Abstract
Proteins comprise the largest soil N reservoir but cannot be taken up directly by microorganisms and plants due to size constraints and stabilization of proteins in organo-mineral associations. Therefore the cleavage of this high molecular weight organic N to smaller soluble compounds as amino acids is a key step in the terrestrial N cycle. In the last years two isotope pool dilution approaches have been successfully established to measure gross rates of protein depolymerization and microbial amino acid uptake in soils. However, both require laborious sample preparation and analyses, which limits sample throughput. Therefore, we here present a novel isotope pool dilution approach based on the addition of 15N-labeled amino acids to soils and subsequent concentration and 15N analysis by the oxidation of α-amino groups to NO2− and further reduction to N2O, followed by purge-and-trap isotope ratio mass spectrometry (PT-IRMS). We applied this method in mesocosm experiments with forest and meadow soils as well as with a cropland soil amended with either organic C (cellulose) or organic N (bovine serum albumin). To measure direct organic N mineralization to NH4+, the latter was captured in acid traps and analyzed by an elemental analyzer coupled to an isotope ratio mass spectrometer (EA-IRMS). Our results demonstrate that the proposed method provides fast and precise measurements of at%15N even at low amino acid concentrations, allows high sample throughput and enables parallel estimations of instantaneous organic N mineralization rates.
Keywords: Isotope pool dilution, Protein depolymerization, Free amino acids, Nitrogen mineralization
1. Introduction
The cleavage of high molecular weight (HMW) organic N to small organic N compounds such as free amino acids (FAA) and amino sugars, which can be taken up directly by soil microorganisms and plants, has been recognized as the rate limiting step in the terrestrial N cycle, also fueling the inorganic N cycle (Jan et al., 2009; Jones et al., 2009; Wanek et al., 2010; Hu et al., 2018). However, soil protein depolymerization is constrained by the pool size of soil extracellular enzymes or by protein substrate availability, or by both (Wanek et al., 2010; Hu et al., 2018). Particularly in organic farming systems, where N is mainly applied by organic fertilizers as manure or crop residues, N availability is the most important yield-limiting factor (Berry et al., 2002; Seufert et al., 2012). Equally important, depolymerization processes of high-molecular weight organic N compounds are also thought to control and limit plant N availability in natural and semi-natural forests and grasslands, thereby controlling primary production and the C sink activity in non-agricultural ecosystems worldwide (LeBauer and Treseder, 2008). To improve our understanding of N transformation processes in soils, in situ measurements of gross protein breakdown and the fate of amino acids are a useful tool providing a mechanistic understanding of soil organic N processes.
Promising approaches to fill this knowledge gap are isotope pool dilution (IPD) experiments which have already been successfully applied for inorganic N processes including organic N mineralization and nitrification (Murphy et al., 2003; Booth et al., 2005) but more rarely applied to organic N depolymerization processes (Wanek et al., 2010; Mooshammer et al., 2012; Hu et al., 2017). The IPD approach relies on the labeling of a target pool (i.e. FAA) with an isotopically enriched tracer. The applied tracer is then diluted over time due to an influx of unlabelled FAA (Fig. 1). Gross protein depolymerization can be inferred from the influx of newly produced free amino acids into the labeled pool. Microbial uptake of FAA is inferred from the efflux from the labeled pool. Gross rates of protein depolymerization and microbial amino acid uptake can be calculated from the decline in the tracer:-tracee ratio of the FAA pool over time and the pool size (Di et al., 2000). Calculations of the gross influx and efflux rates are based on the key assumptions that (i) heavy and light isotopes behave the same in soils, (ii) influx and efflux rates are constant during the measurement period, and (iii) immobilized tracers are not re-mobilized during the observation period (Kirkham and Bartholomew, 1954; Braun et al., 2018).
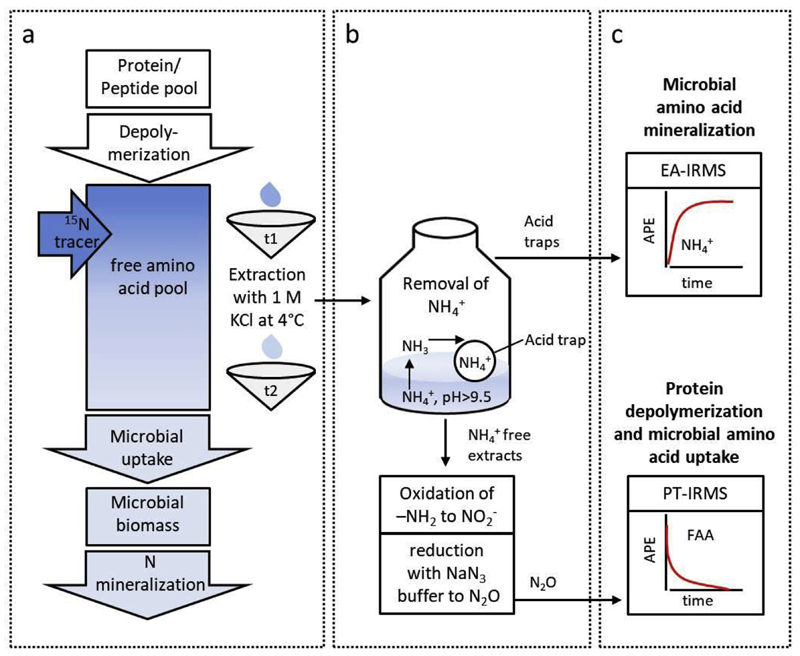
Fig. 1.
Schematic representation of the (a) isotope pool dilution experiment, (b) sample processing and (c) measurements of gross protein depolymerization, gross microbial amino acid uptake and amino acid mineralization rates. (a) The soil free amino acid pool (FAA) is labeled with highly 15N enriched amino acids. Over time, the added tracer is diluted by an influx of unlabelled FAA derived from protein depolymerization. Microbial uptake causes an efflux of FAA from the labeled FAA pool. Further, FAA-N taken up in excess is mineralized and excreted as NH4+ (N mineralization). (b) To measure the changes in isotopic composition and concentration of FAA and NH4+ over time soils are extracted at successive time points (t1 and t2) and NH4+ is removed from the soil extracts by microdiffusion. The NH4+-free extracts are further processed to N2O by oxidation of the α-amino group to NO2- and subsequent reduction of NO2- to N2O. Isotopic composition and concentration are measured by purge-and-trap isotope ratio mass spectrometry (PT-IRMS) for FAA and by an elemental analyzer coupled to an isotope ratio mass spectrometer (EA-IRMS) for NH4+. (c) The atom percent excess (APE) of the FAA pool declines due to the dilution of the tracer over time, whereas the APE of the NH4+ pool increases with time due to FAA mineralization.
Currently available protocols for isotope analysis of FAA are based on compound-specific isotope analysis by gas chromatography-mass spectrometry (GC-MS) of derivatized samples (e.g. Wanek et al., 2010; Zhang and Amelung, 1996; Kvitvang et al., 2011), capillary electrophoresis/mass spectrometry (CE/MS) of underivatized samples (Warren, 2016) or liquid chromatography/mass spectrometry (LC-MS) with hydrophilic interaction chromatography separation (HILIC; Warren, 2014a; Hu et al., 2017).
However, both protocols involve tedious sample preparation and subsequent isotope analysis of single amino acids, and one may only want the integral of FAA production and consumption rates. We therefore sought to simplify the isotope analysis for FAA, and here present a high-throughput method to measure FAA concentrations and δ15N or at%15N in soil extracts, adapted from Zhang and Altabet (2008) and Wanek et al. (2010).
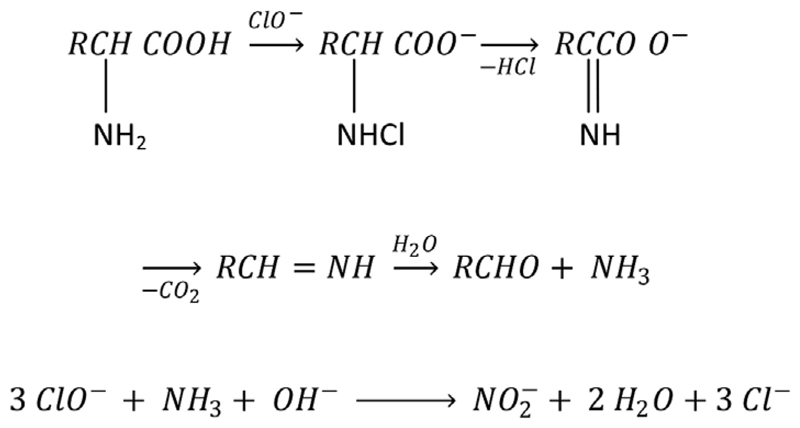
The proposed method for isotope analysis of FAA-N is based on the cleavage and oxidation of the terminal α-amino groups (α-NH2) of FAA to nitrite (NO2−) and subsequent reduction of NO2− to N2O. The N2O produced can then be measured by isotope ratio mass spectrometry coupled to a purge and trap system (PT-IRMS) via He carrier flow (Lachouani et al., 2010).
To evaluate the method proposed by Zhang and Altabet (2008) for application in soil amino acid IPD approaches, we were addressing (i) the precision of the isotopic measurements in soil extracts, (ii) high resolution time kinetics in a forest and in a meadow soil, and (iii) effects of C and N availability on organic N cycling processes in a cropland soil. For the latter we hypothesized (a) higher gross protein depolymerization rates in elevated C treatments and (b) lower rates in elevated N treatments, due to changes in the microbial C:N imbalance (Mooshammer et al., 2012). Further, we expected (c) that the proportion of mineralized amino acid 15N would be higher in the N treatments than in the C treatments, due to enhanced microbial release of excess N as NH4+ (Mooshammer et al., 2014).
2. Methods
2.1. Sampling and basic soil parameters
Mineral soil samples (0–15 cm) used for high resolution time kinetics were collected in triplicate from a temperate meadow and a mixed spruce-beech forest in the vicinity of Vienna (Austria) in spring 2016 (Table 1). All soil samples were sieved to 2 mm, adjusted to 60% water holding capacity (WHC) by addition of ultrapure water, and were allowed to equilibrate for 1 week at 20 °C. Soil pH was measured in ultra-pure water and in 10 mM CaCl2 solution (1:5 (w:v)) using an ISFET pH sensor (Sentron, Leek, The Netherlands). Soil texture was estimated from the SoilGrids project v0.5.1 (ISRIC, 2013). Soil organic C (SOC) and total soil N (TN) were determined by an Elemental analyzer (Carlo Erba 1110, CE Instruments) coupled to a DeltaPlus Isotope Ratio Mass Spectrometer (Finnigan MAT, Germany) via a Conflo III interface (Thermo Fisher, Austria). One day before starting the pool dilution experiment, an aliquot of pre-incubated soil was extracted with 1 M KCl (1:5 (w:v)) for 1 h and free amino acid concentrations were measured by the OPAME fluorescence method (Jones et al., 2002) as modified by Prommer et al. (2014). NH4+ and NO3− were determined colorimetrically in the same extracts (Hood-Nowotny et al., 2010) and dissolved organic C (DOC) and total dissolved N (TDN) were measured by a TOC/TN analyzer (TOC-VCPH/TNM-1, Shimadzu, Austria).
Table 1.
Selected initial soil physicochemical properties of the three soils (forest, meadow, cropland) sampled in Austria for15N isotope pool dilution method development.
| forest | meadow | cropland | |
|---|---|---|---|
| Latitude | 48.27880 | 48.27180 | 47.49656 |
| Longitude | 16.21322 | 16.23062 | 14.10086 |
| Elevation | 336 | 336 | 768 |
| pH (CaCl2) | 3.6 | 5.2 | 5.0 |
| Sand [%] | 37 | 34 | 44 |
| Silt [%] | 42 | 42 | 47 |
| Clay [%] | 21 | 24 | 9 |
| SOC [mg g−1] | 60.0 | 30.0 | 21.6 |
| TN [mg g−1] | 3.0 | 3.5 | 2.1 |
| DOC [μg g−1] | 252 | 107 | 70 |
| DON [μg g−1] | 21.4 | 13.5 | 14.3 |
| NH4+-N [μg g−1] | 4.07 | 1.28 | 0.40 |
| FAA-N [μg g−1] | 12.1 | 6.9 | 6.2 |
| C:N | 17.2 | 9.9 | 9.2 |
To address the effects of the relative availabilities of organic C and organic N on protein depolymerization and microbial FAA uptake in cropland soils, the presented IPD assay was applied to two treatments, manipulating the availabilities of organic C or organic N, as well as to a control treatment. Triplicate mineral soil samples (0–15 cm) were collected from a cropland in Styria (Austria) in June 2016 and sieved to < 2 mm. The soil was cropped by a mixture of vegetables and classified as Eutric Cambisol (WRB, 2015). Soil basic parameters were analyzed as described above. Cellulose or bovine serum albumin were added to the soil accounting for 33% of the respective total organic C or total N pool size. After manipulating the soil C:N ratios the samples were allowed to equilibrate for 3 day at 20 °C.
2.2. Amino acid isotope pool dilution assay
Gross rates of protein depolymerization and microbial amino acid uptake were measured by an isotope pool dilution assay (Fig. 1). Duplicate aliquots of 4 g pre-incubated soil were placed into 50 ml HDPE centrifuge tubes and 0.2 ml of a 98 at% 15N amino acid tracer solution (U-15N-98% 15N amino acid mixture from crude algae protein, Cambridge Isotope Laboratories Europe, Radeberg, Germany) were added drop-wise. Samples were vigorously shaken to promote good mixing of the tracer solution. The amount of tracer added accounted for approximately 20% of the total soil free amino acid pool as determined by the OPAME fluorescence procedure (see above) one day before starting the isotope pool dilution experiment. The applied 15N amino acid tracer contained most of the amino acids measured in soils and soil solutions, closely matching its composition (Table S2, Hu et al., 2017; Fischer et al., 2007; Warren and Taranto, 2010). All pool dilution assays were run in triplicate at 20 °C and 60% WHC. We first ran the pool dilution experiment in the forest and meadow soil to obtain high resolution time kinetics, and reactions were terminated after 15 (t1), 30 (t2), 60 (t3), 180 (t4) and 360 (t5) min in these soils. For the pool dilution experiment with the agricultural soil (C:N experiment) we omitted the last time point (360 min), since we did not observe any changes after 180 min in the previous experiment, and terminated reactions after 15 (t1), 60 (t2), 120 (t3) and 180 (t4) min by extraction with cold (4 °C) 1M KCl. Since NH4+ is also oxidized to NO2− by the proposed method, NH4+ was removed from soil extracts by microdiffusion (Fig. 1b) as described by Lachouani et al. (2010). In brief, acid traps were prepared from cellulose filter disks soaked with 2.5 M KHSO4 and enclosed in commercially available semi-permeable Teflon tape. 10 ml of soil extract were transferred to 20 mL-HDPE vials with 100 mg pre-weighed MgO, and one acid trap was added to each vial. Microdiffusion was run for 2 day at room temperature. Acid traps were dried, folded into tin capsules and N concentration and isotopic composition were measured by an Elemental Analyzer (Carlo Erba 1110, CE Instruments) coupled to a DeltaPlus Isotope Ratio Mass Spectrometer (Finnigan MAT, Germany) via a Conflo III interface (Thermo Fisher, Austria) to address the FAA mineralization process.
To measure concentrations of FAA and at% 15N of FAA in the NH4+-free extracts, we adopted the method proposed by Zhang and Altabet (2008). The proposed mechanism relies on the degradation of amino acids by ClO− in the presence of a base and Br− as catalyst and is known as Strecker degradation (Schonberg and Moubacher, 1952). Thereby the terminal α-NH2 group is cleaved as NH4+ and subsequently oxidized to NO2− (Fig. 1). In a second step NO2− is reduced to N2O by buffered NaN3 (Lachouani et al., 2010). To determine the concentration and isotopic composition of FAA the produced N2O was measured by PT-IRMS.
All chemicals were purchased from Sigma-Aldrich, Merck or Fluka in the highest available purity, and reagent solutions were prepared in ultrapure water (MilliQ, Millipore, Germany). For the cleavage and oxidation of the α-NH2 group to NO2−, aliquots of 2 mL soil extract (1 M KCl, 1:5 (w:v)) were transferred to gas-tight 12 mL glass exetainers with PTFE septa (IVA Analysentechnik, Germany), and atmospheric N2O was removed by flushing the exetainers with He for 10 min 50 μL 60 mM KBr were added as catalyst, followed by addition of 20 μL 10 M NaOH to increase solution pH to 12. As NH4+ is produced as intermediate by the cleavage/oxidation process and might be lost as NH3 at alkaline pH, exetainers were closed tightly before adding 50 μL of 30 mM NaClO with a gas-tight syringe. Samples were placed in a water bath at 50 °C for 30 min. After incubation excess ClO− was deactivated with 100 μL of Na-meta-arsenite (0.4 M, NaAsO2). Prior to starting the reduction of NO2− to N2O, samples were allowed to cool to room temperature. Then 140 μL buffered NaN3 (1:1 mix of 20 mM NaN3 with 100% acetic acid) was added with a gas-tight syringe, and samples were subsequently incubated for 30 min on a shaker at room temperature. The reaction was quenched by addition of 80 μL 6 M NaOH. Finally, sample exetainers were overpressurized by adding 12 mL He, and subsequently 12 mL of the He-sample mixture were transferred into evacuated new exetainers, to avoid blockage of the auto sampler needle by salt or condensates forming on the septa.
Measurements of N2O concentrations and isotopic compositions were done by PT-IRMS, using a Gasbench II headspace analyzer (Thermo Fisher, Bremen, Germany) with cryo-focusing unit, coupled to a Finnigan Delta V Advantage IRMS (Thermo Fisher, Bremen, Germany) as outlined in Lachouani et al. (2010).
Accounting for the variable oxidation yields of different amino acids (Zhang and Altabet, 2008), an equimolar amino acid mix (FAA-mix) containing the 20 proteinogenic amino acids was used for calibration. Yield and precision of the conversion of terminal amino groups to N2O were tested for the FAA-mix against NO2− as reference standard. The NO2− yield by oxidation of valine, alanine, leucine, glycine, lysine and the FAA-mix was examined by photometric NO2− measurements (Hood-Nowotny et al., 2010). The performance of isotope ratio measurements at low concentrations was tested by adding a spike of natural abundance FAA-mix to a 1 μM isotope calibration series (0.366–10 at %15N) to increase the FAA-concentration to 10 μmol L−1.
2.3. Data analyses and statistics
Recoveries of added tracer in the FAA and NH4+ pools were calculated as the percentage of added tracer recovered in the respective pool based on atom percent excess (APE) 15N. Atom percent excess was calculated as at%15N in the sample minus the background 15N abundance (0.366 at%15N) in non-labeled samples. Gross protein depolymerization rates (GD) and gross amino acid uptake rates (GU) were calculated from the means following the equations by Kirkham and Bartholomew (1954) and Wanek et al. (2010):
| (1) |
| (2) |
where Nt1 and Nt2 are the concentrations of FAA-N at the time points t1 and t2. Atom percent excess (APE) in FAA at the time points of termination are expressed as APEt1 and APEt2.
Limits of detection (LOD) were calculated as 3σ/S and limits of quantification (LOQ) as 9σ/S, where σ is the standard deviation of repeated blank measurements and S is the slope of the respective regression line (a(x)) of the N2O peak area against concentration.
Mean residence times (MRT) were calculated as FAA concentration divided by gross amino acid uptake. Errors of gross depolymerization, gross microbial amino acid uptake rate and MRT were propagated by Gaussian error propagation using an online tool (http://julianibus.de, 27.08.2018). Pearson correlation analysis was performed for calibration purposes and to investigate constancy of process rates. Differences between C and N treatments were analyzed by unpaired t-tests or one-way ANOVA followed by Tukey's HSD post-hoc test. Differences between treatments over incubation time were investigated by repeated measures ANOVA. Greenhouse-Geisser correction for departure from sphericity was used if sphericity was not matched (Mauchly's test). All statistical analyses were performed in R (R Development Core Team, 2008).
3. Results
3.1. Method validation
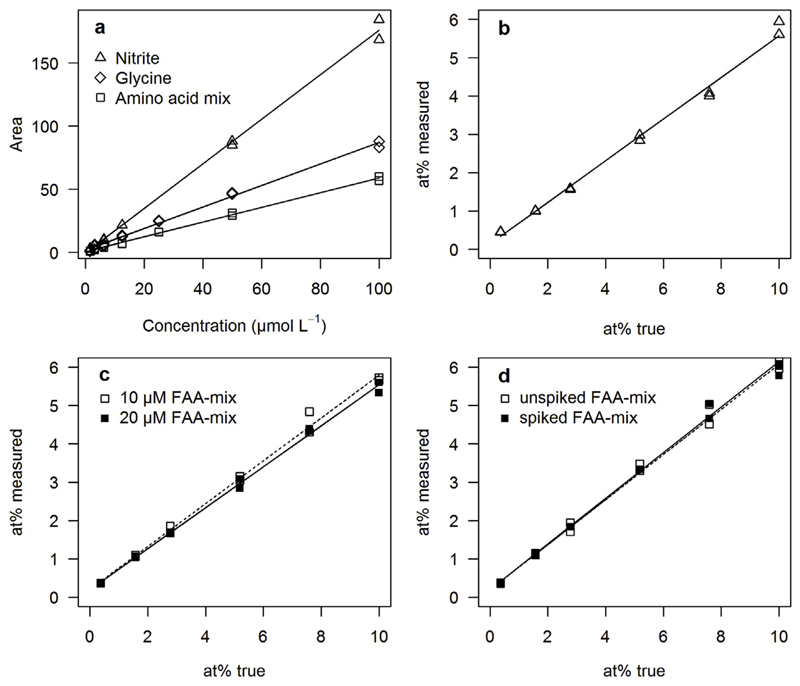
The yield of NO2− from the oxidation of α-NH2 groups ranged from 28 ± 4 to 66 ± 5% for the different FAA (Table 2), with valine > glycine > leucine > alanine ≫ lysine. Oxidation of the FAA-mix obtained a mean yield of 41 ± 6%. Yield and precision of the conversion of the α-NH2 group to N2O were tested for glycine and the FAA-mix against NO2− as a reference standard over a concentration range of 1.56–100 μmol L−1. The increment of N2O peak area with concentration was highly linear over the tested concentration range ( Fig. 2a). The limit of quantification was 0.8 μmol L−1 for glycine and 2.0 μmol L−1 for the FAA-mix (Table 3). The relationship of assigned at%15N and measured at%15N was highly linear with an r2 of 0.994 for NO2− (Figs. 2b), 0.995 for the 10 μM FAA-mix, 0.997 for the 20 μM FAA-mix (Figs. 2c) and 0.995 for the spiked 1 μM FAA-mix (Fig. 2d). The slopes of regression lines were similar for NO2− (0.51) as for the 10 μM FAA-mix (0.56), the 20 μM FAA-mix (0.53) and the spiked 1 μM FAA-mix (0.60). The coefficient of variation (CV) of isotope ratio measurements was less than 0.5% for 0.366 at%15N at 10 μmol L−1 nitrite, glycine or FAA-mix (Table 3). In the FAA-mix the CV increased at higher enrichments to 1.34% for 1.57 at%15N and 2.84% for 10 at%15N.
Table 2.
Effect of concentration of single amino acids or an amino acid mixture (FAA-mix) on the yield of NO2− measured spectrophotometrically. Coefficient of determination (r2), mean yield (± 1sd) and change in yield with increasing concentration (Slopeyield) are presented. Data are from single amino acids (n = 2) and the FAA-mix (n = 4) for concentrations ranging from 10 to 60 μM.
| Compound | r2 | % Yield |
|---|---|---|
| Glycine | 0.992 | 62 ± 8 |
| Alanine | 0.984 | 52 ± 4 |
| Leucine | 0.970 | 58 ± 6 |
| Valine | 0.992 | 66 ± 5 |
| Lysine | 0.900 | 28 ± 4 |
| FAA-mix | 0.996 | 41 ± 6 |
Fig. 2.
Concentration and isotope calibration of the method based on purge-and-trap isotope ratio mass spectrometry (PT-IRMS) method. (a) Increment of N2O peak area (Vs) with concentration of nitrite (slope = 1.76, r2 = 0.997, n = 2), glycine (slope = 0.85, r2 = 0.995, n = 2) and amino acid mix (slope = 0.58, r2 = 0.998, p < 0.001, n = 2) and isotope calibration for (b) nitrite (20 μM, slope = 0.51, r2 = 0.994, n = 2). (c) 10 μM FAA-mix (slope = 0.56, r2 = 0.995, n = 2, dashed line) and 20 μM FAA-mix (0.53, r2 = 0.9974, n = 2, solid line). (d) Isotope calibration for 10 μM FAA-mix (slope = 0.59, r2 = 0.997, n = 2, dashed line) and spiked 1 μM FAA-mix (slope = 0.60, r2 = 0.995, n = 2, solid line).
Table 3.
Determination of PT-IRMS method sensitivity, linearity and isotope precision for glycine and an amino acid mixture (FAA-mix): Limit of detection (LOD) and limit of quantification (LOQ), results of regression analyses for increment of N2O peak area with concentration from 0.78 to 50 μM (slope, y-intercept, r2, p-value), and coefficient of variation (CV) of measured at%15N (at 10 μM and 0.366 at%15N). LOD was calculated as 3σ/S and LOQ as 9σ/S, where σ is the standard deviation of repeated blank measurements and S is the slope of the respective regression line (a(x)).
| Nitrite | Glycine | FAA-mix | |
|---|---|---|---|
| LOD (μM) | 0.2 | 0.3 | 0.7 |
| LOQ (μM) | 0.6 | 0.8 | 2.0 |
| slope | 1.76 | 1.11 | 0.88 |
| y-intercept | 0.48 | 0.62 | 0.43 |
| r2 | 0.997 | 0.998 | 0.990 |
| p | < 0.001 | < 0.001 | < 0.001 |
| CV % (10 μM, 0.366 at%) | 0.21 | 0.11 | 0.43 |
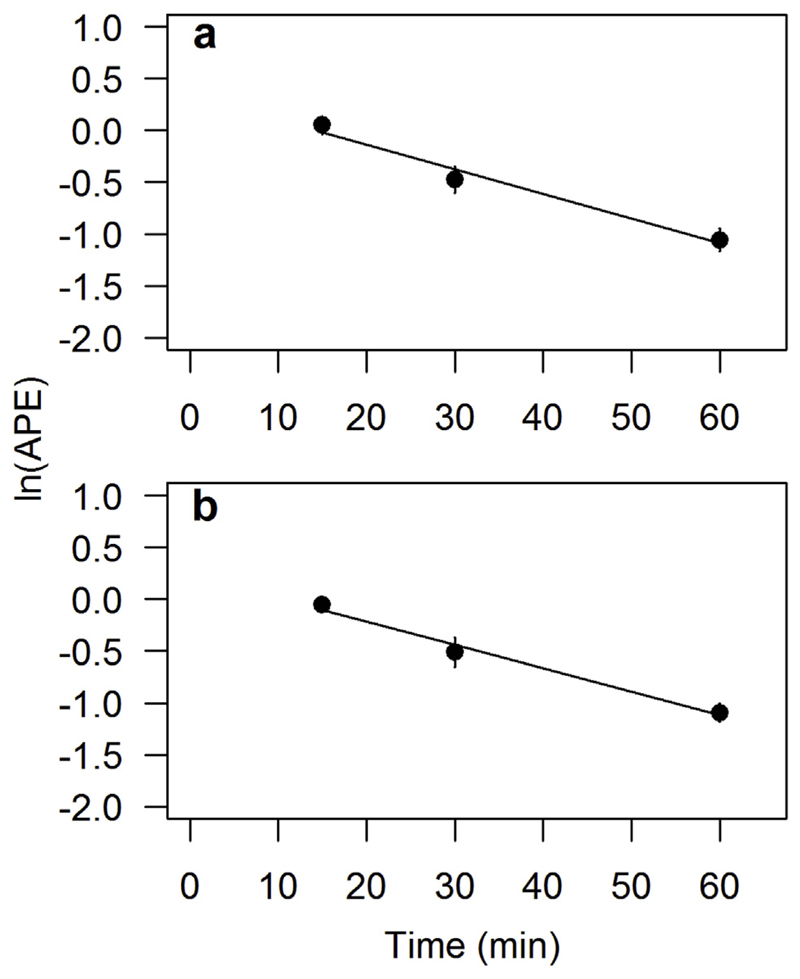
The applicability of the presented 15N amino acid pool dilution assay in soil extracts was tested in a forest and a meadow soil terminated after 15, 30, 60, 180 and 360 min. The recovery of the applied 15N amino acid tracer ranged between 60 ± 5% in meadow soils and 96 ± 22% in forest soils after 15 min incubation period and decreased rapidly with time, particularly between 15 and 60 min. During 120 and 360 min we found no significant changes in 15N-APE (Fig. S1). To test for the consistency of isotope pool dilution rates over time, we plotted the natural logarithm (LN) of APE against time. Constant IPD rates, a prerequisite for applying IPD approaches, are shown by linear decreases in LN (APE) over time over the incubation period. Linear regression analyses showed (almost) constant rates of IPD between 15 and 60 min in forest soils (r2 = 0.861, Fig. 3a) and meadow soils (r2 = 0.841, Fig. 3b). Rates of IPD were slightly higher in forest soils (k = −0.025) than in meadow soils (k = −0.023) during the respective incubation period, where k is the rate constant of isotope pool dilution (i.e. the slopes in the relationship between time and LN(APE) of amino acids). Estimated gross protein depolymerization rates in forest soils (71.3 μgN g−1 d−1) exceeded those in meadow soils (31.5 μg N g−1 d−1) by twofold, due to higher FAA contents in forest soil (Table 1). Gross microbial amino acid uptake rates were also higher in forest soils (118.4 μgN g−1 d−1) than in meadow soil (28.8 μg N g−1 d−1).
Fig. 3.
Time linearity of 15N isotope pool dilution (IPD) in the free amino acid pool over time: Linear regression analysis of the natural logarithm of atom percent excess 15N (ln(APE)) of FAA in (a) forest soils (k = −0.025, r2 = 0.861, p < 0.001) and (b) pasture soils (k = −0.023, r2 = 0.841, p < 0.001) over the incubation time 15−60 min. Constant rates of IPD are shown by linear relationsships between time and ln(APE). Data are given as means ± 1sd (n = 3). Error bars fell within confines of the symbols in some instances.
3.2. Effects of C and N amendment
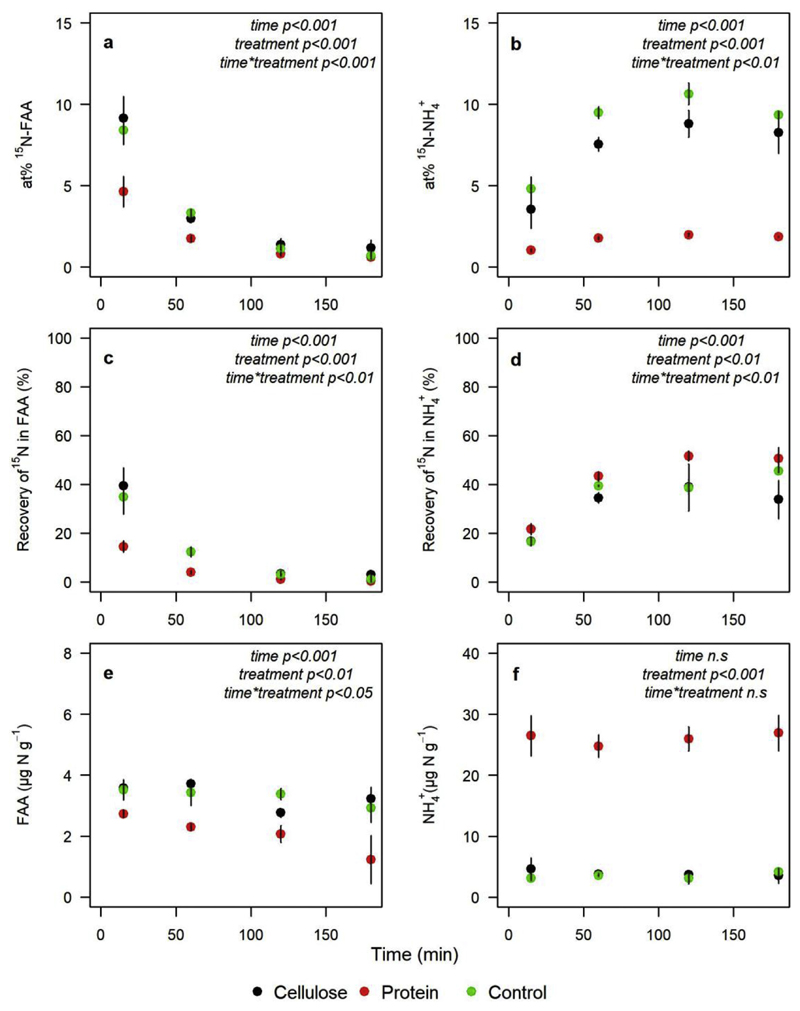
To study the response of organic N transformations to soil C and N availabilities, we conducted a mesocosm experiment with a cropland soil with addition of either organic C (cellulose) or organic N (bovine serum albumin) and with unamended soil as a control treatment. Due to the addition of cellulose the soil C:N ratio increased by 3 units in the C treatment, while C:N ratios in the protein-N treatment decreased by 1.4 units (Table 4). Gross protein depolymerization rates were higher in the C treatment than in the N treatment (p < 0.05) and the control (p < 0.05). We found no significant differences for FAA uptake rates and MRT of FAA across treatments (Table 4). Repeated measures ANOVA analyses showed significant treatment effects for at%15N-FAA, at%15N-NH4+, recovery of 15N in the FAA pool, recovery of 15N in the NH4+ pool, soil FAA concentration and NH4+ concentration (Fig. 4). Effects of incubation time and time*treatment were also significant except for the NH4+ concentration. After 15 min 15N recoveries in the FAA pool were significantly smaller in the N treatment than in the C treatment (p < 0.01) and the control (p < 0.05), and this pattern remained stable during the first 60 min (Table 4, Fig. 4c). On the contrary, 15N recoveries in the NH4+ pool were significantly higher in the N treatment than in the C treatment (p < 0.05) and the control (p < 0.05, Table 4). During the further course of the incubation we found no significant differences between the N treatment and the control, but significantly lower tracer recoveries in the NH4+ pool of the C treatments (p < 0.05, Table 4, Fig. 4d).
Table 4.
Soil C:N ratios and results of the 15N pool dilution assays of the cellulose, protein (BSA) and control treatments of an Austrian cropland soil (mean ± 1 sd). Soil C:N ratios of the cellulose and BSA treatment were calculated from the sum of C or N in the control soil and the amount of C (approx. 44% in cellulose and 53% in BSA) or N (approx. 16% in BSA) added. Gross rates and mean residence times were calculated for the incubation period from 15 to 60 min. Errors of gross depolymerization, gross immobilization and MRT were propagated using Gaussian error propagation (n = 3). Letters indicate significant differences between treatments obtained from unpaired t-tests (protein depolymerization, FAA uptake, MRT) or from one-way ANOVA and Tukey's HSD tests (tracer recoveries).
| Control | Cellulose | BSA | |
|---|---|---|---|
| C:N | 9.2 | 12.3 | 7.8 |
| Gross depolymerization [μg N g−1 d−1] | 112 ± 17b | 143 ± 20a | 91 ± 23b |
| Gross FAA uptake [μg N g−1 d−1] | 114 ± 23 | 138 ± 20 | 105 ± 23 |
| MRT of amino acids [min] | 43 ± 9 | 38 ± 7 | 34 ± 8 |
| 15N tracer recovery in FAA [% after 15 min] | 34.9 ± 7a | 39.4 ± 7.5a | 14.5 ± 2.3b |
| 15N tracer recovery in FAA [% after 60 min] | 12.3 ± 2.1a | 13.3 ± 0.9a | 3.9 ± 0.8b |
| 15N tracer recovery in NH4+ [% after 15 min] | 16.1 ± 1.1a | 16.2 ± 0.8a | 20.9 ± 2.00b |
| 15N tracer recovery in NH4+ [% after 60 min] | 39.4 ± 0.4b | 32.9 ± 3.3a | 42.3 ± 1.0b |
Fig. 4.
Changes in isotopic enrichment, tracer recovery and concentration of free amino acids (FAA) and NH4+in an Austrian cropland soil amended with cellulose or protein, or remaining untreated (control). 15N enrichment (mean at% 15N of the soil FAA (a) and NH4+ (b) pool, mean tracer recovery in FAA (c) and NH4+ (c) pool and soil FAA and NH4+ pool size over the incubation time (mean ± sd, n = 3). Significant effects from repeated-measures ANOVA are shown in italicized text. Error bars fell within confines of the symbols in some instances.
4. Discussion
4.1. Method validation
The observed yields of NO2− produced by oxidation/cleavage of α-NH2 groups was around 60% for glycine, valine, alanine and leucine (Table 2), which is about 10% less than those reported by Zhang and Altabet (2008). Yields obtained by the oxidation/cleavage of lysine's multiple amino groups were lower than those of the other tested amino acids containing only one terminal α-NH2 group (28%, Table 2) but similar to those reported previously by Zhang and Altabet (2008). However, the mechanism which hampers the oxidation of lysine's multiple amino groups by ClO- is not fully understood yet (Zhang and Altabet, 2008). The yield of the oxidation/cleavage step generally depends on the amount of ClO− added (Zhang and Altabet, 2008). The lower efficiency reported here might be triggered by the quality/activity of the used ClO−, which decreases markedly over time. Although only fresh NaClO solutions were used, the actual ClO− activity can fluctuate from batch to batch and between suppliers, and thereby might cause differences in yields between sample batches and laboratories.
Considering the variability of the NO2− yield from different amino acids, a mixed FAA calibration standard was considered more suitable when applying the PT-IRMS method to soil extracts. NO2− yields from the oxidation/cleavage of the FAA-mix were about 40% and showed a high linearity (r2 = 0.994, Table 2) across the range of typical soil FAA concentrations in 1 M KCl extracts (10–60 μmol L−1, Fig. S2). The LOQ of the FAA-mix (2 μmol L−1) obtained from PT-IRMS measurements was almost a magnitude higher than the LOQs reported by Hu et al. (2017) for 22 individual amino compounds (1–75 nM) using LC-MS, and about 6 fold higher than the LOQs for 18 individual amino acids measured by GC-MS (LOQ by PT-IRMS: 0.29 μg per injection for total free amino acids, average LOQ by GC-MS: 0.047 μg per injection for each individual amino acid; Wanek et al., 2010).
However, the precision of LC-MS and GC-MS measurements is affected by the amino acid composition, and comparisons with PT-IRMS need to account for single versus total free amino acids. The precision of the isotope calibrations was excellent even at low concentrations (10 μM, Fig. 2c), and was similar to those obtained by the LC-MS and the GC-MS for single amino (acid) compounds (Wanek et al., 2010; Hu et al., 2017). The lower slopes of the isotope calibration (about 0.5, Fig. 2b and c,d) derive from the reduction of NO2− to N2O by NaN3. The N2O produced contains only one N atom from NO2− from the terminal α-NH2 group, and one originating from NaN3 (Lachouani et al., 2010). However, alternative methods, as the LC-MS method proposed by Hu et al. (2017) using HILIC separation and the GC-MS method by Wanek et al. (2010), cannot handle high salt contents such as 1 M KCl soil extracts. For the LC-MS method, salts have to be removed by methanol extraction before cation-exchange solid phase extraction (Hu et al., 2017). The GC-MS method was optimized for litter and uses 10 mM CaSO4 extracts to circumvent high salt concentrations (Wanek et al., 2010). Alternative derivatization protocols exist that allow direct derivatization of amino acids in salt extracts and analysis by reversed phase LC-MS (Meyer et al., 2008).
In a preliminary experiment with soils from the sampling site in Styria (Austria) we tested the extraction of FAA with different concentrations of KCl (0.1 M, 0.2 M, 0.4 M, 0.6 M, 0.8 M, 1 M) and different extractants (1 M KCl, 10 mM CaSO4, water). The measured amino acid concentrations showed a sigmoidal increase with salt concentration of the extractant, which leveled off at a concentration of 1 M KCl (Fig. S1). In 10 mM CaSO4 extracts the measured amino acid concentrations accounted only for 1–38% of the concentration measured in 1 M KCl (Table S1). Water extracted between 1 and 15% of those in 1 M KCl (Table S1). Similar differences between 1 M KCl and water extracts were also observed by Jones et al. (2002). Since the IRMS method can directly process 1 M KCl soil extracts, it recovers a larger proportion of the soil total free amino acid pool than the GC-MS method, allowing precise measurements in small amounts of sample or at low soil FAA concentrations. The IRMS method allows the quantification of FAA and the respective N isotope ratios in 2 mL of soil extract or 0.4 g fresh mineral soil (1:5 (w:v)), which is 100 fold smaller than reported for the GC-MS method (Wanek et al., 2010). The enhanced precision also allows adding less tracer (only 20% of native pool size) compared to the GC-MS method. Enhancing the pool size has most likely no effect on depolymerization rates but increases FAA efflux by microbial uptake, almost linearly (Wanek et al., 2010). We compared LOD and LOQ of the FAA-mix obtained from the IRMS method to a data compilation of FAA concentrations in 1 M KCl (1:7.5 (w:v)) extracts ranging from Antarctic, tropical, and temperate to subarctic and arctic ecosystems, including grassland, forest, shrubland, tundra and cropland soils. Only 4% of all samples had FAA concentrations < 2 μM and therefore were below the LOQ (Fig. S3). These samples can still be analyzed by IPD for gross protein depolymerization, running the FAA isotope analysis after adding a spike (standard addition) of known amount of a natural 15N abundance FAA-mix to increase the FAA concentration above the LOQ, given the high precision of the PT-IRMS method (Fig. 2d).
In summary, the LC-MS method provides by far the highest precision of all currently available methods for amino acid N isotope analysis, and it offers the great potential to be applied in metabolic flux analyses using uniformly 15N- and double 15N-/13C-labelled compounds. Moreover, compound-specific isotope analysis may help reveal that separate amino acids can have various sources, informing us about the dynamics of amino acid production, and that specific amino acids may be preferentially taken up or undergo different chemical reactions or interactions with the mineral phase. However, laborious data analysis limits the sample throughput and requires extensive training of the operator (Table S3). The analysis of 50 samples requires about 12 working hours for the PT-IRMS method and 14 respectively 101 working hours for the LC-MS and the GC-MS method. Unattended procedural times add to this and increase time requirements to 96, 107 and 230 h, respectively. However, doubling the sample throughput to 100 samples puts constraints on specific parts of the procedures, particularly due to sample numbers that can be processed simultaneously in N2 drying, vacuum drying and analyzer time, which double while other steps can be easily scaled up to 100 samples, like microdiffusion, freeze-drying and sample derivatization. This gives for 100 samples about 1.5 weeks for PT-IRMS, 2.5 weeks for LC-MS and 6 weeks for GC-MS. This clearly demonstrates the trade-off between speed of analysis for total AA fluxes by PT-IRMS and detail of information that is maximized through LC-MS-based compound-specific isotope analysis of single AA fluxes. Therefore, the proposed PT-IRMS method provides a suitable alternative for analyzing larger numbers of samples with high precision. The high time resolution applied in the IPD experiments with forest and meadow soils showed constant 15N dilution rates between 15 and 60 min (Fig. 3), suggesting 15 min and 1 h as optimal incubation periods. Considering the similar isotope pool dilution rates k in both soils (i.e. the slopes in the relationship between time and LN(APE) of amino acids), but significantly higher free amino acid contents in forest soils, this can be translated into higher influx (depolymerization) rates in forest soils (Table 1).
A major limitation of isotope pool dilution experiments is that other potential sources of influx cannot be distinguished from the target process (i.e. protein depolymerization). For example, increasing soil water potential due to liquid 15N-tracer addition might trigger the release of microbial osmolytes such as amino acids and sugars into soil solutions by cell lysis or excretion from microbial biomass (Fierer and Schimel, 2003). However, in the case of soils, release of osmolytes after rewetting has been shown to be negligible (Boot et al., 2013; Kakumanu and Williams, 2014; Warren, 2014a,b; Warren, 2016). Even though addition of 200 μL tracer solution was likely not a major disturbance, as soils were already at 60% WHC, potential osmotic effects should be considered when using dried or air-dried soils. Moreover, protease inhibitor experiments can be included to distinguish between protein depolymerization and other potential amino acid sources (Wanek et al., 2010).
4.2. Effects of C and N availability
Our results on the MRT of FAA suggest rapid turnover of the soil FAA pool (Table 4), which is in accordance with previous observations in a wide range of soils covering different ecosystem and management types (Jones et al., 2005, 2009; Wanek et al., 2010; Hu et al., 2018). In contrast, turnover times of inorganic soil N pools are much longer, ranging between several hours and days for NH4+ and NO3−(Booth et al., 2005; Inselsbacher et al., 2010), indicating that direct uptake of amino acids is energetically favored over the uptake of inorganic N forms (Jones et al., 2005). The addition of cellulose or bovine serum albumin to the studied cropland soil had no effect on the MRT of FAA, but resulted in lower gross protein depolymerization rates in the N than in the C treatment (Table 4). Moreover, relatively more 15N tracer was recovered in the NH4 pool after 15 min in the N treatment than in the C treatment and in the control (Table 4), indicating that a higher proportion of the available FAA-N was rapidly mineralized and not incorporated into microbial biomass. The rapid mineralization might be triggered by the abrupt surplus of available FAA-N due to protein addition. We expect that the system equilibrated over the course of time, as indicated by decreasing mineralization rates in all treatments (Fig. 4d). However, after 180 min organic N mineralization rates of the N treatment were still slightly higher than in the control (Fig. 4d), and the 5-fold higher NH4+ concentrations in the N treatments compared to the C treatment and the control (Fig. 4f) indicate that NH4+ had already accumulated during the pre-incubation period. We suggest that due to the N surplus, soil microbes downregulated their nitrogen use efficiency (NUE), resulting in higher excretion of excess N taken up in organic form as NH4+. These findings are in accordance with Mooshammer et al. (2014), who showed a decrease of microbial NUE with decreasing microbial N limitation, from organic soils and plant litter with wide C:N ratios to mineral soils with low C:N ratios. Moreover, the scarcity of available C in mineral soils was shown to depress microbial growth and consequently microbial NH4+ immobilization, which promotes the accumulation of NH4+ in soils (Jones et al., 2004). Lower depolymerization rates in the N treatment (Table 4) indicate that soil microbes likely invested less C in the production of extracellular proteases and peptidases. As extracellular enzymes, once excreted, are not under direct metabolic control, the adjustment to changes in the C:N imbalance might be much slower than the adjustment of microbial NUE, explaining the less pronounced differences in gross depolymerization rates within the short time frame of this study.
The addition of organic C in the form of cellulose resulted in higher average protein depolymerization rates, although not significantly (Table 4). After 60 min the FAA mineralization was significantly lower than in the N treatment. We assume that the cellulose-C amendment to soils alleviated microbial C limitation. Therefore the N demand for growth and thus microbial NUE increased while FAA mineralization decreased, and simultaneously microbial C (and N) allocation to extracellular enzyme production increased to mobilize N from proteins. A larger sample size (replication) might have increased the statistical power to detect small differences between treatments, but the data obtained suggest that mineralization of organic N was more sensitive to short-term changes in the availability of C or N than extracellular protein depolymerization, as indicated by the stronger effects on 15N tracer recoveries in the NH4+ pool (Fig. 4c and d) than on gross protein depolymerization.
Our work shows that the proposed PT-IRMS method is suitable for 15N amino acid IPD and isotope tracing approaches in soils to measure gross protein depolymerization and microbial amino acid uptake. The new method outperforms other existing methods in terms of time requirement and precision, allowing high sample throughput and low 15N-FAA tracer addition rates. Moreover, parallel measurements of the tracer recovery in the NH4+ pool allow new insights into the linkage between organic and inorganic terrestrial N cycling. This new approach will help to address questions related to the controls of organic N cycling in soils, which will contribute to our knowledge on efficient fertilizer management in organic farming systems, where inorganic N inputs are low, and in (semi)natural ecosystems, where N availability constrains the terrestrial C sink activity.
Supplementary Material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.soilbio.2018.12.005.
Acknowledgements
We thank Theresa Böckle, Claudia Schneider, Josephine Pfeifer and Sebastian Schneider for their help with the C:N-experiment. We are also grateful to Judith Prommer and Yuntao Hu for providing data on the GC-MS and the LC-MS methods. This study was funded by the Austrian Science Fund (FWF, project P-28037-B22).
References
- Berry PM, Sylvester-Bradley R, Philipps L, Hatch DJ, Cuttle SP, Rayns FW, Gosling P. Is the productivity of organic farms restricted by the supply of available nitrogen? Soil Use & Management. 2002;18:248–255. [Google Scholar]
- Boot CM, Schaeffer SM, Schimel JP. Static osmolyte concentrations in microbial biomass during seasonal drought in a California grassland. Soil Biology and Biochemistry. 2013;57:356–361. [Google Scholar]
- Booth MS, Stark JM, Rastetter E. Controls on nitrogen cycling in terrestrial ecosystems: a synthetic analysis of literature data. Ecological Monographs. 2005;75:139–157. [Google Scholar]
- Braun J, Mooshammer M, Wanek W, Prommer J, Walker TWN, Rütting T, Richter A. Full 15N tracer accounting to revisit major assumptions of 15N isotope pool dilution approaches for gross nitrogen mineralization. Soil Biology and Biochemistry. 2018;117:16–26. doi: 10.1016/j.soilbio.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di H, Cameron K, McLaren R. Isotopic dilution methods to determine the gross transformation rates of nitrogen, phosphorus, and sulfur in soil: a review of the theory, methodologies, and limitations. Soil Research. 2000;38:213–230. [Google Scholar]
- Fierer N, Schimel JP. A proposed mechanism for the pulse in carbon dioxide production commonly observed following the rapid rewetting of a dry soil. Soil Science Society of America Journal. 2003;67:798–805. [Google Scholar]
- Fischer H, Meyer A, Fischer K, Kuzyakov Y. Carbohydrate and amino acid composition of dissolved organic matter leached from soil. Soil Biology and Biochemistry. 2007;39:2926–2935. [Google Scholar]
- Hood-Nowotny R, Umana NHN, Inselbacher E, Lachouani P, Wanek W. Alternative methods for measuring inorganic, organic, and total dissolved nitrogen in soil. Soil Science Society of America Journal. 2010;74:1018–1027. [Google Scholar]
- Hu Y, Zheng Q, Wanek W. Flux analysis of free amino sugars and amino acids in soils by isotope tracing with a novel liquid chromatography/high resolution mass spectrometry platform. Analytical Chemistry. 2017;89:9192–9200. doi: 10.1021/acs.analchem.7b01938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zheng Q, Zhang S, Noll L, Wanek W. Significant release and microbial utilization of amino sugars and D-amino acid enantiomers from microbial cell wall decomposition in soils. Soil Biology and Biochemistry. 2018;123:115–125. doi: 10.1016/j.soilbio.2018.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inselsbacher E, Umana NHN, Stange FC, Gorfer M, Schüller E, Ripka K, Zechmeister-Boltenstern S, Hood-Nowotny R, Strauss J, Wanek W. Short-term competition between crop plants and soil microbes for inorganic N fertilizer. Soil Biology and Biochemistry. 2010;42:360–372. [Google Scholar]
- ISRIC. SoilGrids: an Automated System for Global Soil Mapping. 2013.
- IUSS Working Group. World Soil Resources Reports No. 106. FAO; Rome: 2015. World Reference Base for Soil Resources 2014 (Update 2015), International Soil Classification System for Naming Soils and Creating Legends for Soil Maps. [Google Scholar]
- Jan MT, Roberts P, Tonheim SK, Jones DL. Protein breakdown represents a major bottleneck in nitrogen cycling in grassland soils. Soil Biology and Biochemistry. 2009;41:2272–2282. [Google Scholar]
- Jones D, Kielland K, Sinclair F, Dahlgren R, Newsham K, Farrar J, Murphy D. Soil organic nitrogen mineralization across a global latitudinal gradient. Global Biogeochemical Cycles. 2009;23:GB1016. [Google Scholar]
- Jones DL, Owen AG, Farrar JF. Simple method to enable the high resolution determination of total free amino acids in soil solutions and soil extracts. Soil Biology and Biochemistry. 2002;34:1893–1902. [Google Scholar]
- Jones DL, Shannon D, Murphy DV, Farrar J. Role of dissolved organic nitrogen DON in soil N cycling in grassland soils. Soil Biology and Biochemistry. 2004;36:749–756. [Google Scholar]
- Jones DL, Kemmitt SJ, Wright D, Cuttle SP, Bol R, Edwards AC. Rapid intrinsic rates of amino acid biodegradation in soils are unaffected by agricultural management strategy. Soil Biology and Biochemistry. 2005;37:1267–1275. [Google Scholar]
- Kakumanu ML, Williams MA. Osmolyte dynamics and microbial communities vary in response to osmotic more than matric water deficit gradients in two soils. Soil Biology and Biochemistry. 2014;79:14–24. [Google Scholar]
- Kirkham D, Bartholomew W. Equations for following nutrient transformations in soil, utilizing tracer data: I. Soil Science Society of America Journal. 1954;18:33–34. [Google Scholar]
- Kvitvang HF, Andreassen T, Adam T, Villas-Bôas SG, Bruheim P. Highly sensitive GC/MS/MS method for quantitation of amino and nonamino organic acids. Analytical Chemistry. 2011;83:2705–2711. doi: 10.1021/ac103245b. [DOI] [PubMed] [Google Scholar]
- Lachouani P, Frank AH, Wanek W. A suite of sensitive chemical methods to determine the delta15N ammonium, nitrate and total dissolved N in soil extracts. Rapid Communications in Mass Spectrometry. 2010;24:3615–3623. doi: 10.1002/rcm.4798. [DOI] [PubMed] [Google Scholar]
- LeBauer DS, Treseder KK. Nitrogen limitation of net primary productivity in terrestrial ecosystems is globally distributed. Ecology. 2008;89:371–379. doi: 10.1890/06-2057.1. [DOI] [PubMed] [Google Scholar]
- Meyer A, Fischer H, Kuzyakov Y, Fischer K. Improved RP-HPLC and anion-exchange chromatography methods for the determination of amino acids and carbohydrates in soil solutions. Journal of Plant Nutrition and Soil Science. 2008;171:917–926. [Google Scholar]
- Mooshammer M, Wanek W, Schnecker J, Wild B, Leitner S, Hofhansl F, Blöchl A, Hämmerle I, Frank AH, Fuchslueger L. Stoichiometric controls of nitrogen and phosphorus cycling in decomposing beech leaf litter. Ecology. 2012;93:770–782. doi: 10.1890/11-0721.1. [DOI] [PubMed] [Google Scholar]
- Mooshammer M, Wanek W, Hammerle I, Fuchslueger L, Hofhansl F, Knoltsch A, Schnecker J, Takriti M, Watzka M, Wild B, Keiblinger KM, et al. Adjustment of microbial nitrogen use efficiency to carbon:nitrogen imbalances regulates soil nitrogen cycling. Nature Communications. 2014;5:3694–3700. doi: 10.1038/ncomms4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D, Recous S, Stockdale E, Fillery I, Jensen L, Hatch D, Goulding K. Gross nitrogen fluxes in soil: theory, measurement and application of 15 N pool dilution techniques. Advances in Agronomy. 2003;79:69–118. [Google Scholar]
- Prommer J, Wanek W, Hofhansl F, Trojan D, Offre P, Urich T, Schleper C, Sassmann S, Kitzler B, Soja G, Hood-Nowotny R. Biochar decelerates soil organic nitrogen cycling but stimulates soil nitrification in a temperate arable field trial. PLoS One. 2014;9:e86388. doi: 10.1371/journal.pone.0086388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]
- Schonberg A, Moubacher R. The strecker degradation of α-amino acids. Chemical Reviews. 1952;50:261–277. [Google Scholar]
- Seufert V, Ramankutty N, Foley JA. Comparing the yields of organic and conventional agriculture. Nature. 2012;485:229–232. doi: 10.1038/nature11069. [DOI] [PubMed] [Google Scholar]
- Wanek W, Mooshammer M, Blöchl A, Hanreich A, Richter A. Determination of gross rates of amino acid production and immobilization in decomposing leaf litter by a novel 15N isotope pool dilution technique. Soil Biology and Biochemistry. 2010;42:1293–1302. [Google Scholar]
- Warren CR, Taranto MT. Temporal variation in pools of amino acids, inorganic and microbial N in a temperate grassland soil. Soil Biology and Biochemistry. 2010;422:353–359. [Google Scholar]
- Warren CR. Development of liquid chromatography mass spectrometry method for analysis of organic N monomers in soil. Soil Biology and Biochemistry. 2014a;78:233–242. [Google Scholar]
- Warren CR. Response of organic N monomers in a sub-alpine soil to a dry–wet cycle. Soil Biology and Biochemistry. 2014b;77:233–242. [Google Scholar]
- Warren CR. Do microbial osmolytes or extracellular depolymerisation products accumulate as soil dries? Soil Biology and Biochemistry. 2016;98:54–63. [Google Scholar]
- Zhang L, Altabet MA. Amino-group-specific natural abundance nitrogen isotope ratio analysis in amino acids. Rapid Communications in Mass Spectrometry. 2008;22:559–566. doi: 10.1002/rcm.3393. [DOI] [PubMed] [Google Scholar]
- Zhang X, Amelung W. Gas chromatographic determination of muramic acid, glucosamine, mannosamine, and galactosamine in soils. Soil Biology and Biochemistry. 1996;28:1201–1206. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.