Abstract
Protein glycosylation plays a key role in various biological processes and disease-related pathological progression. Mass spectrometry (MS)-based glycoproteomics is a powerful approach that provides a system-wide profiling of the glycoproteome in a high-throughput manner. There have been numerous significant technological advances in this field, including improved glycopeptide enrichment, hybrid fragmentation techniques, emerging specialized software packages, and effective quantitation strategies, as well as more dedicated workflows. With increasingly sophisticated glycoproteomics tools on hand, researchers have extensively adapted this approach to explore different biological systems both in terms of in-depth glycoproteome profiling and comparative glycoproteome analysis. Quantitative glycoproteomics enables researchers to discover novel glycosylation-based biomarkers in various diseases with potential to offer better sensitivity and specificity for disease diagnosis. In this review, we present recent methodological developments in MS-based glycoproteomics and highlight its utility and applications in answering various questions in complex biological systems.
Keywords: Mass spectrometry, Glycoproteomics, N-Glycosylation, O-Glycosylation, Biological samples, Biomarker, Disease
1. Introduction
As one of the most common protein post-translational modi-fications (PTMs), protein glycosylation plays an important role in protein stability, intra- and intercellular signaling, fertilization, embryogenesis, organ development, hormone activity, and immunological regulation [1]. It is estimated that half of the proteins expressed in cells are glycoproteins. There are many types of protein glycosylation, and the most widely studied types are N-linked (amide nitrogen of asparagine residue) and O-linked (hydroxyl oxygen of serine or threonine residue). There is a consensus amino acid sequence (Asn-X-Thr/Ser (X is any amino acid except proline)) that contains the glycosylation site of N-glycoproteins, while no consensus sequence has been found for O-linked glycoproteins yet.
Numerous studies have shown that altered glycosylation played a key role in the pathological process during disease progression. Mass spectrometry (MS)-based glycoproteomics is a powerful, high-throughput approach that enables system-wide screening of glycosylation-based biomarkers. In fact, many current disease biomarkers are glycoproteins, such as CEA for colorectal cancer, CA-125 for ovarian cancer, and AFP for hepatocellular carcinoma etc., and the glycans attached to them have been shown to be altered during oncogenesis [2–4]. With glycosylation being particularly sensitive to malignant transformation, glycosylation-based bio-markers hold great promise to improve the sensitivity and specificity of current protein-based biomarkers and may eventually contribute to disease early diagnosis and better treatment [5]. Therefore, comprehensive profiling of protein glycosylation is prerequisite to better understand its role in these pathological and physiological processes.
Over the past decade, substantial progress has been made to obtain detailed information of protein glycosylation, including glycan structures, glycosylation site and its occupancy, and protein sequence. Traditionally, there are two different approaches to study protein glycosylation: (1) the ‘glycomics’ approach, which focuses on the glycan structures after glycan release from proteins and other sugar-containing moieties and (2) the ‘glycoproteomics’ approach, which examines the localization of glycosylation site and structural elucidation of glycans, as well as protein sequence. In this review, we will focus on MS-based glycoproteomics approach. Substantial advances have been made in this field, and here we highlight recent methodological developments and their applications toward comprehensive understanding of the function of protein glycosylation.
2. Glycopeptide enrichment
Comprehensive profiling of the glycoproteome from a complex biological sample is still challenging due to the wide dynamic range of proteins and the micro- and macro-heterogeneity of glycosylation [6,7]. Isolating glycopeptides from complex samples by an appropriate enrichment method is the most efficient way to reduce the sample complexity and achieve an in-depth glycoproteome analysis.
2.1. Hydrazide chemistry enrichment
Hydrazide chemistry (HC) enrichment is based on the formation of covalent bonds between the NaIO4 oxidized cis-diol on N-and O-linked glycans and the hydrazide groups on the hydrazide beads. The advantage of HC method is its high enrichment spec-ificity, as normally 90% of the enriched peptides are glycopeptides [8]. In the original workflow, the glycopeptides captured on hydrazide beads were deglycosylated and released by the treatment of PNGase F for glycosylation site analysis [9,10]. Recently, the capture and release steps were modified to allow glycopeptides to be released without losing the glycan. Specifically, samples were treated with moderate NaIO4 to selectively oxidize the terminal sialic acid of the glycan to generate an aldehyde while leaving the other parts intact. Next, the sialylated glycopeptides were captured by the hydrazide beads and released through acid hydrolysis of the glycosidic bond of sialic acid by trifluoroacetic acid (TFA) [11,12]. This method enabled sialylated glycopeptides to be analyzed, but, unfortunately, the degree of sialylation information was lost [13]. To preserve the sialylation information, Nishimura et al. employed ice-cold 1M hydrochloride to cleave hydrazone bond between the sialic acid and hydrazide beads, allowing the sialic acid to remain on the glycan [14].
2.2. Lectin affinity chromatography
Lectin affinity chromatography (LAC) is another popular enrichment method for protein glycosylation analysis and has been approved by FDA for cancer glycoprotein biomarker detection [15]. Several well-characterized lectins had been used for selective enrichment of specific type of N- or O-linked glycopeptides, which is based on the affinity of lectins to glycans with specific structure motif. For example, Concanavalin A (Con A) binds to mannose, wheat germ agglutinin (WGA) binds to sialic acid and N-acetyl-glucosamine, Vicia villosa (VVA) binds to N-acetyl-galactosamine, Aleuria aurantia lectin (AAL) binds to fucose, and Ricinus communis Agglutinin (RCA120) captures terminal β-galactose [16–19]. Multiple lectins can be combined to improve the glycoproteome coverage [20–22], and including additional enrichment methods sequentially after LAC enrichment would further improve the enrichment specificity [23]. Moreover, lectin enrichment has also been incorporated in serial online reactors to allow simultaneous online proteolysis and glycopeptide enrichment, which is useful for the glycopeptide analysis where sample amount is very limited [24].
2.3. Hydrophilic interaction chromatography
Hydrophilic interaction chromatography (HILIC) as a universal glycopeptide enrichment method is based upon glycopeptides being more hydrophilic than non-glycopeptides due to the attached N- or O-linked glycans [25,26]. To increase the hydrophilicity difference between glycopeptides and non-glycopeptides, ion pairing reagents like TFA can be used [27]. Compared with the HC and LAC method, the HILIC method is more versatile and thus can provide a more comprehensive glycoproteome profile [28]. The disadvantage of HILIC is its poor enrichment specificity, which calls for the development of new HILIC materials with stronger hydrophilic functional group to improve the specificity [29–33]. The enrichment specificity can also be further improved by combining the HILIC with other enrichment methods such as LAC [34,35]. New devices, which integrate the HILIC materials in micro-column or tip, have been developed to minimize the sample loss during enrichment procedure and facilitate the detection of low abundance glycopeptides [33,36–39].
Other than the three enrichment methods summarized above, methods using boronic acid materials [40–43], titanium dioxide [44,45], responsive smart polymers [46–49], porous graphitized carbon (PGC) [50], acetone precipitation [51,52], size exclusion chromatography [53], and molecular weight cutoff filter [54] for the enrichment of glycopeptides are also in the ascendant. It is worth mentioning that most of current glycopeptide enrichment methods are suitable for both the N- and O-linked glycopeptides; however, due to the relatively high abundance of N-linked glycopeptides, to efficiently analyze O-linked glycopeptides, deglycosylation of N-linked glycopeptides or deep fractionation of sample should be performed.
3. Characterization and quantitation of glycopeptides
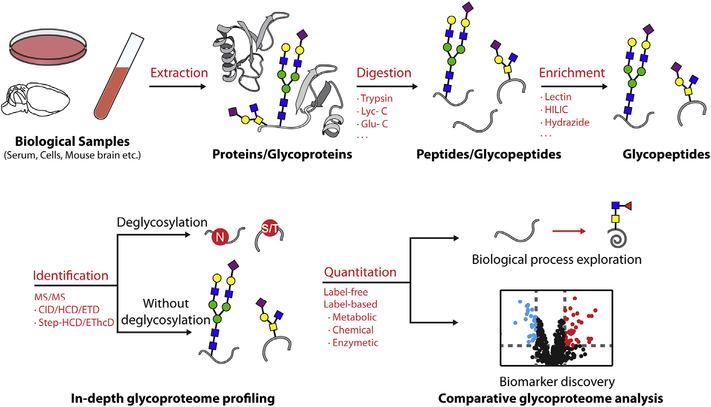
Qualitative characterization of glycopeptides includes two aspects: glycosylation site profiling and site-specific intact glyco-peptide analysis. A typical MS-based glycoproteomics workflow is shown in Fig. 1.
Fig. 1.
A typical workflow for MS-based glycoproteomics in different complex biological samples.
3.1. Glycosylation site profiling
3.1.1. N-glycosylation site profiling
Since N-glycosylation site profiling was originally performed by deglycosylation with peptide-N-glycosidase F (PNGase F) or endo-b-N-acetylglucosaminidase F&H (endo F&H), several studies focused on improving the deglycosylation efficiency. Huang et al. found that when glycosylation site profiling was performed by HC method, glycopeptides with an N-terminal serine/threonine can be oxidized on both the N-termini and glycans; thus, this type of glycopeptides cannot be released by PNGase F treatment due to being covalently coupling to the hydrazide beads through the N-termini. To overcome this problem, they utilized a peptide N-terminal protection strategy to block the primary amine groups on peptides, which avoided the adjacent amino alcohols on peptide N-termini being oxidized. The results showed that this strategy successfully prevented the oxidation of peptide N-termini and significantly improved the coverage of glycoproteome [55]. Recently, the same group found that releasing the glycopeptides captured on hydrazide beads by PNGase F deglycosylation was inefficient due to steric hindrance in the heterogeneous condition. Thus, they developed a hydroxylamine-assisted PNGase F deglycosylation method which used the hydroxylamine to efficiently cleave hydrazone bonds by transamination and release intact glycopeptides. As deglycosylation of the released glycopeptides was performed under homogeneous condition, the recovery rate of deglycosylated peptides was improved significantly [56]. Another study by Weng et al. reported that N-terminal glycosylated peptides are difficult to be deglycosylated due to the limitation of PNGase F enzymatic specificity, which cannot cleave N-glycans attached to N- or C-termini and require the presence of an extra amino acid at the termini. To overcome this drawback, they developed an N-terminal site-selective succinylation strategy by incorporating an amide bond to mimic an amino acid at the peptide N-termini, which greatly improved N-glycosylation site coverage [57].
Other studies involved combining different sample preparation techniques, enrichment methods, and fractionation strategies to improve the glycoproteome coverage. Mann and coworkers developed an N-glyco-FASP sample preparation approach, where the lectin column in conventional method was replaced with ultrafiltration units, to decrease the glycopeptide loss. In this method, the glycopeptides were enriched by binding to lectins on top of a filter, which greatly reduced the sample loss and improved the glycosite coverage. The robustness of this approach was successfully demonstrated in the large-scale glycosylation site profiling in mouse plasma and four different tissues where 6367 N-glycosylation sites were identified. Combining different enrichment methods is also an effective approach to increase the glycosite coverage [58]. Recently, Qian group combined two widely used glycopeptide enrichment methods, HC and HILIC, for N-glycosylation site analysis of the secretome of two human hepatocellular carcinoma (HCC) cell lines. A total of 1212 unique N-glycosylation sites from 611 N-glycoproteins were confidently identified. Overall, the overlap of N-glycosylation sites determined by the two methods was only 28.4% [59]. Zou group performed a similar strategy which combined the click maltose-HILIC and the HC method to comprehensively map the N-glycosylation sites of human liver tissue. Altogether,14,480 N-glycopeptides, corresponding to 2210 N-glycoproteins and 4783 N-glycosylation sites, were identified [60]. As another example, Yang group combined seven protease treatments (trypsin, trypsin coupled with Lys-C (Try + Lys), trypsin coupled with Glu-C (Try + Glu), Lys-C, Glu-C,chymotrypsin and pepsin), four different enrichment techniques (HILIC, ZIC-HILIC, HC, and TiO2 chromatography), and two different fractionation strategies (SCX and high-pH RP), which aided in identifying a total of 13,492 N-glycopeptides, corresponding to 8386 N-glycosylation sites on 3982 proteins in the mouse brain. Considering the efficiency and simplicity, a workflow combining the use of trypsin, Try + Lys and Try + Glu for protein digestion, HILIC and ZIC-HILIC for glycopeptide enrichment, and 1D-RPLCMS/MS for N-glycopeptide detection can also produce a comparable glycosite coverage [61].
3.1.2. O-glycosylation site profiling
Mapping of O-glycosylation is also an active area of research. Among the different types of O-glycosylation, the O-GlcNAcylation and O-GalNAcylation are the most widely studied [62,63]. Profiling of O-glycosylation sites is even more difficult compared to N-glycosylation due to a lack of a consensus sequon and the lack of an enzyme that can effectively deglycosylate the O-linked glycans.
To date, the most successful approach for profiling of OGlcNAcylation has been metabolic and enzymatic labeling, which incorporates an azide-containing group to the O-GlcNAc moiety [64]. Then, the derivatized O-GlcNAc is enriched by an alkynyl biotin or photo-cleavable tag containing alkynyl beads to be analyzed by LC-MS/MS. By using this highly specific strategy, tens to hundreds of O-GlcNAcylation sites can be mapped [65–68]. The drawback of this approach is relatively low labeling efficiency, leading to the limited coverage of O-GlcNAcylation sites [69]. Besides the enzymatic and metabolic labeling methods, Burlingame and coworkers developed a lectin weak affinity chromatography (LWAC) strategy to enrich O-GlcNAc peptides with wheat germ agglutinin (WGA) lectin [70]. The same group recently optimized this LWAC strategy and identified over 1750 sites of O-GlcNAcylation from murine synaptosomes [71]. Due to the particularly low abundance, low hydrophilicity of the O-GlcNAcylation peptides, and severe interference from other N/O-glycopeptides, isolating O-glycopeptides from a complex sample by HILIC enrichment was originally thought to be ineffective. However, after combining PNGase F, sialidase and O-glycosidase to selectively cleave and remove most of the N/O-linked glycans in glycoproteins, Shen et al. were able to eliminate the interference of other N/O-glycopeptides while still preserving the O-GlcNAcylation modified peptides. Benefiting from the improved enrichment specificity of the OGlcNAc peptides, a total of 474 O-GlcNAc peptides from 457 proteins were identified from a human urinary sample. In comparison, performing HILIC enrichment without the deglycosylation step only identified 107 O-GlcNAc proteins, and an immunoprecipitation (IP) approach using an anti-O-GlcNAc antibody only profiled 31 O-GlcNAc proteins [72].
For O-GalNAcylation, the glycan structures are of higher diversity than O-GlcNAcylation. To facilitate the MS identification of these glycopeptide sequences and their attached sites, Medzihradszky et al. utilized exoglycosidase digestion to partially deglycosylate O-GalNAcylation peptides and reduce the complexity of glycan structures and was able to identify 124 O-GalNAcylation sites in 51 O-GalNAcylated proteins from human serum samples [73]. Besides this in vitro approach, Clausen and coworkers developed an alternative method called SimpleCell strategy in vivo, which utilized a zinc-finger nuclease gene targeting to block the O-GalNAcylation elongation pathway to generate short glycan homogenous O-GalNAcylation. This strategy allowed directly enrichment by the LWAC method followed by MS/MS detection of OGalNAcylation peptides from different cell lines [74]. Recently, they extended this approach to characterize samples from 12 human cell lines and profiled almost 3000 O-GalNAcylation sites in over 600 OGalNAcylation glycoproteins, which represented the first map of the human O-glycoproteome [75].
3.2. Site-specific characterization of intact glycopeptides
Along with rapid development of glycosylation site profiling, two major breakthroughs have significantly facilitated the site-specific characterization of intact glycopeptides. These breakthroughs are the advancement of MS/MS dissociation methods towards acquiring both the glycan and peptide backbone fragments and the development of new search engines to decipher the MS/MS spectra of intact glycopeptides.
3.2.1. Comparison of dissociation methods for intact glycopeptides
The dissociation modes for peptide analysis mainly include collision-induced dissociation (CID), beam-type CID (occurs in triple quadrupole (QQQ) and quadrupole time-of-flight (Q-TOF) instruments, and the so-called high-energy collisional dissociation (HCD) in Thermo-Fisher™ instruments), and electron-induced dissociation (ExD, such as electron transfer/capture dissociation, ETD/ECD). Each of these methods alone cannot provide a full picture of the glycopeptide structure [76]. CID prefers to break glycosidic bonds, and it generates strong characteristic ions of peptides bearing different numbers of glycans after the stepwise release of peripheral monosaccharides (Y ions) (Fig. 2). It provides abundant information for deciphering glycan structures but limited information for peptide backbone identification. As for HCD dissociation, in low collision energy, a series of Y ions are preferentially generated, which is similar to CID; while in high collision energy, the peptide backbone fragmentation yielded a decreased intensity of Y ions. ExD mode mainly fragments the peptide backbone while leaving the glycan intact (Fig. 2), which is suitable for the localization of glycosylation sites with a wealth of peptide fragments [77].
Fig. 2.
MS/MS of 3 + charge state precursor ion at m/z 1577.9 of bovine fetuin triantennary N-glycopeptide KLCPDCPLLAPLNDSR (AA 126–141). Alternating between CID/ETD/EThcD resulted in different sets of ions. (a) CID and ETD spectra (inset). Asterisk (*) in the peptide sequence indicates carbamidomethylation. (b) EThcD spectrum. Starred peaks (*) in the spectra were deconvoluted and annotated in the inset. Adapted from Ref. [68] with permission.
3.2.2. Intact glycopeptide analysis by combining different dissociation methods
As no single dissociation method is available to produce a complete picture of intact glycopeptides, combining the complementary fragment information from multiple dissociation modes is an effective strategy to decipher the intact glycopeptides. Larson and coworkers combined CID and ExD to analyze desialylated glycopeptides, where CID-MS2 spectra of glycopeptides were used for the glycan characterization and the subsequent CID-MS3 spectra of selected CID-MS2 fragment ions for peptide sequence identification. Moreover, ExD as a complementary peptide fragmentation mode was used for the characterization of O-glycosylation sites, where 58 N- and 63 O-glycopeptides from 53 glycoproteins were identified and 40 of the 57 putative O-glycosylation sites were accurately localized [78]. However, the requirement of prior knowledge of the targeted peptides to be selected for MS3 and the longer duty cycle due to ExD reaction time in ExD-MS2 limit its capacity compared to HCD- and CID-MS2, and currently only tens to hundreds of intact glycopeptides can be profiled from complex biological samples using this strategy [79].
Sun et al. developed an integrated method that enabled comprehensive characterization of N-linked glycans and glycosite-containing peptides of glycoproteins and generated the glycan and glycosylation site database for spectral interpretation of intact glycopeptides acquired from another enrichment. This strategy allows simultaneous profiling and monitoring of N-linked glycans, glycosites, glycoproteins and site-specific glycosylation in a single experiment [80]. Chen et al. developed an alternative complementary method that enabled analysis of intact glycopeptides by sequentially interrogating the deglycosylated peptides and intact glycopeptides using CID and HCD, respectively. A total of 811 N-glycosylation sites from 567 glycoproteins were identified from HEK293T membrane proteins, and 177 intact N-glycopeptides were also identified by manually integrating the CID and HCD spectra. The number of identified intact glycopeptides was much smaller than the number of identified N-glycosites, which can be attributed to the low ionization efficiency of intact glycopeptides and manual interpretation of the complicated MS/MS spectra [81]. Recently, the same group developed a fully-automated software platform for high-throughput characterization of intact N-glycopeptides. They used the strong correlation of retention time to effectively remove the random matches and were able to control the probability of random matches within 1%. In total, 2249 intact glycopeptides, representing 1769 site-specific N-glycans on 453 glycosylation sites, were identified [82]. Liu et al. developed a similar strategy which profiled 1145 non-redundant glycopeptides from 225 core peptides and 95 glycoproteins from human serum samples [83].
3.2.3. Intact glycopeptide analysis by integrated dissociation methods
Rather than implementing two dissociation methods to obtain the complementary structure information in two separate LC-MS/MS runs, it would be beneficial if hybrid fragmentation spectra were acquired in a single run. To this end, the evolution of several new dissociation approaches offered an effective solution to this problem. Among them, the stepped collision energy HCD (step-HCD), beam-type CID with high energy, and electron-transfer/higher-energy collision dissociation (EThcD) show great potential.
As different HCD collision energies could generate complementary fragments, performing step-HCD (e.g. 30 ± 10%) will give a more complete intact glycopeptide structure information in a single spectrum. Qian and coworkers first applied the step-HCD to analyze partially deglycosylated core-fucosylated glycopeptides in mouse liver tissue and HeLa cell samples and found that the overall performance increased by 7-fold [84]. Recently, this method has been widely used in intact glycopeptide analysis with impressive results being reported [85,86]. Current MS instruments can only provide a three-step collision energy in one spectrum, and more flexible collision energy settings could definitely improve intact glycopeptide analysis.
Under typical beam-type CID conditions, ions produced from dissociation of the peptide backbone are in low abundance. Zaia and coworkers reported that abundant peptide backbone fragments could be generated by increasing the collision energy, along with oxonium ions and intact peptide ions with varying numbers of saccharide units attached. They successfully used this approach for intact glycopeptide analysis from several standard N-glycoproteins [87]. Recently, Sung et al. utilized the same strategy in complex sample analysis and profiled 36 intact glycopeptides of 26 glycoproteins in a HeLa cell sample [88]. Ye & Zou and coworkers continuously explored this strategy in intact O-GalNAcylation peptide analysis and established an automated workflow for O-GalNAcylation peptide MS2 spectral interpretation, enabling identification of 407 intact O-GalNAcylation peptides from 93 glycoproteins in human serum sample [89].
Integrating the HCD and ETD in one spectrum (Fig. 2), EThcD also enables information of both glycan and peptide fragments to be acquired [35,90,91]. Li and coworkers first optimized the parameters of EThcD for intact glycopeptide analysis, and determined that the efficiency of dissociation was greatly improved by using charge-dependent optimized ETD reaction times. Large-scale experiments in rat carotids collected over the course of restenosis progression resulted in over 2000 N-glycopeptide identifications [92]. Qian and coworkers found that EThcD provided more complete fragmentation information on O-GalNAcylation peptides and a more confident site localization of O-GalNAcylation than HCD method. By combining multiple enzyme digestions and multidimensional separation, they identified 173 O-glycosylation sites, 499 non-redundant intact O-glycopeptides, and 6 glycan compositions originating from 49 O-glycoprotein groups from normal human serum [93].
3.2.4. Database search for intact glycopeptide analysis
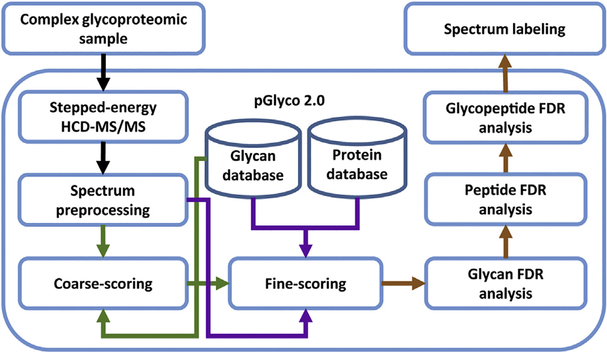
One of the biggest challenges for intact glycopeptide charac terization is the accurate interpretation of the resulting spectra. Based on different strategies, several new search engines have been developed for intact glycopeptide identification, such as Glyco-Master DB [94], GPQuest [95], I-GPA [96], Byonic [97], SweetNET [98], pGlyco [99], pGlyco 2.0 [85], etc. Particularly, pGlyco 2.0 conducted a comprehensive false discovery rates (FDR) evaluation at all three levels of glycans, peptides and glycopeptides, greatly improving the accuracy of intact glycopeptide identification (Fig. 3). Moreover, a quantitative analysis method utilizing 15N/13C meta bolically labeled glycoproteome samples to validate glycopeptide identification was specifically designed [85]. By taking advantage of the optimized step-HCD collision for fragmentation of the HILIC enriched intact glycopeptides and sophisticated algorithm of pGlyco 2.0, the researchers were able to identify 10,009 distinct glycopeptides in five mouse tissues, in site-specific manner, corresponding to 1988 glycosylation and 955 glycoproteins. Some other analytical tools, like MAGIC and SugarQb, which translate the intact glycopeptide spectra and enable them to be analyzed using current peptide search engines, have also been developed [86,88,100]. More details about the development of search engines can be found in several recent reviews [101,102]. Due to space limitations and lack of standard glycopeptide spectral datasets, a fair comparison between different software has not been performed [103]. Comprehensive evaluation of the current software regarding the coverage of the glycoproteome and quality control of the identification results would provide valuable insights for future software design [104]. Large-scale glycoproteomics research would especially benefit greatly from the improvement of automated glycopeptide identification, due to the large volumes of data being generated.
Fig. 3.
Design of a dedicated software pGlyco 2.0 for intact glycopeptide interpretation. Adapted from Ref. [60] with permission.
3.3. Quantitation of glycopeptides
Quantitation of protein glycosylation can be performed at the glycan, glycopeptide or glycoprotein level based on the target molecule, and at relative or absolute quantitation levels based on the strategies used. Absolute quantitation is often conducted by employing targeted MS approach. As the theme of the current review is non-targeted bottom-up glycoproteomics, the quantitation strategy discussed here will focus on quantitation at the glycopeptide level.
3.3.1. Label-free quantitation
The label-free approach has been regularly used in proteomics studies to measure protein abundance changes, and it offers the advantage of a simple workflow, low cost and high proteome coverage [105]. Normalization is needed to overcome the MS response variations in different samples and reliable quantitation results could be obtained by normalizing the data to the total ion abundance [106,107]. However, it could be problematic for glycopeptide analysis due to the low ionization efficiency of glycopeptides, which means small changes of nonglycosylated interferences could lead to large variability in quantitative assays. To overcome this problem, Desaire and coworkers developed a new normalization strategy based on the intensity of all glycopeptides and a twotiered quantitative analysis to discriminate between glycosylation changes of a given protein and glycoprotein’s concentration changes [108]. Additionally, the large volumes of data produced by label-free experiments need rigorous statistical assessment for accurate data processing and interpretation, which requires effective algorithm models and software tools to be developed [109]. Mayampurath et al. developed a novel ANOVA-based mixed effects model for label-free glycopeptide quantitation and demonstrated its effectiveness by applying this method to biomarker discovery in human serum [110]. To facilitate simultaneous identification and label-free quantitation of glycopeptides, Park et al. developed an automated Integrated GlycoProteome Analyzer (I-GPA) platform and successfully quantified 598 N-glycopeptides from human plasma sample [96].
3.3.2. Label-based quantitation
Compared with label-free methods, the greatest advantage of stable isotope labeling is that different samples are mixed together and analyzed simultaneously, which largely reduces instrument time and run-to-run variations. In general, stable isotope labeling can be classified into three major categories: metabolic labeling, chemical labeling and enzymatic labeling.
The most commonly used metabolic labeling in quantitative proteomics is the stable isotope labeling by amino acids in cell culture (SILAC), which incorporates stable isotope-encoded essential amino acids into living cells [111]. The main advantage of SILAC is that it allows different samples to be combined at the intact cell level, minimizing the possible quantitation error introduced by the sample preparation process [112]. By incorporating a glycopeptide enrichment step, the regular SILAC workflow can be easily modified for quantitative glycoproteomics studies. After treatment with PNGase F, the total glycosylation expression changes at each site can be quantified through comparison of light and heavy-labeled deglycopeptides, and a number of studies have successfully utilized this approach to quantify glycosylation changes on hundreds of N-glycosites [113–117]. Furthermore, Parker et al. utilized the SILAC approach to quantify the intact glycopeptides without PNGase F treatment, which enabled the changes in N-glycosylation micro-heterogeneity to be revealed [118]. By combining glycopeptide enrichment using hydrazide chemistry with SILAC, Taga et al. conducted a quantitative analysis of O-glycosylation and showed increased glycosylation of collagen in Osteogenesis Imperfecta [119].
However, the disadvantage of metabolic labeling is that some biological systems are not suited to efficient metabolic labeling and the cost is relatively high [120]. To this end, chemical labeling approaches have been developed to label proteins or peptides extracted from tissues/cells or biofluids with stable isotope-incorporated tags. In early 2003, Zhang et al. used stable isotope labeling by succinic anhydride after glycoprotein capture by hydrazide beads for identification and quantitation of N-glycopep-tides [9]. However, succinic anhydride labeling method requires repeated labeling to achieve reaction completeness and side reactions may happen during the process [121]. To overcome this problem, Sun et al. developed an approach that enables sequential glycopeptide enrichment and dimethyl labeling on hydrazide beads, which showed high quantitation accuracy over two orders of magnitude in dynamic range [122]. However, both succinic anhydride and dimethyl labeling have limited capability for quantitative analysis across different samples; hence, isobaric tags have been developed to allow for multiplexing capability, such as 10-plex tandem mass tag (TMT) [123,124], 8-plex isobaric tags for relative and absolute quantitation (iTRAQ) [125,126], and 12-plex N,N-dimethyl leucine (DiLeu) isobaric tags [127,128] etc. Employing a 6-plex TMT labeling strategy, Kroksveen et al. conducted a quantitative glycoproteomics analysis between 21 subjects in relapsingremitting multiple sclerosis group and 21 subjects in neurological control group, and successfully quantified 1700 deglycopeptides with 235 deglycopeptides showing significant differences between disease group and control group [129]. Notably, Melo-Braga et al. conducted a global comparative proteomic study, as well as changes in N-glycosylation, phosphorylation, and Lys-acetylation with 4-plex iTRAQ tagging scheme [130]. The reason that multiple quantitative PTMs analysis could be conducted in parallel is because both proteome and PTMs analysis shared the same upper stream steps and samples could be split into aliquots and subject to different PTMs-targeted enrichment methods after isobaric labeling. Such capacity allows multiple PTMs to be analyzed from a limited amount of sample and largely facilitates the study of crosstalks between different PTMs. Besides chemical labeling, enzymatic labeling was also developed by incorporating 18O into the peptides during the enzyme-catalyzed digestion process [131]. Later, Liu et al. developed a tandem 18O stable isotope labeling strategy for quantitation of N-Glycoproteome by combining 18O labeling in the C-terminal carboxylic acid during proteolytic process and another 18O labeling in the asparagine residue during deglycosylation process by PNGase F hydrolysis [132].
4. Application of MS-based glycoproteomics in complex biological samples
4.1. In-depth glycoproteome profiling in complex biological samples
4.1.1. Human serum and other human tissues
In-depth glycoproteome profiling has been extensively conducted in different biological systems, including body fluids, cells and tissues, etc. Serving as an indicator of physiological and pathological states alteration in the body system, serum/plasma is the most common clinical specimen for disease diagnosis. The majority of serum proteins are glycosylated as many proteins are secreted in glycosylated form, with an estimated 50% after removing high abundance proteins [133]. To facilitate the detection of low abundance glycoproteins, Sparbier et al. utilized magnetic lectins (ConA, LCA, WGA) beads and boronic acid beads for the enrichment at both protein and peptide levels, resulting in 95 N-glycosylation sites from 193 N-glycoproteins [134]. Nevertheless, the coverage is still not desirable mainly due to the extreme complexity of serum and the wide dynamic range of proteins with concentrations spanning over 10 orders of magnitude [135]. To further decrease sample complexity, various approaches have been applied, including immunoaffinity depletion of high-abundance serum proteins (albumin, IGG etc.), sequential enrichment strategies (lectins, HILIC etc.), off-line fractionation (HpH, SCX) and 2D-LC, which yielded more than 600 N-glycosylation sites from over 300 N-glycoproteins [136,137]. Faced with the challenges of sample complexity brought by various glycoforms, several studies focused on a subset of total glycopeptides such as core-fucosylated peptides which could be enriched by highly specific binding afforded by lectin LcH [138–140]. Park et al. developed a novel automated Integrated GlycoProteome Analyzer (I-GPA) with FDR control for fast and confident intact N-glycopeptide identifications, and successfully identified 619 intact N-glycopeptides with an FDR below 1% from human serum [96]. Compared to N-glycosylation, O-glycosylation in serum is less studied mainly due to its lack of consensus motif and diversity of core structures. Recently, Zhang et al. developed a systematic strategy that combined multiple enzyme digestion, multidimensional separation and EThcD fragmentation, and identified 499 non-redundant intact O-glycopeptides in serum, covering singly, doubly and triply O-glycosylated peptides [93]. Besides serum, in-depth glycoproteome profiling has also been conducted in other human body fluids/tissues such as urine [141–143], liver [60,144] and so on.
4.1.2. Cell culture
In addition to human serum, the glycoproteome of different cell types have also been extensively explored. Cell culture has helped us gain valuable insights into various biological processes and disease-related pathological alterations, and has contributed enormously in drug discovery and development [145]. Adding glycoproteome data to the cellular models would help us gain a better understanding of the inherent complexity in biological systems [146]. Notably, the Clausen group developed a robust SimpleCell approach for an O-GalNAc study [75], and successfully mapped human O-GalNAc glycoproteome with almost 3000 glycosites from over 600 O-glycoproteins in 12 human cell lines from different organs [147]. Although SimpleCell approach has shown extraordinary performance in terms of O-glycosite mapping, it falls short in intact glycopeptide analysis due to glycan truncation during the process. To this end, Bertozzi group developed an IsoTaG strategy for intact glycopeptide characterization [148], and 1375 intact N-glycopeptides and 2159 intact O-glycopeptides were successfully identified from 15 human tissue-derived cell lines [149]. Later, this approach was also applied for human T-cells O-GlcNA-cylation analysis, with over 2000 O-GlcNAcylation peptides identified [150]. Some other studies focused on the glycoproteome on the cell surface, which are crucial for the understanding of cell-cell communication and cell-environment interaction [151,152]. In order to selectively capture surface glycoproteins, in 2009, Wollscheid et al. developed a powerful unbiased cell surface-capturing (CSC) technology through covalently labeling cell surface N-glycan moieties [153]. Since then, this approach has been widely used for cell surface N-glycoproteome profiling including embryonic stem cells [154], induced pluripotent stem cells (iPSCs) [155], gastric adenocarcinoma cells [156], hepatocellular carcinoma cells [157], and hundreds of surface glycoproteins have been identified. Another rich source of glycoproteins come from the secreted proteins, or secretome, as many proteins undergo glycosylation prior to secretion [158]. For secreted glycoprotein analysis, conditioned media from serum-free cell culture is usually collected, followed by extraction of the secreted proteins, and then is subject to a typical glycoproteomics workflow. Li et al. have extensively conducted glycoproteome profiling of hepatocellular carcinoma cell lines [59] and have mapped 1213 unique N-glycosites from 611 N-glycoproteins [159]. Cell component analysis revealed that these N-glyco-proteins were primarily localized to the extracellular space and plasma membrane, indicating important role of N-glycosylation in the secretory pathway. The study of secreted glycoproteome of other commonly used cell lines such as human embryonic kidney (HEK) cells [160], Chinese hamster ovary cells (CHO) and endothelial cells [161], and some microorganisms such as green algae [162] and filamentous fungi [163] have also been conducted, which provide valuable insights into the secretory pathway and their responses to the environmental stimuli.
4.1.3. Animal tissues and plants
Due to their easy accessibility, the glycoproteome of animal tissues and plants have also been comprehensively profiled. Among them, mouse or rat brain is perhaps among the most extensively studied tissue. By employing different enrichment strategies, including lectin, HILIC, hydrazide chemistry and TiO2, Zhang et al. have successfully mapped 3446 unique glycosylation sites from 1597 N-glycoproteins in mouse brain, and 65% of the identified N-glycoproteins are membrane or extracellular proteins [28]. To take a step further, Fang et al. further increased the coverage by optimizing protease treatments and fractionation strategies and identified 8386 glycosylation sites on 3982 N-glycoproteins, representing the largest N-glycosylation site dataset in mouse brain ever reported [61]. Site-specific N-glycoproteome study in rat brain has also been conducted by utilizing a combined glycomics and glycoproteomics approach, resulting in the identifications of 863 unique intact N-glycopeptides [164]. The N-glycosylation site mapping studies in other mouse/rat tissues such as liver, kidney, heart, plasma, stomach, ovary etc. revealed a tissue-specific expression pattern of N-glycosylation, indicating the close relation between glycosylation and the specialized function of different organs/tissues [58,165,166]. Compared with the large number of glycoproteome studies in mammalian, the glycoproteome studies in plants are quite limited, despite an increased interest in deciphering the plants glycoproteome [167–169]. So far, hundreds of N-glycosites have been mapped in rice [170], cereal crop Brachypodium distachyon L. [136], tomato[22], flowering plant Arabidopsis [171] etc., providing valuable insights into the biological role of this ubiquitous protein modi-fication in different plant species.
4.1.4. Microorganisms
As one of the most popular models for basic biological research, yeast has also attracted substantial interest in the glycoscience field. Breidenbach et al. started out mapping the N-glycosites in yeast, yielding a total of 133 N-glycosites spanning 58 glycoproteins, which were mainly distributed in the yeast ER, plasma membrane, vacuole, and cell wall [172]. It has been a puzzle for researchers that O-GlcNAcylation was found in all eukaryotic cells except yeast until Halim et al. discovered and mapped nucleocytoplasmic O-mannose glycoproteome in yeast in 2015, which opened new avenues for the investigation of O-glycosylation based biological events in yeast [173]. Later, Neubert et al. successfully mapped 2300 O-mannosylation sites in 500 O-glycoproteins from whole yeast cell lysates, and one interesting finding was that these O-mannosylation sites were in the proximity of N-glycosylation sites, indicating their potential interplay [174]. The glycoproteome of some common bacteria [175–177] and viruses [178,179] have also been mapped, providing a molecular foundation for further understanding of glycosylation-assisted physiological processes.
4.2. Comparative MS-based glycoproteomics in complex samples
4.2.1. Disease biomarker discovery
Previously, glycosylation-based biomarker studies relied on lectin staining or 2D gel electrophoresis approaches to measure the total glycosylation changes or the total glycoprotein changes, which suffer from low-throughput, limited sensitivity, and limited site-specific glycosylation information [180,181]. With the advancement of glycoproteomics methodologies, glycosylation level changes can be pinpointed on a specific site and further micro-heterogeneity differences can be revealed through intact glycopeptide analysis in a high-throughput manner.
In a quantitative proteome and glycoproteome study of relapsing-remitting multiple sclerosis and neurological controls, Kroksveen et al. identified 96 altered deglycopeptides where their associated protein abundance was not affected, indicating the alterations were due to glycosylation occupancy changes instead of changes at the protein level [129]. Similarly, Zhang and coworkers utilized an integrated proteomics and glycoproteomics approach to explore the mechanism of castration resistance for androgen-deprivation therapy in prostate cancer [182]. This integrated omics approach not only allowed the detection of changes in glycosylation occupancy and microheterogeneity, but also identi-fied associated altered fucosyltransferase and fucosidase expression. To take one step further, after the initial finding of the increased terminal galactosylation and up-regulation of B4GalT5 galactosyltransferases upon TNF-Alpha-Induced insulin resistance in adipocytes through an integrated proteomics and glycoproteomics approach, Parker et al. showed that the knockdown of B4GalT5 down-regulated the terminal galactosylation, confirming the involvement of B4GalT5 in the TNF-alpha-regulated N-glycome [118]. Instead of analyzing the whole glycoproteome, some other studies focused on a specific type of glycopeptides (fucosylated, sialylated etc.) to improve the coverage depth. Tan et al. employed an LCA enrichment approach to selectively enrich core-fucosylated glycopeptides, and were able to identify 613 core-fucosylated peptides and 8 of them exhibited a significant difference between pancreatic cancer and controls [183]. Due to close crosstalks between cells and the extracellular space, the secreted glycoproteins in extracellular space is another rich source for biomarker discovery. Li et al. conducted a glycoproteomics study in the secretome of human hepatocellular carcinoma metastatic (HCC) cell lines, and two glycoproteins FAT1 or DKK3 were proposed as novel prognostic biomarkers of HCC after validation with Western blot and tissue array immunohistochemistry (IHC) [159]. Specifically, extracellular vesicles (EVs) in the secretory system have been exploited as an attractive source for biomarker discovery. Very recently, Chen et al. identified 1453 unique deglycosylated glycopeptides from 556 glycoproteins in plasma-derived EVs, among which 20 were veri-fied to be significantly higher in breast cancer patients [184]. Additionally, 5 of these glycoprotein candidates were later successfully validated in patients and healthy individuals through a novel polymer-based reverse phase glycoprotein array (polyGPA) platform.
4.2.2. Biological process exploration
With glycosylation playing a key role in many biological processes, comparative glycoproteomics could reveal the dynamic changes and further shed light upon its functions along these processes. In a recent study, Kang et al. employed quantitative glycoproteomics approach to explore the molecular mechanism underlying the increased insulin secretion of normal pancreatic islet b-cells (PBCs) in response to elevated blood glucose levels [185]. Their results showed that altered sialylation of surface glycoproteins, such as integrins, integrin ligands, semaphorins and plexins was involved in the process of glucose-stimulated insulin secretion (GSIS). In order to uncover the glyco-markers in the neuronal differentiation process, Tyleckova et al. have successfully quantified hundreds of N-glycoproteins at onset and upon neuronal differentiation, as well as in mature hNT neurons using the cell surface capture (CSC) technology, and validated the glycosylation alterations of several cell adhesion glycoproteins using selected reaction monitoring (SRM) strategy [186]. Glycosylation has been known to affect the development of central nervous system (CNS) and defective glycosylation has also been shown to impair development and neurological function [187]. To this end, Palmisano et al. conducted a glycoproteomics study to monitor the glycosylation changes associated with cell signaling during mouse brain development using the postnatal mice from day 0 until maturity at day 80 [188]. Their results confirmed the role of sialylation in organ development and provided the first extensive global view of dynamic changes in N-glycosylation during mouse brain development. A comprehensive N-glycoproteomics analysis was also conducted to investigate the role of N-glycosylation during the deetiolation process, which is one of the most dramatic developmental processes known in plants [189]. The study has shown 186 N-glycosylation sites from 162 N-glycoproteins were significantly regulated over the course of the 12-h de-etiolation period, indicating the important role of N-glycosylation during deetiolation process. Besides the biological process without disturbance, the biological processes that are the result of environmental stimuli, such as infection, have also been investigated. Melo-Braga et al. explored the modulation of N-glycosylation in grape by Lobesia botrana pathogen infection and demonstrated the importance of N-glycosylation in plant response to biotic stimulus through the glycosylation changes of disease-resistance response glycoprotein DDR206 [130]. In another study into regulation of protein N-glycosylation in human macrophages and their secreted microparticles (MPs) upon Mycobacterium tuberculosis infection, Hare et al. showed an increased complex-type glycosylation and concomitant down-regulation of paucimannosylation of macrophages upon infection [190].
5. Concluding remarks
With rapid advancements in various methodologies, including improved enrichment methods, novel MS/MS fragmentation techniques, powerful workflows, and advanced bioinformatics, MS-based glycoproteomics is gaining more attention and has been increasingly applied to studies of various biological systems. Unprecedented glycoproteome depth has been achieved in different complex samples, providing valuable molecular basis for further structure-function studies of glycosylation. Due to the advances in hybrid fragmentation methods and maturing search engines for intact glycopeptide analysis, site-specific glycoproteomics have become increasingly feasible and will eventually become a routine and practical approach for large-scale glycosylation analysis, which could help decipher the long-time puzzle of glycosylation microheterogeneity. Although great advancements have been made, limitations still exist in the following aspects. The current O-glycopeptide enrichment efficiency is not ideal, and intact O-glycan structure information was often lost in many existing O-glycopeptide enrichment methods. Because of these remaining challenges, the O-glycoproteome information from different complex samples is still quite sparse, with most of the studies focusing on O-glycosite mapping or O-glycoproteome profiling with truncated O-glycans information. More efficient O-glycopeptide enrichment methods that preserve native O-glycan structures are highly desirable to advance the O-glycoproteomics forward. Furthermore, integrated workflows that enable both N-glycosylation and O-glycosylation to be analyzed simultaneously are in great demand. Additionally, the current glycoproteomics methodologies only allow glycan composition and partial glycan structure information to be revealed; although isomer differentiation has been advanced by utilizing PGC columns [191], more diverse tools are needed to improve the resolution for isomer differentiation. Future direction includes improving glycan structural analysis by incorporating isomer differentiation tools such as infrared spectroscopy (IR) or ion mobility (IM) into the workflow.
Acknowledgements
The authors would like to thank Kellen DeLaney, Amanda Buchberger and Jillian Johnson in the Li Lab for critical reading and helpful comments on the early drafts of the manuscript. This research was supported in part by the National Institutes of Health grants U01CA231081, R21AG055377, R01AG052324, and R01DK071801, and a Robert Draper Technology Innovation Fund grant with funding provided by the Wisconsin Alumni Research Foundation (WARF). LL acknowledges a Vilas Distinguished Achievement Professorship and Janis Apinis Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
References
- [1].Ohtsubo K, Marth JD, Glycosylation in cellular mechanisms of health and disease, Cell 126 (2006) 855–867. [DOI] [PubMed] [Google Scholar]
- [2].Jankovic MM, Milutinovic BS, Glycoforms of CA125 antigen as a possible cancer marker, Cancer Biomark 4 (2008) 35–42. [DOI] [PubMed] [Google Scholar]
- [3].van Gisbergen KP, Aarnoudse CA, Meijer GA, Geijtenbeek TB, van Kooyk Y, Dendritic cells recognize tumor-specific glycosylation of carcinoembryonic antigen on colorectal cancer cells through dendritic cellespecific intercellular adhesion molecule-3egrabbing nonintegrin, Cancer Res 65 (2005) 5935–5944. [DOI] [PubMed] [Google Scholar]
- [4].Noda K, Miyoshi E, Uozumi N, Yanagidani S, Ikeda Y, Gao C.x., Suzuki K, Yoshihara H, Yoshikawa M, Kawano K, Gene expression of a1–6 fucosyltransferase in human hepatoma tissues: a possible implication for increased fucosylation of a-fetoprotein, Hepatology 28 (1998) 944–952. [DOI] [PubMed] [Google Scholar]
- [5].Drake PM, Cho W, Li B, Prakobphol A, Johansen E, Anderson NL, Regnier FE, Gibson BW, Fisher SJ, Sweetening the pot: adding glycosylation to the biomarker discovery equation, Clin. Chem 56 (2010) 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chen Z, Glover MS, Li L, Recent advances in ion mobilitye-mass spectrometry for improved structural characterization of glycans and glycoconjugates, Curr. Opin. Chem. Biol 42 (2018) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhong X, Chen Z, Snovida S, Liu Y, Rogers JC, Li L, Capillary electrophoresis-electrospray ionization-mass spectrometry for quantitative analysis of glycans labeled with multiplex carbonyl-reactive tandem mass tags, Anal. Chem 87 (2015) 6527–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Huang G, Sun Z, Qin H, Zhao L, Xiong Z, Peng X, Ou J, Zou H, Preparation of hydrazine functionalized polymer brushes hybrid magnetic nanoparticles for highly specific enrichment of glycopeptides, Analyst 139 (2014) 2199–2206. [DOI] [PubMed] [Google Scholar]
- [9].Zhang H, Li X.-j., Martin DB, Aebersold R, Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry, Nat. Biotechnol 21 (2003) 660–666. [DOI] [PubMed] [Google Scholar]
- [10].Sun B, Ranish JA, Utleg AG, White JT, Yan X, Lin B, Hood L, Shotgun glycopeptide capture approach coupled with mass spectrometry for comprehensive glycoproteomics, Mol. Cell. Proteomics 6 (2007) 141–149. [DOI] [PubMed] [Google Scholar]
- [11].Kurogochi M, Amano M, Fumoto M, Takimoto A, Kondo H, Nishimura S-I, Reverse glycoblotting allows rapid-enrichment glycoproteomics of biopharmaceuticals and disease-related biomarkers, Angew. Chem. Int. Ed 46 (2007) 8808–8813. [DOI] [PubMed] [Google Scholar]
- [12].Nilsson J, Ruetschi U, Halim A, Hesse C, Carlsohn E, Brinkmalm G,Larson G, Enrichment of glycopeptides for glycan structure and attachment site identification, Nat. Methods 6 (2009) 809–811. [DOI] [PubMed] [Google Scholar]
- [13].Halim A, Rüetschi U, Larson G, Nilsson J, LC–MS/MS characterization of O-glycosylation sites and glycan structures of human cerebrospinal fluid glycoproteins, J. Proteome Res 12 (2013) 573–584. [DOI] [PubMed] [Google Scholar]
- [14].Kurogochi M, Matsushista T, Amano M, Furukawa J.-i, Shinohara Y,Aoshima M, Nishimura S-I, Sialic acid-focused quantitative mouse serum glycoproteomics by multiple reaction monitoring assay, Mol. Cell. Proteomics 9 (2010) 2354–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Badr HA, AlSadek DMM, Darwish AA, ElSayed AI, Bekmanov BO, Khussainova EM, Zhang X, Cho WCS, Djansugurova LB, Li C-Z, Lectin approaches for glycoproteomics in FDA-approved cancer biomarkers, Expert Rev. Proteomics 11 (2014) 227–236. [DOI] [PubMed] [Google Scholar]
- [16].Ruiz-May E, Catala C, Rose JK, N-glycoprotein enrichment by lectin affinity chromatography, Methods Mol. Biol 1072 (2014) 633–643. [DOI] [PubMed] [Google Scholar]
- [17].Lee LY, Hincapie M, Packer N, Baker MS, Hancock WS, Fanayan S, An optimized approach for enrichment of glycoproteins from cell culture lysates using native multi-lectin affinity chromatography, J. Sep. Sci 35 (2012) 2445–2452. [DOI] [PubMed] [Google Scholar]
- [18].Wang Y, Wu S.-l., Hancock WS, Approaches to the study of N-linked glycoproteins in human plasma using lectin affinity chromatography and nano-HPLC coupled to electrospray linear ion trapdfourier transform mass spec-trometry, Glycobiology 16 (2006) 514–523. [DOI] [PubMed] [Google Scholar]
- [19].Gbormittah FO, Hincapie M, Hancock WS, Development of an improved fractionation of the human plasma proteome by a combination of abundant proteins depletion and multi-lectin affinity chromatography, Bioanalysis 6 (2014) 2537–2548. [DOI] [PubMed] [Google Scholar]
- [20].Ruiz-May E, Hucko S, Howe KJ, Zhang S, Sherwood RW, Thannhauser TW, Rose JKC, A comparative study of lectin affinity based plant N-glycoproteome profiling using tomato fruit as a model, Mol. Cell. Proteomics 13 (2014) 566–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Totten SM, Kullolli M, Pitteri SJ, Multi-lectin affinity chromatography for separation, identification, and quantitation of intact protein glycoforms in complex biological mixtures, in: Comai L, Katz JE, Mallick P (Editors), Proteomics: Methods and Protocols, Springer New York, New York, NY, 2017, pp. 99–113. [DOI] [PubMed] [Google Scholar]
- [22].Gbormittah FO, Lee LY, Taylor K, Hancock WS, Iliopoulos O, Comparative studies of the proteome, glycoproteome, and N-glycome of clear cell renal cell carcinoma plasma before and after curative nephrectomy, J. Proteome Res 13 (2014) 4889–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li Y, Shah P, De Marzo AM, Van Eyk JE, Li Q, Chan DW, Zhang H, Identification of glycoproteins containing specific glycans using a lectin-chemical method, Anal. Chem 87 (2015) 4683–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yang JS, Qiao J, Kim JY, Zhao L, Qi L, Moon MH, Online proteolysis and glycopeptide enrichment with thermoresponsive porous polymer membrane reactors for nanoflow liquid chromatography-tandem mass spectrometry, Anal. Chem 90 (2018) 3124–3131. [DOI] [PubMed] [Google Scholar]
- [25].Hägglund P, Bunkenborg J, Elortza F, Jensen ON, Roepstorff P, A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation, J. Proteome Res 3 (2004) 556–566. [DOI] [PubMed] [Google Scholar]
- [26].Chen Z, Zhong X, Tie C, Chen B, Zhang X, Li L, Development of a hydro-philic interaction liquid chromatography coupled with matrix-assisted laser desorption/ionization-mass spectrometric imaging platform for N-glycan relative quantitation using stable-isotope labeled hydrazide reagents, Anal. Bioanal. Chem 409 (2017) 4437–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mysling S, Palmisano G, Højrup P, Thaysen-Andersen M, Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics, Anal. Chem 82 (2010) 5598–5609. [DOI] [PubMed] [Google Scholar]
- [28].Zhang C, Ye Z, Xue P, Shu Q, Zhou Y, Ji Y, Fu Y, Wang J, Yang F, Evaluation of different N-glycopeptide enrichment methods for N-glycosylation sites mapping in mouse brain, J. Proteome Res 15 (2016) 2960–2968. [DOI] [PubMed] [Google Scholar]
- [29].Xiong Z, Zhao L, Wang F, Zhu J, Qin H, Wu R.a., Zhang W, Zou H, Synthesis of branched PEG brushes hybrid hydrophilic magnetic nanoparticles for the selective enrichment of N-linked glycopeptides, Chem. Commun 48 (2012) 8138–8140. [DOI] [PubMed] [Google Scholar]
- [30].Xiong Z, Qin H, Wan H, Huang G, Zhang Z, Dong J, Zhang L, Zhang W,Zou H, Layer-by-layer assembly of multilayer polysaccharide coated magnetic nanoparticles for the selective enrichment of glycopeptides, Chem. Commun 49 (2013) 9284–9286. [DOI] [PubMed] [Google Scholar]
- [31].Huang G, Xiong Z, Qin H, Zhu J, Sun Z, Zhang Y, Peng X, ou J, Zou H, Synthesis of zwitterionic polymer brushes hybrid silica nanoparticles via controlled polymerization for highly efficient enrichment of glycopeptides, Anal. Chim. Acta 809 (2014) 61–68. [DOI] [PubMed] [Google Scholar]
- [32].Jiao F, Gao F, Wang H, Deng Y, Zhang Y, Qian X, Zhang Y, Polymeric hydrophilic ionic liquids used to modify magnetic nanoparticles for the highly selective enrichment of N-linked glycopeptides, Sci. Rep 7 (2017) 6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Selman MH, Hemayatkar M, Deelder AM, Wuhrer M, Cotton HILIC SPE microtips for microscale purification and enrichment of glycans and glycopeptides, Anal. Chem 83 (2011) 2492–2499. [DOI] [PubMed] [Google Scholar]
- [34].Sugahara D, Kaji H, Sugihara K, Asano M, Narimatsu H, Large-scale identification of target proteins of a glycosyltransferase isozyme by Lectin-IGOTLC/MS, an LC/MS-based glycoproteomic approach, Sci. Rep 2 (2012) 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen Z, Yu Q, Hao L, Liu F, Johnson J, Tian Z, Kao WJ, Xu W, Li L, Site-specific characterization and quantitation of N-glycopeptides in PKM2 knockout breast cancer cells using DiLeu isobaric tags enabled by electron-transfer/higher-energy collision dissociation (EThcD), Analyst 143 (2018) 2508–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhu J, Wang F, Chen R, Cheng K, Xu B, Guo Z, Liang X, Ye M, Zou H, Centrifugation assisted microreactor enables facile integration of trypsin digestion, hydrophilic interaction chromatography enrichment, and on-column deglycosylation for rapid and sensitive N-glycoproteome analysis, Anal. Chem 84 (2012) 5146–5153. [DOI] [PubMed] [Google Scholar]
- [37].Liu J, Wang F, Lin H, Zhu J, Bian Y, Cheng K, Zou H, Monolithic capillary column based glycoproteomic reactor for high-sensitive analysis of N-glycoproteome, Anal. Chem 85 (2013) 2847–2852. [DOI] [PubMed] [Google Scholar]
- [38].Dedvisitsakul P, Jacobsen S, Svensson B, Bunkenborg J, Finnie C, H€agglund P, Glycopeptide enrichment using a combination of ZIC-HILIC and cotton wool for exploring the glycoproteome of wheat flour albumins,J. Proteome Res 13 (2014) 2696–2703. [DOI] [PubMed] [Google Scholar]
- [39].Jiang H, Yuan H, Qu Y, Liang Y, Jiang B, Wu Q, Deng N, Liang Z, Zhang L,Zhang Y, Preparation of hydrophilic monolithic capillary column by in situ photo-polymerization of N-vinyl-2-pyrrolidinone and acrylamide for highly selective and sensitive enrichment of N-linked glycopeptides, Talanta 146 (2016) 225–230. [DOI] [PubMed] [Google Scholar]
- [40].Xie Y, Liu Q, Li Y, Deng C, Core-shell structured magnetic metal-organic framework composites for highly selective detection of N-glycopeptides based on boronic acid affinity chromatography, J. Chromatogr. A 1540 (2018) 87–93. [DOI] [PubMed] [Google Scholar]
- [41].Xiao H, Chen W, Smeekens JM, Wu R, An enrichment method based on synergistic and reversible covalent interactions for large-scale analysis of glycoproteins, Nat. Commun 9 (2018) 1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chen W, Smeekens JM, Wu R, A universal chemical enrichment method for mapping the yeast N-glycoproteome by mass spectrometry (MS), Mol. Cell. Proteomics 13 (2014) 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bie Z, Chen Y, Li H, Wu R, Liu Z, Off-line hyphenation of boronate affinity monolith-based extraction with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for efficient analysis of glycoproteins/glycopeptides, Anal. Chim. Acta 834 (2014) 1–8. [DOI] [PubMed] [Google Scholar]
- [44].Larsen MR, Jensen SS, Jakobsen LA, Heegaard NH, Exploring the sialiome using titanium dioxide chromatography and mass spectrometry, Mol. Cell. Proteomics 6 (2007) 1778–1787. [DOI] [PubMed] [Google Scholar]
- [45].Yan J, Li X, Yu L, Jin Y, Zhang X, Xue X, Ke Y, Liang X, Selective enrichment of glycopeptides/phosphopeptides using porous titania microspheres, Chem. Commun 46 (2010) 5488–5490. [DOI] [PubMed] [Google Scholar]
- [46].Zhang W, Liu T, Dong H, Bai H, Tian F, Shi Z, Chen M, Wang J, Qin W,Qian X, Synthesis of a highly azide-reactive and thermosensitive bio-functional reagent for efficient enrichment and large-scale identification of O-GlcNAc proteins by mass spectrometry, Anal. Chem 89 (2017) 5810–5817. [DOI] [PubMed] [Google Scholar]
- [47].Li X, Xiong Y, Qing G, Jiang G, Li X, Sun T, Liang X, Bioinspired saccharideesaccharide interaction and smart polymer for specific enrichment of sialylated glycopeptides, ACS Appl. Mater. Interfaces 8 (2016) 13294–13302. [DOI] [PubMed] [Google Scholar]
- [48].Jiang L, Messing ME, Ye L, Temperature and pH dual-responsive core-brush nanocomposite for enrichment of glycoproteins, ACS Appl. Mater. Interfaces 9 (2017) 8985–8995. [DOI] [PubMed] [Google Scholar]
- [49].Bai H, Fan C, Zhang W, Pan Y, Ma L, Ying W, Wang J, Deng Y, Qian X,Qin W, A pH-responsive soluble polymer-based homogeneous system for fast and highly efficient N-glycoprotein/glycopeptide enrichment and identification by mass spectrometry, Chem. Sci 6 (2015) 4234–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Liu J, Wang F, Zhu J, Mao J, Liu Z, Cheng K, Qin H, Zou H, Highly efficient N-glycoproteomic sample preparation by combining C18 and graphitized carbon adsorbents, Anal. Bioanal. Chem 406 (2014) 3103–3109. [DOI] [PubMed] [Google Scholar]
- [51].Takakura D, Harazono A, Hashii N, Kawasaki N, Selective glycopeptide profiling by acetone enrichment and LC/MS, J. Proteomics 101 (2014) 17–30. [DOI] [PubMed] [Google Scholar]
- [52].Mancera-Arteu M, Giménez E, Benavente F, Barbosa J, Sanz-Nebot V, Analysis of O-glycopeptides by acetone enrichment and capillary electrophoresis-mass spectrometry, J. Proteome Res 16 (2017) 4166–4176. [DOI] [PubMed] [Google Scholar]
- [53].Alvarez-Manilla G, Atwood J III, Guo Y, Warren NL, Orlando R, Pierce M, Tools for glycoproteomic analysis: size exclusion chromatography facilitates identification of tryptic glycopeptides with N-linked glycosylation sites,J. Proteome Res 5 (2006) 701–708. [DOI] [PubMed] [Google Scholar]
- [54].Yin H, An M, So P.k., Wong MYM, Lubman DM, Yao Z, The Analysis of Alpha-1-antitrypsin Glycosylation with Direct LC-MS/MS, Electrophoresis, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Huang JF, Qin HQ, Sun Z, Huang G, Mao JW, Cheng K, Zhang Z, Wan H, Yao YT, Dong J, Zhu J, Wang FJ, Ye ML, Zou HF, A peptide N-terminal protection strategy for comprehensive glycoproteome analysis using hydrazide chemistry based method, Sci. Rep 5 (2015) 10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Huang J, Wan H, Yao Y, Li J, Cheng K, Mao J, Chen J, Wang Y, Qin H,Zhang W, Ye M, Zou H, Highly efficient release of glycopeptides from hydrazide beads by hydroxylamine assisted PNGase F deglycosylation for N-glycoproteome analysis, Anal. Chem 87 (2015) 10199–10204. [DOI] [PubMed] [Google Scholar]
- [57].Weng Y, Sui Z, Jiang H, Shan Y, Chen L, Zhang S, Zhang L, Zhang Y, Releasing N-glycan from peptide N-terminus by N-terminal succinylation assisted enzymatic deglycosylation, Sci. Rep 5 (2015) 9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zielinska DF, Gnad F, Wiśniewski JR, Mann M, Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints, Cell 141 (2010) 897–907. [DOI] [PubMed] [Google Scholar]
- [59].Li X, Jiang J, Zhao X, Wang J, Han H, Zhao Y, Peng B, Zhong R, Ying W,Qian X, N-glycoproteome analysis of the secretome of human metastatic hepatocellular carcinoma cell lines combining hydrazide chemistry, HILIC enrichment and mass spectrometry, PloS One 8 (2013) e81921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zhu J, Sun Z, Cheng K, Chen R, Ye M, Xu B, Sun D, Wang L, Liu J, Wang F,Zou H, Comprehensive mapping of protein N-glycosylation in human liver by combining hydrophilic interaction chromatography and hydrazide chemistry, J. Proteome Res 13 (2014) 1713–1721. [DOI] [PubMed] [Google Scholar]
- [61].Fang P, Wang X.-j., Xue Y, Liu M.-q., Zeng W.-f., Zhang Y, Zhang L, Gao X,Yan G.-q., Yao J, Shen H.-l., Yang P.-y., In-depth mapping of the mouse brain N-glycoproteome reveals widespread N-glycosylation of diverse brain proteins, Oncotarget 7 (2016) 38796–38809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Levery SB, Steentoft C, Halim A, Narimatsu Y, Clausen H, Vakhrushev SY, Advances in mass spectrometry driven O-glycoproteomics, Biochim. Biophys. Acta Gen. Subj 1850 (2015) 33–42. [DOI] [PubMed] [Google Scholar]
- [63].Ma J, Hart GW, Analysis of protein O-GlcNAcylation by mass spectrometry, Curr. Protoc. Protein. Sci 87 (2017), 24.10.21–24.10.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Worth M, Li H, Jiang J, Deciphering the functions of protein O-GlcNAcylation with chemistry, ACS Chem. Biol 12 (2017) 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-Wilson LC, Probing the dynamics of OGlcNAc glycosylation in the brain using quantitative proteomics, Nat. Chem. Biol 3 (2007) 339–348. [DOI] [PubMed] [Google Scholar]
- [66].Zaro BW, Yang YY, Hang HC, Pratt MR, Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4–1, Proc. Natl. Acad. Sci. U. S. A 108 (2011) 8146–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Parker BL, Gupta P, Cordwell SJ, Larsen MR, Palmisano G, Purification and identification of O-GlcNAc-Modified peptides using phosphate-based alkyne CLICK chemistry in combination with titanium dioxide chromatography and mass spectrometry, J. Proteome Res 10 (2010) 1449–1458. [DOI] [PubMed] [Google Scholar]
- [68].Alfaro JF, Gong CX, Monroe ME, Aldrich JT, Clauss TRW, Purvine SO, Wang ZH, Camp DG, Shabanowitz J, Stanley P, Hart GW, Hunt DF,Yang F, Smith RD, Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets, Proc. Natl. Acad. Sci. U. S. A 109 (2012) 7280–7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Thompson JW, Griffin ME, Hsieh-Wilson LC, Chapter four - methods for the detection, study, and dynamic profiling of O-GlcNAc glycosylation, in:Imperiali B (Editor), Methods Enzymol, Academic Press, 2018, pp. 101–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA, Schoepfer R, Burlingame AL, O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry, Mol. Cell. Proteomics 5 (2006) 923–934. [DOI] [PubMed] [Google Scholar]
- [71].Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, Burlingame AL, Global identification and characterization of both OGlcNAcylation and phosphorylation at the murine synapse, Mol. Cell. Proteomics 11 (2012) 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Shen B, Zhang W, Shi Z, Tian F, Deng Y, Sun C, Wang G, Qin W, Qian X, A novel strategy for global mapping of O-GlcNAc proteins and peptides using selective enzymatic deglycosylation, HILIC enrichment and mass spectrometry identification, Talanta 169 (2017) 195–202. [DOI] [PubMed] [Google Scholar]
- [73].Darula Z, Sherman J, Medzihradszky KF, How to dig deeper? Improved enrichment methods for mucin core-1 type glycopeptides, Mol. Cell. Proteomics 11 (2012). O111 016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Steentoft C, Vakhrushev SY, Vester-Christensen MB, Schjoldager KTBG,Kong Y, Bennett EP, Mandel U, Wandall H, Levery SB, Clausen H, Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines, Nat. Methods 8 (2011) 977–982. [DOI] [PubMed] [Google Scholar]
- [75].Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KTBG, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L,Gupta R, Paul Bennett E, Mandel U, Brunak S, Wandall HH, Levery SB,Clausen H, Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology, EMBO J 32 (2013) 1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hu H, Khatri K, Klein J, Leymarie N, Zaia J, A review of methods for interpretation of glycopeptide tandem mass spectral data, Glycoconj. J 33 (2016) 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Thaysen-Andersen M, Packer NH, Schulz BL, Maturing glycoproteomics technologies provide unique structural insights into the N-glycoproteome and its regulation in health and disease, Mol. Cell. Proteomics 15 (2016) 1773–1790. 10.1074/mcp.O115.057638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Halim A, Nilsson J, Rüetschi U, Hesse C, Larson G, Human urinary glycoproteomics; attachment site specific analysis of N- and O-linked glycosylations by CID and ECD, Mol. Cell. Proteomics 11 (2012). M111.013649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Mayampurath A, Yu C-Y, Song E, Balan J, Mechref Y, Tang H, Computational framework for identification of intact glycopeptides in complex samples, Anal. Chem 86 (2014) 453e463. [DOI] [PubMed] [Google Scholar]
- [80].Sun S, Shah P, Eshghi ST, Yang W, Trikannad N, Yang S, Chen L, Aiyetan P,Hoti N, Zhang Z, Chan DW, Zhang H, Comprehensive analysis of protein glycosylation by solid-phase extraction of N-linked glycans and glycosite-containing peptides, Nat. Biotechnol 34 (2016) 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Chen R, Seebun D, Ye M, Zou H, Figeys D, Site-specific characterization of cell membrane N-glycosylation with integrated hydrophilic interaction chromatography solid phase extraction and LCeMS/MS, J. Proteomics 103 (2014) 194–203. [DOI] [PubMed] [Google Scholar]
- [82].Cheng K, Chen R, Seebun D, Ye M, Figeys D, Zou H, Large-scale characterization of intact N-glycopeptides using an automated glycoproteomic method, J. Proteomics 110 (2014) 145–154. [DOI] [PubMed] [Google Scholar]
- [83].Liu M, Zhang Y, Chen Y, Yan G, Shen C, Cao J, Zhou X, Liu X, Zhang L,Shen H, Lu H, He F, Yang P, Efficient and accurate glycopeptide identification pipeline for high-throughput site-specific N-glycosylation analysis,J. Proteome Res 13 (2014) 3121–3129. [DOI] [PubMed] [Google Scholar]
- [84].Cao Q, Zhao X, Zhao Q, Lv X, Ma C, Li X, Zhao Y, Peng B, Ying W,Qian X, Strategy integrating stepped fragmentation and glycan diagnostic ion-based spectrum refinement for the identification of core fucosylated glycoproteome using mass spectrometry, Anal. Chem 86 (2014) 6804–6811. [DOI] [PubMed] [Google Scholar]
- [85].Liu MQ, Zeng WF, Fang P, Cao WQ, Liu C, Yan GQ, Zhang Y, Peng C, Wu JQ, Zhang XJ, Tu HJ, Chi H, Sun RX, Cao Y, Dong MQ, Jiang BY, Huang JM, Shen HL, Wong CCL, He SM, Yang PY, pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification, Nat. Commun 8 (2017) 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Bollineni RC, Koehler CJ, Gislefoss RE, Anonsen JH, Thiede B, Large-scale intact glycopeptide identification by Mascot database search, Sci. Rep 8 (2018) 2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Khatri K, Staples GO, Leymarie N, Leon DR, Turiák L, Huang Y, Yip S,Hu H, Heckendorf CF, Zaia J, Confident assignment of site-specific glycosylation in complex glycoproteins in a single step, J. Proteome Res 13 (2014) 4347–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Lynn KS, Chen CC, Lih TM, Cheng CW, Su WC, Chang CH, Cheng CY, Hsu WL, Chen YJ, Sung TY, MAGIC: an automated N-linked glycoprotein identification tool using a Y1-ion pattern matching algorithm and in silico MS(2) approach, Anal. Chem 87 (2015) 2466–2473. [DOI] [PubMed] [Google Scholar]
- [89].Qin H, Cheng K, Zhu J, Mao J, Wang F, Dong M, Chen R, Guo Z, Liang X,Ye M, Zou H, Proteomics analysis of O-GalNAc glycosylation in human serum by an integrated strategy, Anal. Chem 89 (2017) 1469–1476. [DOI] [PubMed] [Google Scholar]
- [90].Frese CK, Altelaar AFM, van den Toorn H, Nolting D, Griep-Raming J, Heck AJR, Mohammed S, Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry, Anal. Chem 84 (2012) 9668–9673. [DOI] [PubMed] [Google Scholar]
- [91].Glover MS, Yu Q, Chen Z, Shi X, Kent KC, Li L, Characterization of intact sialylated glycopeptides and phosphorylated glycopeptides from IMAC enriched samples by EThcD fragmentation: toward combining phosphoproteomics and glycoproteomics, Int. J. Mass Spectrom 427 (2018) 35–42. [Google Scholar]
- [92].Yu Q, Wang B, Chen Z, Urabe G, Glover MS, Shi X, Guo L-W, Kent KC,Li L, Electron-transfer/higher-energy collision dissociation (EThcD)-Enabled intact glycopeptide/glycoproteome characterization, J. Am. Soc. Mass Spec-trom 28 (2017) 1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Zhang Y, Xie X, Zhao X, Tian F, Lv J, Ying W, Qian X, Systems analysis of singly and multiply O-glycosylated peptides in the human serum glycoproteome via EThcD and HCD mass spectrometry, J. Proteomics 170 (2018) 14–27. [DOI] [PubMed] [Google Scholar]
- [94].He L, Xin L, Shan B, Lajoie GA, Ma B, GlycoMaster DB, Software to assist the automated identification of N-linked glycopeptides by tandem mass spectrometry, J. Proteome Res 13 (2014) 3881–3895. [DOI] [PubMed] [Google Scholar]
- [95].Toghi Eshghi S, Shah P, Yang W, Li X, Zhang H, GPQuest: a spectral library matching algorithm for site-specific assignment of tandem mass spectra to intact N-glycopeptides, Anal. Chem 87 (2015) 5181–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Park GW, Kim JY, Hwang H, Lee JY, Ahn YH, Lee HK, Ji ES, Kim KH, Jeong HK, Yun KN, Integrated GlycoProteome analyzer (I-GPA) for automated identification and quantitation of site-specific N-glycosylation, Sci. Rep 6 (2016) 21175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Bern M, Kil YJ, Becker C, Byonic: advanced peptide and protein identification software, Curr. Protoc. Bioinformatics 40 (2012) 1, 13.20.11–13.20.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Nasir W, Toledo AG, Noborn F, Nilsson J, Wang M, Bandeira N, Larson G, SweetNET: a bioinformatics workflow for glycopeptide MS/MS spectral analysis, J. Proteome Res 15 (2016) 2826–2840. [DOI] [PubMed] [Google Scholar]
- [99].Zeng W-F, Liu M-Q, Zhang Y, Wu J-Q, Fang P, Peng C, Nie A, Yan G,Cao W, Liu C, Chi H, Sun R-X, Wong CCL, He S-M, Yang P, pGlyco: a pipeline for the identification of intact N-glycopeptides by using HCD- and CID-MS/MS and MS3, Sci. Rep 6 (2016) 25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Stadlmann J, Taubenschmid J, Wenzel D, Gattinger A, Dürnberger G,Dusberger F, Elling U, Mach L, Mechtler K, Penninger JM, Comparative glycoproteomics of stem cells identifies new players in ricin toxicity, Nature 549 (2017) 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Hu H, Khatri K, Zaia J, Algorithms and design strategies towards automated glycoproteomics analysis, Mass Spectrom. Rev 36 (2017) 475–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Tsai P-L, Chen S-F, A brief review of bioinformatics tools for glycosylation analysis by mass spectrometry, Mass Spectrom 6 (2017). S0064–S0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Lakbub JC, Su X, Zhu Z, Patabandige MW, Hua D, Go EP, Desaire H, Two new tools for glycopeptide analysis researchers: a glycopeptide decoy generator and a large data set of assigned CID spectra of glycopeptides,J. Proteome Res 16 (2017) 3002–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Lee LY, Moh ESX, Parker BL, Bern M, Packer NH, Thaysen-Andersen M, Toward automated N-glycopeptide identification in glycoproteomics,J. Proteome Res 15 (2016) 3904–3915. [DOI] [PubMed] [Google Scholar]
- [105].Li Z, Adams RM, Chourey K, Hurst GB, Hettich RL, Pan C, Systematic comparison of label-free, metabolic labeling, and isobaric chemical labeling for quantitative proteomics on LTQ Orbitrap Velos, J. Proteome Res 11 (2012) 1582–1590. [DOI] [PubMed] [Google Scholar]
- [106].Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG, Comparison of label-free methods for quantifying human proteins by shotgun proteomics, Mol. Cell. Proteomics 4 (2005) 1487–1502. [DOI] [PubMed] [Google Scholar]
- [107].Callister SJ, Barry RC, Adkins JN, Johnson ET, Johnson ET,Johnson ET,Johnson ET, Qian W.-j., Webb-Robertson B-JM, Smith RD, Lipton MS, Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics, J. Proteome Res 5 (2006) 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Rebecchi KR, Wenke JL, Go EP, Desaire H, Label-free quantitation: a new glycoproteomics approach, J. Am. Soc. Mass Spectrom 20 (2009) 1048–1059. [DOI] [PubMed] [Google Scholar]
- [109].Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M,Assadourian G, Lee A, Van Sluyter SC, Haynes PA, Less label, more free: approaches in label-free quantitative mass spectrometry, Proteomics 11 (2011) 535–553. [DOI] [PubMed] [Google Scholar]
- [110].Mayampurath A, Song E, Yu C.-y., Hammoud Z, Mechref Y, Tang H, Label-free glycopeptide quantification for biomarker discovery in human sera, J. Proteome Res 13 (2014) 4821–4832. [DOI] [PubMed] [Google Scholar]
- [111].Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A,Mann M, Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics, Mol. Cell. Proteomics 1 (2002) 376–386. [DOI] [PubMed] [Google Scholar]
- [112].Mann M, Innovations: functional and quantitative proteomics using SILAC, Nat. Rev. Mol. Cell Biol 7 (2006) 952–958. [DOI] [PubMed] [Google Scholar]
- [113].Ji Y, Wei S, Hou J, Zhang C, Xue P, Wang J, Chen X, Guo X, Yang F, Integrated proteomic and N-glycoproteomic analyses of doxorubicin sensitive and resistant ovarian cancer cells reveal glycoprotein alteration in protein abundance and glycosylation, Oncotarget 8 (2017) 13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Yang X, Wang Z, Guo L, Zhu Z, Zhang Y, Proteome-wide analysis of N-glycosylation stoichiometry using SWATH technology, J. Proteome Res 16 (2017) 3830–3840. [DOI] [PubMed] [Google Scholar]
- [115].Smeekens JM, Xiao H, Wu R, Global analysis of secreted proteins and glycoproteins in Saccharomyces cerevisiae, J. Proteome Res 16 (2016) 1039–1049. [DOI] [PubMed] [Google Scholar]
- [116].Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M, Super-SILAC mix for quantitative proteomics of human tumor tissue, Nat. Methods 7 (2010) 383–385. [DOI] [PubMed] [Google Scholar]
- [117].Deeb SJ, Cox J, Schmidt-Supprian M, Mann M, N-linked glycosylation enrichment for in-depth cell surface proteomics of diffuse large B-cell lymphoma subtypes, Mol. Cell. Proteomics 13 (2014) 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Parker BL, Thaysen-Andersen M, Fazakerley DJ, Holliday M, Packer NH, James DE, Terminal galactosylation and sialylation switching on membrane glycoproteins upon TNF-alpha-induced insulin resistance in adipocytes, Mol. Cell. Proteomics 15 (2016) 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Taga Y, Kusubata M, Ogawa-Goto K, Hattori S, Site-specific quantitative analysis of overglycosylation of collagen in osteogenesis imperfecta using hydrazide chemistry and SILAC, J. Proteome Res 12 (2013) 2225–2232. [DOI] [PubMed] [Google Scholar]
- [120].Tolonen AC, Haas W, Chilaka AC, Aach J, Gygi SP, Church GM, Proteome-wide systems analysis of a cellulosic biofuel-producing microbe, Mol. Syst. Biol 7 (2011) 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Wang S, Regnier FE, Proteomics based on selecting and quantifying cysteine containing peptides by covalent chromatography, J. Chromatogr. A 924 (2001) 345–357. [DOI] [PubMed] [Google Scholar]
- [122].Sun Z, Qin H, Wang F, Cheng K, Dong M, Ye M, Zou H, Capture and dimethyl labeling of glycopeptides on hydrazide beads for quantitative glycoproteomics analysis, Anal. Chem 84 (2012) 8452–8456. [DOI] [PubMed] [Google Scholar]
- [123].Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G,Neumann T, Hamon C, Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS, Anal. Chem 75 (2003) 1895–1904. [DOI] [PubMed] [Google Scholar]
- [124].Werner T, Sweetman G, Savitski M.F.l., Mathieson T, Bantscheff M, Savitski MM, Ion coalescence of neutron encoded TMT 10-plex reporter ions, Anal. Chem 86 (2014) 3594–3601. [DOI] [PubMed] [Google Scholar]
- [125].Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S,Khainovski N, Pillai S, Dey S, Daniels S, Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents, Mol. Cell. Proteomics 3 (2004) 1154–1169. [DOI] [PubMed] [Google Scholar]
- [126].Choe L, D’Ascenzo M, Relkin NR, Pappin D, Ross P, Williamson B,Guertin S, Pribil P, Lee KH, 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer’s disease, Proteomics 7 (2007) 3651–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Xiang F, Ye H, Chen R, Fu Q, Li L, N, N-dimethyl leucines as novel isobaric tandem mass tags for quantitative proteomics and peptidomics, Anal. Chem 82 (2010) 2817–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Frost DC, Greer T, Li L, High-resolution enabled 12-plex DiLeu isobaric tags for quantitative proteomics, Anal. Chem 87 (2014) 1646–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Kroksveen AC, Guldbrandsen A, Vaudel M, Lereim RR, Barsnes H, Myhr K-M, Torkildsen Ø, Berven FS, In-depth cerebrospinal fluid quantitative proteome and deglycoproteome analysis: presenting a comprehensive picture of pathways and processes affected by multiple sclerosis, J. Proteome Res 16 (2016) 179–194. [DOI] [PubMed] [Google Scholar]
- [130].Melo-Braga MN, Verano-Braga T, León IR, Antonacci D, Nogueira FC, Thelen JJ, Larsen MR, Palmisano G, Modulation of protein phosphorylation, N-glycosylation and Lys-acetylation in grape (Vitis vinifera) mesocarp and exocarp owing to Lobesia botrana infection, Mol. Cell. Proteomics 11 (2012) 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].López-Ferrer D, Ramos-Fernández A, Martínéz-Bartolome S, García-Ruiz P,Vézquez J, Quantitative proteomics using 16O/18O labeling and linear ion trap mass spectrometry, Proteomics 6 (2006) S4–S11. [DOI] [PubMed] [Google Scholar]
- [132].Liu Z, Cao J, He Y, Qiao L, Xu C, Lu H, Yang P, Tandem 18O stable isotope labeling for quantification of N-glycoproteome, J. Proteome Res 9 (2009) 227–236. [DOI] [PubMed] [Google Scholar]
- [133].Yang Z, Hancock WS, Approach to the comprehensive analysis of glyco-proteins isolated from human serum using a multi-lectin affinity column,J. Chromatogr. A 1053 (2004) 79–88. [PubMed] [Google Scholar]
- [134].Sparbier K, Asperger A, Resemann A, Kessler I, Koch S, Wenzel T, Stein G,Vorwerg L, Suckau D, Kostrzewa M, Analysis of glycoproteins in human serum by means of glycospecific magnetic bead separation and LC-MALDITOF/TOF analysis with automated glycopeptide detection, J. Biomol. Tech.:J. Biochem 18 (2007) 252–258. [PMC free article] [PubMed] [Google Scholar]
- [135].Anderson NL, Anderson NG, The human plasma proteome history, character, and diagnostic prospects, Mol. Cell. Proteomics 1 (2002) 845–867. [DOI] [PubMed] [Google Scholar]
- [136].Zhang M, Chen G-X, Lv D-W, Li X-H, Yan Y-M, N-linked glycoproteome profiling of seedling leaf in Brachypodium distachyon L, J. Proteome Res 14 (2015) 1727–1738. [DOI] [PubMed] [Google Scholar]
- [137].Liu T, Qian W-J, Gritsenko MA, Camp DG, Monroe ME, Moore RJ, Smith RD, Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry, J. Proteome Res 4 (2005) 2070–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Ma C, Zhang Q, Qu J, Zhao X, Li X, Liu Y, Wang PG, A precise approach in large scale core-fucosylated glycoprotein identification with low-and high-normalized collision energy, J. Proteomics 114 (2015) 61–70. [DOI] [PubMed] [Google Scholar]
- [139].Hägglund P, Matthiesen R, Elortza F, Højrup P, Roepstorff P, Jensen ON,Bunkenborg J, An enzymatic deglycosylation scheme enabling identification of core fucosylated N-glycans and O-glycosylation site mapping of human plasma proteins, J. Proteome Res 6 (2007) 3021–3031. [DOI] [PubMed] [Google Scholar]
- [140].Jia W, Lu Z, Fu Y, Wang H-P, Wang L-H, Chi H, Yuan Z-F, Zheng Z-B, Song L-N, Han H-H, A strategy for precise and large scale identification of core fucosylated glycoproteins, Mol. Cell. Proteomics 8 (2009) 913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Kawahara R, Saad J, Angeli CB, Palmisano G, Site-specific characterization of N-linked glycosylation in human urinary glycoproteins and endogenous glycopeptides, Glycoconj. J 33 (2016) 937–951. [DOI] [PubMed] [Google Scholar]
- [142].Yang N, Feng S, Shedden K, Xie X, Liu Y, Rosser CJ, Lubman DM,Goodison S, Urinary glycoprotein biomarker discovery for bladder cancer detection using LC-MS/MS and label-free quantification, Clin. Cancer Res 17 (2011) 3349–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Saraswat M, Joenvaara S, Musante L, Peltoniemi H, Holthofer H,Renkonen R, N-linked (N-) glycoproteomics of urinary exosomes, Mol. Cell. Proteomics 14 (2015) 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Chen R, Jiang X, Sun D, Han G, Wang F, Ye M, Wang L, Zou H, Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry, J. Proteome Res 8 (2009) 651–661. [DOI] [PubMed] [Google Scholar]
- [145].Zhang D, Luo G, Ding X, Lu C, Preclinical experimental models of drug metabolism and disposition in drug discovery and development, Acta Pharm. Sin. B 2 (2012) 549–561. [Google Scholar]
- [146].Breker M, Schuldiner M, The emergence of proteome-wide technologies: systematic analysis of proteins comes of age, Nat. Rev. Mol. Cell Biol 15 (2014) 453–464. [DOI] [PubMed] [Google Scholar]
- [147].Vakhrushev SY, Steentoft C, Vester-Christensen MB, Bennett EP,Clausen H, Levery SB, Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells, Mol. Cell. Proteomics 12 (2013) 932–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Woo CM, Iavarone AT, Spiciarich DR, Palaniappan KK, Bertozzi CR, Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N-and O-glycopeptide discovery and analysis, Nat. Methods 12 (2015) 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Woo CM, Felix A, Byrd WE, Zuegel DK, Ishihara M, Azadi P, Iavarone AT, Pitteri SJ, Bertozzi CR, Development of IsoTaG, a chemical glycoproteomics technique for profiling intact N-and O-glycopeptides from whole cell proteomes, J. Proteome Res 16 (2017) 1706–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Woo CM, Lund PJ, Huang AC, Davis MM, Bertozzi CR, Pitteri S, Mapping and quantification of over 2,000 O-linked glycopeptides in activated human T cells with isotope-targeted glycoproteomics (IsoTaG), Mol. Cell. Proteomics 17 (2018) 764–775 mcp. RA117. 000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Von Heijne G, The membrane protein universe: what’s out there and why bother? J. Intern. Med 261 (2007) 543–557. [DOI] [PubMed] [Google Scholar]
- [152].Wollscheid B, von Haller PD, Yi E, Donohoe S, Vaughn K, Keller A, Nesvizhskii AI, Eng J, Li X.-j, Goodlett DR, Lipid Raft Proteins and Their Identification in T Lymphocytes, Membrane Dynamics and Domains, Springer Publishing, New York, 2004, pp. 121–152. [DOI] [PubMed] [Google Scholar]
- [153].Wollscheid B, Bausch-Fluck D, Henderson C, O’brien R, Bibel M, Schiess R,Aebersold R, Watts JD, Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins, Nat. Biotechnol 27 (2009) 378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Gundry RL, Riordon DR, Tarasova Y, Chuppa S, Bhattacharya S, Juhasz O,Wiedemeier O, Milanovich S, Noto FK, Tchernyshyov I, A cell surfaceome map for immunophenotyping and sorting pluripotent stem cells, Mol. Cell. Proteomics 11 (2012) 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Mallanna SK, Cayo MA, Twaroski K, Gundry RL, Duncan SA, Mapping the cell-surface N-glycoproteome of human hepatocytes reveals markers for selecting a homogeneous population of iPSC-derived hepatocytes, Stem cell reports 7 (2016) 543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Li K, Sun Z, Zheng J, Lu Y, Bian Y, Ye M, Wang X, Nie Y, Zou H, Fan D, In-depth research of multidrug resistance related cell surface glycoproteome in gastric cancer, J. Proteomics 82 (2013) 130–140. [DOI] [PubMed] [Google Scholar]
- [157].Mi W, Jia W, Zheng Z, Wang J, Cai Y, Ying W, Qian X, Surface glycoproteomic analysis of hepatocellular carcinoma cells by affinity enrichment and mass spectrometric identification, Glycoconj. J 29 (2012) 411–424. [DOI] [PubMed] [Google Scholar]
- [158].Tjalsma H, Bolhuis A, Jongbloed JD, Bron S, van Dijl JM, Signal peptide-dependent protein transport inBacillus subtilis: a genome-based survey of the secretome, Microbiol. Mol. Biol. Rev 64 (2000) 515–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Li X, Jiang J, Zhao X, Zhao Y, Cao Q, Zhao Q, Han H, Wang J, Yu Z, Peng B,Ying W, Qian X, In-depth analysis of secretome and N-glycosecretome of human hepatocellular carcinoma metastatic cell lines shed light on metastasis correlated proteins, Oncotarget 7 (2016) 22031–22049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Sugahara D, Tomioka A, Sato T, Narimatsu H, Kaji H, Large-scale identification of secretome glycoproteins recognized by Wisteria floribunda agglutinin: a glycoproteomic approach to biomarker discovery, Proteomics 15 (2015) 2921–2933. [DOI] [PubMed] [Google Scholar]
- [161].Yin X, Bern M, Xing Q, Ho J, Viner R, Mayr M, Glycoproteomic analysis of the secretome of human endothelial cells, Mol. Cell. Proteomics 12 (2013) 956–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Ruiz-May E, Sørensen I, Fei Z, Zhang S, Domozych DS, Rose JK, The secretome and N-glycosylation profiles of the charophycean green alga, penium margaritaceum, resemble those of embryophytes, Proteomes 6 (2018) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Rubio MV, Zubieta MP, Cairo JPLF, Calzado F, Leme AFP, Squina FM, Prade RA, Damásio ARL, Mapping N-linked glycosylation of carbohydrate-active enzymes in the secretome of Aspergillus nidulans grown on ligno-cellulose, Biotechnol. Biofuels 9 (2016) 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Parker BL, Thaysen-Andersen M, Solis N, Scott NE, Larsen MR, Graham ME, Packer NH, Cordwell SJ, Site-specific glycan-peptide analysis for determination of N-glycoproteome heterogeneity, J. Proteome Res 12 (2013) 5791–5800. [DOI] [PubMed] [Google Scholar]
- [165].Tian Y, Kelly-Spratt KS, Kemp CJ, Zhang H, Mapping tissue-specific expression of extracellular proteins using systematic glycoproteomic analysis of different mouse tissues, J. Proteome Res 9 (2010) 5837–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Medzihradszky KF, Kaasik K, Chalkley RJ, Tissue-specific glycosylation at the glycopeptide level, Mol. Cell. Proteomics 14 (2015) 2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Ruiz-May E, Thannhauser TW, Zhang S, Rose JKC, Analytical technologies for identification and characterization of the plant N-glycoproteome, Front. Plant Sci 3 (2012) 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Strasser R, Plant protein glycosylation, Glycobiology 26 (2016) 926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Song W, Henquet MG, Mentink RA, van Dijk AJ, Cordewener JH,Bosch D, America AH, van der Krol AR, N-glycoproteomics in plants: perspectives and challenges, J. Proteomics 74 (2011) 1463–1474. [DOI] [PubMed] [Google Scholar]
- [170].Ying J, Zhao J, Hou Y, Wang Y, Qiu J, Li Z, Tong X, Shi Z, Zhu J, Zhang J, Mapping the N-linked glycosites of rice (Oryza sativa L.) germinating embryos, PloS One 12 (2017) e0173853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Song W, Mentink RA, Henquet MG, Cordewener JH, van Dijk AD,Bosch D, America AH, van der Krol AR, N-glycan occupancy of Arabidopsis N-glycoproteins, J. Proteomics 93 (2013) 343–355. [DOI] [PubMed] [Google Scholar]
- [172].Breidenbach MA, Palaniappan KK, Pitcher AA, Bertozzi CR, Mapping yeast N-glycosites with isotopically recoded glycans, Mol. Cell. Proteomics 11 (2012). M111. 015339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Halim A, Larsen ISB, Neubert P, Joshi HJ, Petersen BL, Vakhrushev SY,Strahl S, Clausen H, Discovery of a nucleocytoplasmic O-mannose glycoproteome in yeast, Proc. Natl. Acad. Sci. U.S.A 112 (2015) 15648–15653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Neubert P, Halim A, Zauser M, Essig A, Joshi HJ, Zatorska E, Larsen ISB,Loibl M, Castells-Ballester J, Aebi M, Mapping the O-mannose glycoproteome in Saccharomyces cerevisiae, Mol. Cell. Proteomics 15 (2016) 1323–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Shaw TI, Zhang M, HIV N-linked glycosylation site analyzer and its further usage in anchored alignment, Nucleic Acids Res 41 (2013) W454–W458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Go EP, Irungu J, Zhang Y, Dalpathado DS, Liao H-X, Sutherland LL, Alam SM, Haynes BF, Desaire H, Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic Epitopesʼ accessibility, J. Proteome Res 7 (2008) 1660–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Bagdonaite I, Nordén R, Joshi HJ, King SL, Vakhrushev SY, Olofsson S, Wandall HH, Global mapping of O-glycosylation of varicella zoster virus, human cytomegalovirus, and Epstein-Barr virus, J. Biol. Chem 291 (2016) 12014–12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Smith GT, Sweredoski MJ, Hess S, O-linked glycosylation sites profiling in Mycobacterium tuberculosis culture filtrate proteins, J. Proteomics 97 (2014) 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Boysen A, Palmisano G, Krogh TJ, Duggin IG, Larsen MR, Møller-Jensen J, A novel mass spectrometric strategy “BEMAP” reveals Extensive O-linked protein glycosylation in Enterotoxigenic Escherichia coli, Sci. Rep 6 (2016) 32016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Fodero LR, Sáez-Valero J, Barquero MS, Marcos A, McLean CA, Small DH, Wheat germ agglutinin-binding glycoproteins are decreased in Alzheimer’s disease cerebrospinal fluid, J. Neurochem 79 (2001) 1022–1026. [DOI] [PubMed] [Google Scholar]
- [181].Hashimoto Y, Taniguchi M, Kitaura M, Nakamura Y, Jimbo D, Urakami K, Alteration of Concanavalin a binding glycoproteins in cerebro spinal fluid and serum of alzheimer’s disease patients, Yonago Acta Med 51 (2008) 1–9. [Google Scholar]
- [182].Shah P, Wang X, Yang W, Eshghi ST, Sun S, Hoti N, Chen L, Yang S,Pasay J, Rubin A, Zhang H, Integrated proteomic and glycoproteomic analyses of prostate cancer cells reveal glycoprotein alteration in protein abundance and glycosylation, Mol. Cell. Proteomics 14 (2015) 2753–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Tan Z, Yin H, Nie S, Lin Z, Zhu J, Ruffin MT, Anderson MA, Simeone DM, Lubman DM, Large-scale identification of core-fucosylated glycopeptide sites in pancreatic cancer serum using mass spectrometry, J. Proteome Res 14 (2015) 1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Chen I-H, Aguilar HA, Paez Paez JS, Wu X, Pan L, Wendt MK, Iliuk AB,Zhang Y, Tao WA, Analytical pipeline for discovery and verification of glycoproteins from plasma-derived extracellular vesicles as breast cancer biomarkers, Anal. Chem 90 (2018) 6307–6313. [DOI] [PubMed] [Google Scholar]
- [185].Kang T, Jensen P, Huang H, Christensen GL, Billestrup N, Larsen MR, Characterization of the molecular mechanisms underlying glucose stimulated insulin secretion from isolated pancreatic β-cells using post-translational modification specific proteomics (PTMomics), Mol. Cell. Proteomics 17 (2018) 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Tyleckova J, Valekova I, Zizkova M, Rakocyova M, Marsala S, Marsala M, Gadher SJ, Kovarova H, Surface N-glycoproteome patterns reveal key proteins of neuronal differentiation, J. Proteomics 132 (2016) 13–20. [DOI] [PubMed] [Google Scholar]
- [187].Freeze HH, Understanding human glycosylation disorders: biochemistry leads the charge, J. Biol. Chem 288 (2013) 6936–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Palmisano G, Parker BL, Engholm-Keller K, Lendal SE, Kulej K, Schulz M,Schwämmle V, Graham ME, Saxtorph H, Cordwell SJ, A novel method for the simultaneous enrichment, identification, and quantification of phosphopeptides and sialylated glycopeptides applied to a temporal profile of mouse brain development, Mol. Cell. Proteomics 11 (2012) 1191–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Bu T.-t., Shen J, Chao Q, Shen Z, Yan Z, Zheng H.-y., Wang B.-c., Dynamic N-glycoproteome analysis of maize seedling leaves during de-etiolation using Concanavalin A lectin affinity chromatography and a nano-LCeMS/MS-based iTRAQ approach, Plant Cell Rep 36 (2017) 1943–1958. [DOI] [PubMed] [Google Scholar]
- [190].Hare NJ, Lee LY, Loke I, Britton WJ, Saunders BM, Thaysen-Andersen M, Mycobacterium tuberculosis infection manipulates the glycosylation machinery and the N-glycoproteome of human macrophages and their micro-particles, J. Proteome Res 16 (2016) 247–263. [DOI] [PubMed] [Google Scholar]
- [191].Huang Y, Zhou S, Zhu J, Lubman DM, Mechref Y, LC-MS/MS isomeric profiling of permethylated N-glycans derived from serum haptoglobin of hepatocellular carcinoma (HCC) and cirrhotic patients, Electrophoresis 38 (2017) 2160–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]