Abstract
The transcription factor, myocyte enhancer factor-2 (MEF2), is required for normal circadian behavior in Drosophila; however, its role in the mammalian circadian system has not been established. Of the four mammalian Mef2 genes, Mef2d is highly expressed in the suprachiasmatic nucleus (SCN) of the hypothalamus, a region critical for coordinating peripheral circadian clocks. Using both conventional and brain-specific Mef2d KO (Mef2d−/−) mouse lines, we demonstrate that MEF2D is essential for maintaining the length of the circadian free-running period of locomotor activity and normal sleep patterns in male mice. Crossing Mef2d−/− with Per2::luc reporter mice, we show that these behavioral changes are achieved without altering the endogenous period of the master circadian oscillator in the SCN. Together, our data suggest that alterations in behavior in Mef2d−/− mice may be the result of an effect on SCN output, rather than an effect on timekeeping within the SCN itself. These findings add to the growing body of evidence that MEF2 proteins play important roles in the brain.
SIGNIFICANCE STATEMENT These studies are the first to show a role for MEF2 proteins in the brain outside of the hippocampus, and our findings suggest that these proteins may play diverse roles in the CNS. It is important to continue to build on our understanding of the roles of proteins acting in the SCN because SCN dysfunction underlies jet lag in humans and influences the response to shift work schedules, which are now known as risk factors for the development of cancer. Our work on MEF2D could be the basis for opening new lines of research in the development and regulation of circadian rhythms.
Keywords: circadian, MEF2D, sleep
Introduction
The circadian clock is essential to the proper timing and regulation of behavior and physiology. In mammals, a master pacemaker in the suprachiasmatic nucleus (SCN) of the ventral hypothalamus coordinates circadian clocks throughout the organism (Welsh et al., 2010; Albrecht, 2012; Herzog et al., 2017). The circadian time-keeping mechanism is composed of a transcriptional autoregulatory feedback loop. At the core of this loop, CLOCK and BMAL1 drive transcription of Period (Per1, Per2) and Cryptochrome (Cry1, Cry2), whose protein products, in turn, dimerize, translocate back into the nucleus, and repress the activity of CLOCK and BMAL1, thereby inhibiting their own expression. In addition, accessory loops interact with the core clock loop, forming a web of modulators contributing to the refinement of the oscillatory network and, ultimately, circadian biological outputs (Shearman et al., 2000; Ueda et al., 2005; Takahashi, 2017).
The transcription factor myocyte enhancer factor-2 (MEF2) has been linked to the maintenance of normal circadian behavior in Drosophila (Blanchard et al., 2010; Sivachenko et al., 2013). Overexpression of Mef2 in clock neurons leads to lengthening of the free-running period of locomotor activity and weak behavioral rhythms in flies. Repressing Mef2 activity has a similar effect, with flies displaying weakly rhythmic or arrhythmic patterns of locomotor activity (Blanchard et al., 2010). However, the role of MEF2 in the mammalian circadian system has not been established.
In vertebrates, there are four Mef2 genes: Mef2a, Mef2b, Mef2c, and Mef2d. These genes display both differential and overlapping expression patterns throughout the body but are most abundantly expressed in muscle (both skeletal and cardiac) (Black and Olson, 1998) and brain (Leifer et al., 1993; Ikeshima et al., 1995; Lyons et al., 1995). Their protein products play vital roles in muscle development and stress- and activity-dependent signaling (Shalizi and Bonni, 2005; Potthoff and Olson, 2007; Dietrich, 2013). In neurons, MEF2 targets a diverse set of genes involved in synapse development and remodeling (Flavell et al., 2006, 2008), whereas loss of MEF2 has been associated with neuronal apoptosis (Mao et al., 1999; Li et al., 2001; Liu et al., 2003; Akhtar et al., 2012). Both in vitro and in vivo studies have shown that MEF2 activation or overexpression can lead to the reduction of excitatory synapses in hippocampal neurons (Barbosa et al., 2008; Pfeiffer et al., 2010). Interestingly, while prenatal loss of Mef2c in the brain has been shown to impair hippocampal-dependent learning in mice (Barbosa et al., 2008; Akhtar et al., 2012; Rashid et al., 2014), postnatal loss of Mef2c had no such behavioral effect (Adachi et al., 2016), and overexpression of MEF2 in discrete regions within the hippocampus disrupts memory formation (Cole et al., 2012; Rashid et al., 2014). These findings suggest a complex role for MEF2 proteins in the development and function of the hippocampus, but the impact of loss of MEF2 on non–hippocampal-dependent behaviors has not been thoroughly investigated.
Global deletion of Mef2a or Mef2c is lethal in mice, whereas Mef2d null mice are viable (Arnold et al., 2007; Kim et al., 2008; Omori et al., 2015) and Mef2d is highly expressed in the SCN (Lein et al., 2007). Therefore, using both a conventional and brain-specific Mef2d KO, we began our investigation of the role of MEF2 proteins in the regulation of mammalian circadian behavior. We demonstrate that MEF2D is essential for maintaining the length of the circadian free-running period of locomotor activity. Loss of MEF2D leads to a lengthening of the behavioral period and to disruption and fragmentation of sleep patterns without altering the endogenous period of the master circadian oscillator. Importantly, these effects are specific to MEF2D, as loss of either MEF2A or MEF2C in the brain had no effect on locomotor free-running period.
Materials and Methods
Animals.
Mef2d−/− mice (Arnold et al., 2007; Kim et al., 2008) and WT littermates on a (129SvEv/C57BL/6J) mixed genetic background were derived from heterozygote crosses. Brain-specific Mef2a, Mef2c, and Mef2d heterozygote and KO mice (and floxed, cre-negative controls) were derived from crossing CaMKIIa -iCre mice (Casanova et al., 2001) with Mef2a/c/dfx/fx triple-floxed mice (Barbosa et al., 2008; Liu et al., 2014; Adachi et al., 2016) and intercrossing to obtain the desired genotypes. For bioluminescence recordings to assess circadian rhythms in the SCN and peripheral tissues, Mef2d−/− mice were crossed with mice carrying the Per2::luc reporter (Yoo et al., 2004). These reporter mice are used to study circadian gene expression in vivo as the Period2 (mPer2) promoter drives luciferase (luc) expression. In all of our studies, Zeitgeber time (ZT) 0 denotes the time of lights on in light/dark cycles (LD), whereas circadian time (CT) 0 denotes start of subjective day and CT12 denotes start of subjective night in constant darkness (DD). Food and water were provided ad libitum. All animal care and procedures were performed in strict accordance with the recommendations of the Guide for the care and use of laboratory animals of the National Institutes of Health and approved by the University of Texas Southwestern Medical Center Institutional Animal Care and Use Committee.
Circadian behavioral analyses.
Adult male mice (8–12 weeks, n ≥ 12/group) were housed individually in running wheel cages, and their activity was recorded continuously as described previously (Shimomura et al., 2001) using ClockLab hardware and software (Actimetrics; RRID:SCR_014309). Phase angle, activity bouts, and free-running period were calculated using ClockLab. Activity onset (psi) was calculated during LD. Bout analyses were calculated after ∼8 weeks in DD, across 14 d of data, with bouts defined by a 10 counts/min threshold and a 20 min maximum gap. Free-running period was calculated after ∼8 weeks in DD, across 14 d of data using a χ2 periodogram. Animals whose data could not be scored for the full time for any measure were excluded from analysis. For behavioral ethograms, mice (n = 2/group) were housed in LD and visualized with a Sony DCR-HC62 camcorder, recording 1 frame/min. Data were recorded over the active (dark) portion of the LD cycle, with recordings extending into the morning hours (∼ZT12-ZT3.5).
Bioluminescence recordings of circadian rhythms.
To investigate circadian rhythms of clock gene expression in the SCN and peripheral tissues of these mice, bioluminescence recordings were performed as previously described (Yoo et al., 2004; Liu et al., 2007). Briefly, adult male Per2::luc (WT and Mef2d−/−) mice (n = 7/group) were killed by rapid cervical dislocation ∼ZT11-Z12 for LD experiments or CT11-CT12 for DD experiments. Tissues were harvested and kept in chilled Hanks' balanced salt solution. SCN was taken from a 300 mm coronal brain section sliced on a vibratome. A heart sample was taken from the exsanguinated atrium. Kidney, liver, and lung were hand sliced into thin samples. Anterior pituitary was cultured whole. Explanted tissues were placed on Millicell culture inserts (Millipore; SCR_015799) in 35 mm culture dishes in tissue culture medium containing luciferin (Promega). Cultures were incubated for at least 6 d under constant conditions (DD, constant temperature of 35°C, and no medium change). The light emitted from each individual culture dish was recorded in real-time with photomultiplier tube detectors using LumiCycle hardware and software (Actimetrics). Bioluminescence data were detrended by subtracting the 24 h running average from the raw data. The detrended datasets were smoothed by taking 2 h running averages. Free-running period was calculated over 5 cycles with LumiCycle software using Levenberg-Marquardt sin fit, and mean periods.
RNA expression.
Tissue was collected from male mice (n = 2 of 3 per group) housed in either LD or DD. Mice were killed at the indicated times (2 time points for LD and 6 time points for DD) by rapid cervical dislocation. RNA isolation and qRT-PCR were performed as previously described (Siepka et al., 2007). Primers for qRT-PCR are shown in Table 1. Relative quantification was calculated using the ΔΔCt method and Gapdh as housekeeping gene.
Table 1.
Primers for qRT-PCR of mRNA expression
| Gene | Primer set |
|---|---|
| Bmal1 | Forward: 5′-CCA CCT CAG AGC CAT TGA TAC A-3′ |
| Reverse: 5′-CGA AGA ACG TCA TGA TGC AG-3′ | |
| Per1 | Forward: 5′-CCC AGC TTT ACC TGC AGA AG-3′ |
| Reverse: 5′-ATG GTC GAA AGG AAG CCT CT-3′ | |
| Per2 | Forward: 5′-TGT GCG ATG ATG ATT CGT GA-3′ |
| Reverse: 5′-GGT GAA GGT ACG TTT GGT TTG C-3′ | |
| Dbp | Forward: 5′-CGA AGA ACG TCA TGA TGC AG-3′ |
| Reverse: 5′-GGT TCC CCA ACA TGC TAA GA-3′ | |
| Nr1d1 (Reverba) | Forward: 5′-AGA CTT CCC GCT TCA CCA AG-3′ |
| Reverse: 5′-AGC TTC TCG GAA TGC ATG TT-3′ | |
| Mef2d | Forward: 5′-ACG AGA GCC GCA CCA ATG CT-3′ |
| Reverse: 5′-GGA ACA GAT GAC CCA TAG CGC CTG-3′ | |
| Gapdh | Forward: 5′-CAA GGA GTA AGA AAC CCT GGA CC-3′ |
| Reverse: 5′-CGA GTT GGG ATA GGG CCT CT-3′ |
Cell culture and in vitro assays.
HEK293A cells were cultured in DMEM (#D5671; Sigma-Aldrich) supplemented with 10% FBS (#F2442; Sigma-Aldrich). Cells were plated 1 d before transfection and Effectene reagent (#301425; QIAGEN) was used to transfect DNA according to the manufacturer's protocol. Approximately 36 h after transfection, cells were harvested for downstream applications. For the luminescence assays, cells were lysed, and luminescence was measured from 20 μl of lysate with the Luciferase Assay System (#E1500; Promega) using a luminometer (AutoLumat Plus; Berthold Technologies). Luminescence is reported relative to Renilla (#E2810; Renilla Luciferase Assay, Promega). For the immunoprecipitation assay, cells were lysed, and immunoprecipitation and Western blotting were performed as previously described (Siepka et al., 2007), using anti-FLAG M2 agarose (#A2220; Sigma-Aldrich). Western blot was performed using anti-HA (Roche Diagnostics; RRID:AB_2687407), anti-CLOCK (generously given by Dr. Seung-Hee Yoo), anti-Flag (Sigma-Aldrich; RRID:AB_796202), and anti-α-tubulin (Santa Cruz Biotechnology). All assays were performed in duplicate and, except for the BMAL::Luc assay, were repeated in 2 or 3 independent transfection experiments.
Sleep recording and analyses.
Sleep recording was performed as previously described (Funato et al., 2010). Briefly, four electrode pins (for EEG) and two flexible wires (for EMG recording) were implanted under ketamine/xylazine anesthesia. EEG electrodes were positioned over the frontal and occipital cortices (AP: 0.5 mm; ML: 1.3 mm; DV: −1.3 mm; and AP: −4.5 mm; ML: 1.3 mm; DV: −1.3 mm; respectively). For recordings, male mice (n = 8/group) were housed individually in LD and tethered to a counterbalance arm (#SMCLA; Instech Laboratories), allowing for free movement. All mice were allowed 14 d of recovery before recording. EEG/EMG signal was recorded continuously for two, consecutive 24 h periods. EEG/EMG signals were amplified using a Grass Model 78 (AstroNova), filtered, digitized at a sampling rate of 250 Hz, and displayed using custom polygraph software. The vigilance state in each 20 s epoch was classified as NREM sleep, REM sleep, or wakefulness by a custom, semiautomated sleep staging software based in MATLAB (The MathWorks). For power density analyses, EEG power density in each frequency bin was expressed as a percentage of the mean total EEG power over all frequency bins and vigilance states.
Experimental design and statistical analyses.
All statistical tests were performed using Prism version 8.0 for for Mac OS X (GraphPad Software; RRID:SCR_002798). For behavioral experiments, sleep studies, and SCN explant experiments, means were calculated for each group for each measure and compared with Student's t tests (conventional KO) or one-way ANOVA (brain-specific KO) to compare across genotypes. Real-time PCR data were analyzed via two-way ANOVA with genotype and time as between and within variables, respectively. In vitro assays were analyzed with one-way ANOVAs with Bonferroni correction for multiple comparisons or unpaired t tests with Welch's correction for differences in variance.
Results
Mef2d KO mice have altered circadian behavior
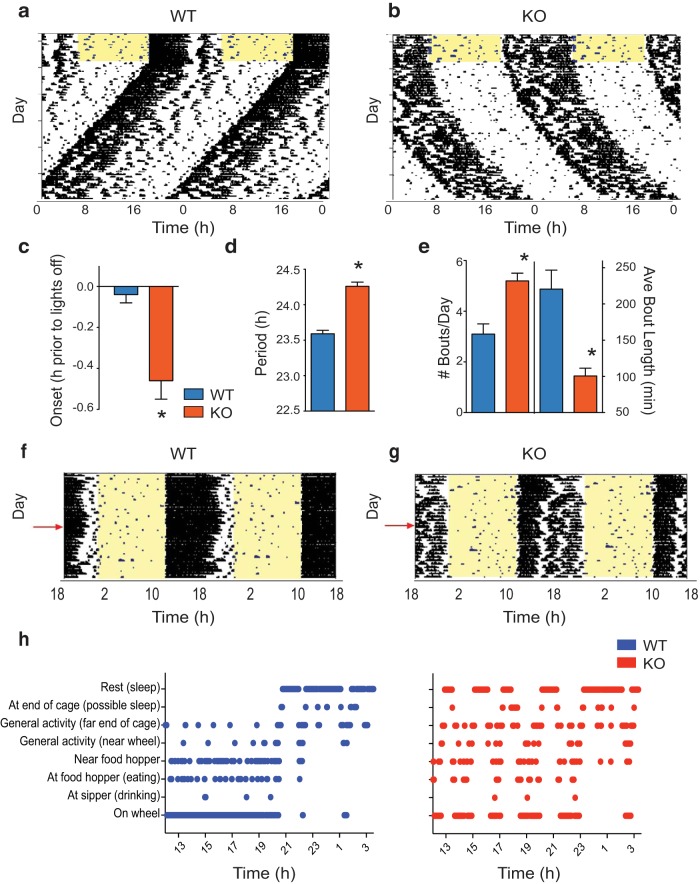
Loss of functional MEF2D had a profound impact on behavioral circadian rhythms (Fig. 1). Mef2d−/− (KO) mice housed with running wheels in 12:12 light/dark cycles with entraining T cycle 24 h (T24, LD 12:12; hereafter referred to as LD) exhibited a delayed phase of activity onset (t(29) = 3.762, *p = 0.0008; t test), beginning their wheel-running ∼25 min later than WT mice (Fig. 1a–c). KO mice also had a substantially lengthened (40 min longer than WT) free-running period (t(38) = 8.340, *p < 0.0001; t test) in DD (Fig. 1a,b,d). Notably, running wheel activity was fragmented in KO mice. KO mice displayed a greater number of activity bouts per day (t(29) = 4.257, *p = 0.0002; t test), with the individual activity bouts being of shorter duration (t(29) = 4.844, *p < 0.0001; t test) compared with WT mice (Fig. 1a,b,e). These findings suggested that MEF2D could play a larger role in the general control of behavior. Therefore, we performed an in-depth analysis of the behavior of KO mice throughout the active portion of the 24 h day. We compiled ethograms from images of individual mice, which were recorded 1×/min throughout the night. These analyses revealed that KO mice spent less consolidated time on the running wheel than WT mice, and wheel-running activity of KO mice was frequently interrupted by intermittent rest events (Fig. 1f–h). No other gross behavioral alterations were observed.
Figure 1.
Global loss of Mef2d lengthens circadian free-running period and alters behavioral activity patterns. Representative actograms of WT (a) and KO (b) mice on running wheels. Actograms are double-plotted, with each horizontal line indicating 48 h of activity. Mice were initially housed in LD12:12, as indicated by the yellow shading (lights on) and results analyzed by Student's t tests. c, Summary data (mean ± SEM) for activity onset in LD (n = 13 WT, 18 KO; t(29) = 3.762, *p = 0.0008). d, Summary data (mean ± SEM) for free-running period in DD (n = 18 WT, 22 KO; t(38) = 8.340, *p < 0.0001). e, Mean ± SEM number (t(29) = 4.257, *p = 0.0002) and duration (t(29) = 4.844, *p < 0.0001) of activity bouts as measured in DD (n = 12 WT, 19 KO). f, g, Representative ethograms from video analyses of WT (f) and KO (g) mice (n = 2/group). Ethograms are double-plotted, with each horizontal line indicating 48 h of activity. Mice were housed in LD12:12, as indicated by the yellow shading (lights on). h, Representative scored ethograms from WT (blue) and KO (red) mice, representing 15.5 h of data beginning at ZT12 (lights-off) on the day indicated by the red arrows in f, g.
Neuronal MEF2D is required for normal circadian behavior
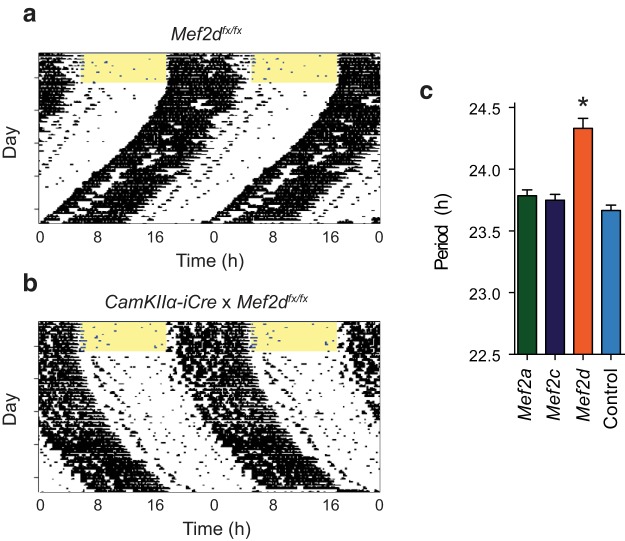
To further investigate whether loss of MEF2D in the brain was responsible for the behavioral phenotype in the global Mef2d KO, we crossed Mef2a/c/d floxed mice with a CaMKIIa-iCre line to generate mice with postnatal brain-specific loss of MEF2 function. Intercrossing for subsequent generations resulted in mice with loss of varying combinations of Mef2 genes in the brain. Only postnatal loss of MEF2D in the brain led to a change in circadian free-running period during DD (F(3,59) = 25.19, *p < 0.0001; one-way ANOVA), as CaMKIIa -iCre × Mef2afx/fx or Mef2cfx/fx mice were indistinguishable from WT controls. In contrast, CaMKIIa -iCre × Mef2dfx/fx mice had a long circadian period of running wheel activity similar to what was observed for the conventional KO (Fig. 2a–c). However, these mice did not differ from controls in their phase-response curves to light or dark pulses (data not shown).
Figure 2.
Mef2d activity in the brain is necessary for normal circadian behavior. CaMKIIα-iCre mice were crossed with Mef2a, Mef2c, and Mef2d floxed lines to produce mice with a varying combination of floxed Mef2 alleles. Representative actograms from (a) Mef2dfx/fx control and (b) CaMKIIα-iCre:Mef2dfx/fx mice. Actograms are double-plotted, with each horizontal line indicating 48 h of activity. Mice were initially housed in LD12:12, as indicated by the yellow shading (lights on). c, Free-running periods in DD (mean ± SEM) from floxed and control mice (n = 14 Mef2a, 4 Mef2c, 13 Mef2d, 32 control; analyzed by one-way ANOVA, F(3,59) = 25.19). *p < 0.0001, different from all other conditions assessed by Tukey post hoc comparisons.
Mef2d KO in the SCN causes light-dependent effects on free-running period
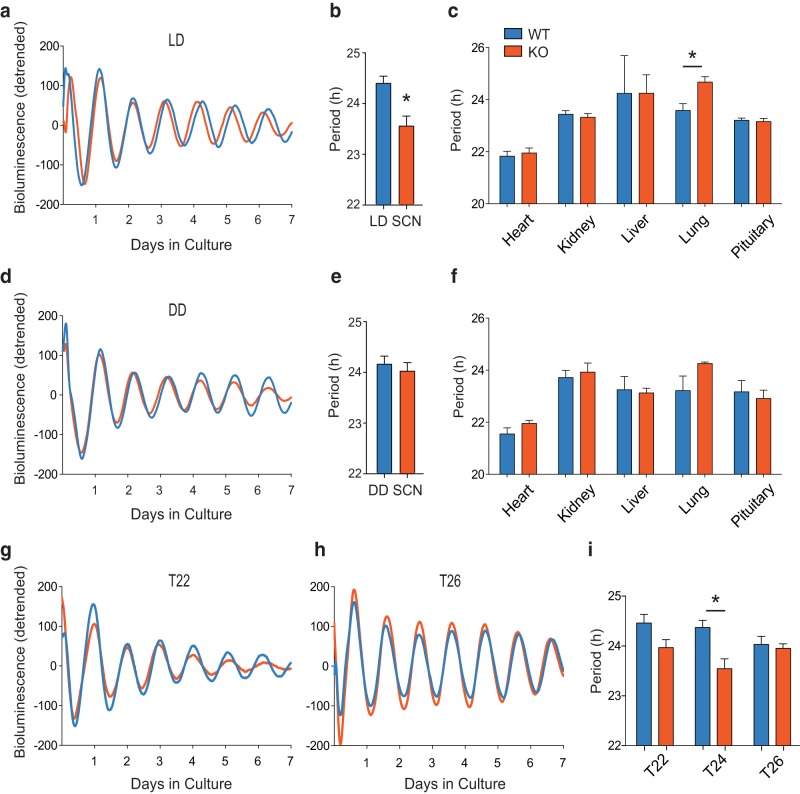
To test the effects of Mef2d on circadian rhythms in the SCN, we crossed Mef2d−/− mice with mice carrying the Per2::luc reporter. When SCNs were harvested from animals housed in LD, the KO SCNs displayed a delayed phase of PER2 bioluminescence from the acute culture (Fig. 3a), recapitulating our behavioral findings. Curiously, PER2 rhythms in the SCN of these animals were ∼50 min shorter in the presence of a T24 entraining cycle (Fig. 3a,b; t(12) = 3.443, *p = 0.0049; t test). PER2::LUC rhythms in other tissues from KO mice were largely unaffected, with the exception of a lengthened period in lungs (Fig. 3c; t(17) = 3.279, *p = 0.0044; t test). The shorter free-running period in KO SCNs harvested from LD led us to hypothesize that the SCNs of these mice may be exhibiting an aftereffect of the LD cycle on the master oscillator. To test this hypothesis, we harvested tissues from animals housed in DD. Interestingly, despite the behavioral differences seen in DD (Fig. 1d), SCNs from mice housed in DD had no observable difference in the rhythm of PER2::LUC (Fig. 3d,e; t(6) = 0.598, p = 0.572; t test). Likewise, no period difference was found in any of the other tissues tested (Fig. 3f), and SCN amplitude did not differ between genotypes in LD or DD (p > 0.05).
Figure 3.
Mef2d KO causes a short free-running period in the SCN. a, Detrended, smoothed, bioluminescence traces from representative WT and KO mouse SCNs. Mice were housed in T24 (LD12:12) before tissue harvest and results analyzed with Student's t tests. b, Periods (mean ± SEM) of LD12:12 SCN explants (n = 7 WT, 7 KO; t(12) = 3.443, *p = 0.0049). c, Periods (mean ± SEM) of LD12:12 peripheral explants (n = 3–10 mice/tissue/genotype; each tissue analyzed separately, t(17) = 3.279, *p = 0.0044). d, Detrended, smoothed, bioluminescence traces from representative WT and KO mouse SCNs. Mice were housed in DD before tissue harvest. e, Periods (mean ± SEM) of DD SCN explants (n = 4 WT, 4 KO; t(6) = 0.598, p = 0.572). f, Periods (mean ± SEM) of DD peripheral tissue explants (n = 2–4 mice/tissue/genotype). g, h, Detrended, smoothed, bioluminescence traces from representative WT and KO mouse SCNs. Mice were housed in T22 (LD 11:11; g) and in T26 (LD 13:13; h) before tissue harvest and results analyzed by two-way ANOVA. i, Periods (mean ± SEM) of SCN explants (T22: n = 6 WT, 9 KO; T24: n = 7 WT, 7 KO, also in b; T26: n = 6 WT, 6 KO; main effect of genotype F(1,35) = 12.24, *p = 0.0013 for T24; no main effect of T length).
Therefore, to explore the impact of LD cycles on the PER2::LUC rhythms of Mef2d−/− SCNs, we housed WT and KO mice in short (T22, LD11:11) and long (T26, LD13:13) LD cycles. SCNs were harvested following at least 3 weeks of acclimation to the altered cycle length. When analyzed by two-way ANOVA, neither a short (T22) nor a long (T26) LD cycle produced a significant change in free-running period length for either genotype (Fig. 3g–i); however, genotype effects were diminished under T26 conditions (Fig. 3i; F(1,35) = 12.24, *p = 0.0013 for T24; two-way ANOVA). Together, the data from Mef2d KO SCNs suggest that the behavioral phenotype is not a result of a gross change in Per2 expression in the SCN, but rather is a result of an interaction between MEF2D and the photic environment.
Clock gene expression is normal in Mef2d KO tissues
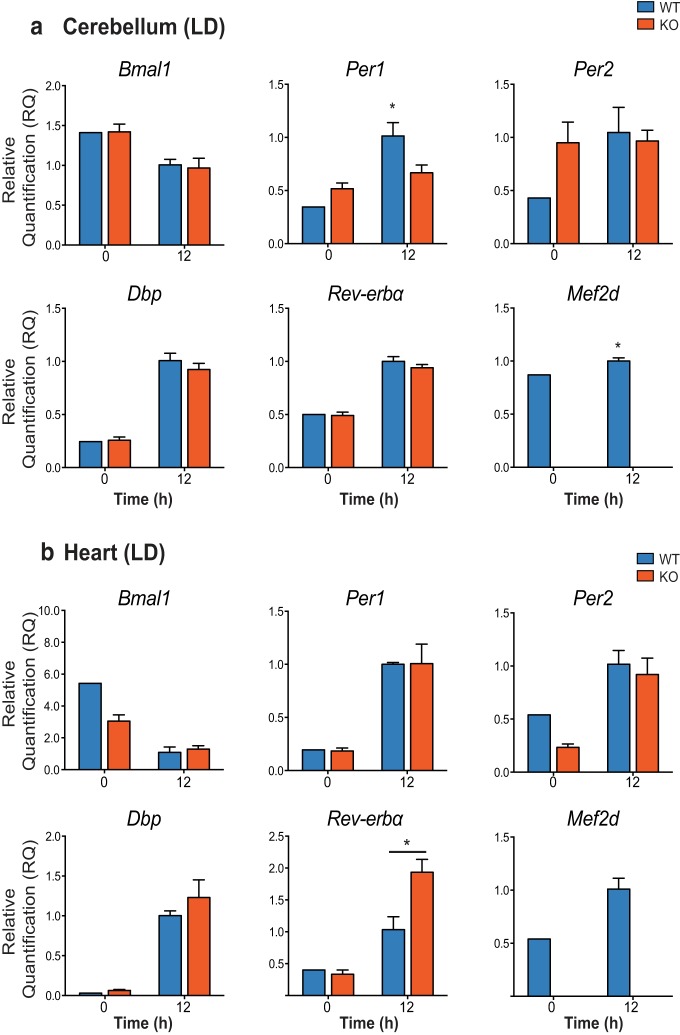
To begin to elucidate the actions of MEF2D on core clock genes, we next measured mRNA expression in cerebellum and heart samples from WT and Mef2d KO mice housed in LD. There was no obvious effect of genotype on clock gene expression in the cerebellum at two different time points (Fig. 4a). However, for Per1 mRNA expression, there was an interaction between genotype and time of day, with a less dramatic increase observed at ZT12 in the cerebellum of KO mice compared with WT mice [interaction (F(1,7) = 8.502, p = 0.0225) and main effect of time (F(1,7) = 21.19, p = 0.0025); *p = 0.0089; two-way ANOVA]. In heart samples, only Rev-erba expression differed between WT and KO mice, with KO mice having higher levels of Rev-erba at ZT12 (Fig. 4b; interaction, F(1,7) = 8.076, p = 0.0250; main effect of time, F(1,7) = 43.10, p = 0.0003; and main effect of genotype, F(1,7) = 6.001, p = 0.0441; *p = 0.0218; two-way ANOVA). Mef2d expression differed significantly with time of day in the cerebellum where it was slightly higher at ZT12 than ZT0 (Fig. 4a; t(3) = 3.296, *p = 0.0459; t test). No differences were observed in clock gene expression or Mef2d expression in either cerebellum or heart tissues from mice housed in DD (data not shown).
Figure 4.
Clock gene expression is not greatly affected by loss of Mef2d. qRT-PCR analysis of clock gene expression in WT and Mef2d KO cerebellum (a) and heart (b) taken from mice housed in T24, LD12:12. Data are presented as relative quantification compared with WT0; n = 2 or 3/group; and analyzed by two-way ANOVA. Per1: interaction (F(1,7) = 8.502, p = 0.0225) and main effect of time (F(1,7) = 21.19, p = 0.0025). *p = 0.0089 WT12 different from WT0. Reverba: interaction (F(1,7) = 8.076, p = 0.0250, main effect of time (F(1,7) = 43.10, p = 0.0003, and main effect of genotype (F(1,7) = 6.001, p = 0.0441). *p = 0.0218 KO12 different from WT12; Mef2d: t(3) = 3.296. *p = 0.0459 via Student's t test.
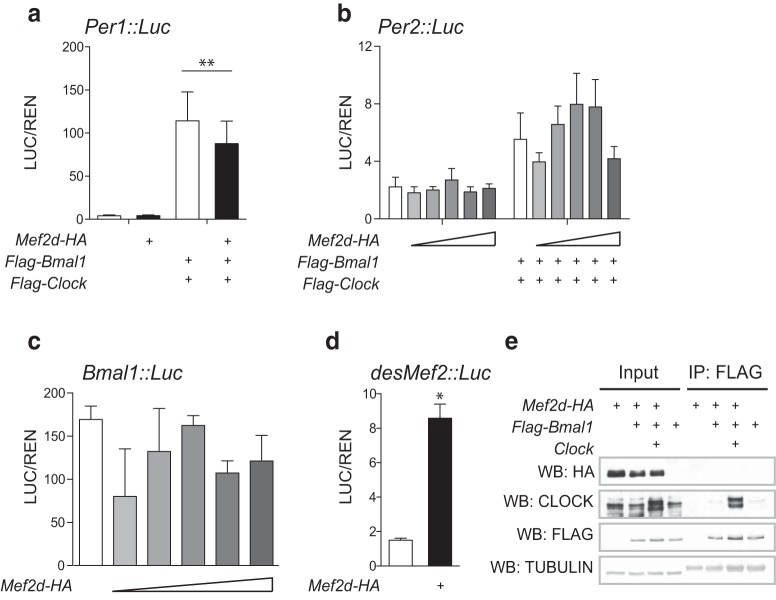
Because such small differences in clock gene expression were observed, we also tested for the ability of MEF2D to transcriptionally regulate clock genes in fibroblasts using a luciferase reporter assay. MEF2D did not activate Per1, Per2, or Bmal1 in our experiments (Fig. 5a–c) but did trans-activate a positive control (desMef2; Fig. 5d; t(3.127) = 8.472; *p = 0.029; t test). Further, we observed no physical interactions between MEF2D and BMAL1 in coimmunoprecipitation experiments (Fig. 5e). Along with our gene expression results, these findings suggest that the robust period difference in the circadian behavior of Mef2d KO mice is not the result of a substantial, direct impact on clock gene or protein expression.
Figure 5.
MEF2D does not directly interact with core clock components. Cotransfection of Mef2d does not lead to activation of (a) Per1::Luc (average of 2 replicates across 3 experiments; 125 ng Mef2d transfected), (b) Per2::Luc (average of 2 replicates across 2 experiments; increasing doses of Mef2d from 5 to 50 ng), or (c) Bmal1::Luc (average of 2 replicates from 1 experiment; increasing doses of Mef2d from 5 to 50 ng) in reporter assays. Both Per1::Luc and Per2::Luc activation is observed in the presence of known transcriptional activators, BMAL1 and CLOCK (Per1: F(3,20) = 10.80; **p = 0.0002; Per2: F(10,33) = 3.42, p = 0.0037, not significant after correction for multiple comparisons). d, Cotransfection of Mef2d activates a Mef2 binding domain fused to a TATA box (average of 2 replicates across 2 experiments; 125 ng Mef2d transfected; t(3.127) = 8.472; *p = 0.029). e, MEF2D is not pulled down with BMAL1 in coimmunoprecipitation experiments, whereas the known binding partner, CLOCK, was successfully coimmunoprecipitated and FLAG (BMAL1) enrichment is observed.
Mef2d KO mice have disrupted sleep patterns
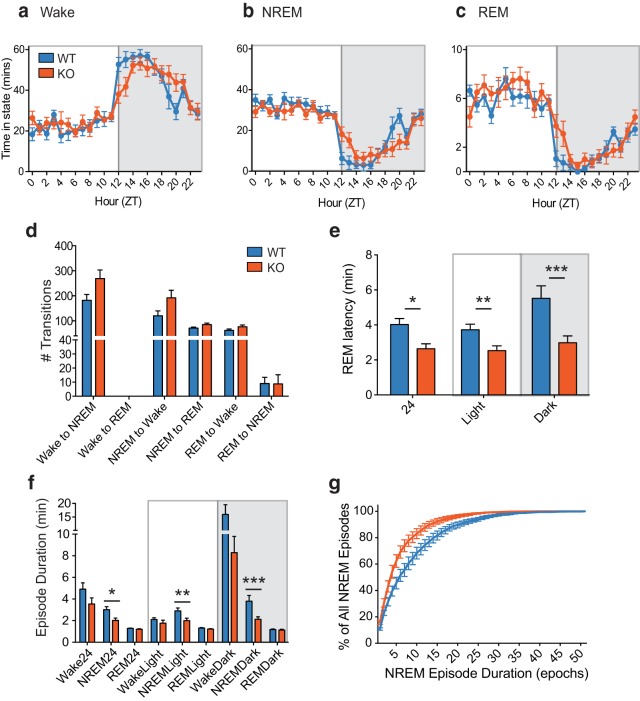
Our findings of disturbed circadian running wheel behavior and increased rest events in Mef2d−/− mice, without corresponding broad changes in the molecular circadian clock, led us to hypothesize these behavioral changes were reflective of abnormal sleep. Thus, as a final behavioral analysis, we recorded sleep patterns from WT and KO mice. Wake and sleep episodes were phase delayed in KO mice (Fig. 6a–c), mirroring what was observed for running wheel behavior (Fig. 1a–c). There were no differences in the EEG power density across sleep stages or the total time spent in any sleep stage (data not shown); however, KO mice tended to have more transitions (Fig. 6d) from wakefulness to NREM sleep (t(14) = 2.077; p = 0.0567; t test) and from NREM sleep to wakefulness (t(14) = 2.015; p = 0.0635; t test) or REM sleep (t(14) = 2.002; p = 0.0651; t test). More in-depth analysis revealed that KO mice had extremely fragmented sleep with significantly shorter REM latency [Fig. 6e; Full day: t(14) = 3.147, *p = 0.0071; Light: t(14) = 2.848, **p = 0.0129; Dark: t(14) = 3.108, ***p = 0.0077 (t tests)] as well as shorter NREM episode duration [Fig. 6f; Full day: t(14) = 2.829, *p = 0.0134; Light: t(14) = 2.557, **p = 0.0228; Dark: t(14) = 2.910, ***p = 0.0114 (t tests)]. Plotting the number of NREM episodes as a function of episode duration (Fig. 6g) revealed that the majority of NREM episodes were shorter in KO mice compared with WT mice. Together, these results suggest that the circadian alterations in activity in Mef2d−/− mice are due, at least in part, to disrupted sleep patterns.
Figure 6.
Loss of Mef2d leads to fragmented sleep. Patterns of wakefulness (a), NREM (b), and REM sleep (c) are shifted toward a later phase in the KO mice, mirroring the delayed phase observed from running wheel behavior. d, Mean ± SEM number of transitions over the full day. Data are presented as mean ± SEM (n = 8 WT, 8 KO) and analyzed by Student's t tests; there were no significant differences between genotypes. However, KO mice tended to have more transitions from wakefulness to NREM sleep (t(14) = 2.077; p = 0.0567) and from NREM sleep to wakefulness (t(14) = 2.015; p = 0.0635) or REM sleep (t(14) = 2.002; p = 0.0651). e, Mean ± SEM REM latency is shorter in Mef2d KO mice (n values same as above; results analyzed with Student's t tests). Data are presented over the full day (24; t(14) = 3.147, *p = 0.0071), the light portion of the LD cycle (Light; t(14) = 2.848, **p = 0.0129), or the dark portion of the LD cycle (Dark; t(14) = 3.108, ***p = 0.0077). f, NREM episode duration is shorter in Mef2d KO mice (n values same as above; analyzed with Student's t tests). Data are presented over the full day (NREM24; t(14) = 2.829, *p = 0.0134), the light portion of the LD cycle (NREMLight; t(14) = 2.557, **p = 0.0228), or the dark portion of the LD cycle (NREMDark: t(14) = 2.910, ***p = 0.0114). g, NREM episode duration as a percentage of all NREM episodes reveals fragmented sleep patterns in KO mice.
Discussion
It is rare for a single-gene mutation/loss to cause a large change in the circadian free-running period. When such changes do occur, they are typically the result of loss of function for a gene directly involved in the core clock feedback loop or a gene that strongly influences the expression or stability of core clock genes (Takahashi et al., 2008). Here we demonstrate that loss of Mef2d results in a 40 min lengthening of the free-running period of locomotor activity. This change in free-running period is not associated with concomitant changes in the levels of core clock components, and thus appears to result from a role of Mef2d downstream of the molecular clock.
In this study, we observed a lengthening of period both in conventional Mef2d−/− and CaMKIIa -iCre × Mef2dfx/fx mice; therefore, it is likely that the loss of Mef2d impacts the circadian system through its actions in the brain. The CaMKIIa -iCre line used expresses at postnatal day 3, with maximal expression occurring by postnatal day 15 (Casanova et al., 2001); thus, our results suggest that the effects of Mef2d on the circadian system are not due to developmental loss of Mef2d. The floxed Mef2d allele does not cause a complete KO, but rather it produces a nonfunctional truncated protein product (Kim et al., 2008). However, these same mice have been crossed with a different Cre-driver line shown to have reduced Mef2d gene expression in the hippocampus (Akhtar et al., 2012). Importantly, the long free-running period observed in Mef2d−/− mice is specific, as we did not observe a change in any circadian parameter in brain-specific Mef2a or Mef2c floxed mice.
MEF2D has been shown to influence synapse elimination, dendritic spine formation, and the expression of neurotrophic factors in hippocampal neurons (Lyons et al., 1995; Akhtar et al., 2012; Dietrich, 2013; Rashid et al., 2014; Chang et al., 2017), so it is possible that the postnatal loss of MEF2D causes reorganization of SCN circuitry. While there is some evidence that alterations in synaptic connectivity lead to changes in circadian outputs in the fly (Fernández et al., 2008; Gilestro et al., 2009; Gorostiza et al., 2014; Krzeptowski et al., 2018), it remains to be established whether this is the case in mammals. Previous studies have concluded that MEF2 in the fly may be necessary for a daily pattern of dendritic remodeling via inhibition of the cell-adhesion molecule Fasciclin2 (Fas2) (Sivachenko et al., 2013), so it is possible that MEF2 may exert its actions through similar mechanisms in mice.
Interestingly, while the behavioral free-running period was lengthened in Mef2d−/− mice, SCNs from animals previously entrained to LD cycles had shorter free-running periods than WT mice. This effect was not observed when SCNs were harvested from mice held in constant DD conditions, suggesting that the short period SCN in Mef2d−/− mice is an aftereffect of the LD cycle and not due to an intrinsic change in the period of the SCN itself. Aftereffects of LD cycles on SCN rhythms have been reported previously, although they are not well understood (Aton et al., 2004; Mickman et al., 2008; Molyneux et al., 2008). When SCNs were taken from mice housed in LD cycles shorter than 24 h (T22; LD11:11), the periods of KO mice remained shorter than those of WT mice, although this difference did not reach statistical significance. It should be noted, however, that in this short LD cycle, the phase of locomotor activity onset was very variable for both genotypes, suggesting that animals may not have been fully entrained in this condition. It has been proposed that normal MEF2 levels may be necessary to maintain synchrony between clock neurons in the fly brain (Blanchard et al., 2010; Sivachenko et al., 2013). In mice, however, we did not observe any evidence via gene expression data that the SCN is strongly affected by the loss of MEF2D. Based on our data, we suggest that the long period of locomotor activity we observe in Mef2d−/− mice may be due to their being pushed to conform to a 24 h LD cycle, and could be the result of an effect on SCN output, rather than an effect on timekeeping within the SCN itself. While we did not directly assess the SCN anatomy or neuronal activity in Mef2d−/− mice, nor did we measure core SCN-driven physiological outputs, these would be important to measure in future studies to test this hypothesis.
Alternatively, it is possible that, rather than affecting SCN outputs, the loss of MEF2D alters SCN inputs from the retina. MEF2D binds to retinal-specific promoters to drive photoreceptor development, and Mef2d−/− mice have progressive retinal degeneration beginning at P30 (Andzelm et al., 2015; Omori et al., 2015). While these studies examined gross changes to rods and cones, intrinsically photosensitive retinal ganglion cells could also be disrupted. Changes in behavioral circadian rhythms have been reported with retinal degeneration or bilateral enucleation (Lupi et al., 1999; Yamazaki et al., 2002; Tosini et al., 2007); therefore, it is possible that the Mef2d−/− mice have altered behavioral rhythms due to decreased retinal signaling. Future studies could examine light induction in the SCN and light-induced phase shifts in behavior in these mice. In addition, while we did not examine aging mice, it would be interesting for future studies to examine the circadian phenotype over extended periods of time to determine whether the progressive retinal degeneration seen in these mice correlates with behavioral changes.
In addition to the circadian effects, loss of MEF2D had a profound effect on sleep/wake patterning. Both activity and sleep were extremely fragmented in Mef2d−/− mice. NREM sleep, in particular, was largely restricted to multiple, very short duration bouts in KOs. This difference in the patterning of NREM sleep occurred without a corresponding change in either total time spent in NREM sleep across the day or power density during any sleep stage. While we did not measure melatonin levels or other physiological markers of sleep patterning, future studies should examine the larger implications of this disrupted sleep pattern on behavioral and physiological function in these animals. In particular, it would be fascinating to determine whether loss of MEF2D influences memory consolidation during sleep. The MEF2 family of transcription factors have been implicated in memory formation and synaptic plasticity (Barbosa et al., 2008; Akhtar et al., 2012; Dietrich, 2013; Rashid et al., 2014; Adachi et al., 2016). Further, a growing body of literature suggests that MEF2 proteins exert neuroprotective effects in response to neuronal inflammation and oxidative stress (Gong et al., 2003; Tang et al., 2005; Smith et al., 2006; She et al., 2011; Ryan et al., 2013; Yang et al., 2015). The present study provides additional evidence for a specific role of MEF2D in maintaining normal function of the mammalian circadian clock and sleep homeostasis. Future studies should focus on understanding the influence of MEF2 on sleep and circadian function and its impacts on neuronal health.
Footnotes
This work was supported by Japan Society for the Promotion of Science FIRST program and Japan's Ministry of Education, Culture, Sports, Science and Technology WPI program to M.Y., National Institutes of Health Grants AR-067294 and HL-130253 and Robert A. Welch Foundation Grant 1-0025 to E.N.O., and National Institutes of Health Grants MH078024 and AG045795 to J.S.T. M.Y. was, and J.S.T. is, an Investigator in the Howard Hughes Medical Institute. We thank Dr. Rhonda Bassel-Duby for contributing the Mef2d KO and floxed mice; and Dr. Robert Greene for suggesting the analysis in Figure 6g.
The authors declare no competing financial interests.
References
- Adachi M, Lin PY, Pranav H, Monteggia LM (2016) Postnatal loss of Mef2c results in dissociation of effects on synapse number and learning and memory. Biol Psychiatry 80:140–148. 10.1016/j.biopsych.2015.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar MW, Kim MS, Adachi M, Morris MJ, Qi X, Richardson JA, Bassel-Duby R, Olson EN, Kavalali ET, Monteggia LM (2012) In vivo analysis of MEF2 transcription factors in synapse regulation and neuronal survival. PLoS One 7:e34863. 10.1371/journal.pone.0034863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U. (2012) Timing to perfection: the biology of central and peripheral circadian clocks. Neuron 74:246–260. 10.1016/j.neuron.2012.04.006 [DOI] [PubMed] [Google Scholar]
- Andzelm MM, Cherry TJ, Harmin DA, Boeke AC, Lee C, Hemberg M, Pawlyk B, Malik AN, Flavell SW, Sandberg MA, Raviola E, Greenberg ME (2015) MEF2D drives photoreceptor development through a genome-wide competition for tissue-specific enhancers. Neuron 86:247–263. 10.1016/j.neuron.2015.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN (2007) MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell 12:377–389. 10.1016/j.devcel.2007.02.004 [DOI] [PubMed] [Google Scholar]
- Aton SJ, Block GD, Tei H, Yamazaki S, Herzog ED (2004) Plasticity of circadian behavior and the suprachiasmatic nucleus following exposure to non-24-hour light cycles. J Biol Rhythms 19:198–207. 10.1177/0748730404264156 [DOI] [PubMed] [Google Scholar]
- Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel-Duby R, Olson EN (2008) MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci U S A 105:9391–9396. 10.1073/pnas.0802679105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BL, Olson EN (1998) Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol 14:167–196. 10.1146/annurev.cellbio.14.1.167 [DOI] [PubMed] [Google Scholar]
- Blanchard FJ, Collins B, Cyran SA, Hancock DH, Taylor MV, Blau J (2010) The transcription factor Mef2 is required for normal circadian behavior in Drosophila. J Neurosci 30:5855–5865. 10.1523/JNEUROSCI.2688-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova E, Fehsenfeld S, Mantamadiotis T, Lemberger T, Greiner E, Stewart AF, Schütz G (2001) A CaMKIIalpha iCre BAC allows brain-specific gene inactivation. Genesis 31:37–42. 10.1002/gene.1078 [DOI] [PubMed] [Google Scholar]
- Chang CW, Wilkerson JR, Hale CF, Gibson JR, Huber KM (2017) Distinct stages of synapse elimination are induced by burst firing of CA1 neurons and differentially require MEF2A/D. Elife 6:e26278. 10.7554/eLife.26278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole CJ, Mercaldo V, Restivo L, Yiu AP, Sekeres MJ, Han JH, Vetere G, Pekar T, Ross PJ, Neve RL, Frankland PW, Josselyn SA (2012) MEF2 negatively regulates learning-induced structural plasticity and memory formation. Nat Neurosci 15:1255–1264. 10.1038/nn.3189 [DOI] [PubMed] [Google Scholar]
- Dietrich JB. (2013) The MEF2 family and the brain: from molecules to memory. Cell Tissue Res 352:179–190. 10.1007/s00441-013-1565-2 [DOI] [PubMed] [Google Scholar]
- Fernández MP, Berni J, Ceriani MF (2008) Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol 6:e69. 10.1371/journal.pbio.0060069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME (2006) Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311:1008–1012. 10.1126/science.1122511 [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME (2008) Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60:1022–1038. 10.1016/j.neuron.2008.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato H, Sato M, Sinton CM, Gautron L, Williams SC, Skach A, Elmquist JK, Skoultchi AI, Yanagisawa M (2010) Loss of goosecoid-like and DiGeorge syndrome critical region 14 in interpeduncular nucleus results in altered regulation of rapid eye movement sleep. Proc Natl Acad Sci U S A 107:18155–18160. 10.1073/pnas.1012764107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilestro GF, Tononi G, Cirelli C (2009) Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science 324:109–112. 10.1126/science.1166673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Tang X, Wiedmann M, Wang X, Peng J, Zheng D, Blair LA, Marshall J, Mao Z (2003) Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron 38:33–46. 10.1016/S0896-6273(03)00191-0 [DOI] [PubMed] [Google Scholar]
- Gorostiza EA, Depetris-Chauvin A, Frenkel L, Pírez N, Ceriani MF (2014) Circadian pacemaker neurons change synaptic contacts across the day. Curr Biol 24:2161–2167. 10.1016/j.cub.2014.07.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog ED, Hermanstyne T, Smyllie NJ, Hastings MH (2017) Regulating the suprachiasmatic nucleus (SCN) circadian clockwork: interplay between cell-autonomous and circuit-level mechanisms. Cold Spring Harb Perspect Biol 9:a027706. 10.1101/cshperspect.a027706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeshima H, Imai S, Shimoda K, Hata J, Takano T (1995) Expression of a MADS box gene, MEF2D, in neurons of the mouse central nervous system: implication of its binary function in myogenic and neurogenic cell lineages. Neurosci Lett 200:117–120. 10.1016/0304-3940(95)12092-I [DOI] [PubMed] [Google Scholar]
- Kim Y, Phan D, van Rooij E, Wang DZ, McAnally J, Qi X, Richardson JA, Hill JA, Bassel-Duby R, Olson EN (2008) The MEF2D transcription factor mediates stress-dependent cardiac remodeling in mice. J Clin Invest 118:124–132. 10.1172/JCI33255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzeptowski W, Hess G, Pyza E (2018) Circadian plasticity in the brain of insects and rodents. Front Neural Circuits 12:32. 10.3389/fncir.2018.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifer D, Krainc D, Yu YT, McDermott J, Breitbart RE, Heng J, Neve RL, Kosofsky B, Nadal-Ginard B, Lipton SA (1993) MEF2C, a MADS/MEF2-family transcription factor expressed in a laminar distribution in cerebral cortex. Proc Natl Acad Sci U S A 90:1546–1550. 10.1073/pnas.90.4.1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176. 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Li M, Linseman DA, Allen MP, Meintzer MK, Wang X, Laessig T, Wierman ME, Heidenreich KA (2001) Myocyte enhancer factor 2A and 2D undergo phosphorylation and caspase-mediated degradation during apoptosis of rat cerebellar granule neurons. J Neurosci 21:6544–6552. 10.1523/JNEUROSCI.21-17-06544.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ 3rd, Takahashi JS, Kay SA (2007) Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129:605–616. 10.1016/j.cell.2007.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cavanaugh JE, Wang Y, Sakagami H, Mao Z, Xia Z (2003) ERK5 activation of MEF2-mediated gene expression plays a critical role in BDNF-promoted survival of developing but not mature cortical neurons. Proc Natl Acad Sci U S A 100:8532–8537. 10.1073/pnas.1332804100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Nelson BR, Bezprozvannaya S, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN (2014) Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc Natl Acad Sci U S A 111:4109–4114. 10.1073/pnas.1401732111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupi D, Cooper HM, Froehlich A, Standford L, McCall MA, Foster RG (1999) Transgenic ablation of rod photoreceptors alters the circadian phenotype of mice. Neuroscience 89:363–374. 10.1016/S0306-4522(98)00353-4 [DOI] [PubMed] [Google Scholar]
- Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN (1995) Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J Neurosci 15:5727–5738. 10.1523/JNEUROSCI.15-08-05727.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME (1999) Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science 286:785–790. 10.1126/science.286.5440.785 [DOI] [PubMed] [Google Scholar]
- Mickman CT, Stubblefield JJ, Harrington ME, Nelson DE (2008) Photoperiod alters phase difference between activity onset in vivo and mPer2::luc peak in vitro. Am J Physiol Regul Integr Comp Physiol 295:R1688–R1694. 10.1152/ajpregu.90510.2008 [DOI] [PubMed] [Google Scholar]
- Molyneux PC, Dahlgren MK, Harrington ME (2008) Circadian entrainment aftereffects in suprachiasmatic nuclei and peripheral tissues in vitro. Brain Res 1228:127–134. 10.1016/j.brainres.2008.05.091 [DOI] [PubMed] [Google Scholar]
- Omori Y, Kitamura T, Yoshida S, Kuwahara R, Chaya T, Irie S, Furukawa T (2015) Mef2d is essential for the maturation and integrity of retinal photoreceptor and bipolar cells. Genes Cells 20:408–426. 10.1111/gtc.12233 [DOI] [PubMed] [Google Scholar]
- Pfeiffer BE, Zang T, Wilkerson JR, Taniguchi M, Maksimova MA, Smith LN, Cowan CW, Huber KM (2010) Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron 66:191–197. 10.1016/j.neuron.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN (2007) MEF2: a central regulator of diverse developmental programs. Development 134:4131–4140. 10.1242/dev.008367 [DOI] [PubMed] [Google Scholar]
- Rashid AJ, Cole CJ, Josselyn SA (2014) Emerging roles for MEF2 transcription factors in memory. Genes Brain Behav 13:118–125. 10.1111/gbb.12058 [DOI] [PubMed] [Google Scholar]
- Ryan SD, Dolatabadi N, Chan SF, Zhang X, Akhtar MW, Parker J, Soldner F, Sunico CR, Nagar S, Talantova M, Lee B, Lopez K, Nutter A, Shan B, Molokanova E, Zhang Y, Han X, Nakamura T, Masliah E, Yates JR 3rd, et al. (2013) Isogenic human iPSC Parkinson's model shows nitrosative stress-induced dysfunction in MEF2-PGC1alpha transcription. Cell 155:1351–1364. 10.1016/j.cell.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi AK, Bonni A (2005) Brawn for brains: the role of MEF2 proteins in the developing nervous system. Curr Top Dev Biol 69:239–266. 10.1016/S0070-2153(05)69009-6 [DOI] [PubMed] [Google Scholar]
- She H, Yang Q, Shepherd K, Smith Y, Miller G, Testa C, Mao Z (2011) Direct regulation of complex I by mitochondrial MEF2D is disrupted in a mouse model of Parkinson disease and in human patients. J Clin Invest 121:930–940. 10.1172/JCI43871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM (2000) Interacting molecular loops in the mammalian circadian clock. Science 288:1013–1019. 10.1126/science.288.5468.1013 [DOI] [PubMed] [Google Scholar]
- Shimomura K, Low-Zeddies SS, King DP, Steeves TD, Whiteley A, Kushla J, Zemenides PD, Lin A, Vitaterna MH, Churchill GA, Takahashi JS (2001) Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res 11:959–980. 10.1101/gr.171601 [DOI] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS (2007) Circadian mutant overtime reveals F box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129:1011–1023. 10.1016/j.cell.2007.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivachenko A, Li Y, Abruzzi KC, Rosbash M (2013) The transcription factor Mef2 links the Drosophila core clock to Fas2, neuronal morphology, and circadian behavior. Neuron 79:281–292. 10.1016/j.neuron.2013.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PD, Mount MP, Shree R, Callaghan S, Slack RS, Anisman H, Vincent I, Wang X, Mao Z, Park DS (2006) Calpain-regulated p35/cdk5 plays a central role in dopaminergic neuron death through modulation of the transcription factor myocyte enhancer factor 2. J Neurosci 26:440–447. 10.1523/JNEUROSCI.2875-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS. (2017) Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18:164–179. 10.1038/nrg.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL (2008) The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet 9:764–775. 10.1038/nrg2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Wang X, Gong X, Tong M, Park D, Xia Z, Mao Z (2005) Cyclin-dependent kinase 5 mediates neurotoxin-induced degradation of the transcription factor myocyte enhancer factor 2. J Neurosci 25:4823–4834. 10.1523/JNEUROSCI.1331-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosini G, Aguzzi J, Bullock NM, Liu C, Kasamatsu M (2007) Effect of photoreceptor degeneration on circadian photoreception and free-running period in the royal college of surgeons rat. Brain Res 1148:76–82. 10.1016/j.brainres.2007.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S (2005) System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet 37:187–192. 10.1038/ng1504 [DOI] [PubMed] [Google Scholar]
- Welsh DK, Takahashi JS, Kay SA (2010) Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol 72:551–577. 10.1146/annurev-physiol-021909-135919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Alones V, Menaker M (2002) Interaction of the retina with suprachiasmatic pacemakers in the control of circadian behavior. J Biol Rhythms 17:315–329. 10.1177/074873040201700405 [DOI] [PubMed] [Google Scholar]
- Yang S, Gao L, Lu F, Wang B, Gao F, Zhu G, Cai Z, Lai J, Yang Q (2015) Transcription factor myocyte enhancer factor 2D regulates interleukin-10 production in microglia to protect neuronal cells from inflammation-induced death. J Neuroinflammation 12:33. 10.1186/s12974-015-0258-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS (2004) PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 101:5339–5346. 10.1073/pnas.0308709101 [DOI] [PMC free article] [PubMed] [Google Scholar]