Abstract
Skp1 is hydroxylated by an O2-dependent prolyl hydroxylase (PhyA) that contributes to O2-sensing in the social amoeba Dictyostelium and the mammalian pathogen Toxoplasma gondii. HO-Skp1 is subject to glycosylation and the resulting pentasaccharide affects Skp1 conformation in a way that influences association of Skp1 with F-box proteins, and potentially the assembly of E3(SCF) ubiquitin ligase complexes that mediate the polyubiquitination of target proteins that are degraded in the 26S-proteasome. To investigate the conservation and specificity of these modifications, we analyzed proteins from the oomycete Pythium ultimum, an important crop plant pathogen. Putative coding sequences for Pythium’s predicted PhyA and first glycosyltransferase in the predicted five-enzyme pathway, a GlcNAc-transferase (Gnt1), predict a bifunctional enzyme (Phgt) that, when expressed in Dictyostelium, rescued a knockout of phyA but not gnt1. Though recombinant Phgt was also unable to glycosylate Dictyostelium HO-Skp1, it could hydrolyze UDP-GlcNAc and modify a synthetic hydroxypeptide from Dictyostelium Skp1. Pythium encodes two highly similar Skp1 isoforms, but only Skp1A was efficiently hydroxylated and glycosylated in vitro. While kinetic analysis revealed no evidence for processive processing of Skp1, the physical linkage of the two activities implies dedication to Skp1 in vivo. These findings indicate a widespread occurrence of the Skp1 modification pathway across protist phylogeny, suggest that both Gnt1 and PhyA are specific for Skp1 and indicate that the second Skp1 provides a bypass mechanism for O2-regulation in Pythium and other protists that conserve this gene.
Keywords: E3 ubiquitin ligase, glycosyltransferase, plant pathogen, prolyl hydroxylase, Pythium ultimum
Introduction
The Skp1/Cullin-1/F-box protein (SCF) family of E3 polyubiquitin ligases is highly conserved throughout eukaryotes (Willems et al. 2004). This enzyme family catalyzes the polyubiquitination and turnover in the 26S-proteasome of a spectrum of cellular proteins that is recognized by the multiplicity of rapidly evolving F-box proteins (FBPs), which serve primarily as substrate receptors. SCF complexes are regulated in many ways, including covalent priming of the substrate, Neddylation of Cullin-1 and competition between Skp1/FBP subcomplexes and Cand1 for binding to Cullin-1 (Deshaies and Joazeiro 2009; Zheng and Shabek 2017). In two protists, the social amoeba Dictyostelium discoideum and the apicomplexan Toxoplasma gondii, a new form of regulation that controls the interaction between Skp1 and FBPs, have been described (West and Blader 2015). This involves posttranslational hydroxylation of a specific Pro residue near the docking interface with the F-box domain of the FBP. Modification of the resulting hydroxyproline (Hyp) with a glycan, up to five sugars in D. discoideum and T. gondii (Figure 1A), affects the conformation of Skp1 in a way that may explain the increased binding to three FBPs observed in in vitro and in vivo interaction studies (Sheikh et al. 2017b). The hydroxylation of Pro143 (in D. discoideum; Pro154 in T. gondii) is catalyzed by a cytoplasmic O2- and α-ketoglutarate (αKG)-dependent prolyl 4-hydroxylase, PhyA (van der Wel et al. 2005; Xu et al. 2012), which is related to the HIFα prolyl hydroxylase involved in O2 and metabolic sensing in animals (Islam et al. 2018). These enzymes distribute O atoms to the 4(trans)-position of the prolyl residue and αKG, which then decomposes into succinate and CO2, and are rate limited by physiological levels of these substrates. In D. discoideum, O2 levels serve as a guide to coordinate starvation-induced fruiting body morphogenesis with arrival of the migrating slug to the surface of the soil (West et al. 2007). While prolyl hydroxylation is the direct O2-sensor, glycosylation of Hyp is required to manifest a biochemical effect on SCF complex formation (Sheikh et al. 2014; Sheikh et al. 2015; West and Kim 2019). Though less well studied, a similar mechanism appears to operate in T. gondii (West and Blader 2015; Rahman et al. 2016; Rahman et al. 2017), a pathogen of warm-blooded animals that is the agent for acute human toxoplasmosis and chronic infections that persist for life (Blader et al. 2015).
Fig. 1.
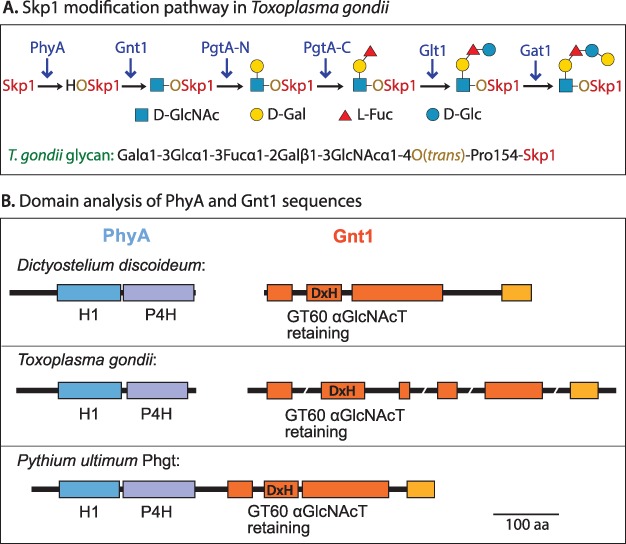
The Skp1 modification pathway. (A) The hydroxylation/glycosylation pathway in T. gondii. A homologous pathway is predicted in P. ultimum. PgtA-N and PgtA-C refer to separate domains of the same protein. (B) Domain diagrams of PhyA and Gnt1 proteins from D. discoideum (van der Wel et al. 2005; van der Wel et al. 2002b), T. gondii (Xu et al. 2012; Rahman et al. 2016) and P. ultimum. The prediction for P. ultimum is based on genomic sequence data and amino acid homology of the translated sequence (Figure S1). Colored boxes refer to regions of substantial homology.
Full glycosylation requires the action of five glycosyltransferase activities, encoded by four genes in T. gondii: gnt1, pgtA, glt1 and gat1 (Rahman et al. 2016; Rahman et al., 2017), as illustrated in Figure 1A. Each gene encodes a single glycosyltransferase activity, with the exception of pgtA, which encodes a 2-domain diglycosyltransferase (Rahman et al. 2016) that, in D. discoideum, mediates the successive addition of βGal and αFuc in processive manner (van der Wel et al. 2002a). The glycosyltransferases are sugar nucleotide dependent and exist as soluble proteins that can be isolated from cytosolic fractions, and thus represent a well-defined example of eukaryotic cytoplasmic glycosylation (West and Hart 2017). In D. discoideum, glt1 and gat1 are replaced by agtA, which alone catalyzes addition of a distinct nonreducing-end disaccharide, Galα1,3Galα,1-, at the 3-position of the α-l-Fuc residue (Wang et al. 2009).
Pythium ultimum is an oomycete protist that is a major necrotrophic plant pathogen causing damping off and root rot diseases in dozens of diverse plant hosts including corn, soybean, potato, wheat, fir and many ornamental species (Lévesque et al. 2010). The P. ultimum genome possesses sequences predicted to encode all six of the enzymatic activities known to modify T. gondii Skp1, and PuGat1 was recently found to catalyze addition of the nonreducing terminal αGal in 3-linkage to Glc in vitro, as does its counterpart in T. gondii (Mandalasi et al., unpublished). Remarkably, sequences homologous to phyA and gnt1 appear to be represented within a single open reading frame that encodes a two-domain protein (Figure 1B, Figure S1), referred to here as PuPhgt.
The physical association of the predicted prolyl 4(trans)-hydroxylase and αGlcNAc-transferase activities of PuPhgt suggests that they act in concert to modify their substrate, as observed for multidomain lysyl hydroxylase/glycosyltransferase enzymes that modify collagen and collagen-like proteins in mammals and a virus (Luther et al. 2011; Wang et al. 2002). This is relevant to a key question regarding the substrate specificity of these enzymes. There is substantial evidence that Gnt1 and the other glycosyltransferases are absolutely specific for Skp1 in D. discoideum and T. gondii, based on enzyme specificity assays (Zhang et al. 2012), and genetic evidence for the role of Skp1 abundance and its Pro143 residue, as well as the modification enzymes, in O2 sensing in D. discoideum (Wang et al. 2011). However, genetic evidence indicates more severe phenotypes in D. discoideum and T. gondii in phyA-KO vs. gnt1-KO strains (Zhang et al. 2012; Rahman et al. 2016), suggesting that i) hydroxylation alone affects Skp1, ii) additional unknown substrates of PhyA might contribute to O2-sensing or iii) PhyA has a second activity such as binding Skp1. Because of the large phylogenetic distance between P. ultimum and T. gondii or D. discoideum, and the unusual fusion of P. ultimum’s predicted phyA and gnt1 coding sequences, we investigated the predicted enzymatic activity of PuPhgt toward both ectopic and endogenous Skp1s. Our analysis of recombinant proteins confirmed the predicted prolyl hydroxylase and GlcNAc-transferase activities of PuPhgt, and revealed an unexpected species-specific selectivity toward endogenous PuSkp1A relative to PuSkp1B, suggesting that P. ultimum has evolved a mechanism to bypass posttranslational control via Skp1 isoform selection.
Results
Complementation of Dictyostelium mutants with PuPhgt
We first tested whether Puphgt could complement absence of the corresponding genes in D. discoideum. Synthetic FLAG-Puphgt cDNA was expressed in phyA-KO and phyA/gnt1-double KO strains under control of the constitutive actin 15 promoter from an extrachromosomal vector, and Skp1 modification was analyzed by western blotting using isoform specific antibodies (Table I). In phyA− cells (Figure 2, lane 1), Skp1 migrates as a single band that is recognized by pAb UOK87 (panel A), which is selective for unmodified Skp1, and by the isoform nonselective mAb 4E1 (panel C). When FLAG-PuPhgt is expressed (lane 2), as evidenced by western blotting with anti-FLAG (panel E), Skp1 migrates as a doublet when probed with mAb 4E1 (panel C), with only the lower band being reactive with pAb UOK87 (panel A). The more slowly migrating band shows that, once endogenous Skp1 is hydroxylated, it becomes available for further modification by the Skp1 glycosyltransferases resulting in retarded mobility. Thus PuPhgt is likely to modify Skp1 P143 in the same manner as endogenous PhyA, forming a 4(trans)-hydroxyproline (van der Wel et al. 2011). The efficiency of hydroxylation by PuPhgt is evidently lower than that of DdPhyA (Sheikh et al. 2014), which may reflect different thresholds for sensing O2 and αKG, or reduced ability to recognize DdSkp1, which is 49% identical and 66% similar to PuSkp1A (Figure S4).
Table I.
Antibodies employed
| Name | Typea | Immunogen | Specificity | Reference |
|---|---|---|---|---|
| 4E1 | mAb | His10DdSkp1A | All Dd isoforms | Kozarov et al. 1995 |
| UOK75 | pAb | TgSkp1 | All Tg isoforms | Xu et al. 2012 |
| UOK77 | pAb | DdSkp1 | All Dd isoforms | Sheikh et al. 2015 |
| UOK87 | pAb | Dd Pro-peptide | Skp1 | Sheikh et al. 2014 |
| UOK85 | pAb | Dd Hyp-peptide | HO-Skp1 | Sheikh et al. 2014 |
| 1C9 | mAb | Dd GlcNAc-peptide | GlcNAc-Skp1 | Wang et al. 2009 |
| 4H2 | mAb | Dd Hyp-peptide | Skp1, HO-Skp1 | West et al. 2015 |
| M2 | mAb | FLAG epitope | FLAG epitope | Srila and Yamabhai 2013 |
amAb = mouse monoclonal antibody; pAb = rabbit polyclonal antibody.
Fig. 2.
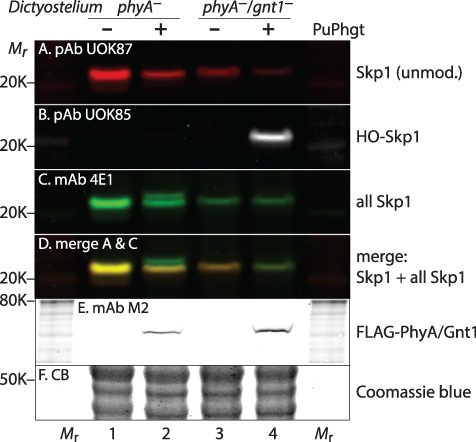
PuPhgt is a Skp1 prolyl hydroxylase. Strains of D. discoideum that are phyA− (lane 1) or phyA−/gnt1− (lane 3) were transfected with synthetic cDNA encoding FLAG-PuPhgt (lanes 2 and 4, respectively), and soluble (S100) extracts from a clone of each transfectant were analyzed for Skp1 modification status using isoform specific antibodies (Table I). (A) Polyclonal antibody (pAb) UOK87, selective for unmodified Skp1. (B) pAb UOK85, specific for prolyl-hydroxylated Skp1 (HO-Skp1). (C) mAb 4E1, which is pan-specific for all glycoforms of Skp1. Skp1 migrates as a doublet in lane 2. (D) A merge of panels A and C, showing that the more slowly migrating band in lane 2 is unreactive with UOK87 or UOK85 as expected for fully glycosylated Skp1 (GGFGGn-Skp1). (E) mAb M2 specific for FLAG-PuPhgt. (F) Coomassie blue stain of gel after western blotting for panel E, confirming similar loading.
To test the activity of PuPhgt to glycosylate HO-Skp1, PuPhgt was expressed in a phyA−/gnt1− double mutant. As expected, the level of unmodified Skp1 was decreased (compare lane 4 with lane 3). However, a second band of slower mobility failed to appear when probed with mAb 4E1. Instead, Skp1 was labeled with pAb UOK85 (panel B), which is selective for HO-Skp1. Thus, PuPhgt was unable to modify HO-Skp1 in such a manner that it could be extended by the other endogenous Skp1 glycosyltransferases (Figure 1A).
Evidence of GlcNAcT activity of PuPhgt in vitro
To examine why PuPhgt did not exhibit expected GlcNAcT activity in D. discoideum cells, FLAG-PuPhgt was purified from D. discoideum using anion exchange chromatography and mAb M2 affinity chromatography, which generated a highly purified preparation of anti-FLAG reactive protein (Figure 3A) at the expected Mr position for PuPhgt (71,400). Activity was initially characterized as an ability to hydrolyze UDP-GlcNAc, its predicted donor substrate, in the absence of acceptor substrate, which is a common characteristic of glycosyltransferases (Sheikh et al. 2017a) including members of the CAZy GT60 family (Heise et al. 2009). As shown in Figure 3B, PuPhgt exhibited robust UDP-GlcNAc hydrolysis activity, as measured by generation of UDP, which was highly specific to UDP-GlcNAc relative to other common sugar nucleotides. Thus failure to allow DdSkp1 glycosylation in vivo was not due to utilization of a different sugar nucleotide that would not be recognized by the next glycosyltransferase (PgtA).
Fig. 3.
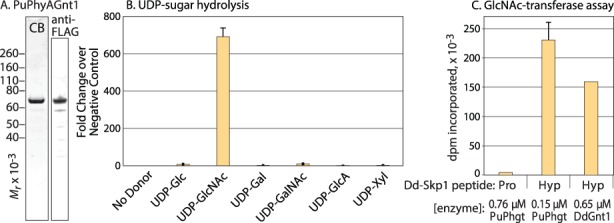
PuPhgt is a GlcNAc-transferase. (A) SDS-PAGE gel of purified FLAG-PuPhgt, after staining with Coomassie Blue, or after western blotting with mAb M2 (anti-FLAG). (B) Sugar nucleotide hydrolysis. Purified PuPhgt was incubated with the indicated sugar nucleotide and, after 17 h, the reaction was quenched and the concentration of UDP generated was measured using the UDP-Glo assay. (C) GlcNAc-transferase assays using DdSkp1 peptides. A total of 1 mM Pro-peptide(133–155) or Hyp-peptide(133–155) was incubated with PuPhgt or DdGnt1 in the presence of 1 μM UDP-[3H] GlcNAc and absence of αKG. Incorporation was measured by liquid scintillation counting of the peptide band excised from an SDS-PAGE gel. Error bars represent standard deviation of three technical replicates of a single trial; similar results were obtained using a separate enzyme preparation.
To test whether PuPhgt was able to transfer GlcNAc to Skp1 from UDP-GlcNAc in vitro, we first utilized a previously described assay based on a 23-mer peptide from DdSkp1 (Teng-umnuay et al. 1999). As shown in Figure 3C, PuPhgt exhibited robust ability to transfer [3H] GlcNAc from UDP-[3H] GlcNAc to the Hyp-form of the peptide, similar to DdGnt1 as a positive control, but not to the Pro-form. This indicated that PuPhgt was indeed capable of GlcNAcylating the Hyp residue of at least an isolated peptide of DdSkp1.
PuPhgt efficiently modifies PuSkp1A, but not PuSkp1B, DdSkp1 or TgSkp1 in vitro
Because of the evolutionary distance between P. ultimum and D. discoideum or T. gondii, we tested the in vitro activity of PuPhgt toward its homologous Skp1. Inspection of the P. ultimum genome revealed two Skp1-like sequences, which are named Skp1A and Skp1B and described in Figures S2 and S3. PuSkp1A is encoded from three exons, whereas PuSkp1B appears to be a single exon, based on est sequences. The proteins are 66% identical and 83% similar to one another at the amino acid level, and each has the equivalent of Pro154 (for T. gondii or Pro143 for D. discoideum). PuSkp1A and PuSkp1B were each expressed as their native predicted sequences from synthetic cDNAs in Escherichia coli, and partially purified over anion exchange and hydrophobic interaction columns (Figure 4A). We compared the substrate activities of the Pythium Skp1’s with equivalent preparations of Skp1 from D. discoideum and T. gondii, and compared the activities of PuPhgt with PhyA and Gnt1 from D. discoideum. While T. gondii expresses only a single Skp1, D. discoideum also expresses a second Skp1, which, however, varies only by a single poorly conserved amino acid (S39 vs. A39) in a disordered region near the N-terminus. This specific variation is not conserved in other cellular slime molds, and there is no evidence for a functional difference in genetic studies (Wang et al., 2011). DdSkp1A was utilized for the in vitro reactions.
Fig. 4.
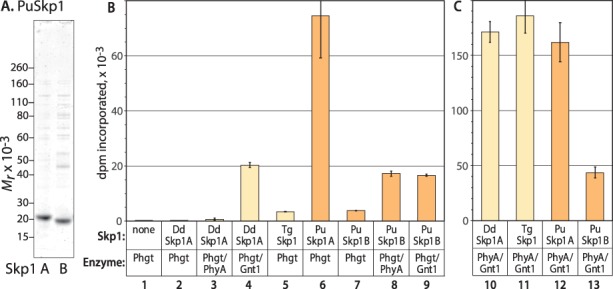
PuPhgt preferentially modifies PuSkp1A relative to PuSkp1B in vitro. (A) The native protein sequences of PuSkp1A and PuSkp1B were expressed in E. coli, purified by conventional chromatographic methods and analyzed by SDS-PAGE with staining by Coomassie blue for total protein. (B) PuPhgt, supplemented as indicated with DdPhyA or DdGnt1, was incubated in the presence of atmospheric O2 and αKG with the indicated type of 1.8 μM Skp1 and 2 μM UDP-[3H] GlcNAc for 2 h. Incorporation of radioactivity into Skp1 was determined after precipitation with trichloroacetic acid. (C) Similarly, a mixture of DdPhyA and DdGnt1 was incubated with the indicated type of Skp1. Error bars represent standard deviation of two technical replicates of a single trial; similar results were obtained using separate protein preparations.
In accord with the complementation studies (Figure 2), PuPhgt was essentially inactive toward DdSkp1A in a tandem reaction that included substrates for both PhyA (αKG, O2) and Gnt1 (UDP-[3H]GlcNAc) and was monitored based on incorporation of 3H into Skp1 (Figure 4B, lane 2). Activity could be partially restored by the addition of DdGnt1 (lane 4) but not DdPhyA (lane 3) to PuPhgt, consistent with the complementation data that showed a lack of Gnt1 activity. The relatively weak P4H activity of PuPhgt toward DdSkp1A (lane 4) vs. PuSkp1A (lane 6) correlates with the incomplete hydroxylation of DdSkp1 in the complementation assay (Figure 2, lane 2). TgSkp1 was also essentially inactive in the tandem assay (lane 5). In comparison, the D. discoideum enzymes were robustly active toward both DdSkp1A and TgSkp1 (Figure 4C, lanes 10 and 11).
In contrast, the homologous substrate PuSkp1A was an excellent substrate for both the P4H and GlcNAcT activities of PuPhgt (Figure 4B, lane 6). Similarly, PuSkp1A was a good substrate for a mixture of DdPhyA and DdGnt1 (Figure 4C, lane 12).
However, PuSkp1B was a poor substrate for PuPhgt in the same in vitro analysis (Figure 4B, lane 7). This occurred despite its substantial activity toward the mixture of DdPhyA and DdGnt1 (Figure 4C, lane 13). To dissect out which activity was deficient, the reactions were supplemented with DdPhyA or DdGnt1. Each supplement incrementally and similarly improved the activity of the combined reaction (lanes 8 and 9), suggesting that PuSkp1B was a weak substrate for each activity.
To confirm these findings, the Skp1 reaction products were analyzed by mass spectrometry. Parallel reactions conducted in the absence of detergent at a PuPhgt:PuSkp1 molar ratio of 1:10 for 4 h were resolved on a C4 nLC column and eluted directly into a Q-Exactive+ Orbitrap MS. As shown by examples in Figure S6A and summarized in Table II, the expected masses of unmodified PuSkp1A and PuSkp1B were observed prior to incubation. The reaction of PuSkp1A with PuPhgt yielded a mass increase corresponding to complete conversion to a singly hydroxylated and GlcNAcylated derivative. In contrast, the reaction of PuSkp1B yielded evidence for partial hydroxylation and no evidence for GlcNAcylation, despite the high relative concentration of enzyme and prolonged reaction time.
Table II.
PuSkp1 Mr measurements
| Monoisotopic mass (MH+) | |||||||
|---|---|---|---|---|---|---|---|
| untreated | 240 min | ||||||
| Expecteda | Obs. | Intens.b | Δppm | Obs. | Intens. | Δppm | |
| Skp1A | 20022.7 | 20022.7 | 3 × 106 | −1.8 | n.d. | ||
| HO-Skp1 | 20038.7 | n.d.c | n.d. | ||||
| Gn-Skp1A | 20241.7 | n.d. | 20239.7 | 2 × 106 | −96 | ||
| Skp1B | 19703.9 | 19703.8 | 2 × 105 | −0.9 | 19703.8 | 2 × 105 | −0.8 |
| HO-Skp1 | 19719.9 | n.d. | 19718.0 | 3 × 105 | −95 | ||
| Gn-Skp1B | 19922.9 | n.d. | n.d. | ||||
aFull length minus N-terminal Met.
bIntensity, from Xtract algorithm.
cNot detected.
Interdependence of the P4H and GlcNAcT activities of PuPhgt
Time course studies were analyzed using isoform-specific Abs to examine the relative rates of the separate P4H and GlcNAcT reactions toward PuSkp1A. At an enzyme: PuSkp1A ratio of 1:10 in the presence of substrates for both enzyme activities, the great majority of Skp1 was converted to Gn-Skp1 by 60 min (Figure 5A). In the absence of UDP-GlcNAc, nearly all of PuSkp1A was converted to HO-Skp1 within this time frame. The isoform specific nature of Ab recognition was conserved between the species, providing additional evidence for the formation of a GlcNAcα1–4(trans)-Pro-Skp1 linkage as observed in DdSkp1 and TgSkp1.
Fig. 5.
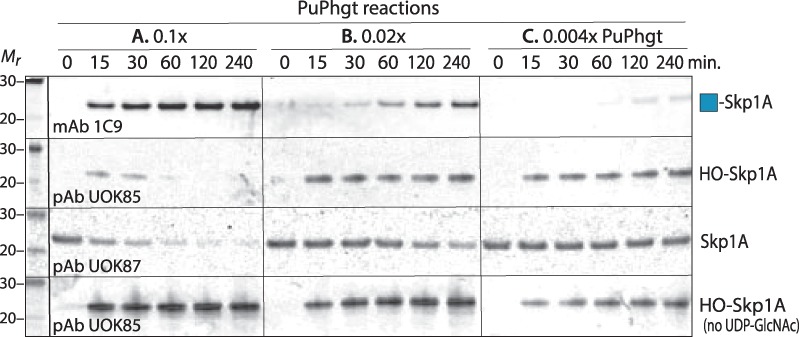
Hydroxylation and GlcNAcylation of PuSkp1A by PuPhgt are uncoupled in vitro. PuSkp1A (5 μM) was incubated with purified FLAG-PuPhgt at the indicated molar ratios (panels A–C) in the presence of atmospheric O2, αKG, and ±UDP-GlcNAc, without catalase, for the indicated periods of time. The reaction products (containing 0.1 μg Skp1) were subjected to SDS-PAGE and western blotting with the indicated Abs specific for Gn-Skp1 (top row), HO-Skp1 (second and bottom rows) and unmodified Skp1 (third row). Reactions in the bottom row were conducted in the absence of UDP-GlcNAc to assay P4H activity alone. Data are representative of two independent trials.
At a lower enzyme: Skp1 ratio of 0.02 (panel B), it was evident that formation of Gn-Skp1A lagged substantially behind formation of HO-Skp1A. This difference was even more evident at an enzyme: Skp1 ratio of 1:250 (panel C). At this ratio, progress of the P4H reaction was similar regardless of the presence of UDP-GlcNAc to support the GlcNAcT reaction. These observations are most consistent with a model in which the enzymatic reactions are independent, and that progress of the GlcNAcT reaction depends on accumulation of the product of the first reaction, to sufficient concentrations to drive it.
Discussion
PuPhgt is a bifunctional enzyme that combines, in one protein, the first two steps of Skp1 modification that were previously shown to involve separate proteins in the social amoeba D. discoideum and the apicomplexan parasite T. gondii. The modification generated by PuPhgt appears to be the same GlcNAcα1-4(trans)-Hyp structure catalyzed by the separate enzyme proteins in the other protists, based on i) the ability of DdGnt1 to modify the product of the PuPhgt P4H reaction, ii) cross-reactivity of the isoform specific Abs generated against D. discoideum HO-Skp1 and Gn-Skp1 with the products of the respective PuPhgt reactions, iii) specificity of PuPhgt’s GlcNAcT activity toward the DdSkp1 Hyp-peptide and its utilization of UDP-GlcNAc and iv) the property of all known CAZy GT60 and related GT27 family members to be retaining GTs.
The findings confirm predictions from genomics studies that the Skp1 modification pathway is present in oomycetes, which evolved within the stramenopiles that are distantly related to the alveolates, of which apicomplexa including T. gondii are members, as part of the larger SAR (stramenopiles/alveolates/rhizaria) group (Figure 6). The P. ultimum genome contains coding sequences related to each of the four other Skp1 glycosyltransferase activities that assemble a linear pentasaccharide on TgSkp1 (Figure 1A), including the bifunctional PgtA, Glt1 and Gat1 (unpublished studies). Numerous other oomycetes, including Phytophthora spp., and other stramenopiles, including diatoms and brown algae, also possess Skp1 and Skp1 modification gene candidates (asterisked in Figure 6). These genes potentially contribute to O2- or metabolite sensing, physiological processes that have received negligible attention in these organisms.
Fig. 6.
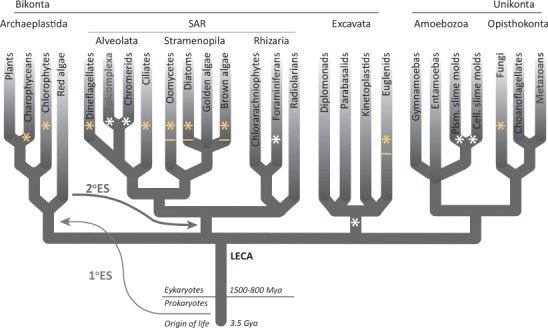
Distribution of separate and joined PhyA- and Gnt1-like sequences in eukaryotic phylogeny. In this tree, adapted from White and Suvorova (2018) with permission of the publisher, phylogenetic groups with at least one member possessing genomic sequences predicted to encode PhyA and Gnt1 are marked with an asterisk. Those groups containing genomes with Phgt-like sequences, i.e., PhyA- and Gnt1-encoded within the same open reading frame, are asterisked in yellow. See Figure S5 for species names. Those groups that include species with a second Skp1 that is predicted to be resistant to modification are underlined. See Figure S4 for species names. LECA refers to last eukaryotic common ancestor; ES refers to endosymbiotic incorporations.
P. ultimum expresses two Skp1 genes, each possessing the equivalent of the target Pro (Figures S2–S4). The second Skp1, Skp1B, is however a very poor substrate for PuPhgt in vitro. This is despite a rather high degree of sequence similarity with PuSkp1A (Figure S4), and its being a substrate for DdPhyA and DdGnt1 (Figure 4, lane 13). Supplementing the reaction of PuSkp1B and PuPhgt with either DdPhyA or DdGnt1 indicated that PuSkp1B was a weak substrate for both activities (Figure 4, lanes 7–9). Skp1’s from D. discoideum and T. gondii were also poor substrates for PuPhgt, especially its GlcNAcT activity (Figures 2, 4). This selectivity was unexpected, because DdPhyA and TgPhyA, and DdGnt1 and TgGnt1, readily modify each other’s Skp1’s in vitro and in vivo, and DdPhyA efficiently hydroxylates Skp1A from C. elegans and even a mutant human Skp1 in which the equivalent of Pro143 is restored (van der Wel et al. 2011). In addition, D. discoideum and T. gondii PhyA and Gnt1 genes readily modify PuSkp1A. Thus the specificity of PuPhgt for PuSkp1A over PuSkp1B is a property of both the enzyme and PuSkp1B. However, we cannot currently exclude the possibility that PuSkp1B can be modified in vivo as a result of, e.g., a posttranslational modification or another protein factor. Since DdSkp1 readily spontaneously refolds after denaturation (Sheikh et al. 2014), it is less likely that PuSkp1B is simply misfolded when expressed in E. coli.
To address whether the expression of a second poorly modifiable Skp1 is a conserved phenomenon, we queried the genomes of a range of protists for related sequences using BLASTp and tBLASTn algorithms. As shown by the alignments of Figure S4 and elsewhere (Sheikh et al. 2017b), the occurrence of two closely related Skp1 genes is a common occurrence and occur in divergent clades of protists (underlined asterisks in Figure 6). Several differences occur in the vicinity of the target Pro residue that are expected to contribute to enzyme recognition and F-box binding. In other oomycetes, including several Phytophthora spp., and the diatom Fragilariopsis cylindrus, the second Skp1 lacks the target Pro so obviously cannot be modified. In other stramenopiles, including other diatoms and a phaeophyte, as well as representatives from other groups including cryptophytes (Guillardia theta), excavates (Naegleria gruberi), and even distantly related social amoebae (Sheikh et al. 2017b), substitutions occur in highly conserved positions near the target Pro that are similar to the differences between PuSkp1B and PuSkp1A. In contrast, the two Skp1’s in D. discoideum and closely related social amoebae, which vary only by a single (or few) changes in a poorly conserved region of Skp1 associated with Cul1-binding, exhibit no indication of different properties (West et al. 1997; Wang et al. 2011). Based on the example of PuSkp1B, it is tempting to speculate that various evolutionary events have converged to render the second Skp1 in these highly varied protists to be resistant to regulation by O2-dependent glycosylation. Expression of a nonmodifiable Skp1 may represent a general bypass mechanism for Skp1 regulation of all FBPs, or the nonmodifiable Skp1 might be dedicated to a subset of FBPs that are therefore not subject to this regulation.
The evolutionary origin of the fusion of the P4H and GlcNAcT activities appears to be ancient given its widespread occurrence across protist phylogeny (Figure S5), where it is found not only in other stramenopiles but also alveolates, cryptophytes, haptophytes, green algae and excavates (yellow asterisks in Figure 6). Evidently the enzymes do not need to act in concerted fashion because they are often expressed as separate proteins and, in D. discoideum and T. gondii, there is no indication that they physically associate. Our analysis of the relative rates of sequential hydroxylation and GlcNAcylation of PuSkp1A by PuPhgt in vitro are most consistent with a model in which two enzyme activities act independently. Time course studies suggest that the reaction occurs rapidly and the GlcNAcT reaction occurs slowly (Figure 5), as expected if the P4H product dissociates from the enzyme and must build up in concentration to drive the GlcNAcT reaction mediated by the other domain. Thus there is no evidence that PuPhgt acts processively to accomplish both reactions, in contrast to the next enzyme in the pathway, which in D. discoideum processively mediates the successive additions of βGal and αFuc in vitro (van der Wel et al. 2002a). Nevertheless, the physical linkage evidently offers a selective advantage for P. ultimum, and can be expected to support a higher likelihood of GlcNAcylation of hydroxylated Skp1 in the crowded environment of the cytoplasm or nucleus where these proteins reside, but also allow for independent regulation of the GlcNAcylation reaction. Follow-on GlcNAcylation of the P4H product implies that both enzymes are specific for the same original substrate, PuSkp1A. This in turn supports the model that the entire modification pathway, including PuPhgt, acts solely on Skp1 in the cytoplasm, which is consistent with the complementary O2-sensing effects that result from genetic modulation of Skp1 levels in Dictyostelium (Wang et al. 2011).
Materials and methods
Bioinformatic studies
Searches for homologs of D. discoideum PhyA, Gnt1 and Skp1 were initially conducted by applying the BLASTp algorithm to the nonredundant NCBI database (December 2017), and gene candidates were further assembled using tBLASTn in NCBI Whole Genome Shotgun databases. Multiple examples of predicted open reading frames and protein models containing sequences homologous to both PhyA and Gnt1 were found, as partially enumerated in Figure S5. The example from P. ultimum is represented by the protein model PYU1_T004073 at www.pythiumdb.org and is referred to as Phgt; the corresponding genomic sequence was found in DNA contig ADO01001287 at NCBI (Figure S1). Two highly similar Skp1-like sequences, referred to as Skp1A and Skp1B, were found in P. ultimum. Skp1A is from protein model PYU1_T013675 at www.pythiumdb.org; the nucleotide sequence is from genomic DNA ADOS01001474.1 (Figure S2). Skp1B is from protein model PYU1_T011629 at www.pythiumdb.org; the nucleotide sequence is from genomic DNA ADOS01001682.1 at NCBI wgs database (Figure S3). Other Skp1-like sequences from organisms with Phgt-like sequences are enumerated together with reference Skp1 sequences in Figure S4.
Genetic complementation studies
The predicted cDNA for PuPhgt was codon optimized for recombinant expression in E. coli and D. discoideum, and synthesized by GenScript (Figure S1). PuPhgt cDNA was cloned into the BglII and SpeI sites of D. discoideum transient expression vector pDM320 (Veltman et al. 2009), which was expected to yield an N-terminally FLAG-tagged fusion protein of Mr 71,367 (average mass). The plasmid was electroporated into D. discoideum strains HW288 (phyA−/bsR) (van der Wel et al. 2005) and HW512 (phyA−/gnt1−/bsR/hygR) (Zhang et al. 2012), and grown in HL-5 axenic media in the presence of 120 μg/mL G418 without cloning as described (Sheikh et al. 2014). Cytosolic fractions (S100) were prepared by filter lysis and centrifugation at 100,000 × g for 60 min, and subjected to SDS-PAGE followed by western blotting with anti-Skp1 Abs to assess the modification status of Skp1 (Sheikh et al. 2014).
Recombinant protein expression and purification
The Phgt expression plasmid (pDM320-PuPhgt) was electroporated into D. discoideum strain Ax3 as above. FLAG-PuPhgt was purified from an S100 fraction from a 4-l culture by successive chromatography DEAE-FastFlow Sepharose and immobilized M2-antibody (anti-FLAG) columns, exactly as described previously for DdPgtA (Sheikh et al. 2014). FLAG-PuPhgt elution was monitored by western blotting with mAb M2, and purity was assessed by staining SDS-PAGE gels with Coomassie blue-R250. Attempts to express FLAG-PuPhgt in E. coli, from a pET15TEV vector (Bochkareva et al. 1998), generated abundant protein that was, however, insoluble.
The predicted cDNA for P. ultimum Skp1A (PuSkp1A) was codon optimized for recombinant expression in E. coli, synthesized by GenScript (Figure S2), cloned into the NcoI and SpeI sites of pET15TEV and expressed in E. coli Gold cells at 22°C after overnight induction with 0.5 mM IPTG in LB media supplemented with 100 μg/mL ampicillin. The same method was used for PuSkp1B except that the synthetic cDNA (Figure S3) was cloned into the BspH1 and SpeI sites of pET15TEV. The expressed Skp1 proteins were harvested and purified by successive chromatography on DEAE-FastFlow Sepharose and Phenyl-High-Sub-FastFlow Sepharose columns, as described (Sheikh et al. 2014). PuSkp1A was detected using mAb 4H2, which was generated against an unmodified D. discoideum Skp1-peptide (selective for unmodified and hydroxylated isoforms; West et al. 2015); PuSkp1B was detected using pAb UOK75 generated against unmodified T. gondii Skp1 (pan-specific against all isoforms; Xu et al. 2012). See Table I for antibodies employed. Phenyl fractions containing PuSkp1 were concentrated by centrifugal ultrafiltration and buffer exchanged using PD-10 desalting columns (GE Healthcare) into 25 mM Tris-HCl (pH 7.5), 2 mM DTT, 5 mM MgCl2, 10 μg/mL aprotinin and leupeptin. Purity was assessed by staining SDS-PAGE gels as above.
DdSkp1A and TgSkp1 were expressed as native sequences in E. coli and purified as previously described (Sheikh et al. 2014; Xu et al. 2012). D. discoideum expresses two Skp1 isoforms, from separate genes (West et al. 1997) that are functionally indistinguishable (Wang et al. 2011). DdSkp1A, which differs from DdSkp1B by only a single poorly conserved amino acid (S39A), is analyzed here.
Sugar nucleotide hydrolysis assays
FLAG-PuPhgt was incubated in the presence of 50 mM HEPES-NaOH, pH 7.4, 50 mM NaCl, 1 mM MgCl2, 1 mM MnCl2, 1 mM DTT, 50 μM UDP-sugar for 17 h at 37°C, and generation of UDP was assayed using the UDP-Glo assay (Promega) as previously described (Sheikh et al. 2017a).
Glycosyltransferase assay
GlcNAcT activity toward peptides was assayed as previously described (Teng-umnuay et al. 1999). Briefly, a preparation of purified FLAG-PuPhgt (0.15–0.76 μM) lacking divalent cations was incubated in the presence of 1 mM DdSkp1 peptide(133–155) or Pro-peptide(133–155), and 1 μM UDP-3H-GlcNAc (81,400 dpm/pmol), in 35 μL of 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 0.2% Tween-20, 0.3–11 mM MgCl2 for 2 h at 29°C, and an additional 2 h at 37°C. Incorporation was assayed by resolving the reaction mixture on an SDS-PAGE gel, excising the Coomassie blue stained peptide from the gel, and measuring the radioactivity in a liquid scintillation counter, as described (Rahman et al. 2016).
Tandem prolyl hydroxylase/GlcNAc-transferase assay
The one-step tandem assay using full-length Skp1 as the substrate was conducted as previously described (van der Wel et al. 2011). Briefly, the reaction buffer consisted of 30–50 μL of 50 mM Tris-HCl (pH 7.5), 2.5–5 mM DTT, 5 mM MgCl2, ±2 mM MnCl2, 0.1% Tween-20, 5 μM FeSO4, 1 mM ascorbate, 1.5 mM αKG, 10 μg/mL aprotinin and 10 μg/mL leupeptin, ±0.2 mg/mL catalase. As indicated, reactions contained 0.045–0.72 μM PuPhgt, 1–5.4 μM DdSkp1A, TgSkp1, PuSkp1A or PuSkp1B and 1–2 μM UDP-[3H] GlcNAc or 1–50 μM unlabeled UDP-GlcNAc, and were incubated for 0–18 h at 29°C. UDP-GlcNAc, MgCl2 and MnCl2 were omitted when assaying P4H activity alone. The reaction product was detected based on western blotting with isoform-specific anti-Skp1 Abs (Sheikh et al. 2014) or, for reactions containing UDP-[3H] GlcNAc, trichloroacetic acid precipitation followed by liquid scintillation counting.
Mass spectrometry
Mass analysis of full length Skp1 was performed approximately as described (Xu et al. 2018). Samples were prepared as above, except in the absence of Tween-20, with 0.5 μM PuPhgt and 5 μM Skp1, buffer exchanged into 2% (v/v) acetonitrile using a 10-kDa cut-off Amicon Ultra-05 centrifugal filter (UFC901096, Millipore), diluted to 20 ng/μL Skp1 and brought to 0.05% (v/v) trifluoroacetic acid. A total of 5 μL (100 ng of Skp1) was injected into a Acclaim PepMap C4 trap cartridge (300 μm × 5 mm) equilibrated with 0.05% trifluoroacetic acid, 2% acetonitrile, ramped up with an increasing gradient to 0.1% (v/v) formic acid, 25% acetonitrile and introduced into a Acclaim PepMap analytical C4 column (75 μm × 15 cm, 5 μm pore size) maintained at 35°C in an Ultimate 3000 RSLC (ThermoFisher Scientific/Dionex), which was coupled to a ThermoScientific QE+ Orbitrap mass spectrometer. After equilibrating the analytical column in 98% LC-MS Buffer A (water, 0.1% formic acid) for 10 min and 6-min ramp up to 27% LC-MS Buffer B (90% acetonitrile, 0.1% formic acid), separation was achieved using a linear gradient from 27% to 98% Buffer B over 20 min at a flow rate of 300 nl/min. The column was regenerated after each run by maintaining it at 98% Buffer B for 5 min. The effluent was introduced into the mass spectrometer by nanospray ionization in positive ion mode via a stainless-steel emitter with spray voltage set to 1.9 k, capillary temperature set at 250°C and probe heater temperature set at 350°C. The MS method consisted of collecting Full ITMS (MS1) scans (400–2000 m/z) at 140,000 resolution in intact protein mode (default gas P set to 0.2). Skp1 species eluting between 18 and 20 min (60–68% acetonitrile) were processed with Xcalibur Xtract deconvolution software to generate monoisotopic masses from the multicharged, protonated ion series. Parameters including fit, remainder and signal to noise were varied to ensure detection of all products. Average masses were calculated manually for verification. Ion intensities of isoforms in mixtures were generated by the deconvolution software.
Supplementary Material
Acknowledgments
We are grateful M. Osman Sheikh for conducting the UDP-Glo assays, Hyun Woo Kim for protein purification, and Julia Clark for assistance in preparing the expression plasmids.
Funding
National Institutes of Health (grants R01-GM084383 and R01-GM037539).
Conflict of interest statement
None declared.
Abbreviations
αKG, α-ketoglutarate;
FBP, F-box protein;
GlcNAcT, polypeptide UDP-GlcNAc:Hyp-Skp1 GlcNAc-transferase;
GT, glycosyltransferase;
Hyp, (2S,4R)-4-hydroxy-l-proline;
mAb, monoclonal antibody;
P4H, prolyl 4-hydroxylase;
pAb, polyclonal antibody;
SCF, Skp1/Cullin-1/F-box protein subcomplex of E3 Cullin-RING-1 ubiquitin ligases
References
- Blader IJ, Coleman BI, Chen CT, Gubbels MJ. 2015. Lytic cycle of Toxoplasma gondii: 15 years later. Annu Rev Microbiol. 69:463–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E, Frappier L, Edwards AM, Bochkarev A. 1998. The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J Biol Chem. 273:3932–3936. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 78:399–434. [DOI] [PubMed] [Google Scholar]
- Heise N, Singh D, van der Wel H, Sassi SO, Johnson JM, Feasley CL, Koeller CM, Previato JO, Mendonça-Previato L, West CM. 2009. Molecular analysis of a UDP-GlcNAc:polypeptide alpha-N-acetylglucosaminyltransferase implicated in the initiation of mucin-type O-glycosylation in Trypanosoma cruzi. Glycobiology. 19:918–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MS, Leissing TM, Chowdhury R, Hopkinson RJ, Schofield CJ. 2018. 2-oxoglutarate-dependent oxygenases. Annu Rev Biochem. 87:585–620. [DOI] [PubMed] [Google Scholar]
- Kozarov E, van der Wel H, Field M, Gritzali M, Brown RD Jr, West CM. 1995. Characterization of FP21, a cytosolic glycoprotein from Dictyostelium. J Biol Chem. 270:3022–3030. [DOI] [PubMed] [Google Scholar]
- Lévesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, Raffaele S, Robideau GP, Thines M, Win J et al. 2010. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 11:R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther KB, Hülsmeier AJ, Schegg B, Deuber SA, Raoult D, Hennet T. 2011. Mimivirus collagen is modified by bifunctional lysyl hydroxylase and glycosyltransferase enzyme. J Biol Chem. 286:43701–43709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman K, Zhao P, Mandalasi M, van der Wel H, Wells L, Blader IJ, West CM. 2016. The E3 ubiquitin ligase adaptor protein Skp1 is glycosylated by an evolutionarily conserved pathway that regulates protist growth and development. J Biol Chem. 291:4268–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman K, Mandalasi M, Zhao P, Sheikh MO, Taujale R, Kim HW, van der Wel H, Matta K, Kannan N, Glushka JN et al. 2017. Characterization of a cytoplasmic glucosyltransferase that extends the core trisaccharide of the Toxoplasma Skp1 E3 ubiquitin ligase subunit. J Biol Chem. 292:18644–18659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh MO, Schafer CM, Powell JT, Rodgers KK, Mooers BH, West CM. 2014. Glycosylation of Skp1 affects its conformation and promotes binding to a model F-box protein. Biochemistry. 53:1657–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh MO, Xu Y, van der Wel H, Walden P, Hartson SD, West CM. 2015. Glycosylation of Skp1 promotes formation of Skp1/Cullin-1/F-box protein complexes in Dictyostelium. Mol Cell Proteomics. 14:66–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh MO, Halmo SM, Patel S, Middleton D, Takeuchi H, Schafer CM, West CM, Haltiwanger RS, Avci FY, Moremen KW et al. 2017a. Rapid screening of sugar-nucleotide donor specificities of putative glycosyltransferases. Glycobiology. 27:206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh MO, Thieker D, Chalmers G, Schafer CM, Ishihara M, Azadi P, Woods RJ, Glushka JN, Bendiak B, Prestegard JH et al. 2017b. O2 sensing–associated glycosylation exposes the F-box–combining site of the Dictyostelium Skp1 subunit in E3 ubiquitin ligases. J Biol Chem. 292:18897–18915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srila W, Yamabhai M. 2013. Identification of amino acid residues responsible for the binding to anti-FLAG™ M2 antibody using a phage display combinatorial peptide library. Appl Biochem Biotechnol. 171:583–589. [DOI] [PubMed] [Google Scholar]
- Teng-umnuay P, van der Wel H, West CM. 1999. Identification of a UDP-GlcNAc:Skp1-hydroxyproline GlcNAc-transferase in the cytoplasm of Dictyostelium. J Biol Chem. 274:36392–36402. [DOI] [PubMed] [Google Scholar]
- van der Wel H, Fisher SZ, West CM. 2002a. A bifunctional diglycosyltransferase forms the Fucalpha1,2Galbeta1,3-disaccharide on Skp1 in the cytoplasm of Dictyostelium. J Biol Chem. 277:46527–46534. [DOI] [PubMed] [Google Scholar]
- van der Wel H, Morris HR, Panico M, Paxton T, Dell A, Kaplan L, West CM. 2002b. Molecular cloning and expression of a UDP-N-acetylglucosamine (GlcNAc):Hydroxyproline polypeptide GlcNAc-transferase that modifies Skp1 in the cytoplasm of Dictyostelium. J Biol Chem. 277:46328–46337. [DOI] [PubMed] [Google Scholar]
- van der Wel H, Ercan A, West CM. 2005. The Skp1 prolyl hydroxylase from Dictyostelium is related to the hypoxia-inducible factor-alpha class of animal prolyl 4-hydroxylases. J Biol Chem. 280:14645–14655. [DOI] [PubMed] [Google Scholar]
- van der Wel H, Johnson JM, Xu YC, Karunaratne CV, Wilson KD, Vohra Y, Boons GJ, Taylor CM, Bendiak B, West CM. 2011. Requirements for Skp1 processing by cytosolic prolyl 4(trans)-hydroxylase and alpha-N-acetylglucosaminyltransferase enzymes involved in O2 signaling in Dictyostelium. Biochemistry. 50:1700–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman DM, Akar G, Bosgraaf L, Van Haastert PJM. 2009. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid. 61:110–118. [DOI] [PubMed] [Google Scholar]
- Wang C, Risteli M, Heikkinen J, Hussa AK, Uitto L, Myllyla R. 2002. Identification of amino acids important for the catalytic activity of the collagen glucosyltransferase associated with the multifunctional lysyl hydroxylase 3 (LH3). J Biol Chem. 277:18568–18573. [DOI] [PubMed] [Google Scholar]
- Wang ZA, van der Wel H, Vohra Y, Ercan A, Feasley CL, Buskas T, Boons G-J, West CM. 2009. Role of a cytoplasmic dual-function glycosyltransferase in O2-regulation of development in Dictyostelium. J Biol Chem. 284:28896–28904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZA, Singh D, van der Wel H, West CM. 2011. Prolyl hydroxylation- and glycosylation-dependent functions of Skp1 in O2-regulated development of Dictyostelium. Dev Biol. 349:283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West CM, Kim HW. 2019. Nucleocytoplasmic O-glycosylation in protists. Curr Opin Struct Biol. 56:204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West CM, Kozarov E, Teng-umnuay P. 1997. The cytosolic glycoprotein FP21 of Dictyostelium discoideum is encoded by two genes resulting in a polymorphism at a single amino acid position. Gene. 200:1–10. [DOI] [PubMed] [Google Scholar]
- West CM, van der Wel H, Wang ZA. 2007. Prolyl 4-hydroxylase-1 mediates O2 signaling during development of Dictyostelium. Development. 134:3349–3358. [DOI] [PubMed] [Google Scholar]
- West CM, Blader IJ. 2015. Oxygen sensing by protozoans: How they catch their breath. Curr Opin Microbiol. 26:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West CM, van der Wel H, Chinoy Z, Boons GJ, Gauthier TJ, Taylor CM, Xu Y. 2015. Generating isoform-specific antibodies: lessons from the nucleocytoplasmic glycoprotein Skp1 In: Taniguchi N, Endo T, Hart G, Seeberger P, Wong C-H, editors. Glycoscience: Biology and Medicine. Japan: Springer; p. 927–934. [Google Scholar]
- West CM, Hart GW. 2017. Nucleocytoplasmic glycosylation In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH et al., editors. Essentials of Glycobiology [Internet], 3rd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; Chapter 18. [Google Scholar]
- White MW, Suvorova ES. 2018. Apicomplexa cell cycles: Something old, borrowed, lost, and new. Trends Parasitol. 34:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems AR, Schwab M, Tyers M. 2004. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 1695:133–170. [DOI] [PubMed] [Google Scholar]
- Xu Y, Brown KM, Wang ZA, van der Wel H, Teygong C, Zhang D, Blader IJ, West CM. 2012. The Skp1 protein from Toxoplasma is modified by a cytoplasmic prolyl 4-hydroxylase associated with oxygen sensing in the social amoeba Dictyostelium. J Biol Chem. 287:25098–25110PMC3408163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Eletsky A, Sheikh MO, Prestegard JH, West CM. 2018. Glycosylation promotes the random coil to helix transition in a region of a protist Skp1 associated with F-box binding. Biochemistry. 57:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, van der Wel H, Johnson JM, West CM. 2012. Skp1 prolyl 4-hydroxylase of Dictyostelium mediates glycosylation-independent and -dependent responses to O2 without affecting Skp1 stability. J Biol Chem. 287:2006–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Shabek N. 2017. Ubiquitin ligases: Structure, function, and regulation. Annu Rev Biochem. 86:14.1–14.29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


