Abstract
Gene regulatory networks and the dynamic responses they produce offer a wealth of information about how biological systems process information about their environment. Recently, researchers interested in dissecting these networks have been outsourcing various parts of their experimental workflow to computers. Here we review how, using microfluidic or optogenetic tools coupled with fluorescence imaging, it is now possible to interface cells and computers. These platforms enable scientists to perform informative dynamic stimulations of genetic pathways and monitor their reaction. It is also possible to close the loop and regulate genes in real time, providing an unprecedented view of how signals propagate through the network. Finally, we outline new tools that can be used within the framework of cell-machine interfaces.
Introduction
Deciphering the inner workings of regulatory networks within cells is arguably one of the most daunting reverse engineering problems today. Through billions of years of evolution, organisms have been optimized in a massively parallelized process to adapt to a plethora of changing environments. Dissecting the dynamics of these complex gene regulatory networks is an important step in our comprehension of living systems. However, even long after Jacob and Monod’s foundational discovery on cellular regulation of lactose metabolism [1], tools for dissecting such networks remained primarily limited to gene knock-outs. In addition to requiring methods for chromosomal modification, knock-outs do not allow experimenters to follow the effect of their perturbation dynamically, requiring researchers to deduce the connections within gene networks from static, steady state measurements. Decades later, the emergence of chemically or heat responsive genetic induction systems ushered in a new wave of experiments that formed the foundation for the field of synthetic biology [2–4]. With those inducible systems, it was now possible to perturb gene regulatory networks and track their reaction over time. Over the next few years, synthetic biologists set out to develop genetic circuits that would allow them to precisely control gene expression or cellular processes to study genetic networks (for an excellent comprehensive review see Ref. [5]). In addition, the emergence of optogenetic tools [6–9] to steer gene expression has expanded the toolbox that scientists can use to dissect the regulatory mechanisms of the cell.
In this review, we focus on a new paradigm that has appeared with the emergence and refinement of these tools: systems and synthetic biologists have started outsourcing the signal processing part of their genetic circuits to computers. By using microfluidics or computer-controlled light signals on the one hand and automated microscopy or flow cytometry on the other, they have implemented cell-machine interfaces in which a computer is able to both perturb and monitor expression of genes in living cells in a completely automated process. These advances have off-loaded intricate and often burdensome genetic circuit design to easily-configurable and rewritable software, which greatly speeds up the design-build-test cycle for implementing new perturbations. Here, we review this nascent and evolving field, the classes of scientific questions it can help investigate, and how it might expand.
Time-varying stimulations
Gene regulation networks process signals. While static perturbations like gene knock-outs can give some insight into their operation, time-varying stimulations provide more information about how the system adapts to changes. With time-varying perturbations, the experimenter can observe both the resting state of gene regulatory networks and their reaction to a stimulus. Without measuring these transient effects, one might miss critical nodes or connections in the network that would not appear to be impacted in steady state measurements [10,11]. For this reason, scientists have developed chemically inducible systems that can deliver well-defined dynamic inputs to their circuit.
By combining chemical inputs with computer-controlled time-lapse fluorescence microscopy, researchers have started assembling cell-machine interfaces that can extract an increasing amount of information about their system in an entirely automated process. In 2008, pioneering studies in the model organism Saccharomyces cerevisiae demonstrated the potential of this approach [12–14]. Oscillatory fluctuations in the nutritive or osmotic environment of the cells not only gave insights into the frequency-response of essential cell adaptation pathways, but also unearthed key regulatory interactions within those pathways. An attractive aspect of these automated experimental platforms is the relative ease of scaling up and massively parallelizing the technique: a few years later Dénervaud and colleagues [15] conducted a proteome-wide screen of dynamic responses to chemical and UV stresses in S. cerevisae by simultaneously monitoring over one thousand separate experiments per microfluidic chip.
Albeit less heavily parallelized, a similar approach using 96 independent experiments was adopted by Sorre et al. to look into signal processing in embryogenesis regulation in mammalian cells [16]. By using step changes, ramps, and trains of pulses in the concentration of the TGF-β morphogen in the cell’s environment, the authors demonstrated that the differentiation pathway they studied is mostly responsive to the rate of change in the morphogen concentration rather than its absolute value. This led them to propose a new mechanism for embryogenic cell-fate determination: While gradients of morphogens do direct spatial organization, the rate of change in the morphogen concentration varies throughout the gradient, and gene regulation networks involved in cell differentiation might respond to the time-derivative of the establishment of the gradient. The study of this mechanism would have been near impossible had the authors not been able to dynamically alter the environment of their cells. In a similar spirit, Young and coworkers used microfluidics to inject chemical stressors in the environment of Bacillus subtilis cells, ramping the concentration up at various rates [17]. The authors showed that induction under the σB sigma factor, which targets over 200 genes in response to stresses, is adaptive, meaning that the rate of stress introduction will influence the intensity of activation. More recently, a commanding study of stress response in yeast [18] used tools from information theory to decipher how environmental information is encoded into gene regulation dynamics using single-cell microscopy acquisitions under time-varying inputs. The authors analyzed the dynamics of ten transcription factors known to be related to stress response, and demonstrated how their temporal response profile encoded not only the nature of the stresses that the cells were subjected to, but also their magnitude.
In the past decade, a number of optogenetic induction systems have been developed for bacteria [6,9], yeast [7], and mammalian cells [8]. Light is an attractive signal in that it is relatively straightforward to program and apply in time-varying inputs, in contrast to chemically inducible signals that typically require some kind of microfluidic apparatus. Harnessing the tight and precise control offered by optogenetics, Olson and co-workers used pre-determined sequences of light inputs to precisely drive gene expression over time [19]. They then used their method as a “function generator” to characterize the behavior of a synthetic inverter circuit. Using a light-activated enzyme, Stewart-Orstein and colleagues investigated the protein kinase A (PKA) pathway [20]. This approach allowed them to directly perturb the pathway internally, with a temporal precision and resolution that identified new dynamical relationships within this pathway that adapt on the order of minutes. Wilson and co-workers recently used optogenetics to control the Ras/Erk pathway, which is a fundamental network in cell division and growth in mammalian cells [21,22]. They also monitored expression of downstream genes and followed the regulatory activity of the pathway at the signaling, transcriptional, and translational levels simultaneously. By applying various time-varying profiles as inputs, they identified frequency-dependent processing of the optogenetic input as well as logic gating between the processed optogenetic input and other environmental cues, and modeled the pathway’s behavior based on their experimental data. Taken together, these examples demonstrate the wealth of information that can be extracted about a gene regulatory network by applying time-varying perturbations. In many of these cases, the knowledge gathered about the networks would have been impossible to obtain without some kind of dynamic stimulus.
Cell-machine interfaces and computer-based feedback
Although microfluidics and optogenetics can be used to manipulate the environment of cells or induce gene expression, reverse engineering often requires precise and robust manipulation of elements within the system itself. In recent years, this has led several teams to close the loop between perturbation and monitoring by developing cell-machine interfaces that implement computer-based feedback to steer the system towards a desired, potentially informative state. Synthetic biologists have genetically encoded feedback mechanisms within synthetic circuits [23–25], and recent developments in the field promise ever finer control [26,27]. However the flexibility offered by running the algorithmic part of the feedback loop through a computer or microcontroller offers the possibility to both rapidly iterate over different feedback parameters or strategies, and to implement complex feedback algorithms from modern control theory.
Toettcher et al. [28] used the optogenetic Phy-PIF system [29] to precisely control recruitment of both fluorescent proteins and enzymes at the membrane of human HeLa cells on the timescale of seconds to minutes. They used a relatively simple proportional-integral control scheme to achieve this. Proportional-integral control is a ubiquitous algorithm in traditional engineering fields that has been around for over a century. Despite its relative mathematical simplicity, it has not yet been successfully implemented in synthetic genetic circuits. Out-sourcing the feedback control to a computer can allow for implementation of algorithms that are not easily achievable with biological parts.
In another example, Milias-Argeitis et al. were able to control gene expression in S. cerevisiae over periods of hours [30]. The authors also used the Phy-PIF optogenetic system to control fluorescence levels in a batch liquid culture by periodically measuring cells state in a flow cytometer. They used a feedback strategy called model predictive control, in which a minimal mathematical model of the genetic system is used to infer the optimal inputs that will drive the cells to the desired fluorescence level. This framework is particularly well-adapted to the control of non-linear, noisy systems, but its algorithmic complexity far exceeds the current capabilities of synthetic circuit engineering. A study by Uhlendorf et al. focused on single-cell control in S. cerevisiae by driving the expression level of a promoter downstream of the endogenous osmolarity-responsive HOG signaling pathway via microfluidics [31]. The authors also used model predictive control and were able to manipulate the fluorescence level of a single cell for over sixteen hours.
Since then, the field has been expanding, both in the variety of systems investigated and finesse of the control. Days-long control of gene expression in mammalian cell lines has been reported using a variety of chemically-inducible promoters with microfluidics [32,33]. Chait and coworkers [34] combined microfluidics and optogenetics to control fluorescence levels in several single Escherichia coli cells independently, reducing cell-to-cell variation at the population level. They then used their platform to implement cell-to-cell communication through the computer, paving the way for the study of group behavior through single-cell control. Beyond gene expression level manipulation, feedback systems also make it possible to control other measurable properties of the cells. For example, an interesting tool for cell physiology studies has been implemented by Milias-Argeitis and coworkers [35]. By placing an essential synthase under the control of the CcaS/R bacterial optogenetic system [9,36] they were able to precisely control the growth rate of batch cultures of E. coli cells [35].
In the last two years, studies have appeared using computer-based feedback and synthetic biology methods to investigate questions relevant to systems biology. By studying the controllability of a bistable genetic toggle switch [37], an architecture common in cellular decision-making circuits, researchers serendipitously discovered a dynamic stabilization mechanism that can explain how pluripotency is maintained prior to cell fate determination [38]. In another study, Rullan et al. [39] used a single-cell optogenetic feedback interface to control transcriptional activity. By automatically detecting nascent RNA transcripts in single cells, they quantitively investigated transcriptional bursting in yeast, and proposed a mechanistic model describing the relationship between transcription factor binding and bursting dynamics that hints at the potential effect of histones. Finally, the traditional approach to disentangling feedback in a regulatory network typically involves deleting a potential regulator and identifying the nodes in the network that are perturbed. However this yields no information regarding the dynamics of those feedback loops. In a remarkable effort to quantify and parameterize such feedback loops in the yeast pheromone response MAPK pathway, Harrigan et al. replaced endogenous pathway regulators with light-inducible ones, and used model predictive control to try to compensate for the lost feedback loops [40]. Although the first attempts sometimes failed, the information gathered allowed them to re-fit their models until they achieved satisfactory compensation, thus inferring dynamical models for three feedback loops in the MAPK network. This interplay between iterative parameter inference and computer-based feedback control is a promising avenue for fast, automated regulatory network analysis.
Optimal experimental design
Because precise and dynamically complex genetic perturbations are now possible, and finely-resolved data can be extracted out of experiments, the refinement of our analysis and mathematical models of gene regulatory networks keeps increasing. But investigating and dissecting these networks in ever more details calls for more informative experiments. Moreover, with dynamic perturbations, the parameter space for designing experiments increases dramatically. Optimal experimental design is a powerful tool to minimize the number of experiments necessary to investigate a gene regulatory network. Given a system to reverse engineer, the core concept of optimal experimental design is to calculate what the optimal series of experiments would be to perform on that system in order to produce data that would best constrain its modeling. Theoretical studies describing its potential use in systems biology arose early [41–44]. At the time, because both perturbations and observations were less dynamically refined, the studies focused on static genetic manipulations to infer network structure. Since then, with technical advances, a body of work has been building up on optimal time-varying inputs to maximize dynamic understanding of gene regulation networks [45–47]. However, these studies have remained predominantly theoretical, and few experimenters actually incorporate optimal experimental design into their workflow. A notable exception is the work by Bandara and colleagues in which an ordinary differential equation model describing the phosphatidylinositol 3,4,5-trisphosphate (PIP3) lipid signaling dynamics in mammalian cells was fitted to the results of iteratively-designed experiments with time-varying chemical inputs [48]. In a remarkable testament to the power of optimal experimental design, the study showed that only two iterative experiments suffice to reduce the uncertainty on the parameter estimates by over 60-fold, where previous “intuitively designed” experiments failed to constrain the fit. Another notable study reports the optimal experimental design-driven characterization of an optogenetic system in yeast [49]. After the authors demonstrate that, again, only two optimally-designed experiments were necessary to constrain the parameters of the model, they used their fitted model to perform computer-based, model predictive feedback control on both the average fluorescence level of their population and its variance. While experimental implementation of optimal experimental design is still somewhat lacking, we expect that it will combine synergistically with advanced dynamic stimulation frameworks and cell-machine interfaced feedback control to play an increasing role in gene regulatory network analysis.
Current limitations and new tools for controlling gene regulation networks
Cell-machine interfaces arose thanks to technical advances in, among other things, molecular biology, microfluidics, optogenetics, and image processing. Naturally, their capabilities are constrained by the current limitations of these underlying technologies. For example, long term single-cell control of gene expression in mammalian cells is a major challenge because it requires near perfect on-the-fly image segmentation. Recently however, a revolutionary deep learning-based segmentation framework, U-Net [50], has been streamlined for use in biomedical acquisitions [51] and has shown outstanding results for mammalian cell segmentation. Another limitation of these platforms is the difficulty in measuring gene expression deep within tissues in real time, which confines the interfaces to the study of either monolayers of single cells or liquid cultures that can be sampled from. An exciting potential solution that could bring cell-machine interfaces into animal models is photoacoustic tomography [52]. This technique can exploit the photoacoustic properties of reporter proteins to measure gene expression in a non-invasive way, at depths of several millimeters to centimeters.
Regardless of the organism, experimenters are limited to the study of gene regulation networks that can be perturbed, which restricts them to either systems that naturally respond to environmental stimuli, or to potentially disruptive genetic modifications. Fortunately, technological solutions for better and easier manipulation of cellular processes are emerging. CRISPR-derived genome-targeting tools, especially photo-activatable ones [53–56], are a clear example. CRISPR activation/inhibition systems can be used to target and perturb endogenous pathways within their physiologically relevant ranges and with minimal off-target interference. These tools offer the exciting potential for time-varying dCas9-mediated perturbations to investigate gene regulation dynamics. Photoactivatable recombinases offer another potential solution [57]. With this system, not only can a gene of interest be turned completely off by excising its DNA, it can also be turned from off to on with appropriate arrangement of the recombinase’s recognition sites. This tool allows the experimenter to investigate the effect of complete gene knock-outs or knock-ins dynamically. The emergence of frameworks for automated circuit design and cloning [58] could also lead researchers to study cellular processes more extensively and systematically. By automating the time-consuming and often repetitive genetic modifications required to interface the machine with different parts of a gene regulation network, studying complex networks with an otherwise impractically large number of genes may become possible. An exciting potential development would be to integrate the design and construction of new strains into an optimal experimental design workflow, suggesting both the genetic modifications and the dynamic experiments that would yield the most information.
Increasing the number of inputs and outputs in the network also offers the potential to increase performance. Optogenetic actuation and fluorescence sensing quickly run into spectral crowding issues, especially when used simultaneously. As an alternative approach, the development of microfluidic chips integrating valves and pumps, like the one used by Sorre et al. [16], makes it possible to use several chemically inducible systems to perturb gene regulation networks. The renewed interest in fluorescent probe development that followed the super-resolution breakthrough [59,60] could help extract more information out of dynamic experiments. By expanding the toolbox of available biosensors, protein engineers have made it possible to investigate, and possibly control, gene regulation networks in more refined ways. These advances can also enable tracking of more nodes simultaneously in a network, as illustrated by the signaling-transcription-translation monitoring described in Wilson et al. [22]. Exciting studies have shown how the use of photoswitchable fluorescent reporters with modulated light signals can discriminate between spectrally similar reporters [61,62]. Such techniques would make it possible to monitor many nodes simultaneously in gene networks. Finally, further development of periodic sampling platforms would allow cell-machine interfaces to be integrated with other, more holistic cell biology techniques like RNA-seq to contextualize the knowledge acquired through dynamic stimulation of an isolated gene regulation network.
Conclusions
Following the same trend that more traditional engineering fields followed decades before, synthetic biologists have started interacting with the physical processes they intend to perturb and analyze through the digital realm. Much like electromechanical actuators and sensors, microfluidic and optogenetic tools and biosensors allow researchers to submit genetic systems to programmed, time-varying inputs and to monitor their reaction over time. By closing the loop between sensing and actuation, experimenters have now started to precisely and robustly steer nodes within gene regulatory networks. While the domain is still young and constantly evolving, we expect this approach to spread as standards and protocols are established. We also anticipate that recent progress in the field of optimal experimental design can be used in the framework of cell-machine interfaces to drive on-line experimentation. In conjunction with new minimally-invasive chemical or light-inducible actuators and fluorescent biosensors, autonomous and automated investigation of the dynamics of gene regulatory networks, possibly in a massively parallelized fashion, has the potential to revolutionize our approach to systems biology.
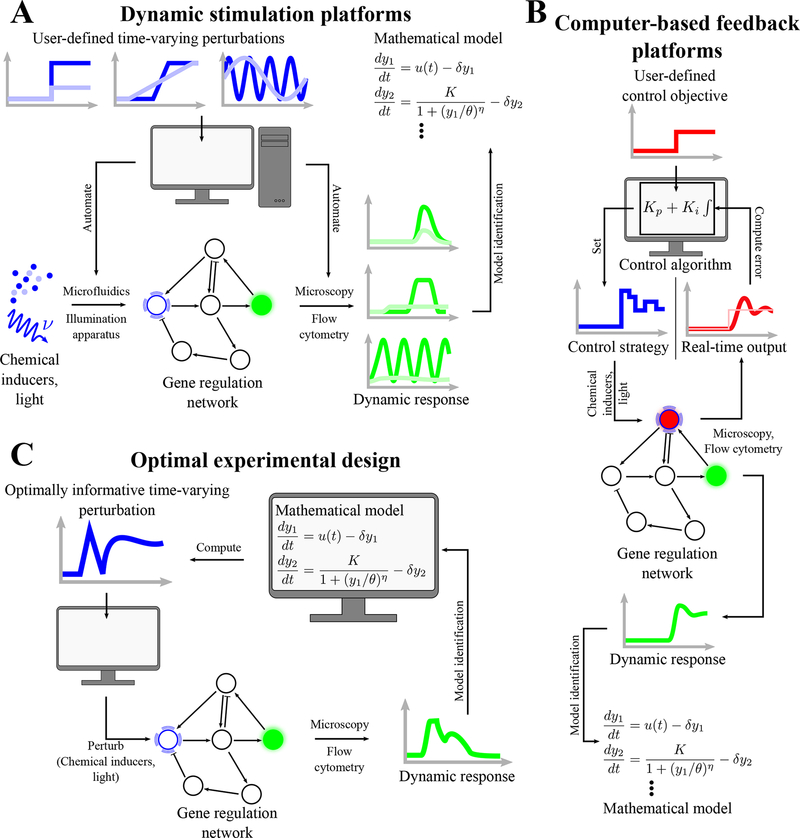
Figure 1.
Computer-based study of gene regulatory networks. (A) Computers are used to subject cells to dynamic stimulations using chemical inducers or light. Automated microscopy or flow cytometry is used to monitor the dynamic response of the gene regulatory network in real time, and the data is then analyzed to infer a mathematical model capturing the dynamics of the system. (B) Automated microscopy or flow cytometry can also be used to implement feedback control through the computer. The state of one or more nodes of the regulatory network is measured and compared to a reference objective that the experimenter chooses a priori. This error is fed into a control algorithm that will modify the chemical or light input in real-time to steer the controlled gene or genes towards the objective. One or more other nodes can also be monitored to study their reaction to this internal perturbation. (C) A model of the system can be used to infer what dynamic perturbation would yield the most information to improve understanding and modeling of the system. The process can be iterated, with the improved model being used to design new perturbations.
Highlights.
Cell-machine interfaces are increasingly used to perturb gene regulatory networks and study their dynamics.
By implementing computer-based feedback loops, researchers can precisely and robustly manipulate the inner workings of genetic pathways.
Optimal experimental design can help identify and model the structure of gene regulatory networks.
Acknowledgements
We thank Dr. Imane El Meouche, Nathan Tague, and Michael B. Sheets for helpful comments on the manuscript. This work was supported by the National Institutes of Health grant R21AI137843.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jacob F, Monod J: Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 1961, 3:318–356. [DOI] [PubMed] [Google Scholar]
- 2.Lutz R, Bujard H: Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 1997, 25:1203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skerra A: Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene 1994, 151:131–135. [DOI] [PubMed] [Google Scholar]
- 4.Guzman LM, Belin D, Carson MJ, Beckwith J: Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 1995, 177:4121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bashor CJ, Collins JJ: Understanding Biological Regulation Through Synthetic Biology. Annu Rev Biophys 2018, 47:399–423. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z, Zhang J, Jin J, Geng Z, Qi Q, Liang Q: Programming Bacteria With Light—Sensors and Applications in Synthetic Biology. Front Microbiol 2018, 9:2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salinas F, Rojas V, Delgado V, Agosin E, Larrondo LF: Optogenetic switches for light-controlled gene expression in yeast. Appl Microbiol Biotechnol 2017, 101:2629–2640. [DOI] [PubMed] [Google Scholar]
- 8.Beyer HM, Naumann S, Weber W, Radziwill G: Optogenetic control of signaling in mammalian cells. Biotechnol J 2015, 10:273–283. [DOI] [PubMed] [Google Scholar]
- 9.Ong NTX, Tabor JJ: A miniaturized E. coli green light sensor with high dynamic range. ChemBioChem 2018, doi: 10.1002/cbic.201800007. [DOI] [PubMed] [Google Scholar]
- 10.Locke JCW, Elowitz MB: Using movies to analyse gene circuit dynamics in single cells. Nat Rev Microbiol 2009, 7:383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett MR, Hasty J: Microfluidic devices for measuring gene network dynamics in single cells. Nat Rev Genet 2009, 10:628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mettetal JT, Muzzey D, Gomez-Uribe C, van Oudenaarden A: The Frequency Dependence of Osmo-Adaptation in Saccharomyces cerevisiae. Science (80- ) 2008, 319:482–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett MR, Pang WL, Ostroff N a, Baumgartner BL, Nayak S, Tsimring LS, Hasty J: Metabolic gene regulation in a dynamically changing environment. Nature 2008, 454:1119–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hersen P, McClean MN, Mahadevan L, Ramanathan S: Signal processing by the HOG MAP kinase pathway. Proc Natl Acad Sci 2008, 105:7165–7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dénervaud N, Becker J, Delgado-Gonzalo R, Damay P, Rajkumar AS, Unser M, Shore D, Naef F, Maerkl SJ: A chemostat array enables the spatio-temporal analysis of the yeast proteome. Proc Natl Acad Sci 2013, 110:15842–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorre B, Warmflash A, Brivanlou AH, Siggia E: Encoding of Temporal Signals by the TGF-β Pathway and Implications for Embryonic Patterning. Dev Cell 2014, 30:334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young JW, Locke JCW, Elowitz MB: Rate of environmental change determines stress response specificity. Proc Natl Acad Sci 2013, 110:4140–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granados AA, Pietsch JMJ, Cepeda-Humerez SA, Farquhar IL, Tkačik G, Swain PS: Distributed and dynamic intracellular organization of extracellular information. Proc Natl Acad Sci 2018, 115:6088–6093.•• The authors submit trapped single Saccharomyces cerevisiae cells in a microfluidic device to various chemical stresses. By monitoring the activity of several genes connected to stress response, they break down the temporal encoding of stresses in the dynamics of those genes and identify specific response strategies.
- 19.Olson EJ, Hartsough LA, Landry BP, Shroff R, Tabor JJ: Characterizing bacterial gene circuit dynamics with optically programmed gene expression signals. Nat Methods 2014, 11:449–455. [DOI] [PubMed] [Google Scholar]
- 20.Stewart-Ornstein J, Chen S, Bhatnagar R, Weissman JS, El-Samad H: Model-guided optogenetic study of PKA signaling in budding yeast. Mol Biol Cell 2017, 28:221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toettcher JE, Weiner OD, Lim WA: Using Optogenetics to Interrogate the Dynamic Control of Signal Transmission by the Ras/Erk Module. Cell 2013, 155:1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson MZ, Ravindran PT, Lim WA, Toettcher JE: Tracing Information Flow from Erk to Target Gene Induction Reveals Mechanisms of Dynamic and Combinatorial Control. Mol Cell 2017, 67:757–769.e5.• The authors optogenetically perturb signaling through the Ras/Erk pathway and monitor transcription and translation of target genes. They uncover dynamic and combinatorial gating.
- 23.Nevozhay D, Adams RM, Murphy KF, Josic K, Balazsi G: Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc Natl Acad Sci 2009, 106:5123–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bashor CJ, Helman NC, Yan S, Lim WA: Using Engineered Scaffold Interactions to Reshape MAP Kinase Pathway Signaling Dynamics. Science (80- ) 2008, 319:1539–1543. [DOI] [PubMed] [Google Scholar]
- 25.Del Vecchio D, Dy AJ, Qian Y: Control theory meets synthetic biology. J R Soc Interface 2016, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Briat C, Gupta A, Khammash M: Antithetic Integral Feedback Ensures Robust Perfect Adaptation in Noisy Bimolecular Networks. Cell Syst 2016, 2:15–26. [DOI] [PubMed] [Google Scholar]
- 27.Agrawal DK, Tang X, Westbrook A, Marshall R, Maxwell CS, Lucks J, Noireaux V, Beisel CL, Dunlop MJ, Franco E: Mathematical Modeling of RNA-Based Architectures for Closed Loop Control of Gene Expression. ACS Synth Biol 2018, 7:1219–1228. [DOI] [PubMed] [Google Scholar]
- 28.Toettcher JE, Gong D, Lim W a, Weiner OD: Light-based feedback for controlling intracellular signaling dynamics. Nat Methods 2011, 8:837–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levskaya A, Weiner OD, Lim WA, Voigt CA: Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 2009, 461:997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milias-Argeitis A, Summers S, Stewart-Ornstein J, Zuleta I, Pincus D, El-Samad H, Khammash M, Lygeros J: In silico feedback for in vivo regulation of a gene expression circuit. Nat Biotechnol 2011, 29:1114–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlendorf J, Miermont A, Delaveau T, Charvin G, Fages F, Bottani S, Batt G, Hersen P: Long-term model predictive control of gene expression at the population and single-cell levels. Proc Natl Acad Sci 2012, 109:14271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fracassi C, Postiglione L, Fiore G, di Bernardo D: Automatic Control of Gene Expression in Mammalian Cells. ACS Synth Biol 2016, 5:296–302. [DOI] [PubMed] [Google Scholar]
- 33.Postiglione L, Napolitano S, Pedone E, Rocca DL, Aulicino F, Santorelli M, Tumaini B, Marucci L, di Bernardo D: Regulation of gene expression and signaling pathway activity in mammalian cells by automated microfluidics feedback control. ACS Synth Biol 2018, doi: 10.1021/acssynbio.8b00235. [DOI] [PubMed] [Google Scholar]
- 34.Chait R, Ruess J, Bergmiller T, Tkačik G, Guet CC: Shaping bacterial population behavior through computer-interfaced control of individual cells. Nat Commun 2017, 8:1535.• By trapping Escherichia coli cells in a “Mother machine” microfluidic device, the authors are able to shine spatially structured light on single cells over hours. They are able to control gene expression in each cell independently using an optogenetic system.
- 35.Milias-Argeitis A, Rullan M, Aoki SK, Buchmann P, Khammash M: Automated optogenetic feedback control for precise and robust regulation of gene expression and cell growth. Nat Commun 2016, 7:12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabor JJ, Levskaya A, Voigt CA: Multichromatic Control of Gene Expression in Escherichia coli. J Mol Biol 2011, 405:315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins JJ, Gardner TS, Cantor CR: Construction of a genetic toggle switch in Escherichia coli. Nature 2000, 403:339–342. [DOI] [PubMed] [Google Scholar]
- 38.Lugagne J-B, Carillo SS, Kirch MM, Köhler A, Batt G, Hersen P, Sosa Carrillo S, Kirch MM, Köhler A, Batt G, et al. : Balancing a genetic toggle switch by real-time feedback control and periodic forcing. Nat Commun 2017, 8:1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rullan M, Benzinger D, Schmidt GW, Milias-argeitis A, Khammash M: An optogenetic platform for real-time , single-cell interrogation of stochastic transcription regulation. Mol Cell 2018, 70:745–756.e6.• Using spatially structured light, the authors implement computer-based feedback of gene expression in single Saccharomyces cerevisiae cells. They monitor and transcriptional bursting by detecting nascent mRNAs in real-time.
- 40.Harrigan P, Madhani HD, El-Samad H: Real-Time Genetic Compensation Defines the Dynamic Demands of Feedback Control. Cell 2018, 175:877–886.e10.•• The authors remove key signaling proteins in the MAPK pathway in yeast and them with photo-activatable ones. They use model predictive control to implement dynamically-correct replacements for the deleted genes, sometimes by iteratively fitting their model experiments. By doing so, they discover new dynamic regulatory elements of the pathway.
- 41.Ideker TE, Thorsson V, Karp RM: Discovery of regulatory interactions through perturbation: inference and experimental design. Pac Symp Biocomput 2000, 5:305–16. [DOI] [PubMed] [Google Scholar]
- 42.Tegner J, Yeung MKS, Hasty J, Collins JJ: Reverse engineering gene networks: Integrating genetic perturbations with dynamical modeling. Proc Natl Acad Sci 2003, 100:5944–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinke F, Seeger M, Tsuda K: Experimental design for efficient identification of gene regulatory networks using sparse Bayesian models. BMC Syst Biol 2007, 1:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagen DR, White JK, Tidor B: Convergence in parameters and predictions using computational experimental design. Interface Focus 2013, 3:20130008–20130008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nandy P, Unger M, Zechner C, Koeppl H: Optimal Perturbations for the Identification of Stochastic Reaction Dynamics. IFAC Proc Vol 2012, 45:686–691. [Google Scholar]
- 46.Zechner C, Nandy P, Unger M, Koeppl H: Optimal variational perturbations for the inference of stochastic reaction dynamics In 2012 IEEE 51st IEEE Conference on Decision and Control (CDC). . IEEE; 2012:5336–5341. [Google Scholar]
- 47.Ruess J, Milias-Argeitis A, Lygeros J: Designing experiments to understand the variability in biochemical reaction networks. J R Soc Interface 2013, 10:20130588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bandara S, Schlöder JP, Eils R, Bock HG, Meyer T: Optimal Experimental Design for Parameter Estimation of a Cell Signaling Model. PLoS Comput Biol 2009, 5:e1000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruess J, Parise F, Milias-Argeitis A, Khammash M, Lygeros J: Iterative experiment design guides the characterization of a light-inducible gene expression circuit. Proc Natl Acad Sci 2015, 112:8148–8153.• By optimally designing dynamic experiments, the authors rapidly infer the parameters for a model of the optogenetic system they are using. They then use that model to conduct predictive control experiments and demonstrate better control accuracy than with other models.
- 50.Ronneberger O, Fischer P, Brox T: U-Net: Convolutional Networks for Biomedical Image Segmentation In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Edited by Navab N, Hornegger J, Wells WM, Frangi AF. Springer; 2015:234–241. [Google Scholar]
- 51.Falk T, Mai D, Bensch R, Çiçek Ö, Abdulkadir A, Marrakchi Y, Böhm A, Deubner J, Jäckel Z, Seiwald K, et al. : U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods 2018, doi: 10.1038/s41592-018-0261-2. [DOI] [PubMed] [Google Scholar]
- 52.Brunker J, Yao J, Laufer J, Bohndiek SE: Photoacoustic imaging using genetically encoded reporters: a review. J Biomed Opt 2017, 22:070901. [DOI] [PubMed] [Google Scholar]
- 53.Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M: CRISPR-Cas9-based Photoactivatable Transcription System. Chem Biol 2015, 22:169–174. [DOI] [PubMed] [Google Scholar]
- 54.Polstein LR, Gersbach CA: A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 2015, 11:198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shao J, Wang M, Yu G, Zhu S, Yu Y, Heng BC, Wu J, Ye H: Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proc Natl Acad Sci 2018, 115:E6722–E6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nihongaki Y, Kawano F, Nakajima T, Sato M: Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol 2015, 33:755–760. [DOI] [PubMed] [Google Scholar]
- 57.Kawano F, Okazaki R, Yazawa M, Sato M: A photoactivatable Cre–loxP recombination system for optogenetic genome engineering. Nat Chem Biol 2016, 12:1059–1064. [DOI] [PubMed] [Google Scholar]
- 58.Appleton E, Madsen C, Roehner N, Densmore D: Design Automation in Synthetic Biology. Cold Spring Harb Perspect Biol 2017, 9:a023978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodriguez EA, Campbell RE, Lin JY, Lin MZ, Miyawaki A, Palmer AE, Shu X, Zhang J, Tsien RY: The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem Sci 2017, 42:111–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Z, Sharma A, Qi J, Peng X, Lee DY, Hu R, Lin D, Qu J, Kim JS: Super-resolution fluorescent materials: an insight into design and bioimaging applications. Chem Soc Rev 2016, 45:4651–4667. [DOI] [PubMed] [Google Scholar]
- 61.Quérard J, Le Saux T, Gautier A, Alcor D, Croquette V, Lemarchand A, Gosse C, Jullien L: Kinetics of Reactive Modules Adds Discriminative Dimensions for Selective Cell Imaging. ChemPhysChem 2016, 17:1396–1413. [DOI] [PubMed] [Google Scholar]
- 62.Quérard J, Zhang R, Kelemen Z, Plamont M-A, Xie X, Chouket R, Roemgens I, Korepina Y, Albright S, Ipendey E, et al. : Resonant out-of-phase fluorescence microscopy and remote imaging overcome spectral limitations. Nat Commun 2017, 8:969. [DOI] [PMC free article] [PubMed] [Google Scholar]