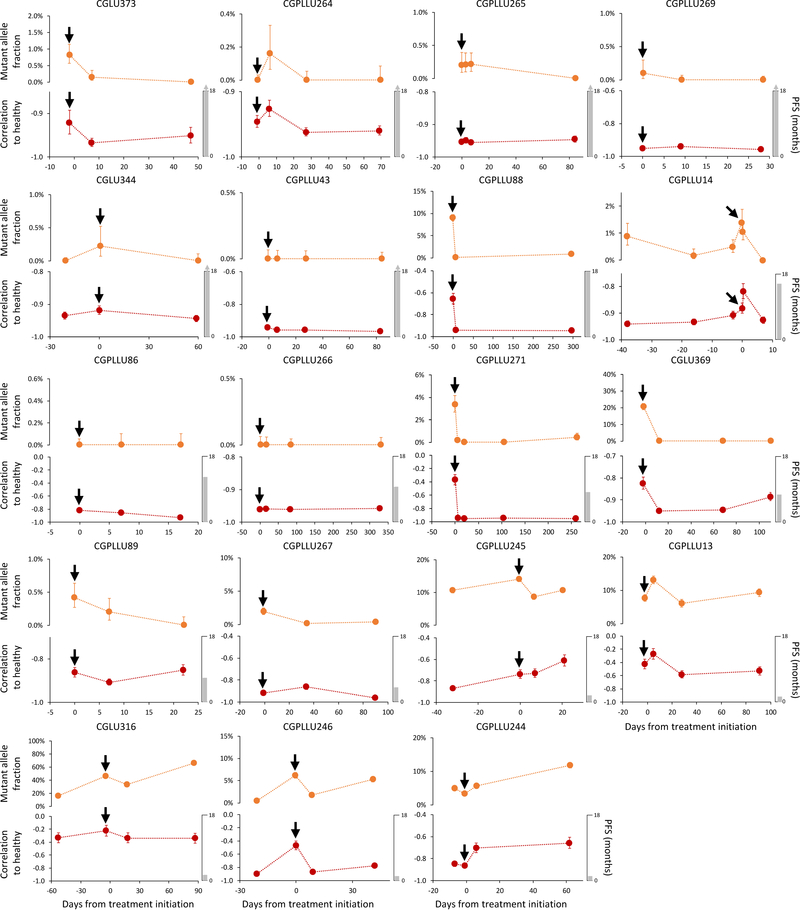
Extended Data Fig. 4. cfDNA fragmentation profiles and sequence alterations during therapy.
Detection and monitoring of cancer in serial blood draws from NSCLC patients (n=19) undergoing treatment with targeted tyrosine kinase inhibitors (black arrows) was performed using targeted sequencing (top) as previously reported29 and genome-wide fragmentation profiles (bottom). For each case, the vertical axis of the lower panel displays −1 times the Pearson correlation of each sample to the median healthy cfDNA fragmentation profile. Error bars depict confidence intervals from binomial tests for mutant allele fractions and confidence intervals calculated using Fisher transformation for genome-wide fragmentation profiles. Although the approaches analyze different aspects of cfDNA (whole genome compared to specific alterations) the targeted sequencing and fragmentation profiles were similar for patients responding to therapy as well as those with stable or progressive disease. As fragmentation profiles reflect both genomic and epigenomic alterations, while mutant allele fractions only reflect individual mutations, mutant allele fractions alone may not reflect the absolute level of correlation of fragmentation profiles to healthy individuals.