Abstract
Objective:
Radiomics pipelines have been developed to extract novel information from radiological images, which may help in phenotypic profiling of tumours that would correlate to prognosis. Here, we compared two publicly available pipelines for radiomics analyses on head and neck CT and MRI in nasopharynx cancer (NPC).
Methods and materials:
100 biopsy-proven NPC cases stratified by T- and N-categories were enrolled in this study. Two radiomics pipeline, Moddicom (v. 0.51) and Pyradiomics (v. 2.1.2) were used to extract radiomics features of CT and MRI. Segmentation of primary gross tumour volume was performed using Velocity v. 4.0 by consensus agreement between three radiation oncologists. Intraclass correlation between common features of the two pipelines was analysed by Spearman’s rank correlation. Unsupervised hierarchical clustering was used to determine association between radiomics features and clinical parameters.
Results:
We observed a high proportion of correlated features in the CT data set, but not for MRI; 76.1% (51 of 67 common between Moddicom and Pyradiomics) of CT features and 28.6% (20 of 70 common) of MRI features were significantly correlated. Of these, 100% were shape-related for both CT and MRI, 100 and 23.5% were first-order-related, 61.9 and 19.0% were texture-related, respectively. This interpipeline heterogeneity affected the downstream clustering with known prognostic clinical parameters of cTN-status and GTVp. Nonetheless, shape features were the most reproducible predictors of clinical parameters among the different radiomics modules.
Conclusion:
Here, we highlighted significant heterogeneity between two publicly available radiomics pipelines that could affect the downstream association with prognostic clinical factors in NPC
Advances in knowledge:
The present study emphasized the broader importance of selecting stable radiomics features for disease phenotyping, and it is necessary prior to any investigation of multicentre imaging datasets to validate the stability of CT-related radiomics features for clinical prognostication.
Background
Nasopharyngeal carcinoma (NPC) is a malignant head and neck cancer that is endemic in the Southeastern parts of Asia and North Africa.1 This tumour is characterized by its sensitivity to radiation and platinum-based chemotherapy, and as such survival has improved substantially with the advent of radiotherapy advancement and combination chemoradiotherapy in locally advanced cases.2 Nonetheless, combination therapies also contribute to incremental treatment-related toxicities, and thus there is a push to tailor treatment intensity based on more precise clinical risk stratification. To this end, there is a keen interest to explore novel biomarkers derived from molecular profiling of tumours that could inform on prognosis. More recently, radiomics, which aims to use algorithms to extract deep radiological features not visible to the naked eye, has been explored as a tool to characterize novel radiological phenotypes that are correlated with prognosis in a number of tumour types.3–6 Several radiomics pipelines have been developed, most of which are agnostic to the type of imaging modality and anatomical site from which the region of interest (ROI) is localized.7 It is thus possible that differences in image acquisition and reconstruction methods contributing to variation in image quality, among other factors including accuracy of ROI segmentation, coding variation and baseline clinical characteristics, could affect the downstream feature extraction process in radiomics. Collectively, these caveats motivated a global collaborative effort to harmonize the radiomics analytical processes,8–10 and the first undertaking of the group was the standardization of some publicly available radiomics pipelines.
Among them, Pyradiomics is the most widely reported radiomics tool in the literature. It provides a flexible analytical platform with both a simple and convenient front-end interface in 3-dimensional (3D) Slicer—a free open-source platform for medical image computing.11 It regards the voxel-to-voxel relationship in a 3D manner (default software option), and outputs several radiomics indices relating to first-order, texture modules, and shape. Next, Moddicom is another in-house developed radiomics pipeline, which is able to handle DICOM/DICOM-RT objects; unlike Pyradiomics, it considers two-dimensional voxel-to-voxel relationships for each transverse slice within the ROI, and then generates an aggregated score by using the mean of all the slices for the textural radiomics indices. It was developed primarily to interrogate fractal radiomics in MRIs.12 Both radiomics tools have been investigated in several human cancers, including oesophageal, lung and head and neck cancers, etc. and specific radiomics features have been highlighted to correlate with prognosis.5,13,14 All of the common features defined in both radiomics tools are International Image Biomarker Standardization Initiative-compliant.11,12 However, while both pipelines share common radiomics features and definitions, as aforementioned, the algorithms implementations underlying the feature extraction process differ between them. It is therefore not known if such interpipeline variations could affect the association with clinical parameters.
In this background, we investigated the utility of Pyradiomics and Moddicom as radiomics tools to analyze CT and MRI datasets in a cohort of NPC cases. In addition, we included a third radiomics pipeline—Computational Environment for Radiological Research (CERR),15 for comparison with Pyradiomics and Moddicom. We included tumours of different T-, N-categories, and gross tumour volumes (GTV), and observed significant heterogeneity even for the same features between both tools that affected the correlation with these known prognostic clinical parameters.
Methods and materials
Study cohort
We utilized a data set of 100 patients with biopsy-proven NPC from a single academic institution. All patients fulfilled the following criteria: (1) differentiated or undifferentiated non-keratinizing NPC based on the WHO classification; (2) absence of distant metastasis; (3) and were treated with intensity-modulated radiotherapy (IMRT). Patient demographics, including age, gender and baseline comorbidities were collected. Tumour characteristics including T-, N-category, and GTV of the primary tumour (GTVp) were recorded; all patients were restaged according to the American Joint Committee on Cancer seventh edition/International Union Against Cancer (2010) stage classification system. For the purpose of this study, which was primarily to investigate the implications of inter pipeline heterogeneity, we included equal numbers of patients with T1-4 status NPC. Ethical approval for the study was obtained from the SingHealth Centralised Institutional Review Board (protocol no. 2018/2352). Informed consent was obtained from all living patients.
Treatment strategies
All patients underwent IMRT as primary treatment of NPC. The IMRT planning and treatment protocol were as previously reported.16 Briefly, GTVp and clinically involved nodes were outlined, followed by high-risk and low-risk clinical target volumes (CTVs) in the primary tumour region and uninvolved nodal levels. 70, 60 and 54 Gy, delivered as simultaneous boost technique in 33 fractions, were prescribed to the GTV, high- and low-risk CTV, respectively. Dose constraints to critical organs at risk were determined by the standard threshold doses. Additionally, for patients with Stage III–IVb disease, IMRT was given in combination with concurrent chemotherapy of cisplatin (either 40 mg/m2 weekly or 100 mg/m2 3-weekly), along with either neoadjuvant or adjuvant platinum-based chemotherapy regimens.
Imaging protocol
CT perfusion (CTP) scans of the nasopharynx were performed using a 120 kVp 64-slice multidetector scanner (Somatom AS, Siemens Medical Solutions, Forchhein, Germany). It has a field of view (FOV) of 50 cm, slice thickness of 2 mm, and matrix of 512 × 512. The reconstruction kernel used is B40s. 60 ml of non-ionic low-osmolar contrast material was administered intravenously at an injection rate of 1.2 ml s−1. It was performed after a 62 s injection delay, using following parameters; 120 kVp, 100 mAs, and 2.5 mm contiguous sections with ongoing injection of 30 ml of saline boost at flow rate 1.3 ml s−1 with total scan time of 72 s.
All patients underwent nasopharynx and cervical region contrast-enhanced MR examination using head and neck coils with 1.5 T MR scanners (GE HDxt 1.5T, GE Healthcare, Chicago, IL). The MR scanner has a FOV of 23 cm, number of signal average (NSA) of 2 and Acquisition matrix of 320 by 224. T1 weighted (T1W) fast spin-echo images in the axial plane (spacing between slices = 3 mm), T2 weighted (T2W) fast spin echo MR images in the axial plane (spacing between slices = 3 mm) were obtained before contrast was administrated. After bolus injection of contract, axial T1 weighted fast spin echo sequences were performed (with the same parameters as before contrast).
GTVp segmentation
Manual segmentation was performed on all CT and MRI images by three experienced radiation oncologists (ZL, LL, and MC). Final GTVp ROI was decided by consensus agreement between the outlined contours. NPC often has a irregularly shaped contour due to its propensity to infiltrate adjacent anatomical barriers in the cranium and parapharynx which results in a high degree of interindividual heterogeneity in tumour outlining. Hence, the consensus agreement between the radiation oncologists in tumour delineation represents a more pragmatic approach towards extracting robust feature values. Delineation on CT was performed using standardised window settings: –99, –60, 88 Hounsfield units (HU; tumour); –132, –176, 257 HU (soft tissue); –99, –176, 1440 HU (bone).
Radiomics feature extraction
Following anonymization of DICOM images, Pyradiomics (v. 2.1.2)11 and Moddicom (v. 0.51)12 were applied for feature extraction from both contrast-enhanced CT and MRI images; only MRI T2W images were considered for this study to ensure consistency in the GTVp segmentation and feature extraction processes. For both pipelines, features from the following five radiomics modules were extracted: (1) first-order, (2) grey level co-occurrence matrix (GLCM), (3) grey level run length matrix (GLRLM), (4) grey level size zone matrix (GLSZM) and (5) shape. In addition, two other modules [grey level difference matrix (GLDM) and neighbouring grey tone difference matrix (NGTDM)] were extractable by Pyradiomics. In addition, CERR, as another open-source platform for developing computational radiomics,15 was also used for feature extraction from MRI T2 images.
Statistical considerations
Common radiomics features between Pyradiomics and Moddicom were analysed using Spearman’s rank correlation. Correlated features were defined as p-value ≤ 0.05. Unsupervised hierarchical clustering of radiomics features of the two pipelines against known prognostic clinical variables of T-, N- categories, and GTVp was performed in R (using ward.D2 as the agglomeration method).17–19 All statistical analyses were performed on the R statistical package (v. 3.5.2, https://www.r-project.org). A two-sided p ≤ 0.05 was set as the cut-off for statistical significance. The use of the above analysis method instead of conventional regression analysis or random forest for model building is due to the small sample size used in this study (100 patients). Additionally, common radiomics features between Pyradiomics and CERR were also analysed using Spearman’s rank correlation for MRI images.
Results
Patient characteristics
Clinical characteristics of our study cohort are listed in Table 1. Corresponding N-status, GTVp and treatment parameters are summarized for each T-category. Overall, 72 patients were male and 28 were female; median GTVp was 20.5 (IQR = 13.0–34.2) cc. 29 and 71 patients received IMRT and chemo-IMRT, respectively, as treatment for their disease.
Table 1. .
Clinical characteristics of 100 nasopharynx cancer patients who wereincluded in the present study.
| Clinical Parameters | Number of patients | T1 | T2 | T3 | T4 |
| Gender | |||||
| Male | 72 | 18 | 20 | 17 | 17 |
| Female | 28 | 7 | 5 | 8 | 8 |
| Age, year | |||||
| Median (IQR) | 52 (44–61) | 56 (45.5–62.5) | 52 (39.5–63.5) | 54 (40–60) | 51 (47.5–59) |
| T-category | |||||
| T1 | 25 | ||||
| T2 | 25 | ||||
| T3 | 25 | ||||
| T4 | 25 | ||||
| N-category | |||||
| N0 | 14 | 5 | 3 | 1 | 5 |
| N1 | 27 | 6 | 9 | 7 | 5 |
| N2 | 43 | 11 | 7 | 16 | 9 |
| N3 | 16 | 3 | 6 | 1 | 6 |
| TNM-stage | |||||
| Ⅰ | 5 | 5 | - | - | - |
| Ⅱ | 18 | 6 | 3 | - | - |
| Ⅲ | 42 | 11 | 7 | 24 | - |
| ⅣA-B | 35 | 3 | 6 | 1 | 25 |
| Treatment | |||||
| Chemo-IMRT | 71 | 15 | 14 | 21 | 21 |
| IMRT alone | 29 | 10 | 11 | 4 | 4 |
| GTV, cc | |||||
| median (IQR) | 20.5 (13.0–34.2) | 9.8 (8.2–16.9) | 20.7 (14.9–25.3) | 27.8 (14.7–39.8) | 44.1 (22.4–74.5) |
GTV, gross tumour volume; IMRT, Intensity modulated radiotherapy; IQR, Inter-quartile range.
CT and MRI radiomics features in NPC
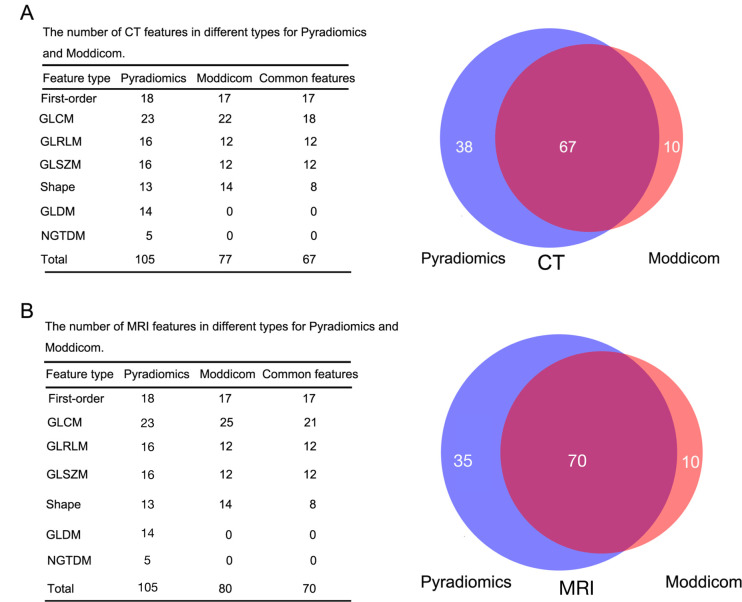
The extracted radiomics features from the CT and MRI data sets are summarized in Supplementary Table 1. In the CT data set, 105 features were extracted using Pyradiomics; 88 features were extracted using Moddicom, of which 11 features were excluded, as these features yielded an infinite value. Between them, 67 common features were identified: 17 first-order, 18 GLCM, 12 GLRLM, 12 GLSZM and 8 shape (Figure 1A). 64 patients had available MRI at baseline (3 patients did not have paired MRI images and feature extraction failed in 33 patients). 105 features were extracted using Pyradiomics; 88 features from Moddicom, of which 8 were excluded. This yielded 70 common features: 17 first-order, 21 GLCM, 12 GLRLM, 12 GLSZM, and 8 shape (Figure 1B). The numerical symbols of radiomics features extracted by Moddicom and Pyradiomics are summarized in Supplementary Table 2.
Figure 1.
The number of CT and MRI features in each type for Pyradiomics and Moddicom. A: CT data set; B: MRI data set. The blue color represents Pyradiomics, while the red colour represents Moddicom. The cross-areas indicate the common features. GLCM, grey level co-occurrence matrix; GLRLM, grey level run length matrix; GLSZM, grey level size zonematrix; GLDM, grey level difference matrix; NGTDM, neighbouring grey tone difference matrix.
Interpipeline variation for radiomics features
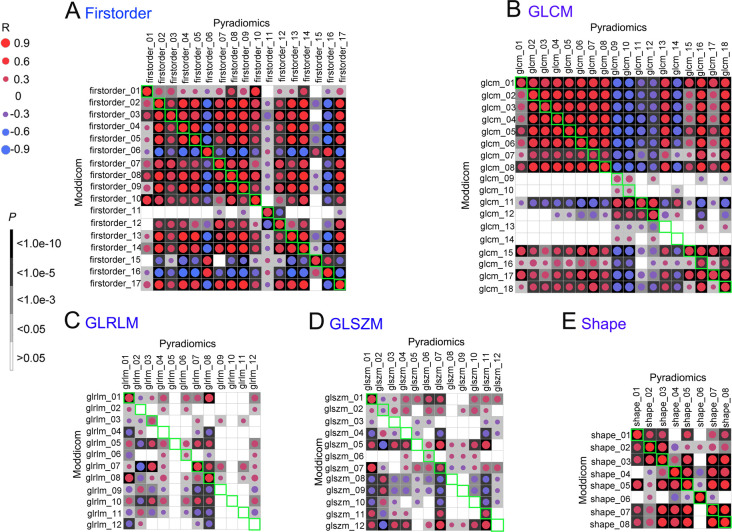
For the CT data set, Spearman’s rank correlation analyses between Pyradiomics and Moddicom revealed that 51 of 67 common CT features met the p-value ≤ 0.05 (Supplementary figure 1). Among them, all the shape-class features were correlated between the pipelines (Figure 2), indicating that the calculation algorithm for this feature class is comparable between the two pipelines. First-order- and GLCM-class features also showed a high proportion of correlation between the common features, while a substantial proportion of GLRLM- and GLSZM-class features showed discordance between the pipelines (Figure 2). Percentages of correlated features for all the feature classes in descending order were as follows: 100% (8/8) for shape,100% (17/17) for first-order, 88.9% (16/18) for GLCM, 41.7% (5/12) for GLSZM and 41.7% (5/12) for GLRLM.
Figure 2.
Correlations of 67 common CT features in each type between Pyradiomics and Moddicom. A: First-order; B: GLCM; C: GLRLM; D: GLSZM; E: Shape. The green areas showed the Spearman correlation of the common features. Blank means no correlation, red circle means positive correlation, blue circle means negative correlation. The darker of the background or larger of the circle, more relevant the correlation of the common features. GLCM, grey level co-occurrence matrix; GLRLM, grey level run length matrix; GLSZM, grey level size zone matrix.
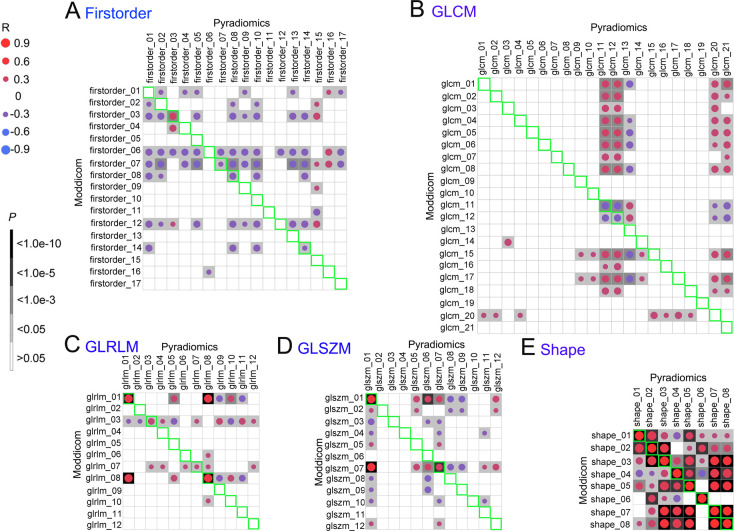
In contrast, only 20 of the 70 common MRI features met the p-value ≤ 0.05 (Supplementary figure 2). Interestingly, only shape-related features were fully concordant between the pipelines, but not for the other modules (Figure 3). Percentages of correlated features for all the feature classes in descending order were as follows: 100% (8/8) for shape, 33.3% (4/12) for GLRLM, 23.5% (4/17) for first-order, 16.7% (2/12) for GLSZM and 9.5% (2/21) for GLCM.
Figure 3.
Correlations of 70 common MRI features in each type between Pyradiomics and Moddicom. A: First-order; B: GLCM; C: GLRLM; D: GLSZM; E: Shape. The green areas showed the Spearman correlation of the common features. Blank means no correlation, red circle means positive correlation, blue circle means negative correlation. The darker of the background or larger of the circle, more relevant the correlation of the common features. GLCM, grey level co-occurrence matrix; GLRLM, grey level run length matrix; GLSZM, grey level size zone matrix.
Additionally, spearman’s rank correlation analyses between Pyradiomics and CERR were also performed for the MRI data set. 33 of the 69 common MRI features met the p-value ≤ 0.05 (Supplementary figure 3). Shape and first-order related features were both fully concordant between the two pipelines. Percentages of correlated features for all the feature classes in descending order were as follows: 100% (16/16) for first-order, 100% (8/8) for shape, 33.3% (4/12) for GLRLM, 23.5%, 19.0% (4/21) for GLCM and 8.3% (1/12) for GLSZM.
Impact of interpipeline heterogeneity on clustering of clinical variables
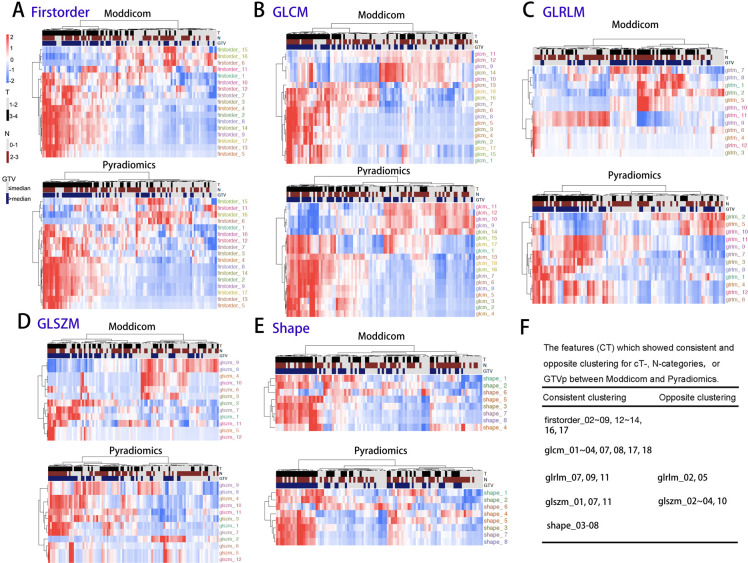
Next, we interrogated the effects of the inter pipeline variation in the extracted CT and MRI features on their association with known prognostic clinical variables of T-, N-categories and GTVp. For the CT features, we observed consistent clustering with clinical parameters for the feature classes that showed high proportion of interpipeline consistency (first-order, GLCM and shape; Figure 4A, B and E), whereas it was unsurprising that clustering patterns differed between pipelines for GLRLM and GLSZM features (Figure 4C and D). To summarize, 13 for first-order (firstorder_02–09,12-14,16,17), 8 for GLCM (glcm_01–04,07,08,17,18), 3 for GLRLM (glrlm_07,09,11), 3 for GLSZM (glszm_01,07,11) and 6 for shape (shape_03–08) of the common features showed the consistent clustering for cT-category and GTVp, while 2 for GLRLM (glrlm_02,05), and 4 for GLSZM (glszm_02–04,10) showed the opposite clustering for cT-category and GTVp. No common shape features showed similar clustering with cN-category (Figure 4F). First-order and shape feature classes were highly correlated (100%) between pipelines, which explains the higher number of consistently clustered features and lower numbers of oppositely clustered features. Textural features (GLRLM and GLSZM) are generally poorly correlated (Figure 2) which results in a lower numbers of consistently clustered features and higher number of oppositely clustered features.
Figure 4.
The interpipeline heterogeneity and clustering of cT-, cN-categories, and GTV of CT features of each type. A: First-order; B:GLCM; C:GLRLM; D: GLSZM; E: Shape; F: The features (CT) which showed consistent and opposite clustering for cT-, N-categories, or GTVp between Moddicom and Pyradiomics. GLCM, grey level co-occurrence matrix; GLRLM, grey level run length matrix; GLSZM, grey level size zone matrix.
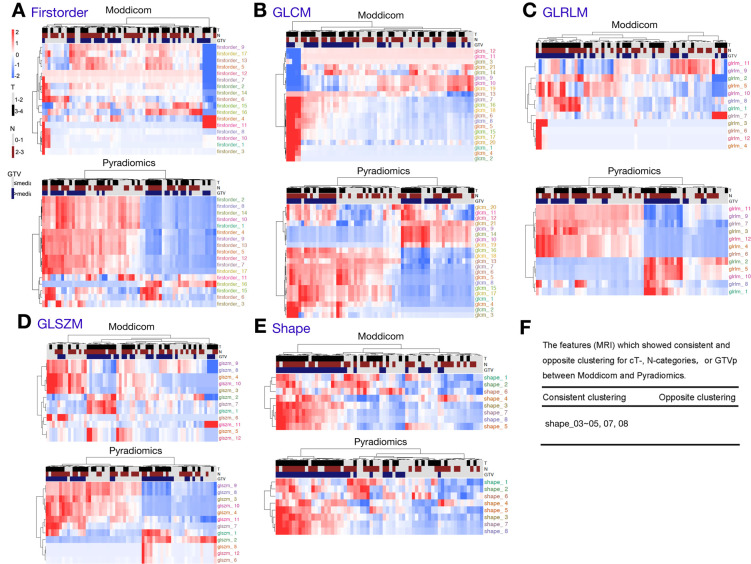
For the MRI features, we observed consistent clustering with clinical parameters only for shape-related features, which was expected given that this class of features showed the least inter pipeline variation (Figure 5E). However, for the other classes of features that showed significant inter pipeline heterogeneity, clustering patterns differed between Moddicom and Pyradiomics for first-order, GLCM, GLRLM and GLSZM features (Figure 5A–D). Clustering outcomes were as such for MRI; five common shape (shape_03–05, 07, 08) showed the same clustering for cT-status, cN-status and GTVp (Figure 5F). In contrast to the clustering result of CT features, only shape features (which give perfect correlation between pipeline) give a higher numbers of consistently clustered features. The rest of the features are poorly correlated (Figure 3) and results in little consistently clustered features.
Figure 5.
The interpipeline heterogeneity and clustering of cT-, cN-categories, and GTV of MRI features. A: First-order; B:GLCM; C:GLRLM; D: GLSZM; E: Shape; F: The features (MRI) which showed consistent and opposite clustering for cT-, N-categories, or GTVp between Moddicom and Pyradiomics. GLCM, grey level co-occurrence matrix; GLRLM, grey level run length matrix; GLSZM, grey level size zone matrix; GTV, gross tumour volume; GLDM, grey level difference matrix; NGTDM, neighbouring grey tone difference matrix.
Discussion
Radiomics refers to the comprehensive quantification of radiological phenotypes using data characterization algorithms. Through this scientific method, we potentially harbour a new paradigm of interrogating imaging data sets, which allows us to add information beyond quantification of tumour volume, number and locality. The latter are conventional indices that could potentially inform on tumour aggression, but nonetheless, few, if any, of these factors are being used to guide treatment in the clinic. It is therefore envisioned that characterization of deeper radiological phenotypes would help to enhance the prediction of tumour biology and improve clinical stratification of cancer patients. Moreover, its appeal stems from the advantage that radiomics relies on profiling of images as opposed to molecular profiling of biopsied tumour specimens, and thus offers a non-invasive method for surveillance of tumour response to treatment. On this note, Pyradiomics and Moddicom are published open-source platforms for radiomics analyses, which have been investigated in several human cancers.11,12 While several signatures of tumour aggression and treatment response have been reported to date, the community remains cynical due to uncertainty on the reproducibility of these radiomics signatures. Several queries, not limited to data input, image processing, feature extraction, and ultimately sensitivity of the radiomics workflow to inter population heterogeneity have not been addressed; all of which can influence the robustness of radiomics as a clinical tool. To partly address this conundrum, we embarked on an important study to compare the output of Pyradiomics and Moddicom on CT and MRI imaging datasets in NPC, which is a common viral-associated head and neck cancer in East and South-Eastern Asia.20 There are other works on comparison of other radiomics tools,21,22 but this is the first work comparing Pyradiomics and Moddicom in NPC context. We made several key observations: (1) significantly more radiomics features extracted from CT data sets were comparable between Pyradiomics and Moddicom compared to MRI (76.1% vs 28.6%); (2) consequently, CT-based radiomics features were significantly more stable and pipeline-agnostic in terms of association with clinical parameters; and (3) finally, it is interesting that that among the different features classes, several shape-related features were associated with GTVp (Figure 4E). These findings are crucial in instructing the workflow for future radiomics work in NPC.
The results on clustering and Spearman correlation show that first-order and shape features are more robust to interpipeline heterogeneity compared to textural features such as GLCM, GLRLM and GLSZM. This is because textural features calculations are sensitive to pre-processing steps: (1) interpolation methods (important when having data with different slice and pixel spacings); (2) two-dimensional against 3D methods for textural features extractions; (3) aggregation method for obtaining scalar value from textural matrices; (4) quantization of voxel values for textural matrix computation. The poor correlation between pipelines for MR-based features (especially first-order) can be explained by noting that Moddicom12 performed a re-scaling of voxel values to account for variation in physics acquisition settings for MRI sequences. The results on interpipeline correlation in this work are also mirrored in our additional study with a different radiomics software—CERR15(Supplementary figure 3), where first-order and shape features tend to be a more robust features compared to textural one. Hence, this work shows that apart from features calculation being International Image Biomarker Standardization Initiative-compliant, it is important to understand and perhaps standardisze the pre-processing method prior to features extraction to achieve robust radiomics phenotyping.
Perhaps, it is interesting to note that in our cohort, CT-derived images were more stable and reproducible than MRI images. This may be in part due to the complexity of the image acquisition protocols between the imaging modalities. MRI image quality is dependent on spin echo sequences of electromagnetic waves, as opposed to CT that relies on the photoelectric effect of kV-strength ionizing radiation passing through tissues of differing atomic number. The discrepancy in our study perhaps highlights the importance of image processing prior to data input in the radiomics workflow. At present, it is not known if different normalization protocols are needed for radiomics analyses of CT and MRI data sets, and much work is needed in this domain going forward.
Next, we observed that several shape features were correlated with known clinical prognostic variables in NPC such as cT- and cN-status and GTVp, which would suggest that various aspects of the tumour shape geometry and surface irregularities may be linked to the tumour burden. For example, shape_03 ~ 08 of CT, and shape_03 ~ 05, 07, 08 of MRI showed the same direction of correlation with GTVp in both Pyradiomics and Moddicom. Previous studies also showed shape features might correlate with GTVp of breast cancer and glioblastoma.14,23 The reason why shape features are stable may be that few differences existed for the parameters of the shape information between CT and MRI regardless of pipelines. Shape_03 (shape_LeastAxis) yields the smallest axis length of the ROI-enclosing ellipsoid while shape_07 (shape_surfaceArea) yields the surface area of the ROI which appears to be consistent as the tumour volume or ROI is similar between CT and MRI. As such, these two features should be observed in future studies to confirm clinical prognostic correlations.
Finally, we acknowledge that a main limitation of this present study relates to the small sample size of our cohort. Nonetheless, we contest that such a preliminary analysis is necessary prior to any large-scale multicohort radiomics study in NPC. Though there exist more commonly used machine learning techniques (lasso, regression, support vector machine, random forest) in predicting the end points, our motivation for going with the unsupervised clustering method is to reduce the number of tested features and yet circumvent the problem of limited sample size in analysis. We will compare between both statistical (supervised against unsupervised) approaches in a larger cohort going forward. In addition, the fact that significant inter pipeline variation is detectable for a proportion of the feature classes and across imaging modalities, even in a limited cohort of 100 patients support our hypothesis that radiomics phenotyping is highly heterogeneous, and feature reproducibility is crucial for clinical prediction. Hence, our next phase of study will include interrogation of multi centre imaging data sets to validate the stability of CT-related radiomics features for clinical prognostication. In addition, we aim to investigate for the biological correlates of these radiomics indices, as previously described in non-small cell lung cancer.24
Conclusion
Here, we report on the significant heterogeneity in radiomics phenotyping between two publicly available feature extraction tools in NPC. The degree of inter pipeline variation differs by feature classes and imaging modality. This has a downstream impact on association with prognostic clinical parameters in NPC such as tumour volume and extent of infiltration. Collectively, our findings emphasize the broader importance of selecting stable radiomics features for disease phenotyping, so as to lead to the development of a robust radiomics-based biomarker for clinical implementation.
Funding Statement
This work is funded by the National Research Foundation Clinical Research Programme (NRF-CRP17-2017-05) and the Duke-NUS Oncology Academic Clinical Programme Proton Radiation Physics Fund (08/FY2018/EX(SL)/48-A89). MC is supported by National Medical Research Council (NMRC) Clinician Scientist Award (NMRC/CSA-INV/0027/2018), and the Duke-NUS Oncology Academic Clinical Programme Proton Research Fund. ZL is supported by the International Communication of Guangxi Medical University Graduate Education in 2018. This work is also partly supported by a structured research agreement with Varian for use of the Velocity v4.0 software.
Footnotes
Acknowledgment: We thank the members of the joint Soo and Chua laboratory for the valuable scientific inputs and comments on the manuscript. We thank Prof Paul C. Boutros (UCLA) for his constructive comments on the manuscript.
Conflict of interests and disclosures:There is no conflict of interest for the submitted work. MC reports personal fees from Astellas, personal fees from Janssen, grants and personal fees from Ferring, non-financial support from Astrazeneca, personal fees and non-financial support from Varian, grants from Sanofi Canada, grants from GenomeDx Biosciences, non-financial support from Medlever, outside the submitted work.
Competing interests: There is no conflict of interest for the submitted work. MC reports personal fees from Astellas, personal fees from Janssen, grants and personal fees from Ferring, non-financial support from Astrazeneca, personal fees and non-financial support from Varian, grants from Sanofi Canada, grants from GenomeDx Biosciences, non-financial support from Medlever, outside the submitted work.
Contributors: Data collection, analysis and interpretation: all authors
Initiated the project: MC, YS
Supervised research: MC
Wrote the first draft of manuscript: ZL, FZ, LT, MC
Approved the manuscript: all authors
ZHONG-GUO LIANG and HONG QI TAN are co-first authors
Ethics approval: Ethical approval for the study was obtained from the SingHealth Centralised Institutional Review Board (protocol no. 2018/2352).
Patient consent: Informed consent was obtained from all living patients.
Contributor Information
Ying Sun, Email: melvin.chua.l.k@singhealth.com.sg;melvin.chua.09@ucl.ac.uk.
Melvin Lee Kiang Chua, Email: melvin.chua.l.k@singhealth.com.sg; melvin.chua.09@ucl.ac.uk.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, et al. . Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Lee AWM, Ma BBY, Ng WT, Chan ATC, BB M. Management of nasopharyngeal carcinoma: current practice and future perspective. JCO 2015; 33: 3356–64. doi: 10.1200/JCO.2015.60.9347 [DOI] [PubMed] [Google Scholar]
- 3.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016; 278: 563–77. doi: 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdollahi H, Mofid B, Shiri I, Razzaghdoust A, Saadipoor A, Mahdavi A, et al. . Machine learning-based radiomic models to predict intensity-modulated radiation therapy response, Gleason score and stage in prostate cancer. Radiol Med 2019; 124: 555–67. doi: 10.1007/s11547-018-0966-4 [DOI] [PubMed] [Google Scholar]
- 5.Tan X, Ma Z, Yan L, Ye W, Liu Z, Liang C. Radiomics nomogram outperforms size criteria in discriminating lymph node metastasis in resectable esophageal squamous cell carcinoma. Eur Radiol 2019; 29: 392–400. doi: 10.1007/s00330-018-5581-1 [DOI] [PubMed] [Google Scholar]
- 6.Yang L, Dong D, Fang M, Zhu Y, Zang Y, Liu Z, et al. . Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur Radiol 2018; 28: 2058–67. doi: 10.1007/s00330-017-5146-8 [DOI] [PubMed] [Google Scholar]
- 7.Verma V, Simone CB, Krishnan S, Lin SH, Yang J, Hahn SM. The rise of Radiomics and implications for oncologic management. J Natl Cancer Inst 2017; 10901 07 2017. doi: 10.1093/jnci/djx055 [DOI] [PubMed] [Google Scholar]
- 8.Nie K, Al-Hallaq H, Li XA, Benedict SH, Sohn JW, Moran JM, et al. . NCTN assessment on current applications of Radiomics in oncology. Int J Radiat Oncol Biol Phys 2019; 104: 302–15. doi: 10.1016/j.ijrobp.2019.01.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keek SA, Leijenaar RT, Jochems A, Woodruff HC. A review on radiomics and the future of theranostics for patient selection in precision medicine. Br J Radiol 2018; 91: 20170926. doi: 10.1259/bjr.20170926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. arXiv preprint arXiv:1612.07003; 2016. [Google Scholar]
- 11.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. . Computational Radiomics system to decode the radiographic phenotype. Cancer Res 2017; 77: e104–7. doi: 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinapoli N, Alitto AR, Vallati M, Gatta R, Autorino R, Boldrini L, et al. . Moddicom: a complete and easily accessible library for prognostic evaluations relying on image features. Conf Proc IEEE Eng Med Biol Soc 2015; 2015: 771–4. doi: 10.1109/EMBC.2015.7318476 [DOI] [PubMed] [Google Scholar]
- 13.Welch ML, McIntosh C, Haibe-Kains B, Milosevic MF, Wee L, Dekker A, et al. . Vulnerabilities of radiomic signature development: the need for safeguards. Radiotherapy and Oncology 2019; 130: 2–9. doi: 10.1016/j.radonc.2018.10.027 [DOI] [PubMed] [Google Scholar]
- 14.Sanghani P, Ang BT, King NKK, Ren H. Overall survival prediction in glioblastoma multiforme patients from volumetric, shape and texture features using machine learning. Surg Oncol 2018; 27: 709–14. doi: 10.1016/j.suronc.2018.09.002 [DOI] [PubMed] [Google Scholar]
- 15.Apte AP, Iyer A, Crispin-Ortuzar M, Pandya R, van Dijk LV, Spezi E, et al. . Technical note: extension of CERR for computational radiomics: a comprehensive Matlab platform for reproducible radiomics research. Med Phys 2018; 45: 3713–20. doi: 10.1002/mp.13046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tham IW-K, Hee SW, Yeo RM-C, Salleh PB, Lee J, Tan TW-K, et al. . Treatment of nasopharyngeal carcinoma using intensity-modulated Radiotherapy—The National cancer centre Singapore experience. Int J Radiat Oncol Biol Phys 2009; 75: 1481–6. doi: 10.1016/j.ijrobp.2009.01.018 [DOI] [PubMed] [Google Scholar]
- 17.OuYang P-Y, You K-Y, Zhang L-N, Xiao Y, Zhang X-M, Xie F-Y. External validity of a prognostic nomogram for locoregionally advanced nasopharyngeal carcinoma based on the 8th edition of the AJCC/UICC staging system: a retrospective cohort study. Cancer Commun 2018; 38: 55. doi: 10.1186/s40880-018-0324-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang X-L, Wang Y, Liang S-B, He S-S, Chen D-M, Chen H-Y, et al. . Comparison of the seventh and eighth editions of the UICC/AJCC staging system for nasopharyngeal carcinoma: analysis of 1317 patients treated with intensity-modulated radiotherapy at two centers. BMC Cancer 2018; 18: 606. doi: 10.1186/s12885-018-4419-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y-H, Huang T-L, Chien C-Y, Chen H-C, Hsu H-C, Huang E-Y, et al. . Pretreatment prognostic factors of survival and late toxicities for patients with nasopharyngeal carcinoma treated by simultaneous integrated boost intensity-modulated radiotherapy. Radiat Oncol 2018; 13: 45. doi: 10.1186/s13014-018-0990-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. The Lancet 2016; 387: 1012–24. doi: 10.1016/S0140-6736(15)00055-0 [DOI] [PubMed] [Google Scholar]
- 21.Bogowicz M, Leijenaar RTH, Tanadini-Lang S, Riesterer O, Pruschy M, Studer G, et al. . Post-radiochemotherapy PET radiomics in head and neck cancer – the influence of radiomics implementation on the reproducibility of local control tumor models. Radiotherapy and Oncology 2017; 125: 385–91. doi: 10.1016/j.radonc.2017.10.023 [DOI] [PubMed] [Google Scholar]
- 22.Foy JJ, Robinson KR, Li H, Giger ML, Al-Hallaq H, Armato SG. Variation in algorithm implementation across radiomics software. J. Med. Imag. 2018; 5: 1. doi: 10.1117/1.JMI.5.4.044505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgiou H, Mavroforakis M, Dimitropoulos N, Cavouras D, Theodoridis S. Multi-scaled morphological features for the characterization of mammographic masses using statistical classification schemes. Artif Intell Med 2007; 41: 39–55. doi: 10.1016/j.artmed.2007.06.004 [DOI] [PubMed] [Google Scholar]
- 24.Grossmann P, Stringfield O, El-Hachem N, Bui MM, Rios Velazquez E, Parmar C, et al. . Defining the biological basis of radiomic phenotypes in lung cancer. eLife 2017; 6. doi: 10.7554/eLife.23421 [DOI] [PMC free article] [PubMed] [Google Scholar]