Abstract
The period (per) and timeless (tim) genes encode interacting components of the circadian clock. Levels and phosphorylation states of both proteins cycle with a circadian rhythm, and the proteins drive cyclic expression of their RNAs through a feedback mechanism that is, at least in part, negative. We report here that a hypophosphorylated mutant PER protein, produced by creating a small internal deletion, displays increased stability and low-amplitude oscillations, consistent with previous reports that phosphorylation is required for protein turnover. In addition, this protein appears to be defective in feedback repression because it is associated with relatively high levels of RNA and high levels of TIM. Transgenic flies carrying the mutant PER protein display a temperature-dependent shortening of circadian period and are impaired in their response to light, particularly to pulses of light in the late night that normally advance the phase of the rhythm. Interestingly, per RNA is induced by light in these flies, most likely because of the removal of the light-sensitive TIM protein, thus implicating a more direct role for TIM in transcriptional inhibition. These data have relevance for mechanisms of feedback repression, and they also address existing models for the differential behavioral response to light at different times of the night.
Keywords: circadian rhythms, Drosophila, per, tim, entrainment to light, feedback
In Drosophila, the period (per) and timeless (tim) genes are interacting components of the circadian clock (Sehgal et al., 1996). Mutations in these genes affect the periodicity of eclosion and locomotor activity rhythms, and a lack of either gene product results in complete loss of circadian rhythms (Konopka and Benzer, 1971; Konopka et al., 1994;Sehgal et al., 1994; Rutila et al., 1996; Ousley et al., 1998). RNA and protein levels of per and tim cycle with a circadian period, and the two proteins form a heterodimer that negatively regulates the synthesis of both mRNAs (Hardin et al., 1990; Zerr et al., 1990; Sehgal et al., 1995; Hunter-Ensor et al., 1996; Myers et al., 1996; Zeng et al., 1996). The feedback loop thus generated is thought to constitute the molecular basis of behavioral rhythms. Other components of this feedback loop are the products of thedclock (clk) and cycle(cyc) genes, both of which are required for transcriptional activation of per and tim (Allada et al., 1998;Bae et al., 1998; Darlington et al., 1998; Rutila et al., 1998), and the double-time (DBT) kinase that phosphorylates PER (Kloss et al., 1998; Price et al., 1998). Both PER and TIM are cyclically phosphorylated (Edery et al., 1994; Zeng et al., 1996), a modification that appears to be required for protein turnover (Price et al., 1998;Naidoo et al., 1999).
Levels of TIM are rapidly decreased by light treatment, thereby providing a mechanism for photic entrainment of this circadian clock (Hunter-Ensor et al., 1996; Myers et al., 1996; Zeng et al., 1996). Pulses of light in the early night, which delay the phase of the overt rhythm, as well as those in the latter half of the night, which advance the phase, reduce TIM levels (Suri et al., 1998; Yang et al., 1998). PER levels are unchanged, although PER phosphorylation is delayed in the former case and advanced in the latter (Lee et al., 1996). A number of models have been proposed to explain how a unidirectional response of a clock protein (TIM) is converted to a bidirectional effect on the overt rhythm. One possible explanation is that delays versus advances are determined by the level of tim RNA, such that high RNA levels produce delays because they can replace the degraded protein, and low RNA levels produce advances because the protein is not replaced (Myers et al., 1996; Zeng et al., 1996). Other models invoke the phosphorylation state of the proteins or perhaps their subcellular localization (Lee et al., 1996).
We report here that deletion of a segment of PER that is downstream of the PAS protein–protein interaction domain affects the rescue of rhythms in transgenic per01 flies. In addition to aberrant circadian periodicity, transgenic flies show altered responses to pulses of light. The molecular deficits underlying these behavioral phenotypes include an increase in protein stability and a defect in negative feedback. Apart from identifying an important functional domain in PER, these studies suggest novel aspects of feedback regulation and shed light on possible resetting mechanisms.
MATERIALS AND METHODS
Activity assays. Three to 7-d-old flies were entrained on a 12 hr light/dark cycle at 25°C for 3 d, after which time they were placed in constant darkness (DD) at the appropriate temperature (20, 25, or 27°C) to monitor locomotor activity. Locomotor activity was monitored using the TriKinetics Inc. (Waltham, MA) system (Konopka et al., 1994). Periodogram analysis was performed on 7 consecutive days of data using the tau analysis package from Mini-Mitter Co. (Sunriver, OR) (Sehgal et al., 1994). For temperature compensation assays, some animals were monitored for 1 week each at both 20 and 27°C, whereas others were monitored at one temperature only. Data from both procedures were pooled into a 20°C pool and 27°C pool, and statistical comparison of means was performed using Student's t test.
Resetting experiments. Three to 7-d-old flies were entrained on a 12 hr light/dark cycle for 3 d. An Aschoff TypeII phase response curve (PRC) was used to facilitate comparison between animals of different period (Aschoff, 1965). In this modification of the standard PRC, light pulses normally delivered during the subjective night [CT12–CT0 (CT, or circadian time, denotes time of day under free-running conditions)] are instead delivered during the last night of light–dark conditions (LD) [ZT12–ZT0 (ZT, or zeitgeber time, refers to the entrainment regimen with ZT0 being lights on and ZT12 being lights off in a 12 hr light/dark cycle)]. Subjective day pulses were delivered during the first day of DD. At the times tested, 23 flies were treated with light (2000 lux, 10 min) and placed into constant darkness at 27°C for monitoring of locomotor activity (described above). Seven consecutive days of activity records were analyzed for period and activity offsets using the Circadia software package (Mistlberger et al., 1996). Flies that survived 7 d and were deemed rhythmic by χ2periodogram analysis were used to compute phase shifts; the number of such flies varied between 14 and 20, except the ΔC2population pulsed at ZT6 of which 11 flies were used. Phase shifts were computed as the difference in average activity offset between light-treated populations and an unpulsed control. Statistical significance of phase shifts was determined by comparing average activity offsets of light-treated populations with an unpulsed control using Student's t test. For the experiment shown in Figure6, the temperature throughout the experiment was kept at 27°C to facilitate comparison between the two genotypes because the period difference is smaller at that temperature (see Table 2). However, a similar PRC for both genotypes was generated at 25°C.
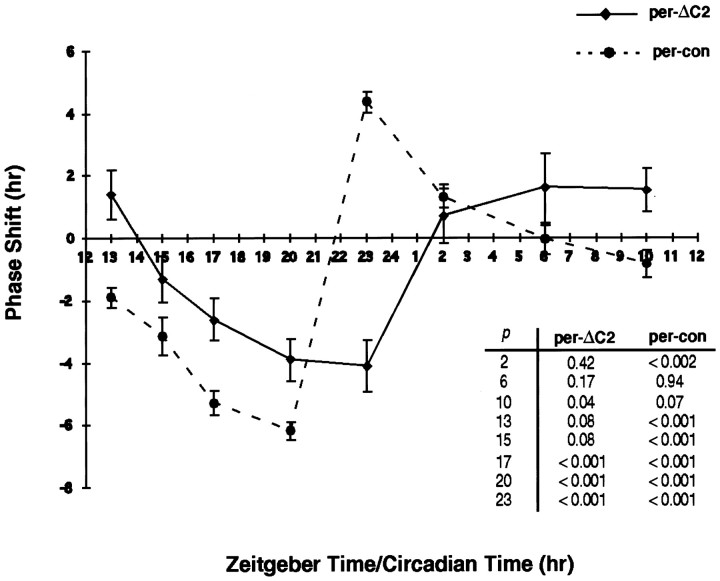
Fig. 6.
Behavioral response of per-ΔC2flies to light. per-con and per- ΔC2flies were exposed to light at the times indicated, and activity rhythms were assayed using the Aschoff Type II PRC (Aschoff, 1965). The experiment was performed at 27°C to facilitate the comparison of the two genotypes because the period difference is smaller at that temperature (Table 2). Phase shifts were computed as the difference in average activity offsets between light-treated populations and untreated controls. Note that per-ΔC2 flies are deficient in advances at all times tested, with the exception of ZT10. Student's t test was used to compare average activity offsets of light-treated populations with untreated controls (table ofp values at right). Similar PRCs for both genotypes were obtained at 25°C.
Table 2.
Temperature compensation in ΔC2 flies
| Genotype (transgene copy #) | Period (hr ± SEM)2-a | % Rhythmic | n2-b | Δτ2-c | |||
|---|---|---|---|---|---|---|---|
| 20°C | 27°C | 20°C | 27°C | 20°C | 27°C | ||
| 1 Canton S | 23.0 ± 0.2 | 23.9 ± 0.1 | 96 | 88 | 24 | 17 | 0.9 ± 0.2 |
| 2 ywper0 per-con (1) | 28.3 ± 0.4 | 29.4 ± 0.2 | 85 | 85 | 13 | 13 | 1.1 ± 0.3 |
| 3 ywper0 per-con (2) | 24.8 ± 0.1 | 26.7 ± 0.1 | 60 | 91 | 87 | 88 | 1.9 ± 0.1 |
| 4 ywper0 ΔC2.2c (1) | 24.7 ± 0.1 | 26.9 ± 0.1 | 78 | 95 | 69 | 75 | 2.2 ± 0.1 |
| 5 ywper0 ΔC2.2c (2) | 22.4 ± 0.1 | 28.0 ± 0.2 | 73 | 92 | 80 | 75 | 5.6 ± 0.2 |
| 6 ywper0 ΔC2.3f (2) | 22.9 ± 0.3 | 27.7 ± 0.2 | 102-e | 59 | 89 | 68 | 4.8 ± 0.3 |
| 7 yw ΔC2 (1) | 22.8 ± 0.5 | 22.8 ± 0.4 | 88 | 88 | 16 | 16 | 0.0 ± 0.4 |
| 8 yw ΔC2 (2) | NA2-d | 22.9 ± 0.2 | 72-e | 50 | 15 | 15 | NA |
The degree of temperature compensation was determined by one of two methods, and the results were combined. In method 1, locomotor activity of each animal was recorded for 1 week each at both 20 and 27°C; only flies surviving both weeks were counted. In method 2, locomotor activity of each animal was recorded for 1 week at 20 or 27°C only.
Number of individuals tested.
Within genotype difference in mean period at 20 and 27°C. The differences in Δτ between genotypes 2 and 4, 3 and 5, and 3 and 6 were compared using Student's t test (p < 0.001 for all).
NA, Not applicable. Only 1 animal of 15 was rhythmic at 20°C.
Two of the per-ΔC2 lines showed significant loss of rhythmicity at 20°C. This may indicate an effect of the mutation on the temperature limits of rhythmicity. However, there was also some loss of rhythmicity at 20°C in theper-con (2) line; thus, the specificity of the effect is questionable.
Generation of constructs. A 159 bp in-frame deletion (per-ΔC2) was made by excising sequences between restriction sites EcoRV (at bp 4572) andXbaI (at bp 4630) of the per gene. AnXba-Xho fragment containing this deletion was cloned in the context of a genomic per construct in CasPer 4 (Chen et al., 1998)
Western analysis. Three to 7-d-old flies were placed in bottles containing a 5% sucrose–2% agar medium, entrained for 3 d at 25°C on a 12 hr light/dark cycle, and collected at the indicated times on dry ice (see Figs. 2, 3). Heads were fractionated on stacked, liquid N2-chilled sieves, and homogenized in lysis buffer (150 mm NaCl, 50 mm Tris-HCl, pH7.6, 10 mmEDTA, 0.1% Triton X-100, 10 mm DTT, and protease inhibitors) with a Kontes motorized pestle. Homogenates were spun two times at 4°C, 15,000 × g for 5 min. Supernatant protein concentrations were assayed using the Bio-Rad (Hercules, CA) DC protein assay. Total protein (50 μg) from each sample were run on a 6% SDS-PAGE gel and transferred to a nitrocellulose membrane. Immunological staining was visualized using the ECL chemiluminescence detection system (Amersham, Arlington Heights, IL) according to the manufacturer's instructions. For detection of the PER and PER-ΔC2 proteins, we used a rabbit anti-PER antibody kindly provided by M. Rosbash (Brandeis University, Waltham, MA) at a dilution of 1:20,000. For detection of TIM, a rat anti-TIM antibody was used at a dilution of 1:500 (Hunter-Ensor et al., 1996).
Fig. 2.
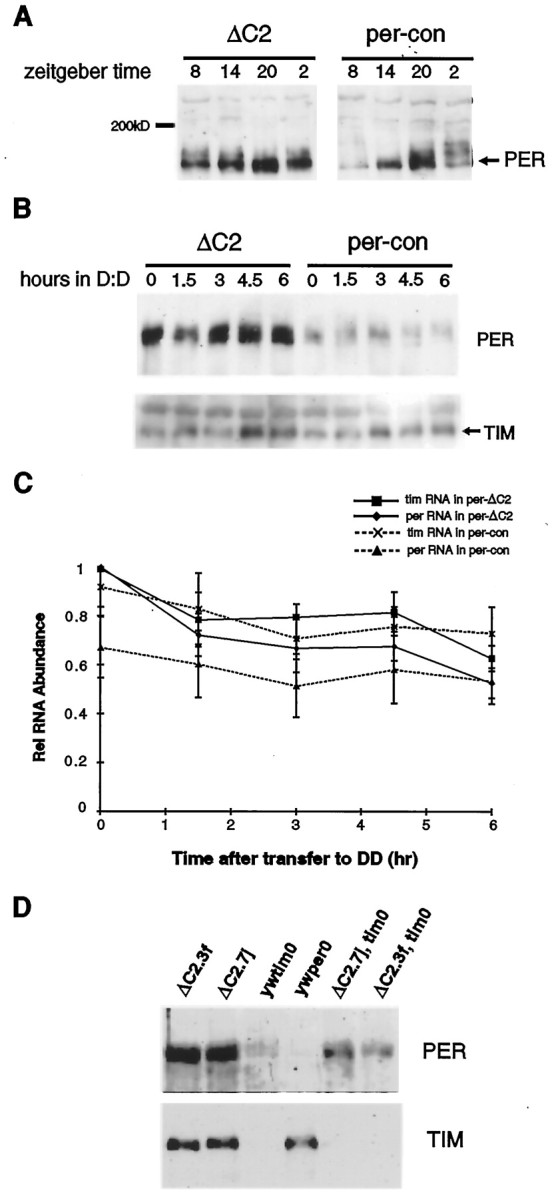
Effect of the ΔC2 mutation on PER expression in LD and LL. A, Western blot of head protein extracts from per-con and per-ΔC2 flies collected at 6 hr intervals during a 12 hr light/dark cycle. The blot was probed with an anti-PER antibody (see Materials and Methods). Thenumbers at the top of thelanes denote ZT. PER-ΔC2 is hypophosphorylated and cycles with reduced amplitude compared with wild-type PER. Samples were run on the same gel to allow comparison of relative mobilities. Note that we do not observe a significant difference in mobility between control and mutant PER because of the high molecular weight of PER compared with that of the deleted region. B, After 3 d in LL, per-con and per-ΔC2flies were transferred to DD. PER expression was assayed before the transfer to DD and at 1.5 hr intervals thereafter. The Western blot shown here indicates that PER-ΔC2 is more abundant than PER in LL (top). Stripping the blot and staining with an anti-TIM antibody revealed that TIM levels are equivalent in the wild-type and mutant backgrounds (bottom). Thus, theΔC2 mutation increases PER stability. Alllanes were equivalently loaded as determined by measuring protein concentrations using the Bio-Rad DC assay and also as reflected by nonspecific bands that cross-react with both antibodies (see top band in A and also thetop band in the TIM blot in B). These bands were also present in per0 andtim0 controls that were loaded on the gels shown in A and B, respectively (data not shown). C, Flies were collected as inB and treated for RNase protection assays.D, Two ΔC2 transgenic lines, ΔC2.7j and ΔC2.3f, were placed in atim0 background, and head extracts were Western blotted and stained with an anti-PER antibody. PER is more abundant in thetim0,ΔC2 lines than in the yw;tim0 line, further supporting an effect of the ΔC2 mutation on PER stability.
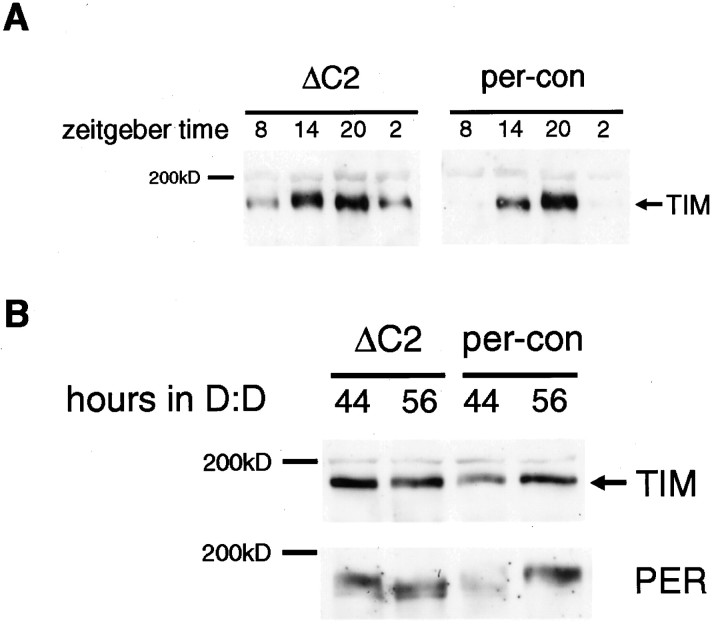
Fig. 3.
Effect of the ΔC2 mutation on TIM expression in LD and DD. A, TIM expression was assayed at the time points indicated in per-ΔC2 andper-con flies. The blot used here was the same as that probed with an anti-PER antibody in Figure 2A. As seen for PER, the amplitude of TIM oscillations is reduced, and it cycles around a higher level. B, per-conand per-ΔC2 flies were entrained to a 12 hr light/dark cycle for 3 d, then transferred to DD, and collected at 44 and 56 hr after transfer. The Western analysis indicates that TIM continues to be expressed at high levels in per-ΔC2 flies in DD. The blot was stripped and probed with an anti-PER antibody (bottom). PER-ΔC2 is high at 44 and 56 hr, consistent with its increased stability, and yet TIM is also expressed at high levels, suggesting a defect in feedback. Equivalent loading was based on the criteria mentioned in the legend to Figure 1.
RNase protection assays. Three to 7-d-old flies were placed in bottles containing a 5% sucrose–2% agar medium, entrained for 3 d at 25°C on a 12 hr light/dark cycle, and collected at the indicated times on dry ice (see Figs. 4, 5). Heads were fractionated on stacked, liquid N2-chilled sieves and homogenized in RNA extraction buffer (50 mm Tris-HCl, pH 9, 1% SDS, 150 mm NaOAc, 5 mmEDTA, and 0.5 vol of Tris-HCl-saturated phenol, pH 9) at 60°C. Extracts were spun at 15,000 × g for 10 min at 4°C, and the supernatants were removed to another tube. The supernatants were phenol–chloroform extracted, then chloroform extracted, and finally ethanol precipitated. RNase protection assays were performed as described by Sehgal et al. (1995). per RNA levels were assayed with a 32P-labeled riboprobe antisense to nucleotides 123–438 of per, and tubulin RNA levels were assayed with a32P-labeled riboprobe antisense to nucleotides 1–142 of d-tubulin. Total head RNA (10 μg) was used per RNase protection assay. Protected bands were quantitated with a phosphorimager (Molecular Dynamics, Sunnyvale, CA).
Fig. 4.
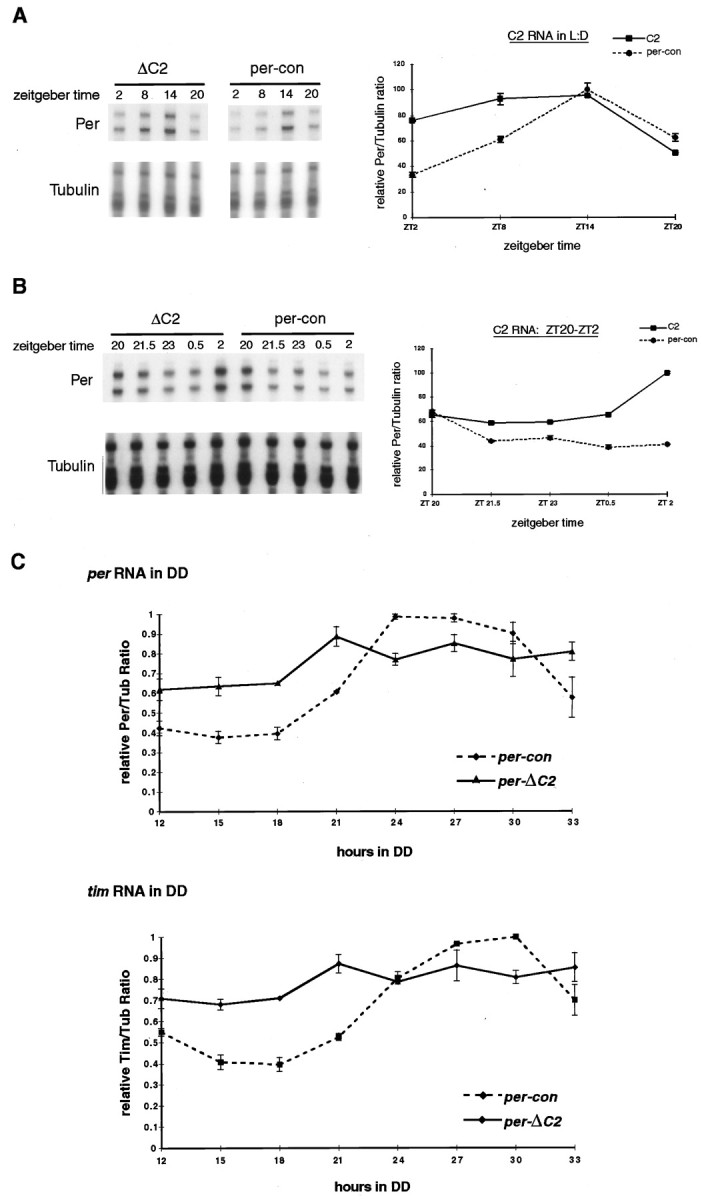
Effect of the ΔC2mutation on per RNA expression. A, RNase protection assay of per RNA from fly heads collected at 6 hr intervals during a 12 hr light/dark cycle. A probe for tubulin RNA was used for normalization of per RNA. Bands were quantitated on a phosphorimager, and per/tubulin ratios were expressed as a percentage of the maximum value (which corresponds to the ZT20 time point in per-con) and graphed (bottom). Note that per-ΔC2 RNA cycles with reduced amplitude compared with per RNA fromper-con flies. B, RNase protection assay of per RNA from fly heads collected at 1.5 hr intervals from ZT20 to ZT2 during a 12 hr light/dark cycle. Procedures and data analysis were identical to those in Figure 4A. Note that per-ΔC2 RNA levels increase rapidly between ZT0.5 and ZT2. C, RNase protection assay ofper and tim RNA under free-running conditions. per-ΔC2 and per-con flies were collected at 3 hr intervals starting 12 hr after transfer to constant darkness. This was selected as the first time point because the first 12 hr of DD correspond to the dark phase of the last LD cycle. Levels of per and tim RNA were assayed as indicated in A and B. All values are expressed relative to the maximum per/tubulin (top) or tim/tubulin (bottom) ratio in per-con flies. Statistical analysis of these data indicate the RNA values cycle. One-way ANOVA performed on per-ΔC2 mRNA values yieldsp = 0.0327 and F = 4.108. Furthermore, post hoc planned comparison analysis shows significant differences between the following time points (in DD): 12 versus 21, 27, and 33 (p < 0.006, 0.012, and 0.03, respectively); 15 versus 21, 27, and 33 (p < 0.008, 0.016, and 0.041, respectively); 18 versus 21 and 27 (p < 0.011 and 0.023, respectively). Additionally, planned comparison analysis shows no significant differences between 21, 24, 27, 30, and 33 in any combination. This indicates a trend toward lower RNA in the night and higher RNA in the subjective day, i.e., the RNA cycles.
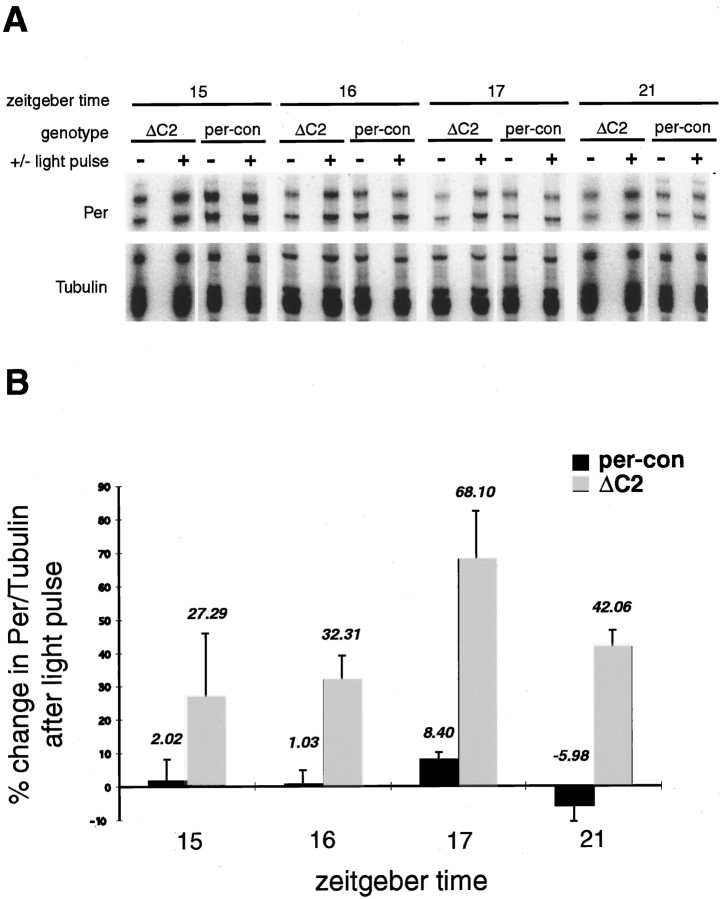
Fig. 5.
Response of per RNA to light in per-ΔC2 flies. A, A composite image of RNase protection assays of per RNA from light-treated flies. per-con andper-ΔC2 flies were treated with light of 2000 lux intensity for 30 min at the zeitgeber times indicated. Pulsed and unpulsed flies were collected 2 hr after the initiation of the light pulse. Probes used were identical to those in Figure 4.B, The gels shown in A were quantitated on a phosphorimager, and the data were graphed as percent induction by light compared with unpulsed controls. As in Figure 4,per RNA was normalized relative to tubulin. Significant induction of per RNA in per-ΔC2 flies was seen at ZT16, ZT17, and ZT21, whereas induction at ZT15 was not reliable, resulting in the large error shown. Error bars represent one SE.
RESULTS
A mutated C domain (ΔC2) affects function of aper transgene
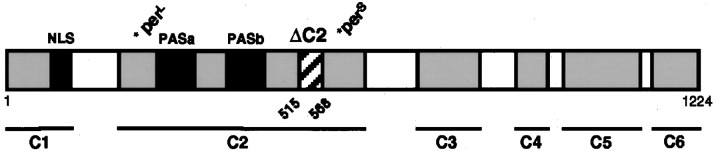
Based on the analysis of different Drosophila species,per was found to contain six conserved regions (C1–C6) (Colot et al., 1988) (Fig. 1). Of these, the C2 region contained the largest block of conserved sequence and was also well conserved in silkmoth and mammalian per homologs (Reppert et al., 1994; Albrecht et al., 1997; Shearman et al., 1997;Sun et al., 1997; Tei et al., 1997; Takumi et al., 1998; Zylka et al., 1998). The C2 region contains the PAS domain, which consists of two repeats of 51 amino acids each: the cytoplasmic localization domain (amino acids 448–512), which retains PER in the cytoplasm in the absence of TIM, and thepers domain, which when mutated produces short period rhythms (Baylies et al., 1992; Saez and Young, 1996). The perl mutation also maps to this region (Baylies et al., 1987; Yu et al., 1987; Hamblen et al., 1998). In addition, a large segment of the C2 region called the C domain (amino acids 524–686) binds the PAS domain through an intramolecular interaction that was proposed to account for temperature-compensation of circadian period (Huang et al., 1995b).
Fig. 1.
Map of PER protein showing the region deleted in this study. The map shows Drosophilid-conserved regions C1–C6, the PAS A and PAS B repeats, and the nuclear localization signal (NLS). The positions of persand perl mutations are also indicated. Transgenes with and without the ΔC2 region (amino acids 515–568) were made in a full-length per genomic construct (Baylies et al., 1992). The constructs included 4 kb of upstream promoter sequence and several hundred base pairs downstream of the transcription stop site.
To address possible functions of the C2 region, we made small deletions in a full-length per genomic construct containing theper promoter (Chen et al., 1998). Resulting constructs were introduced into per01 flies and tested for their ability to rescue behavioral rhythms. As one might predict, based on its requirement for the interaction of PER with TIM, a deletion in the PAS domain failed to rescue rhythms (data not shown). We focused instead on a per transgene carrying a deletion of amino acids 515–568, including 11 serine and threonine residues (per-ΔC2) (Fig. 1). This transgene rescued rhythms at ambient temperature with periods that were shorter than those produced by the full-length control per construct (per-con) (Table 1). Data in Table 1 were obtained from four per-ΔC2 transgenic lines (4a and 4e represent lines that were maintained independently but derived from the injection of a single egg) and one per-conline. However, consistent data from three per-con lines, including the one shown here, have been reported previously (Chen et al., 1998).
Table 1.
Free-running locomotor activity of ΔC2 flies at 25°C
| Genotype (transgene copy#) | na | % Rhythmic | Period (±SEM) | p1-b |
|---|---|---|---|---|
| ywper0per-con (2) | 25 | 70 | 26.7 ± 0.2 | |
| ywper0per-con (1) | 45 | 76 | 27.2 ± 0.2 | |
| ywper0ΔC2.2c (2) | 72 | 85 | 24.3 ± 0.2 | <0.001 [vs per-con (2)] |
| ywper0ΔC2.3f (2) | 75 | 401-c | 25.6 ± 0.2 | <0.001 [vs per-con (2)] |
| ywper0ΔC2.4a (1) | 32 | 281-c | 26.4 ± 0.8 | 0.15 [vs per-con (1)] |
| ywper0ΔC2.4e (1) | 32 | 441-c | 25.2 ± 0.4 | <0.001 [vs per-con (1)] |
| ywper0ΔC2.7j (1) | 32 | 75 | 24.5 ± 0.3 | <0.001 [vsper-con (1)] |
Number of individuals tested.
Comparison of mean period of mutant genotype tested with that of wild-type control using Student's ttest.
Some per-ΔC2 lines (3f, 4a, and 4e) also displayed a low percent rhythmicity. However, the percent rhythmicity in lines ΔC2.2c andΔC2.7j is greater than or equal to that ofper-con. The reason for this variability is not clear, although it could depend on the expression level of the transgene.
per-ΔC2 transgenic flies fail to compensate circadian period with alterations in temperature
Because the C2 domain is included in the larger region implicated previously in temperature compensation (Huang et al., 1995b), we testedper-ΔC2 flies for activity rhythms at different temperatures. As shown in Table 2, these flies displayed an increase in period of as much as 5 hr when moved from 20 to 27°C compared with a 0.9 hr increase in Canton S flies. The effect was especially pronounced (5 hr) with flies that carried two copies of the mutant transgene. For comparison, the period increased by ∼2 hr in control per transgenics carrying two copies of the transgene, whereas there was no significant difference in period inper-con flies carrying one copy of the transgene. The small loss of temperature compensation in two-copy per-con flies may reflect the consequence of increasing doses of the pergene (the endogenous per gene is present although it does not make protein). However, the ΔC2 mutant flies are significantly impaired compared with controls in their ability to maintain period length at different temperatures. In a wild-type background, the per-ΔC2 transgene does not affect temperature compensation, but it does shorten period (Table 2).
PER-ΔC2 is hypophosphorylated and cycles with reduced amplitude
To determine the molecular basis for the behavioral phenotype ofper-ΔC2 flies, we assayed levels of PER protein at different times of day. In per-con flies, levels of PER cycle as they do in wild-type flies, with peak levels reached in the middle–end of the night. Thus, levels are high at ZT20 and low at ZT8 (Fig. 2A). Inper-ΔC2 flies, PER cycles with reduced amplitude, such that trough levels (at ZT8 and even ZT2) are higher than those seen inper-con flies. Consistent with higher trough levels, immunocytochemistry experiments indicated that PER is expressed in nuclei at all times in per-ΔC2 flies as opposed to its cyclic expression in wild-type and per-con flies (data not shown).
We also noticed that, although the PER protein in per-conflies underwent mobility shifts over the course of the day/night cycle, it did not do so in per-ΔC2 flies. These mobility shifts are characteristic of wild-type PER (and TIM) and are the result of cyclic phosphorylation (Edery et al., 1994; Zeng et al., 1996). Alterations of these shifts is routinely taken as a measure of altered phosphorylation (Rutila et al., 1996; Dembinska et al., 1997; Price et al., 1998). In the per-ΔC2 mutant, the maximally phosphorylated forms of PER that appear at the end of the night–beginning of the day are absent (Fig. 2A, compare ZT2 in per-con and per-ΔC2). Given that the deletion removes 11 serines and threonines, this is perhaps not surprising and could account for lower amplitude cycling of PER (see below). We cannot preclude a constant, hyperphosphorylated state (which is also consistent with the absence of gel shifts in PER-ΔC2), but we believe this is unlikely given the increased stability that mimics thedbt phenotype (see below), as well as the fact that multiple serines and threonines were deleted. PER expression data were based on the analysis of two independent lines (per-ΔC2.2cand per-ΔC2.3f), which gave the same results. All subsequent characterization was done in the per-ΔC2.2cline, which contained a high percentage of rhythmic flies (Table2).
The ΔC2 mutation increases the stability of PER
Phosphorylation of PER by the DBT protein renders it unstable in the absence of TIM (Price et al., 1998). Increased stability of the hypophosphorylated PER-ΔC2 protein could accelerate its rate of accumulation and delay its disappearance, thereby producing low-amplitude oscillations. To test the rate of accumulation of PER-ΔC2, we first placed flies under constant light conditions (LL) for 3 d to promote the turnover of TIM and PER (light degrades TIM, and PER is unstable in the absence of TIM). After 3 d of LL, we transferred the flies to DD and assayed PER levels at regular intervals (Fig. 2B).
In per-con flies, levels of PER were extremely low at the end of the constant light treatment (Fig. 2B). This is similar to what is seen in wild-type flies (Price et al., 1995). However, the PER-ΔC2 protein continued to be expressed at high levels (Fig. 2B). To confirm that the light treatment had been effective in decreasing expression of TIM, we stripped and reprobed the blot with an anti-TIM antibody. As expected, TIM levels were equivalent at the light-to-dark transition in bothper-con and per-ΔC2 flies. per andtim mRNA levels were also measured to address the possibility that the PER-ΔC2 protein is higher in constant light because of elevated mRNA levels (Fig.2C). The difference in mRNA levels was not sufficient to explain the elevated PER-ΔC2 protein.
To further test the effect of the ΔC2 mutation on PER stability, we assayed PER-ΔC2 levels in a tim0background. Because all of our per-ΔC2 transgene insertions map to the same chromosome as tim, we used meiotic recombination to place the per-ΔC2 transgene in atim0 background. Head extracts from these lines were Western blotted and stained with an anti-PER antibody (Fig. 2D). Both the ΔC2.7j and ΔC2.3f lines show increased PER abundance in atim0 background compared with theper+,tim0control in which PER levels are low because of the absence of TIM (we were unable to generate a ΔC2.2c,tim0 recombinant line). Thus, the PER-ΔC2 protein is less dependent on TIM for stability.
TIM is expressed at high levels and cycles with reduced amplitude in per-ΔC2 flies
If PER-ΔC2 levels are high and nuclear at all times, one would expect increased feedback repression, resulting in lower levels of RNA and, therefore, lower levels of TIM. To determine whether this was the case, we examined the expression of TIM in per-ΔC2 flies. Figure 3A demonstrates that these flies actually express high levels of TIM. In addition, as seen for PER, TIM cycling is of reduced amplitude with peak levels equivalent to those in wild-type flies but with higher trough levels.
dbtP flies, likeper-ΔC2 flies, express a highly stable form of PER (Price et al., 1998). In these flies, TIM expression oscillates in the presence of light/dark cycles because cyclic feedback is driven, to some extent, by light effects on PER–TIM oscillations. However, in constant darkness, TIM levels are greatly reduced (undetectable by immunocytochemistry) because of constitutive feedback from the stabilized PER (Price et al., 1998). To determine whether high levels of TIM persist in per-ΔC2 flies in the absence of light, we transferred flies to DD and assayed TIM expression at 44 and 56 hr (second day of DD). Under these conditions, TIM levels were still high in per-ΔC2 flies, and the oscillation was further dampened such that levels were equivalent at 44 and 56 hr. The increased dampening is typical for flies kept in constant dark and also occurs inper-con flies, although an oscillation is still evident in these flies (Fig. 3B). The higher levels inper-ΔC2 suggest that expression of tim RNA, and ultimately TIM protein, is not being effectively repressed by the more abundant mutant PER (Fig. 3B).
The ΔC2 mutation decreases the repression ofper RNA
The higher levels of TIM in per-ΔC2 flies could not be explained by increased PER stability because TIM does not depend on PER for stability. In addition, increased stability of the PER-ΔC2 protein was not sufficient to explain the behavioral phenotype ofper-ΔC2 flies. The prediction for a more stable PER would be a longer, not a shorter, period than per-con. If the increased stability occurred only during the rising phase of the protein profile, i.e., in the early night, perhaps a shorter period would be generated. However, this is clearly not the case for this mutant. Its expression persists well into the day and even after several days in constant light (Fig. 2B). As mentioned above, the most likely explanation was that feedback was affected in the per-ΔC2 mutant.
To address the issue of feedback, we studied the temporal expression pattern of per RNA in per-ΔC2 flies and compared it with that of per-con flies using RNase protection assays. In per-con flies, the RNA cycled as expected, peaking at ZT14, falling off at ZT20, and decreasing still further until ZT2 (Fig.4A). Inper-ΔC2 flies, on the other hand, the RNA was low as expected at ZT20, but it rose again after that so it was close to peak levels at ZT2. Throughout this time, the PER-ΔC2 protein is still expressed at high levels (Fig. 2A), suggesting that the protein is ineffective in repressing its own transcription. We examined the phase of feedback repression in these flies more closely by determining RNA levels at 1.5 hr intervals. As shown in Figure4B, the RNA levels are never reduced to the same extent as in wild type, and they start to rise again at ZT0.5, with a dramatic increase seen at ZT2.
We also assayed the temporal profiles of per andtim RNA in per-con and per-ΔC2 flies in constant darkness. Entrained flies were collected every 3 hr on the first day of DD, and head RNA was processed for RNase protection assays in which levels of per and tim RNA were assessed simultaneously. As reported previously , oscillations of both RNAs dampened (Hardin et al., 1990; Sehgal et al., 1995) in both genotypes, although the amplitude of the oscillation was significantly lower inper-ΔC2 flies (Fig. 4C). Consistent with the LD experiments, trough levels in per-ΔC2 flies were higher than those in per-con flies. However, unlike the increase at ZT2 in the LD data, there was no increase of RNA levels at the 15 hr DD time point (which would correspond to CT3 for a 24 hr rhythm) or even at the 18 hr point. Note that, because these flies have a longer period than 24 hr under free-running conditions, we would have expected the ZT2 equivalent rise to occur a little later in DD.
per RNA is induced by light inper-ΔC2 flies
We were intrigued by the relatively large increase inper RNA levels at ZT2 in ΔC2 flies. The DD data argued against a clock-controlled event that coincided with the advent of light, suggesting instead an induction of per RNA by light, reminiscent of the mPer1 and mPer2 RNA induction seen in mammals (Albrecht et al., 1997; Shearman et al., 1997; Shigeyoshi et al., 1997). To test this idea, we treatedper-con and per-ΔC2 flies with bright light (∼2000 lux, 30 min) at ZT15, ZT16, ZT17, and ZT21 and assayedper RNA levels 2 hr after the initiation of the light pulse. At all these times, we observed an induction of per RNA inper-ΔC2 flies with light treatment (Fig.5). The unpulsed controls in Figure 5were collected at the same time as the light-treated populations. This was done to exclude any clock-regulated changes within the 2 hr period.
The highest level of induction was seen at ZT17, with slightly less at ZT16 and ZT21 (note that in each case the samples are being examined 2 hr later, i.e., at ZT18 and ZT23). As indicated by the size of the error bars in the ZT15 sample, the data at this time point were quite variable. This temporal pattern of RNA induction correlates with the phase of feedback repression, suggesting that light is perhaps relieving transcriptional inhibition (further discussed in Discussion). The variability at ZT15, therefore, may reflect the fact that it is on the cusp between active transcription and repression. Small changes from one experiment to another, such as the time of collection, or even some period differences between individual flies could account for this variability. Experiments at ZT21, which actually reflect a collection at ZT23, lie at the end of the repression phase in per-ΔC2flies and also shows less RNA induction. Finally, we examined the response of tim mRNA to light at ZT16, ZT17, and ZT21 and found that it was induced in a parallel manner, which further supports the hypothesis that light removes a transcriptional repressor (data not shown).
per-ΔC2 alters the PRC of behavioral rhythms
As discussed earlier, several models have been proposed to account for the differential behavioral response to light at different times of the night. Because the ΔC2 mutation affected some of the parameters that were thought to be critical for resetting (phosphorylation and RNA levels), we reasoned that this mutation could be informative in addressing current resetting models. Thus, we generated a PRC for this line using 10 min pulses of light and again compared it with that of per-con flies. The resetting assays were performed as described previously (Yang et al., 1998), except that they were conducted at 27°C. This was done because per-conand per-ΔC2 flies have similar periods at this temperature (Table 2), thus reducing period-influenced changes in the PRC.
As shown in Figure 6, per-conflies displayed a PRC that one would predict for flies with 27 hr periods. In wild-type flies, relatively little resetting occurs in response to pulses during subjective day, delays occur in the first half of the night, and advances occur in the second half. Inper-con flies, consistent with their longer period, the switch from delay to advance occurs later in the night and advances occur up to CT2. Thus, delays are observed at ZT15, ZT17, and ZT20 and advances at ZT22 and CT2. per-ΔC2 flies display a very different PRC despite the similarity of their periods. Delays do not occur until ZT17 (the delay at ZT15 is statistically insignificant), and advances do not occur at all at the times predicted by theper-con PRC (ZT22 and CT2). In fact, a pulse at ZT22 elicits a large delay in per-ΔC2 flies but produces the maximal advance in per-con flies. Advances of very low magnitude are observed in per-ΔC2 flies during the day, although only one of these (ZT10) was statistically significant.
DISCUSSION
Although per has been the subject of intense research for ∼14 years, major aspects of its regulation and function remain unresolved. Available data indicate that cyclic expression of PER can be achieved in the absence of per RNA cycling (Frisch et al., 1994; Cheng and Hardin, 1998), but RNA cycling requires the protein, suggesting that the feedback loop is driven primarily by the cycling of the protein (Hardin et al., 1990; Sehgal et al., 1995). This makes it all the more important to address the mechanisms that control protein turnover and the effects of protein on transcription. Traditional methods to dissect the regulation of a protein involve structure–function studies, which allow specific functions to be assigned to distinct domains. In the case of PER, these studies have been limited. We show here that a deletion within the conserved C2 region in PER does not prevent rescue by a pertransgene but affects the properties of the behavioral rhythm. Behavioral phenotypes of the per-ΔC2 flies have their basis in specific underlying molecular defects that we discuss below.
We believe that the direct effects of the ΔC2 mutation are twofold: (1) to increase PER stability and (2) to decrease its ability to effectively feedback repress transcription. Thus, the effect on RNA cycling is probably secondary and caused by inadequate feedback by the protein. A direct effect on RNA stability is unlikely given that the deletion is in coding regions and has a similar effect on the cycling of tim transcripts as it does on the transgenic RNA (Fig.4C). Consistent with this idea, the effect on RNA cycling is to reduce amplitude via an increase in trough levels, i.e., decrease repression during the trough. An effect on feedback is also supported by the fact that the protein itself is expressed at high levels and is constitutively nuclear. Normal negative feedback should lead to a reduction of RNA expression and, ultimately, protein expression, making this phenotype (of high RNA and high protein levels) highly unlikely.
The question then is the following: what causes the increased stability and decreased feedback associated with the protein? The increased protein stability is almost certainly attributable to the lack of phosphorylation events. A hypophosphorylated, more stable protein is also produced by a truncated form of per fused to β-galactosidase (Dembinska et al., 1997). The latter protein retains the region deleted in this study, indicating that more than one region of PER can confer this phenotype. Either one or both regions may be targets of the DBT kinase, which phosphorylates and destabilizes PER monomers. If the DBT sites are also responsible for the reduced feedback in per-ΔC2 flies, then one would predict that DBT mutants themselves must be defective in feedback. This does not appear to be the case because, although PER does not cycle in larvae of strongly hypomorphic dbt mutants, tim RNA and protein do so in a robust manner (although absolute levels could not be quantitated) in the presence of LD cycles (Price et al., 1998). Moreover, expression of tim gene products in dbtmutants is greatly reduced in DD compared with wild type, which is inconsistent with reduced feedback. In contrast, tim RNA, as well as TIM, continue to be expressed at high levels in DD inper-ΔC2 flies (Figs. 3B, 4C). We propose that the effect of the PER-ΔC2 protein on feedback is caused by an additional defect, such as in DBT-independent phosphorylation sites or in the region per se.
The current model for how PER and TIM feedback to inhibit their own transcription is that they bind transcriptional activators dCLK and CYC, both of which contain PAS domains, as does per (Allada et al., 1998; Darlington et al., 1998; Lee et al., 1998; Rutila et al., 1998). Although homotypic interactions between PAS-containing proteins are common, the PAS domain can also engage in heterotypic interactions. For instance, the PAS A repeat in PER binds a part of TIM through a heterotypic association (Gekakis et al., 1995). In addition, a larger region encompassing the ΔC2 sequence was shown previously to interact with the PAS domain (Huang et al., 1995a). Although it was thought to engage in an intramolecular interaction, association with a different molecule, possibly a different protein, cannot be excluded. Thus, this domain may be required for the interaction of PER with a PAS-containing transcription factor.
The defect in feedback can explain the short periodicity displayed byper-ΔC2 flies. As discussed earlier, increased stability of the protein, in particular during the falling phase after lights are turned on, is inconsistent with a shorter period. In fact, thedbtL allele, which reduces PER phosphorylation and increases PER stability, produces a long period (Price et al., 1998). Moreover, thepers mutant, which accelerates decay of the protein in a day/night cycle (Marrus et al., 1996), shortens circadian period. Thus, feedback is truncated and the RNA levels rise earlier. In case of the per-ΔC2 flies, the protein remains but is impaired in feedback inhibition. Note that the phenotype of these flies ranges from short periods to arrhythmia. Reduction of feedback below a certain threshold level could result in arrhythmia. Relevant to this issue is the phenotype of flies kept in altered light/dark cycles. When the length of the night is reduced to <8 hr, flies are arrhythmic (Qiu and Hardin, 1996). Thus, a minimum night length is critical to maintain circadian cycles, perhaps by ensuring adequate feedback. Note that the defect in feedback inper-ΔC2 flies also elevates levels of tim RNA and protein (Fig. 3A), thereby leading to accelerated accumulation of PERΔC2–TIM dimers and contributing to a shorter circadian cycle.
The response of the per-ΔC2 flies to pulses of light is intriguing. Induction of per and tim RNA by light has never been seen in wild-type flies, although it is a characteristic of some of the mammalian per homologs (Albrecht et al., 1997; Shearman et al., 1997; Shigeyoshi et al., 1997). In case of theΔC2 mutant, it is most likely because of the removal of transcriptional repression, supported by the fact that best induction is observed at times of maximal feedback. Because PER appears to be defective in feedback inhibition in per-ΔC2 flies, we believe that repression is mediated primarily by the light-sensitive TIM protein. We examined the response of PER and TIM proteins in response to light at ZT21, using the same pulse that induced the two RNAs, and found, as expected, that TIM levels were reduced, but PER levels were constant (data not shown). The apparent increased dependence on TIM in per-ΔC2 flies may indicate that TIM compensates for the defect in PER or it may reflect the normal role of TIM, which is usually masked by PER, i.e., in wild-type flies PER continues to repress after TIM is degraded by light. In either event, these data suggest that TIM can participate in feedback repression. Note that TIM can bind the PAS domain of PER (Gekakis et al., 1995) and was shown recently to interact with dCLK in a per null background (Lee et al., 1998). The hypothesis that TIM can mediate feedback is also consistent with the report of tim mRNA and protein cycling in dbtP flies in LD but not DD (Price et al., 1998). Light-driven cycling of timgene products in dbtP flies indicates that light-dependent TIM degradation can drive rhythmic feedback, even when PER does not cycle.
The behavioral response to resetting pulses of light is also altered. A model that incorporates increased stability of PER-ΔC2, together with reduced feedback, can account for these resetting defects (Fig.7). We propose that the lack of advances is attributable to the high RNA levels in per-ΔC2 flies, which are further increased in response to light. As proposed (Myers et al., 1996), these could replace the TIM protein lost by light and thereby delay the cycle instead of advancing it to the following dawn (Fig. 7). The reduced delays can be explained on the basis of the RNA induction, as well as by the generally elevated levels oftim RNA, both of which allow rapid resynthesis of TIM. The large pool of stable PER monomers may also contribute by allowing PER–TIM heterodimers to accumulate rapidly after a light pulse degrades TIM. Thus, the time required to get back to the same point of the cycle is shortened.
Fig. 7.
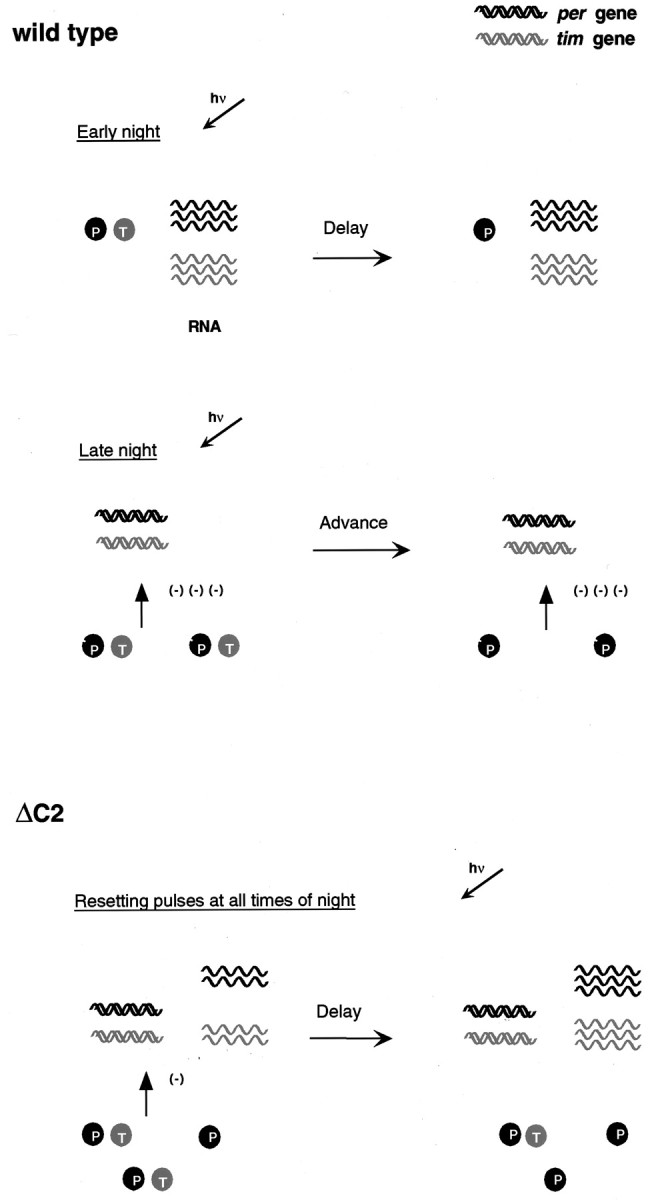
Model for the resetting response in wild-type and per-ΔC2 flies. In wild-type flies, delays occur in response to light in the early part of the night when both genes are being actively transcribed and RNA levels are high. In the second half of the night, highly phosphorylated forms of PER–TIM repress transcription and RNA levels are low. Advances are produced at this time. In per-ΔC2 flies, delays are produced at almost all times, thus only a single situation is shown. These delays correlate with reduced feedback and relatively high levels of RNA.
Although the effect of the PER-ΔC2 protein on temperature compensation is interesting, particularly given that the deleted sequence falls in the C domain, a region implicated previously in temperature compensation (Huang et al., 1995b), it may be premature to conclude that this specific sequence is important for the compensatory property of clocks. In fact, the data presented here run contrary to the model of Huang et al., which would predict an increase in intermolecular over intramolecular PER-ΔC2 interactions and hence a shortening of period with increasing temperature. Other mutations ofper also affect temperature compensation (Huang et al., 1995a; Sawyer et al., 1997; Hamblen et al., 1998), and at the present time it is difficult to reconcile all these data with a single molecular model.
Footnotes
This work was supported by National Institutes of Health Grant NS35703 and American Cancer Society Grant DB-140. A.S. is an assistant investigator of the Howard Hughes Medical Institute. We thank Michael Young for useful comments and suggestions, Michael Rosbash for the anti-PER antibody, Jeffrey Field for critical comments on this manuscript, and other members of the laboratory for helpful discussions.
Correspondence should be addressed Amita Sehgal at the above address. E-mail: amita@mail.med.upenn.edu.
Dr. Hunter-Ensor's present address: Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139.
REFERENCES
- 1.Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mPer1 and mPer2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 2.Allada R, White NE, Venus So W, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 3.Aschoff J. Response curves in circadian periodicity. North Holland; Amsterdam: 1965. [Google Scholar]
- 4.Bae K, Lee C, Sidote D, Chuang KY, Edery I. Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol Cell Biol. 1998;18:6142–6151. doi: 10.1128/mcb.18.10.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baylies MK, Bargielli TA, Jackson FR, Young MW. Changes in abundance or structure of the per gene product can alter periodicity of the Drosophila clock. Nature. 1987;326:390–392. doi: 10.1038/326390a0. [DOI] [PubMed] [Google Scholar]
- 6.Baylies MK, Vosshall LB, Sehgal A, Young MW. New short period mutations of the Drosophila clock gene per. Neuron. 1992;9:575–581. doi: 10.1016/0896-6273(92)90194-i. [DOI] [PubMed] [Google Scholar]
- 7.Cheng Y, Hardin PE. Drosophila photoreceptors contain an autonomous circadian oscillator that can function without period mRNA cycling. J Neurosci. 1998;18:741–750. doi: 10.1523/JNEUROSCI.18-02-00741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Hunter-Ensor M, Schotland P, Sehgal A. Alterations of per RNA in noncoding regions affect periodicity of circadian behavioral rhythms. J Biol Rhythms. 1998;13:364–379. doi: 10.1177/074873098129000192. [DOI] [PubMed] [Google Scholar]
- 9.Colot HV, Hall JC, Rosbash M. Interspecific comparison of the period gene of Drosophila reveals large blocks on non-conserved coding DNA. EMBO J. 1988;7:3929–3937. doi: 10.1002/j.1460-2075.1988.tb03279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darlington TK, Wagner-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TDL, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: clock-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 11.Dembinska ME, Stanewsky R, Hall JC, Rosbash M. Circadian cycling of a period-lacZ fusion protein in Drosophila: evidence for cyclic degradation. J Biol Rhythms. 1997;12:157–172. doi: 10.1177/074873049701200207. [DOI] [PubMed] [Google Scholar]
- 12.Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frisch B, Hardin PE, Hamblen CM, Rosbash M, Hall JC. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 14.Gekakis N, Saez L, Delahaye BA, Myers MP, Sehgal A, Young MW, Weitz CJ. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 15.Hamblen MJ, White NE, Emery PTJ, Kaiser K, Hall JC. Molecular and behavioral analysis of four period mutants in Drosophila melanogaster encompassing extreme short, novel long, and unorthodox arrhythmic types. Genetics. 1998;149:165–178. doi: 10.1093/genetics/149.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 17.Huang ZJ, Curtin KD, Rosbash M. PER protein interactions and temperature compensation of a circadian clock in Drosophila. Science. 1995a;267:1169–1172. doi: 10.1126/science.7855598. [DOI] [PubMed] [Google Scholar]
- 18.Huang Y, Baker RT, Fischer-Vize JA. Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science. 1995b;270:1828–1831. doi: 10.1126/science.270.5243.1828. [DOI] [PubMed] [Google Scholar]
- 19.Hunter-Ensor M, Ousley A, Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84:677–686. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- 20.Kloss B, Price JP, Saez L, Blau J, Rothenflugh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 21.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konopka RJ, Hamblen-Coyle MJ, Jamison CF, Hall JC. An ultrashort clock mutation at the period locus of Drosophila melanogaster that reveals some new features of the fly's circadian system. J Biol Rhythms. 1994;9:189–216. doi: 10.1177/074873049400900303. [DOI] [PubMed] [Google Scholar]
- 23.Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- 24.Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- 25.Marrus SB, Zeng H, Rosbash M. Effect of constant light and circadian entrainment of perS flies: evidence for light-mediated delay of the negative feedback loop in Drosophila. EMBO J. 1996;15:6877–6886. [PMC free article] [PubMed] [Google Scholar]
- 26.Mistlberger RE, Marchant EG, Sinclair SV. Nonphotic phase-shifting and the motivation to run: cold exposure reexamined. J Biol Rhythms. 1996;11:208–215. doi: 10.1177/074873049601100303. [DOI] [PubMed] [Google Scholar]
- 27.Myers MP, Wager-Smith K, Rothenflugh A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 28.Naidoo N, Song W, Hunter-Ensor M, Sehgal A. A role for the proteasome in the light response of the timeless clock protein. Science. 1999;285:1737–1741. doi: 10.1126/science.285.5434.1737. [DOI] [PubMed] [Google Scholar]
- 29.Ousley A, Zafarullah K, Chen Y, Emerson M, Hickman L, Sehgal A. Conserved regions of the timeless (tim) clock gene in Drosophila analyzed through phylogenetic and functional studies. Genetics. 1998;148:815–825. doi: 10.1093/genetics/148.2.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price JL, Dembinska ME, Young MW, Rosbash M. Suppression of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless. EMBO J. 1995;14:4044–4049. doi: 10.1002/j.1460-2075.1995.tb00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price JL, Blau J, Rothenflugh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 32.Qiu J, Hardin PE. per mRNA cycling is locked to lights-off under photoperiodic conditions that support circadian feedback loop function. Mol Cell Biol. 1996;16:4182–4188. doi: 10.1128/mcb.16.8.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reppert SM, Tsai T, Roca AL, Sauman I. Cloning of a structural and functional homolog of the circadian clock gene period from the giant silkmoth Antheraea pernyi. Neuron. 1994;13:1167–1176. doi: 10.1016/0896-6273(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 34.Rutila JE, Zeng H, Le M, Curtin KD, Hall JC, Rosbash M. The timSL mutant of the Drosophila rhythm gene timeless manifests allele-specific interactions with period gene mutants. Neuron. 1996;17:921–929. doi: 10.1016/s0896-6273(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 35.Rutila JE, Suri V, Le M, Venus So W, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 36.Saez L, Young MW. Regulation of nuclear entry of the Drosophila clock proteins Period and Timeless. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 37.Sawyer LA, Hennesy JM, Peixoto AA, Rosato E, Parkinson H, Costa R, Kyriacou CP. Natural variation in a Drosophila clock gene and temperature compensation. Science. 1997;278:2117–2120. doi: 10.1126/science.278.5346.2117. [DOI] [PubMed] [Google Scholar]
- 38.Sehgal A, Price JL, Man B, Young MW. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- 39.Sehgal A, Rothenfluh-Hilfiker A, Hunter-Ensor M, Chen Y, Myers MP, Young MW. Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science. 1995;270:808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- 40.Sehgal A, Ousley A, Hunter-Ensor M. Control of circadian rhythms by a two-component clock. Mol Cell Neurosci. 1996;7:165–172. doi: 10.1006/mcne.1996.0013. [DOI] [PubMed] [Google Scholar]
- 41.Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF, Reppert SM. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 42.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 43.Sun ZS, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee CC. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 44.Suri V, Zuwei Q, Hall JC, Rosbash M. Evidence that the TIM light response is relevant to light-induced phase shifts in Drosophila melanogaster. Neuron. 1998;21:225–234. doi: 10.1016/s0896-6273(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 45.Takumi T, Taguchi K, Miayake S, Sakakida Y, Takashima N, Matsubara C, Maebayashi Y, Okumura K, Takekida S, Yamamoto S, Yagita K, Yan L, Young MW, Okamura H. A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 1998;17:4753–4759. doi: 10.1093/emboj/17.16.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 47.Yang Z, Emerson M, Su HS, Sehgal A. Response of the timeless protein to light correlates with behavioral entrainment and suggests a non-visual pathway for circadian photoreception. Neuron. 1998;21:215–223. doi: 10.1016/s0896-6273(00)80528-0. [DOI] [PubMed] [Google Scholar]
- 48.Yu Q, Jacquier AC, Citri Y, Hamblen M, Hall JC, Rosbash M. Molecular mapping of point mutations in the period gene that stop or speed up clocks in Drosophila melanogaster. Proc Natl Acad Sci USA. 1987;84:784–788. doi: 10.1073/pnas.84.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng H, Qian Z, Myers MP, Rosbash M. A light entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 50.Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian rhythms of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]