Abstract
Immune repertoire is a collection of enormously diverse adaptive immune cells within an individual. As the repertoire shapes and represents immunological conditions, identification of clones and characterization of diversity are critical for understanding how to protect ourselves against various illness such as infectious diseases and cancers. Over the past several years, fast growing technologies for high throughput sequencing have facilitated rapid advancement of repertoire research, enabling us to observe the diversity of repertoire at an unprecedented level. Here, we focus on B cell receptor (BCR) repertoire and review approaches to B cell isolation and sequencing library construction. These experiments should be carefully designed according to BCR regions to be interrogated, such as heavy chain full length, complementarity determining regions, and isotypes. We also highlight preprocessing steps to remove sequencing and PCR errors with unique molecular index and bioinformatics techniques. Due to the nature of massive sequence variation in BCR, caution is warranted when interpreting repertoire diversity from error-prone sequencing data. Furthermore, we provide a summary of statistical frameworks and bioinformatics tools for clonal evolution and diversity. Finally, we discuss limitations of current BCR-seq technologies and future perspectives on advances in repertoire sequencing.
Keywords: Antibody, BCR, Genomics, NGS, Repertoire
INTRODUCTION
Adaptive immunity is a specialized protective immune system in vertebrates against a variety of pathogens by utilizing B and T lymphocytes (1). Acquired immune responses can be initialized and achieved through highly specific recognition of antigen with cell surface receptors including B cell receptor (BCR) and T cell receptor (TCR) (2). Undoubtedly, antigens are extremely diverse spanning from infectious particles to self-peptides. Receptors that can recognize these antigens with high binding affinity are selected out of a huge receptor pool prepared by random mutations. As a result, this immunological selection mechanism can lead to a highly personalized collection of lymphocytes in every individual which is called immune repertoire. In particular, B lymphocytes play pivotal role in humoral immunity through secreting BCR molecules, also known as antibodies and immunoglobulins, into body fluid. In this review, we will focus on BCR repertoire and interchangeably use both terms, BCR and antibody (3).
In recent years, research on BCR repertoire is not only an intriguing area in natural sciences, but also in pharmaceutical industries with significant impact. In immunology, emergence of powerful high-throughput methods such as parallel sequencing and mass spectrometry has allowed us to deeply investigate BCR repertoires. The repertoire analysis has expanded our horizons of understanding unseen diversity and complexity of the adaptive immune system. In clinical field, immunotherapy is leading a new paradigm in oncology. Thus, monoclonal antibody market has been rapidly enlarged (4). Examination of in vivo repertoire provides candidates of antigen-pecific monoclonal antibodies that can help us discover antibody drugs (5, 6). Furthermore, the repertoire of in vitro phage antibody library can be explored by deep sequencing to accelerate antibody discovery without conventional screening (7).
Major challenge in BCR repertoire analysis arises from difficulties in interrogating astronomical diversity. Heterogeneous clonality in BCR repertoire is derived from the fact that recombination of V, D, J gene segment occurs at DNA levels independently in every B cell (8). Additionally, somatic hyper-mutation (SHM) and insertion and deletion (INDEL) of nucleotides at V-D-J junctions can tremendously increase the junctional diversity (3, 8). The region translated from the junction determines antigen specificity. Such region is called complementary determining region 3 (CDR3). The highest throughput technology in genomics is currently next generation sequencing (NGS). The advent and improvement of NGS technology has revolutionized the scope to investigate repertoires (9). In this review, we will discuss B cell receptor sequencing (BCR-seq), a genomics approach to analyze BCR repertoire. In particular, library preparation, initial process of NGS, and the downstream analysis are emphasized.
OVERVIEW OF BCR-SEQ LIBRARY CONSTRUCTION
To study BCR repertoire, a process of separating B cells from diverse cell populations is the first step. B cells in peripheral blood, spleen, lymph node, and even in tumor tissue can be purified by surface markers. Sorted B cells are sequenced in bulk and also at single cell level after additional isolation step that is performed mostly using microfluidic devices (Fig. 1A). Sequencing B cells in bulk costs less but produces higher throughput that allows for identification of rare V(D)J recombination. However, it is unable to distinguish a unique pair of light chain and heavy chain in a B cell owing to lysis of pooled numerous cells. On the other hand, single cell sequencing resolves this limitation by tracing a single cell with a molecular barcode that can maintain heavy and light chain pair information and correct experimental bias and errors (10). However, since current methods of single cell transcriptome sequencing cover under a million cells that are insufficient to fully represent a huge BCR repertoire, single cell sequencing for repertoire should be conducted at high cost in order to obtain comprehensive repertoire with sufficient depth. As a result, BCR-seq is typically defined as a high throughput sequencing of only BCR regions, not transcriptome levels. In this review, BCR-seq as BCR region specific sequencing will be discussed.
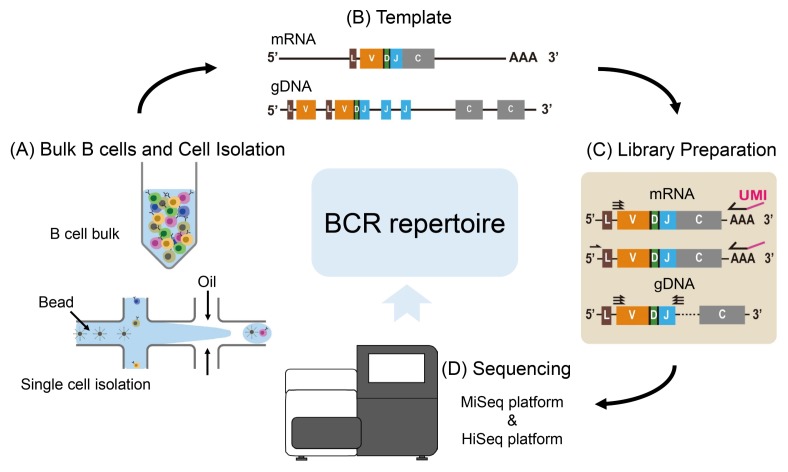
Fig. 1.
Experimental workflow of repertoire sequencing. (A) Sorted B cells by cell surface markers are prepared for sequencing in bulk or single cell state with further isolation by droplet microfluidics. (B) Templates available for BCR repertoire analysis are both gDNA and cDNA. gDNA contains intron sequences and both V and J genes that are not taken part in V(D)J recombination, resulting in far genomic distance between V region and C region in particular B cell clones. In contrast, only rearranged V(D)J segments exist in cDNA. They are juxtaposed with the C region. (C) For both gDNA and cDNA, a library is constructed by using PCR with multiplex primers targeting multiple V segments. Alternatively, in order to prevent primer bias from a large number of primer sets, a universal forward priming site is attached to the 5′ RACE region by template switching. (D) Once library preparation is completed, NGS platform is chosen considering the depth and length of BCR to be examined.
Before library construction, it is important to carefully consider suitable templates and genomic regions depending on purpose of the study because a choice of DNA or RNA and genomic regions to be examined provides different biological interpretation in the long run after customized analysis. First of all, genomic DNA (gDNA) or mRNA needs to be selected to construct a library (Fig. 1B). mRNA commonly used as BCR-seq template has already undergone V(D)J recombination and class switching that would allow a constant region to juxtapose with recombined variable region in one read of NGS. In the course of cDNA synthesis from mRNA on beads, cDNA can be barcoded with reverse transcription (RT) primers that include short random nucleotide sequences called Unique Molecular Identifiers (UMI) (Fig. 1C) (11). UMIs that are 8–12 nucleotide long can differentiate individual molecules and provide information to correct bias and errors generated during PCR amplification and sequencing (12). One important consideration when establishing a library with mRNA is where to prime for PCR amplification. Because mRNA undertakes not only V(D)J recombination, but also somatic hypermutation (SHM), it is difficult to choose priming sites around highly variable regions. Multiplex PCR with a mixture of primers near the 5′ end of variable regions can be a possible solution. However, the amplicon suffers significant primer bias owing to SHM (13). In order to reduce primer bias, universal priming sites can be attached by using template switching technique at 5′ end of cDNA for 5′ rapid amplification of cDNA ends (5′ RACE) (14, 15). Mostly, 5′ RACE universal primers are paired with backward primers aligning to constant regions to amplify the whole variable regions, suggesting that isotype-specific repertoire can be explored with corresponding primers. In the meanwhile, CDR3 is another target region to be sequenced because this region confers capability of antigen recognition with the most diverse sequences. Since CDR3 region spans only 50–100bp upstream constant region, amplicons can be deeply sequenced by short read technology.
gDNA can also be used to construct a library for BCR repertoire, indicating that BCR-seq does not necessarily mean RNA sequencing (Fig. 1B, C). In order to prepare gDNA library, a mixture of primers aligning to all V segments is selected as forward primers and a few primers targeting J segments are designed as backward primers. Different from mRNA template, priming constant regions is not preferred when using gDNA as template because constant regions are located several kb away from variable regions before class switching. Multiplex PCR is then performed to amplify heavy chain and light chain variable regions (VH and VL) (16). Unlike mRNA, gDNA has one copy per cell. It consists of intron sequences and V segments that do not participate in V(D)J recombination. Due to these properties of BCR gDNA, sequencing of gDNA enables us to infer and quantify clones in a less biased manner, although it requires more amplification for those with fewer copies than RNA. Moreover, B cells prior to class switching can retain extra DNA segments compared to BCR mRNA, resulting in both productive and nonproductive V(D)J sequences in the library. Detection of nonproductive recombination may lead to ambiguity in the analysis when the objective of BCR-seq is to identify full-length antibody sequences. Nonetheless, sequences of nonproductive and even aberrant recombination can help examine particular types of B cells (17). In summary, taking advantages of both gDNA and mRNA can yield better results and comprehensive interpretation through complementary information.
Finally, sequencing platform is critical in that applications vary in throughput, read length, and error rate. As HiSeq and NovaSeq of Illumina can easily produce a few hundreds of millions (M) of reads at less than a thousand dollars, these platforms are optimal to investigate comprehensiveness of repertoire. However, these short-read technologies (up to 150 bp) are of limited use. Although they are applicable to sequencing of CDR3, they are not applicable to full-length sequencing of variable regions of VH or VL. In order to sequence full-length variable regions, ranging from 400–600 bp, of VH and VL separately, MiSeq is the best platform as it is capable of producing reads of length 2 × 300 bp. However, its throughput is only up to 25M. Oxford Nanopore sequencing and Single Molecule Real-Time (SMRT) sequencing of PacBio are suitable for long gDNA library and VH:VL pairing library in principle. However, they are rarely applied to repertoire sequencing because bases of high quality are indispensable to discriminate true diversity from errors.
PREPROCESSING FOR DECONVOLUTION OF BCR-SEQ
Emergence of erroneous variants in BCR-seq is inevitable during PCR amplification and sequencing steps, although NGS technologies have been significantly improved over the past decade (18–20). Base calling errors originated from experiments and analyses can cause seriously biological mis-interpretation in repertoire analysis (21, 22). It is notable that SHM and INDEL on CDR3 occur at much higher rate than other somatic mutations in cancers and tissues, indicating that misleading identification of BCR sequences is highly sensitive to erroneous base calls in BCR-seq. To avoid identification of artefactual sequences bearing accumulated errors, various molecular and bioinformatics strategies such as quality control for base quality, assembly of paired-end reads, UMI, and clustering can be used.
The first error correction step for raw data is to check base quality that is optically measured by NGS machine. The best performance of NGS is currently at an error rate of 0.1% for ≥ 90% of bases per read. However, still about 5% of the data have errors occurring at 0.1%. Intrinsic NGS errors can be accurately computed based on fluorescence blurriness of clusters on a flow cell. They can be easily eliminated because base quality scores are recorded in FASTQ file as Phred score (23). In most cases, bases with error rate > 0.1% are trimmed using bioinformatics tools such as Trimmomatic at the beginning of analyses (24). Additional method to remove errors using base quality score is to choose bases with higher scores at overlapped positions when paired-end reads are assembled for fragments short enough to be stitched (Fig. 2B). For accurate reads assembly, at least 10 overlapping nucleotides are needed for a short read assembly program called FLASH (25).
Fig. 2.
Procedure of error correction and clonotyping. (A) Through UMIs attached to each read, the same sequences with the same UMI are considered as duplicates to be eliminated. Different sequences with the same UMI are produced by errors during PCR and sequencing. These sequences are collapsed based on UMI. (B) Read 1 and read 2 of paired-end reads from V(D)J full-length are assembled into a single sequence when reads have overlapping regions at their 3′ ends. Assembled sequences with high similarity are then clustered into the same clone in order to infer the origin of these sequences. Individual counts of collapsed sequences are summed to quantitate clonal abundance. Red squares in lines represent PCR and sequencing errors that can be corrected by UMI and/or clustering. Sky blue indicates bases with poor quality in paired-end reads.
More challenging problem is to correct errors introduced by PCR during target enrichment and NGS library preparation. An experimental way to eliminate these errors is to use UMIs encoded within PCR primers so that they could be added into molecules during RT (Fig. 2A). Since a random UMI sequence is configured differently for each molecule, it can serve as a unique ID of a molecule. After amplification and sequencing, a number of reads with the same UMI are produced. They are originated from one molecule. As a result, identical sequence of reads with different UMIs indicates detection of independent transcripts expressed from the same gene rather than by PCR bias (12, 26). Moreover, when two different types of UMIs are designed for a molecule and a cell, respectively, it is easy to tell whether the same sequences are stemmed from different transcripts in the same cells or from the same sequence transcript of different cells. Therefore, UMI strategy helps accurate detection and quantification of B cell clones by correcting PCR errors and tracking single cell simultaneously. Upon wide and common use of UMI, recently developed tools often implement utilities for de-multiplexing UMIs (27). It should be noted that UMI is not a perfect method because sequencing errors in UMI sequence are not ignorable in long UMI design that aims to resolve sequence saturation issue of short UMIs. Thus, long UMI sequences have high chance to include errors in themselves, resulting in the same molecule that has different UMIs (28, 29).
The next process is to detect and define distinct B cell clones from large and complex sequencing data containing sequence variants derived from recombination, SHM, and INDEL. Clonality can be identified by clustering sequences. This is called as clonotyping (Fig. 2B). Clonotyping starts with grouping reads based on sequence similarity to infer a shared ancestor without SHM or INDEL. In general, homology comparison is conducted for clonotyping using sequences of CDR3, the most variable region in BCR, and under the highest selection pressure during affinity maturation. Multiple thresholds of homology around 90–100% are applied. The threshold should be cautiously chosen owing to the fact that relaxed parameters can underestimate clone diversity whereas strict parameters might define sequences with errors as distinct B cell clones. In this sense, clonotyping process has also function as a remover of PCR and sequencing errors because nucleotide variants sparsely distributed at low frequency can be regarded as errors from highly abundant consensus sequences in a cluster. As pairwise comparison and clustering for large repertoire-size data are computationally intensive, UCLUST and CD-Hit are the most widely used programs that employ a strategy of matching short words in common among sequences for rapid identification of correct hits (30–32). Although clustering analysis is widespread, it is not necessary in all repertoire analyses. Alternative approaches are also available. For example, in order to study clonal abundance of B cells elicited by pathogens or cancer, UMI can be used for clonotyping, error correction, and quantification of BCR. In contrast, antibody engineering requires sequence clustering for clonotyping. Research on novel antibody discovery should identify error-free full-length sequences, suggesting that clonotyping and error correction by clustering is an efficient method beyond UMI (Fig. 2B).
DOWNSTREAM ANALYSES
Once identification of clones is completed, further downstream analyses are performed to understand clonal evolution and diversity at a system-wide level. Clonal evolution of BCR repertoire follows the principle of evolution (i.e., genetic diversity is followed by clonal selection through competition). Likewise, BCR repertoire is an evolving ecological system within our bodies as clonal selection keeps occurring among numerous unique B cells in a population generated by accumulation of genetic variation. Therefore, analyses of clonality from evolutionary point of view allow us to understand how BCR repertoire has been initially built and then adapted.
One of the very first steps in evolution is genetic diversification within a population. Thus, recombination and SHM in BCR should be measured before studying systematic evolution of repertoire (Fig. 3A). For both analyses of gene usage and SHM, BCR-seq data are aligned onto germline BCR database such as the international ImMunoGeneTics information system (IMGT) (33). For annotation step, caution is warranted regarding the choice of annotation tools as it can result in varying alignment outputs, similar to other NGS read mapping processes. An advantage of these annotation tools such as MIXCR, IgBlast, and IMGT/V-QUEST is that they can provide information for calculating gene usage and SHM rates (34–36). In general, SHM is calculated by simply counting the total number of mutations and then dividing it by the length of DNA regions. Statistical modeling is also utilized for SHM calculation. For example, SHazaM infers SHM through building the targeting model that takes neighboring sequences into account (37).
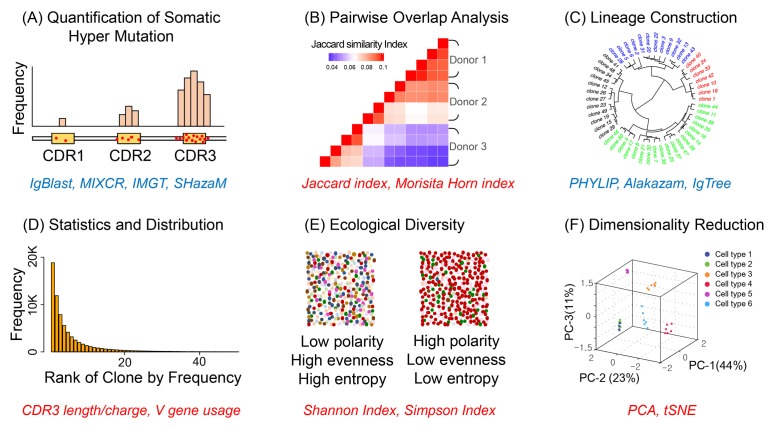
Fig. 3.
Bioinformatics analysis for clonal evolution and diversity. (A) Red dots represent mutations on CDR3 of BCR. CDR3 has the highest mutation rate that enables us to define clonotypes of B cells. (B) Due to high diversity by random SHM, shared repertoire between unrelated individuals is small. (C) Phylogenetic tree of B cell lineage reveals history of clonal evolution and origins. (D) Statistical distribution of BCR characteristics (e.g. CDR3 length, amino acid composition) shows the shape of diverse repertoires. (E) Diversity indices quantitate size and evenness of repertoires. BCR population is considered as an ecology within a body. Ecological statistics are then applied. (F) Dimensionality reduction algorithm projects repertoires in 2D or 3D. An axis represents a variance explained. Reduced dimension visualizes distance between repertoires. Italicized blue text indicates names of software and statistical tools. Red text represents concepts or indicators.
After clonotyping, comparison at repertoire level is performed by overlapping clones that are selected by affinity maturation after stochastic diversification from each naïve repertoire (Fig. 3B). Due to random selection in repertoire, the number of shared clones is small among individuals. Common clones are called public repertoire. For quantitative measurement of overlap, Morisita-Horn index is commonly computed in that this index considers different sizes of two populations that often occur in repertoire analysis (Fig. 3B) (38). The ratio of overlapping intersection to union called Jaccard index is another way to show a degree of overlap (39). Practically, package vegan in R provides various methods to calculate dissimilarity indices. Computing multiple indices is recommended to avoid mis-interpretation by a single value (40). Utilizing a unique sequence per clone as another critical evolutionary analysis can construct a lineage of B cell clones (Fig. 3C). Phylogenetic tree also exhibits evolutionary relationship among sequences with accumulated variants on variable regions or CDR3, one of which can be used for building the dendrogram. In other words, the tree enables us to trace B cells before and after SHM in germinal center of lymph nodes. PHYLIP is a general phylogenetic tree construction tool that can enable visualization of BCR lineage from multiple alignments of sequences. In addition, repertoire specific lineage tools such as IgPhyML, IgTree, and Alakazam implement maximum likelihood framework or minimal mutation assumption (41–43).
Analysis of clonal evolution explained above can be understood in great detail through interrogating clonal diversity. Basic methods to examine the diversity is to conduct statistical analyses on frequencies of biochemical and biophysical properties in variable regions of BCR (Fig. 3D). Frequencies of CDR3 length, charge, and hydrophobicity are informative to learn the profile of a repertoire under immunological conditions. For investigation at amino acid level, Package Peptides in R is useful for summarizing biochemical characteristics of amino acid sequences. The frequency of V gene usage is also of interest because its usage frequency can discriminate B cell types within individuals (44). Furthermore, the distribution of clonal frequency provides the shape of a repertoire upon external immunological stimulations regarding how much a few clones dominate and how the distribution is skewed (45). Since frequency statistics is a relatively straightforward computation, no particular tool is available. However, repertoire pipeline tools (e.g., IGGalaxy and VDJServer) are often implemented to report the statistics (46, 47). Next, quantitative estimate of repertoire diversity is crucial as repertoire diversity is closely associated with immune responses (Fig. 3E). Comprehensive investigation of diversity has been started recently due to the emergence of NGS. Thus, the concept of diversity indices is adapted from ecology, an academic field with much longer history (48). Richness and evenness represent the number of clones and a degree of equal distribution, respectively. Entropy and polarity are other parameters that simultaneously quantitate the size and distribution of clones in a repertoire profile. For instance, Simpson index is a probability that two randomly chosen sequences belong to the same clone and Shannon index is −∑Piln Pi, where Pi is the proportional abundance of clones. Like overlap index, the package vegan in R can be utilized to easily perform computation of multiple diversity indices.
Lastly, dimensionality reduction is a technique that can dissect the repertoire complexity and visualize multi-dimensional data in 2- or 3-dimensional space (Fig. 3F). Defining clones in repertoire generates high order multi-dimensional data. It is noteworthy that dimensions of repertoire profile are not the same as those of gene expression profile in single cell sequencing. In single cell RNA-sequencing data analysis, each dimension represents an expressed gene. Thus, a cell is a data point in the multi-dimensional space before and after reduction. In contrast, a clonotype in a repertoire corresponds to a dimension, indicating that the size of original space is highly variable across repertories. Therefore, feature selection is necessary before reduction for repertoires. Principal Component Analysis (PCA) and t-distributed Stochastic Neighbor Embedding (t-SNE) are two commonly used machine learning techniques. PCA is performed to maximally explain the variance of complex data in reduced dimensions by computing eigenvectors from the covariance matrix, whereas t-SNE is a nonlinear dimensionality reduction method through constructing probability distributions of similar data points and then minimizing the divergence between probability distributions. From a low dimensional space, we can visually inspect which repertoires are closely located in 2D or 3D space. Both PCA and t-SNE are implemented in R packages. Python machine learning tool, scikit-learn (49), is also freely available and useful owing to its great detailed documentations and graphical examples.
PERSPECTIVE
In BCR repertoire analysis, it would be ideal to obtain BCR repertoire of error-free full-length VH:VL sequences with single cell tracking at high throughput of more than a million cells. This aim has not been accomplished yet, although fundamental technologies for individual sub-aims have been already developed. For example, widely used single cell sequencing protocols such as Drop-seq and 10X Genomics platform can track single cells with UMIs. However, neither of them can identify full-length sequence of VH and VL (50). This is because reads from both methods cover only about a hundred nucleotides at the 3′ end and UMI at the 5′ end of cDNA. These sequences can be utilized only for clonotyping and repertoire diversity analysis. In contrast, another powerful single cell sequencing protocol, SMART-seq, allows us to sequence whole transcriptome and identify a combination of VH and VL in a single B cell through cell isolation by fluorescence-activated cell sorting (FACS) into 96- or 384-well plate (51). As the number of wells on a plate is the number of cells to be sequenced, a low throughput of SMART-seq is an obvious caveat to be overcome, although such small scale full-length sequencing is particularly useful for antibody discovery and engineering. Taken together, there might be two possible solutions to sequence full-length of BCR from a great number of cells: 1) To use SMART-seq to sequence whole transcriptome for a high number of plates, 2) To use Drop-seq or 10X Genomics platform with long read technologies. The first solution is not practical because it has unnecessarily high cost. It is noteworthy that enrichment of BCR sequences by either hybridization-based capture or PCR amplification is cost-effective by removing unrelated mRNA species to repertoire. The second solution is not realistic for repertoire either because long read sequencing technology does not satisfy the need of accuracy to identify highly variable sequences. Error-free long read is particularly crucial for BCR-seq as it can improve and broaden applications. For example, the library size of VH:VL pairs is around 1 kb, meaning that PacBio and Nanopore machines are needed to sequence the full-length whereas the longest read methodology of Illumina, 2 × 300 MiSeq platform, cannot fully cover these regions. However, error rates of PacBio and Nanopore platforms are still too high to accurately infer clones with SHM, although these platforms have been significantly improved to identify genomic variants in general. At present, the most realistic solution to obtain high throughput data of full-length BCR at single cell level might be as follows: i) To interrogate over a million B cells by flow-focusing microfluidics, ii) To enrich VH and VL by PCR, and iii) To sequence VH and VL separately and together through short read technologies with low error rate and high throughput (52). Therefore, innovation for the future of BCR-seq requires further advances in next generation sequencing technologies.
Lastly, a drawback of BCR-seq comes from the fact that genomic sequences and mRNA abundance of B cells in peripheral blood neither perfectly represent amino acid sequences of serological antibodies nor correlate with expression levels of antibodies. Furthermore, as post-translational modification determines functions of antibodies, genomic approaches to BCR repertoire are not optimal for isolating functional antibodies (53). One of solutions to these issues is to converge BCR-seq with proteomics (45). Mass spectrometry can be applied to obtain spectra of trypsinized peptides from serological antibodies after antigen enrichment. These spectra are then searched against the database generated by BCR-seq. Identified peptides, especially CDR3 peptides, are valuable molecular signatures that respond to antigens such as pathogens in serum. This in vivo screening of antigen-specific antibody can be applied in pharmaceutical industries in order to identify functional serological antibodies at protein levels. Nevertheless, limitations of proteomics approaches to BCR repertoire are their low sensitivity to detect low expressed antibodies and the requirement of personal search database. The future of proteomic antibody repertoire would be de novo peptide sequencing by using nanopore or single molecule imaging with high sensitivity in the absence of search database (54, 55).
ACKNOWLEDGEMENTS
This work was supported by a new faculty research fund of Ajou University and by a grant (No. 2019R1C1C1008181) of the National Research Foundation of Korea (NRF) to D.P.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Bonilla FA, Oettgen HC. Adaptive immunity. J Allergy Clin Immunol. 2010;125:S33–40. doi: 10.1016/j.jaci.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 2.Bishop GA, Haxhinasto SA, Stunz LL, Hostager BS. Antigen-specific B-lymphocyte activation. Crit Rev Immunol. 2003;23:149–197. doi: 10.1615/CritRevImmunol.v23.i3.10. [DOI] [PubMed] [Google Scholar]
- 3.Murphy K, Weaver C. Janeway’s immunobiology. 9th ed. Garland Science; 2016. pp. 410–411. [Google Scholar]
- 4.Weiner GJ. Building better monoclonal antibody-based therapeutics. Nat Rev Cancer. 2015;15:361–370. doi: 10.1038/nrc3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X, Zhou T, Zhu J, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doria-Rose NA, Schramm CA, Gorman J, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glanville J, Zhai W, Berka J, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A. 2009;106:20216–20221. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakano H, Kurosawa Y, Weigert M, Tonegawa S. Identification and nucleotide sequence of a diversity DNA segment (D) of immunoglobulin heavy-chain genes. Nature. 1981;290:562–565. doi: 10.1038/290562a0. [DOI] [PubMed] [Google Scholar]
- 9.Rouet R, Jackson KJL, Langley DB, Christ D. Next-generation sequencing of antibody display repertoires. Front Immunol. 2018;9:118. doi: 10.3389/fimmu.2018.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wardemann H, Busse CE. Novel approaches to analyze immunoglobulin repertoires. Trends Immunol. 2017;38:471–482. doi: 10.1016/j.it.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Vollmers C, Sit RV, Weinstein JA, Dekker CL, Quake SR. Genetic measurement of memory B-cell recall using antibody repertoire sequencing. Proc Natl Acad Sci U S A. 2013;110:13463–13468. doi: 10.1073/pnas.1312146110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith T, Heger A, Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan TA, Friedensohn S, Gorter de Vries AR, Straszewski J, Ruscheweyh HJ, Reddy ST. Accurate and predictive antibody repertoire profiling by molecular amplification fingerprinting. Sci Adv. 2016;2:e1501371. doi: 10.1126/sciadv.1501371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matz M, Shagin D, Bogdanova E, et al. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 1999;27:1558–1560. doi: 10.1093/nar/27.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waltari E, Jia M, Jiang CS, et al. 5′ Rapid amplification of cDNA ends and illumina MiSeq reveals B cell receptor features in healthy adults, adults with chronic HIV-1 infection, cord blood, and humanized mice. Front Immunol. 2018;9:628. doi: 10.3389/fimmu.2018.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calis JJ, Rosenberg BR. Characterizing immune repertoires by high throughput sequencing: strategies and applications. Trends Immunol. 2014;35:581–590. doi: 10.1016/j.it.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chovanec P, Bolland DJ, Matheson LS, et al. Unbiased quantification of immunoglobulin diversity at the DNA level with VDJ-seq. Nat Protoc. 2018;13:1232–1252. doi: 10.1038/nprot.2018.021. [DOI] [PubMed] [Google Scholar]
- 18.Robasky K, Lewis NE, Church GM. The role of replicates for error mitigation in next-generation sequencing. Nat Rev Genet. 2014;15:56–62. doi: 10.1038/nrg3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kircher M, Heyn P, Kelso J. Addressing challenges in the production and analysis of illumina sequencing data. BMC Genomics. 2011;12:382. doi: 10.1186/1471-2164-12-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shagin DA, Shagina IA, Zaretsky AR, et al. A high-throughput assay for quantitative measurement of PCR errors. Sci Rep. 2017;7:2718. doi: 10.1038/s41598-017-02727-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X, Chockalingam SP, Aluru S. A survey of error-correction methods for next-generation sequencing. Brief Bioinform. 2013;14:56–66. doi: 10.1093/bib/bbs015. [DOI] [PubMed] [Google Scholar]
- 22.Friedensohn S, Khan TA, Reddy ST. Advanced methodologies in high-throughput sequencing of immune repertoires. Trends Biotechnol. 2017;35:203–214. doi: 10.1016/j.tibtech.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. doi: 10.1101/gr.8.3.186. [DOI] [PubMed] [Google Scholar]
- 24.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kivioja T, Vaharautio A, Karlsson K, et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods. 2011;9:72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- 27.Vander Heiden JA, Yaari G, Uduman M, et al. pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics. 2014;30:1930–1932. doi: 10.1093/bioinformatics/btu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He L, Sok D, Azadnia P, et al. Toward a more accurate view of human B-cell repertoire by next-generation sequencing, unbiased repertoire capture and single-molecule barcoding. Sci Rep. 2014;4:6778. doi: 10.1038/srep06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egorov ES, Merzlyak EM, Shelenkov AA, et al. Quantitative profiling of immune repertoires for minor lymphocyte counts using unique molecular identifiers. J Immunol. 2015;194:6155–6163. doi: 10.4049/jimmunol.1500215. [DOI] [PubMed] [Google Scholar]
- 30.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 31.Li W, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 32.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 33.Lefranc MP, Giudicelli V, Duroux P, et al. IMGT(R), the international ImMunoGeneTics information system(R) 25 years on. Nucleic Acids Res. 2015;43:D413–422. doi: 10.1093/nar/gku1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolotin DA, Poslavsky S, Mitrophanov I, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 35.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giudicelli V, Chaume D, Lefranc MP. IMGT/V-QUEST an integrated software program for immunoglobulin and T cell receptor V-J and V-D-J rearrangement analysis. Nucleic Acids Res. 2004;32:W435–440. doi: 10.1093/nar/gkh412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui A, Di Niro R, Vander Heiden JA, et al. A Model of somatic hypermutation targeting in mice based on high-throughput Ig sequencing data. J Immunol. 2016;197:3566–3574. doi: 10.4049/jimmunol.1502263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ritvo PG, Saadawi A, Barennes P, et al. High-resolution repertoire analysis reveals a major bystander activation of Tfh and Tfr cells. Proc Natl Acad Sci U S A. 2018;115:9604–9609. doi: 10.1073/pnas.1808594115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas N, Best K, Cinelli M, et al. Tracking global changes induced in the CD4 T-cell receptor repertoire by immunization with a complex antigen using short stretches of CDR3 protein sequence. Bioinformatics. 2014;30:3181–3188. doi: 10.1093/bioinformatics/btu523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oksanen J, Kindt R, Legendre P, et al. The vegan package. Community Ecology Package. 2007;10:631–637. [Google Scholar]
- 41.Hoehn KB, Lunter G, Pybus OG. A phylogenetic codon substitution model for antibody lineages. Genetics. 2017;206:417–427. doi: 10.1534/genetics.116.196303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barak M, Zuckerman NS, Edelman H, Unger R, Mehr R. IgTree: creating Immunoglobulin variable region gene lineage trees. J Immunol Methods. 2008;338:67–74. doi: 10.1016/j.jim.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Gupta NT, Vander Heiden JA, Uduman M, Gadala-Maria D, Yaari G, Kleinstein SH. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeKosky BJ, Lungu OI, Park D, et al. Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc Natl Acad Sci U S A. 2016;113:E2636–2645. doi: 10.1073/pnas.1525510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J, Boutz DR, Chromikova V, et al. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat Med. 2016;22:1456–1464. doi: 10.1038/nm.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moorhouse MJ, van Zessen D, IJspeert H, et al. ImmunoGlobulin galaxy (IGGalaxy) for simple determination and quantitation of immunoglobulin heavy chain rearrangements from NGS. BMC Immunol. 2014;15:59. doi: 10.1186/s12865-014-0059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christley S, Scarborough W, Salinas E, et al. VDJServer: A cloud-Based analysis portal and data commons for immune repertoire sequences and rearrangements. Front Immunol. 2018;9:976. doi: 10.3389/fimmu.2018.00976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hurlbert SH. The nonconcept of species diversity: A critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 49.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine learning in python. J Machine Learning Res. 2011;12:2825–2830. [Google Scholar]
- 50.Rajan S, Kierny MR, Mercer A, et al. Recombinant human B cell repertoires enable screening for rare, specific, and natively paired antibodies. Commun Biol. 2018;1:5. doi: 10.1038/s42003-017-0006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu YL, Stubbington MJ, Daly M, Teichmann SA, Rada C. Intrinsic transcriptional heterogeneity in B cells controls early class switching to IgE. J Exp Med. 2017;214:183–196. doi: 10.1084/jem.20161056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDaniel JR, DeKosky BJ, Tanno H, Ellington AD, Georgiou G. Ultra-high-throughput sequencing of the immune receptor repertoire from millions of lymphocytes. Nat Protoc. 2016;11:429–442. doi: 10.1038/nprot.2016.024. [DOI] [PubMed] [Google Scholar]
- 53.Jung ST, Reddy ST, Kang TH, et al. Aglycosylated IgG variants expressed in bacteria that selectively bind FcgammaRI potentiate tumor cell killing by monocyte-dendritic cells. Proc Natl Acad Sci U S A. 2010;107:604–609. doi: 10.1073/pnas.0908590107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piguet F, Ouldali H, Pastoriza-Gallego M, Manivet P, Pelta J, Oukhaled A. Identification of single amino acid differences in uniformly charged homopolymeric peptides with aerolysin nanopore. Nat Commun. 2018;9:966. doi: 10.1038/s41467-018-03418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swaminathan J, Boulgakov AA, Hernandez ET, et al. Highly parallel single-molecule identification of proteins in zeptomole-scale mixtures. Nat Biotechnol. 2018;36:1076–1082. doi: 10.1038/nbt.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]