Abstract
Ets-1 is a prototype of the ETS protein family. Members of the ETS protein family contain a unique ETS domain. Ets-1 is associated with cancer progression and metastasis in many types of cancer. Many studies have shown a link between elevated expression of Ets-1 in cancer biopsies and poor survival. CCR7 is a chemokine that binds to specific ligand CCL21/CCL19. CCR7 expression is associated with tumor metastasis and infiltration into lymph nodes. The objective of this study was to test whether Ets-1 could regulate CCR7 expression and enhance tumor metastasis. Our data showed that CCR7 expression was downregulated in Ets-1-deficient T cells upon T-cell stimulation. Overexpression of Ets-1 increased CCR7 expression in breast cancer cell lines. In contrast, knockdown of Ets-1 reduced CCR7 expression. Ets-1 could directly bind to CCR7 promoter and mediate CCR7 expression in luciferase reporter assays and chromatin immunoprecipitation assays. Transactivation activity of Ets-1 was independent of the Pointed domain of Ets-1. Ets-1 could also enhance NF-κB and CBP transactivation of CCR7 promoter. Our results also showed that Ets-1 could modulate cancer cell transmigration by altering CCR7 expression in transwell assay and wound healing assay. Taken together, our data suggest that Ets-1 can enhance CCR7 expression and contribute to tumor cell migration.
Keywords: CCR7, Ets-1, PNT domain, Tumor migration
INTRODUCTION
Chemokines are a family of small chemotactic cytokines that direct the homing of immune cells and control their homeostasis (1). In addition to their roles in the immune system, chemokines have also been implicated in the pathogenesis of cancer. Several studies have shown that elevated expression of chemokines is correlated with cancer progression and metastasis (2, 3).
CCR7 is a CC chemokine family member, the ligand of which is CCL21/CCL19. In the immune system, CCL19/CCL21 expressed on fibroblastic reticular cells (FRCs) in secondary lymphoid organs can help CCR7-expressing cells move to lymphoid organs. Several studies, including those on breast cancer, melanoma, gastric cancer, and head and neck cancer, have shown that tumor cells also use this interaction to establish lymph node metastasis (4–7). In a mouse model, anti-CCR7 treatment could cause significant delay in tumor growth and metastasis (8). CCR7 expression is controlled by transcription factors, miRNAs, and epigenetic mechanisms. Transcription factors such as NF-AT1, AP-1, NF-κB, and SP-1 can bind to CCR7’s promoter region and control its mRNA transcription (4, 9, 10). However, whether these transcription factors are involved in tumor metastasis has not been elucidated yet.
Ets-1 is a transcription factor that contains a unique ETS domain with DNA binding activity. The ETS domain is a winged helix-turn-helix structure that recognizes GGAA/T core motif of DNA. Ets-1 has been shown to play a critical role in regulating the differentiation and function of T helper (Th) cells (11). Deficiency in Ets-1 has a profound impact on Th1 immune responses (12). Pointed (PNT) domain located on the N-terminus of Ets-1 can serve as a protein-interacting domain. Among interacting proteins, ERK2 has been shown to bind to the PNT domain and phosphorylate threonine residue T38 at the N-terminus. Phosphorylation of T38 increases the ability of Ets-1 to induce reporter gene expression in transient expression assay. This effect is closely related to the recruitment of coactivator CBP/p300 (10, 11). Ets-1 has been linked to progression of many types of cancer. Elevated expression of Ets-1 in cancer biopsies is associated with poor survival in many studies (13). However, how Ets-1 enhances tumor metastasis remains unknown.
The objective of the present study was to determine whether Ets-1 could regulate CCR7 expression and enhance tumor metastasis. We found that Ets-1 could bind to CCR7 promoter and enhance CCR7 expression in a concentration-dependent manner. Ets-1 increased CCR7 expression in a PNT domain-independent manner. In addition, Ets-1 cooperated with NF-κB and CBP to enhance CCR7 expression. Overexpression of Ets-1 enhanced tumor cell migration through CCR7/CCL21 interaction.
RESULTS
CCR7 expression is upregulated during T-cell activation
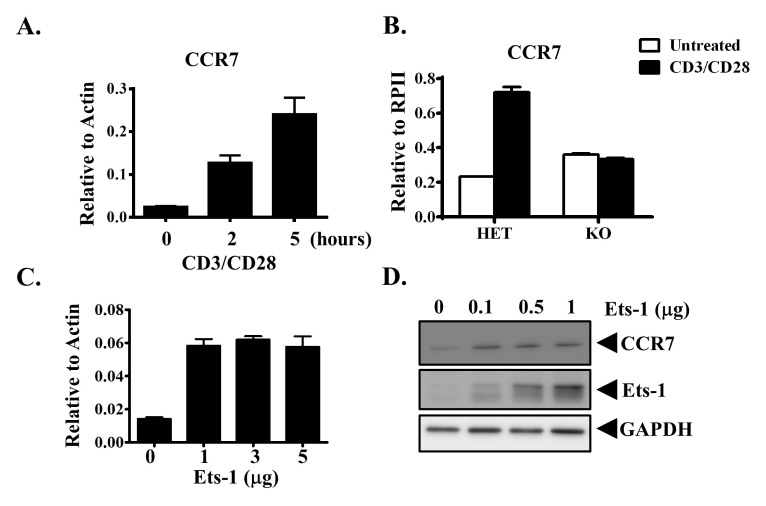
Previous studies have shown that CCR7 expression is upregulated in T cells upon T-cell stimulation (14). To determine whether Ets-1 regulated CCR7 expression, we first determined expression level of CCR7 upon T-cell activation. Splenocytes from wild-type (WT) mice were activated with CD3 and CD28. CCR7 expression was then measured with quantitative RT-PCR (Q-PCR). Similar to previous results, CCR7 expression levels were increased by at least 10-fold after cells were stimulated for 5 hours (Fig. 1A). To determine whether Ets-1 regulated CCR7 expression during T-cell activation, we examined CCR7 expression in Ets-1-deficient cells. Splenocytes from Ets-1 heterozygous (Het) and knockout (KO) mice were activated with CD3 and CD28 for 5 hours. CCR7 expression was then measured with Q-PCR. Ets-1 protein deficiency did not affect CCR7 expression in splenocytes without stimulation (Fig. 1B). This suggests that Ets-1 does not control CCR7 expression under resting conditions. However, CCR7 expression was upregulated in Het splenocytes, but not in Ets-1-deficient splenocytes, upon stimulation (Fig. 1B). The expression level of CCR7 in KO cells was approximately 50% of that in Het cells after activation. These data suggest that Ets-1 controls CCR7 expression upon T-cell activation.
Fig. 1.
CCR7 expression is regulated by Ets-1. (A) Splenocytes from B6 mice were activated with CD3/CD28 for the indicated time periods. Cells were harvested and the CCR7 expression level was examined. (B) Splenocytes from Ets-1 heterozygous (Het) or knockout (KO) mice were left untreated (white bar) or activated with CD3/CD28 (black bar) for 5 hours. Cells were harvested and the CCR7 expression level was examined. MDA-MB-231 cells were transfected with indicated amounts of Ets-1 DNA. These cells were harvested after 24 hours and CCR7 mRNA (C) and protein (D) expression levels are shown.
Ets-1 enhances CCR7 expression through promoter binding and upregulates transcriptional activity in cancer cells
Expression of CCR7 in some tumor cells plays a role in tumor metastasis to lymph nodes. Overexpression of Ets-1 in basal-like tumor cells has been linked to tumor metastasis (15). We hypothesized that Ets-1 might promote tumor cell metastasis by upregulating CCR7 expression. To test this hypothesis, we overexpressed Ets-1 in MDA-MB-231 breast cancer cells and examined CCR7 expression. Overexpression of Ets-1 increased CCR7 mRNA expression (Fig. 1C). CCR7 protein expression in MDA-MB-231 cells was also increased upon Ets-1 overexpression. Its expression was increased further with higher levels of Ets-1 transfection (Fig. 1D). These data suggest that Ets-1 can upregulate CCR7 expression.
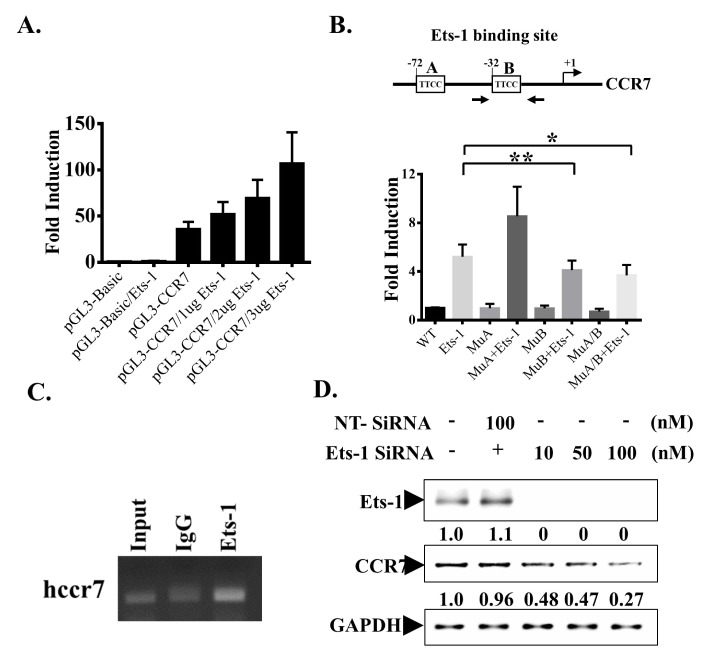
To test whether Ets-1 could bind to CCR7 promoter and regulate CCR7 expression, we searched for putative Ets-1 binding sites in the 1000 bp of CCR7 promoter region using multiTF and Mulan software. There were at least two potential Ets-1 binding sites in the CCR7 promoter region. These two putative Ets-1 binding sites were located at −72 bp and −32 bp upstream of the transcription start site of the CCR7 gene. Sequences of these two potential binding sites showed conservation in both human and mouse CCR7 promoter regions. The promoter region of human CCR7 was then cloned into a luciferase reporter plasmid and co-transfected with different concentrations of Ets-1 into MDA-MB-231 cells. Transfection of the 1 kb CCR7 promoter region caused a 50-fold increase in promoter activity compared to that caused by the control plasmid. The promoter activity was further increased by co-transfection with different concentrations of Ets-1. The promoter activity was increased by up to 120-fold when cells were transfected with 3 μg of Ets-1 expression plasmid (Fig. 2A). These results indicate that CCR7 is constitutively expressed in MDA-MB-231 cells and that overexpression of Ets-1 further upregulates CCR7 promoter activity.
Fig. 2.
Ets-1 binds to CCR7 promoter and controls its expression. (A) MDA-MB-231 cells were transfected with indicated plasmids. Cells were harvested after 24 hours and the promoter activity was examined by luciferase assay. (B) 2 Ets-1 binding sites on the CCR7 promoter region are shown. These sites were either mutated separately (MuA or MuB) or together (both mutated, MuA/B). The promoter activity was examined by luciferase assay and shown in the lower panel. (C) MDA-MD-231 cells were subjected to ChIP analysis. Chromatin was precipitated with an anti-Ets-1 antibody or an IgG antibody as a negative control. Precipitated DNA was amplified using primers (Fig. 2B, arrows) specific for a 220 bp fragment of the CCR7 promoter spanning the −32 Ets-1 binding sites. As a positive control, total DNA was diluted 1:100 and subjected to PCR (input). (D) MDA-MB-231 cells were transfected with indicated amounts of Ets-1 siRNA and harvested after 24 hours. Whole-cell lysates were prepared and subjected to Western blot analyses with indicated antibodies.
We then mutated these two binding sites through site-directed mutagenesis and tested Ets-1 transactivation activity. While Ets-1 co-transfected with WT promoter increased CCR7 promoter activity by 5-fold, co-transfection with Ets-1 having a mutation in the binding site at the −72 region did not affect luciferase activity (Fig. 2B). Ets-1 transcriptional activity was decreased by approximately 30% when cells were transfected with Ets-1 having a mutation in the binding site at the −32 region (Fig. 2B). Its transcriptional activity also showed a 30% reduction when both binding sites were mutated. These data suggest that Ets-1 can bind to the −32 region of the CCR7 promoter region and enhances CCR7 promoter activity. To determine whether Ets-1 can bind to the CCR7 promoter region in vivo, we performed a chromatin immunoprecipitation (ChIP) assay to immunoprecipitate the CCR7 promoter region with an Ets-1 antibody using normal rabbit Ig as a background control. These samples were subjected to Q-PCR using primers to specifically amplify the −32 Ets-1 binding region. Results showed an enrichment in anti-Ets-1-immunoprecipitated DNA containing the −32 Ets-1 binding site (Fig. 2C).
To confirm the role of Ets-1 in contributing to CCR7 expression, we knocked down Ets-1 expression in MDA-MB-231 cells with siRNA. Ets-1 expression in cells was completely abolished upon transfection with 25 nM Ets-1 siRNA. However, CCR7 expression was reduced by approximately 70% after transfection with 100 nM Ets-1 siRNA which also caused a complete knockdown of Ets-1 expression. These data suggest that a complete knockdown of Ets-1 expression does not abolish CCR7 expression (Fig. 2D). Taken together, these data suggest that Ets-1 is sufficient but not necessary for CCR7 expression.
Ets-1 regulates CCR7 expression through a PNT domain-independent mechanism
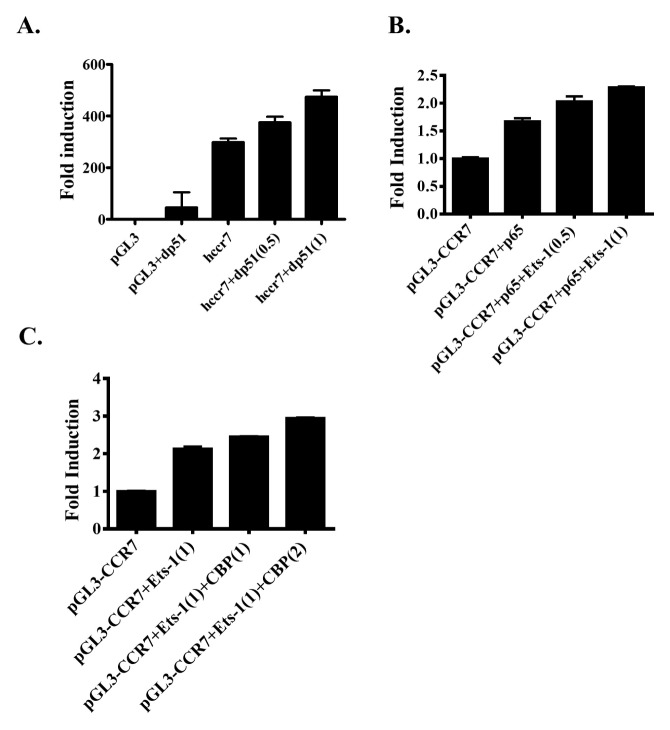
To examine the role of the PNT domain in CCR7 promoter activity, we deleted the PNT domain of Ets-1 (dp51 Ets-1) and tested its transcriptional activity. Similar to the effect of full-length Ets-1, co-transfection of dp51 Ets-1 enhanced CCR7 promoter activity. The enhancement of CCR7 promoter activity by dp51 Ets-1 was also concentration dependent (Fig. 3A). These data suggest that Ets-1 can enhance CCR7 promoter through a PNT domain-independent mechanism.
Fig. 3.
Control of CCR7 expression by Ets-1. (A) MDA-MB-231 cells were transfected with a reporter plasmid containing wild-type human CCR7 promoter or an empty vector plasmid (pGL3). These cells were co-transfected with various concentrations of plasmid containing Ets-1 with the Pointed domain deleted (dp51). The promoter activity was examined by luciferase assay. (B) MDA-MB-231 cells were transfected with a reporter plasmid containing wild-type human CCR7 promoter and a NF-κB expression plasmid (p65). Additionally, these cells were transfected with various amounts of Ets-1 expression plasmids. The promoter activity was examined by luciferase assay. (C) MDA-MB-231 cells were transfected with a reporter plasmid containing the wild-type human CCR7 promoter and an Ets-1 expression plasmid. Additionally, cells were transfected with various amounts of CBP expression plasmids. The promoter activity was examined by luciferase assay.
Among proteins involved in CCR7 expression, Ets-1 has been shown to interact with NF-κB protein (10). We then tested whether Ets-1 together with NF-κB could enhance the transcriptional activity of CCR7 promoter. Similar to results of a previous report (10), transfection with NF-κB enhanced CCR7 promoter activity by 1.5-fold. Co-transfection of NF-κB with Ets-1 enhanced CCR7 promoter activity in a concentration-dependent manner. The promoter activity was increased by up to 2-fold after transfection with 1 μg of Ets-1 (Fig. 3B). These data indicate that Ets-1 can interact with NF-κB to regulate CCR7 expression.
Previous studies have shown that CBP interacts with and enhances Ets-1 transcriptional activity (16). We tested whether CBP could enhance Ets-1 transcriptional activity on CCR7 promoter. Transfection with 1 μg of CBP expression vector and Ets-1 resulted in 2.5-fold enhancement of the transactivation of the CCR7 promoter. Furthermore, co-expression of 2 μg of CBP led to a 2.8-fold enhancement in CCR7 promoter activity (Fig. 3C). These data suggest that CBP can further enhance Ets-1 transcriptional activity on CCR7 promoter.
Ets-1 modulates tumor cell migration
MDA-MB-231 cells were co-transfected with Ets-1 and GFP expression plasmids. Migration activity of MDA-MB-231 cells was then measured. GFP was used to mark Ets-1-overexpressing cells. Transfected cells were placed in the upper chamber of a transwell plate and GFP+ cells in the lower chamber representing migrated cells were then counted. The number of GFP+ cells was increased by 4-fold in the lower chamber when cells were transfected with the Ets-1 expression plasmid. The number of migrating cells was increased when CCR7 ligand CCL21 was added to the lower chamber. The number of GFP+ cells was increased 10-fold when 3 μg of Ets-1 was transfected (Fig. 4A). We also examined the migration activity in a wound healing assay. The migration activity of MDA-MD-231 cells in CCL21 environment was decreased when Ets-1 expression was knocked down (Fig. 4B). These data clearly suggest that the expression level of Ets-1 affects tumor cell migration in a CCL21-containing environment.
Fig. 4.
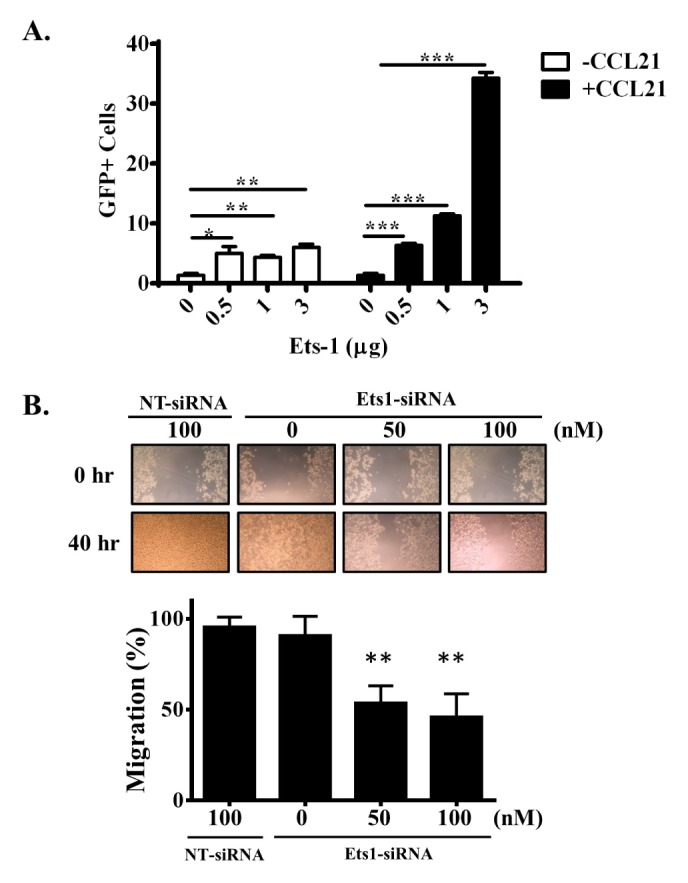
Ets-1 modulates tumor metastasis. (A) MDA-MB-231 cells were co-transfected with Ets-1 and GFP. After transfection, cells were added to the upper chamber of a transwell plate. The lower chamber was filled with medium without CCL21 (white bar) or with CCL21 (black bar). After 48 hours, GFP+ cells in the lower chamber were quantified. (B) MDA-MB-231 cells were transfected with indicated siRNA. Cells were then treated with CCL21 and subjected to wound healing assay. Pictures were acquired at 40 hours. Migration area was calculated with MRI wound healing tool of Image J software and expressed as percentage change of Ets-1 siRNA vs. Non-target control (NT-SiRNA).
DISCUSSION
In this study, we examined the role of Ets-1 in the regulation of CCR7 expression. We found that Ets-1 could directly bind to and transactivate CCR7 promoter activity. Mutation at Ets-1 binding sites on CCR7 promoter or knockdown of Ets-1 expression in a tumor cell line altered CCR7 expression. Functionally, overexpression of Ets-1 enhanced cancer cell transmigration activity based on tumor cell migration assay. Taken together, our data suggest that Ets-1 may enhance CCR7 expression in tumor cells and help tumor cells infiltrate lymph nodes.
Our results demonstrated that Ets-1 directly activated CCR7 promoter activity based on luciferase promoter assay. However, mutations in two conserved Ets-1 binding sites in the CCR7 promoter reduced CCR7 expression only by approximately 30%. The reduction in activity was similar when we mutated the −32 Ets-1 binding site (B site in Fig. 2B), suggesting that the −32 site might be the major Ets-1 binding site in controlling CCR7 expression. These data are consistent with results of Ets-1 knockdown experiment which showed that complete knockdown of Ets-1 expression only partially affected CCR7 protein expression. We also found a similar regulation of CCR7 expression in T cells. CCR7 expression was unaffected in unstimulated Ets-1-deficient T cells. However, CCR7 was downregulated in Ets-1-deficient T cells upon T-cell stimulation (Fig. 1). Collectively, these data suggest that Ets-1 may not play a role in basal CCR7 expression.
Our data showed that Ets-1 could control CCR7 expression in MDA-MB-231 cells. MDA-MDA-MB-231 is a highly metastatic and triple negative (ER−, PR− and HER2-negative) basal type breast cancer cell line (17, 18). Constitutive expression of Ets-1 in this cell line is well correlated with CCR7 expression and a highly metastatic phenotype. Our data also showed that the overexpression of Ets-1 in MDA-MB-231 cells further enhanced CCR7 expression and cell migration activity (Fig. 1D and 4). Ets-1 is an oncogene that plays a role in epithelial-to-mesenchymal transition (EMT) (19, 20). Transwell migration assay showed that overexpression of Ets-1 in tumor cells also enhanced tumor migration to an environment containing CCL21. In summary, our data suggest that Ets-1 plays multiple roles during cancer development. Overexpression of Ets-1 during cancer development may contribute to EMT and CCR7 overexpression which aids in tumor cell migration. Downregulation of Ets-1 in tumor therapy might be a promising strategy to block EMT and tumor metastasis.
MATERIALS AND METHODS
Mice
Ets-1 KO mice were described previously (12). Heterozygous littermates were used as controls. All animals were housed under specific pathogen-free conditions. Experiments were performed in accordance with institutional guidelines for animal care at E-Da University using approved protocols (IACUC-105001).
Quantitative RNA analysis
Total RNA was purified using a TRIzol Plus RNA Purification Kit (Invitrogen). First-strand cDNA synthesis was performed using 200 ng of total RNA and a QuantiTect Reverse Transcription Kit (QIAGEN). Gene expression levels were determined by real-time PCR analysis using a PowerUp SYBR Green Q-PCR Kit (Applied Biosystem) according to the manufacturer’s protocol on a 7500 Real Time PCR system (Applied Biosystem) using the following cycling conditions: denaturation at 95°C for 30 s, annealing at 56°C for 60 s, and extension at 72°C for 30 s. Primer sets were designed using Primer3 web utility. mRNA levels were adjusted for differences in actb (β-actin) or RPII (RNA polymerase II) expression.
Transwell migration assay
CytoSelectTM 24-well cell migration assay (Cell Biolab, San Diego, CA, USA) was used to examine cell migration activity. A total of 1 × 105 MDA-MB-231 cells transfected with pcDNA4/GFP or Est-1/GFP vectors in 200 μl of serum-free medium were placed in the upper chamber and the insert was placed in the lower well which held 500 μl of complete medium containing 10% fetal bovine serum with or without 100 ng/ml CCL21. After incubation for 48 hours, nonmigratory cells were removed from the upper surface of the membrane by scrubbing with wet cotton-tipped swabs 2–3 times. GFP-positive cells that migrated through the polycarbonate membrane and those climbing the bottom side were photographed. Four random images of each insert were acquired using fluorescence microscopy at 40× magnification. Migrated GFP-positive cells in each image were counted. Experiments were performed in triplicate.
Wound healing assay
Migratory capacity of MDA-MB-231 cells was examined using Culture-Insert 2 Well (80209; Ibidi) according to the manufacturer’s instructions. Briefly, 2.1 × 104 cells were suspended in 70 μl medium and incubated in each chamber overnight. After cell attachment, the culture insert was gently removed by tweezers, leaving a cell-free 500 μm gap. Fresh DMEM medium with CCL-21 (100 ng/ml) was added. Migration of cells was monitored and pictures were acquired after 40 hours. Pictures were analyzed and migration area was calculated with MRI Wound Healing Tool (http://dev.mri.cnrs.fr/projects/imagej-macros/wiki/Wound_Healing_Tool) in ImageJ.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed using Chromatin Immuno-precipitation Kit (#17-10085, EMD Millipore, Billerica, MA, USA) according to the protocol of the Magna ChIPTM A/G. For each sample, 1 × 107 cells were fixed in 1% freshly prepared formaldehyde for 10 min at room temperature and glycine (125 mM) was added to quench unreacted formaldehyde for 5 min. Cells were then washed twice with ice-cold phosphate-buffered saline (PBS, pH 7.2) and lysed in cell lysis buffer (5 mM PIPES, pH 8.0; 85 mM KCl; 0.5% NP-40) for 15 min on ice. After centrifugation, the cell pellet was resuspended in nuclear lysis buffer (50 mM Tris-Cl, pH 8.1; 10 mM EDTA; 1% SDS). Cross-linked DNA was sheared with a Misonix Sonicator® 3000 (Qsonica, Newtown, CT, USA) in six 15 s pulses with 50 s rest between pulses and a power setting of 6. Sheared chromatin was separated from debris by centrifugation at 10,000 × g for 10 min at 4°C. A total of 1 × 106 cell equivalents of chromatin was used for immunoprecipitation. Chromatin was well mixed with 1 μg of the indicated antibody and protein A/G magnetic beads and incubated for 4 hours at 4°C with rotation. The chromatin-antibody-protein A/G complex was separated with a magnetic separator and washed. The complex was then digested with proteinase K at 62°C for 2 hours and beads were separated with a magnetic separation device. DNA was purified with a spin column for PCR.
Plasmids, transfection, and luciferase assay
An approximately 2 kb genomic fragment containing nucleotides +28 to −1964 in relation to the transcriptional start site of human CCR7 gene was amplified and cloned into BglII site of pGL3-Basic Vector (Promega). In all luciferase assays, cells were transfected with 2 μg of luciferase reporter and 0.5 μg of pTK-Renilla with Lipofectamine 2000 (Invitrogen). Luciferase activity was determined in duplicate with a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Firefly luciferase activity obtained from each sample was normalized to Renilla luciferase activity from the same sample.
Western blot analysis and antibodies
In each sample, cells were lysed with freshly prepared whole-cell lysis buffer. Cell lysate was separated from debris by centrifugation at 13,000 rpm for 10 min. The lysate was loaded onto 10% polyacrylamide gels and transferred onto PVDF membranes (PolyScreen, PerkinElmer). These membranes were subsequently blocked in 5% milk and probed with an anti-Ets-1 or anti-CCR7 antibody. Proteins were visualized using an enhanced chemiluminescence (ECL) kit (PerkinElmer).
Statistical analysis
All statistical analyses were performed with Student’s t-test. Statistical significance was defined at P < 0.05 and indicated with asterisk (*P < 0.05; **P < 0.01; ***P < 0.001).
ACKNOWLEDGEMENTS
This study was supported by research grants (EDAHT-103030, EDAHT-104046, EDPJ-106084 and EDAHI-106001) from E-Da Hospital, Taiwan.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Vela M, Aris M, Llorente M, Garcia-Sanz JA, Kremer L. Chemokine receptor-specific antibodies in cancer immunotherapy: achievements and challenges. Front Immunol. 2015;6:12. doi: 10.3389/fimmu.2015.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zlotnik A. Chemokines in neoplastic progression. Semin Cancer Biol. 2004;14:181–185. doi: 10.1016/j.semcancer.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Chen K, Bao Z, Tang P, Gong W, Yoshimura T, Wang JM. Chemokines in homeostasis and diseases. Cell Mol Immunol. 2018;15:324–334. doi: 10.1038/cmi.2017.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al Akoum C, Akl I, Rouas R, et al. NFAT-1, Sp-1, Sp-3, and miR-21: New regulators of chemokine C receptor 7 expression in mature human dendritic cells. Hum Immunol. 2015;76:307–317. doi: 10.1016/j.humimm.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Muller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Xi L, Hunt JL, et al. Expression pattern of chemokine receptor 6 (CCR6) and CCR7 in squamous cell carcinoma of the head and neck identifies a novel metastatic phenotype. Cancer Res. 2004;64:1861–1866. doi: 10.1158/0008-5472.CAN-03-2968. [DOI] [PubMed] [Google Scholar]
- 7.Ma H, Gao L, Li S, et al. CCR7 enhances TGF-beta1-induced epithelial-mesenchymal transition and is associated with lymph node metastasis and poor overall survival in gastric cancer. Oncotarget. 2015;6:24348–24360. doi: 10.18632/oncotarget.4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Somovilla-Crespo B, Alfonso-Pérez M, Cuesta-Mateos C, et al. Anti-CCR7 therapy exerts a potent anti-tumor activity in a xenograft model of human mantle cell lymphoma. J Hematol Oncol. 2013;6:89. doi: 10.1186/1756-8722-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moran TP, Nakano H, Kondilis-Mangum HD, Wade PA, Cook DN. Epigenetic Control of Ccr7 Expression in Distinct Lineages of Lung Dendritic Cells. J Immunol. 2014;193:4904–4913. doi: 10.4049/jimmunol.1401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mburu YK, Egloff AM, Walker WH, et al. Chemokine receptor 7 (CCR7) gene expression is regulated by NF-kappaB and activator protein 1 (AP1) in metastatic squamous cell carcinoma of head and neck (SCCHN) J Biol Chem. 2012;287:3581–3590. doi: 10.1074/jbc.M111.294876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrett-Sinha LA. Review of Ets1 structure, function, and roles in immunity. Cell Mol Life Sci. 2013;70:3375–3390. doi: 10.1007/s00018-012-1243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grenningloh R, Kang BY, Ho IC. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J Exp Med. 2005;201:615–626. doi: 10.1084/jem.20041330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol. 2015;35:20–38. doi: 10.1016/j.semcancer.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Schwendemann J, Choi C, Schirrmacher V, Beckhove P. Dynamic differentiation of activated human peripheral blood CD8+ and CD4+ effector memory T cells. J Immunol. 2005;175:1433–1439. doi: 10.4049/jimmunol.175.3.1433. [DOI] [PubMed] [Google Scholar]
- 15.Bosman JD, Yehiely F, Evans JR, Cryns VL. Regulation of alphaB-crystallin gene expression by the transcription factor Ets1 in breast cancer. Breast Cancer Res Treat. 2010;119:63–70. doi: 10.1007/s10549-009-0330-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C, Shapiro LH, Rivera M, Kumar A, Brindle PK. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol Cell Biol. 1998;18:2218–2229. doi: 10.1128/MCB.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Betapudi V, Licate LS, Egelhoff TT. Distinct roles of nonmuscle myosin II isoforms in the regulation of MDA-MB-231 breast cancer cell spreading and migration. Cancer Res. 2006;66:4725–4733. doi: 10.1158/0008-5472.CAN-05-4236. [DOI] [PubMed] [Google Scholar]
- 18.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Wang Z, Chen Y, et al. Transcriptional silencing of ETS-1 abrogates epithelial-mesenchymal transition resulting in reduced motility of pancreatic cancer cells. Oncol Rep. 2015;33:559–565. doi: 10.3892/or.2014.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilles C, Polette M, Birembaut P, Brunner N, Thompson EW. Expression of c-ets-1 mRNA is associated with an invasive, EMT-derived phenotype in breast carcinoma cell lines. Clin Exp Metastasis. 1997;15:519–526. doi: 10.1023/A:1018427027270. [DOI] [PubMed] [Google Scholar]