural progenitor markers, were increased. Confirming neural differentiation, neuronal and glial gene and protein makers were upregulated over the 14 day time course. While it is challenging to directly compare timepoints between mESC lines, differentiation techniques, and media types, N-cadherin bound surfaces demonstrated neurites within 10 days of differentiation of mESC ST1 line and Nanog-GFP expressing miPS cell line, yet had low expression of TUBB3 at day 4, indicating a similar progression to our study.[68] Overall, the laminin-substrate, along with the differentiation medium, encouraged both the commitment and differentiation toward neurons and glia over 14 days.
We next investigated the effect of fibers on the same process. Electrospun nanofibers have been shown to enhance differentiation of embryonic stem cells into various types of cell lineages compared to flat surfaces,[69] including osteogenic differentiation,[70] cardiomyocytes,[71] adipocytes,[72] and neurons.[15, 73] Ultimately, interactions between nanofibers and neural stem cells, mesenchymal stem cells, or other cells have been broadly studied, yet little has been done to investigate the direct interactions between nanofibers and pluripotent stem cells for neural differentiation.[74] As each cell type offers unique challenges, much of our focus of comparison from the literature is to mESCs and nanofibers, and the induction, commitment, and differentiation to neural cells. Synthetic nanofibers offer many advantages in processing and manufacture, yet, in general, synthetic electrospun fibers do not offer bioactive sites for mESCs with which to interact, which is a significant drawback. The need to have bioactive components adsorbed or bound to the fiber surface to exert its influence has limited studies seeking to avoid costly protein or xenogenic components. While some studies have not utilized an adsorbed protein, these protocols employed embryoid body to induce differentiation prior to cell seeding on the fibers. For example, mESC neural differentiation was examined on uncoated PLLA microfibers after induction by embryoid body formation.[21] By post-electrospinning placement of bioactive components, our fibers offered an advantage over others by preventing peptides from possible degradation during electrospinning and focusing them on the surfaces that could be in contact with cells. Our simple method of nanofiber post-electrospinning modification with peptide involving strain-promoted azide-alkyne cycloaddition provided easier characterization of the surface peptide concentration. Our previous work on YIGSR-functionalized PLLA fibers demonstrated early commitment of mESCs on aligned over random nanofibers without embryoid body formation,[26] however, did not compare results for time points after 3 days or to whole protein. To fully investigate the effects of the topography or the peptide, a more stringent comparison over longer times to the whole protein is necessary.
The topography of the substrate is defined by both the fiber diameter and alignment. While fiber diameter has been found to play a role in neural differentiation, the results are mixed. For example, fiber diameter played a role in rat neural stem cell differentiation on laminin-adsorbed polyethersulfone fibers in comparison to laminin-adsorbed tissue culture polystyrene, where at 5 days of culture, cells had higher TUBB3 protein expression on 749 nm fibers compared to gelatin-coated plastic and 283 nm nanofibers. In contrast, oligodendrocytes were found in higher numbers on 283 nm fibers over larger fibers and gelatin-coated plastic.[16] In this study, we found few oligodendrocytes on 212 nm fibers, and high expression of β-III tubulin both in protein and gene expression, high Nes in gene expression, and little detectable GFAP by both gene and protein expression by day 7. These results were more similar to previous work on PLLA fibers with tethered GYIGSR[26] and neat PLLA fibers, where the rate of neural stem cell differentiation was higher on nanofibers (250–300 nm) than on microfibers (1.25–1.5μm).[18] The differences noted between each study can be due to surface/protein/peptide functionality, fiber density,[75] or potential topography influences. While we did not study fiber diameter as a variable, this variable could be studied in the future to potentially drive one cell population over another.
As noted above, the alignment of the fibers has played a role in the resulting differentiation, with aligned topographies demonstrating increased neuronal differentiation. After 10 days on PLGA fibers, mESCs (mESC1 and mESC5) on aligned scaffolds had statistically higher Nes gene expression than gelatin-coated substrates or random fibers, but not Tubb3 or Pax6.[21] In addition, PLLA fibers (350 nm) with bound YIGSR had increased neural differentiation after 3 days over similar fibers that were randomly aligned, or aligned or random unfunctionalized fibers.[26] This trend is similar with adult neural stem cells.[76] Therefore, we selected to only study aligned fibers, and compare those results to laminin-coated substrates, which are a typical platform for neural differentiation. In this comparison at early time points, YIGSR-aligned fibers had higher expression of neural progenitor and neuronal genes Sox1, Tubb3, Cdh2, Gap43, Syp, and earlier NES, TUBB3, GAP43 and MAP2 protein expression on synthetic nanofiber substrates. Thus, YIGSR-aligned fibers were a suitable substrate for neural differentiation, producing an increased rate of differentiation compared to whole-protein coated glass substrates. After 14 days, any differences were gone, implying a more mature population on both substrates. Future work, over longer time frames, can continue to determine if further differences exist after differentiation on a peptide-modified scaffold with different topographies.
GYIGSR-tethered substrates were shown to promote neural differentiation previously for neural stem cells on membranes,[77] embryonic hippocampal neurons on YIGSR-modified substrates,[78] and human mesenchymal stem cells on silk fibroin films[46] compared to laminin-coated surfaces. The role of GYIGSR in cell adhesion has been well established,[77] and we demonstrated that surface-tethered GYIGSR peptide increased mESC adhesion over non-functionalized fibers. However, the numbers of cells adhering to the functionalized nanofibers was far less than those adhering to the flat laminin-coated substrate. As activity of the substrate has been found to be dependent upon the surface concentration of peptide, we calculated that the GYIGSR peptide on laminin-coated surfaces would be approximately 1.2 pmol/cm2.[14] Since laminin has other bioactive sites, a higher surface concentration of GYIGSR peptide on synthetic substrates was used (17.3 ± 6.6 pmol/cm2) than on laminin-coated glass, but this concentration was still lower than previous studies on PLLA fibers (57.3 pmol/cm2).[26] While the concentration of GYIGSR was theoretically lower on the laminin-coated substrates, adhesion of the cells to a surface can rely on multiple cell-substrate binding sites, which is demonstrated here by the increased adhesion to whole protein over peptide-functionalized fibers. Interestingly, the reduction in cell number would typically be thought of as a negative influence on differentiation, yet, we saw increased rates of differentiation on the fiber substrates compared to the laminin substrates. This result provides even further evidence of the potential power in using topography and peptides in the differentiation process.
The authors could not find another single-peptide system that increased the rate of mESC commitment and differentiation over protein-coated substrates. Our nanofiber scaffolds, which combine topographical properties with bioactive binding sites, represent a versatile biomaterial platform that can be used for differentiation of other cell lines for neural lineages. Similar to our study, neural progenitor cells had a higher rate of differentiation on peptide (IKVAV)-functionalized peptide nanofibers than on laminin-coated substrates, where a higher percentage of cells expressed TUBB3 and fewer cells expressed GFAP on fibers than on laminin.[14] However, the concentration of peptide on the nanofiber surface was higher by a factor of 103 than on laminin, where our concentration was only 10-fold higher. As our previous study was 50-fold higher GYIGSR concentration and a slightly higher fiber diameter (~338 nm), yet demonstrated similar results, we hypothesize that the faster differentiation is related to both the peptide specificity and alignment of the fibers, compared to protein-coated glass substrates. But by day 7 and day 14, cells on both substrates are at the same differentiation stage, suggesting that GYIGSR sequence plays a more important role at the early stages of neural commitment and differentiation than at the late stages. However, at later stages, the fiber topography played a more significant role in directing the neurite extensions.[15, 18, 76, 79–82] Our work demonstrated that aligned GYIGSR-functionalized scaffolds provided this contact guidance for the extension of neurites along the fiber direction and showed increased total neurite length. Previous results using bioactive species demonstrated improved neurite extensions on bioactive fibers compared to neat fibers.[79] This directional guidance could be useful in the end applications, but the exact role of fiber alignment in the neural induction or differentiation process is still unclear and would require further study.
Even though most of our results indicate faster differentiation on YISGR-modified fibers than on laminin, better cell performance on laminin substrates was also noted, including higher adhesion, higher expression of the Syp by day 14, and earlier and more mature glial and oligodendrocyte markers. These results, in particular the high level of initial cell adhesion, are likely related to multiple bioactive sites working synergistically as a consequence of laminin having a native ECM.[54] Therefore, in order to fully mimic the cell-laminin interaction it would be interesting to investigate nanofiber substrates with multiple functionalized bioactive cues, with ability to balance each of their concentrations to promote target cell behavior. This functionality could be easily achieved by introducing functional monomers into the PCL chain with the possibility for easy post-electrospinning modification with multiple factors.[53] These results, and the possibility of future modifications, demonstrate the versatility of our substrates, which could be used for culturing or differentiation of other cell lines by tethering other bioactive factors.
Conclusions
The present study describes fabrication of a versatile nanofiber platform, combining topographical features and surface-tethered bioactive species, and its application as a substrate for mESC neural differentiation. Detailed analysis of gene and protein expression results reveal s that even with fewer adherent cells, GYIGSR-functionalized fibers promoted similar neural differentiation of D3 mESCs when compared to laminin-coated glass, and induced faster differentiation times on functional nanofibers (higher expression of neural progenitor and neuronal genes Sox1, Tubb3, Cdh2, Gap43, Syp at early time points, and earlier NES, TUBB3, GAP43 and MAP2 protein expression on synthetic nanofiber substrates). These results indicated that functional nanofiber substrates could promote even faster differentiation than laminin. The aligned nanofibers can also be used as substrates to guide neurite extension. Aligned nanofibers and post-electrospinning surface modification with bioactive species can be combined to produce translationally relevant xeno-free substrates for stem cell therapy. Future development efforts are focused on additional bioactive species that are able to function as surrogates for other xenogenic factors found in differentiation media.
Supplementary Material
In this paper, we report the use of GYIGSR-functionalized poly(ԑ-caprolactone) aligned nanofibers as a tool to accelerate the neural lineage commitment and differentiation of D3 mouse embryonic stem cells. The results indicate that functional nanofiber substrates promote faster differentiation than laminin coated substrates. The data suggest that aligned nanofibers and post-electrospinning surface modification with bioactive species can be combined to produce translationally relevant xeno-free substrates for stem cell therapy. Future development efforts are focused on additional bioactive species that are able to function as surrogates for other xenogenic factors found in differentiation media.
Acknowledgments
This work is funded by the National Institutes of Health (R15-GM113155) and National Science Foundation (CBET BME 1603832). RKW acknowledges support from the Margaret F. Donovan Endowed Chair for Women in Engineering and MLB acknowledges support from the W. Gerald Austen Endowed Chair in Polymer Science and Polymer Engineering via the John S. and James L. Knight Foundation. OM would like to thank the National Science Foundation REU Program in Polymer Science and Engineering at the University of Akron (DMR# 1359321).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kleinman HK, Martin GR, Matrigel: basement membrane matrix with biological activity, Semin Cancer Biol 15(5) (2005) 378–386. [DOI] [PubMed] [Google Scholar]
- [2].Kohen NT, Little LE, Healy KE, Characterization of Matrigel interfaces during defined human embryonic stem cell culture, Biointerphases 4(4) (2009) 69–79. [DOI] [PubMed] [Google Scholar]
- [3].Fridman R, Giaccone G, Kanemoto T, Martin GR, Gazdar AF, Mulshine JL, Reconstituted Basement-Membrane (Matrigel) and Laminin Can Enhance the Tumorigenicity and the Drug-Resistance of Small-Cell Lung-Cancer Cell-Lines, Proc. Natl. Acad. Sci 87(17) (1990) 6698–6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mullen P, Ritchie A, Langdon SP, Miller WR, Effect of Matrigel on the tumorigenicity of human breast and ovarian carcinoma cell lines, Int J Cancer 67(6) (1996) 816–820. [DOI] [PubMed] [Google Scholar]
- [5].Brafman DA, Chang CW, Fernandez A, Willert K, Varghese S, Chien S, Long-term human pluripotent stem cell self-renewal on synthetic polymer surfaces, Biomaterials 31(34) (2010) 9135–9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Higuchi A, Ling QD, Chang Y, Hsu ST, Umezawa A, Physical cues of biomaterials guide stem cell differentiation fate, Chem. Rev 113(5) (2013) 3297–3328. [DOI] [PubMed] [Google Scholar]
- [7].Seidlits SK, Khaing ZZ, Petersen RR, Nickels JD, Vanscoy JE, Shear JB, Schmidt CE, The effects of hyaluronic acid hydrogels with tunable mechanical properties on neural progenitor cell differentiation, Biomaterials 31(14) (2010) 3930–3940. [DOI] [PubMed] [Google Scholar]
- [8].Sharifi F, Patel BB, Dzuilko AK, Montazami R, Sakaguchi DS, Hashemi N, Polycaprolactone Microfibrous Scaffolds to Navigate Neural Stem Cells, Biomacromolecules 17(10) (2016) 3287–3297. [DOI] [PubMed] [Google Scholar]
- [9].McNamara LE, McMurray RJ, Biggs MJ, Kantawong F, Oreffo RO, Dalby MJ, Nanotopographical control of stem cell differentiation, J Tissue Eng 2010(1) (2010) 120623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cherry JF, Bennett NK, Schachner M, Moghe PV, Engineered N-cadherin and L1 biomimetic substrates concertedly promote neuronal differentiation, neurite extension and neuroprotection of human neural stem cells, Acta Biomater 10(10) (2014) 4113–4126. [DOI] [PubMed] [Google Scholar]
- [11].Gonzalez R, Garitaonandia I, Abramihina T, Wambua GK, Ostrowska A, Brock M, Noskov A, Boscolo FS, Craw JS, Laurent LC, Snyder EY, Semechkin RA, Deriving dopaminergic neurons for clinical use. A practical approach, Sci. Rep 3((2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yim EK, Pang SW, Leong KW, Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage, Exp Cell Res 313(9) (2007) 1820–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kim H, Kim I, Choi HJ, Kim SY, Yang EG, Neuron-like differentiation of mesenchymal stem cells on silicon nanowires, Nanoscale 7(40) (2015) 17131–17138. [DOI] [PubMed] [Google Scholar]
- [14].Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI, Selective differentiation of neural progenitor cells by high-epitope density nanofibers, Science 303(5662) (2004) 1352–1355. [DOI] [PubMed] [Google Scholar]
- [15].Xie J, Willerth SM, Li X, Macewan MR, Rader A, Sakiyama-Elbert SE, Xia Y, The differentiation of embryonic stem cells seeded on electrospun nanofibers into neural lineages, Biomaterials 30(3) (2009) 354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Christopherson GT, Song H, Mao HQ, The influence of fiber diameter of electrospun substrates on neural stem cell differentiation and proliferation, Biomaterials 30(4) (2009) 556–564. [DOI] [PubMed] [Google Scholar]
- [17].Patel S, Kurpinski K, Quigley R, Gao H, Hsiao BS, Poo MM, Li S, Bioactive nanofibers: synergistic effects of nanotopography and chemical signaling on cell guidance, Nano Lett. 7(7) (2007) 2122–2128. [DOI] [PubMed] [Google Scholar]
- [18].Yang F, Murugan R, Wang S, Ramakrishna S, Electrospinning of nano/micro scale poly (L-lactic acid) aligned fibers and their potential in neural tissue engineering, Biomaterials 26(15) (2005) 2603–2610. [DOI] [PubMed] [Google Scholar]
- [19].Mahairaki V, Lim SH, Christopherson GT, Xu L, Nasonkin I, Yu C, Mao HQ, Koliatsos VE, Nanofiber matrices promote the neuronal differentiation of human embryonic stem cell-derived neural precursors in vitro, Tissue Eng Part A 17(5–6) (2011) 855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Corey JM, Lin DY, Mycek KB, Chen Q, Samuel S, Feldman EL, Martin DC, Aligned electrospun nanofibers specify the direction of dorsal root ganglia neurite growth, J Biomed Mater Res A 83(3) (2007) 636–645. [DOI] [PubMed] [Google Scholar]
- [21].Sperling LE, Reis KP, Pozzobon LG, Girardi CS, Pranke P, Influence of random and oriented electrospun fibrous poly(lactic-co-glycolic acid) scaffolds on neural differentiation of mouse embryonic stem cells, J Biomed Mater Res A 105(5) (2017) 1333–1345. [DOI] [PubMed] [Google Scholar]
- [22].Li D, Xia Y, Electrospinning of Nanofibers: Reinventing the Wheel?, Adv. Mater. 16(14) (2004) 1151–1170. [Google Scholar]
- [23].Doshi J, Reneker DH, Electrospinning Process and Applications of Electrospun Fibers, J. Electrost 35(2–3) (1995) 151–160. [Google Scholar]
- [24].Park JH, Rutledge GC, 50th Anniversary Perspective: Advanced Polymer Fibers: High Performance and Ultrafine, Macromolecules 50(15) (2017) 5627–5642. [Google Scholar]
- [25].Agarwal S, Wendorff JH, Greiner A, Use of electrospinning technique for biomedical applications, Polymer 49(26) (2008) 5603–5621. [Google Scholar]
- [26].Callahan LA, Xie S, Barker IA, Zheng J, Reneker DH, Dove AP, Becker ML, Directed differentiation and neurite extension of mouse embryonic stem cell on aligned poly(lactide) nanofibers functionalized with YIGSR peptide, Biomaterials 34(36) (2013) 9089–9095. [DOI] [PubMed] [Google Scholar]
- [27].Xie J, MacEwan MR, Schwartz AG, Xia Y, Electrospun nanofibers for neural tissue engineering, Nanoscale 2(1) (2010) 35–44. [DOI] [PubMed] [Google Scholar]
- [28].Ingavle GC, Leach JK, Advancements in electrospinning of polymeric nanofibrous scaffolds for tissue engineering, Tissue Eng Part B Rev 20(4) (2014) 277–293. [DOI] [PubMed] [Google Scholar]
- [29].Luo CJ, Stride E, Edirisinghe M, Mapping the Influence of Solubility and Dielectric Constant on Electrospinning Polycaprolactone Solutions, Macromolecules 45(11) (2012) 4669–4680. [Google Scholar]
- [30].Li D, Wang YL, Xia YN, Electrospinning of polymeric and ceramic nanofibers as uniaxially aligned arrays, Nano Lett. 3(8) (2003) 1167–1171. [Google Scholar]
- [31].Liu T, Teng WK, Chan BP, Chew SY, Photochemical crosslinked electrospun collagen nanofibers: synthesis, characterization and neural stem cell interactions, J Biomed Mater Res A 95(1) (2010) 276–282. [DOI] [PubMed] [Google Scholar]
- [32].Zhang Y, Ouyang H, Lim CT, Ramakrishna S, Huang ZM, Electrospinning of gelatin fibers and gelatin/PCL composite fibrous scaffolds, J Biomed Mater Res B Appl Biomater 72(1) (2005) 156–165. [DOI] [PubMed] [Google Scholar]
- [33].Ghasemi-Mobarakeh L, Prabhakaran MP, Morshed M, Nasr-Esfahani M-H, Ramakrishna S, Electrospun poly (ɛ-caprolactone)/gelatin nanofibrous scaffolds for nerve tissue engineering, Biomaterials 29(34) (2008) 4532–4539. [DOI] [PubMed] [Google Scholar]
- [34].Neal RA, McClugage SG III, Link MC, Sefcik LS, Ogle RC, Botchwey EA, Laminin nanofiber meshes that mimic morphological properties and bioactivity of basement membranes, Tissue Engineering Part C: Methods 15(1) (2008) 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zeugolis DI, Khew ST, Yew ES, Ekaputra AK, Tong YW, Yung LY, Hutmacher DW, Sheppard C, Raghunath M, Electro-spinning of pure collagen nano-fibres - just an expensive way to make gelatin?, Biomaterials 29(15) (2008) 2293–2305. [DOI] [PubMed] [Google Scholar]
- [36].Teo WE, He W, Ramakrishna S, Electrospun scaffold tailored for tissue-specific extracellular matrix, Biotechnol J 1(9) (2006) 918–929. [DOI] [PubMed] [Google Scholar]
- [37].Liu X, Holzwarth JM, Ma PX, Functionalized synthetic biodegradable polymer scaffolds for tissue engineering, Macromol Biosci 12(7) (2012) 911–919. [DOI] [PubMed] [Google Scholar]
- [38].Murphy AR, Laslett A, O’Brien CM, Cameron NR, Scaffolds for 3D in vitro culture of neural lineage cells, Acta Biomater 54((2017) 1–20. [DOI] [PubMed] [Google Scholar]
- [39].Zhao L, Ma S, Pan Y, Zhang Q, Wang K, Song D, Wang X, Feng G, Liu R, Xu H, Zhang J, Qiao M, Kong D, Functional Modification of Fibrous PCL Scaffolds with Fusion Protein VEGF-HGFI Enhanced Cellularization and Vascularization, Adv Healthc Mater 5(18) (2016) 2376–2385. [DOI] [PubMed] [Google Scholar]
- [40].Kleinman HK, Cannon FB, Laurie GW, Hassell JR, Aumailley M, Terranova VP, Martin GR, DuBois-Dalcq M, Biological activities of laminin, J. Cell. Biochem 27(4) (1985) 317–325. [DOI] [PubMed] [Google Scholar]
- [41].Aumailley M, Smyth N, The role of laminins in basement membrane function, J Anat 193 ( Pt 1)(1) (1998) 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Terranova VP, Rohrbach DH, Martin GR, Role of laminin in the attachment of PAM 212 (epithelial) cells to basement membrane collagen, Cell 22(3) (1980) 719–726. [DOI] [PubMed] [Google Scholar]
- [43].Yamada Y, Kleinman HK, Functional domains of cell adhesion molecules, Curr Opin Cell Biol 4(5) (1992) 819–823. [DOI] [PubMed] [Google Scholar]
- [44].Graf J, Ogle RC, Robey FA, Sasaki M, Martin GR, Yamada Y, Kleinman HK, A pentapeptide from the laminin B1 chain mediates cell adhesion and binds the 67,000 laminin receptor, Biochemistry 26(22) (1987) 6896–6900. [DOI] [PubMed] [Google Scholar]
- [45].Graf J, Iwamoto Y, Sasaki M, Martin GR, Kleinman HK, Robey FA, Yamada Y, Identification of an amino acid sequence in laminin mediating cell attachment, chemotaxis, and receptor binding, Cell 48(6) (1987) 989–996. [DOI] [PubMed] [Google Scholar]
- [46].Manchineella S, Thrivikraman G, Basu B, Govindaraju T, Surface-Functionalized Silk Fibroin Films as a Platform To Guide Neuron-like Differentiation of Human Mesenchymal Stem Cells, Acs Applied Materials & Interfaces 8(35) (2016) 22849–22859. [DOI] [PubMed] [Google Scholar]
- [47].Beatty KE, Fisk JD, Smart BP, Lu YY, Szychowski J, Hangauer MJ, Baskin JM, Bertozzi CR, Tirrell DA, Live-cell imaging of cellular proteins by a strain-promoted azide-alkyne cycloaddition, Chembiochem 11(15) (2010) 2092–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ning X, Guo J, Wolfert MA, Boons GJ, Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast huisgen cycloadditions, Angew. Chem. Int. Ed. Engl 47(12) (2008) 2253–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sletten EM, de Almeida G, Bertozzi CR, A homologation approach to the synthesis of difluorinated cycloalkynes, Org. Lett 16(6) (2014) 1634–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].de Almeida G, Townsend LC, Bertozzi CR, Synthesis and reactivity of dibenzoselenacycloheptynes, Org. Lett 15(12) (2013) 3038–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zheng J, Liu K, Reneker DH, Becker ML, Post-assembly derivatization of electrospun nanofibers via strain-promoted azide alkyne cycloaddition, J. Am. Chem. Soc 134(41) (2012) 17274–17277. [DOI] [PubMed] [Google Scholar]
- [52].Zheng J, Xie S, Lin F, Hua G, Yu T, Reneker DH, Becker ML, 4-Dibenzocyclooctynol (DIBO) as an initiator for poly(ε-caprolactone): copper-free clickable polymer and nanofiber-based scaffolds, Polym. Chem 4(7) (2013) 2215–2218. [Google Scholar]
- [53].Zheng JK, Hua G, Yu JY, Lin F, Wade MB, Reneker DH, Becker ML, Post-Electrospinning “Triclick” Functionalization of Degradable Polymer Nanofibers, Acs Macro Letters 4(2) (2015) 207–213. [DOI] [PubMed] [Google Scholar]
- [54].Zheng J, Kontoveros D, Lin F, Hua G, Reneker DH, Becker ML, Willits RK, Enhanced Schwann cell attachment and alignment using one-pot “dual click” GRGDS and YIGSR derivatized nanofibers, Biomacromolecules 16(1) (2015) 357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Calabrese J, Cushing MA, Ittel SD, Sterically Hindered Magnesium Aryloxides, Inorg. Chem 27(5) (1988) 867–870. [Google Scholar]
- [56].Wilson JA, Hopkins SA, Wright PM, Dove AP, ‘Immortal’ ring-opening polymerization of ω-pentadecalactone by Mg(BHT)2(THF)2, Polym. Chem 5(8) (2014) 26912694. [Google Scholar]
- [57].Zheng J, Smith Callahan LA, Hao J, Guo K, Wesdemiotis C, Weiss RA, Becker ML, Strain-Promoted Crosslinking of PEG-based Hydrogels via Copper-Free Cycloaddition, ACS Macro Lett 1(8) (2012) 1071–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Jung ME, Mossman AB, Lyster MA, Direct synthesis of dibenzocyclooctadienes via double ortho Friedel-Crafts alkylation by the use of aldehyde-trimethylsilyl iodide adducts, The Journal of Organic Chemistry 43(19) (1978) 3698–3701. [Google Scholar]
- [59].Abràmoff MD, Magalhães PJ, Ram SJ, Image processing with ImageJ, Biophotonics international 11(7) (2004) 36–42. [Google Scholar]
- [60].Liu ZQ, Scale space approach to directional analysis of images, Appl. Opt 30(11) (1991) 1369–1373. [DOI] [PubMed] [Google Scholar]
- [61].Wojdyr M, Fityk: a general-purpose peak fitting program, J. Appl. Crystallogr 43(5) (2010) 1126–1128. [Google Scholar]
- [62].Grandjean C, Boutonnier A, Guerreiro C, Fournier JM, Mulard LA, On the preparation of carbohydrate-protein conjugates using the traceless Staudinger ligation, J. Org. Chem 70(18) (2005) 7123–7132. [DOI] [PubMed] [Google Scholar]
- [63].Janz R, Sudhof TC, Hammer RE, Unni V, Siegelbaum SA, Bolshakov VY, Essential roles in synaptic plasticity for synaptogyrin I and synaptophysin I, Neuron 24(3) (1999) 687–700. [DOI] [PubMed] [Google Scholar]
- [64].Gazina EV, Morrisroe E, Mendis GDC, Michalska AE, Chen J, Nefzger CM, Rollo BN, Reid CA, Pera MF, Petrou S, Method of derivation and differentiation of mouse embryonic stem cells generating synchronous neuronal networks, J Neurosci Methods 293((2018) 53–58. [DOI] [PubMed] [Google Scholar]
- [65].Meade KA, White KJ, Pickford CE, Holley RJ, Marson A, Tillotson D, van Kuppevelt TH, Whittle JD, Day AJ, Merry CL, Immobilization of heparan sulfate on electrospun meshes to support embryonic stem cell culture and differentiation, J. Biol. Chem 288(8) (2013) 5530–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wongpaiboonwattana W, Stavridis MP, Neural differentiation of mouse embryonic stem cells in serum-free monolayer culture, J Vis Exp 99) (2015) e52823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ding K, Wang Y, Wang H, Yuan L, Tan M, Shi X, Lyu Z, Liu Y, Chen H, 6-Osulfated chitosan promoting the neural differentiation of mouse embryonic stem cells, ACS Appl Mater Interfaces 6(22) (2014) 20043–20050. [DOI] [PubMed] [Google Scholar]
- [68].Haque A, Yue XS, Motazedian A, Tagawa Y, Akaike T, Characterization and neural differentiation of mouse embryonic and induced pluripotent stem cells on cadherin-based substrata, Biomaterials 33(20) (2012) 5094–5106. [DOI] [PubMed] [Google Scholar]
- [69].Meade K, Holley R, Merry C, Cell culture systems for stem cell research. In Electrospinning for Tissue Regeneration, Elsevier: 2011; pp 372–396. [Google Scholar]
- [70].Smith LA, Liu X, Hu J, Wang P, Ma PX, Enhancing osteogenic differentiation of mouse embryonic stem cells by nanofibers, Tissue Eng Part A 15(7) (2009) 1855–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Fromstein JD, Zandstra PW, Alperin C, Rockwood D, Rabolt JF, Woodhouse KA, Seeding bioreactor-produced embryonic stem cell-derived cardiomyocytes on different porous, degradable, polyurethane scaffolds reveals the effect of scaffold architecture on cell morphology, Tissue Eng Part A 14(3) (2008) 369–378. [DOI] [PubMed] [Google Scholar]
- [72].Kang XH, Xie YB, Powell HM, Lee LJ, Belury MA, Lannutti JJ, Kniss DA, Adipogenesis of murine embryonic stem cells in a three-dimensional culture system using electrospun polymer scaffolds, Biomaterials 28(3) (2007) 450–458. [DOI] [PubMed] [Google Scholar]
- [73].Carlberg B, Axell MZ, Nannmark U, Liu J, Kuhn HG, Electrospun polyurethane scaffolds for proliferation and neuronal differentiation of human embryonic stem cells, Biomed Mater 4(4) (2009) 045004. [DOI] [PubMed] [Google Scholar]
- [74].Holzwarth JM, Enhancing the Neuronal Differentiation of Mouse Embryonic Stem Cells Using Biomaterials, (2015).
- [75].Liu L, Yuan Q, Shi J, Li X, Jung D, Wang L, Yamauchi K, Nakatsuji N, Kamei K, Chen Y, Chemically-defined scaffolds created with electrospun synthetic nanofibers to maintain mouse embryonic stem cell culture under feeder-free conditions, Biotechnol. Lett 34(10) (2012) 1951–1957. [DOI] [PubMed] [Google Scholar]
- [76].Lim SH, Liu XY, Song H, Yarema KJ, Mao HQ, The effect of nanofiber-guided cell alignment on the preferential differentiation of neural stem cells, Biomaterials 31(34) (2010) 9031–9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Li YC, Liao YT, Chang HH, Young TH, Covalent bonding of GYIGSR to EVAL membrane surface to improve migration and adhesion of cultured neural stem/precursor cells, Colloids Surf B Biointerfaces 102(Supplement C) (2013) 53–62. [DOI] [PubMed] [Google Scholar]
- [78].Tong YW, Shoichet MS, Enhancing the neuronal interaction on fluoropolymer surfaces with mixed peptides or spacer group linkers, Biomaterials 22(10) (2001) 1029–1034. [DOI] [PubMed] [Google Scholar]
- [79].Koh HS, Yong T, Chan CK, Ramakrishna S, Enhancement of neurite outgrowth using nano-structured scaffolds coupled with laminin, Biomaterials 29(26) (2008) 3574–3582. [DOI] [PubMed] [Google Scholar]
- [80].Kim YT, Haftel VK, Kumar S, Bellamkonda RV, The role of aligned polymer fiber-based constructs in the bridging of long peripheral nerve gaps, Biomaterials 29(21) (2008) 3117–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Xie J, MacEwan MR, Li X, Sakiyama-Elbert SE, Xia Y, Neurite outgrowth on nanofiber scaffolds with different orders, structures, and surface properties, ACS Nano 3(5) (2009) 1151–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bini TB, Gao SJ, Wang S, Ramakrishna S, Poly(l-lactide-co-glycolide) biodegradable microfibers and electrospun nanofibers for nerve tissue engineering: an in vitro study, J. Mater. Sci 41(19) (2006) 6453–6459. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.