Abstract
We have performed nonradioactive double in situhybridization to study the expression of glutamic acid decarboxylase and GluR6 or GluR5 subunits in hippocampal slices. Our results indicate that although GluR6 is primarily expressed by pyramidal cells and dentate granule neurons and GluR5 is prominently expressed in nonpyramidal cells, there is a significant population of GABAergic interneurons that coexpress the two glutamate receptor subunits. To assess whether the two subunits could coassemble to form heteromeric receptors, we studied the electrophysiological responses when both subunits were coexpressed in HEK293 cells. Responses evoked by rapid application of either glutamate, (RS)-α-amino-3-hydroxy-5-tert-butyl-4-isoxazolepropionic acid (ATPA) the selective agonist of GluR5 receptors), and AMPA in cells cotransfected with GluR6(R) and GluR5(Q) presented a similar degree of outward rectification. This can only be attributed to the fact that all receptors have at least one GluR6(R) subunit in their structure, conferring outward rectification, and at least one GluR5(Q) subunit to confer sensitivity to ATPA and AMPA. More than 80% of the receptors expressed by a single cell were found to be GluR5/R6 heteromers, presenting different desensitization and gating properties to homomeric R6 receptors. These results lead us to believe that a population of interneurons in the hippocampus express receptors made up of both GluR5 and GluR6 subunits and provide evidence for a greater diversity of kainate receptors in the brain than previously thought, that may account for a higher functional complexity.
Keywords: heteromeric kainate receptors, heteromeric glutamate receptor, ATPA, coassembly, desensitization, hippocampus, interneurons, coexpression
Within the glutamate receptor system, a number of roles have been established for NMDA and AMPA receptors in brain physiology, whereas the role of kainate receptors has remained elusive over the years (Lerma, 1997). High-affinity kainate receptors probably account for the kainate-dependent excitotoxicity and susceptibility to epilepsy of hippocampal circuits (Nadler, 1981; Coyle, 1983; Ben-Ari, 1985). Moreover, recent experiments suggest a role for kainate receptors as transducers and modulators of synaptic transmission (Castillo et al., 1997; Clarke et al., 1997; Rodriguez-Moreno et al., 1997; Vignes and Collingridge, 1997; Cossart et al., 1998; Frerking et al., 1998;Rodriguez-Moreno and Lerma, 1998; Kidd and Isaac, 1999).
Kainate receptors appear to be made up of five different subunits, namely GluR5, GluR6, GluR7, KA1, and KA2 (for review, see Lerma, 1999). Based on sequence homology, this family has been subdivided into two branches, GluR5–7 and KA1-KA2 subunits (Egebjerg et al., 1991;Bettler et al., 1992; Sommer et al., 1992). Both GluR5, GluR6 and GluR7 form homomeric functional channels when expressed in heterologous systems (for review, see Lerma, 1999). In contrast, KA1 and KA2 do not yield functional channels in the same systems but generate new functional receptors when combined with GluR5, GluR6, or GluR7, showing different properties from the homomeric channels (Herb et al., 1992;Sakimura et al., 1992). Such a functional analysis of recombinant subunits has fueled the idea of a heteromeric model for kainate receptors. The model implies the pairing of members of each branch of the kainate receptor subunits, i.e., GluRn/KAx, where n andx are equivalent to 5–7 and 1–2, respectively. The typical patterns of spatial distribution of the subunits have also supported this idea. For instance, GluR6, KA1, and K2 are expressed in the CA3 pyramidal cells, whereas GluR6 and KA2 transcripts are more abundant in the CA1 pyramids. Purkinje cells contain GluR5 and KA1, whereas the granule cells of the cerebellum express GluR6 and KA2 (Wisden and Seeburg, 1993; Bahn et al., 1994). However, expression of GluR5 has also been detected in a small population of hippocampal interneurons and cerebellar granule cells (Wisden and Seeburg, 1993), and single-cell RT-PCR from hippocampal cultures demonstrated coexpression of both GluR5 and GluR6 in a percentage of the cells analyzed (Mackler and Eberwine, 1993; Ruano et al., 1995).
In the present work we have reexamined the distribution of two kainate receptor subunits, GluR5 and GluR6, in the hippocampus by nonradioactive in situ hybridization. We have performed double labeling for GluR5 or GluR6 and glutamic acid decarboxylase (GAD) as a means to identify the GABA interneurons expressing these subunits. Our results indicate that a population of GABAergic neurons in the hippocampus may coexpress both GluR5 and GluR6 subunits. We have also tested the hypothesis that GluR5 and GluR6 could combine to form heteromeric channels. Our results demonstrate the assembly of both subunits to form functional receptor channels with pharmacological properties similar to GluR5 and biophysical properties similar to GluR6 homomeric receptors. Thus, we provide evidence for a greater diversity of native kainate receptors in the brain.
MATERIALS AND METHODS
In situ hybridization in hippocampal slices.Brains were dissected from anesthetized 14-d-old rats and fixed overnight in 4% paraformaldehyde at 4°C. After fixing, the brains were washed three times in PBS and embedded in 7.5% low-melting point agarose in PBS. The brains were sectioned in a vibratome to obtain 100 μm sagittal slices. The agarose was removed from the slices, which were again fixed overnight, again in 4% paraformaldehyde at 4°C. APstI–SalI fragment (649 bp) and aPstI–XbaI fragment (595 bp) from theGluR6 and GluR5 plasmids, respectively, were subcloned into pBluescript-KS. The corresponding fragments were chosen to contain the most divergent region between two cDNA sequences, with an average similarity of ∼60% at the nucleotide level. At the stringency used in this protocol, no cross-hybridization is possible between sequences with <90% similarity. Digoxigenin-labeled antisense riboprobes were synthesized from these plasmids and from a plasmid containing the full-length cDNA sequence forGAD65 (kindly provided by Drs. A. Tobin and N. Tillakaratne, University of California at Los Angeles, Los Angeles, CA). In situ hybridization was performed in free-floating slices as described for whole embryos (Nieto et al., 1996). Double-labeling experiments were performed by simultaneous hybridization with two probes. The GluR5 andGluR6 probes were labeled with digoxigenin-UTP, and theGAD probe was labeled with fluorescein-UTP from Boehringer Mannheim (Mannheim, Germany). After hybridization, the slices were subsequently incubated with alkaline phosphatase-conjugated anti-digoxigenin and anti-fluorescein antibodies. The alkaline phosphatase activity was detected by incubation with the following substrates: nitro blue tetrazolium-5-bromo-4-chloro-3-indolyl phosphate (BCIP) for GluR5 and GluR6, and 2-[4-iodophenyl]-3-[4-nitrophenyl]-5-phenyl tetrazolium chloride (INT)/BCIP for GAD (both substrates from Boehringer Mannheim) following manufacturer's instructions. Subsequently, sections were cleared in 50% glycerol in PBS and mounted in the same solution containing 0.02% sodium azide. Sense probes failed to give any signal. The slices were photographed with a Leica (Nussloch, Germany) DMR microscope under Nomarski optics.
cDNA transient transfection. Plasmids encoding the VCR or VCQ versions of GluR6, and the Q- or R-form ofGluR5–2a were a gift of Dr. P. H. Seeburg (Max-Planck Institute, Heidelberg, Germany). The plasmid encoding the green fluorescent protein (GFP) was kindly supplied by Dr. T. Hughes (Yale University). GluR5 and/or GluR6 plasmids were cotransfected with a GFP expression vector (10 μg ofGluR5, 1 μg of GluR6, and 1 μgGFP) in HEK 293 cells by electroporation (Gene pulser; Bio-Rad, Hercules, CA). GluR6 or GluR5 andKA2 plasmids were cotransfected at 1:1 ratio. Afterwards, cells were seeded in Petri dishes in DMEM supplemented with 10% fetal calf serum and antibiotics, and maintained in a humidified incubator at 37°C and 5% CO2. Highly fluorescent cells (Marshall et al., 1995) were identified and selected for recording.
Electrophysiological recordings and perfusion procedures.Electrophysiological experiments were performed 1 d after electroporation. Membrane currents were recorded at a given membrane potential, using the whole-cell configuration of the patch-clamp technique using a List EPC-7 amplifier. Series resistance was compensated by 50–60%. Patch electrodes were fabricated from borosilicate glass and had a resistance of 5–10 MΩ. Currents were filtered at 1 kHz (two pole Butterworth filter; −12 dB/octave) and transferred at a sampling rate of 1–5 kHz into a personal computer for analysis and display purposes using pClamp software (Axon Instruments, Foster City, CA). All experiments were performed at room temperature (22–25°C). The cells were rapidly perfused using a linear array of eight glass tubes placed 200–300 μm from the cell body. Ringer's solution with and without agonist flowed from adjacent barrels, and solution changes were achieved by laterally displacing the whole perfusion array using a motorized device under the control of a personal computer (Lerma, 1992). To test the speed of this perfusion system, the open tip potential of a patch pipette was monitored after jumping it into a low-strength solution. The current relaxed after a single exponential with a time constant of <1 msec, and thus, in all cases total replacement of the solution around the tip of the patch electrode was achieved in <10 msec (usually 5–6 msec). However, for receptor activation during whole-cell recordings, diffusion may be limited by multiple barriers, which may slow down perfusion speed.
Concentration–response relationships were constructed for glutamate, and sometimes for (RS)-α-amino-3-hydroxy-5-tert-butyl-4-isoxazolepropionic acid (ATPA), and fitted with the logistic equation by using nonlinear regression procedures (SigmaPlot; Jandel Scientific, San Rafael, CA). For activation curves, peak currents were normalized and pooled for fitting with the equation I = Imax/(1 + (EC50/[agonist])n), where EC50 is the concentration of agonist producing half-maximal activation. For steady-state inactivation, the pooled data were fitted by the equation I = Imax/(1 + ([agonist]/IC50)n), where IC50 is the concentration of agonist at which receptors were 50% inactivated. In both equations, nis the slope factor (i.e., the Hill coefficient). All results are expressed as the mean ± SEM.
Experimental solutions. The normal external solution was (in mm) NaCl 160, KCl 2.5, CaCl2 1.8, MgCl2 2, glucose 10, and HEPES 10, pH 7.4. Pipettes were filled with (in mm) cesium methanesulfonate 140, CsCl 5, CaCl2 0.5, MgCl2 5, EGTA 10, and HEPES 10, pH 7.4. Spermine (100 μm) was routinely included in the pipette solution to prevent washout of inward rectification of unedited receptors (Bowie and Mayer, 1995; Kamboj et al., 1995; Koh et al., 1995). Osmolarity was adjusted, when necessary, to 330 and 314 mOsm for extracellular and intracellular solutions, respectively, by adding sucrose. Agonists were dissolved in distilled water at 50 mm stock solutions or directly prepared in normal external solution. Kainate was obtained from either Sigma (St. Louis, MO) or Tocris Cookson (Bristol, UK), and AMPA was obtained from Tocris. Disodium glutamate and ConA were from Sigma, and other salts were obtained from either Sigma or Merck (Darmstadt, Germany). ATPA was a gift of Dr. J. Drejer (NeuroSearch, Ballerup, Denmark).
RESULTS
Expression of GluR5 and GluR6 mRNAs in hippocampal neurons
Radioactive in situ hybridization studies have shown that GluR6 is heavily expressed by hippocampal pyramidal neurons and dentate granule cells (Wisden and Seeburg, 1993; Bahn et al., 1994). Similarly, it has been shown that a population of interneurons in the hippocampus, mainly in the stratum oriens, expresses the GluR5 subunits (Bahn et al., 1994). We have performed nonradioactive in situ hybridization studies to allow single-cell resolution, semiquantitative detection of expression of GluR6 and GluR5 subunits, and to check whether both might be expressed by the same type of cells in the hippocampus. As can be seen in Figure1, GluR6 was abundantly expressed in pyramidal cells and dentate granule neurons, whereasGluR5 was prominently expressed by nonpyramidal cells. However, many neurons laying outside the pyramidal cell layer also expressed GluR6, and there were some cells within the pyramidal cell layer that contained GluR5 transcripts. This was observed not only in the CA1 but also in the CA3 field (Fig.1D–G). Consequently, the possibility exists that both subunits are expressed by both pyramidal cells and interneurons.
Fig. 1.
Expression pattern of GluR5(A), GluR6 (B), andGAD65 (C) mRNA in the hippocampus. Whereas GluR5 is predominantly expressed in interneurons distributed over the whole hippocampus (A),GluR6 is mainly expressed in the principal cells (B). The boxes in Aand B indicate the areas shown in D–G.D, A high-power view of the CA1 layer reveals a high expression of GluR5 in the stratum oriens (SO) and stratum radiatum (SR) interneurons (arrow and white arrowhead, respectively). Note that, in addition, many cells within the pyramidal cell layer (Pyr) are also labeled (black arrowhead). E, A similar zone of the CA1 area showing high expression of GluR6 in pyramidal neurons and interneurons both in the stratum oriens and stratum radiatum (arrow and white arrowhead, respectively). F, High-power view of the CA3 field showing expression of GluR5 in several cells both within and outside the pyramidal cell layer (black andwhite arrowheads, respectively). G, A high-power view of the CA3 field shows GluR6 expression in numerous interneurons (white arrowheads). For simplicity, only a few cells have been highlighted in each panel.
To demonstrate the coexpression of GluR5 andGluR6, we used double in situ hybridization. However, the relatively low level of expression of both mRNAs, in particular that of GluR5, prevented the reliable determination of their coexpression because the first signal developed was attenuated during the second development process.GAD65 mRNA was abundantly expressed by interneurons throughout the hippocampus (Fig. 1C) and gave a signal intensity enough to allow double detection. Therefore, we performed double in situ hybridization with eitherGluR5 or GluR6 andGAD65 as a means to determine whether these kainate receptor subunits were expressed in pyramidal or GABAergic interneurons.
Figure 2 shows the results obtained from double in situ hybridization of GluR5 andGAD mRNA. As can be seen, many neurons expressed both transcripts throughout the hippocampus (Fig. 2A). To estimate the percentage of each type of hippocampal neuron expressingGluR5, we computed in a double-blind test those neurons appearing red (GAD+,GluR5−), blue (GAD−,GluR5+), and brown (GAD+,GluR5+). Cells were counted from the available hippocampal slices (six to eight for each hippocampus; 2500–2900 neurons computed) by at least two different people. The scores given by the different observers (for each color in a given slice) were similar (often differing in <1%), and the mean value was taken. GluR5 was expressed by approximately half of the interneurons (53.6%; 539 of 1004 neurons) in the stratum oriens of CA1. This value dropped to one-third (28.4%) in the pyramidal layer, and only 13.6% of the stratum radiatum interneurons (n= 767) expressed GluR5 mRNA. However, most of the neurons expressing GluR5 mRNA were GABA interneurons in the CA1 field. Thus, 86% of those neurons expressing GluR5 (1025 neurons) also express GAD (885 neurons) (Fig.2F), indicating that although in a minority, some non-GABA neurons express GluR5 subunits. Such a value was estimated to be slightly larger in the stratum oriens than in the other layers. A similar pattern was also found in the CA3 field (Fig.2F).
Fig. 2.
Coexpression of GluR5 subunit andGAD mRNAs in hippocampal neurons. A, Composition of low-power pictures showing the hippocampus from the subiculum (SUB) to the CA3 field. Boxesin A from left to rightare presented enlarged in B–E, respectively.GAD expression is labeled red(white arrowheads), whereas GluR5 mRNA is labeled blue (black arrowheads). Coexpression appears as a brown precipitate (black arrows). F, Cells appearingblue, red, and brown were counted in the stratum oriens (SO), pyramidale (SP), and radiatum of CA1 or lucidum of CA3 (SR/L). Theleft histogram represents the fraction of interneurons (GAD-expressing cells) expressing GluR5 subunits, whereas theright histogram represents the fraction of GluR5-expressing cells that are GABA neurons. The histograms are based on 2500–2900 neurons counted in six to eight slices by at least two different people.
GluR6 was abundantly expressed by principal cells in the hippocampus, and a few interneurons in CA1 stratum oriens (6%) and stratum radiatum (3.3%) contained GluR6 transcripts. However, most of the GABA interneurons lying in the pyramidal cell layer also expressed GluR6 (96.2%; Fig.3B). This was also true in CA3, where 93.3% of the GABA cells lying in the pyramidal cell layer were labeled (240 of 257 neurons counted; Fig. 3D,F). As could be seen in single in situ hybridizations (Fig.1B), numerous cells, probably interneurons, in the stratum lucidum of CA3 contain GluR6 transcripts. This result was corroborated by double in situ labeling, demonstrating that a large number (85%; Fig. 3E) ofGAD-expressing cells also expressed GluR6 (573 of 671 neurons counted). Although only one-half of theGluR6-expressing cells in the CA1 stratum oriens were interneurons (65%), in the stratum oriens of CA3 the majority of the GluR6-positive cells could be classified as interneurons (93%; 227 of 242 neurons). Similarly, virtually all of the GluR6-containing cells in the stratum lucidum were interneurons (97%), although in the CA1 stratum radiatum approximately one-half of GluR6-expressing cells also expressed GAD (48%; Fig. 3F).
Fig. 3.
Coexpression of GluR6 subunit andGAD mRNAs in hippocampal neurons. A, Composition of low-power pictures showing the hippocampus from the subiculum (SUB) to the CA3 field. B–E, Details of CA1 pyramidal cell layer (B), stratum radiatum (C), CA3 pyramidal cell layer (D), and stratum lucidum (E). F, Cells appearingblue, red, and brown were counted in the stratum oriens (SO), pyramidal (SP), and radiatum of CA1 or lucidum of CA3 (SR/L). GAD expression is labeled red (white arrowheads), whereas GluR6 mRNA is labeled blue (black arrowheads). Coexpression appears as abrown precipitate (black arrows). Theleft histogram represents the fraction of interneurons (GAD-expressing cells) expressing the GluR6 subunits, whereas theright histogram represents the fraction of GluR6-expressing cells that are GABA neurons. The histograms are based on 1100–1800 neurons counted in six to eight slices by at least two different people. Calculation of the fraction of GluR6-expressing cells also expressing GAD was not possible (asterisk) because all pyramidal cells were labeled blue.
Taken together, these results indicate that GluR5 and GluR6 subunits may coexist in a population of hippocampal interneurons, possibly 24–27% of GABA interneurons in the pyramidal cell layer (both CA1 and CA3) and stratum oriens of CA3 express both subunits. However, it should be noted that the spread of blue reaction caused by the massive expression of GluR6 in the pyramidal layer may induce an overestimation of the number of brown cells in this field. Although coexpression ofGluR5 and GluR6 by interneurons of stratum oriens in CA1 is less likely, both transcripts appear to be present in at least 17% of the stratum radiatum (CA1) and stratum lucidum (CA3) interneurons.
GluR5 and GluR6 combine to form functional receptors
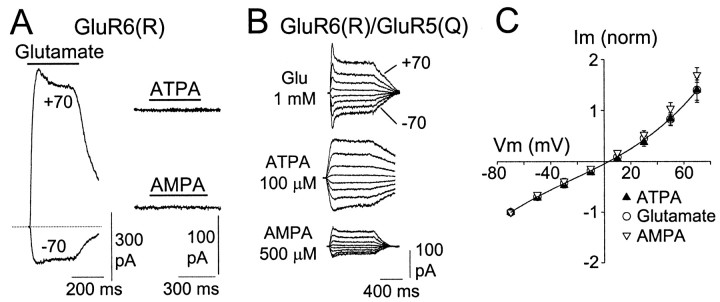
A question that arises as a result of the mRNA expression data is whether or not GluR5 and GluR6 may combine to form heteromeric channels. To address this question, we cotransfected GluR5and GluR6 in HEK293 cells and studied the agonist sensitivity of the receptors. Figure4A shows that homomeric GluR6 receptors are activated by glutamate but are completely insensitive to AMPA and ATPA (Lauridsen et al., 1985), the specific agonist of GluR5 receptors. In our experiments, transfection with theGluR5 plasmid produced barely detectable currents when the cells were challenged by glutamate, kainate, or ATPA (data not shown). However, when GluR6 was cotransfected with GluR5, large currents were generated by the three agonists (Fig.4B). It is known that these subunits hold a site in the second membrane segment that is regulated by mRNA editing (for review, see Seeburg, 1996). Edited forms (R forms) give rise toI–V relationships with outward rectification (Fig.4A), whereas unedited (Q) forms generate receptors that present strong inward rectification. Because R forms are phenotypically dominant (Schoepfer et al., 1994), we looked at theI–V relationship for responses induced by ATPA and AMPA on heteromeric GluR6(R)/GluR5(Q) receptors. As can be seen in Figure 4,B and C, the responses evoked by either glutamate, ATPA, or AMPA in cells cotransfected with both subunits presented a similar degree of outward rectification. Indeed, the index of rectification (IR), calculated as the ratio of conductance at 60 and −60 mV, was 1.17 ± 0.05 (n = 13), 1.16 ± 0.04 (n = 13), and 1.5 ± 0.06 (n= 3) for ATPA, glutamate, and AMPA, respectively. This can only be attributed to the fact that all receptors have at least one GluR6(R) subunit in their structure, which would confer outward rectification, and at least one GluR5(Q), which would confer sensitivity to ATPA and AMPA. We wanted to know whether GluR6(Q) can combine with edited GluR5(R) subunits. Thus, we cotransfected both plasmids and looked at the I–V relationships of the expressed receptors. Homomeric GluR6(Q) generated I–V relationships with a strong inward rectification (IR < 0.1) and were insensitive to ATPA. Cotransfection with GluR5(R) not only abolished inward rectification (IR = 0.84 ± 0.05; n = 6 for glutamate-induced responses) but also conferred sensitivity to ATPA, which presented an I–V relationship with a similar degree of rectification (0.7 ± 0.14; n = 5). These cells were treated with ConA to remove rapid desensitization (Partin et al., 1993) and thus facilitate measurements.
Fig. 4.
GluR6 and GluR5 subunits coassemble to generate heteromeric kainate receptors with new pharmacological properties.A, Typical responses induced by the rapid perfusion of glutamate (1 mm) in HEK293 cells voltage-clamped at the indicated Vm after being transfected with GluR6(R) plasmid. Cells were treated with ConA to remove desensitization. Homomeric GluR6 receptors were not activated by ATPA (100 μm) or S-AMPA (500 μm).B, In cells cotransfected with GluR6(R) and GluR5(Q), currents were activated after perfusion of glutamate, AMPA, and ATPA. Responses activated by either agonist presented a similar degree of outward rectification, indicating the dominance of the R forms (outward-rectifying) over the Q forms (inward-rectifying).C, Current–voltage relationship for these three agonists from cells transfected with GluR5(Q) and GluR6(R). Points are mean ± SEM from eight (ATPA), seven (glutamate), and three (AMPA) cells.
In cells not treated with ConA, 100 μm ATPA induced responses that largely but not totally desensitized (11.6 ± 1% of the peak current remained; n = 21). To determine the population of heteromeric receptors formed after cotransfection of both plasmids, we looked at the efficiency of ATPA to cross-desensitize glutamate-induced response. Figure5A shows that the response to glutamate was largely reduced by a previous pulse of ATPA. In the 17 cells in which GluR5(Q) and GluR6(R) were coexpressed, a prepulse of ATPA depressed the subsequent glutamate-induced peak current by 81.7% (Fig. 5B). In contrast, ATPA did not induce a depression of the glutamate-induced response in cells expressing GluR6 only. The results were similar when GluR5(R) and GluR6(Q) were cotransfected (84.5 ± 2.7%; n = 5 of the glutamate-induced response was cross-desensitized). This result indicates that a large fraction of receptors expressed in these experiments included both GluR5 and GluR6 subunits and that the editing site has no influence on subunit assemblage.
Fig. 5.
In cells cotransfected withGluR5 and GluR6, most of the receptors formed are heteromeric receptors. A, ATPA cross-desensitized most of the response activated by glutamate (1 mm; arrows) in cells cotransfected with both subunits and not treated with ConA, indicating the existence of a largely homogeneous population of heteromeric receptors.B, Average data for the glutamate-induced current remaining after a prepulse of ATPA (100 μm) in cells transfected with GluR6(Q) (black bar), with GluR5(Q) and GluR6(R) (light gray bar), or with GluR5(R) and GluR6(Q) (dark gray bar). The white bar represents the remaining steady-state response in ATPA-induced responses (St. St.; i.e., nondesensitizing).Numbers in parentheses correspond to the number of cells studied.
Activation–inactivation properties of kainate receptors
Because most of the receptors susceptible to glutamate activation in cells cotransfected with GluR5 and GluR6 were heteromeric, we wanted to further characterize this new type of kainate receptor. Therefore, the glutamate dose–response relationship for the peak current was calculated and compared with homomeric GluR6 receptors (Fig. 6). Data from different cells were normalized and pooled for computer fitting with the logistic equation. GluR5/R6 receptors expressed in HEK293 cells did not differ from homomeric GluR6 receptors in their activation or steady-state desensitization in response to glutamate (Fig. 6). The curve that best fitted the dose–response data from GluR5/R6 receptors presented an EC50 value of 1.34 mm for glutamate, with a slope factor of 0.6. The inactivation curve was constructed as previously described (Paternain et al., 1998; Fig.6A). Briefly, after a period of incubation of at least 10 sec, enough time to attain desensitization in a steady-state situation, this solution was rapidly replaced by another one containing a saturating concentration of kainate (Fig. 6A). The peak response was measured and compared to that obtained when no glutamate was included in the conditioning pulse. The fitted curve revealed a half-maximal inactivation at a submicromolar concentration of glutamate (0.33 μm) with a slope factor of 1. These values were close to those observed for GluR6 homomeric receptors in these (Fig. 6) and previous experiments (Heckmann et al., 1996; Paternain et al., 1998).
Fig. 6.

Activation–inactivation properties of heteromeric GluR5/R6 kainate receptors. A, To calculate the steady-state inactivation curve, cells were exposed to different concentrations of glutamate for 10 sec before jumping to a solution containing kainate. The effect of 1 μm of glutamate on a test response evoked by kainate (300 μm; thin trace) is shown (thick trace). B,Activation and inactivation curves of homomeric (dotted) and heteromeric (solid) kainate receptors. The lines through the points represent the best least-squares fit of the Hill equation to pooled data and presented a half-saturated concentration (EC50) of 898 and 1338 μm for GluR6 and GluR5/R6 receptor activation, respectively, and 0.34 and 0.33 μm for inactivation (IC50). Points are the mean ± SEM of data collected from three to seven cells.
The onset and recovery of desensitization was also determined for heteromeric GluR5/R6 receptors. In the continuous presence of agonist, the current desensitization followed a single exponential time course (Fig. 7A). It was observed that the onset of desensitization was slightly faster in cells transfected with GluR5 and GluR6 than withGluR6 alone. As for GluR6 receptors, the time constant of desensitization was concentration-dependent, decreasing as the agonist concentration increased. To calculate the actual rate of the onset of receptor desensitization, we measured the time constant of current decay at several concentrations of agonist. The concentration versus desensitization rate (i.e., 1/τonset) plot reveals a saturating process, which was well described by a rectangular hyperbolic function. The asymptotic value of this function reflects the rate of desensitization when the binding rate of glutamate is no longer a limiting step (Paternain et al., 1998). This value was significantly different in heteromeric and homomeric channels (126 and 92 sec−1, respectively; p < 0.05; normal distribution comparison), indicating that heteromeric GluR5/R6 receptors desensitize slightly but significantly faster than homomeric GluR6 formations.
Fig. 7.
Desensitization kinetics in GluR6 and GluR5/R6 kainate receptors. A, Onset of desensitization was faster in heteromeric than in homomeric receptors. The superimposed lines represent single exponential fits to the desensitization process with the indicated time constants (τDes).B, The time constant value for onset of desensitization was measured in several cells at different concentrations of agonist, normalized and pooled for fitting. The following rectangular hyperbolic function was fitted to the data points: y = a · x/(b + x), where y is the onset rate, measured as 1/τDes at any concentration of glutamate (x), a is the asymptote of the function, and b is the agonist concentration at which rate is 50% of the maximum. The asymptotic values obtained for each receptor type are indicated and were significantly different (p < 0.05). Data points are the mean ± SEM of five or six cells. C, The desensitization recovery of heteromeric receptors was slower than that of homomeric assemblies. Receptors were completely desensitized by applying a 0.6 sec pulse of glutamate (1 mm). Conditioning pulses were followed by test pulses of the same duration delivered at different intervals. The peak amplitude was measured and normalized with respect to the average of all conditioning pulses. This value (recovery) was plotted against the interval between applications (D). The data collected from two (asterisks) to four cells were pooled and fitted with single exponential functions (dotted and solid lines for GluR6 and GluR5/R6 receptors, respectively) with the illustrated time constants of recovery (τrec) which significantly differed (p < 0.05) when normal distributions were statistically compared. Data points are the mean ± SEM.
We then investigated whether the rate of recovery from the desensitized state was different in heteromeric and homomeric receptors. Thus, we applied test pulses of glutamate at different intervals after the receptors were desensitized by a conditioning pulse of agonist (Fig.7C). Peak responses were measured and normalized to permit the comparison of several cells. Homomeric GluR6 receptors recovered from desensitization after an exponential time course with a time constant of 2.2 ± 0.1 sec (cf. Paternain et al., 1998). In cells expressing heteromeric GluR5/R6 receptors, the time course of recovery was also well fitted by a single exponential function but showed a slower time constant (3.2 ± 0.4 sec; Fig. 7D). These two values differed statistically (p < 0.05; normal distribution comparison)
These results indicate that coassembly of GluR5 and GluR6 subunits alters the kinetics of both onset and recovery of desensitization such that heteromeric GluR5/R6 receptors enter more rapidly into the desensitized state and leave it more slowly.
We wanted to further analyze how the addition of new subunits into receptors made of GluR5 (plus KA2 or GluR6) or GluR6 subunits (plus KA2 or GluR5) modifies the activation of the functional receptor. In Figure8, the response patterns induced by rapid application of the three different agonists are compared in receptors expressed after the cotransfection of the three different subunits. Glutamate induced similar responses when activating receptors of either configuration. In contrast, AMPA activated the three heteromeric receptors with remarkably different patterns. Although this agonist desensitized rapidly and completely GluR5/KA2 receptors, it produced currents with GluR6/KA2 that inactivated partially and slowly. Something similar happened when the AMPA derivative ATPA was applied. This agonist did not desensitize heteromeric GluR6/KA2 formations, whereas it produced a rapid and strong desensitization in GluR5/KA2 receptors. Interestingly, as described above, ATPA rapidly desensitized heteromeric GluR5/R6, but left some steady-state current. This distinctive behavior in terms of desensitization makes it possible to differentiate this type of receptor from the other formations. In addition, to generate responses with different properties, ATPA presented a higher affinity for heteromeric GluR5/R6 (EC50 = 21 ± 2.8 μm) than for GluR6/KA2 (EC50 = 84 ± 9.7 μm). As for glutamate and kainate, the treatment of kainate receptors with ConA to remove desensitization drastically increased the apparent affinity (Paternain et al., 1998) because the EC50 for ATPA was decreased by 19-fold (EC50 = 1.1 ± 0.08 μm; Fig.8C).
Fig. 8.
Activation patterns of heteromeric kainate receptors by different agonists. A, HEK cells were cotransfected with either GluR6 and KA2, GluR5 and KA2, or GluR6 and GluR5, and the current response to rapid applications of glutamate (Glu, 300 μm), AMPA (500 μm), and ATPA (100 μm, except where otherwise indicated) were studied. Note that glutamate evoked responses with similar shapes in the three receptor types. However, GluR6/KA2 responded to AMPA and ATPA differently than the other receptors. ATPA differentiated GluR5/KA2 from GluR5/GluR6 in that it did not completely desensitize GluR6-containing GluR5 receptor. Except for the responses induced by ATPA in GluR5/GluR6 receptors, the illustrated recordings were collected from the same cell in each case. B, Dose–response curve for ATPA activation of heteromeric GluR6/KA2 receptors. The current amplitudes were expressed as a percentage of the current induced by 500 μm AMPA. The solid line is the result of fitting the Hill equation with an EC50 of 84 μm and a Hill coefficient of 1.7. Points are the mean ± SEM of data collected from 2 (asterisk) to 26 cells. C, Dose–response curve for ATPA in heteromeric GluR5/GluR6 receptors. Solid lines are the result of fitting the Hill equation with EC50 of 21 and 1.1 μm in cells untreated or treated with ConA, respectively, to remove desensitization. Points are the mean ± SEM of data collected from three to seven cells.
It is noteworthy that ATPA indeed behaves as a partial agonist of GluR6-containing receptors when these also contain the KA2 or GluR5 subunits. In heteromeric GluR6/KA2 receptors, and unlike glutamate or AMPA, ATPA induced nondesensitizing responses (<5% of current attenuation in a 20 sec pulse at 100 μm), and ATPA-induced currents always were smaller than the peak response induced by glutamate (1 mm) or S-AMPA (500 μm). On average, the current induced by 100 μm ATPA was ∼30% of the peak current induced by 500 μmS-AMPA. Similarly, in GluR6/R5 receptors, the peak response to 100 μmATPA was 25 ± 1.5% (n = 51) of that induced by glutamate (1 mm), of which only a fraction desensitized.
These results indicate that ATPA can no longer be considered as a specific agonist of GluR5-containing receptors. Although in heteromeric GluR6/KA2 receptors, the ATPA-induced response would be a fraction of the amplitude of glutamate-induced rapidly desensitizing response, it is worth noting that ATPA induces nondesensitizing currents, and in terms of steady-state activation, it should be much more effective than kainate and the endogenous ligand.
DISCUSSION
In the present work, we have studied the coexpression and coassembly of two of the most abundantly expressed kainate receptor subunits, GluR5 and GluR6, in the hippocampus. Our results indicate that both subunits are able to form heteromeric receptors and that both colocalize in a population of GABA neurons in the hippocampus, implicating these heteromeric receptors in the transduction machinery for glutamate signaling in inhibitory interneurons.
In contrast to other authors' observations (Sommer et al., 1992;Swanson and Heinemann, 1998), under our experimental situation, we were unable to obtain expression of homomeric GluR5 receptors after transfection in HEK293 cells. This was not attributable to unsuccessful plasmid transfection or a failure in cDNA transcription or RNA translation because when the same construct was contransfected withKA2 (a subunit unable to form functional channels by itself), responses were induced in 100% of the cases. We have no obvious explanation for this phenomenon, but it is possible that the functional state of the cell line used (e.g., passage number, time in culture, etc.) might influence the assembly of GluR5 subunits into functional receptors, giving different results in different laboratories. In addition, it is well known that only the shortest splice variant of GluR5 (i.e., GluR5–2a) is able to form functional homomeric channels (Sommer et al., 1992), yet with low efficiency when compared to that of GluR6. Interestingly, most of the receptors assembled after expression of GluR5 and GluR6 subunits were heteromeric. Cross-desensitization experiments allowed us to estimate that >80% of the functional receptors expressed after cotransfection of both cDNAs contained enough GluR5 subunits to confer ATPA sensitivity. This analysis was based on the rationale that whereas glutamate could activate the whole population of receptors (homomeric and heteromeric), ATPA would only be able to activate and desensitize those receptors containing GluR5 subunits because it was unable to activate homomeric GluR6. Together, these results indicate that cotransfection of GluR5 and GluR6 induces a large proportion of heteromers present in the membrane, allowing us to further characterize them.
Responses induced by the endogenous agonist in heteromeric and homomeric GluR6 receptors did not differ in that the current was rapidly activated, and receptors rapidly and almost completely desensitized. However, R5/R6 heteromeric constructions desensitized 30% faster and recovered from desensitization slower (∼40%) than homomeric GluR6. It is worth noting that desensitization rate constants were estimated from the asymptote of fitted curves (Fig.7A), i.e., when the rate becomes independent of agonist concentration and therefore independent of the perfusion speed (Paternain et al., 1998). The desensitization rate measured in this way should be fairly accurate, making the observed differences reliable. Indeed, such a difference in the onset rate of desensitization would in principle account for the difference in the EC50for glutamate from dose–response curves (Fig. 6), because rapid desensitization may curtail activation at even high concentrations of agonist. Therefore, we conclude that heteromeric R5/R6 and homomeric R6 have similar apparent affinities for glutamate. Previous work has reported lower EC50 values for homomeric GluR6 as well as faster desensitization rates (Heckmann et al., 1996; Swanson et al., 1997; Traynelis and Wahl, 1997). Although the slower perfusion speed attained by us could account for larger EC50 values, in the mentioned works fast perfusion was achieved by excising patches or by lifting the cells from the bottom of the dish. It is presently unclear how these manipulations may affect desensitization kinetics and receptor affinity. Indeed, with our perfusion system we have been able to resolve GluR6-mediated responses in excised patches with desensitization time constants of ≤2 msec. Whatever the explanation, it is unlikely that the small changes detected by us in desensitization kinetics and affinity would establish relevant differences in physiological responses mediated by this kind of heteromeric kainate receptor, as compared to homomeric GluR6.
Conversely, the gating properties of heteromeric receptors for agonists other than glutamate presented remarkable differences. GluR6/KA2 and GluR5/KA2 were both sensitive to ATPA and AMPA. However, although responses were poorly or nondesensitizing in the former, currents rapidly faded in the latter (Fig. 8). These same agonists activated heteromeric GluR5/GluR6 with a different pattern. In particular, ATPA desensitized these receptors rapidly but only partially, because a clear steady current remained at all concentrations. In addition, the affinity of heteromeric GluR5/GluR6 receptors for ATPA was greater than heteromeric GluR6/KA2 formations (21 vs 84 μm in EC50, respectively) and as such, more similar to that of native kainate receptors expressed by DRG cells and human homomeric GluR5 receptors (Clarke et al., 1997). However, many other receptor types would predictably be activated by ATPA, because the presence of additional receptor subunits in the hippocampus, such as KA1 and KA2, leaves open the possibility that GluR5/KAx, GluR6/KAx, and GluR5/GluR6/KAx receptors could also exist.
The pharmacological distinction between heteromeric kainate receptors is not limited to the combination of one of the GluR5–7 subunits with one of the KA subunits, an assumption that surprisingly has been the overall accepted scheme for the assembly of kainate receptors (for review, see Lerma, 1999). This assumption may be, at least in part, responsible for a number of discrepancies observed between recombinant and native kainate receptors. For instance, by using single-cell RT-PCR of electrophysiologically characterized neurons, we found that ∼20% of the hippocampal embryonic cells, maintained in culture for a few days, expressed GluR5 and GluR6 mRNAs (Ruano et al., 1995). Interestingly, in those cells we could never observe responses such as those obtained in recombinant systems or DRG neurons, indicating the presence of homomeric GluR5 receptors. This observation, surprising at that time, can be now explained if heteromeric GluR5/R6 receptors were indeed present in the membrane of those cells: the response induced by kainate would not have differed from that obtained in neurons expressing only GluR6. Furthermore, Vignes et al. (1997)observed that two new compounds, LY293558 and LY294486, that antagonize human GluR5 receptors, also antagonize the depolarization induced by kainate in the CA3 region as well as the synaptic response induced by high-frequency stimulation of mossy fibers, which has been unequivocally identified as GluR6-mediated (Mulle et al., 1998). Our demonstration of the assembly of GluR5 and GluR6 subunits reconciles both observations and indicates that the presence of a population of receptors including both subunits should be taken into account. It is worth noting, however, that the presence of heteromeric GluR5/GluR6 receptors may not be restricted to the hippocampus (Mackler and Eberwine, 1993) but may also occur in other parts of the brain (e.g., striatal cells; Ghasemzadeh et al., 1996; Porter et al., 1998).
Finally, what is the role of heteromeric R5/R6 receptors? Is there any advantage in forming heteromeric R5/R6 over homomeric receptors? At the present time, when the functional role of kainate receptors is still largely ignored, it is difficult to define a role for a heteromeric kainate receptor. However, it is likely that the different subunits may confer different targeting capabilities to the corresponding receptors. For instance, it has been reported that GluR6 subunits interact with PSD95/SAP90 proteins through binding to their PDZ1 domain. In contrast, the KA2 C-terminal domain interacts with SH3 and GK domains of the same protein (Garcia et al., 1998). Obviously, the clustering capabilities of heteromeric GluR6/KA2 receptors performed by this kind of structural proteins are wider than those of homomeric GluR6, as would be the case for their coupling to signaling proteins. It is not yet known which proteins are able to bind and cluster GluR5 subunits, but the existence of three different C-terminal variants indicates multiple possibilities for specific receptor targeting. A given binding motif may serve to associate the receptor to a particular signal transduction cascade and/or to localize it to particular regions of the cell (axon terminal vs dendritic spine), implying different functional roles. Although all these possibilities should be studied in detail, the unique pharmacological and kinetic properties described here for heteromeric receptors should prove useful to identify native kainate receptors. The possibility of coassembly of subfamily members provides evidence to indicate that the molecular diversity of kainate receptors in the brain is larger than previously thought and adds new excitement into the study of this receptor type, a receptor that is just beginning to be understood.
During the review process of this paper, a report by Cui and Mayer (1999) appeared demonstrating coassembly of GluR5, GluR6, and GluR7 in HEK 293 cells. The conclusion from this study is essentially the same as ours in that, similar to AMPA receptors, kainate receptor subunits exhibit promiscuous coassembly. This study also revealed that heteromeric GluR5/R6 receptors showed reduced desensitization with enhanced steady-state current in response to kainate. In keeping with this, we also observed some enhanced steady-state current in kainate-induced responses after coexpression of both subunits (steady state of 8 ± 2%; n = 8 of peak amplitude; data not shown) as compared to homomeric GluR6 receptors (<2% of the peak current remained). However, any additional alteration of activation–inactivation kinetic properties of heteromeric receptors is not comparable in both studies because, unlike Cui and Mayer (1999), we evaluated heteromeric receptor properties for the endogenous ligand glutamate.
Footnotes
This work was supported in part by grants to J.L. from the Dirección General de Enseñanza Superior e Investigación Científica (P.M.96/0008) and to J.L. and M.A.N. from the Community of Madrid (08.5/0042/1998). M.T.H. held a fellowship from the Instituto de Cooperación Iberoamericana. We thank Dr. P. H. Seeburg for the cytomegalovirus expression plasmids encoding GluR6, GluR5, and KA2, Drs. A. Tobin and N. Tillakaratne for the GAD65 plasmid, Dr. T. E. Hughes for the plasmid encoding green fluorescent protein (GFP), Dr. J. Drejer for the gift of ATPA, Dr. M. Sefton for proofreading, and D. Guinea for technical assistance.
Correspondence should be addressed to Dr. J. Lerma, Instituto Cajal, Consejo Superior de Investigaciones Científicas, Avenue Doctor Arce 37, 28002 Madrid, Spain. E-mail: lerma@cajal.csic.es.
REFERENCES
- 1.Bahn S, Volk B, Wisden W. Kainate receptor gene expression in the developing rat brain. J Neurosci. 1994;14:5525–5547. doi: 10.1523/JNEUROSCI.14-09-05525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Ari Y. Limbic seizure and brain damage produced by kainic acid: mechanisms and relevance to human temporal lobe epilepsy. Neuroscience. 1985;14:375–403. doi: 10.1016/0306-4522(85)90299-4. [DOI] [PubMed] [Google Scholar]
- 3.Bettler B, Egebjerg J, Sharma G, Pecht G, Hermans-Borgmeyer I, Moll C, Stevens CF, Heinemann S. Cloning of a putative glutamate receptor: a low affinity kainate-binding subunit. Neuron. 1992;8:257–265. doi: 10.1016/0896-6273(92)90292-l. [DOI] [PubMed] [Google Scholar]
- 4.Bowie D, Mayer ML. Inward rectification of both AMPa and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron. 1995;15:453–462. doi: 10.1016/0896-6273(95)90049-7. [DOI] [PubMed] [Google Scholar]
- 5.Castillo PE, Malenka RC, Nicoll RA. Kainate receptors mediate a slow postsynaptic current in hippocampal CA3 neurons. Nature. 1997;388:182–186. doi: 10.1038/40645. [DOI] [PubMed] [Google Scholar]
- 6.Clarke VRJ, Ballyk BA, Hoo KH, Mandelzys A, Pellizari A, Bath CP, Thomas J, Sharpe EF, Davies CH, Ornstein PL, Schoepp DD, Kamboj RK, Collingridge GL, Lodge D, Bleakman D. A hippocampal GluR5 regulating inhibitory synaptic transmission. Nature. 1997;389:599–602. doi: 10.1038/39315. [DOI] [PubMed] [Google Scholar]
- 7.Cossart R, Esclapez M, Hirsch JC, Bernard C, Ben-Ari Y. GluR5 kainate receptor activation in interneurons increases tonic inhibition of pyramidal cells. Nat Neurosci. 1998;1:470–478. doi: 10.1038/2185. [DOI] [PubMed] [Google Scholar]
- 8.Coyle JT. Neurotoxic actions of kainic acid. J Neurochem. 1983;41:1–11. doi: 10.1111/j.1471-4159.1983.tb11808.x. [DOI] [PubMed] [Google Scholar]
- 9.Cui C, Mayer ML. Heteromeric kainate receptors formed by the coassembly of GluR5, GluR6, and GluR7. J Neurosci. 1999;19:8281–8291. doi: 10.1523/JNEUROSCI.19-19-08281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egebjerg J, Bettler B, Hermans-Borgmeyer I, Heinemann S. Cloning of a cDNA for a glutamate receptor subunit activated by kainate but not by AMPA. Nature. 1991;351:745–748. doi: 10.1038/351745a0. [DOI] [PubMed] [Google Scholar]
- 11.Frerking Malenka RC, Nicoll RA. Synaptic activation of kainate receptors on hippocampal interneurons. Nat Neurosci. 1998;1:479–486. doi: 10.1038/2194. [DOI] [PubMed] [Google Scholar]
- 12.Garcia EP, Mehta S, Blair LAC, Wells DG, Shang J, Fukushima T, Fallon JR, Garner CC, Marshall J. SAP90 binds and clusters kainate receptors causing incomplete desensitization. Neuron. 1998;21:727–739. doi: 10.1016/s0896-6273(00)80590-5. [DOI] [PubMed] [Google Scholar]
- 13.Ghasemzadeh MB, Sharma S, Surmeier DJ, Eberwine JH, Chesselet MF. Multiplicity of glutamate receptor subunits in single striatal neurons: an RNA amplification study. Mol Pharmacol. 1996;49:852–859. [PubMed] [Google Scholar]
- 14.Heckmann M, Bufler J, Franke C, Dudel J. Kinetics of homomeric GluR6 glutamate receptor channels. Biophys J. 1996;71:1743–1750. doi: 10.1016/S0006-3495(96)79375-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herb A, Burnashev N, Werner P, Sakmann B, Wisden W, Seeburg PH. The KA-2 subunit of excitatory amino acid receptors shows widespread expression in brain and forms ion channels with distantly related subunits. Neuron. 1992;8:775–785. doi: 10.1016/0896-6273(92)90098-x. [DOI] [PubMed] [Google Scholar]
- 16.Kamboj SK, Swanson GT, Cull-Candy SG. Intracellular spermine confers rectification on rat calcium-permeable AMPA and kainate receptors. J Physiol (Lond) 1995;486:297–303. doi: 10.1113/jphysiol.1995.sp020812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kidd FL, Isaac JTR. Developmental and activity dependent regulation of kainate receptors at thalamocortical synapses. Nature. 1999;400:569–573. doi: 10.1038/23040. [DOI] [PubMed] [Google Scholar]
- 18.Koh DS, Burnashev N, Jonas P. Block of native Ca2+-permeable AMPA receptors in rat brain by intracellular polyamines generates double rectification. J Physiol (Lond) 1995;486:305–312. doi: 10.1113/jphysiol.1995.sp020813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauridsen J, Honore T, Krogsgaard-Larsen P. Ibotenic acid analogues. Synthesis, molecular flexibility, and in vitro activity of agonists and antagonists at central glutamic acid receptors. J Med Chem. 1985;28:668–672. doi: 10.1021/jm50001a022. [DOI] [PubMed] [Google Scholar]
- 20.Lerma J. Spermine regulates N-methyl-d-aspartate receptor desensitization. Neuron. 1992;8:343–352. doi: 10.1016/0896-6273(92)90300-3. [DOI] [PubMed] [Google Scholar]
- 21.Lerma J. Kainate reveals its targets. Neuron. 1997;19:1155–1158. doi: 10.1016/s0896-6273(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 22.Lerma J. Kainate receptors. In: Jonas P, Monyer H, editors. Handbook of experimental pharmacology, Vol 141, Ionotropic glutamate receptors in the CNS. Springer; Berlin: 1999. pp. 275–307. [Google Scholar]
- 23.Mackler SA, Eberwine JH. Diversity of glutamate receptor subunits mRNA expression within live hippocampal CA1 neurons. Mol Pharmacol. 1993;44:308–315. [PubMed] [Google Scholar]
- 24.Marshall J, Molloy R, Moss GWJ, Howe JR, Hughes TE. The Jellyfish green fluorescent protein: a new tool for studying ion channels expression and function. Neuron. 1995;14:211–215. doi: 10.1016/0896-6273(95)90279-1. [DOI] [PubMed] [Google Scholar]
- 25.Mulle C, Sailer A, Perez-Otaño I, Dickinson-Anson H, Castillo PE, Bureau I, Maron C, Gage FH, Mann JR, Bettler B, Heinemann SF. Altered synaptic physiology and reduced susceptibility to kainate-induced seizures in GluR6-deficient mice. Nature. 1998;392:601–605. doi: 10.1038/33408. [DOI] [PubMed] [Google Scholar]
- 26.Nadler J. Kainic acid as a tool for the study of temporal lobe epilepsy. Life Sci. 1981;29:2031–2042. doi: 10.1016/0024-3205(81)90659-7. [DOI] [PubMed] [Google Scholar]
- 27.Nieto MA, Patel K, Wilkinson DG. In situ hybridisation analysis of chick embryos in whole mount and in tissue sections. Methods Cell Biol. 1996;51:219–235. doi: 10.1016/s0091-679x(08)60630-5. [DOI] [PubMed] [Google Scholar]
- 28.Partin KM, Patneau DK, Winters CA, Mayer ML, Buonanno A. Selective modulation of desensitization at AMPA versus kainate receptors by cyclothiazide and concanavalin A. Neuron. 1993;11:1069–1082. doi: 10.1016/0896-6273(93)90220-l. [DOI] [PubMed] [Google Scholar]
- 29.Paternain AV, Rodríguez-Moreno A, Villarroel A, Lerma J. Activation and desensitization properties of native and recombinant kainate receptors. Neuropharmacology. 1998;37:1249–1259. doi: 10.1016/s0028-3908(98)00098-7. [DOI] [PubMed] [Google Scholar]
- 30.Porter JT, Cauli B, Staiger JF, Lambolez B, Rossier J, Audinat E. Properties of bipolar VIPergic interneurons and their excitation by pyramidal neurons in the rat neocortex. Eur J Neurosci. 1998;10:3617–3628. doi: 10.1046/j.1460-9568.1998.00367.x. [DOI] [PubMed] [Google Scholar]
- 31.Rodríguez-Moreno A, Lerma J. Kainate receptor modulation of GABA release involves a metabotropic function. Neuron. 1998;20:1211–1218. doi: 10.1016/s0896-6273(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez-Moreno A, Herreras O, Lerma J. Kainate receptors presynaptically downregulate GABAergic inhibition in the rat hippocampus. Neuron. 1997;19:893–901. doi: 10.1016/s0896-6273(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 33.Ruano D, Lambolez B, Rossier J, Paternain AV, Lerma J. Kainate receptors subunits expressed in single cultured hippocampal neurons: molecular and functional variants by RNA editing. Neuron. 1995;14:1009–1017. doi: 10.1016/0896-6273(95)90339-9. [DOI] [PubMed] [Google Scholar]
- 34.Sakimura K, Morita T, Kushiya E, Mishina M. Primary structure and expression of the γ2 subunit of the glutamate receptor channel selective for kainate. Neuron. 1992;8:267–274. doi: 10.1016/0896-6273(92)90293-m. [DOI] [PubMed] [Google Scholar]
- 35.Schoepfer R, Monyer H, Sommer B, Wisden W, Sprengel R, Kuner T, Lomeli H, Herb A, Kohler M, Burnashev N, Günther W, Ruppersberg P, Seeburg P. Molecular biology of glutamate receptors. Prog Neurobiol. 1994;42:353–357. doi: 10.1016/0301-0082(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 36.Seeburg PH. The role of RNA editing in controlling glutamate receptor channel properties. J Neurochem. 1996;66:1–5. doi: 10.1046/j.1471-4159.1996.66010001.x. [DOI] [PubMed] [Google Scholar]
- 37.Sommer B, Burnashev N, Verdoorn TA, Keinänen K, Sakmann B, Seeburg PH. A glutamate receptor channel with high affinity for domoate and kainate. EMBO J. 1992;11:1651–1656. doi: 10.1002/j.1460-2075.1992.tb05211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanson GT, Heinemann SF. Heterogeneity of homomeric GluR5 kainate receptor desensitization expressed in HEK293 cells. J Physiol (Lond) 1998;15:639–646. doi: 10.1111/j.1469-7793.1998.639ba.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanson GT, Gereau RW, Green T, Heinemann SF. Identification of amino acid residues that control functional behaviour in GluR5 and GluR6 kainate receptors. Neuron. 1997;19:913–926. doi: 10.1016/s0896-6273(00)80972-1. [DOI] [PubMed] [Google Scholar]
- 40.Traynelis SF, Wahl P. Control of rat GluR6 glutamate receptor open probability by protein kinase A and calcineurin. J Physiol (Lond) 1997;503:513–531. doi: 10.1111/j.1469-7793.1997.513bg.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vignes M, Collingridge GL. The synaptic activation of kainate receptors. Nature. 1997;388:179–182. doi: 10.1038/40639. [DOI] [PubMed] [Google Scholar]
- 42.Vignes M, Bleakman D, Lodge D, Collingridge GL. The synaptic activation of the GluR5 subtype of kainate receptor in area CA3 of the rat hippocampus. Neuropharmacology. 1997;36:1477–1481. doi: 10.1016/s0028-3908(97)00158-5. [DOI] [PubMed] [Google Scholar]
- 43.Wisden W, Seeburg PH. A complex mosaic of high-affinity kainate receptors in rat brain. J Neurosci. 1993;13:3582–3598. doi: 10.1523/JNEUROSCI.13-08-03582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]