Abstract
There is evidence that human activities are reducing the population genetic diversity of species worldwide. Given the prediction that parasites better exploit genetically homogeneous host populations, many species could be vulnerable to disease outbreaks. While agricultural studies have shown the devastating effects of infectious disease in crop monocultures, the widespread nature of this diversity–disease relationship remains unclear in natural systems. Here, we provide broad support that high population genetic diversity can protect against infectious disease by conducting a meta-analysis of 23 studies, with a total of 67 effect sizes. We found that parasite functional group (micro- or macroparasite) affects the presence of the effect and study setting (field or laboratory-based environment) influences the magnitude. Our study also suggests that host genetic diversity is overall a robust defence against infection regardless of host reproduction, parasite host range, parasite diversity, virulence and the method by which parasite success was recorded. Combined, these results highlight the importance of monitoring declines of host population genetic diversity as shifts in parasite distributions could have devastating effects on at-risk populations in nature.
Keywords: genetic diversity, host–parasite interactions, monoculture effect, meta-analysis, microparasite
1. Introduction
Most natural populations are genetically diverse [1]. Given there is often specificity between hosts and parasites [2], host population genetic diversity is thought to increase the chance that one or more individuals is resistant to infection. The likelihood of a parasite encountering a susceptible host is thus reduced [3]. Genetically homogeneous host populations are conversely predicted to be more vulnerable to infection, given the uniformity of host susceptibility. This negative relationship between host genetic diversity and parasite success is often referred to as the ‘monoculture effect’ [4].
The study of the monoculture effect in agricultural settings is extensive [5–7]. A recent meta-analysis showed that with increased diversity in intraspecific cultivar mixtures, disease presence is reduced and crop yields increased [7]. However, crop plants are under artificial selection for high yield, and may therefore exhibit less genetic polymorphism than hosts in the wild. We consequently know little of the extent to which low genetic diversity influences parasite success across species and environments beyond agricultural contexts.
Threats to within-species genetic diversity are on the rise. There is evidence that habitat alterations, pollution and global temperature changes, as well as the restriction of species geographical ranges, may lead to increased genetic drift and reduced population genetic diversity [8,9]. Impacts of humans on local species biodiversity, however, remain controversial [10,11]. Populations with reduced genetic diversity might suffer diminished evolutionary potential [12] and increased inbreeding depression [13,14]. Knowing whether there is an additional threat of outbreaks in these populations is crucial for disease management and species conservation approaches.
Theory has illuminated the dynamics of parasite spread [4,15–18] in diverse host populations as well as examined the level of diversity required to stop transmission [19,20]. However, whether population genetic diversity can impact parasite success in nature more broadly remains unclear for several reasons. First, given that parasite transmission can be determined by host density [3], the relative effects of density versus host genetic diversity need to be elucidated [20]. Shrinking habitats, for example, can result in higher population densities (and lower resource availability) where parasites can transmit better due to more contact between hosts [21,22]. Second, even when focusing on host genetic diversity alone, there is great variation across systems in the conditions under which infection and diversity are measured. In comparison to diverse populations, genetically homogeneous bumblebee (Bombus terrestris L.) populations, the microsporidian Nosema bombi has higher success, but the trypanosomelid Crithidia bombi does not [23]. In other cases, we see an increase in parasite success on the homogeneous host populations when multiple parasite species infect [23–26] but not always between one host–parasite species pair [27,28]. Third, because parasite success is measured differently across studies, and even within systems, there is the potential that the relevant measure of parasite success is not used. For example, in honeybee (Apis mellifera) host populations, genetic diversity has a negative impact on parasite success when infection prevalence or parasite load is measured, but not always when host survival is calculated [29]. Host survival might be less informative because the interplay of virulence, force of infection and the timing of infection might determine the overall spread of pathogens in host populations [30]. It is therefore unclear whether the effect of low host genetic diversity on parasite success is relevant to host–parasite interactions in non-agricultural systems across the tree of life.
We tested the effect of host population genetic diversity on parasite success with a formal meta-analysis across a range of host–parasite systems. We searched the published literature for all publicly available data sources and compared the effects of low and high host genetic diversity on parasite success using Hedges's effect size g (with positive values indicating an effect of low host genetic diversity on parasite success) with a nested random mixed effects meta-analysis model. We also tested whether biological traits associated with the species in the interaction, as well as study settings and measures, could explain variation in the effects of genetic diversity on parasite success.
2. Material and methods
(a). Literature search
In July 2019, the literature was searched using keyword searches on Web of Knowledge, Google Scholar and PubMed, with a subset of the terms ‘host genetic diversity’, ‘low versus/and high host genetic diversity’, ‘heterogeneous versus/and homogenous host populations', ‘monoculture effect’, ‘disease spread’ and ‘parasite prevalence’ to investigate the effect of low versus high host population diversity on parasite disease impact (see electronic supplementary material, figure S1 for PRISMA flowchart [31] summarizing study collection process). We gathered data of parasite success in host populations of varying genetic diversity. We define ‘parasite success’ as any measure of a parasite's ability to proliferate within a host population reported in a given study. As parasite presence within a host population is measured differently across studies, the following terms were included as measurements of parasite success: parasite load, parasite virulence, parasite abundance, host mortality rate, viral concentrations, viral load, infection rate and infection intensity. We also checked reference lists for other potential papers. Studies were also searched for and extracted from review papers.
Papers were included in this study if they met the following inclusion criteria:
-
(i)
The study was published in a peer-reviewed academic journal.
-
(ii)
The study collected parasite success data from two distinct comparable host population groups with any measured difference in diversity, such as low versus high genetic diversity, inbred versus outbred, and monoculture versus polyculture.
-
(iii)
In the study, both host population groups contained the same species.
-
(iv)
The study measured genetic diversity at the host population level and not community diversity or individual-level genetic heterozygosity.
-
(v)
The study was not conducted in an agricultural system.
-
(vi)
The study did not interfere with parasite or host life cycle, as in passaging manipulations.
We excluded agricultural studies as a recent meta-analysis had already demonstrated the benefits of intraspecific diversity to crop yields (and thus host fitness) in the presence of infectious disease [7].
(b). Statistical analysis
We calculated Hedges's g from studies using the method described in Hedges [32]. This is a standard and widely used method of calculating effect sizes in meta-analyses which takes into account small sample sizes [33,34]. To calculate effect size g, the mean parasite measurements and their standard deviation for each treatment were extracted in the order of low host population diversity and high host population diversity. We extracted data from either paper figures, reported statistics in the text, or raw data received from authors. Where means and standard deviations in each group were not available (2 out of 23 studies), t-values and degrees of freedom were extracted.
We calculated the standard mean differences using the escalc function in the package metafor in R v. 3.6.0 (R Development Core Team) before performing a nested random mixed effects meta-analysis model using the rma.mv function. We chose this model to account for the fact that we collected several effect sizes per study, where some studies shared the same host species, which has the potential for pseudo-replication and phylogenetic non-independence. Estimates of effect size g were extracted from the model. We first tested for an overall relationship between host population genetic diversity and parasite success using the entire dataset. We then tested whether the magnitude of the relationship was dependent on the following moderator variables: study setting, parasite success measure, host reproduction, parasite functional group, parasite's host range, parasite diversity and ability of parasite to cause host death (see electronic supplementary material, table S1 for variable definitions). The measure of heterogeneity of moderator variables was reported as Cochran's Q test, where Q is the weighted sum of squares about the fixed effect estimate between subgroups [35].
We tested for an effect of both study setting (field or laboratory-based environments) and parasite success measure on the relationship between host genetic diversity and parasite success. For the latter, we separated measures into three groups based on those used in studies included in the meta-analysis: parasite prevalence, parasite load and host mortality (electronic supplementary material, table S1). Studies looking at overall parasite presence in a host population were placed under the category ‘parasite prevalence’. Where measures of parasite propagules per host were taken, studies were placed under ‘parasite load’. Measures of mortality within a population were placed under ‘host mortality’. In order to incorporate studies publishing survival data, measures of host mortality were taken as the inverse of published survival measures.
We then focused on the impact of host and parasite biological traits on variation in the magnitude and direction of effect sizes. We first considered host reproductive mode, given sexual and asexual strategies can generate disparate levels of population genetic diversity. However, one study was placed under a separate reproduction group as the host (Daphnia magna) had undergone both sexual and asexual reproduction in the study. Second, we looked at infection by parasite functional group (micro- or macroparasites) as the former tends to be associated with higher mortality [36], and third, the parasite's host range (1 host species or greater than 1 host species), as this factor has been shown to have an impact in crop studies [37,38] due to the reduced genetic specificity between hosts and multi-host parasites. Fourth, we separated studies into three categories—one genotype of one parasite species (1 genotype), multiple parasite genotypes of one parasite species (greater than 1 genotypes), and multiple parasite species (greater than 1 species)—to determine whether the diversity–disease relationship was dependent on parasite diversity. Higher levels of parasite diversity might increase the pool of susceptible hosts in a diverse population. Lastly, we tested whether effect sizes were dependent on the parasite's ability to cause host death. Compared to less harmful parasites, virulent parasites could select for greater levels and variation of resistance in the host population.
(c). Assessing for potential publication bias
Studies that report larger effects are more likely to get published in comparison to studies reporting smaller effects [34]. To check for publication bias, we visualized the spread of our effect sizes by creating a funnel plot (electronic supplementary material, figure S2). We then performed a fail-safe n analysis to calculate the number of additional studies needed to reduce the significance level of the weighted average effect size [39].
3. Results
We found 32 unique host–parasite interactions in 23 papers containing data that followed the inclusion criteria. Papers often included results from multiple experiments or exposures to multiple parasite species. A total of 67 effect sizes were retrieved from this dataset, covering a diverse range of host and parasite species (table 1).
Table 1.
Summary of the literature on the effect of host population genetic diversity on measures of parasite success across host–parasite systems.
| source paper | paper number | host | measure of host diversity | parasite | host range (ref.) | parasite type | infection measure | data source | data extracted | n effect sizes |
|---|---|---|---|---|---|---|---|---|---|---|
| Altermatt & Ebert [40] | 1 | Daphnia magna | 1 versus 10 genotypes | Octosporea bayeri | 1 host species [40] | fungus | parasite load | fig. 2, raw data | mean ± s.d. | 2 |
| Baer & Schmid-Hempel [23] | 2 | bumblebee (B. terrestris) | queens inseminated with sperm from 1 versus 2 or 4 males | Crithidia bombi, Nosema bombi | >1 host species [66] | protozoa, fungus | parasite load | fig.ure 1, raw data | mean ± s.e. | 4 |
| Baer & Schmid-Hempel [24] | 3 | bumblebee (B. terrestris) | queens inseminated with sperm from 1 versus 2 or 4 males | Crithidia bombi | >1 host species [66] | protozoa | parasite load, parasite prevalence | fig. 1, raw data | mean ± s.d. | 4 |
| Baer & Schmid-Hempel [25] | 4 | bumblebee (B. terrestris) | queens inseminated with sperm from 1 versus 2 or 4 males | Crithidia bombi | >1 host species [66] | protozoa | parasite load, parasite prevalence | raw data | mean ± s.d. | 4 |
| Calleri et al. [42] | 5 | termite (Zootermopsis angusticollis) | inbred versus outbred | Metarhizium anisopliae | >1 host species [62] | fungus | parasite load | in text | mean ± s.d. | 1 |
| Desai & Currie [29] | 6 | honeybee (A. mellifera L.) | queens inseminated with sperm from 1 versus 12 drones |
Varroa destructor, deformed wing virus, black queen cell virus, Israeli acute paralysis virus |
>1 host species [57,69,70] | mite, virus, virus, virus |
parasite load, host mortality, parasite prevalence |
figs. 1, 2, 4, 5, 7 and 8 | mean ± s.e. | 11 |
| Ganz & Ebert [47] | 7 | Daphnia magna | 1 versus 10 genotypes |
Glugoides intestinalis, Ordospora colligate, Microsporidium sp. (undescribed species) |
1 host species [79] and >1 host species [79] | fungus, fungus, fungus |
parasite prevalence | fig. 2 | mean ± s.e. | 3 |
| Hale & Briskie [49] | 8 | New Zealand robin (Petroica australis) | bottleneck versus source population | hippoboscid flies (Ornithomya spp. and Ornithoica spp.), feather mite |
>1 host species [67] | fly, mite |
parasite load | fig. 1 | mean ± s.d. | 2 |
| Hughes & Boomsma [51] | 9 | ant (Acromyrmex echinatior) | 1 patriline versus 3 patrilines | Metarhizium anisopliae (strain KVL 02–73) | >1 host species [62] | fungus | host mortality | fig. 4 | mean ± s.e. | 2 |
| Liersch and Schmid-Hempel [52] | 10 | bumblebee (B. terrestris) | full sister workers versus mixed workers |
Crithidia bombi, Nosema bombi, Apicystis (Mattesia) bombi |
>1 host species [66,76] | protozoa, fungus, protozoa |
parasite prevalence, parasite load |
fig. 1 | mean + CI | 2 |
| Manlik et al. [54] | 11 | bumblebee (B. terrestris) | island versus land population | Nosema bombi | >1 host species [66] | fungus | parasite prevalence | in text | mean ± s.e. | 1 |
| Pearman & Garner [55] | 12 | italian agile frog (Rana latastei) | low versus high population genetic variability | Ranavirus (frog virus 3) | >1 host species [55] | virus | host mortality | fig. 2, raw data | mean ± s.d. | 3 |
| Reber et al. [27] | 13 | ant (Formica selysi) | monogynous versus polygynous colonies | Metarhizium anisopliae | >1 host species [62] | fungus | host mortality | figs. 1 and 2 | mean ± s.e. | 3 |
| Schmidt et al. [28] | 14 | ant (Monomorium pharaonis) | inbred versus mixed colonies | Beauveria bassiana | >1 host species [62] | fungus | host mortality | fig. 3 | mean + CI | 3 |
| Seeley & Tarpy [56] | 15 | honeybee (A. mellifera L.) | queens inseminated with sperm from 1 versus 10 drones | American foulbrood (Paenibacillus larvae) | >1 host species [80] | bacteria | parasite prevalence | fig. 2, raw data | mean ± s.d. | 2 |
| Shykoff & Schmid-Hempel [58] | 16 | bumblebee (B. terrestris) | 1 genotype versus 3 genotypes | Crithidia bombi | >1 host species [66] | protozoa | parasite prevalence | fig. 2 | t-value | 2 |
| Smallbone et al. [59] | 17 | guppy (Poecilia reticulata) | inbred versus outbred | Gyrodactylus turnbulli (strain Gt3) | >1 host species [77] | worm | parasite load | fig. 2 | mean ± s.e. | 1 |
| Strauss et al. [61] | 18 | Daphnia dentifera | 1 genotype versus 3 genotypes | Metschnikowia bicuspidata | >1 host species [79] | fungus | parasite prevalence | fig. S1B | mean ± s.e. | 1 |
| Tarpy [63] | 19 | honeybee (A. mellifera L.) | queens inseminated with sperm from 1 versus multiple drones | chalkbrood disease (Acosphaera apis) | >1 host species [83] | fungus | parasite prevalence | fig. 2 | mean ± s.d. | 1 |
| Tarpy & Seeley [65] | 20 | honeybee (A. mellifera L.) | queens inseminated with sperm from 1 versus 10 drones | sacbrood (Iflavirus genus), chalkbrood disease (Acosphaera apis), European foulbrood (Melissococcus plutonius), American foulbrood (Paenibacillus larvae) |
>1 host species [78,80,83,84] | virus, fungus, bacteria, bacteria |
parasite prevalence | in text | t-value | 4 |
| van Houte et al. [68] | 21 |
Pseudomonas aeruginosa, Streptococcus thermophilus |
1 genotype versus 6, 8, 12, 24, and 48 genotypes and 1 genotype versus 44 genotypes | bacteriophage (DMS3), bacteriophage (2972) |
1 host species [81,82] | virus, virus |
parasite prevalence | fig. 2, raw data | mean ± s.d. | 5 |
| Wargo et al. [71] | 22 | rainbow trout (Oncorhynchus mykiss) | inbred versus outbred | infectious haematopoietic necrosis virus (IHNV) isolates: 220 : 90 (HV), WRAC 039-82 (LV), FF020-91 (B), FF030-91(C) |
1 host species [71] | virus | parasite prevalence | fig. 2, raw data |
mean ± s.e. | 4 |
| Whiteman et al. [26] | 23 | Galapagos hawk (Buteo galapagoensis) | inbred versus outbred |
Colpocephalum turbinatum, Degeerialla regalis |
>1 host species [48] | louse, louse |
parasite load | fig. 2, raw data | mean ± s.d. | 2 |
After the construction of a funnel plot, we find no indication of a publication bias in this meta-analysis dataset, with the majority of points falling symmetrically within the plot (electronic supplementary material, figure S1). The unusual shape of the plot can be explained by the fact that small sample sizes were predominantly found in laboratory studies, whereas large sample sizes were predominantly found in field studies. Consequently, studies with large sample sizes had higher errors than those with small sample sizes explaining the shape of the plot (we highlight this by colourising the plot by study setting). Rosenberg's fail-safe n analysis showed that an additional 604 studies would need to be added to reduce the significance level of this meta-analysis.
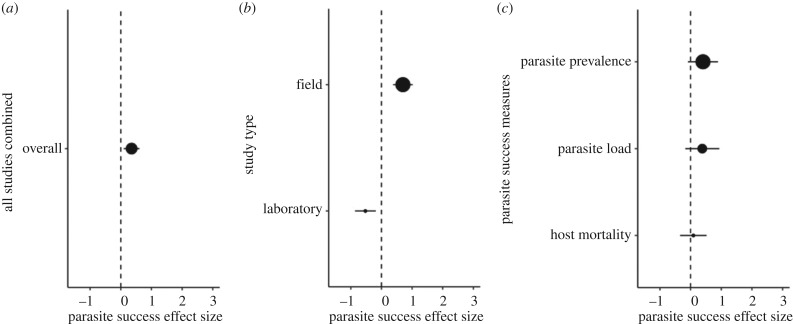
Our results are consistent with the hypothesis that low host genetic diversity results in higher parasite success (g = 0.3527, p < 0.0001; figure 1a). We found that the effect size is influenced by study setting (Q = 9.2111, d.f. = 1, p = 0.0024; figure 1b), where the magnitude of the effect size is significantly greater for field studies (g = 0.7003) in comparison to laboratory studies (g = −0.5249). Parasite success measures used in the studies do not significantly influence the effect size (Q = 2.6526, d.f. = 2, p = 0.2655; figure 1c).
Figure 1.
Impact of study setting on the effect of host genetic diversity on parasite success. Positive values indicate that low host genetic diversity has an impact on parasite success (i.e. a negative association between genetic diversity and parasite success). Negative values represent the opposite relationship. At an effect size of zero (dashed line), there is no relationship between host genetic diversity and parasite success. (a) Overall effect size (n = 67). (b) Moderator analysis of study type between field (n = 36) and laboratory (n = 31) studies. (c) Moderator analysis of parasite success measures between parasite load (n = 19), parasite prevalence (n = 35) and host mortality (n = 13). The size of the dot corresponds to the sample size. Effect sizes are shown with 95% confidence intervals.
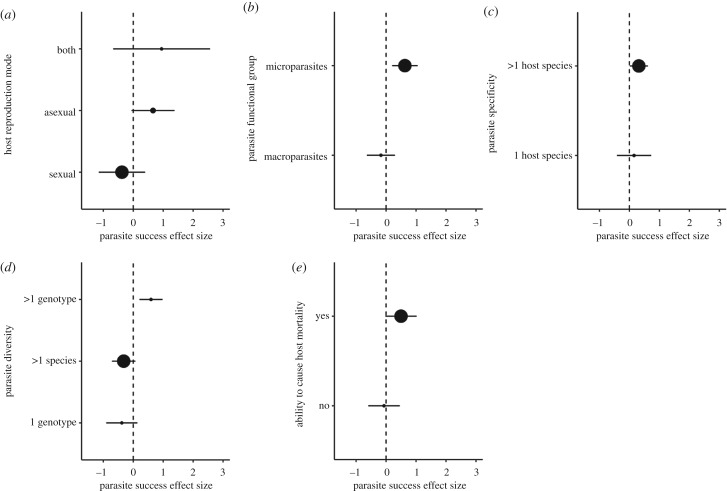
We found no evidence of an effect of host reproduction on the direction or magnitude of the effect size (Q = 4.0711, d.f. = 2, p = 0.1306; figure 2a), even when we excluded the Daphnia study by Altermatt & Ebert [40] (Q = 0.9147, d.f. = 1, p = 0.3389). Conversely, we found that the effect size was dependent on parasite functional group (Q = 8.3621, d.f. = 1, p = 0.0038, figure 2b). The success of microparasites (g = 0.6277), and not macroparasites (g = −0.1725), was limited by high host population genetic diversity. Neither the direction nor magnitude of the effect size was influenced by host range (Q = 0.2864, d.f. = 1, p = 0.5925; figure 2c), parasite diversity (Q = 3.1047, d.f. = 2, p = 0.2118; figure 2d) or whether parasites caused host mortality (Q = 3.5504, d.f. = 1, p = 0.0595; figure 2e).
Figure 2.
Impact of host and parasite characteristics on the effect of host genetic diversity on parasite success. Positive values indicate that low host genetic diversity has an impact on parasite success (i.e. a negative association between genetic diversity and parasite success). Negative values represent the opposite relationship. The dashed line (effect size of zero) represents no relationship between host genetic diversity and disease spread. Moderator analysis of (a) host reproduction mode: asexual (n = 5), both (n = 2) and sexual (n = 60) effect sizes, (b) parasite functional group between microparasite (n = 57) and macroparasite (n = 10) effect sizes, (c) host range between 1 ‘host species’ (n = 13) and greater than 1 ‘host species’ (n = 54) effect sizes, (d) parasite diversity, greater than 1 genotype (n = 15), greater than 1 species (n = 37) and 1 genotype (n = 15) effect sizes, and (e) the ability of a parasite to cause host death, displayed as yes (n = 57) and no (n = 10) effect sizes. The size of the dot corresponds to the sample size. Effect sizes are shown with 95% confidence intervals.
4. Discussion
Our meta-analysis shows that host population genetic diversity reduces parasite success across multiple natural systems. In particular, we find that host population genetic diversity is effective at limiting microparasite infection success, with little to no effect on the macroparasites tested, and the protection is stronger when measured in the field. Our findings additionally highlight the potential damage that emerging infectious diseases may have on genetically homogeneous host populations.
The parasites included in our meta-analysis were highly variable in terms of their host range. However, we found no evidence that a parasite's host range affected its success in host populations of low or high genetic diversity. Indeed, we see evidence of resistance in more diverse populations involving highly specialized interactions [40,68,71], in broad-spectrum interactions at the genotypic level [55] and in those that cross host species boundaries [25,26,72]. That host range is not a factor here is in contrast with those results found in crop studies. For example, in rusts and powdery mildews, disease severity is driven by a pathogen's host specificity [6]. The mirroring of parasite virulence genes to host resistance genes means that crop mixtures need to contain both susceptible and resistant cultivars to avoid a monoculture effect. When there is a lack of host specificity, mixed cultivar populations are just as susceptible as monocultures. For example, mixed cultivar populations have been observed to be slightly more susceptible to infection [37] or completely susceptible [38] in comparison to monocultures to the fungal pathogen Mycosphaerella graminicola. These findings suggest that the threat to crops from generalist parasites is greater than specialist parasites.
Given that host range did not influence whether parasite success was reduced by host genetic diversity, it is possible that novel parasites, just as well-adapted parasites, could have high success in host populations with low genetic diversity. Essentially, homogeneous populations could be vulnerable to outbreaks with spill-over or emerging infectious diseases which are less likely to be host specific [73], but for which there is clearly genetic variation for resistance. The resistance to emerging parasites in these cases could be due to historical contact or similar mechanisms of infection applied by parasites with an evolutionary history to the host [8]. Nevertheless, this result is concerning from a conservation perspective as global climate change has the potential to reduce within-species genetic diversity [74] and alter host population ranges [9,41]. Natural movement of individuals between populations has always served to bolster host diversity [9], and introducing new genotypes is an approach applied by conservation biologists to improve population viability [14]. While adding individuals to a population could increase diversity and reduce inbreeding [43], a risk may be that new individuals, new species and changes in ecological opportunities bring in new parasites to the population [44,45]. There is potential here for an increased overlap between host populations with low genetic diversity and novel infections. Given that we found a stronger effect in field studies, these consequences are of real concern.
The difference in parasite success between diverse and homogeneous host populations was more pronounced in field studies, compared to laboratory studies, despite the additional environmental noise data collection in nature might involve. One reason could be that less diverse populations in the wild are more susceptible to infection than they are in the laboratory for reasons unrelated to genetic diversity. Hosts on islands as well as social insects, such as bees [65], ants [51] and termites [42], live in tight proximities to each other making parasite transmission easier in homogeneous populations. The stronger effect in field studies highlights the importance of the maintenance of diversity in natural populations.
In our meta-analysis, the success of macroparasites was not impeded by genetic heterogeneity in host populations. The macroparasites in the studies included herein were all ectoparasites, and their biology may explain our result. Ectoparasite transmission is often dependent on host-to-host contact [46,48], and thus host density is probably a critical factor in parasite success [46]. Host density may play a more important role than host genetic diversity here such that similarly aggregated populations varying in diversity might be equally susceptible to infection. It has been shown that the clustering of captive animal populations restricted by movement or wild animal populations restricted by ranges are highly vulnerable to ectoparasites [44,50]. Moreover, host social behaviours, such as grooming [29] or preening [26], can reduce ectoparasite success. In fact, in populations where social grooming is correlated with relatedness, ectoparasite load is dramatically reduced in highly related individuals [53]. Taken together, host diversity on its own does not always explain a reduction in parasite success, particularly in the case of ectoparasites.
Understanding the impact of host population genetic diversity on parasite infection outside of agricultural systems is crucial because of anthropogenic threats to the diversity of wild populations. This meta-analysis reveals that the susceptibility conferred by low host genetic diversity is a widespread phenomenon in nature, with microparasites most likely to encounter resistance in diverse host populations. Indeed, these broad patterns show that genetic diversity is a robust weapon against infection, similar to the effects of species biodiversity [60]. Our findings suggest that further erosion of within-species genetic diversity could drive outbreaks of both coevolving and emerging infectious diseases. Conservation efforts should focus on preserving population genetic diversity in vulnerable populations to improve their ability to fight off infections.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We are grateful to P. B. Pearman, A. R. Wargo, P. Schmid-Hempel, N. K. Whiteman, T. D. Seeley, D. R. Tarpy, F. Altermatt, S. van Houte and E. Westra for sharing their raw data with us. We also thank C. M. Lively for comments on our manuscript.
Data accessibility
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.c856930 [75].
Authors' contributions
A.K.E.E. and K.C.K. conceived and designed the study. A.K.E.E. gathered the data and performed the statistical analysis with C.R.-M. A.K.E.E. and K.C.K. wrote the paper.
Competing interests
The authors have no competing interests.
Funding
A.K.E.E. acknowledges funding from Natural Environment Research Council (grant no. NE/L002612/1). K.C.K. thanks funding from the Leverhulme Trust (grant no. RPG-2015-165) and European Research Council (COEVOPRO 802242).
References
- 1.Haldane JBS. 1949. Suggestions as to quantitative measurement of rates of evolution. Evolution 3, 51–56. ( 10.1111/j.1558-5646.1949.tb00004.x) [DOI] [PubMed] [Google Scholar]
- 2.Agrawal A, Lively CM. 2002. Infection genetics: gene-for-gene versus matching-alleles models and all points in between. Evol. Ecol. Res. 4, 79–90. [Google Scholar]
- 3.Anderson RM, May RM. 1982. Coevolution of hosts and parasites. Parasitology 85, 411 ( 10.1017/S0031182000055360) [DOI] [PubMed] [Google Scholar]
- 4.Elton CS. 1958. The ecology of invasions by animals and plants. Chicago, IL: University of Chicago Press. [Google Scholar]
- 5.Smithson JB, Lenné JM. 1996. Varietal mixtures: a viable strategy for sustainable productivity in subsistence agriculture. Ann. Appl. Biol. 128, 127–158. ( 10.1111/j.1744-7348.1996.tb07096.x) [DOI] [Google Scholar]
- 6.Mundt CC. 2002. Use of multiline cultivars and cultivar mixtures for disease management. Annu. Rev. Phytopathol. 40, 381–410. ( 10.1146/annurev.phyto.40.011402.113723) [DOI] [PubMed] [Google Scholar]
- 7.Reiss ER, Drinkwater LE. 2018. Cultivar mixtures: a meta-analysis of the effect of intraspecific diversity on crop yield. Ecol. Appl. 28, 62–77. ( 10.1002/eap.1629) [DOI] [PubMed] [Google Scholar]
- 8.Altizer S, Harvell D, Friedle E. 2003. Rapid evolutionary dynamics and disease threats to biodiversity. Trends Ecol. Evol. 18, 589–596. ( 10.1016/j.tree.2003.08.013) [DOI] [Google Scholar]
- 9.Pauls SU, Nowak C, Bálint M, Pfenninger M. 2013. The impact of global climate change on genetic diversity within populations and species. Mol. Ecol. 22, 925–946. ( 10.1111/mec.12152) [DOI] [PubMed] [Google Scholar]
- 10.Vellend M, et al. 2017. Estimates of local biodiversity change over time stand up to scrutiny. Ecology 98, 583–590. ( 10.1002/ecy.1660) [DOI] [PubMed] [Google Scholar]
- 11.McGill BJ, Dornelas M, Gotelli NJ, Magurran AE. 2015. Fifteen forms of biodiversity trend in the Anthropocene. Trends Ecol. Evol. 30, 104–113. ( 10.1016/j.tree.2014.11.006) [DOI] [PubMed] [Google Scholar]
- 12.Frankham R, Ballou JD, Ralls K, Eldridge M, Dudash MR, Fenster CB, Lacy RC, Sunnucks P. 2017. Genetic management of fragmented animal and plant populations. Oxford, UK: Oxford University Press. [Google Scholar]
- 13.Spielman D, Brook BW, Briscoe DA, Frankham R. 2004. Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv. Genet. 5, 439–448. ( 10.1023/B:COGE.0000041030.76598.cd) [DOI] [Google Scholar]
- 14.Charlesworth D, Willis JH. 2009. The genetics of inbreeding depression. Nat. Rev. Genet. 10, 783–796. ( 10.1038/nrg2664) [DOI] [PubMed] [Google Scholar]
- 15.Schmid-Hempel P. 1998. Parasites in social insects. Princeton, NJ: Princeton University Press. [Google Scholar]
- 16.Springbett AJ, MacKenzie K, Woolliams JA, Bishop SC. 2003. The contribution of genetic diversity to the spread of infectious diseases in livestock populations. Genetics 165, 1465–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keesing F, Holt RD, Ostfeld RS. 2006. Effects of species diversity on disease risk. Ecol. Lett. 9, 485–498. ( 10.1111/j.1461-0248.2006.00885.x) [DOI] [PubMed] [Google Scholar]
- 18.Nath M, Woolliams JA, Bishop SC. 2008. Assessment of the dynamics of microparasite infections in genetically homogeneous and heterogeneous populations using a stochastic epidemic model. J. Anim. Sci. 86, 1747–1757. ( 10.2527/jas.2007-0615) [DOI] [PubMed] [Google Scholar]
- 19.Lively CM. 2010. The effect of host genetic diversity on disease spread. Am. Nat. 175, E149–E152. ( 10.1086/652430) [DOI] [PubMed] [Google Scholar]
- 20.King KC, Lively CM. 2012. Does genetic diversity limit disease spread in natural host populations? Heredity 109, 199–203. ( 10.1038/hdy.2012.33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.May RM. 1988. Conservation and disease. Conserv. Biol. 2, 28–30. ( 10.1111/j.1523-1739.1988.tb00332.x) [DOI] [Google Scholar]
- 22.Loehle C. 1995. Social barriers to pathogen transmission in wild animal populations. Ecology 76, 326–335. ( 10.2307/1941192) [DOI] [Google Scholar]
- 23.Baer B, Schmid-Hempel P. 1999. Experimental variation in polyandry affects parasite loads and fitness in a bumble-bee. Nature 397, 151–154. ( 10.1038/16451) [DOI] [Google Scholar]
- 24.Baer B, Schmid-Hempel P. 2001. Unexpected consequences of polyandry for parasitism and fitness in the bumblebee, Bombus terrestris. Evolution 55, 1639–1643. ( 10.1111/j.0014-3820.2001.tb00683.x) [DOI] [PubMed] [Google Scholar]
- 25.Baer B, Schmid-Hempel P. 2003. Bumblebee workers from different sire groups vary in susceptibility to parasite infection. Ecol. Lett. 6, 106–110. ( 10.1046/j.1461-0248.2003.00411.x) [DOI] [Google Scholar]
- 26.Whiteman NK, Matson KD, Bollmer JL, Parker PG. 2006. Disease ecology in the Galápagos Hawk (Buteo galapagoensis): host genetic diversity, parasite load and natural antibodies. Proc. R. Soc. B 273, 797–804. ( 10.1098/rspb.2005.3396) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reber A, Castella G, Christe P, Chapuisat M. 2008. Experimentally increased group diversity improves disease resistance in an ant species. Ecol. Lett. 11, 682–689. ( 10.1111/j.1461-0248.2008.01177.x) [DOI] [PubMed] [Google Scholar]
- 28.Schmidt AM, Linksvayer TA, Boomsma JJ, Pedersen JS. 2011. No benefit in diversity? The effect of genetic variation on survival and disease resistance in a polygynous social insect. Ecol. Entomol. 36, 751–759. ( 10.1111/j.1365-2311.2011.01325.x) [DOI] [Google Scholar]
- 29.Desai SD, Currie RW. 2015. Genetic diversity within honey bee colonies affects pathogen load and relative virus levels in honey bees, Apis mellifera L. Behav. Ecol. Sociobiol. 69, 1527–1541. ( 10.1007/s00265-015-1965-2) [DOI] [Google Scholar]
- 30.Wells K, Hamede RK, Kerlin DH, Storfer A, Hohenlohe PA, Jones ME, McCallum HI. 2017. Infection of the fittest: devil facial tumour disease has greatest effect on individuals with highest reproductive output. Ecol. Lett. 20, 770–778. ( 10.1111/ele.12776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moher D, Liberati A, Tetzlaff J, Altman DG. 2009. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann. Intern. Med. 151, 264 ( 10.7326/0003-4819-151-4-200908180-00135) [DOI] [PubMed] [Google Scholar]
- 32.Hedges LV. 1981. Distribution theory for glass's estimator of effect size and related estimators. J. Educ. Stati. 6, 107–128. ( 10.3102/10769986006002107) [DOI] [Google Scholar]
- 33.Field AP, Gillett R. 2010. How to do a meta-analysis. Br. J. Math. Stat. Psychology 63, 665–694. ( 10.1348/000711010X502733) [DOI] [PubMed] [Google Scholar]
- 34.Borenstein M, Hedges LV, Higgins JPT, Rothstein HR. 2011. Introduction to meta-analysis. Chichester, UK: John Wiley & Sons. [Google Scholar]
- 35.Schwarzer G, Carpenter JR, Rücker G. 2015. Fixed effect and random effects meta-analysis. In Meta-analysis with R (eds Schwarzer G, Carpenter JR, Rücker G), pp. 21–53. Cham, Switzerland: Springer International Publishing. [Google Scholar]
- 36.Anderson RM, May RM. 1986. The invasion, persistence and spread of infectious diseases within animal and plant communities. Phil. Trans. R. Soc. Lond. B 314, 533–570. ( 10.1098/rstb.1986.0072) [DOI] [PubMed] [Google Scholar]
- 37.Gigot C, Saint-Jean S, Huber L, Maumené C, Leconte M, Kerhornou B, de Vallavieille-Pope C. 2013. Protective effects of a wheat cultivar mixture against splash-dispersed septoria tritici blotch epidemics. Plant Pathol. 62, 1011–1019. ( 10.1111/ppa.12012) [DOI] [Google Scholar]
- 38.Cowger C, Mundt CC. 2002. Effects of wheat cultivar mixtures on epidemic progression of septoria tritici blotch and pathogenicity of Mycosphaerella graminicola. Phytopathology 92, 617–623. ( 10.1094/PHYTO.2002.92.6.617) [DOI] [PubMed] [Google Scholar]
- 39.Rosenberg MS. 2005. The file-drawer problem revisited: a general weighted method for calculating fail-safe numbers in meta-analysis. Evolution 59, 464 ( 10.1111/j.0014-3820.2005.tb01004.x) [DOI] [PubMed] [Google Scholar]
- 40.Altermatt F, Ebert D. 2008. Genetic diversity of Daphnia magna populations enhances resistance to parasites. Ecol. Lett. 11, 918–928. ( 10.1111/j.1461-0248.2008.01203.x) [DOI] [PubMed] [Google Scholar]
- 41.Bálint M, Domisch S, Engelhardt CHM, Haase P, Lehrian S, Sauer J, Theissinger K, Pauls SU, Nowak C. 2011. Cryptic biodiversity loss linked to global climate change. Nat. Clim. Change. 1, 313–318. ( 10.1038/nclimate1191) [DOI] [Google Scholar]
- 42.Calleri DV, Reid EM, Rosengaus RB, Vargo EL, Traniello JFA. 2006. Inbreeding and disease resistance in a social insect: effects of heterozygosity on immunocompetence in the termite Zootermopsis angusticollis. Proc. R. Soc. B 273, 2633–2640. ( 10.1098/rspb.2006.3622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frankham R. 1995. Conservation genetics. Annu. Rev. Genet. 29, 305–327. ( 10.1146/annurev.ge.29.120195.001513) [DOI] [PubMed] [Google Scholar]
- 44.Altizer S, Bartel R, Han BA. 2011. Animal migration and infectious disease risk. Science 331, 296–302. ( 10.1126/science.1194694) [DOI] [PubMed] [Google Scholar]
- 45.Wells K, Clark NJ. 2019. Host specificity in variable environments. Trends Parasitol. 35, 452–465. ( 10.1016/j.pt.2019.04.001) [DOI] [PubMed] [Google Scholar]
- 46.Viney M, Cable J. 2011. Macroparasite life histories. Curr. Biol. 21, 767–774. ( 10.1016/j.cub.2011.07.023) [DOI] [PubMed] [Google Scholar]
- 47.Ganz H, Ebert D. 2010. Benefits of host genetic diversity for resistance to infection depend on parasite diversity. Ecology 91, 1263–1268. ( 10.1890/09-1243.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whiteman NK, Parker PG. 2004. Effects of host sociality on ectoparasite population biology. J. Parasitol. 90, 939–947. ( 10.1645/GE-310R) [DOI] [PubMed] [Google Scholar]
- 49.Hale KA, Briskie JV. 2007. Decreased immunocompetence in a severely bottlenecked population of an endemic New Zealand bird. Animal conservation 10, 2–10. ( 10.1111/j.1469-1795.2006.00059.x) [DOI] [Google Scholar]
- 50.Scott ME. 1988. The impact of infection and disease on animal populations: implications for conservation biology. Conserv. Biol. 2, 40–56. ( 10.1111/j.1523-1739.1988.tb00334.x) [DOI] [Google Scholar]
- 51.Hughes WOH, Boomsma JJ. 2004. Genetic diversity and disease resistance in leaf-cutting ant societies. Evolution 58, 1251–1260. ( 10.1111/j.0014-3820.2004.tb01704.x) [DOI] [PubMed] [Google Scholar]
- 52.Liersch S, Schmid-Hempel P. 1998. Genetic variation within social insect colonies reduces parasite load. Proc. R. Soc. B 265, 221–225. ( 10.1098/rspb.1998.0285) [DOI] [Google Scholar]
- 53.Frumhoff PC, Schneider S. 1987. The social consequences of honey bee polyandry: the effects of kinship on worker interactions within colonies. Anim. Behav. 35, 255–262. ( 10.1016/S0003-3472(87)80231-2) [DOI] [Google Scholar]
- 54.Manlik O, et al. 2017. Parasite infection of specific host genotypes relates to changes in prevalence in two natural populations of bumblebees. Infect. Genet. Evol. 56, 125–132. ( 10.1016/j.meegid.2017.11.019) [DOI] [PubMed] [Google Scholar]
- 55.Pearman PB, Garner TWJ. 2005. Susceptibility of Italian agile frog populations to an emerging strain of Ranavirus parallels population genetic diversity. Ecol. Lett. 8, 401–408. ( 10.1111/j.1461-0248.2005.00735.x) [DOI] [Google Scholar]
- 56.Seeley TD, Tarpy DR. 2007. Queen promiscuity lowers disease within honeybee colonies. Proc. R. Soc. B 274, 67–72. ( 10.1098/rspb.2006.3702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenkranz P, Aumeier P, Ziegelmann B. 2010. Biology and control of Varroa destructor. J. Invertebr. Pathol. 103, 96–119. ( 10.1016/j.jip.2009.07.016) [DOI] [PubMed] [Google Scholar]
- 58.Shykoff JA, Schmid-Hempel P. 1991. Parasites and the advantage of genetic variability within social insect colonies. Proc. R. Soc. B 243, 55–58. ( 10.1098/rspb.1991.0009) [DOI] [Google Scholar]
- 59.Smallbone W, van Oosterhout C, Cable J. 2016. The effects of inbreeding on disease susceptibility: Gyrodactylus turnbulli infection of guppies, Poecilia reticulata. Exp. Parasitol. 167, 32–37. ( 10.1016/j.exppara.2016.04.018) [DOI] [PubMed] [Google Scholar]
- 60.Civitello DJ, et al. 2015. Biodiversity inhibits parasites: broad evidence for the dilution effect. Proc. Natl Acad. Sci. USA 112, 8667–8671. ( 10.1073/pnas.1506279112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strauss AT, et al. 2017. Rapid evolution rescues hosts from competition and disease but—despite a dilution effect—increases the density of infected hosts. Proc. R. Soc. B 284 ( 10.1098/rspb.2017.1970) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyling NV, Eilenberg J. 2007. Ecology of the entomopathogenic fungi Beauveria bassiana and Metarhizium anisopliae in temperate agroecosystems: potential for conservation biological control. Biol. Control. 43, 145–155. ( 10.1016/j.biocontrol.2007.07.007) [DOI] [Google Scholar]
- 63.Tarpy DR. 2003. Genetic diversity within honeybee colonies prevents severe infections and promotes colony growth. Proc. R. Soc. B 270, 99–103. ( 10.1098/rspb.2002.2199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ebert D. 2008. Host–parasite coevolution: insights from the Daphnia–parasite model system. Curr. Opin Microbiol. 11, 290–301. ( 10.1016/j.mib.2008.05.012) [DOI] [PubMed] [Google Scholar]
- 65.Tarpy DR, Seeley TD. 2006. Lower disease infections in honeybee Apis mellifera colonies headed by polyandrous vs monandrous queens. Naturwissenschaften 93, 195–199. ( 10.1007/s00114-006-0091-4) [DOI] [PubMed] [Google Scholar]
- 66.Cordes N, Huang W-F, Strange JP, Cameron SA, Griswold TL, Lozier JD, Solter LF. 2012. Interspecific geographic distribution and variation of the pathogens Nosema bombi and Crithidia species in United States bumble bee populations. J. Invertebr. Pathol. 109, 209–216. ( 10.1016/j.jip.2011.11.005) [DOI] [PubMed] [Google Scholar]
- 67.Jovani R, Tella JL, Sol D, Ventura D. 2001. Are hippoboscid flies a major mode of transmission of feather mites? J. Parasitol. 87, 1187–1189. ( 10.1645/0022-3395(2001)087[1187:AHFAMM]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 68.van Houte S, et al. 2016. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388. ( 10.1038/nature17436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang X, He SY, Evans JD, Pettis JS, Yin GF, Chen YP. 2012. New evidence that deformed wing virus and black queen cell virus are multi-host pathogens. J. Invertebr. Pathol. 109, 156–159. ( 10.1016/j.jip.2011.09.010) [DOI] [PubMed] [Google Scholar]
- 70.Singh R, et al. 2010. RNA viruses in Hymenopteran pollinators: evidence of inter-taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS ONE 5, e14357 ( 10.1371/journal.pone.0014357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wargo AR, Kell AM, Scott RJ, Thorgaard GH, Kurath G. 2012. Analysis of host genetic diversity and viral entry as sources of between-host variation in viral load. Virus Res. 165, 71–80. ( 10.1016/j.virusres.2012.01.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hale KA, Briskie JV. 2006. Decreased immunocompetence in a severely bottlenecked population of an endemic New Zealand bird. Anim. Conserv. 10, 2–10. ( 10.1111/j.1469-1795.2006.00059.x) [DOI] [Google Scholar]
- 73.Brooks DR, Hoberg EP. 2007. How will global climate change affect parasite–host assemblages? Trends Parasitol. 23, 571–574. ( 10.1016/j.pt.2007.08.016) [DOI] [PubMed] [Google Scholar]
- 74.Hoffmann AA, Sgrò CM. 2011. Climate change and evolutionary adaptation. Nature 470, 479–485. ( 10.1038/nature09670) [DOI] [PubMed] [Google Scholar]
- 75.Ekroth AKE, Rafaluk-Mohr C, King KC. 2019. Data from: Host genetic diversity limits parasite success beyond agricultural systems: a meta-analysis Dryad Digital Repository. ( 10.5061/dryad.c856930) [DOI] [PMC free article] [PubMed]
- 76.Durrer S, Schmid-Hempel P. 1995. Parasites and the regional distribution of bumblebee species. Ecography 18, 114–122. ( 10.1111/j.1600-0587.1995.tb00331.x) [DOI] [Google Scholar]
- 77.King TA, Cable J. 2007. Experimental infections of the monogenean Gyrodactylus turnbulli indicate that it is not a strict specialist. Int. J. Parasitol. 37, 663–672. ( 10.1016/j.ijpara.2006.11.015) [DOI] [PubMed] [Google Scholar]
- 78.Li J, et al. 2019. The phylogeny and pathogenesis of sacbrood virus (SBV) infection in European honey bees, Apis mellifera. Viruses. 11, 1–17. ( 10.3390/v11010061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ebert D. 2005. Ecology, epidemiology, and evolution of parasitism in Daphnia. Bethesda, MD: National Center for Biotechnology Information. [Google Scholar]
- 80.Yoshiyama M, Kimura K. 2009. Bacteria in the gut of Japanese honeybee, Apis cerana japonica, and their antagonistic effect against Paenibacillus larvae, the causal agent of American foulbrood. J. Invertebr. Pathol. 102, 91–96. ( 10.1016/j.jip.2009.07.005) [DOI] [PubMed] [Google Scholar]
- 81.Essoh C, et al. 2015. Investigation of a large collection of Pseudomonas aeruginosa bacteriophages collected from a single environmental source in Abidjan, Côte d'Ivoire. PLoS ONE 10, e0130548 ( 10.1371/journal.pone.0130548) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duplessis M, Moineau S. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41, 325–336. ( 10.1046/j.1365-2958.2001.02521.x) [DOI] [PubMed] [Google Scholar]
- 83.Chen D, Guo R, Xiong C, Zheng Y, Hou C, Fu Z. 2018. Morphological and molecular identification of chalkbrood disease pathogen Ascosphaera apis in Apis cerana cerana. J. Apicult. Res. 57, 516–521. ( 10.1080/00218839.2018.1475943) [DOI] [Google Scholar]
- 84.Singh RB, Mohan RK, Chakravarty SK, Katna S. 2012. Characterization of Melissococcus plutonius causing European foulbrood disease in Apis cerana F. J. Apicult. Res. 51, 306–311. ( 10.3896/IBRA.1.51.4.03) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Ekroth AKE, Rafaluk-Mohr C, King KC. 2019. Data from: Host genetic diversity limits parasite success beyond agricultural systems: a meta-analysis Dryad Digital Repository. ( 10.5061/dryad.c856930) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.c856930 [75].