Abstract
Background
Interferon alpha (IFNalpha) exerts its anti-proliferative effect on many human cancers. Among the 13 subtypes of human IFNalpha, IFNalpha-1 subtype has 2 variants, named IFNalpha-1a and IFNalpha-1b, that differ from each other in only 1 amino acid, at residue 114. However, the mechanism by which IFNalpha-1a mediates growth inhibition is still unclear.
Material/Methods
Human laryngeal carcinoma HEp2 cells were treated with IFNalpha-1a by either transient transfection or exogenous delivery. Western blot and RT-PCR analysis were carried out to assess apoptotic pathways active in IFNalpha-1a-treated HEp2 cells. Microarray analysis was conducted to uncover the differential gene expressions after IFNalpha-1a treatment. KEGG pathway enrichment analysis was also performed.
Results
IFNalpha-1a markedly inhibited the proliferation and significantly promoted the apoptosis of HEp-2 cells. Mechanistic studies indicate that IFNalpha-1a-mediated cell apoptosis is directly linked to intrinsic and endoplasmic reticulum (ER) stress-related apoptosis, but is independent of extrinsic apoptosis. The top 40 differentially expressed genes discovered by microarray analysis included 20 upregulated genes (e.g., IFI6, IFI27, IFI44L, and MIR548X) and 20 downregulated genes (e.g., PRKDC, HIST1H3B, DYNC1H1, and HIST1H2AM). KEGG pathway enrichment analysis revealed that 4 out of 6 pathways are TP53-related.
Conclusions
We demonstrated a detailed mechanism involved in IFNalpha-1a-mediated anti-proliferation activity in human laryngeal carcinoma cells.
MeSH Keywords: Apoptosis, Interferon-alpha, Laryngeal Neoplasms
Background
Laryngeal carcinoma, of which the majority is laryngeal squamous cell carcinoma, is the second most common cancer of the human head and neck worldwide. About 60% of patients with this disease were in advanced stage upon diagnosis, with 5-year survival around 65% [1]. Tobacco use and alcohol consumption are the 2 major risk factors linearly associated with the occurrence of laryngeal carcinoma [2,3]. Accumulating evidence shows that the larynx is one of the sites most vulnerable to infection by human papillomavirus (HPV) [4,5]. Therefore, HPV infection could be an emerging risk factor for the development of laryngeal carcinoma [6,7]. However, Xu et al. recently showed that HPV infection in Asian patients did not account for the occurrence of laryngeal carcinoma, indicating that patients’ racial disparities could be another determinant of HPV-mediated laryngeal carcinoma [8]. Traditionally, surgical therapy plus radiotherapy are the main treatments for patients with laryngeal carcinoma [9]. Potential voice loss and extensive rehabilitation after oncologic larynx surgery may be detrimental to a patient’s quality of life. Thus, novel therapeutic approach to effectively restrict the neoplasm growth are urgently needed by patients with laryngeal carcinoma, especially for those with earlier diagnosis.
Interferon (IFN) is a cytokine with potent antiviral, anti-proliferative, and immuno-modulatory capacity [10]. The human type I IFN includes 17 members: 13 alpha subtypes, as well as 1β, 1ɛ, 1κ, and 1ω, which are all located on human chromosome 9 [11–13]. All these IFNalpha subtypes recognize the same receptor molecule, the type I IFN receptor (IFNAR), indicating that the distinct function of each subtype is retained during evolution. The binding of the IFN molecule to its specific receptor activates the intracellular signaling pathway, especially the JAK/STAT pathway, which can drive hundreds of downstream interferon response gene (ISG) expressions. It has been shown that the anti-proliferative effect of IFNalpha could result in cell apoptosis. However, the mechanism is still not well defined. Some studies have shown that a number of the ISGs can promote cell apoptosis by upregulating the expressions of the key pro-apoptotic genes [14–16]. However, IFNalpha-mediated cell apoptosis is not observed in every tissue examined, indicating that IFNalpha-mediated cell apoptosis may function in a tissue-specific manner.
There are 2 well-known pathways responsible for cell apoptosis: the intrinsic and extrinsic cell death pathways. The intrinsic mitochondrial pathway is controlled by the Bcl-2 family proteins, with pro-apoptotic and anti-apoptotic properties. A ‘point of no return’ occurs when an arrived death signal enhances the permeabilization of the mitochondrial outer membrane, which in turn promotes the release of cytochrome c into the cytosol. Activation of the extrinsic apoptotic pathway is marked by cleavage of caspase 8 and caspase 10 in the cytosol. The intrinsic and the extrinsic cell death pathways converge on the activation of the final executioners, caspase 3 and 7. Subsequently, the activated caspase 3 inactivates the highly reliable apoptotic biomarker, PARP1, by cleaving it between Asp214 and Gly215 to further promote apoptosis. In addition, endoplasmic reticulum (ER) stress is a normal cellular response to unfolded proteins by different exogenous and endogenous stimulators. However, prolonged ER stress can induce cell apoptosis due to the accumulations of unfolded or mis-folded proteins. The hallmark of ER stress-mediated apoptosis is the activation of caspase 12 in mice or the activation of caspase 4 in humans, which can subsequently induce the cleavage of caspase 3/7 [17,18].
In this study, we first demonstrate that the anti-proliferative effect of IFNalpha-1a on human laryngeal carcinoma HEp-2 cells is directly linked to the IFNalpha-1a-mediated cell apoptosis. Our mechanism study demonstrates that IFNalpha-1a markedly inhibits cell proliferation by activating the intrinsic and ER stress-mediated apoptotic pathways. In addition, microarray and pathway analysis revealed the top differential genes and the major pathways that respond to IFNalpha-1a treatment. Our study reveals a previously undefined role for IFNalpha-1a in promoting apoptosis of laryngeal carcinoma cells.
Material and Methods
Antibodies and reagents
Anti-HA antibody was from Huaxingbio (Beijing, China); anti-caspase3 antibody was from Cell Signaling Technology (Danvers, MA, USA); anti-caspase 8, anti-caspase 4, anti-cleaved-caspase 3, anti-Bcl-XL, anti-cleaved caspase 8, anti-cytochrome C, anti-CHOP, anti-PARP1, and anti-GRP78 antibodies were all purchased from Proteintech Group, Inc. (Rosemont, IL, USA); anti-caspase 10 antibody was from Bioworld Technology, Inc. (St. Louis Park, MN, USA); anti-β-actin antibody was from Transgen Biotechnology (Beijing, China); anti-GAPDH antibody was from Affinity Biosciences (Cincinnati, OH, USA). Human recombinant IFNalpha-1a was purchased from ProSpec-Tany TechnoGene (Ness Ziona, Israel). Cell counting kit-8 was from Dojindo (Kumamoto, Japan).
The isolation of IFNalpha-1a cDNA and the construction of IFNalpha-1a expression plasmid
The full length of IFNalpha-1a coding sequence was isolated from the total RNAs of human embryonic brain tissue using the forward primer 5′-TTAAGCTTATGGCC TCGCCCTTTGCTTTA-3′ and the reverse primer 5′-TTGAATTCCTAAGCGTAGTC TGGGACGTCGTATGGGTATTCCTTCCTCCTTAATCTTT-3′. After reverse transcription, the reaction product was amplified by polymerase chain reaction (PCR) as follows: denaturation at 94°C for 5 min, then amplification at 94°C/30 s, 55°C/30 s, and 72°C/2 min for 35 cycles. The reaction product was finally extended at 72°C for 10 min. The PCR products were separated and extracted from agarose gel. The IFNalpha-1a cDNA that carries the coding sequence of influenza hemagglutinin (HA) epitope tag was first subcloned into pMD18T-Simple vector to form pMD18T-Simple-IFNalpha-1a-HA construct. After confirmation by sequencing analysis, IFNalpha-1a-HA was released from pMD18T-Simple-IFNalpha-1a-HA by EcoRI/HindIII double-digestion and subsequently subcloned into the EcoRI/HindIII sites of pcDNA3.0 to form pcDNA3.0-IFNalpha-1a-HA construct.
Cell culture, transient transfection, and cell treatment
The human laryngeal squamous cancer cell line HEp-2 cells, human embryonic kidney HEK293T cells, human hepatocellular carcinoma HepG2 cells, and human lung carcinoma A549 cells were obtained from the Cell Culture Center of the Basic Institute of Medical Sciences, Chinese Academy of Medical Sciences (Beijing, China). Hep-2 cells were grown in DMEM (Gibco) supplemented with 10% fetal bovine serum at 37°C with 5% CO2. At about 80% confluency, Hep-2 cells were transiently transfected with increasing doses of plasmid pcDNA3.0-IFNalpha-1a-HA DNAs by NeoFect (Neofect biotech Co, Beijing, China) according to the instruction manual. Hep-2 cells were also treated with increasing doses (0, 50, or 200 ng/mL) of IFNalpha-1a for 48 h before harvesting.
Cell proliferation assays
Cell proliferation potentials were evaluated by 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and cell counting kit-8 (CCK-8) tests. The HEp-2 cells grown on a 12-well culture plate were either transiently transfected with pcDNA3.0-IFNalpha-1a plasmid DNAs or treated with IFNalpha-1a for 48 h. For CCK-8 assay, the transfected HEp2 cells were treated with CCK8 reagent for 2 h before detection using a microplate reader at 490 nm. Cell proliferation potentials were assessed by CCK-8 based on the standard curve of optical density and cell number. For MTT assay, the transfected Hep-2 cells seeded onto 96-well microtiter plates were treated with IFNalpha-1a before testing. After addition of MTT reagent, HEp-2 cells continued to culture for 2 h. Then, the culture medium was discarded and the purple crystal formazan was dissolved by DMSO. Finally, the optical density was detected using a microplate reader at 450 nm.
Apoptosis assay
Cell apoptosis was detected using an Annexin V-FITC apoptosis detection kit (Biotool, Beijing, China). Briefly, the transiently transfected or IFNalpha-1a-treated HEp-2 cells were harvested after 48-h incubation. The cell pellet was resuspended with 100 μL of 1×binding buffer, followed by adding 5 μL Annexin V-FITC and 5 μL propidium iodide (PI). The reaction mixture was incubated away from light for 15 min at room temperature. Then, another 400 μL 1X binding buffer was added into the mixture. The reaction product was subjected to flow cytometric analysis using an Accuri C6 device (BD Biosciences, San Jose, CA, USA).
Western blot analysis
The transiently transfected or IFNalpha-1a-treated HEp-2 cells were lysed with ice-cold lysis buffer (1% NP-40, 50 mM Tris-HCl (pH 7.5), 120 mM NaCl, 200 μM NaVO4, 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 1 μM PMSF) for 30 min at 4°C. About 10 μg of cell lysates was separated on 12% SDS-PAGE. The resolved proteins were transferred to nitrocellulose membranes (Amersham Biosciences, Freiburg, Germany). The membrane was incubated with a primary antibody followed by a horseradish peroxidase-conjugated secondary antibody. The protein bands were developed with Novex ECL Western Blotting substrate (Invitrogen, Thermo Fisher Scientific, USA). The Chemiluminescence image was captured om a Tanon Luminescent imaging workstation (YPH-Bio, Beijing, China).
Microarray analysis
Total RNA was extracted from HEp2 cells using Trizol reagent before being assessed in the microarray experiment. Affymetrix GeneChip® Human Transcriptome 2.0 Arrays analysis was conducted using the GeneChip® WT Plus Reagent Kit according to the manufacturer’s instructions (GeneChip® WT PLUS Reagent Kit Manual Target Preparation for GeneChip® Whole Transcript (WT) Expression Arrays P/N 703174 Rev. 2, 2013). Briefly, the first-strand cDNA was synthetized from 500 ng of RNA using Superscript II reverse transcriptase primed with a poly(T) oligomer that was driven by T7 promoter. The second-strand cDNA synthesis was followed by in vitro transcription amplification. The obtained cRNA was used as a template for a second cDNA synthesis cycle with dUTPs incorporated into the new strand. Uracil-DNA glycosylase and purin-pyrimidin endonuclease were used to fragment the cDNA. The fragments were biotin-labeled and then hybridized against arrays. The arrays were stained, washed, and scanned after 16 h of hybridization. TAC was used to analyze the chip data. Screening of differentially expressed genes was by multiple differential method (Fold change=2experiment group_NS−control group_NS) based on the fold change (FC) ≥2, or fold change ≤–2 (p<0.05).
Reverse transcription-polymerase chain reaction (RT-PCR)
Both semi-quantitative reverse transcription-polymerase chain reaction (RT-PCR) and quantitative real-time RT-PCR (qRT-PCR) were employed to validate the selected gene expressions of IFNalpha-treated Hep2 cells. Total RNAs were extracted from the cultured cells with TRIzol (Invitrogen, Carlsbad, CA, USA). The purified total RNAs were treated with DNase I (Qiagen, Dusseldorf, Germany). All primers used in RT-PCR and qRT-PCR are summarized in Table 1. The RT-PCR reaction was carried with a PrimeScript One-Step RT-PCR Kit (Takara Biotechnology, Dalian, China) by following the manual instructions. The RT-PCR was carried out in DNA Thermal Cycle (Applied Biosystems, Carlsbad, CA, USA) as follows: 50°C for 35 min for reverse transcription; 94°C for 5 min for denaturing; PCR condition was 94°C for 30 s, 50°C for 30 s, 72°C for 50 s, and repeated for 20 cycles; the reaction was extended at 72°C for 10 min before being stored at 4°C. One-step real-time quantitative RT-PCR (qRT-PCR) (Takara Biotechnology, Dalian, China) was also performed according to the instruction manual with the CFX real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA) as follows: 42ºC for 5 min and 95ºC for 10 s; 95ºC for 5 s, and 60ºC for 10 s, and repeated for 40 cycles. The dissociation of the reaction products was conducted at 55–95ºC, as the temperature rose at 0.2ºC per 10 s.
Table 1.
Primer sequences.
| Name | Forward 5′-3′ | Reverse 5′-3′ |
|---|---|---|
| DYNC1H1 | AAGATGCAGATGTTGGAGGA | GCCTTCACCTTTAGCTCCTT |
| *q-DYNC1H1 | AAGATGCAGATGTTGGAGGA | ATATTCTCCACGGTGCGCTT |
| DNA-PKc | AAAGACATTCTCCCCTGCCT | AACGTCCGCTTATAGAGCTG |
| q-DNA-PKc | AAAGACATTCTCCCCTGCCT | AAGAGCTGACACTTCCCAGT |
| ATF6 | AGATATTAAGGCAGAACCCC | CCGACATTTCTCATGGTACT |
| q-ATF6 | AGATATTAAGGCAGAACCCC | TAAGGAAAGGGGAGACATCT |
| IFI6 | TGCTTCTCTTCTCTCCTCCA | AAGGAAGAAGAGGTTCTGGG |
| q-IFI6 | TGCTTCTCTTCTCTCCTCCA | GAGCTCTCCGAGCACTTTTT |
| IFI27 | TTGTGATTGGAGGAGTTGTG | TGGGAAGAGTTGCAACAATT |
| q-IFI27 | TTGTGATTGGAGGAGTTGTG | GGACATCATCTTGGCTGCTA |
| IFI44L | AAGCCTGATCTAACCCCTAG | GCCTATTTCTGTGCTCTCTG |
| q-IFI44L | AAGCCTGATCTAACCCCTAG | ATAGTACATCCCTGACGGCT |
| Actin | CACACTGTGCCCATCTACGA | CTCAGGAGGAGCAATGATCT |
| q-Actin | TCCATCATGAAGTGTGACGT | CTCAGGAGGAGCAATGATCT |
“q” stands for “quantitative RT-PCR”.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
KEGG pathways enrichment analysis is based on the gene and genomic encyclopedia of molecular interaction, reaction, and relation networks [19]. The Database for Annotation, Visualization and Integrated Discovery (DAVID) [20] bioinformatics web (http://david.abcc.ncifcrf.gov/summary.jsp) was utilized to identify KEGG pathways that are enriched with IFNalpha-1a treatment. The DAVID 6.8 program was used to analyze differentially expressed genes at a significance level.
Statistical analysis
Data are presented as mean ± standard deviation (SD) from at least 3 independent experiments and subject to 2-tailed, unpaired t test. A p value less than 0.05 was considered statistically significant.
Results
IFNalpha-1a inhibits the proliferation potential of laryngeal carcinoma Hep-2 cells
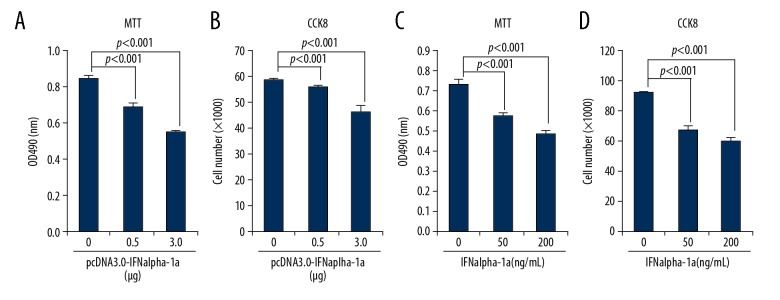
It has been known for many years that IFNalpha can serve as a therapeutic agent for the treatment of human laryngeal carcinoma [21]. However, how IFNalpha exerts its anti-proliferative effect on human laryngeal carcinoma is largely unclear. To address this issue directly, HEp-2 cells were used as a test model of laryngeal cancer for IFNalpha-1a treatment. We hypothesize that IFNalpha-1a might induce the anti-proliferative effect on HEp-2 cells. Two strategies were employed to test this hypothesis: one uses transient transfection approach and the other uses the exogenous delivery of recombinant human IFNalpha-1a into HEp-2 cells. The full-length of coding sequence (cDNA) of IFNalpha-1a was cloned from human fetal brain mRNAs by RT-PCR analysis. After sequencing confirmation, IFNalpha-1a cDNA was subcloned into pcDNA 3.0 to form eukaryotic expression vector pcDNA 3.0-IFNalpha-1a. The plasmid pcDNA 3.0-IFNalpha-1a DNAs were then transiently transfected into HEp-2 cells. Both MTT and CCK-8 results (Figure 1A, 1B) demonstrate that the cell proliferative potentials of HEp-2 cells were significantly inhibited by pcDNA 3.0-IFNalpha-1a. To further consolidate the result, HEp-2 cells were also treated with exogenously delivered recombinant IFNalpha-1a. Figure 1C and 1D demonstrate that the increased delivery of the recombinant IFNalpha-1a into HEp-2 cells markedly inhibited cell proliferation, further confirming that IFNalpha-1a has an anti-proliferative effect on human laryngeal carcinoma cells.
Figure 1.
IFNalpha-1a inhibits the proliferation of laryngeal carcinoma HEp-2 cells. (A, C) 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) analysis of the proliferation of HEp-2 cells. HEp-2 cells were either transiently transfected with increasing doses (0, 0.5, and 3 ug) of pcDNA3.0-IFNalpha-1a (A) or treated with increasing doses (0, 50, and 200 ng/mL) of recombinant human IFNalpha-1a (C). After 48-h incubation, the treated HEp-2 cells were collected and analyzed with MTT assay. The reaction products were measured at 490 nm with a microplate reader. Each value is represented as mean ±SD from 3 independent experiments. The results were considered to be significant if p≤0.05. (B, D) Cell counting kit 8 (CCK-8) analysis of cell proliferation. HEp-2 cells were either transiently transfected with increasing doses (0, 0.5, and 3 ug) of pcDNA3.0-IFNalpha-1a (B) or treated with increasing doses (0, 50, and 200 ng/mL) of recombinant human IFNalpha-1a (D). After 48-h incubation, the treated HEp-2 cells were collected and analyzed by CCK-8 assay. The reaction products were measured at 450 nm with a microplate reader. The number of viable cells for each dose was calculated against the standard curve. Each value is represented as mean ±SD from 3 independent experiments. The results were considered to be significant if p≤0.05.
IFNalpha-1a induces apoptosis of laryngeal carcinoma HEp-2 cells
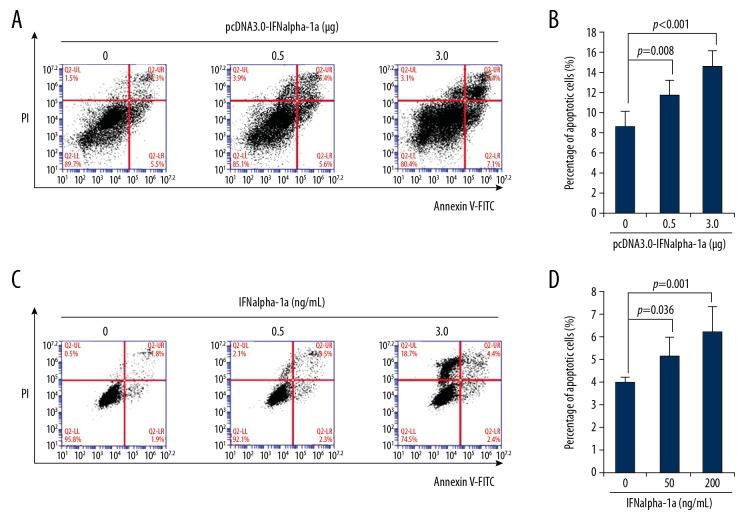
Sustained inhibition of cell proliferation leads to apoptosis of targeted cells. IFNalpha-1a-mediated anti-proliferation of HEp2 cells may induce cell apoptosis. To test whether IFNalpha-1a promotes apoptosis of HEp-2 cells, flow cytometric analysis by Annexin V/PI double staining was employed. Significant apoptosis was detected as the transfected dose of pcDNA 3.0-IFNalpha-1a increased (Figure 2A, 2B). To further confirm the results, HEp-2 cells were also treated with exogenous recombinant human IFNalpha-1a. Consistent with the results of the transient transfection experiment, the increased delivery of the recombinant IFNalpha-1a into HEp-2 cells dramatically promoted apoptosis of the targeted cells (Figure 2C, 2D), indicating that IFNalpha-1a induces apoptosis of human laryngeal carcinoma HEp-2 cells.
Figure 2.
IFNalpha-1a promotes the apoptosis of laryngeal carcinoma HEp-2 cells. (A) Flow cytometric analysis of the apoptosis of HEp-2 cells that were transiently transfected with pcDNA3.0-IFNalpha-1a. HEp-2 cells were first seeded onto a 12-well culture plate and transiently transfected with increasing doses (0, 0.5, and 3 ug) of pcDNA3.0-IFNalpha-1a. After 48-h incubation, the transfected HEp-2 cells were subjected to Annexin V/Propidium iodide (PI) double staining followed by flow cytometric analysis. (B) Quantitation of the apoptosis of the HEp-2 cells after transiently transfecting with pcDNA3.0-IFNalpha-1a. The transfected HEp2 cells in (A) were subjected to Annexin V/PI double staining followed by flow cytometric analysis. Each value is represented as mean ±SD from 3 independent experiments. After statistical analysis, results were considered to be significant if p≤0.05. (C) Flow cytometric analysis of the apoptosis of HEp-2 cells that were treated with recombinant human IFNalpha-1a. Hep-2 cells were seeded onto a 12-well culture plate and treated with increasing doses (0, 50, and 200 ng/mL) of recombinant human IFNalpha-1a. After 48-h culture, the cells were harvested and subjected to Annexin V/PI double staining followed by flow cytometric analysis. (D) Quantitation of the apoptosis of the HEp-2 cells after IFNalpha-1a treatment. The IFNalpha-1a treated HEp-2 cells were prepared as described in (C) and subjected to Annexin V/PI double staining followed by flow cytometric analysis. Each value is represented as mean ±SD from 3 independent experiments. After statistical analysis, results were considered to be significant if p≤0.05.
The effect of IFNalpha-1a on the apoptosis of other cell types
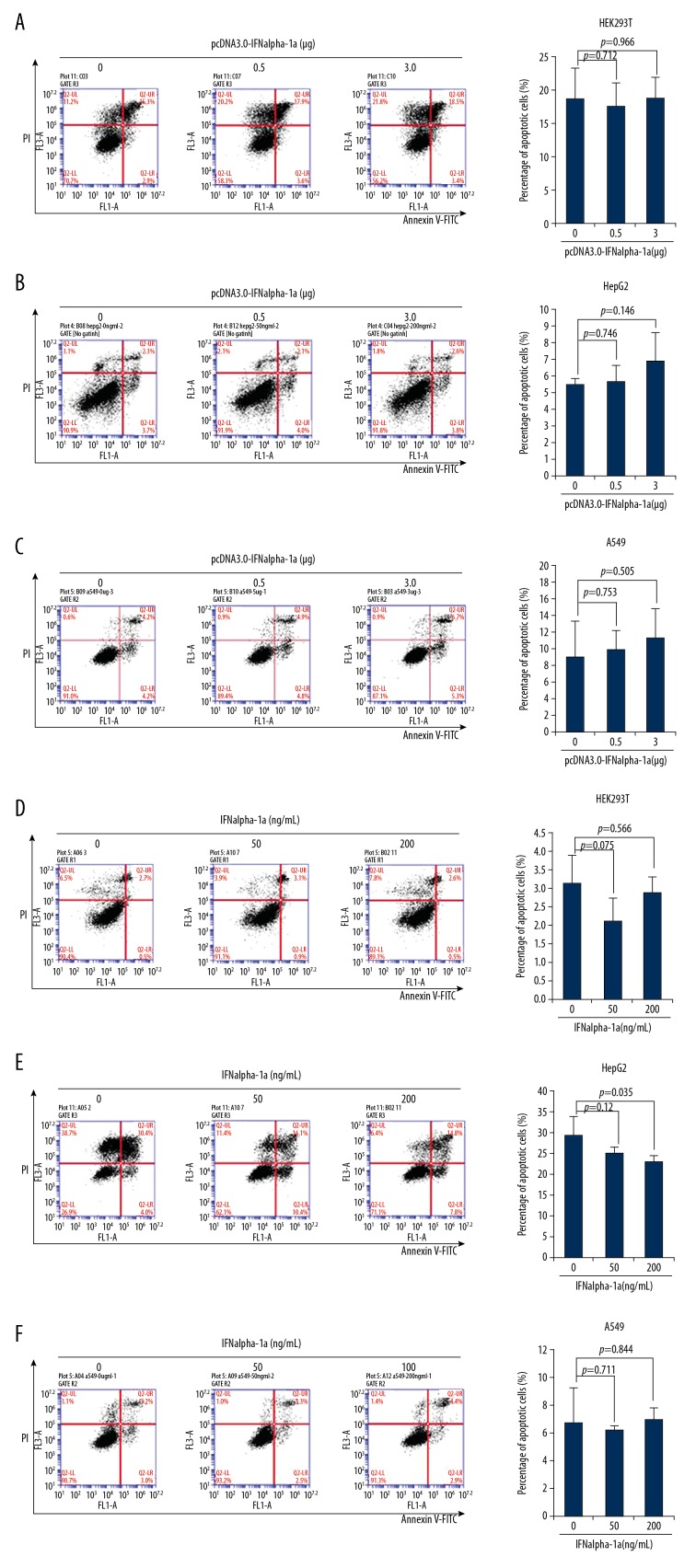
The next question that we attempted to ask was whether IFNalpha-1a treatment leads to apoptosis of other cell types. To address this question directly, 3 human cell lines (transformed human embryotic kidney HEK293T cells, human hepatocellular carcinoma HepG2 cells, and human lung carcinoma A549 cells) were selected for the treatment of IFNalpha-1a. Flow cytometric analysis by Annexin V/PI double staining was performed. The increased delivery of pcDNA 3.0-IFNalpha-1a into HEK293T (Figure 3A), HepG2 (Figure 3B), and A549 (Figure 3C) by transient transfection did not induce significant apoptosis in the respective cell lines. IFNalpha-1a-mediated apoptosis failed to be induced after the exogenous delivery of recombinant IFNalpha-1a into the cell culture medium of HEK293T (Figure 3D) and A549 (Figure 3F) but not HepG2 cells (Figure 3E), indicating that IFNalpha-1a-mediated apoptosis might function in a cell/tissue-specific manner.
Figure 3.
The responses to IFNalpha-1a by other cell lines. (A–C) Quantitation of the apoptosis of the HEK293T cells (A), HepG2 cells (B), and A549 cells (C) after transiently transfecting with increasing doses (0, 0.5, and 3 μg) of pcDNA3.0-IFNalpha-1a. The transfected cells were subjected to Annexin V/PI double staining followed by flow cytometric analysis. Each value is represented as mean ±SD from 3 independent experiments. After statistical analysis, results were considered to be significant if p≤0.05. (D–F) Quantitation of the apoptosis of the HEK293T cells (D), HepG2 cells (E), and A549 cells (F) after treatment with increasing doses (0, 50, and 200 ng/mL) of human recombinant IFNalpha-1a. The treated cells were subjected to Annexin V/PI double staining followed by flow cytometric analysis. Each value is represented as mean ±SD from 3 independent experiments. After statistical analysis, results were considered to be significant if p≤0.05.
IFNalpha-1a activates the intrinsic but not extrinsic apoptotic pathway
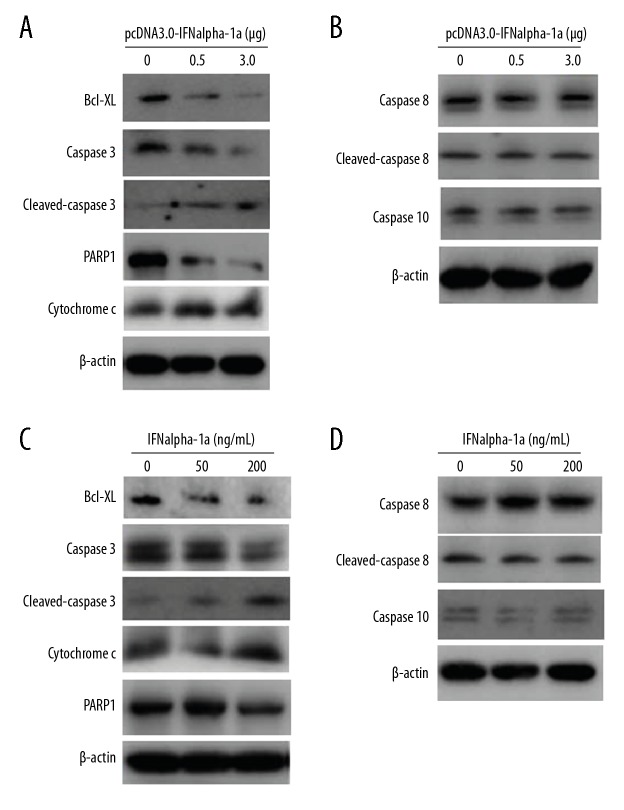
The intrinsic and extrinsic apoptotic pathways are 2 well-defined apoptotic pathways in mammal cells. To detect which pathway is activated by IFNalpha-1a, Western blot analysis was performed to examine the activation status of the key mediators in each pathway. First, HEp-2 cells were transiently transfected with increasing doses (0, 0.5, and 3.0 μg) of pcDNA 3.0-IFNalpha-1a. Overexpression of IFNalpha-1a dramatically inhibited the expression of the anti-apoptotic protein B cell lymphoma-XL (Bcl-XL), increased the cytoplasmic distribution of cytochrome c, and promoted the cleavages of caspase 3 and poly(ADP-Ribose) polymerase 1 (PARP1) dose-dependently (Figure 4A), indicating that the cell-intrinsic apoptotic pathway was activated. In contrast, increased delivery of pcDNA 3.0-IFNalpha-1a into HEp-2 cells failed to promote cleavages of caspase 8 and 10 (Figure 4B), indicating that IFNalpha-1a-mediated apoptosis is independent of the extrinsic apoptotic pathway. To further confirm the above results, HEp-2 cells were also treated with increasing doses (0, 50, and 200 ng/mL) of recombinant IFNalpha-1a. In agreement with the results of transient transfection, the addition of exogenous IFNalpha-1a decreased Bcl-XL expression, enhanced the expression of cytochrome c, and activated the cleavage of caspase 3 and PARP1 (Figure 4C), but had no effects on the expressions of caspase 8 and caspase 10 (Figure 4D), demonstrating that IFNalpha-1a indeed activates the intrinsic but not extrinsic apoptotic pathway.
Figure 4.
IFNalpha-1a-mediated cell apoptosis in HEp-2 cells is associated with the intrinsic but not extrinsic apoptotic pathway. (A, C) Western blot analysis on the expressions of the key mediators involved in the intrinsic apoptotic pathway. HEp-2 cells were either transiently transfected with increasing doses (0, 0.5. and 3ug) of pcDNA3.0-IFNalpha-1a (A) or treated with increasing doses (0, 50, and 200 ng/mL) of recombinant human IFNalpha-1a (C). After 48-h incubation, the whole-cell lysates were prepared and probed with anti-Bcl-XL, anti-cytochrome c, anti-caspase 3, anti-cleaved caspase 3, and anti-PARP1 antibodies. The reaction products were subjected to Western blot analysis. β-actin gene expression served as internal control. (B, D) IFNalpha-1a does not activate the extrinsic apoptotic pathway. Hep-2 cells were either transiently transfected with increasing doses (0, 0.5, and 3 ug) of pcDNA3.0-IFNalpha-1a (B) or treated with increasing doses (0, 50, and 200 ng/mL) of recombinant human IFNalpha-1a (D). After 48-h incubation, the whole-cell lysates were prepared and probed with anti-caspase 8, anti-cleaved caspase 8, and anti-caspase 10 antibodies. The reaction products were subjected to Western blot analysis. β-actin gene expression served as an internal control.
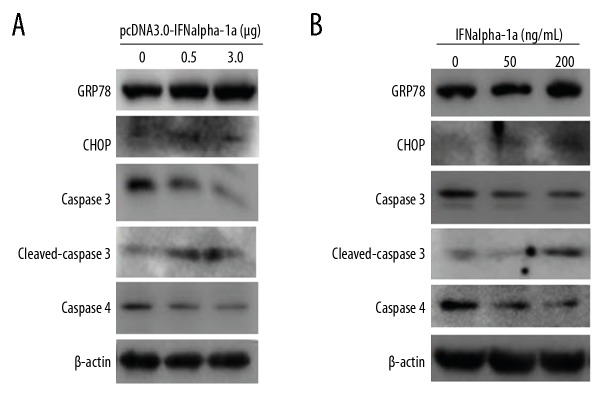
IFNalpha-1a induces endoplasmic reticulum stress-related apoptosis
In addition to intrinsic and extrinsic apoptotic pathways, we investigated a third apoptotic pathway named endoplasmic reticulum (ER) stress-associated apoptosis. Sustained ER stress usually triggers cells to undergo apoptosis. During ER stress, the expression of ER stress response genes (GRP78, CHOP and ATF4) is upregulated, and the activation of human caspase 4 is directly associated with ER stress-mediated cell apoptosis. To detect whether ER stress-mediated apoptosis can be induced by IFNalpha-1a, HEp-2 cells were either transiently transfected with pcDNA 3.0-IFNalpha-1a or treated with exogenous recombinant human IFNα-1a. Figure 5A and 5B show that the increased delivery of either pcDNA 3.0-IFNalpha-1a or recombinant IFNalpha-1a dramatically enhanced the expressions of GRP78 and CHOP and markedly promoted the activation of caspase 3 and caspase 4, indicating that IFNalpha-1a indeed promotes ER stress-mediated apoptosis.
Figure 5.
IFNalpha-1a activates the endoplasmic reticulum stress-induced apoptosis. (A) Western blot analysis of the expressions of the key mediators associated with the endoplasmic reticulum stress-induced apoptosis. HEp-2 cells were either transiently transfected with increasing doses (0, 0.5, and 3 μg) of pcDNA3.0-IFNalpha-1a (A) or treated with increasing does (0, 50, and 200 ng/mL) of human recombinant IFNalpha-1a (B). After 48-h incubation, the whole-cell lysates (WCL) were prepared and probed with anti-caspase4, anti-GRP78, anti-CHOP, anti-caspase 3, and anti-cleaved caspase 3 antibodies. The reaction products were subjected to Western blot analysis. β-actin gene expression served as an internal control.
Microarray and pathway enrichment analysis of the gene expression profiles of IFNalpha-1a-treated HEp-2 cells
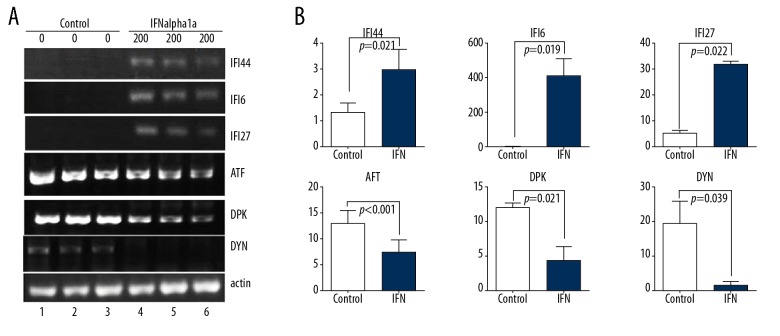
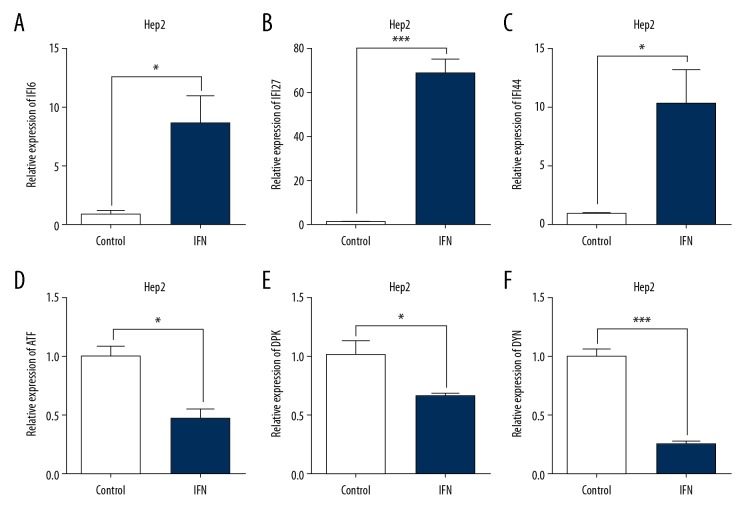
The results of microarray revealed that, under stringent criteria, 1236 upregulated and 531 downregulated probe sets displayed differential expressions by at least 1.5-fold. The top 40 differentially expressed genes that have been characterized are shown in Table 2. Among them, several IFN-alpha-regulated genes were dramatically upregulated, such as interferon alpha-inducible protein 6 (IFI6, 1228 folds), IFI27 (236 folds) and interferon-induced protein 44-like (IFI44L, by 18.25-fold), while the top down-regulated genes include DNA-activated catalytic polypeptide (PRKDC, −8.86 folds), histone cluster 1 H3b (HIST1H3B, by −6.51 fold), dynein cytoplasmic 1 heavy chain 1 (DYNC1H1, by −4.57 fold), histone cluster 1, H2am (HIST1H2AM, by −4.56 fold), and activating transcription factor 6 (ATF6, by −4.51 fold). The results of microarray analysis were validated with both semi-quantitative RT-PCR (Figure 6) and qRT-PCR (Figure 7) approaches on 3 selected upregulated genes (IFI6, IFI27, and IFI44L) and 3 selected downregulated genes (PRKDC, DYNC1H1, and ATF6).
Table 2.
Top 40 differentially expressed mRNAs in HEp2 cells after IFNalpha1a treatment.
| Gene symbol | Description | Fold change | p Value | Regulation | Chromosome | GenBank No. |
|---|---|---|---|---|---|---|
| IFI6 | Interferon, alpha-inducible protein 6 | 1228 | 0.017151 | Up | chr1 | BC015603.2 |
| FI27 | Interferon, alpha-inducible protein 27 | 236 | 0.032977 | Up | chr14 | BC015492.1 |
| IFI44L | Interferon-induced protein 44-like | 18.25 | 0.038627 | Up | chr1 | NM_006820 |
| MIR548X | microRNA 548x | 3.26 | 0.04887 | Up | chr21 | NR_109925.1 |
| EIF2AK2 | Eukaryotic translation initiation factor 2-alpha kinase 2 | 2.56 | 0.037189 | Up | chr2 | NM_002759 |
| RNA5SP473 | RNA, 5S ribosomal pseudogene 473 | 2.54 | 0.003723 | Up | chr19 | NC_000019 |
| DDX60L | DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like | 2.15 | 0.036241 | Up | chr4 | KR632542 |
| IFITM4P | Interferon-induced transmembrane protein 4 pseudogene | 2.05 | 0.029556 | Up | chr6 | NR_001590 |
| RNY4P11 | RNA, Ro-associated Y4 pseudogene 11 | 1.99 | 0.020822 | Up | chr20 | NG_016612 |
| IFITM3 | Interferon-induced transmembrane protein 3 | 1.96 | 0.023115 | Up | chr11 | NM_021034 |
| RNU6-1141P | RNA, U6 small nuclear 1141, pseudogene | 1.90 | 0.021328 | Up | chr10 | NG_043954 |
| TNFSF10 | Tumor necrosis factor (ligand) superfamily, member 10 | 1.89 | 0.022176 | Up | chr3 | NM_003810 |
| FAM218A | Family with sequence similarity 218, member A | 1.78 | 0.020452 | Up | chr4 | NM_153027 |
| RNA5SP238 | RNA, 5S ribosomal pseudogene 238 | 1.76 | 0.041771 | Up | chr7 | NG_033316 |
| RNU6ATAC36P | RNA, U6atac small nuclear 36, pseudogene | 1.66 | 0.023507 | Up | chr5 | NG_044158 |
| RP11-124O11.2 | novel transcript | 1.63 | 0.027078 | Up | chr10 | AL365199 |
| RNA5SP354 | RNA, 5S ribosomal pseudogene 354 | 1.58 | 0.027011 | Up | chr12 | NG_033499 |
| ZZEF1 | Zinc finger, ZZ-type with EF-hand domain 1 | 1.57 | 0.044929 | Up | chr17 | NM_015113 |
| HIST2H2BC | Histone cluster 2, H2bc (pseudogene) | 1.56 | 0.012514 | Up | chr1 | NR_036461 |
| MIR548D2 | microRNA 548d-2; microRNA 548aa-2 | 1.55 | 0.033455 | Up | chr17 | NR_036461 |
| PRKDC | DNA-activated, catalytic polypeptide | −8.86 | 0.003455 | Down | chr8 | NM_006904 |
| HIST1H3B | Histone cluster 1, H3b | −6.51 | 0.039809 | Down | chr6 | NM_003537 |
| DYNC1H1 | Dynein, cytoplasmic 1, heavy chain 1 | −4.57 | 0.044701 | Down | chr14 | NM_001376 |
| HIST1H2AM | Histone cluster 1, H2am | −4.56 | 0.041334 | Down | chr6 | NM_003514 |
| ATF6 | Activating transcription factor 6 | −4.51 | 0.038825 | Down | chr1 | AB015856 |
| HIST2H2AC | Histone cluster 2, H2ac | −4.03 | 0.027939 | Down | chr1 | NM_003517 |
| RPS3AP47 | Ribosomal protein S3a pseudogene 47 | −3.76 | 0.021051 | Down | chr15 | NG_010693 |
| MYOF | Myoferlin | −3.60 | 0.046976 | Down | chr10 | AF182316 |
| HUWE1 | HECT, UBA and WWE domain containing 1 | −3.04 | 0.013349 | Down | chrX | NM_031407 |
| HNRNPA1P3 | Heterogeneous nuclear ribonucleoprotein A1 pseudogene 33 | −2.86 | 0.015543 | Down | chr20 | NC_000020 |
| HNRNPA1P33 | Heterogeneous nuclear ribonucleoprotein A1 pseudogene 33 | −2.86 | 0.015543 | Down | chr10 | NR_003277 |
| HIST1H2AB | Histone cluster 1, H2ab | −2.78 | 0.012627 | Down | chr6 | NM_003513 |
| DST | Dystonin | −2.74 | 0.006619 | Down | chr6 | NM_183380 |
| HIST1H4J | Histone cluster 1, H4j | −2.62 | 0.040169 | Down | chr6 | NM_021968 |
| PTPRS | Protein tyrosine phosphatase, receptor type, S | −2.35 | 0.024097 | Down | chr19 | NM_130854 |
| KIF5B | Kinesin family member 5B | −2.29 | 0.030942 | Down | chr10 | NM_004521 |
| ANXA2 | Annexin A2; annexin A2 pseudogene 2 | −2.24 | 0.008898 | Down | chr15 | NM_001002858 |
| HIST1H2BI | Histone cluster 1, H2bi | −2.15 | 0.014486 | Down | chr6 | NM_003525 |
| NNT | Nicotinamide nucleotide transhydrogenase | −2.14 | 0.018575 | Down | chr5 | NM_012343 |
| HNRNPA1P8 | Heterogeneous nuclear ribonucleoprotein A1 pseudogene 8 | −2.11 | 0.028259 | Down | chr7 | NG_006966 |
Figure 6.
Confirmation of microarray analysis of IFNalpha-1a-responded gene expressions in HEp-2 cells by semi-quantitative RT-PCR analysis. (A) RT-PCR analysis of total RNAs from HEp-2 cells treated with or without IFNalpha-1a. The expressions of the 3 selected upregulated genes (IFI6, IFI27, and IFI44L) and 3 selected downregulated genes (ATF6 (ATF), PRKDC (DPK), and DYNC1H1 (DYN)) were subjected to RT-PCR analysis. The expression of β-actin gene was assayed as a loading control. (B) Quantitation of the results in (A). The relative band intensity was quantitated with Image J program in comparison with β-actin. The triplicated results of both IFNalpha-1a treated or untreated samples were subjected to statistical analysis. The results were considered to be significant if p≤0.05.
Figure 7.
Confirmation of microarray analysis of IFNalpha-1a-responded gene expressions in HEp-2 cells by real-time qRT-PCR analysis. Real-time qRT-PCR was employed to detect the mRNA levels of IFI6 (A), IFI27 (B), IFI44L (C), ATF6 (ATF, D), PRKDC (DPK, E), and DYNC1H1 (DYN, F) on the samples isolated in Figure 6. Each value represents the mean ±SD from 3 reactions. The triplicated results of both IFNalpha-1a-treated or -untreated samples were subjected to statistical analysis. The symbols * and *** represent p values less than 0.05 and 0.001, respectively. The results were considered to be significant if p≤0.05.
Pathway enrichment analysis is an effective approach to identify the potential key cellular pathways for high-throughput data. Here, the identified top 40 genes were formatted and subjected to analysis with the DAVID Bioinformatics program. The results of the KEGG pathway data set revealed that IFNalpha-1a-treated HEp2 cells were enriched in at least 6 KEGG pathways (Table 3). Four out of 6 pathways are associated with TP53: they are pathways in cancer (p=0.011), transcriptional mis-regulation in cancer (p=0.020), MAPK signaling pathway (p=0.032), and small cell lung cancer (p=0.046). The 2 TP53 unrelated pathways are calcium signaling pathway (p=0.027) and signaling pathways regulating pluripotency of stem cells (p=0.043). Therefore, the results also suggest that the TP53-related pathways might be involved in IFNalpha-1a-mediated HEp-2 apoptosis.
Table 3.
Enriched KEGG pathways in IFNalpha-1a treated HEp2 cells.
| KEGG pathways | p Value | Related genes |
|---|---|---|
| Pathways in cancer | 0.011 | BCL2, GNA12, WNT2B, WNT6, AGTR1, ERBB2, FGF3, NOS2, PIK3R1, TP53 |
| Transcriptional mis-regulation in cancer | 0.020 | CCND2, ELANE, MPO, MEF2C, RUNX2, TP53 |
| Calcium signaling pathway | 0.027 | HTR4, AGTR1, ERBB2, GRIN2A, NOS2, PDE1C |
| MAPK signaling pathway | 0.032 | GNA12, DUSP9, FGF3, MAPK8IP1, MEF2C, NTRK2, TP53 |
| Signaling pathways regulating pluripotency of stem cells | 0.043 | WNT2B, WNT6, DUSP9, INHBC, PIK3R1 |
| Small cell lung cancer | 0.046 | BCL2, NOS2, PIK3R1, TP53 |
Discussion
Laryngeal cancer is one of the most common head and neck carcinomas. Surgical therapy and chemo-radiation treatment are the 2 major therapeutic choices for laryngeal cancer patients. However, both treatments can be associated with some severe adverse effects such as voice loss [9] and toxicity [22]. Thus, novel therapeutic strategies are urgently needed [10]. Our current results demonstrate that overexpression of IFNalpha-1a cDNA or addition of recombinant IFNalpha-1a significantly promotes both intrinsic and ER-stress-mediated apoptosis in HEp-2 cells, indicating that IFNalpha-1a may provide a therapeutic alternative for laryngeal cancer patients in the future.
Interferon exerts its antineoplastic property by either serving as an immunological modulator to promote cancer-specific immune responses or functioning as an anti-proliferative agent to block tumor growth. Xu et al. showed that using mesenchymal stem cells (MSC) as a carrier to deliver IFNalpha (MSC-IFNalpha) into tumor tissue failed to induce an anti-tumor effect in immunodeficient mice, but MSC-IFNalpha-mediated anti-tumor activity could be dramatically enhanced in immune-competent mice, indicating the immuno-dependent character of MSC-IFNalpha treatment in vivo [23]. IFN has been regarded as an indirect mediator that exerts its anti-proliferative effect by upregulating the expressions of pro-apoptotic ISGs [24]. For example, inositol hexosephosphate kinase 2 (IP6K2)/interferon-induced death (RID) is an IFNβ-stimulated gene that promotes apoptosis of ovarian cancer cells [25]. Subsequent studies showed that the upregulation of IP6K2/RID expression is directly linked to p53-mediated apoptosis through the DNA-PK/ATM-p53 cell death axis [26,27]. Our study revealed the mechanisms of IFNalpha-1a-mediated apoptosis in HEp-2 cells by which both intrinsic and ER-stress-mediated apoptotic pathways are activated. However, it remains unclear how IFNalpha-1a stimulates these 2 apoptotic pathways in HEp-2 cells. IFN-induced cell apoptosis has been shown to occur indirectly through apoptotic mediators such as ISGs [24,28]. Nevertheless, the pro-apoptotic ISGs specifically induced by IFNalpha-1a are still unknown and need to be further investigated. Therefore, we cannot exclude the possibility that IFNalpha-1a promotes intrinsic and ER-stress-mediated cell apoptosis through a direct mechanism.
In 1986, recombinant IFNalpha-2a and IFNalpha-2b were the first 2 cytokines to be licensed for the treatment of hairy cell leukemia in the United States [29]. Since then, IFNalpha-2 has been primarily used for the treatment of various blood cancers such as chronic myelogenous leukemia [30], non-Hodgkin lymphomas [31,32], and multiple myeloma [31–33], as well as some virus infections such as chronic hepatitis B viral (HBV) infection [34] and chronic hepatitis C viral (HCV) infection [35]. IFNalpha-2 has become the preferred therapeutic molecule approved by the US Food and Drug Administration (FDA) for treatment of certain cancers and infectious diseases [10,36,37]. The reason for this preferred use of IFNalpha-2 subtype in clinic is elusive. One possibility might be that IFNalpha-2a and 2b were the first 2 IFN molecules to be cloned and characterized with higher activity and potency – 10-fold more potent than IFNalpha-1b, as measured by antiviral activity [38]. In 1997, the recombinant IFNalpha-1b was approved for treatment of chronic hepatitis B and C in China [38]. Although the clinical effect and toxicity of IFNalpha-1b have been evaluated in cancer patients [38], IFNalpha-1 has not been approved for use as a therapeutic drug to treat human cancers in clinical practice. It has been shown that IFNalpha-1 has lower anti-proliferative and antiviral activities compared with other IFNalpha subtypes [39,40]. However, our functional study demonstrated that IFNalpha-1a can induce cell apoptosis in laryngeal carcinoma HEp-2 cells. This may be partly due to the stable ternary complex formed upon the binding of IFNalpha-1a to IFNalpha receptors on HEp-2 cells, which might be critical for the generation of certain pro-apoptotic ISGs in HEp-2 cells.
Microarray analysis reveals several differential genes that are dramatically upregulated after IFNalpha-1a treatment in HEp-2 cells. The 3 most upregulated genes – IFI6, IFI27, and IFI44L – are typical interferon alpha-inducing genes. Studies indicate that IFI6 might be double-edged sword to regulate cell growth versus apoptosis. Earlier reports have defined IFI6 as an anti-apoptotic gene that promotes melanoma development [39] and facilitates viral infection [40]. In contrast, Li et al. recently found that a short spliced variant of CTCF (CTCF-s) could up-regulate IFI-6 expression by competing with CTCF binding at the IFI6 promoter region, which subsequently induces cell apoptosis [41]. The effect of CTCF-s has great similarity to that of IFNalpha-1a. This is because, in addition to IFI6, CTCF-s like IFNalpha-1a can also upregulate IFI27 and ISG15 expressions [41]. IFI27 has been shown to stimulate cell apoptosis by promoting mitochondrial membrane destabilization [42], while IFI44L has a tumor-suppressor effect [43]. Therefore, upregulating IFI6, IFI27, and IFI44L gene expressions should facilitate IFNalpha-1a-mediated cell apoptosis. When cells are undergoing apoptosis, apoptosis-related genes can be induced, but growth-related genes need to be repressed. The downregulated genes discovered by our microarray analysis are indeed associated with cell survival and proliferation. For example, both PRKDC [44] and HIST1H3B [45] have been shown to play a regulatory role in apoptotic inhibition and tumor growth. Therefore, downregulating these gene expressions by IFNalpha-1a should promote the apoptosis of HEp-2 cells.
Conclusions
Our study elucidates a previously undefined mechanism for IFNalpha-1a, which could be used to induce apoptosis of certain cancer cells, such as laryngeal carcinoma HEp-2 cells, via both intrinsic and ER-stressed apoptotic pathways. However, more research is needed to determine how the IFNalpha-1 molecule can serve as effective inducer of apoptosis.
Footnotes
Source of support: The study is supported by grant from CAMS Innovation Fund for Medical Sciences (2017-I2M-3-007)
Conflicts of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Kuper H, Boffetta P, Adami HO. Tobacco use and cancer causation: Association by tumour type. J Intern Med. 2002;252:206–24. doi: 10.1046/j.1365-2796.2002.01022.x. [DOI] [PubMed] [Google Scholar]
- 3.Boffetta P, Hashibe M. Alcohol and cancer. Lancet Oncol. 2006;7:149–56. doi: 10.1016/S1470-2045(06)70577-0. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez Barrueco A, Gonzalez Galan F, Lora Pablos D, et al. HPV in larynx squamous cell carcinoma: New serotypes and survival study within 10-year follow-up. Otolaryngol Head Neck Surg. 2017;156:677–82. doi: 10.1177/0194599817695545. [DOI] [PubMed] [Google Scholar]
- 5.Vietia D, Liuzzi J, Avila M, et al. Human papillomavirus detection in head and neck squamous cell carcinoma. Ecancermedicalscience. 2014;8:475. doi: 10.3332/ecancer.2014.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsimplaki E, Argyri E, Sakellaridis A, et al. Oropharyngeal and laryngeal but not oral cancers are strongly associated with high-risk human papillomavirus in 172 Greek patients. J Med Virol. 2017;89:170–76. doi: 10.1002/jmv.24614. [DOI] [PubMed] [Google Scholar]
- 7.Weiss D, Heinkele T, Rudack C. Reliable detection of human papillomavirus in recurrent laryngeal papillomatosis and associated carcinoma of archival tissue. J Med Virol. 2015;87:860–70. doi: 10.1002/jmv.24124. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, Liu S, Yi H, et al. Human papillomavirus infection in 674 Chinese patients with laryngeal squamous cell carcinoma. PLoS One. 2017;9:e115914. doi: 10.1371/journal.pone.0115914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steuer CE, El-Deiry M, Parks JR, et al. An update on larynx cancer. Cancer J Clin. 2017;67:31–50. doi: 10.3322/caac.21386. [DOI] [PubMed] [Google Scholar]
- 10.Antonelli G, Scagnolari C, Moschella F, Proietti E. Twenty-five years of type I interferon-based treatment: A critical analysis of its therapeutic use. Cytokine Growth Factor Rev. 2015;26:121–31. doi: 10.1016/j.cytogfr.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardy MP, Owczarek CM, Jermiin LS, et al. Characterization of the type I interferon locus and identification of novel genes. Genomics. 2004;84:331–45. doi: 10.1016/j.ygeno.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 12.van Pesch V, Lanaya H, Renauld JC, Michiels T. Characterization of the murine alpha interferon gene family. J Virol. 2004;78:8219–28. doi: 10.1128/JVI.78.15.8219-8228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zwarthoff EC, Mooren AT, Trapman J. Organization, structure and expression of murine interferon alpha genes. Nucleic Acids Res. 1985;13:791–804. doi: 10.1093/nar/13.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, Liu S, Xu F, et al. Inhibition of proteasome activity induces aggregation of IFIT2 in the centrosome and enhances IFIT2-induced cell apoptosis. Int J Biol Sci. 2017;13:383–90. doi: 10.7150/ijbs.17236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stawowczyk M, Van Scoy S, Kumar KP, Reich NC. The interferon stimulated gene 54 promotes apoptosis. J Biol Chem. 2011;286:7257–66. doi: 10.1074/jbc.M110.207068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuno T, Mejido J, Zhao T, et al. BID is a critical factor controlling cell viability regulated by IFN-alpha. J Immunother. 2012;35:23–31. doi: 10.1097/CJI.0b013e3182372dcf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hitomi J, Katayama T, Eguchi Y, et al. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death. J Cell Biol. 2004;165:347–56. doi: 10.1083/jcb.200310015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakagawa T, Zhu H, Morishima N, et al. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–61. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang dW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haglund S, Lundquist PG, Cantell K, Strander H. Interferon therapy in juvenile laryngeal papillomatosis. Arch Otolaryngol. 1981;107:327–32. doi: 10.1001/archotol.1981.00790420001001. [DOI] [PubMed] [Google Scholar]
- 22.Wiegand S. Evidence and evidence gaps of laryngeal cancer surgery. GMS Curr Top Otorhinolaryngol Head Neck Surg. 2016;15 doi: 10.3205/cto000130. Doc03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu C, Lin L, Cao G, et al. Interferon-alpha-secreting mesenchymal stem cells exert potent antitumor effect in vivo. Oncogene. 2014;33:5047–52. doi: 10.1038/onc.2013.458. [DOI] [PubMed] [Google Scholar]
- 24.Kotredes KP, Gamero AM. Interferons as inducers of apoptosis in malignant cells. J Interferon Cytokine Res. 2013;33:162–70. doi: 10.1089/jir.2012.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison BH, Bauer JA, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 mediates growth suppressive and apoptotic effects of interferon-beta in ovarian carcinoma cells. J Biol Chem. 2001;276:24965–70. doi: 10.1074/jbc.M101161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koldobskiy MA, Chakraborty A, Werner JK, Jr, et al. p53-mediated apoptosis requires inositol hexakisphosphate kinase-2. Proc Natl Acad Sci USA. 2010;107:20947–51. doi: 10.1073/pnas.1015671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao F, Cha J, Xu J, et al. Inositol pyrophosphates mediate the DNA-PK/ATM-p53 cell death pathway by regulating CK2 phosphorylation of Tti1/Tel2. Mol Cell. 2014;54:119–32. doi: 10.1016/j.molcel.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Jamal SM, Taylor EB, Abd Elmageed ZY, et al. Interferon gamma-induced apoptosis of head and neck squamous cell carcinoma is connected to indoleamine-2,3-dioxygenase via mitochondrial and ER stress-associated pathways. Cell Div. 2016;11:11. doi: 10.1186/s13008-016-0023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Golomb HM, Ratain MJ, Mick R, Daly K. Interferon treatment for hairy cell leukemia: An update on a cohort of 69 patients treated from 1983–1986. Leukemia. 1992;6:1177–80. [PubMed] [Google Scholar]
- 30.Talpaz M, Kantarjian HM, McCredie KB, et al. Clinical investigation of human alpha interferon in chronic myelogenous leukemia. Blood. 1987;69:1280–88. [PubMed] [Google Scholar]
- 31.Borden EC. Innovative treatment strategies for non-Hodgkin’s lymphoma and multiple myeloma. Semin Oncol. 1994;21:14–22. [PubMed] [Google Scholar]
- 32.Steis RG, Foon KA, Longo DL. Current and future uses of recombinant interferon alpha in the treatment of low-grade non-Hodgkin’s lymphoma. Cancer. 1987;59:658–63. doi: 10.1002/1097-0142(19870201)59:3+<658::aid-cncr2820591315>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 33.Oken MM, Kyle RA, Kay NE, et al. Interferon in the treatment of refractory multiple myeloma: An Eastern Cooperative Oncology Group Study. Leuk Lymphoma. 1990;1:95–100. doi: 10.3109/10428199009042465. [DOI] [PubMed] [Google Scholar]
- 34.Rakela J, Wood JR, Czaja AJ, et al. Long-term versus short-term treatment with recombinant interferon alfa-2a in patients with chronic hepatitis B: A prospective, randomized treatment trial. Mayo Clin Proc. 1990;65:1330–35. doi: 10.1016/s0025-6196(12)62144-2. [DOI] [PubMed] [Google Scholar]
- 35.Zein NN, Rakela J. Interferon therapy in hepatitis C. Semin Gastrointest Dis. 1995;6:46–53. [PubMed] [Google Scholar]
- 36.Degasperi E, Vigano M, Aghemo A, et al. PegIFN-alpha2a for the treatment of chronic hepatitis B and C: A 10-year history. Expert Rev Anti Infect Ther. 2013;11:459–74. doi: 10.1586/eri.13.37. [DOI] [PubMed] [Google Scholar]
- 37.Di Trolio R, Simeone E, Di Lorenzo G, et al. The use of interferon in melanoma patients: A systematic review. Cytokine Growth Factor Rev. 2015;26:203–12. doi: 10.1016/j.cytogfr.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Paul F, Pellegrini S, Uze G. IFNA2: The prototypic human alpha interferon. Gene. 2015;567:132–37. doi: 10.1016/j.gene.2015.04.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta R, Forloni M, Bisserier M, Dogra SK, et al. Interferon alpha-inducible protein 6 regulates NRASQ61K-induced melanomagenesis and growth. Elife. 2016;5 doi: 10.7554/eLife.16432. pii: e16432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi Y, Li Y, Zhang Y, et al. IFI6 inhibits apoptosis via mitochondrial-dependent pathway in dengue virus 2 infected vascular endothelial cells. PLoS One. 2015;10:e0132743. doi: 10.1371/journal.pone.0132743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Huang K, Hu G, et al. An alternative CTCF isoform antagonizes canonical CTCF occupancy and changes chromatin architecture to promote apoptosis. Nat Commun. 2019;10:1535. doi: 10.1038/s41467-019-08949-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheriyath V, Leaman DW, Borden EC. Emerging roles of FAM14 family members (G1P3/ISG 6-16 and ISG12/IFI27) in innate immunity and cancer. J Interferon Cytokine Res. 2011;31:173–81. doi: 10.1089/jir.2010.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang WC, Tung SL, Chen YL, et al. IFI44L is a novel tumor suppressor in human hepatocellular carcinoma affecting cancer stemness, metastasis, and drug resistance via regulating met/Src signaling pathway. BMC Cancer. 2018;18:609. doi: 10.1186/s12885-018-4529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stronach EA, Chen M, Maginn EN, et al. DNA-PK mediates AKT activation and apoptosis inhibition in clinically acquired platinum resistance. Neoplasia. 2011;13:1069–80. doi: 10.1593/neo.111032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcussen M, Sønderkær M, Bødker JS, et al. Oral mucosa tissue gene expression profiling before, during, and after radiation therapy for tonsil squamous cell carcinoma. PLoS One. 2018;13:e0190709. doi: 10.1371/journal.pone.0190709. [DOI] [PMC free article] [PubMed] [Google Scholar]