Abstract
Histone methylation can occur at various sites in histone proteins, primarily on lysine and arginine residues, and it can be governed by multiple positive and negative regulators, even at a single site, to either activate or repress transcription. It is now apparent that histone methylation is critical for almost all stages of development, and its proper regulation is essential for ensuring the coordinated expression of gene networks that govern pluripotency, body patterning and differentiation along appropriate lineages and organogenesis. Notably, developmental histone methylation is highly dynamic. Early embryonic systems display unique histone methylation patterns, prominently including the presence of bivalent (both gene-activating and gene-repressive) marks at lineage-specific genes that resolve to monovalent marks during differentiation, which ensures that appropriate genes are expressed in each tissue type. Studies of the effects of methylation on embryonic stem cell pluripotency and differentiation have helped to elucidate the developmental roles of histone methylation. It has been revealed that methylation and demethylation of both activating and repressive marks are essential for establishing embryonic and extra-embryonic lineages, for ensuring gene dosage compensation via genomic imprinting and for establishing body patterning via HOX gene regulation. Not surprisingly, aberrant methylation during embryogenesis can lead to defects in body patterning and in the development of specific organs. Human genetic disorders arising from mutations in histone methylation regulators have revealed their important roles in the developing skeletal and nervous systems, and they highlight the overlapping and unique roles of different patterns of methylation in ensuring proper development.
DNA is packaged inside eukaryotic nuclei by being wrapped around histone proteins, and this assembly of DNA and histones, together with associated non-histone proteins and RNA, comprises chromatin. DNA and histones can be modified by the attachment or removal of small chemical groups such as methyl or acetyl, which can regulate gene activation or repression. During development, these modification marks control the recruitment of transcription factors and/or RNA polymerase to ensure the proper expression of a highly orchestrated set of gene networks, as cells transition from the pluripotent state through multiple progenitor states to their final differentiated cell fate. Errors in establishing or maintaining proper chromatin modifications are often lethal during embryogenesis. Histone methylation has emerged as a particularly important modification during development, one involved in both gene activation and repression. Although much progress has been made in understanding the multiple roles of histone methylation during development, many of the precise mechanisms by which histone methylation regulates developmental events in response to intracellular and extracellular signals remain incompletely understood.
In this Review, we discuss the effects of histone methylation on gene activity and the factors that regulate histone methylation during development (for a discussion of DNA methylation, see REF.1). The overall effects of histone methylation regulators on different stages of embryogenesis and their roles in promoting the development of specific organ systems are reviewed. We also cover how histone methylation affects genomic imprinting and the regulation of HOX genes. Embryonic stem cells (ESCs) have provided valuable models for studying the effects of histone methylation on development, and the roles of different histone methylation regulators in promoting the maintenance of pluripotency and in driving differentiation are discussed. Finally, we discuss human genetic disorders caused by mutations in histone methylation regulators, which provide further insight into the critical roles of histone methylation in organismal development.
Histone methylation and gene activity
Early models of chromatin function hypothesized a histone ʻcodeʼ or ʻlanguageʼ whereby combinations of different histone modifications — occurring either sequentially or simultaneously — would determine the activity of the associated gene2. Genomic techniques held promise for cracking the code by surveying a large number of modifications in a large number of contexts. Although a one-to-one correspondence between modifications and gene expression did not emerge, such studies3–6 established general themes for the effects of histone modifications on gene expression.
The methylation of proteins involves the attachment of a methyl group to nitrogen atoms in amino acid side chains and/or at the amino termini. In histones, lysine (Lys or K) and arginine (Arg or R) residues serve as the most common acceptor sites of methylation marks, which have varying effects on gene activity depending on the specific residues that are modified, the degree and pattern of methylation, and the genomic context in which the methylation occurs (that is, the exact location of the modified nucleosome in the genome) (FIG. 1). Histone H3 is the primary site of histone methylation, although the other core histones display methylations as well.
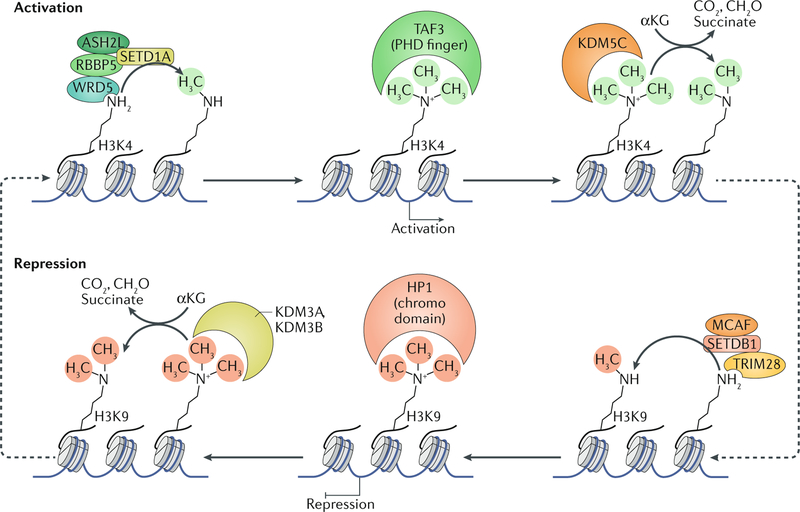
Fig. 1 |. Examples of regulation of gene expression by histone methyltransferases and demethylases.
SETD1A methyltransferase complex (comprising SETD1A (a catalytic SET-domain subunit), together with the binding partners ASH2L, RBBP5, WDR5 and other complex-specific subunits not shown) deposits the gene-activating H3 Lys4 tri-methyl (H3K4me3) mark at the promoters of various genes. H3K4me3 is recognized by PHD finger domains in proteins such as TAF3, which bind to methylated Lys. Gene activation can be reversed through the removal of this modification by the demethylase KDM5C, which utilizes α-ketoglutarate (αKG) as a cofactor. Gene-repressive states can be established by the deposition of H3K9me3 by the SETDB1 histone methyltransferase complex (including the catalytic subunit SETDB1 together with a regulator, MCAF (also known as ATF7IP) and a reader protein, TRIM28). H3K9me3 is recognized by the chromodomain in HP1 proteins and can be removed by the KDM3A and/or KDM3B demethylase in the presence of αKG as a cofactor, to allow for gene activation.
Histone Lys methylation can exist in one of three states: mono-, di- or tri-methylation. Di- and tri-methylation at H3K4, H3K36 and H3K79 are typically gene-activating, with H3K4 tri-methylation (H3K4me3) marking promoters3,7,8, and H3K36 and H3K79 methylations occurring primarily over gene bodies9,10. Mono-methylation of H3K4 is an activating mark unique to enhancers11. H3K9 and H3K27 methylations are generally gene-repressive3,8 but serve unique functions. H3K27me3 is considered easily reversible12 and marks dynamically regulated genes, and thus is especially important in development, when genes need to be switched on and off in a highly dynamic fashion in accordance with developmental signals. H3K9me3 is characteristic of heterochromatin13–15, whereas H3K9me2 is found more commonly at silent or lowly expressed genes in euchromatin13,15.
Generally, methylations at Arg show a greater complexity than do those at Lys, owing to the multiple nitrogen atoms in Arg. Three major forms of methyl-Arg have been identified in mammals, of which omega-NG,N´G-symmetric di-methyl-Arg (SDMA) is found on a small percentage of nuclear and cytoplasmic proteins, whereas omega-NG-mono-methyl-Arg (MMA) and asymmetric di-methyl-Arg (ADMA) are more ubiquitous16,17 (for an expanded discussion of Arg protein methylation, see REF.18). On mammalian histones, MMA and ADMA are the prevailing Arg methylations at sites H3R2, H3R17, H3R26 and H4R3 (REF.17). The association of histone Arg methylation marks with gene expression is poorly understood. Symmetric H3R8me2 (H3R8me2s) and H4R3me2s are generally associated with transcription repression19, but emerging evidence suggests that these modifications might influence certain genomic loci differentially, with both activating and neutral effects of these methylations in specific settings20,21.
Although many other aspects of chromatin structure contribute to its cumulative gene-activating or gene-repressive properties22–24, the genomic studies mentioned above have established histone methylations as important activating and repressing chromatin marks (FIG. 1).
Histone methylation regulators
Histone modifications are regulated by chromatin ʻwritersʼ (methyltransferases, for methylation), which add modifications; ʻerasersʼ (demethylases, for methylation), which remove modifications; and ʻreadersʼ (for example, chromodomain and bromodomain proteins for methylation), which recognize modifications and influence gene expression; all of these have important roles in governing the modifications present at each genetic locus and for their translation into gene-activating or gene-repressing events (FIG. 1). For a comprehensive review of all known histone modifications and their regulators, see REF.25.
Multiple histone methyltransferases with varying activities have now been identified. Some generate multiple degrees or types of methylation, whereas others catalyse a single species. The catalytic activity of histone lysine methyltransferases often resides in the SET domain, first identified in the human and mouse H3K9 methyltransferase SUV39H1 (REF.14), and subsequently in many other histone Lys methyltransferases26–30. Notably, the H3K79 methyltransferase DOT1 and its homologues are the only histone Lys methyltransferases to lack a SET domain31–33. Most methyltransferases show a strong preference for specific sites; for example, the SETD1/MLL family places gene-activating H3K4 methylation marks34, whereas the PRC2 complex35,36 places repressive H3K27 methylation marks37,38. The H3K9 methyltransferases show varying specificities for genomic regions: SUV39H1 and SUV39H2 regulate H3K9me3 at pericentric heterochromatin, whereas G9a (also known as EHMT2) and GLP (also known as EHMT1) regulate H3K9me1 and H3K9me2 in euchromatin15,39. Arg methyltransferases, in addition to showing site specificity, also show specificity for the type of methylation. Protein arginine methyltransferase 1 (PRMT1), the founding member of the class, ubiquitously catalyses the formation of MMA and ADMA in mouse tissues on histone and non-histone proteins16,40 and accounts for the majority of mammalian Arg methylation17. SDMA is found on a small percentage of nuclear and cytoplasmic proteins and is catalysed by PRMT5 (REF.41). The third and least understood family of PRMTs is thought to catalyse the formation of only MMA and, so far, has only one putative member, PRMT7.
The specificities of many methyltransferases have been determined using antibody-based methods. Although in many cases the results agree with mass spectrometry data, in some cases contradictory results have emerged, possibly arising from nonspecific antibody binding42–44. Even then, antibody-based methods are preferred, due to their relative ease of use, ability to be used in single-cell imaging studies and ability to probe modification sites missed during trypsin cleavage for mass spectrometry-based detection of Arg and Lys methylation. The varying specificities of enzymes for different substrates, as well as the possible nonspecific binding of antibodies that recognize histone modifications, have made it challenging to decipher the precise functions of these enzymes and their products — the specific patterns of histone methylation — in vivo.
Histone methylation was considered stable until the report of lysine-specific demethylase 1 (LSD1) in 2004. LSD1 was initially found to demethylate mono-methylated and di-methylated H3K4 (REF.45), and later to act on mono-methylated and di-methylated H3K9, as well46 (because of the chemical mechanism LSD1 employs, it cannot act on tri-methylated Lys). Soon after the report of LSD1, the jumonji domain class of demethylases was found to act on mono-methylated, di-methylated and tri-methylated Lys47–50. Eighteen demethylases of this class have now been identified51. Like methyltransferases, demethylases show specificity for the Lys residues they act on, although several show activity towards two or more substrates; for example, LSD1 demethylates H3K4 and H3K9 (REFS45,46), and KDM4A demethylates H3K9 and H3K36 (REFS48,50). To date, no Arg demethylases have been conclusively identified.
In vivo, histone methyltransferases and demethylases often operate in large protein complexes, and their genomic targets are often influenced by the presence of reader proteins or domains, which recognize various histone modifications. Although the members of a family of readers can recognize different degrees of methylation at different sites, each individual reader typically recognizes a single or a few closely related methylation marks. The plant homeodomain (PHD) fingers represent the largest class of readers, recognizing unmethylated or methylated Lys residues52–56. The Tudor domain shows greater versatility, binding both methylated Lys and Arg residues57. Reader domains can occur in writers and erasers or in their binding partners, and they can recognize the products or substrates of the enzymes present together in the complex or marks generated by other enzymes that help target the complexes to appropriate genomic locations. For instance, BHC80 (also known as PHF21A), a member of the LSD1 demethylase complex, recognizes the reaction product of LSD1, unmethylated H3K4 (H3K4me0), and helps maintain LSD1 at H3K4me0 sites to protect H3K4 from re-methylation52. The SETD1 family recognizes its target sites, unmethylated Cpg islands, via the binding partner CFP1 (REF.58). Some chromatin regulatory complexes contain multiple readers that facilitate the integration of multiple histone marks59–61. In other cases, a single reader domain provides the desired specificity62,63. Thus, readers are essential for ensuring that initial chromatin modifications are translated into additional structural changes, and eventually into the activity of functional effectors to mediate and reinforce the appropriate gene expression networks25.
Histone methylation in development
Histone methylation orchestrates developmental gene expression programmes beginning before fertilization and continuing into the postnatal period. Defects in histone methylation affect various developmental processes and can result in developmental arrest and lethality at different stages or lead to specific deficits in organ function in mature animals, depending on the nature and cell-type specificity of the methylation defect. Histone methylation regulators are generally ubiquitously expressed during development64,65. However, knockout experiments in mice and expression profiling studies have revealed that there are certain cell-type-specific and tissue-specific differences in the activity of histone methylation regulators: the tissues that are most affected in knockouts often show moderate to high expression of the regulator during normal development.
Analysis of individual methylation marks in early embryonic systems has revealed their unique distribution and functions in gene regulation. Notably, H3K4 methylation was widely present at transcription start sites in human ESCs66, zebrafish67 and mouse embryos68, but it showed varying degrees of correlation with gene expression. In human ESCs, 80% of H3K4me3-marked genes were expressed66, whereas in early zebrafish embryos (before the mid-blastula transition (MBT)), minimal expression of H3K4-methylated genes was observed67 (likely because embryonic transcription is globally repressed by other mechanisms in zebrafish embryos at this stage). However, the H3K4me3-marked genes in zebrafish were enriched among the set of genes expressed following the MBT, suggesting that early deposition of H3K4me3 poised these genes for later expression67. Histone methylation patterns are clearly associated with the earliest stage of differentiation: the formation of the trophectoderm and the inner cell mass. In mouse embryos, H3K27 methylation was found at distinct sets of genes in these tissues, whereas H3K4 methylation sites were largely common between the trophectoderm and inner cell mass68. Neither the H3K4 nor H3K27 methylation patterns correlated especially well with gene expression, suggesting the presence of more complex regulatory mechanisms in vivo than those emerging from studies of differentiated cells68.
During development, cells commit to specific lineages and must silence genes that promote pluripotency as well as those that determine alternative fates. Both H3K27 and H3K9 methylations contribute to this silencing. Comparison of H3K27me3 in ESCs and in differentiated cells revealed a broadening of H3K27me3 domains in the differentiated cells69,70. Genes silenced in this manner included those encoding pluripotency factors, developmental factors and lineage-specific transcription factors69. Similarly, differentiated liver and brain cells harboured expanded H3K9me3 tracts relative to ESCs71. Genes silenced by H3K9 methylation in differentiated cells were additionally maintained in an inactive state by physical association with the nuclear lamina70, which was shown to depend on H3K9 methyltransferase G9a in Caenorhabditis elegans embryos72. These data suggest the occurrence of progressive heterochromatinization during development and lineage specification. However, a recent study using mouse embryos showed few changes in the overall numbers of H3K9me3-marked genes when comparing the inner cell mass, the three germ layers at gastrulation and differentiated cells73. Instead, H3K9me3 was dynamically altered during development, such that lineage-specific genes became activated by the loss of H3K9me3, while pluripotency genes and those pertaining to other lineages gained this mark73. This study indicates that developmentally programmed heterochromatin reorganization, rather than an overall increase in heterochromatin, accompanies the progression of development. Although the exact mechanisms that govern the acquisition and readout of these methylation patterns remain to be fully determined, these studies cumulatively demonstrate the importance of repressive H3K9 and H3K27 methylations in promoting differentiation and lineage specification during development.
A unique aspect of developmental systems related to histone methylation is the presence of genes bivalently marked with both activating H3K4me3 and repressive H3K27me3 marks, which partially explains the weak correlations between gene expression and single H3K4 or H3K27 methylation marks discussed above. These marks typically occur in the promoters of lowly expressed genes in early embryos — before lineage commitment — that often encode developmental transcription factors such as the SOX, PAX and POU families4,5. During differentiation and lineage specification, cells lose one of the two marks in specific regions, resulting in gene activation or repression that is appropriate for the fate the cell will acquire. For example, neuronal differentiation led to the loss of H3K27me3 from neuronal gene promoters4, whereas mouse embryonic fibroblasts retained this mark at these promoters but lost the activating H3K4me3 modification5. Genomic studies have revealed enrichments of bivalent marks at developmental genes in zebrafish67, human ESCs66,74 and mouse embryos68. By contrast, the genes with reduced H3K4me3 and H3K27me3 marks encoded proteins with functions in physiological responses, such as receptors and other proteins that respond to environmental stimuli68,74. Surprisingly, bivalently marked genes were not found in very early embryogenesis (in pre-MBT zebrafish and in pre-implantation mouse embryos)75,76. In mouse embryos, H3K27 marks, which were initially present in gametes, disappeared after fertilization and were re-established only post-implantation77. By contrast, maternal chromosomes in pre-implantation embryos contained broad, non-canonical tracts of H3K4me3 (REF.77), which in humans were correlated with open chromatin78. The non-canonical H3K4me3 tracts in mouse embryos were paradoxically associated with gene silencing77. These studies hint at the existence of a mechanism separate from bivalency to suppress gene expression in early embryos, but how such a mechanism operates and possibly interacts with bivalent marks remains elusive. Bivalent histone marks have also been documented at enhancers. Active enhancers display H3K4me1 together with H3K27 acetylation, whereas poised enhancers harbour H3K27me3 and H3K4me1 (primed enhancers lack H3K27 modifications but retain H3K4me1)79,80.
Beyond the regulation of lineage fate decisions in early embryos, bivalent methylation marks also have other specialized roles during development. For example, several pluripotency-associated genes expressed in ESCs were silenced during differentiation by acquiring bivalent marks associated with their promoters66. Similarly, most bivalently marked genes in haematopoietic progenitor cells were shown to lose H3K4me3 and to become silenced upon differentiation. However, in progenitors destined to become erythrocytes, some bivalently labelled genes lost H3K27me3 and were activated upon differentiation, which correlated with the presence of additional histone marks: H3K9me1, H3K27me1 and H4K20me1 at the promoters and gene bodies. This led the authors to conclude that these marks confer activation potential to bivalent genes and that cell fate was predetermined before the onset of differentiation and could be predicted on the basis of the histone modification patterns present in progenitor cells81. Similar predictions about future gene expression could be made on the basis of histone methylation patterns in zebrafish embryos before zygotic gene activation67,82. Thus, although bivalently marked genes were initially thought to be a unique feature of early embryonic systems, it is likely that they play special roles in later stages of development and in specialized stem cell populations in adults.
Importance for animal development
The importance of histone methylation is conserved across animal development. Accordingly, as has been indicated by many studies throughout the years, the removal of various histone methylation regulators has profound effects on early embryogenesis, body patterning and organ development.
Histone methylation in whole-body development and body patterning.
The first indications of the importance of histone methylation in embryonic development came from studies in Drosophila melanogaster. Early genetic screens in D. melanogaster identified numerous genes required for embryo development — in particular, body patterning — many of which were later found to regulate histone methylation. Mutations in these components typically led to homeotic transformations (changes of one body segment into another). For example, mutations in Trithorax (Trx) — later shown to be an H3K4 methyltransferase83 — resulted in transformations of the first and third thoracic segments towards the second84, whereas heterozygous mutants of the Polycomb group (PcG) complex — the fly orthologue of the H3K27 methyltransferase complex PRC2 — developed extra sex combs on the limbs85, which was linked to aberrant homeotic gene regulation86,87. Various PcG components were later identified in screens for body pattern regulation88–90. Surprisingly, demethylases were not found in these genetic screens, possibly because demethylases function redundantly (for example, KDM4A can substitute for KDM4B, and vice versa91) and/or because their depletion has a less pronounced impact on early development, owing to maternal contributions of RNA or protein (as seen, for example, for LSD1 (REF.92)), thereby resulting in milder phenotypes. Such maternal effects have also been observed for histone demethylases in mammals93–95. It is also worth noting that not all histone methylation regulators are required for D. melanogaster development. For example, loss of Su(var)3–9, an H3K9 methyltransferase14,96, had minimal effects on fertility and embryogenesis, but it was required for position effect variegation97, a process known to depend on chromatin structure.
Similar to their roles in D. melanogaster, many histone methylation regulators have critical roles in mammalian development (FIG. 2a), as revealed by extensive knockout studies in mice. These studies have largely focused on analysing the effects of embryonically expressed regulators; fewer studies have analysed the maternal contributions of these regulators, but those that have suggest that many regulators are indispensable for very early stages of development94,98. Notably, the timing, nature and extent of defects associated with embryonic knockouts are largely uncorrelated with the affected methylation site or whether the mark is activating or repressive. Overall, the effects of the removal of histone methylation regulators have complex aetiologies and likely arise from misregulation of specific sets of developmental genes and/or perturbations of other aspects of chromatin regulation (BOX 1).
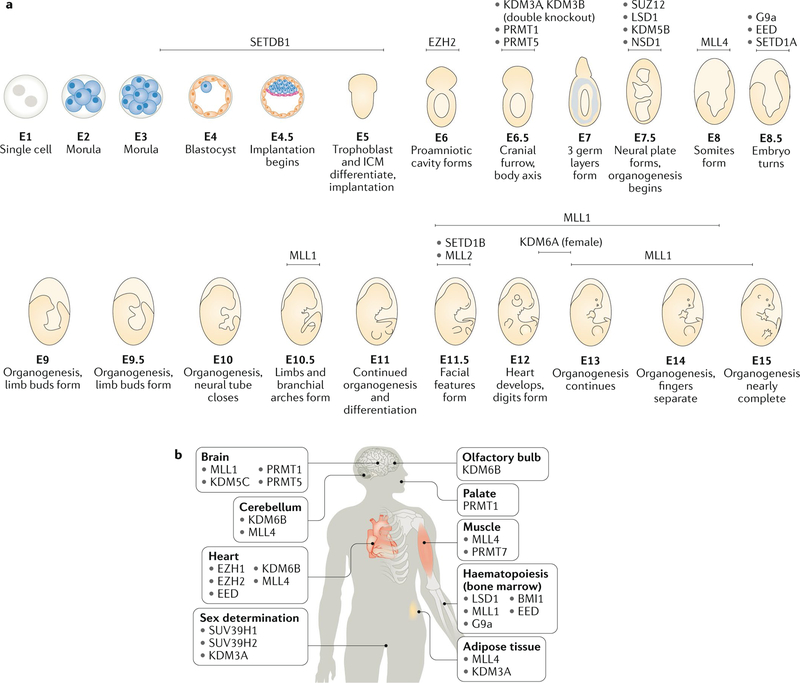
Fig. 2 |. The importance of histone methylation regulators in mammalian development and organogenesis.
a | Mouse developmental stages at which null alleles of the indicated histone methylation regulators exhibit embryonic lethality. Loss of some histone methylation regulators causes very early lethality, before or during implantation (for example, SETDB1), whereas other regulators are required at later stages of organogenesis, with the majority exhibiting lethality between embryonic day 7 (E7) and E12. For some regulators (MLL1, SETDB1), lethality was observed at different stages, depending on the report, in which case all reports are shown. For references to the relevant reports, see Supplementary Table 1. b | Tissue-specific sites of action of histone methylation regulators, as revealed by conditional knockout analysis or by analysis of viable systemic knockouts in mice. Many regulators are essential for neurodevelopment and cardiac development, whereas others regulate myogenesis, adipogenesis and haematopoiesis. For references to the relevant reports, see Supplementary Table 2. ICM, inner cell mass.
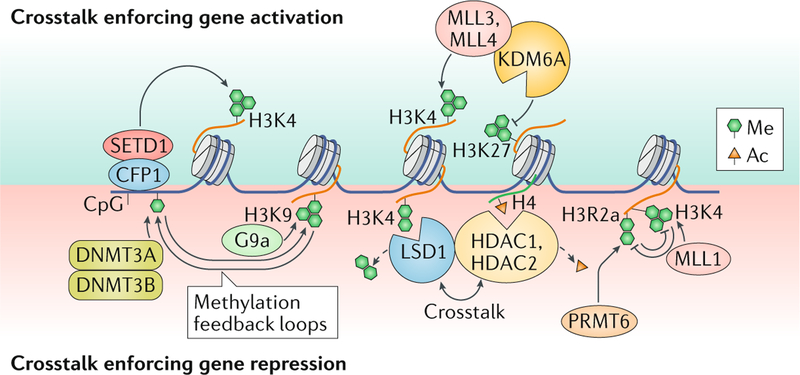
Box 1 |. crosstalk between chromatin marks.
Chromatin is typically marked by multiple modifications, and it is essential that these marks work in concert to achieve a coordinated cellular response (see figure). in some cases, the presence or absence of one modification can stimulate or inhibit deposition of another. For example, during neuronal differentiation, PRMt6-mediated catalysis of asymmetric arginine di-methylation at histone H3 (H3r2me2a) — by affecting recruitment of the relevant methyltransferases — precluded H3 methylation at lysine 4 (H3K4) at promoters, but stimulated it at enhancers, which was required for proper induction of neuronal gene networks227. reciprocally, H3K4 tri-methylation (H3K4me3) precluded PRMt6-mediated H3r2 methylation228. a similar interplay was observed with histone methylation (Me) and acetylation (ac): demethylation of H3K4 by LSD1 was stimulated by the removal of acetylation by histone deacetylase 1 (HDaC1) and HDaC2, which are LSD1 binding partners229,230; reciprocally, deacetylation was stimulated by H3K4 demethylation229. this positive feedback loop was shown to be required for maintaining pluripotency in embryonic stem cells (ESCs)231.
several chromatin regulators have important catalytically independent roles in setting chromatin modifications, often by recruiting other enzymes. PRC2 can be recruited by both PRMt6 (REF.232) and SETDB1 (REF.233) to appropriately govern cell-type-specific gene repression programmes. similarly, LSD1 was recruited to enhancers by MLL4 in a catalytically independent manner to suppress pluripotency genes234. the H3K27 demethylase KDM6a forms important associations with multiple chromatin regulators. For example, KDM6a–MLL4 complex235,236 led to a coordinated increase in H3K4 methylation and decrease in H3K27me3 at certain HOX loci237. Notably, H3K4me3 preceded the H3K27 demethylation, suggesting the importance of temporal coordination of these events237. in other cases, KDM6a had a purely scaffolding role, recruiting MLL4 and p300 to activate enhancers in ESCs with H3K4me1 and H3K27 acetylation238, or recruiting chromatin remodellers to cardiac enhancers in order to drive cardiac differentiation of ESCs119,172. the latter function could be partially supplied by the H3K27 demethylase KDM6C182. similar roles in recruiting chromatin remodellers were reported for KDM6a and KDM6B, which were required for proper gene expression in mature T cells239.
Histone methylations are also highly coordinated with DNA methylation through multiple feedback loops. in ESCs, unmethylated H3K4 acted to promote gene repression by providing a binding site for the recruitment of DNA methyltransferase 3a2 (DNMt3A2)240. Conversely, unmethylated DNA at CpG islands activated genes via the recruitment of CFP1, a member of the SETD1 complexes that deposit gene-activating H3K4 methylation58. similarly, H3K9 and DNA methylation show strong cooperation. in ESCs, the H3K9me3 reader HP1 (REF.241) recruited DNMT3B242, while MBD1, a DNA methylation reader, recruited the H3K9 methyltransferase SETDB1 (REF.243), thereby coordinating the two methylation marks to enforce gene silencing. accordingly, proper DNA methylation and the suppression of aberrant gene expression programmes were shown to require the H3K9 methyltransferases SUV39H1, SUV39H2 (REF.242) and G9a244.
Loss-of-function mutation of the methyltransferase Setdb1 responsible for H3K9 tri-methylation was associated with the earliest embryonic lethality of all methyltransferase knockouts, occurring before embryo implantation (between embryonic day 3.5 (E.3.5) and E5.5 (REF.99). Loss of many other methyltransferases led to lethality at various stages of organogenesis (E6–E15), suggesting that these methyltransferases may have critical roles in promoting the development of specific organs (see also below). Notably, the loss of enzymes that regulate the same mark can have differential consequences. For example, in contrast to the early embryonic lethality associated with Setdb1 mutations, double knockouts of a pair of related H3K9 methyltransferases, SUV39H1 and SUV39H2, did not display a fully penetrant requirement for unperturbed embryonic development, and these mice generated viable progeny at sub-Mendelian ratios39 (TABLE 1). These results suggest that the SUV39 and SETDB1 methyltransferases regulate distinct sets of genes during development, but the nature of these differences has not been elucidated. Several other histone methylation regulators have also been shown to be dispensable for normal development (TABLE 1). Nevertheless, loss of these regulators still led to reduced viability after birth or to tissue-specific defects, suggesting tissue-context-specific functions of the different regulators and distinct roles for histone methylation marks in different tissues.
Table 1 |.
Histone methylation regulators not essential for development
| Regulator gene | Phenotype | Refs |
|---|---|---|
| Nsd2 | Lethal at P1 | 267 |
| Kdm3b | Lethal at P1 | 268 |
| Prmt4 | Partial embryonic lethality, fully lethal at P1 | 100,101 |
| Kdm6b | Lethal by P18 | 173 |
| Bmi1 (encodes PRC2 complex component) | 50% lethal between P1 and P3, with further lethality at 3–20 weeks | 113 |
| Kdm6A (males) | Reduced viability after birth, runted body | 171 |
| Suv39h1 and Suv39h2 (double knockout) | Viable at sub-Mendelian ratios | 39 |
| Kdm3a | Viable, obese | 131 |
| Prmt7 | Viable, reduced muscle-to-fat ratio | 226 |
| Prmt6 | Viable | 269 |
| Kdm5c | Viable, neurological deficits | 123 |
| Ezh1 (encodes PRC2 catalytic subunit) | Viable | 270 |
P, postnatal day.
The roles of histone Arg methylation during mammalian development have been more challenging to decipher than those of Lys methylation, largely because of the broad specificity of PRMTs, which target histone and non-histone proteins (BOX 2) (see also REF.18). Some embryonic lethality was observed around E18.5 and E19.5 in Prmt4−/− embryos, which were significantly smaller than their wild-type counterparts100. At birth, all the Prmt4−/− pups showed reduced levels of H3R17 and p300 methylation and died due to a failure to breathe101. Prmt5 (REF.102) and Prmt1 (REF.103) homozygous mutant mice showed much earlier embryonic lethality, at E6.5. However, in Prmt1 mutant cells, global protein methylation was markedly reduced103, obscuring any contributions of histone methylation to this phenotype. Direct examination of changes in histone Arg methylation, rather than relying on the analysis of methyltransferase mutants, will thus be required to unravel the chromatin-specific effects of Arg methylation in development.
Box 2 |. Non-histone protein methylation in development.
Although rich in lysine (Lys) and arginine (Arg) residues, histone proteins form a small fraction of the known protein methylome. soon after the discovery of histone Lys methylation245, numerous groups reported the presence of methylated Lys and Arg residues in non-histone proteins involved in transcription regulation, signalling pathways, DNA damage response and RNA processing246,247. in the first example of methylation-dependent regulation of non-histone proteins, SET7 was shown to mono-methylate transcription initiation factor TFIID subunit 10 (TAF10) and p53, with transcription-stimulatory effects248,249. the p53 protein is required for the proper development of organs in the renal, musculoskeletal and nervous systems250–254, but how methylation regulates the activity of p53 in the development of these systems has not been investigated. since then, multiple transcription factors have been shown to be regulated through methylation, with some having direct effects on mammalian development. EZH2-mediated methylation of GATA4 at Lys299 attenuates the activity of GATA4 as a transcription factor and is essential for proper cardiogenesis255. similarly, SMYD1 methylates the putative kinase TRB3, which activates its function as a transcription co-repressor to downregulate anti-proliferative and autophagy-related genes in embryonic cardiomyocytes256.
Importantly, complete knockout of methylation regulators often fails to delineate the contributions of histone versus non-histone protein methylation towards developmental programmes. in analysing histone versus non-histone modifications, it has to be considered that many chromatin readers also recognize non-histone substrates, which could have important roles in development. For example, tudor domain proteins are involved in many aspects of cytoplasmic and nuclear RNA regulation, and they are critical for gamete formation and other stages of development (reviewed in REF.257). recent genetic studies in Drosophila melanogaster revealed that mutations in some histone residues that are targeted by methyltransferases, such as histone H3 Lys36 (H3K36) and H3K27, closely mimicked the phenotypes caused by mutations in their methyltransferases258, pointing to a pivotal role of these methylation sites in embryogenesis. By contrast, other sites, such as H4K20, were shown to be dispensable for fly development, unlike the H4K20 methyltransferase PR-set7 (SET8)259, suggesting essential non-histone targets of this enzyme. in a different approach, introducing mutated elongation factor 1 alpha 1 (EF1α1) in which lysines were substituted for alanines in chick embryos elucidated the importance of EF1α1 methylation in neural crest development260.
Large-scale mass spectrometry studies have shown that the scope and complexity of Arg methylation is greater than that of Lys methylation, and that non-histone Arg methylation affects the activity, localization and protein–protein interactions of several signalling proteins involved in development18,247,261. the first example of the role of non-histone arg methylation in transcription regulation was provided in 2001, when it was shown that methylation of the transcription co-activator CBP by CARM1 reduces the association of CBP with its binding partner, caMP-dependent transcription factor CreB, which results in a decrease in caMP-dependent gene expression262. so far, more than 100 substrates of CARM1 have been identified, many of which are involved in RNA processing263. For example, methylation of p54nrb (also known as NONO) by CARM1 is critical for decreasing its binding to mRNAs containing inverted-repeat Alu elements, thereby reducing their nuclear retention264. recent studies in mouse embryos suggest that this methylation event, along with CARM1-mediated methylation of H3r26, is critical for the establishment of the embryonic and extra-embryonic lineages during early embryogenesis265. these studies corroborate and provide mechanistic insights into the small size and perinatal death that was observed decades earlier in CARM1 knockout embryos or embryos containing a catalytically dead knock-in of CARM1 (REFS101,266).
Despite these insights, our understanding of the contributions of hundreds of non-histone protein methylation events to mammalian development still remains elementary. Future studies utilizing gene-editing techniques to install site-specific mutations targeting methylated residues should help elucidate the molecular basis of this versatile modification in mammalian development.
Histone methylation in organogenesis.
Histone methylation regulators have many established roles in the development of specific organs (FIG. 2b). These roles have been identified by observing mouse full-body knockouts or tissue-specific knockouts designed to circumvent embryonic lethality at earlier stages (see above). Full-body knockouts revealed important roles for H3K4 methylation in embryonic haematopoiesis. Reports of MLL1 loss have documented a plethora of effects, including specific deficiencies in myeloid and lymphoid lineages104–107, reduced overall haematopoiesis104 and reduced haematopoietic stem cell (HSC) function105. LSD1 was also reported to be required at multiple stages of haematopoietic differentiation, and its activity was shown to repress critical targets of the haematopoietic transcription factors GFI-1 and GFI-1b108,109. Proposed functions of LSD1 include promoting granulocytic differentiation108, impairing both the self-renewal and differentiation of HSCs110 and enhancing production of precursor cell populations for specific lineages111. Additionally, H3K9 methylation mediated by G9a was shown to be involved in silencing pluripotency genes to promote HSC differentiation to mature lineages112, consistent with the observed roles for H3K9 in inactivating pluripotency-associated gene expression programmes during lineage specification (see above). Although the H3K27 methyltransferase EZH2 was required for embryogenesis at a very early stage (FIG. 2a), a role for H3K27 methylation in haematopoiesis was suggested by knockouts of two PRC2 complex members: BMI1 in embryos, which were viable until birth but showed reduced haematopoietic cells113, and EED in adults, which led to bone marrow failure114. At later stages of haematopoiesis, the recombinase RAG2, which contains a PHD finger reader domain recognizing the H3K4me3 and H3R2me2s marks62,63, is essential for DNA recombination and the maturation of adaptive immune cells115. Although the exact roles of H3K4 methylation regulators are yet to be elucidated, it is clear that dynamic H3K4 methylation is required for successful haematopoiesis.
Dynamic regulation of H3K27 methylation has a role in cardiac development. Cardiomyocyte-specific knockouts of PRC2 components, which circumvented the requirement for PRC2 in haematopoiesis113,114, led to multiple defects in cardiac morphology, including defects in separation of the heart chambers and myocardial hypoplasia, and were accompanied by the expression of non-cardiac genes116,117. Just as opposing H3K4 methylation and demethylation were required for haematopoiesis, the H3K27 demethylase KDM6B was required along with methyltransferase PRC2 for cardiac development, as shown in zebrafish, where the loss of KDM6B led to a lack of cardiomyocyte proliferation late in development118. Interestingly, loss of KDM6A led to much earlier defects in embryogenesis, during embryo patterning, suggesting non-redundant roles for these related H3K27 demethylases in organismal development119. Cardiac development was also highly sensitive to changes in H3K4 methylation, with the H3K4 mono-methyltransferase SETD7 being required for proper cardiac morphology64 and the expression of cardiac-specific genes65, suggesting roles for both activating and repressive methylation marks in forming cardiac structures.
Roles for multiple types of histone methylation have been observed in neurodevelopment. KDM6B promoted the differentiation of neuronal precursors in both the cerebellum120 and olfactory bulb121, and PRMT1 was required in neural crest cells for palate development122. Full knockout of the H3K4me3 demethylase KDM5C in some genetic backgrounds resulted in mice with neuro-developmental deficits and impaired cortical development123,124. Deletion of MLL1 in sub-ventricular zone neural stem cells (NSCs) did not affect embryonic development but did lead to postnatal lethality resulting from a reduced numbers of neurons125. NSC-specific knockout of both Prmt1 and Prmt5 also led to postnatal lethality126,127, although the underlying mechanisms of their action differed greatly. PRMT5 was shown to control the differentiation and proliferation of NSCs by generating H4R3me2s to downregulate specific pro-mitotic genes128 and promote proper mRNA splicing126, resulting in neural progenitor cell depletion and decreased neuronal numbers, whereas Prmt1 loss caused a large reduction in the number of mature oligodendrocytes, resulting in severe hypomyelination in the central nervous system127. Brain-specific knockout of Mll1 early in development led to changes in H3K4 methylation patterns at superenhancers, resulting in increased proliferation of cells in the cerebellum and increased susceptibility to the development of medulloblastoma, which was traced to the role of MLL4 in activating superenhancers to stimulate the expression of tumour suppressor genes129. Thus, neurodevelopment is governed by various histone methylations that employ both transcriptional and post-transcriptional mechanisms to ensure the expression of appropriate developmental programmes.
The development of the reproductive system is also regulated by histone methylation. Knockout of the H3K9 demethylase Kdm3a in mice showed no effects on mortality130,131 but caused a fraction of XY mice to develop into females132, which can be linked to defects in spermatogenesis arising from misexpression of the sex determination gene Sry upon KDM3A loss of funcion133. Similarly, Suv39h1 and Suv39h2 double knockout mice also displayed deficits in spermatogenesis39. Finally, the founding member of the Tudor family of readers (encoded by tud), which recognizes H3R2me2s marks, was first identified in D. melanogaster more than three decades ago, on the basis of defective germ cell development in the progeny of mutant mothers134.
Overall, it is now well established that haematopoiesis, cardiac development, neurodevelopment and reproduction are all importantly controlled by histone methylation regulators. This indicates that the establishment of precise patterns of histone methylation is required for the development of these different tissues, and it is likely that histone methylation is important for nearly all aspects of organogenesis. The molecular mechanisms by which the different histone methylations act and their functional outcomes are poorly conserved across tissues. Essentially, the same methylation type or enzyme can have vastly different effects depending on the context, which precludes predictions of the roles of each regulator and its associated mark in development. Recently, meta-analyses combined with machine-learning algorithms have provided a new approach for predicting the developmental functions of histone methylation marks135.
Roles in developmental processes
It is now clear that histone methylation has a vast impact on animal development, but in many cases the exact underlying mechanisms are elusive. Here we will discuss three notable examples of developmental processes controlled by histone methylation in early embryonic systems: genomic imprinting, HOX gene expression and the regulation of pluripotency and differentiation programmes.
Genomic imprinting.
One of the earliest steps in development is the establishment of embryonic and extra-embryonic lineages. Genomic imprinting, which leads to mono-allelic expression of a gene by silencing one of the parental copies, is critical for this stage, by ensuring proper dosage of each gene product. This process is especially important for genes on the X chromosome, which exist in two copies in females and one in males. In females, gene expression from one copy of the X chromosome is silenced in cis by the long non-coding RNA Xist, which is expressed from the otherwise silenced X chromosome and recruits PRC2 to deposit gene-inactivating H3K27me3 marks136. On the active chromosome, Xist itself is silenced by PRC2-mediated H3K27me3 deposition137. Consequently, mice with mutated Eed — which encodes the essential PcG protein EED — were defective in X inactivation in the extra-embryonic lineages138 and in pre-implantation embryos139. Genes on other chromosomes are also imprinted, and the prevailing model has been that one allele is silenced by DNA methylation1, with or without co-occurring H3K9 methylation. DNA methylation and H3K27 methylation are mutually exclusive at one imprinted gene, Rasgrf1 (REF.140). However, H3K27 methylation was recently shown to mediate imprinting at sites with low levels of DNA methylation141, demonstrating the existence of two independent imprinting pathways (FIG. 3a). Approximately half of the active chromatin on the paternal DNA showed DNA hypomethylation on the maternal allele and contained H3K27me3 tracts141. Genes imprinted by this mechanism included Sfmbt2 (REFS141,142), a PRC2 component required for the development of extra-embryonic lineages143. In the future, it will be important to understand how H3K27-mediated imprinting contributes to organ development and lineage specification in the embryo.
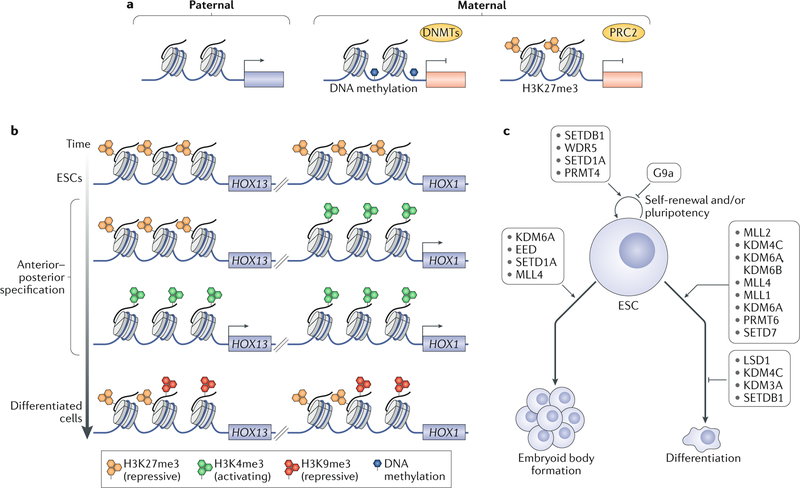
Fig. 3 |. Developmental processes regulated by histone methylation.
a | Genomic imprinting is mediated by both histone and DNA methylation. Paternally expressed genes display undetectable or very low levels of repressive DNA and H3K27 methylation marks. The maternal allele is silenced either by DNA methylation, introduced by DNA methyltransferases (DNMTs), or, in regions of hypomethylated DNA, by tri-methylation of Lys27 of histone H3 (H3K27me3), introduced by the PRC2 complex. The predominant mechanism for the silencing of imprinted genes varies by the locus. b | Regulation of HOX genes by histone methylation. In embryonic stem cells (ESCs), histone tri-methylation at H3K27 represses all HOX genes. At later stages of differentiation, early HOX genes are activated along the anterior–posterior axis by removal of H3K27me3 and addition of H3K4me3. Subsequently, late HOX genes are activated in caudal regions by a similar mechanism. In differentiated cells, HOX genes are again repressed by tri-methylation at H3K9 and H3K27. It is not known whether these methylation marks occur on the same or separate nucleosomes. c | Effects of histone methylation regulators on ESC self-renewal and differentiation. Although some regulators directly influence ESC self-renewal (SETDB1, WDR5, SETD1A, G9a), most affect the ability of ESCs to differentiate (as assessed by using embryoid body formation assays, which indicate the ability to form the three germ layers, or by various differentiation-inducing protocols (including those that direct ESCs towards a particular lineage)). In cases where different studies have indicated divergent roles, all outcomes are listed. For references to the relevant reports, see Supplementary Table 3.
Regulation of HOX genes.
One critical and well-conserved role for histone methylation in development is in regulating expression of HOX genes. These genes are arranged in linear arrays, where the position of the gene controls its spatiotemporal expression patterns: genes at the distal ends of the clusters are typically expressed later in development and are restricted to caudal regions of the embryo144,145. Progressive activation of HOX genes corresponds to removal of H3K27me3 and appearance of H3K4me3 (REF.146). Later in development, HOX genes are silenced by methylation at H3K27 and H3K9 (REF.69) (FIG. 3b). Interestingly, recent studies have suggested that HOX genes are briefly devoid of H3K27 methylation after fertilization, but it is not known how they are kept inactive during this stage76.
The effects of many genes discovered in the early D. melanogaster embryogenesis screens for developmental defects were traced to their roles in regulating HOX gene expression. Trx mutant embryos showed reduced expression of the HOX gene Ultrabithorax (Ubx) concomitant with thoracic transformations. Overexpression of Ubx rescued these transformations84, leading to the conclusion that Trx was a Hox activator84,147. PcG mutants, by contrast, led to expanded expression of Hox genes in embryonic regions in which they are normally not expressed148,149, indicating a negative role for PcG in restricting Hox expression to specific body regions149. Combined mutations of PcG and Trx restored some of the body patterning defects seen in the single mutants, suggesting that the two regulators act in opposite ways147. Of note, later studies uncovered more complex interactions between Trx and PcG group proteins, in which the double mutants failed to completely rescue Hox expression in all regions of the embryo. The wing discs of double mutants expressed Ubx at elevated levels comparable to those in single PcG mutants, leading the authors to conclude that Trx proteins act as ‘anti-repressors’ to suppress PcG-mediated repression of gene expression, and that Trx is not otherwise required for Hox gene activation in all contexts150. Although the biochemical functions of Trx and PcG proteins were not understood at the time, these studies have demonstrated essential roles for both gene-activating (H3K4) and gene-repressing (H3K27) methylation in ensuring that appropriate Hox genes are expressed in each body segment.
Despite the variable effects of different types of histone methylation on mammalian development, their effects on HOX gene expression typically align with the activating or repressive features of each mark. Mouse embryos lacking the H3K4 methyltransferase MLL1, the homologue of D. melanogaster Trx, displayed embryonic lethality between E10.5 and E15 (REFS104,106,151), with defects in body patterning — abnormal branchial arches, mixed rostral–caudal neural patterning152 and caudalization of Hox gene expression153 — similar to its effects in flies154. Knockout of LSD1, the H3K4 demethylase, showed effects opposite to those of MLL1 at the molecular level, leading to premature or excessive expression of Hoxb7 and Hoxd8 (REF.155) and embryonic lethality at E7.5 (REF.95). Thus, it is clear that a balance of histone methylation and demethylation is also required for proper Hox gene expression and successful embryogenesis in mammals. However, in mammals, more complex roles for H3K4 methylation have been seen, owing to the presence of six H3K4 methyltransferases: SETD1A, SETD1B and MLL1–MLL4. These enzymes were individually required at various stages of embryogenesis, resulting in lethality between E8 and E15 in the individual knockouts99,104–106,151,156–158. Their distinct embryonic phenotypes supported their non-overlapping methylation patterns, including global H3K4 tri-methylation (SETD1A and SETD1B159), promoter-specific methylation (MLL1 (REF.160)) and mono-methylation at enhancers (MLL4 (REF.161)). Currently, the detailed mechanisms by which these locus-specific methylations ultimately govern HOX genes and other developmental regulators remain poorly understood. As in D. melanogaster, methylation dynamics at H3K27 are also required in mammals for successful embryogenesis and HOX gene regulation. Accordingly, mutations in the components of the PRC2 complex — which is the mammalian PcG homologue, consisting of the methyltransferases EZH1 and EZH2, along with the other regulatory components BMI1, EED, SUZ12, RBAP46 and RBAP48 (REF.35) — resulted in body-patterning defects in mice. Conditional knockout of Ezh2 in specific cell types later in development — to circumvent early embryonic lethality in Ezh2-null embryos162,163 by E6.5 (before body patterning) — led to mispatterned (anteriorized)164 or generally increased165 expression of Hox genes. Both Eed and Suz12 knockout mutations were lethal by E8.5 and E7.5, respectively, with major body-patterning defects such as reduced mesoderm and misexpression of key developmental regulators (brachyury, Evx1)166–168. Bmi1 knockout mice, despite clear alterations in the proper establishment of Hox gene expression boundaries169, were viable until birth, but died shortly afterwards with impaired haematopoiesis and subtle skeletal, neurological and haematopoietic defects113. These variable results suggest that PRC2 subunits may perform some functions independently of their roles in the methyltransferase complex, and/or that the complex may retain some functions during development in the absence of some subunits. As expected, the H3K27 demethylases KDM6A and KDM6B displayed effects opposite to PRC2 on Hox gene expression170. In zebrafish, loss of KDM6A resulted in posterior patterning defects and reduced expression of multiple Hox genes119. In mice, female embryos lacking KDM6A died before birth with defects in cardiac development and neural tube closure; however, the requirement for KDM6A for unperturbed development was partially compensated for in males by KDM6C (also known as UTY)171, a catalytically inactive homologue of the enzyme present on the Y chromosome119,172, which allowed survival until birth. The critical roles of KDM6A may involve the recruitment of other chromatin regulators instead of direct H3K27 demethylation (BOX 1), a function that KDM6C is expected to be able to fulfil. Loss of the catalytic activity of KDM6B delayed expression of posterior Hox genes and led to the anteriorization of Hox boundaries, with resulting skeletal abnormalities; however, the embryos were viable until birth, albeit with postnatal loss of viability173. Overall, the distinct phenotypes resulting from loss of biochemically equivalent H3K27 methylation regulators suggest that they likely have distinct genomic targets, and therefore act on non-overlapping sets of essential developmental genes.
Regulation of cell fate and specification.
The embryonic defects arising at the onset of organogenesis from aberrant histone methylation suggest that histone methylation potently affects cell fate decisions. To circumvent the obstacles of embryonic lethality and to study cell fate decisions directly, many studies have analysed the self-renewal or differentiation of ESCs in vitro. An emerging theme from these studies is that the regulators that are required early in development have roles in maintaining ESC pluripotency, whereas those acting during organogenesis or later tend to suppress or enhance ESC differentiation (FIG. 3c; see also FIG. 2). For example, SETDB1, an H3K9 methyltransferase that shows the earliest requirement during embryogenesis, was required for the maintenance of pluripotency in ESCs99. By contrast, regulators that are required later during embryonic development, such as the methyltransferases MLL1 (REFS174,175), MLL2 (REFS174,176,177), SETD1A178, G9a179 and PRC2 (REF.180), or demethylases such as KDM6A and KDM6B181, were largely not required for the maintenance of pluripotency, but their loss compromised differentiation in vitro174,177,179,182. Loss of the demethylase LSD1 (REF.183) or KDM3A184 promoted ESC differentiation, in accordance with their antagonistic relationships to the H3K4 and H3K9 methyltransferases. These results suggest that the stage at which a regulator acts during embryogenesis may be inferred from in vitro models of ESC maintenance and differentiation. Notably, however, not all methylation regulators showed this direct correspondence between embryonic and ESC phenotypes. For example, PRMT4 (REF.100) supported pluripotency maintenance in human and mouse ESCs, likely by directly promoting ESC self-renewal, rather than by regulating pluripotency versus differentiation programmes as such100,185,186. A study separate from REF.178 reported a similar role for SETD1A187 in supporting ESC pluripotency. Also included in this group are regulators that promote ESC differentiation when they are lost (for example, LSD1 (REF.183)). These regulators shift the cell fate decision away from self-renewal, and thus appear to be required for the maintenance of pluripotency via affecting differentiation programmes, which recapitulates their embryonic roles. KDM4C, whose embryonic phenotype has not been investigated, was reported in separate studies to both promote188 and inhibit184 differentiation, a discrepancy that has not been resolved. Thus, although ESC-based studies have been very powerful in elucidating the developmental functions of chromatin regulators, such exceptions, along with discrepancies in histone methylation patterns between ESCs and pre-implantation embryos76, highlight the imperfect correspondence between in vitro and in vivo studies.
Another complication in studying the role of histone methylation in cell fate determination stems from the fact that individual histone marks and their regulatory enzymes often show distinct effects on ESC differentiation towards different lineages. For example, MLL4 loss led to deficits in differentiation towards endoderm, an increase in mesodermal markers, but relatively few effects on differentiation into embryoid bodies and ectoderm189. As another example, PRMT6 overexpression induced endodermal markers but had varying effects on mesodermal and ectodermal markers, whereas its depletion led to varying increases in markers of all three lineages190. Furthermore, these effects on differentiation due to changes in the expression of methylation regulators in many cases do not correspond to the effects observed for other related and/or seemingly functionally equivalent enzymes. For example, both KDM4C and KDM3A remove repressive H3K9 methylations (tri-methylation and di-methylation, respectively), yet the gene expression programmes in knockout ESCs showed distinct differences, which corresponded to the differentiation towards different lineages: endodermal in Kdm4c knockout cells, and mesodermal in Kdm3a knockout cells184. In addition, an independent study showed no deficits at all in self-renewal of Kdm3a knockout ESCs191. These differences may arise from localization of the KDM4C and KDM3A substrates to different loci, with H3K9me3 localizing to pericentric heterochromatin and H3K9me2 localizing to euchromatin13,15. The details of how these distinct marks lead to differences in lineage specification remain to be elucidated. Similarly, SETD1A and MLL2 both methylate H3K4, but only SETD1A appeared to promote methylation during ESC differentiation, in part by regulating the Hoxa locus178 via the methylation of two enhancers187. The loss of WDR5, which forms a complex with SET1D1A (and SET1D1B), showed effects opposite to those of MLL2 loss, leading to impaired ESC self-renewal192 in contrast to impaired differentiation. Finally, MLL2 was required for establishing methylation at bivalent genes in ESCs, whereas the highly related MLL1 showed a weaker requirement174. Although the finding is in contrast to the late-stage lethality of MLL2 knockout embryos (FIG. 2a), the involvement of MLL2 in promoting bivalent methylation marks in ESCs is consistent with observations that embryos lacking maternal MLL2 arrest by the two-cell stage98, whereas MLL1 is required for development only much later, at the onset of haematopoiesis104,151. In summary, owing to these complex effects of methylation marks and the enzymes that regulate them, a description of a unifying developmental histone code has been so far inaccessible.
Relevance to human genetic disorders
Human disease-associated mutations in genes encoding histone methylation regulators (TABlE 2) have highlighted the importance of dynamic regulation of histone methylation in skeletal and neurodevelopment. Mutations in the H3K4 methyltransferase genes SETD1B and MLL1–4 have all been implicated in syndromes characterized by skeletal and facial abnormalities and intellectual disability, typically involving reduced body size and microcephaly193–199. Mutations in the H3K4 demethylase-encoding genes KDM5C200–202 and LSD1 (REFS203,204) or in the gene encoding the LSD1 binding partner PHD finger protein 21A (BHC80; also known as PHF21A)52,205 resulted in similar syndromes, characterized by intellectual disability and skeletal defects. Notably, mutations in both MLL4 and its antagonist LSD1 have been associated with Kabuki syndrome197,198,203,204, revealing that opposing activities at a molecular level can cause similar outcomes at an organismal level. Dynamic methylation at H3K9 is also required for proper skeletal and neural development. Mutations in both the H3K9 demethylase-encoding PHF8 (REFS206,207) and the methyltransferase-encoding GLP were associated with craniofacial defects and intellectual disability in both humans208–211 and animal models212,213.
Table 2 |.
Histone methylation regulators associated with inherited genetic disorders in humans
| Gene | Mutation | Syndrome | Refs |
|---|---|---|---|
| MLL1 | Point mutations | Wiedemann–Steiner syndrome | 193 |
| MLL2 | Point mutations | Dystonia, skeletal abnormalities, intellectual disability | 194 |
| MLL3 | Point mutations | Kleefstra syndrome, autism spectrum disorder | 195 |
| MLL4 | Point mutations | Kabuki syndrome | 197,198 |
| SETD1B | Point mutations | Seizures, developmental delay, intellectual disability | 199 |
| LSD1 | Point mutations | Kabuki syndrome | 203,204,224 |
| BHC80 (also known as PHF21A) | Deletiona, translocation | Potocki–Schaeffer | 205 |
| GLP (also known as EHMT1) | Translocation, microdeletiona | Kleefstra syndrome | 211,271 |
| KDM5C | Point mutations | Skeletal abnormalities, X-linked intellectual disability | 200–202 |
| PHF8 | Point mutations | Craniofacial abnormality, X-linked intellectual disability | 208–210 |
| EZH2 | Point mutations | Weaver syndrome | 214 |
| NSD1 | Point mutations | Weaver syndrome | 215,216 |
| Deletiona, point mutation | Sotos syndrome | 215,216 | |
| Microinsertion or microdeletiona | Beckwith–Wiedemann syndrome | 217 | |
| NSD2 | Deletiona | Wolf–Hirschhorn syndrome | 218 |
| PRMT7 | Point mutations | Skeletal abnormalities, intellectual disability | 222 |
Haploinsufficient deletions.
In contrast to the above mutations that cause reduced skeletal and brain size, mutations in other histone methylation regulators can lead to overgrowth syndromes. For example, mutations in EZH2 and NSD1 — which encode H3K27 and H3K36 methyltransferases, respectively — lead to overgrowth pathologies typical of Weaver, Sotos, and Beckwith–Wiedemann syndromes214–217. However, mutations in NSD2, another H3K36 methyltransferase-encoding gene, led to growth delays, indicating non-redundant, or even opposing, roles for these related enzymes. Beyond the differences between the enzymes, it should also be considered that germline and somatic mutations can result in divergent phenotypes. This seems to be the case for EZH2 and NSD1, which are often overexpressed in cancers218–220, despite their loss of function in embryos being associated with overgrowth as discussed above, and even in cancer predisposition221.
In the only known example of a clinical phenotype associated with genetic disruption of Arg methyltransferases, loss-of-function mutations in PRMT7 were associated with mild intellectual disability along with obesity and poor development of bones222. A recent large-scale sequencing study showed many mutations in histone methyltransferases and demethylases linked to developmental delays223, suggesting that the full scope of involvement of these enzymes in embryonic and postnatal development in humans has yet to be explored.
Analyses of disease-associated mutations in histone methylation regulators have provided limited insights into the molecular mechanisms underlying the associated pathologies. Many of the reported mutations disrupt or reduce the activity of the corresponding enzymes193,194,197,205,214,224, interfere with their binding to complex members or to DNA194,214, or reduce their expression at the RNA or protein level205,225. It is not known whether truncated or catalytically inactive forms of the mutant proteins have functional roles, although the variations in disease presentation caused by different mutations (for example, in NSD1 (REFS215–217)) suggest that these mutant proteins may have non-catalytic effects. In some cases, phenotypes have been linked to misregulation of a few critical genes. For example, MLL2 mutations led to reduced expression of intellectual disability-associated genes such as THAP1 and TOR1 (REF.194). A mouse model of Kleefstra syndrome, driven by Glp mutation, showed increased rather than decreased H3K9me3, as would be expected on the basis of the function of GLP as an H3K9 methyltransferase. It was then proposed that a concomitant reduction in expression of protocadherins underlies some of the phenotypes of Glp mutants213. Although mouse knockouts have served as faithful models of disease in some cases213,226, many human genetic disorders are not recapitulated in mouse models. For example, the loss of H3K4 methyltransferases in mouse results in haematopoietic rather than skeletal or neural disorders, although one report showed that heterozygous Mll4 knockouts display smaller body size, similar to human patients with MLL mutations198. Further characterization of the functions of histone methylation regulators in various organs and cell types in humans — for example, using organoid systems — may begin to uncover the molecular mechanisms by which their deregulation causes disease.
Concluding remarks
Studies in embryonic model systems laid the foundational basis for the field of epigenetics five decades ago, yet many mysteries still remain regarding the roles and regulation of epigenetic marks during embryonic development. It is clear that histone methylations have important functions throughout development in nearly all cell and tissue types. Key roles for histone methylation in promoting body patterning via HOX gene regulation are well-conserved across species, and histone methylations additionally regulate other developmental gene networks. However, different chromatin regulators often have distinct roles during development even when acting on the same modification; thus, obtaining a full understanding of their functions will require integrating their effects at different genomic loci — for example, promoters, enhancers, heterochromatin and individual genes — and in different cellular contexts with the phenotypic effects on development. Analyses of chromatin in specific embryonic tissues have begun to shed light on some of these differences. In addition, ESCs have provided powerful alternative models for studying the effects of chromatin regulators on maintaining pluripotency and promoting differentiation along diverse lineages.
Although genomic studies have defined overarching histone methylation patterns during development, direct causal relationships between methylation events and the formation of embryonic structures or mature organs have been difficult to define. In many cases, methylation has critical effects at multiple genetic loci, making classical reverse genetic studies focused on the regulation of individual genes difficult to interpret. Furthermore, bulk analyses can mask the heterogeneity of methylation patterns and their outcomes observed across cell types and developmental stages. Whereas imaging analysis can provide useful single-cell information, further advances in molecular techniques will be needed in order to define how methylation patterns influence gene expression in a tissue-specific manner. Determining the relative contributions of promoter and enhancer regulation by methylation, as well as how histone methylations interact with other aspects of chromatin structure, such as nucleosome positioning and 3D genome architecture, will help to further define the molecular mechanisms by which histone methylations operate in development. Studies of early embryos in mouse and zebrafish have suggested the existence of unique mechanisms of gene regulation. For example, zygotic transcription in zebrafish is suppressed despite the presence of activating marks at some genes67, and mouse Hox genes transiently lose repressive H3K27me3 marks after fertilization yet maintain gene silencing76. Determining how these alternative modes of gene repression operate and how they interact with histone methylations will be important for fully understanding the regulatory mechanisms of gene expression in embryos. Finally, although much progress has been made in understanding the roles of histone Lys methylation, our understanding of histone Arg methylation lags behind. Comprehensive mapping of Arg methylation across developmental stages and tissue types will be an essential first step towards understanding its role in development.
Overall, dissecting the roles of various histone methylation regulators at each stage of embryogenesis, using conventional genetics combined with conditional knockout studies and analyses of ESCs, will help us understand the overlapping and unique features of human genetic disorders involving aberrant histone methylation, and eventually will allow the developmental histone code to be cracked.
Supplementary Material
Genomic imprinting
Monoallelic expression of a gene specifically from either the maternal or paternal copy.
HOX genes
Genes encoding homeodomain-containing developmental transcription factors that are arranged in linear arrays and expressed in a spatially and temporally regulated manner corresponding to their one-dimensional arrangement along the chromosome.
Chromodomain
Protein domain that binds methylated lysine.
Bromodomain
Protein domain that binds acetylated lysine.
SET domain
Protein domain that typically harbours catalytic methyltransferase activity.
Plant homeodomain (PHD) fingers
Zinc-finger-containing domain involved in recognizing histone modifications.
Tudor domain
Protein domain that recognizes methylated lysines and arginines.
CpG islands
Regions of the genome with elevated frequency of Cpg dinucleotide, often occurring in gene regulatory regions and often displaying DNA hypomethylation on cytosine.
Mid-blastula transition
Embryonic stage at which cells in the blastula switch from rapid cycling between S and M phases to lengthened cell cycles including G1 and G2 phases.
Trophectoderm
Cells forming the outer layer of the mammalian embryo that later give rise to the placenta.
Inner cell mass
Cluster of undifferentiated cells in the mammalian embryo that give rise to the fetus.
Poised enhancers
enhancers that contain H3K4me1 and H3K27me3 marks and are unable to activate gene expression but that remain capable of activation in the future.
Primed enhancers
An enhancer state that is intermediate between poised and active states. Such enhancers are characterized by H3K4me1 marks and DNA hypomethylation but are unable to activate gene expression.
Zygotic gene activation
Activation of transcription from the genome of the embryo, accompanied by clearance of maternal transcripts.
Sex combs
In Drosophila melanogaster, a set of male-specific bristles on the leg.
Position effect variegation
Variation in gene expression levels based on the surrounding genomic context of the gene.
Neural crest cells
embryonic cells that arise from the ectoderm and give rise to multiple tissues, including craniofacial structures and peripheral nerves.
Sub-ventricular zone
Region of the brain lining the ventricles that generates neural and glial precursors.
Medulloblastoma
Paediatric brain tumour, believed to originate from primitive (undifferentiated) neuro-ectodermal cells, that most commonly arises in the cerebellum during the first decade of life and accounts for approximately 10% of primary brain tumours in children.
Alu elements
Short stretches of DNA containing the Alui restriction site that are repeated millions of times throughout the human genome.
Embryoid bodies
Aggregates of pluripotent cells that contain cells differentiating towards each of the three germ layers.
Microcephaly
Reduced head circumference.
Protocadherins
Family of cell adhesion proteins important for the development of neurons.
Acknowledgements
Research in the Shi lab is supported by grants from the US National Institutes of Health (RO1 GM117264, RO1 MH096066, R35 CA210104). Y.S. is an American Cancer Society Research Professor.
Footnotes
Competing interests
Y.S. is a cofounder of Constellation Pharmaceuticals and Athelas Therapeutics, as well as a consultant for Active Motif, Inc. A.J. and A.D. declare no competing interests.
Peer review information
Nature Reviews Molecular Cell Biology thanks C. Lu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at https://doi.org/10.1038/s41580-019-0151-1.
References
- 1.Greenberg M. & Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol In the press. [DOI] [PubMed]
- 2.Strahl BD & Allis CD The language of covalent histone modifications. Nature 403, 41–45 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Barski A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Bernstein BE et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).An early paper demonstrating bivalent histone methylation marks and their functions.
- 5.Mikkelsen TS et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Arensbergen J. et al. Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing beta-cells to adopt a neural gene activity program. Genome Res 20, 722–732 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein BE et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl Acad. Sci. USA 99, 8695–8700 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein BE et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120, 169–181 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Bannister AJ et al. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem 280, 17732–17736 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Wang Z. et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet 40, 897–903 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heintzman ND et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet 39, 311–318 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Trojer P. & Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol. Cell 28, 1–13 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Peters AH et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Rea S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000).First demonstration of the histone methyltransferase activity of the SET domain.
- 15.Rice JC et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12, 1591–1598 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Lin WJ, Gary JD, Yang MC, Clarke S. & Herschman HR The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem 271, 15034–15044 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Tang J. et al. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem 275, 7723–7730 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Guccione E. & Richard S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell .Biol 10.1038/s41580-019-0155-x (2019). [DOI] [PubMed]
- 19.Majumder S. et al. Methylation of histone H3 and H4 by PRMT5 regulates ribosomal RNA gene transcription. J. Cell. BioChem 109, 553–563 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dacwag CS, Ohkawa Y., Pal S., Sif S. & Imbalzano AN The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol. Cell. .Biol 27, 384–394 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girardot M. et al. PRMT5-mediated histone H4 arginine-3 symmetrical dimethylation marks chromatin at G + C-rich regions of the mouse genome. Nucleic Acids Res 42, 235–248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egger G., Liang G., Aparicio A. & Jones PA Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Dixon JR et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieberman-Aiden E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y. & Garcia BA Comprehensive catalog of currently documented histone modifications. Cold Spring Harb. Perspect. .Biol 7, a025064 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L. et al. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene 21, 148–152 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Tachibana M., Sugimoto K., Fukushima T. & Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem 276, 25309–25317 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Wang H. et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 8, 1207–1217 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Nishioka K. et al. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev 16, 479–489 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishioka K. et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 9, 1201–1213 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Feng Q. et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. .Biol 12, 1052–1058 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Ng HH et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev 16, 1518–1527 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Leeuwen F., Gafken PR & Gottschling DE Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109, 745–756 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell .Biol 20, 341–348 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P. & Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev 16, 2893–2905 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao R. et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Muller J. et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197–208 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Lee TI et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters AH et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Chen D. et al. Regulation of transcription by a protein methyltransferase. Science 284, 2174–2177 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Wang M., Xu RM & Thompson PR Substrate specificity, processivity, and kinetic mechanism of protein arginine methyltransferase 5. Biochemistry 52, 5430–5440 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Miranda TB, Miranda M., Frankel A. & Clarke S. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J. Biol. Chem 279, 22902–22907 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Lee JH et al. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J. Biol. Chem 280, 3656–3664 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Zurita-Lopez CI, Sandberg T., Kelly R. & Clarke SG Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming omega-NG-monomethylated arginine residues. J. Biol. Chem 287, 7859–7870 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi Y. et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004).First demonstration of enzymatic demethylase activity.
- 46.Metzger E. et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Cloos PA et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442, 307–311 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Klose RJ et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442, 312–316 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Tsukada Y. et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Whetstine JR et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467–481 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Franci G., Ciotta A. & Altucci L. The Jumonji family: past, present and future of histone demethylases in cancer. Biomol. Concepts 5, 209–224 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Lan F. et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 448, 718–722 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H. et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pena PV et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442, 100–103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi X. et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442, 96–99 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wysocka J. et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86–90 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Chen C., Nott TJ, Jin J. & Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nat. Rev. Mol. Cell .Biol 12, 629–642 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Thomson JP et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464, 1082–1086 (2010).This paper shows how crosstalk is established between DNA methylation and H3K4 methylation deposited by the SETD1 complex.
- 59.Laue K. et al. The multidomain protein Brpf1 binds histones and is required for Hox gene expression and segmental identity. Development 135, 1935–1946 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiu Y. et al. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes Dev 26, 1376–1391 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vezzoli A. et al. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat. Struct. Mol. .Biol 17, 617–619 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Ramon-Maiques S. et al. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc. Natl Acad. Sci. USA 104, 18993–18998 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matthews AG et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450, 1106–1110 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim JD et al. Proper activity of histone H3 lysine 4 (H3K4) methyltransferase is required for morphogenesis during zebrafish cardiogenesis. Mol. Cells 38, 580–586 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee J. et al. SETD7 drives cardiac lineage commitment through stage-specific transcriptional activation. Cell Stem Cell 22, 428–444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao XD et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 1, 286–298 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Lindeman LC et al. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev. Cell 21, 993–1004 (2011).Demonstration that inactive genes can be ‘pre-marked’ for expression later in development.
- 68.Dahl JA, Reiner AH, Klungland A., Wakayama T. & Collas P. Histone H3 lysine 27 methylation asymmetry on developmentally-regulated promoters distinguish the first two lineages in mouse preimplantation embryos. PLOS ONE 5, e9150 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hawkins RD et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6, 479–491 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu J. et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell 152, 642–654 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wen B., Wu H., Shinkai Y., Irizarry RA & Feinberg AP Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet 41, 246–250 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Towbin BD et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150, 934–947 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Nicetto D. et al. H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science 363, 294–297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pan G. et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1, 299–312 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Zhang B. et al. Widespread enhancer dememorization and promoter priming during parental-to-zygotic transition. Mol. Cell 72, 673–686 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Zheng H. et al. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol. Cell 63, 1066–1079 (2016). [DOI] [PubMed] [Google Scholar]
- 77.Zhang B. et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Wu J. et al. Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature 557, 256–260 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Calo E. & Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol. Cell 49, 825–837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Whyte WA et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221–225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cui K. et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4, 80–93 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lindeman LC et al. Chromatin states of developmentally-regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos. Int. J. Dev. .Biol 54, 803–813 (2010). [DOI] [PubMed] [Google Scholar]
- 83.Byrd KN & Shearn A. ASH1, a Drosophila trithorax group protein, is required for methylation of lysine 4 residues on histone H3. Proc. Natl Acad. Sci. USA 100, 11535–11540 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ingham PW & Whittle R. Trithorax: a new homoeotic mutation of Drosophila melanogaster causing transformations of abdominal and thoracic imaginal segments. Mol. Gen. Genet 179, 607–614 (1980). [Google Scholar]
- 85.Slifer EH A mutant stock of Drosophila with extra sex-combs. J. Exp. Zool 90, 31–40 (1942). [Google Scholar]
- 86.Lewis EB A gene complex controlling segmentation in Drosophila. Nature 276, 565–570 (1978). [DOI] [PubMed] [Google Scholar]
- 87.Struhl G. A gene product required for correct initiation of segmental determination in Drosophila. Nature 293, 36–41 (1981). [DOI] [PubMed] [Google Scholar]
- 88.Jurgens G., Wieschaus E., Nusslein-Volhard C. & Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster: II. Zygotic loci on the third chromosome. Wilehm Roux Arch. Dev. .Biol 193, 283–295 (1984). [DOI] [PubMed] [Google Scholar]
- 89.Nusslein-Volhard C., Wieschaus E. & Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster: I. Zygotic loci on the second chromosome. Wilehm Roux Arch. Dev. .Biol 193, 267–282 (1984). [DOI] [PubMed] [Google Scholar]
- 90.Wieschaus E., Nusslein-Volhard C. & Jurgens G. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster: III. Zygotic loci on the X-chromosome and fourth chromosome. Wilehm Roux Arch. Dev. .Biol 193, 296–307 (1984). [DOI] [PubMed] [Google Scholar]
- 91.Tsurumi A., Dutta P., Shang R., Yan SJ & Li WX Drosophila Kdm4 demethylases in histone H3 lysine 9 demethylation and ecdysteroid signaling. Sci. Rep 3, 2894 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Di Stefano L., Ji JY, Moon NS, Herr A. & Dyson N. Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development. Curr. .Biol 17, 808–812 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sankar A. et al. Maternal expression of the histone demethylase Kdm4a is crucial for pre-implantation development. Development 144, 3264–3277 (2017). [DOI] [PubMed] [Google Scholar]
- 94.Ancelin K. et al. Maternal LSD1/KDM1A is an essential regulator of chromatin and transcription landscapes during zygotic genome activation. eLife 5, e08851 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang J. et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446, 882–887 (2007). [DOI] [PubMed] [Google Scholar]
- 96.Tschiersch B. et al. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3–9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J 13, 3822–3831 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reuter G., Dorn R., Wustmann G., Friede B. & Rauh G. Third chromosome suppressor of position-effect variegation loci in Drosophila melanogaster. Mol. Gen. Genet 202, 481–487 (1986). [Google Scholar]
- 98.Andreu-Vieyra CV et al. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLOS .Biol 8, e1000453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dodge JE, Kang YK, Beppu H., Lei H. & Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. .Biol 24, 2478–2486 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Torres-Padilla ME, Parfitt DE, Kouzarides T. & Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445, 214–218 (2007).Seminal study showing that different amounts of Arg methylation within four-cell blastomeres can account for their different cell fates and potencies during early development.
- 101.Yadav N. et al. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl Acad. Sci. USA 100, 6464–6468 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tee WW et al. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev 24, 2772–2777 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pawlak MR, Scherer CA, Chen J., Roshon MJ & Ruley HE Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol. Cell. .Biol 20, 4859–4869 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hess JL, Yu BD, Li B., Hanson R. & Korsmeyer SJ Defects in yolk sac hematopoiesis in Mll-null embryos. Blood 90, 1799–1806 (1997). [PubMed] [Google Scholar]
- 105.Jude CD et al. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell 1, 324–337 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McMahon KA et al. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell 1, 338–345 (2007). [DOI] [PubMed] [Google Scholar]
- 107.Ernst P. et al. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev. Cell 6, 437–443 (2004). [DOI] [PubMed] [Google Scholar]
- 108.Saleque S., Kim J., Rooke HM & Orkin SH Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol. Cell 27, 562–572 (2007).Identification of a role and mechanism for LSD1 in regulating haematopoietic differentiation.
- 109.Thambyrajah R. et al. GFI1 proteins orchestrate the emergence of haematopoietic stem cells through recruitment of LSD1. Nat. Cell .Biol 18, 21–32 (2016). [DOI] [PubMed] [Google Scholar]
- 110.Sprussel A. et al. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia 26, 2039–2051 (2012). [DOI] [PubMed] [Google Scholar]
- 111.Kerenyi MA et al. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. eLife 2, e00633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ugarte F. et al. Progressive chromatin condensation and H3K9 methylation regulate the differentiation of embryonic and hematopoietic stem cells. Stem Cell Rep 5, 728–740 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van der Lugt NM et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev 8, 757–769 (1994). [DOI] [PubMed] [Google Scholar]
- 114.Ikeda K. et al. Maintenance of the functional integrity of mouse hematopoiesis by EED and promotion of leukemogenesis by EED haploinsufficiency. Sci. Rep 6, 29454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Oettinger MA, Schatz DG, Gorka C. & Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248, 1517–1523 (1990). [DOI] [PubMed] [Google Scholar]
- 116.Ai S. et al. Divergent requirements for EZH1 in heart development versus regeneration. Circ. Res 121, 106–112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ai S. et al. EED orchestration of heart maturation through interaction with HDACs is H3K27me3-independent. eLife 6, e24570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Akerberg AA, Henner A., Stewart S. & Stankunas K. Histone demethylases Kdm6ba and Kdm6bb redundantly promote cardiomyocyte proliferation during zebrafish heart ventricle maturation. Dev. .Biol 426, 84–96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lan F. et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449, 689–694 (2007). [DOI] [PubMed] [Google Scholar]
- 120.Wijayatunge R. et al. The histone demethylase Kdm6b regulates a mature gene expression program in differentiating cerebellar granule neurons. Mol. Cell. Neurosci 87, 4–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Park DH et al. Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep 8, 1290–1299 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gou Y. et al. Protein arginine methyltransferase PRMT1 is essential for palatogenesis. J. Dent. Res 97, 1510–1518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Iwase S. et al. A mouse model of X-linked intellectual disability associated with impaired removal of histone methylation. Cell Rep 14, 1000–1009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Scandaglia M. et al. Loss of Kdm5c causes spurious transcription and prevents the fine-tuning of activity-regulated enhancers in neurons. Cell Rep 21, 47–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lim DA et al. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458, 529–533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bezzi M. et al. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev 27, 1903–1916 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hashimoto M. et al. Severe hypomyelination and developmental defects are caused in mice lacking protein arginine methyltransferase 1 (PRMT1) in the central nervous system. J. Biol. Chem 291, 2237–2245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chittka A., Nitarska J., Grazini U. & Richardson WD Transcription factor positive regulatory domain 4 (PRDM4) recruits protein arginine methyltransferase 5 (PRMT5) to mediate histone arginine methylation and control neural stem cell proliferation and differentiation. J. Biol. Chem 287, 42995–43006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dhar SS et al. MLL4 is required to maintain broad H3K4me3 peaks and super-enhancers at tumor suppressor genes. Mol. Cell 70, 825–841 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Inagaki T. et al. Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice. Genes Cells 14, 991–1001 (2009). [DOI] [PubMed] [Google Scholar]
- 131.Tateishi K., Okada Y., Kallin EM & Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 458, 757–761 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kuroki S. et al. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science 341, 1106–1109 (2013). [DOI] [PubMed] [Google Scholar]
- 133.Okada Y., Scott G., Ray MK, Mishina Y. & Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 450, 119–123 (2007). [DOI] [PubMed] [Google Scholar]
- 134.Boswell RE & Mahowald AP tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell 43, 97–104 (1985). [DOI] [PubMed] [Google Scholar]
- 135.Capra JA Extrapolating histone marks across developmental stages, tissues, and species: an enhancer prediction case study. BMC Genomics 16, 104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Plath K. et al. Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131–135 (2003).Demonstration of the mechanism of X-chromosome inactivation via recruitment of PRC2 by Xist RNA.
- 137.Inoue A., Jiang L., Lu F. & Zhang Y. Genomic imprinting of Xist by maternal H3K27me3. Genes Dev 31, 1927–1932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang J. et al. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat. Genet 28, 371–375 (2001). [DOI] [PubMed] [Google Scholar]
- 139.Inoue A., Chen Z., Yin Q. & Zhang Y. Maternal Eed knockout causes loss of H3K27me3 imprinting and random X inactivation in the extraembryonic cells. Genes Dev 32, 1525–1536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lindroth AM et al. Antagonism between DNA and H3K27 methylation at the imprinted Rasgrf1 locus. PLOS Genet 4, e1000145 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Inoue A., Jiang L., Lu F., Suzuki T. & Zhang Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424 (2017).Demonstration of a histone-based mechanism of imprinting by H3K27 methylation.
- 142.Kuzmin A. et al. The PcG gene Sfmbt2 is paternally expressed in extraembryonic tissues. Gene Expr. Patterns 8, 107–116 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Miri K. et al. The imprinted polycomb group gene Sfmbt2 is required for trophoblast maintenance and placenta development. Development 140, 4480–4489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Harding K., Wedeen C., McGinnis W. & Levine M. Spatially regulated expression of homeotic genes in Drosophila. Science 229, 1236–1242 (1985). [DOI] [PubMed] [Google Scholar]
- 145.Kondo T. & Duboule D. Breaking colinearity in the mouse HoxD complex. Cell 97, 407–417 (1999). [DOI] [PubMed] [Google Scholar]
- 146.Soshnikova N. & Duboule D. Epigenetic temporal control of mouse Hox genes in vivo. Science 324, 1320–1323 (2009). [DOI] [PubMed] [Google Scholar]
- 147.Ingham PW Differential expression of Bithorax complex genes in the absence of the Extra sex combs and Trithorax genes. Nature 306, 591–593 (1983).First demonstration that Trithorax and Polycomb genes have opposing effects on Hox gene expression.
- 148.Simon J., Chiang A. & Bender W. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 114, 493–505 (1992). [DOI] [PubMed] [Google Scholar]
- 149.Struhl G. & Akam M. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J 4, 3259–3264 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Klymenko T. & Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep 5, 373–377 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yagi H. et al. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood 92, 108–117 (1998). [PubMed] [Google Scholar]
- 152.Yu BD, Hanson RD, Hess JL, Horning SE & Korsmeyer SJ MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl Acad. Sci. USA 95, 10632–10636 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yu BD, Hess JL, Horning SE, Brown GA & Korsmeyer SJ Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378, 505–508 (1995).Initial link between MLL1 and HOX gene expression during mammalian embryogenesis.
- 154.Breen TR & Harte PJ Molecular characterization of the trithorax gene, a positive regulator of homeotic gene expression in Drosophila. Mech. Dev 35, 113–127 (1991). [DOI] [PubMed] [Google Scholar]
- 155.Foster CT et al. Lysine-specific demethylase 1 regulates the embryonic transcriptome and CoREST stability. Mol. Cell. .Biol 30, 4851–4863 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ang SY et al. KMT2D regulates specific programs in heart development via histone H3 lysine 4 di-methylation. Development 143, 810–821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bledau AS et al. The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development 141, 1022–1035 (2014). [DOI] [PubMed] [Google Scholar]
- 158.Glaser S. et al. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 133, 1423–1432 (2006). [DOI] [PubMed] [Google Scholar]
- 159.Wu M. et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol. Cell. .Biol 28, 7337–7344 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Milne TA et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10, 1107–1117 (2002). [DOI] [PubMed] [Google Scholar]
- 161.Hu D. et al. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell. .Biol 33, 4745–4754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.O’Carroll D. et al. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. .Biol 21, 4330–4336 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Huang XJ et al. EZH2 is essential for development of mouse preimplantation embryos. Reprod. Fertil. Dev 26, 1166–1175 (2014). [DOI] [PubMed] [Google Scholar]
- 164.Wyngaarden LA, Delgado-Olguin P., Su IH, Bruneau BG & Hopyan S. Ezh2 regulates anteroposterior axis specification and proximodistal axis elongation in the developing limb. Development 138, 3759–3767 (2011). [DOI] [PubMed] [Google Scholar]
- 165.Schwarz D. et al. Ezh2 is required for neural crest-derived cartilage and bone formation. Development 141, 867–877 (2014). [DOI] [PubMed] [Google Scholar]
- 166.Faust C., Lawson KA, Schork NJ, Thiel B. & Magnuson T. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development 125, 4495–4506 (1998). [DOI] [PubMed] [Google Scholar]
- 167.Faust C., Schumacher A., Holdener B. & Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. Development 121, 273–285 (1995). [DOI] [PubMed] [Google Scholar]
- 168.Pasini D., Bracken AP, Jensen MR, Lazzerini Denchi E. & Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23, 4061–4071 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.van der Lugt NM, Alkema M., Berns A. & Deschamps J. The Polycomb-group homolog Bmi-1 is a regulator of murine Hox gene expression. Mech. Dev 58, 153–164 (1996). [DOI] [PubMed] [Google Scholar]
- 170.Agger K. et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734 (2007). [DOI] [PubMed] [Google Scholar]
- 171.Shpargel KB, Sengoku T., Yokoyama S. & Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLOS Genet 8, e1002964 (2012).This paper shows a catalytically independent role of KDM6A in recruiting chromatin regulators that can be substituted for by the catalytically inactive KDM6C.
- 172.Hong S. et al. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl Acad. Sci. USA 104, 18439–18444 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Naruse C. et al. New insights into the role of Jmjd3 and Utx in axial skeletal formation in mice. FASEB J 31, 2252–2266 (2017). [DOI] [PubMed] [Google Scholar]
- 174.Denissov S. et al. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development 141, 526–537 (2014). [DOI] [PubMed] [Google Scholar]
- 175.Zhang H. et al. MLL1 inhibition reprograms epiblast stem cells to naive pluripotency. Cell Stem Cell 18, 481–494 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lubitz S., Glaser S., Schaft J., Stewart AF & Anastassiadis K. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol. Biol. Cell 18, 2356–2366 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Glaser S. et al. The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis. Epigenetics Chromatin 2, 5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sze CC et al. Histone H3K4 methylation-dependent and -independent functions of Set1A/COMPASS in embryonic stem cell self-renewal and differentiation. Genes Dev 31, 1732–1737 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tachibana M. et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16, 1779–1791 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Obier N. et al. Polycomb protein EED is required for silencing of pluripotency genes upon ESC differentiation. Stem Cell Rev 11, 50–61 (2015). [DOI] [PubMed] [Google Scholar]
- 181.Jiang W., Wang J. & Zhang Y. Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway. Cell Res 23, 122–130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lee S., Lee JW & Lee SK UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev. Cell 22, 25–37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Adamo A. et al. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat. Cell .Biol 13, 652–659 (2011). [DOI] [PubMed] [Google Scholar]
- 184.Loh YH, Zhang W., Chen X., George J. & Ng HH Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev 21, 2545–2557 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wu Q. et al. CARM1 is required in embryonic stem cells to maintain pluripotency and resist differentiation. Stem Cells 27, 2637–2645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Xu Z. et al. MicroRNA-181 regulates CARM1 and histone arginine methylation to promote differentiation of human embryonic stem cells. PLOS ONE 8, e53146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cao K. et al. SET1A/COMPASS and shadow enhancers in the regulation of homeotic gene expression. Genes Dev 31, 787–801 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tomaz RA et al. Jmjd2c facilitates the assembly of essential enhancer-protein complexes at the onset of embryonic stem cell differentiation. Development 144, 567–579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wan X. et al. Mll2 controls cardiac lineage differentiation of mouse embryonic stem cells by promoting H3K4me3 deposition at cardiac-specific genes. Stem Cell Rev 10, 643–652 (2014). [DOI] [PubMed] [Google Scholar]
- 190.Lee YH et al. Protein arginine methyltransferase 6 regulates embryonic stem cell identity. Stem Cells Dev 21, 2613–2622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ma DK, Chiang CH, Ponnusamy K., Ming GL & Song H. G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem Cells 26, 2131–2141 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ang YS et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145, 183–197 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Jones WD et al. De novo mutations in MLL cause Wiedemann–Steiner syndrome. Am. J. Hum. Genet 91, 358–364 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Meyer E. et al. Mutations in the histone methyltransferase gene KMT2B cause complex early-onset dystonia. Nat. Genet 49, 223–237 (2017). [DOI] [PubMed] [Google Scholar]
- 195.Kleefstra T. et al. Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am. J. Hum. Genet 91, 73–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.De Rubeis S. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hannibal MC et al. Spectrum of MLL2 (ALR) mutations in 110 cases of Kabuki syndrome. Am. J. Med. Genet. A 155A, 1511–1516 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ng SB et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet 42, 790–793 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hiraide T. et al. De novo variants in SETD1B are associated with intellectual disability, epilepsy and autism. Hum. Genet 137, 95–104 (2018). [DOI] [PubMed] [Google Scholar]
- 200.Abidi FE et al. Mutations in JARID1C are associated with X-linked mental retardation, short stature and hyperreflexia. J. Med. Genet 45, 787–793 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Goncalves TF et al. KDM5C mutational screening among males with intellectual disability suggestive of X-Linked inheritance and review of the literature. Eur. J. Med. Genet 57, 138–144 (2014). [DOI] [PubMed] [Google Scholar]
- 202.Jensen LR et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet 76, 227–236 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chong JX et al. Gene discovery for Mendelian conditions via social networking: de novo variants in KDM1A cause developmental delay and distinctive facial features. Genet. Med 18, 788–795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tunovic S., Barkovich J., Sherr EH & Slavotinek AM De novo ANKRD11 and KDM1A gene mutations in a male with features of KBG syndrome and Kabuki syndrome. Am. J. Med. Genet. A 164A, 1744–1749 (2014). [DOI] [PubMed] [Google Scholar]
- 205.Kim HG et al. Translocations disrupting PHF21A in the Potocki-Shaffer-syndrome region are associated with intellectual disability and craniofacial anomalies. Am. J. Hum. Genet 91, 56–72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Horton JR et al. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat. Struct. Mol. .Biol 17, 38–43 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Feng W., Yonezawa M., Ye J., Jenuwein T. & Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me½ demethylation. Nat. Struct. Mol. .Biol 17, 445–450 (2010). [DOI] [PubMed] [Google Scholar]
- 208.Abidi F., Miano M., Murray J. & Schwartz C. A novel mutation in the PHF8 gene is associated with X-linked mental retardation with cleft lip/cleft palate. Clin. Genet 72, 19–22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Koivisto AM et al. Screening of mutations in the PHF8 gene and identification of a novel mutation in a Finnish family with XLMR and cleft lip/cleft palate. Clin. Genet 72, 145–149 (2007). [DOI] [PubMed] [Google Scholar]
- 210.Laumonnier F. et al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J. Med. Genet 42, 780–786 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kleefstra T. et al. Disruption of the gene euchromatin histone methyl transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J. Med. Genet 42, 299–306 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Qi HH et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466, 503–507 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Iacono G. et al. Increased H3K9 methylation and impaired expression of Protocadherins are associated with the cognitive dysfunctions of the Kleefstra syndrome. Nucleic Acids Res 46, 4950–4965 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Gibson WT et al. Mutations in EZH2 cause Weaver syndrome. Am. J. Hum. Genet 90, 110–118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Douglas J. et al. NSD1 mutations are the major cause of Sotos syndrome and occur in some cases of Weaver syndrome but are rare in other overgrowth phenotypes. Am. J. Hum. Genet 72, 132–143 (2003).Describes non-overlapping sets of point mutations or deletions in NSD1 that are associated with two distinct syndromes.
- 216.Rio M. et al. Spectrum of NSD1 mutations in Sotos and Weaver syndromes. J. Med. Genet 40, 436–440 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Baujat G. et al. Paradoxical NSD1 mutations in Beckwith–Wiedemann syndrome and 11p15 anomalies in Sotos syndrome. Am. J. Hum. Genet 74, 715–720 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bennett RL, Swaroop A., Troche C. & Licht JD The role of nuclear receptor-binding SET domain family histone lysine methyltransferases in cancer. Cold Spring. Harb. Perspect. Med 7, a026708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Laugesen A., Hojfeldt JW & Helin K. Role of the polycomb repressive complex 2 (PRC2) in transcriptional regulation and cancer. Cold Spring Harb. Perspect. Med 6, a026575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Michalak EW, Burr ML, Bannister AJ & Dawson MA The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell .Biol 10.1038/s41580-019-0143-1 (2019). [DOI] [PubMed]
- 221.Rahman N. Mechanisms predisposing to childhood overgrowth and cancer. Curr. Opin. Genet. Dev 15, 227–233 (2005). [DOI] [PubMed] [Google Scholar]
- 222.Akawi N. et al. Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families. Nat. Genet 47, 1363–1369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Faundes V. et al. Histone lysine methylases and demethylases in the landscape of human developmental disorders. Am. J. Hum. Genet 102, 175–187 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Pilotto S. et al. LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors. Hum. Mol. Genet 25, 2578–2587 (2016). [DOI] [PubMed] [Google Scholar]
- 225.Brookes E. et al. Mutations in the intellectual disability gene KDM5C reduce protein stability and demethylase activity. Hum. Mol. Genet 24, 2861–2872 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Blanc RS, Vogel G., Chen T., Crist C. & Richard S. PRMT7 preserves satellite cell regenerative capacity. Cell Rep 14, 1528–1539 (2016). [DOI] [PubMed] [Google Scholar]
- 227.Bouchard C. et al. Genomic location of PRMT6-dependent H3R2 methylation is linked to the transcriptional outcome of associated genes. Cell Rep 24, 3339–3352 (2018). [DOI] [PubMed] [Google Scholar]
- 228.Guccione E. et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449, 933–937 (2007).Describes antagonism between MLL methyltransferases and PRMT6 in depositing H3K4 and H3R2 methylation, respectively.
- 229.Lee MG et al. Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell. .Biol 26, 6395–6402 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Shi YJ et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19, 857–864 (2005). [DOI] [PubMed] [Google Scholar]
- 231.Yin F. et al. LSD1 regulates pluripotency of embryonic stem/carcinoma cells through histone deacetylase 1-mediated deacetylation of histone H4 at lysine 16. Mol. Cell. .Biol 34, 158–179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Stein C., Notzold RR, Riedl S., Bouchard C. & Bauer UM The arginine methyltransferase PRMT6 cooperates with Polycomb proteins in regulating HOXA gene expression. PLOS ONE 11, e0148892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Fei Q. et al. SETDB1 modulates PRC2 activity at developmental genes independently of H3K9 trimethylation in mouse ES cells. Genome Res 25, 1325–1335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Cao K. et al. An Mll4/COMPASS-Lsd1 epigenetic axis governs enhancer function and pluripotency transition in embryonic stem cells. Sci. Adv 4, eaap8747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Cho YW et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem 282, 20395–20406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Issaeva I. et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. .Biol 27, 1889–1903 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lee MG et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318, 447–450 (2007). [DOI] [PubMed] [Google Scholar]
- 238.Wang SP et al. A UTX-MLL4-p300 transcriptional regulatory network coordinately shapes active enhancer landscapes for eliciting transcription. Mol. Cell 67, 308–321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Miller SA, Mohn SE & Weinmann AS Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell 40, 594–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ooi SK et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lachner M., O’Carroll D., Rea S., Mechtler K. & Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001). [DOI] [PubMed] [Google Scholar]
- 242.Lehnertz B. et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. .Biol 13, 1192–1200 (2003). [DOI] [PubMed] [Google Scholar]
- 243.Sarraf SA & Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell 15, 595–605 (2004). [DOI] [PubMed] [Google Scholar]
- 244.Epsztejn-Litman S. et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat. Struct. Mol. .Biol 15, 1176–1183 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Murray K. The occurrence of epsilon-N-methyl lysine in histones. Biochemistry 3, 10–15 (1964). [DOI] [PubMed] [Google Scholar]
- 246.Falnes PO, Jakobsson ME, Davydova E., Ho A. & Malecki J. Protein lysine methylation by seven-beta-strand methyltransferases. Biochem. J 473, 1995–2009 (2016). [DOI] [PubMed] [Google Scholar]
- 247.Larsen SC et al. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal 9, rs9 (2016). [DOI] [PubMed] [Google Scholar]
- 248.Chuikov S. et al. Regulation of p53 activity through lysine methylation. Nature 432, 353–360 (2004). [DOI] [PubMed] [Google Scholar]
- 249.Kouskouti A., Scheer E., Staub A., Tora L. & Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell 14, 175–182 (2004). [DOI] [PubMed] [Google Scholar]
- 250.Armstrong JF, Kaufman MH, Harrison DJ & Clarke AR High-frequency developmental abnormalities in p53-deficient mice. Curr. .Biol 5, 931–936 (1995). [DOI] [PubMed] [Google Scholar]
- 251.Kaufman MH et al. Analysis of fused maxillary incisor dentition in p53-deficient exencephalic mice. J. Anat 191, 57–64 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Rinon A. et al. p53 coordinates cranial neural crest cell growth and epithelial-mesenchymal transition/ delamination processes. Development 138, 1827–1838 (2011). [DOI] [PubMed] [Google Scholar]
- 253.Sah VP et al. A subset of p53-deficient embryos exhibit exencephaly. Nat. Genet 10, 175–180 (1995). [DOI] [PubMed] [Google Scholar]
- 254.Saifudeen Z., Dipp S. & El-Dahr SS A role for p53 in terminal epithelial cell differentiation. J. Clin. Invest 109, 1021–1030 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.He A. et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev 26, 37–42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Rasmussen TL et al. Smyd1 facilitates heart development by antagonizing oxidative and ER stress responses. PLOS ONE 10, e0121765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Pek JW, Anand A. & Kai T. Tudor domain proteins in development. Development 139, 2255–2266 (2012). [DOI] [PubMed] [Google Scholar]
- 258.Pengelly AR, Copur O., Jackle H., Herzig A. & Muller J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339, 698–699 (2013). [DOI] [PubMed] [Google Scholar]
- 259.McKay DJ et al. Interrogating the function of metazoan histones using engineered gene clusters. Dev. Cell 32, 373–386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Vermillion KL, Lidberg KA & Gammill LS Cytoplasmic protein methylation is essential for neural crest migration. J. Cell .Biol 204, 95–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Boisvert FM, Chenard CA & Richard S. Protein interfaces in signaling regulated by arginine methylation. Sci. STKE 2005, re2 (2005). [DOI] [PubMed]
- 262.Xu W. et al. A transcriptional switch mediated by cofactor methylation. Science 294, 2507–2511 (2001). [DOI] [PubMed] [Google Scholar]
- 263.Shishkova E. et al. Global mapping of CARM1 substrates defines enzyme specificity and substrate recognition. Nat. Commun 8, 15571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Hu SB et al. Protein arginine methyltransferase CARM1 attenuates the paraspeckle-mediated nuclear retention of mRNAs containing IRAlus. Genes Dev 29, 630–645 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Hupalowska A. et al. CARM1 and paraspeckles regulate pre-implantation mouse embryo development. Cell 175, 1902–1916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kim D. et al. Enzymatic activity is required for the in vivo functions of CARM1. J. Biol. Chem 285, 1147–1152 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Nimura K. et al. A histone H3 lysine 36 trimethyltransferase links Nkx2–5 to Wolf–Hirschhorn syndrome. Nature 460, 287–291 (2009). [DOI] [PubMed] [Google Scholar]
- 268.Kuroki S. et al. Combined loss of JMJD1A and JMJD1B reveals critical roles for H3K9 demethylation in the maintenance of embryonic stem cells and early embryogenesis. Stem Cell Rep 10, 1340–1354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Neault M., Mallette FA, Vogel G., Michaud-Levesque J. & Richard S. Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res 40, 9513–9521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ezhkova E. et al. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 25, 485–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Yatsenko SA et al. Deletion 9q34.3 syndrome: genotype-phenotype correlations and an extended deletion in a patient with features of Opitz C trigonocephaly. J. Med. Genet 42, 328–335 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.