Abstract
Proteins constitute the single largest soil organic nitrogen (SON) reservoir and its decomposition drives terrestrial N availability. Protein cleavage by extracellular enzymes is the rate limiting step in the soil organic N cycle and can be controlled by extracellular enzyme production or protein availability/stabilization in soil. Both controls can be affected by geology and land use, as well as be vulnerable to changes in soil temperature and moisture/O2. To explore major controls of soil gross protein depolymerization we sampled six soils from two soil parent materials (calcareous and silicate), where each soil type included three land uses (cropland, pasture and forest). Soil samples were subjected to three temperature treatments (5, 15, 25 °C at 60% water-holding capacity (WHC) and aerobic conditions) or three soil moisture/O2 treatments (30 and 60% WHC at 21% O2, 90% WHC at 1% O2, at 20 °C) in short-term experiments. Samples were incubated for one day in the temperature experiment and for one week in the moisture/O2 experiment. Gross protein depolymerization rates were measured by a novel 15N isotope pool dilution approach. The low temperature sensitivity of gross protein depolymerization, the lack of relationship with protease activity and strong effects of soil texture and pH demonstrate that this process is constrained by organo-mineral associations and not by soil enzyme content. This also became apparent from the inverse effects in calcareous and silicate soils caused by water saturation/O2 limitation. We highlight that the specific soil mineralogy influenced the response of gross depolymerization rates to water saturation/O2 limitation, causing (I) increasing gross depolymerization rates due to release of adsorbed proteins by reductive dissolution of Fe- and Mn-oxyhydroxides in calcareous soils and (II) decreasing gross depolymerization rates due to mobilization of coagulating and toxic Al3+ compounds in silicate soils.
Keywords: Protein depolymerization, Amino acid uptake, Organic nitrogen, Temperature, Moisture
1. Introduction
Between 50 and 90% of soil N occurs in the form of high molecular weight (HMW) peptidic N, comprising (glyco)proteins and peptide side chains in peptidoglycan (Martens and Loeffelmann, 2003), the rest deriving from amino sugar polymers (e.g. chitin and glycan strands in peptidoglycan), aromatic N compounds (e.g. nucleic acids) and to date uncharacterized organic N compounds (Schulten and Schnitzer, 1997). These HMW-organic N compounds represent the largest soil N reservoir but are not directly available for microorganisms and plants due to molecular size constraints on cellular uptake mechanisms (Schimel and Bennett, 2004). Depolymerization of these complex organic N compounds is therefore considered the rate limiting step fueling the terrestrial N cycle and allowing organisms to utilize soil organic N compounds (Schimel and Bennett, 2004; Jan et al., 2009; Jones et al., 2009; Hu et al., 2017). The breakdown of the largest fraction of HMW-organic N, proteinaceous substances, to low molecular weight organic compounds such as oligopeptides and amino acids is catalyzed by extracellular proteases and peptidases, secreted into the soil by micro-organisms and plants (Adamczyk et al., 2008; Fuka et al., 2008; Vranova et al., 2013). The products of this breakdown represent a highly dynamic and easily available pool of organic N in soils with half-life times of minutes to hours (Jones et al., 2009; Wanek et al., 2010). In decomposing litter and in soils, protein depolymerization rates by far exceed those of inorganic N processes such as organic N mineralization and nitrification i.e. by > 8 fold (Wanek et al., 2010; Prommer et al., 2014; Wild et al., 2015), showing that organic N cycling exceeds inorganic N cycling in the terrestrial N cycle. Moreover, isotope tracer studies showed that uptake of intact amino acids and small peptides contributes significantly to the N nutrition of soil microorganisms and plants (Gallet-Budynek et al., 2009; Hill et al., 2011; Farrell et al., 2012), short-cutting the soil inorganic N cycle. However, despite its importance and first insights into the process of gross protein depolymerization in soils, we still lack a thorough mechanistic understanding of major controls on this process.
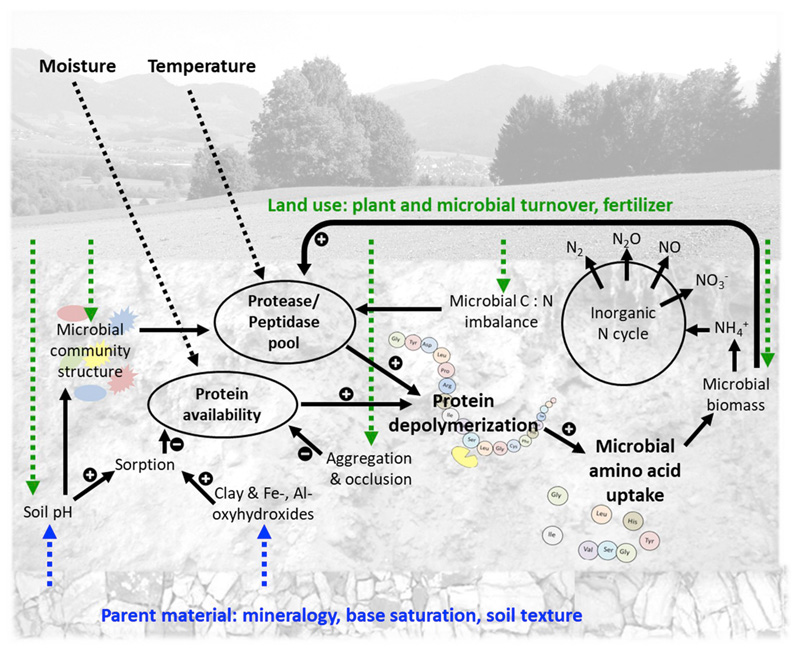
In principle enzyme-catalyzed depolymerization processes in soils can be controlled either by extracellular enzyme content (enzyme limitation) or by resource availability (substrate limitation, Wallenstein and Weintraub (2008)). Both are strongly affected by environmental conditions, such as by soil temperature and moisture controlling substrate diffusion and catalytic rates, and by soil physicochemical factors that influence protein availability and microbial community structure (Fig. 1).
Fig. 1.
Schematic of potential controls on gross protein depolymerization in soils including effects of parent material (blue), land use (green), and soil temperature and moisture/O2 (grey). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Soil extracellular enzyme content, measured as potential enzyme activities at substrate saturation and optimal pH, depends on (i) microbial community structure and activity and (ii) the microbial N and C demand, both affecting the production of extracellular enzymes, and (iii) sorption of enzymes/protein substrates in soils changing their activity and decreasing proteolytic decomposition (Quiquampoix, 2000). All these parameters are affected by land use and parent material (Fig. 1). The set of excreted proteases is shaped by the microbial community structure (Fuka et al., 2008). Soil peptidases are produced by archaea, bacteria, and fungi, belonging to ∼150 peptidase subfamilies, though the occurrence and abundance of the genes coding for secreted peptidases varies across the three microbial kingdoms (Nguyen et al., 2017). Land use is known to be an important factor influencing soil microbial community structure and bacterial to fungal biomass ratios (Fierer and Jackson, 2006; Jangid et al., 2008; Lauber et al., 2009). Moreover, land-use related N inputs by synthetic fertilizers or manure may further suppress (inorganic N) or stimulate (organic N) soil protease production (Geisseler and Horwath, 2008). The production and potential activity of extracellular proteases is therefore driven by substrate supply (Brzostek and Finzi, 2011), litter quality (Wickings et al., 2012) and consequently microbial C and N demands (Geisseler and Horwath, 2008). Moreover, soil parent material shapes soil physicochemical properties such as soil pH and texture which in turn affect microbial community structure and the excreted enzyme complement (Fierer and Jackson, 2006; Kallenbach et al., 2016; Rousk et al., 2010), but also determine the number of sorption sites for enzymes and protein substrates.
Soil protein availability depends (i) on the input of labile proteins by plant litter, microbial turnover and manure, and (ii) on stabilization mechanisms (sorption/desorption equilibria) and protein accessibility (aggregate formation), both of which are strongly affected by soil parent material and land-use. Land-use and the respective plant communities affect litter quality and quantity (Fanin and Bertrand, 2016) and thereby the labile protein input into the soil ecosystem. Microbial C limitation due to reduced litter input in cropland soils promotes organic N mineralization as microbes decompose available organic N compounds to meet their energy demands and excrete excess N as NH4+ (Mooshammer et al., 2014). This is further enhanced by tillage or other disturbances in cropland soils which promote microbial decomposition of soil organic matter (Six and Jastrow, 2002). Microbial turnover in soils also adds proteins from lysing cells (Schimel and Bennett, 2004) and turnover varies with vegetation type, soil depth and fertilizer regime (Spohn et al., 2016; Spohn and Widdig, 2017). Though proteinaceous N makes up the bulk of N in soils and litter, in soils protein availability is strongly constrained by organo-mineral associations (Kögel-Knabner et al., 2008), rendering proteins inaccessible to proteolytic attack (Quiquampoix, 2000).
Fe- and Al- oxides and phyllosilicates are assumed to be the main sorption sites of soil organic matter and to form stable organo-mineral complexes by ligand exchange or electrostatic interactions (Kaiser and Guggenberger, 2003). Both, the net charge of sorbent (mineral phase) and adsorbent (protein) are determined by soil pH, due to protonation of functional groups with decreasing pH. Adsorption of proteins on mineral surfaces is highest at soil pH close to the isoelectric point, where the net charge of a protein is zero (Quiquampoix and Ratcliffe, 1992; Gu et al., 1994). Protein sorption capacity/strength of the soil matrix therefore depends on soil texture, soil mineral composition and soil pH and thereby is strongly affected by soil parent material. Moreover, Mn-oxides, like birnessite catalyze the abiotic hydrolysis of peptide bonds and thus could either increase depolymerization rates by abiotic cleavage of proteins or decrease them by fragmentation and deactivation of proteolytic enzymes (Reardon et al., 2016). We therefore expect strong effects of parent material and land-use on the protein depolymerization process, but rigorous tests thereof are lacking.
Moreover, so far no studies have tested the effects of soil temperature and soil moisture on gross protein depolymerization rates. With increasing soil temperature, catalytic rates of extracellular peptidases increase exponentially with Q10 values of ∼2, while temperature effects are weaker on diffusion, sorption and desorption processes of soil organic matter and protein substrates (Q10 ∼1, Conant et al., 2011). Recent studies have shown that extracellular peptidases are less temperature sensitive than C or P cycling enzymes, resulting in a decoupling of protein depolymerization from C and P acquisition at increasing temperatures (Brzostek and Finzi, 2012; Steinweg et al., 2018). Depending on enzyme- or substrate-limitation of gross protein depolymerization, we therefore would expect rates to increase with Q10 reflecting extracellular enzyme responses or to remain unaltered, respectively (Steinweg et al., 2012).
Soil moisture is also a strong driver of in situ decomposition processes. Diffusion of catalytic enzymes and substrates throughout soil is determined by the soil water content (Moldrup et al., 2001) and modified by soil texture. Soil water content also directly affects soil microbial life. At low soil water content, soil microorganisms are exposed to osmotic stress and consequently reduce their internal water potential by accumulation of osmolytes like amino compounds (Csonka, 1989; Witteveen and Visser, 1995; Schimel et al., 2007). In contrast, at high soil water contents, close to field capacity, oxygen becomes a limiting factor causing a shift from aerobic to anaerobic respiration and fermentation. Since, microbial activity is constrained by the lower energetic efficiency of anaerobic respiration and fermentation compared to aerobic respiration (Ponnamperuma, 1972; Picek et al., 2000), this leads to reduced microbial SOM decomposition in water-logged soils in the long term (Kögel-Knabner et al., 2010; Schädel et al., 2016). However, in the short term reductive dissolution of Fe- and Mn- oxyhydroxides causes a release of stabilized SOM and thereby increases decomposition (Kögel-Knabner et al., 2010).
Here we present the first comprehensive study on the above cited potential controls of the protein depolymerization process, based on two incubation experiments, assessing temperature and soil moisture/O2 effects on gross protein depolymerization and amino acid uptake rates in soils differing in soil parent material (calcareous versus silicate) and land use (forests, pastures and croplands). To assess short-term temperature and moisture effects on gross protein depolymerization (and amino acid uptake) rates, we conducted short-term incubation experiments at three levels of temperature (5, 15 and 25 °C at 60% WHC) and moisture (30 and 60% WHC at 21% O2, and 90% WHC at 21% O2, all at 20 °C). We hypothesized, that (i) gross protein depolymerization is not enzyme but mainly substrate limited and that protein availability and protein sorption is strongly affected by soil parent material and land use. Second (ii) if protein depolymerization were enzyme limited, higher temperatures will induce higher gross rates due to increased enzyme activity while if substrate-limitation prevails, the constraint by substrate availability will suppress the temperature sensitivity of protein depolymerization. Finally, we hypothesized that (iii) an increase in soil moisture will cause increased protein depolymerization rates due to enhanced diffusion of substrates and enzymes throughout the soil. In contrast, at high soil water contents in association with low oxygen concentrations, we expected that protein depolymerization would be less substrate limited due to dissolution of organo-mineral complexes and microbial amino acid uptake will decrease due to reduced microbial activity under suboxic conditions.
2. Materials and methods
2.1. Site description and soil sampling
Soil samples were collected in June 2016 in the Enns valley in the Eastern Alps (Austria) at 700m a.s.l. Mean annual air temperature and precipitation are respectively 7 °C and 1014 mm (Starz et al., 2011). Two different soil parent materials were selected: (1) calcareous glacial deposits covered by loess in Moarhof, Trautenfels-Pürg (47° 30′ N, 14° 4′ E, 708 m a.s.l.) and (2) siliceous bedrock dominated by phyllite in Raumberg-Gumpenstein (G, 47° 29′ N, 14° 6′ E, 690 m a.s.l.). Soils developed on calcareous and loess deposits are classified as Luvisols and soils developed on siliceous bedrock are classified as Cambisols (WRB, 2015). On each site soils from three land use types were sampled: forests, pastures and croplands. Forests were dominated by Norway spruce (Picea abies). Pastures were extensively managed and regularly grazed by cattle at the calcareous site and by sheep at the silicate site. Cropland soils at the calcareous site were cultivated with cereal crops (barley, wheat and oat) and at the silicate site with mixed vegetables (cabbage, onion, potato, beans). At each of the six sites four replicate soil samples were taken with a root corer (diameter 7 cm) to 15 cm soil depth. Prior to soil sampling vegetation and organic horizons were removed. The four replicates were sampled to cover heterogeneities in site topography (hilltop, upper and lower slope, bottom of slope) and were kept and analyzed separately. All soils were first sieved to 4 mm, roots and stones were removed by hand, then homogenized by sieving to 2 mm and stored at 4 °C until further analyses.
2.2. Basic soil parameters and microbial biomass
Soil texture, carbonate content, base saturation (BS), effective cation-exchange capacity (CECeff), and exchangeable Na+, K+, Mg2+, Ca2+, Fe3+, Al3+ and H+ were determined by the soil analysis laboratory of the Federal Office for Food Safety (AGES) in Vienna, Austria in air dried soil samples. All analyses were carried out according to Austrian and European standards (ÖNORM). To determine soil water content, sieved soils were dried at 60 °C for 48 h. Soil water-holding capacity (WHC) was determined by repeatedly saturating 10 g field-moist soil with deionized water and draining in between for 2.5 h in a funnel with an ash-free cellulose filter paper. Soil pH was measured in ultra-pure water and in 10 mM CaCl2 solution (soil:solution ratio = 1:5 (w:v)) using an ISFET pH sensor (Sentron, Leek, The Netherlands). For total soil organic C (SOC) and total soil N (TN), carbonate was removed in soil aliquots with 2 M HCl, subsequently samples were oven-dried, ground in a ball mill (MM 200, Retsch, Germany) and analyzed by an Elemental Analyzer (Carlo Erba 1110, CE Instruments) coupled to a DeltaPlus Isotope Ratio Mass Spectrometer (Finnigan MAT, Germany) via a Conflo III interface (Thermo Fisher, Austria). For dissolved C and N pools, 4 g aliquots of field-moist soil were extracted with 1 M KCl (1:5 w:v) for 1 h, filtered through ash-free cellulose filters and stored at −20 °C until analysis. Dissolved organic C (DOC) and total dissolved N (TDN) were measured by a TOC/TN analyzer (TOC-VCPH/TNM-1, Shimadzu, Austria). Ammonium and nitrate concentrations were measured in the same extracts by colorimetric assays as described by Hood-Nowotny et al. (2010). Dissolved organic N (DON) was calculated as TDN minus ammonium and nitrate. Total primary amines (hereafter, free amino acid concentration (FAA)) were measured fluorimetrically using the OPAME assay (Jones et al.,2002; Darrouzet-Nardi et al., 2013) as modified by Prommer et al. (2014). Interference of NH4+ was corrected using the NH4+ concentrations determined via colorimetric assays, and N compounds other than amino acids (mainly free amino sugars) contributed only between 3 and 15% to primary amine-N in the same soils (Hu et al., 2018). Microbial biomass C and N were estimated using the chloroform-fumigation extraction method (Brookes et al., 1985). Soils were fumigated with chloroform for 48 h and extracted (1:5 (w:v)) with 1 M KCl. Soil total P (TP) and soil total inorganic P (TIP) were determined in 0.5 M H2SO4 extracts of ignited (450 °C, 4 °C, Kuo (1996)) and untreated soil aliquots by malachite green measurements of reactive phosphate (Lajtha et al., 1999). Total soil organic P (TOP) was calculated as the difference of TP – TIP. Microbial P was estimated using the chloroform-fumigation extraction method (Brookes et al., 1985). Soils were fumigated with chloroform for 48 h, extracted (1:5 (w:v)) with 0.5 M NaHCO3 for 1 h and filtered through ash-free cellulose filters. Dissolved inorganic P (DIP, Olsen P) was measured in 0.5 M NaHCO3 extracts (1:7.5 (w:v), pH 8.5) by the malachite green method. Total dissolved P was measured following acid persulfate digestion (Lajtha et al., 1999) and dissolved organic P (DOP) was calculated as the difference of P concentration in non-digested and digested samples. Soil microbial community composition was analyzed by phospholipid fatty acids (PLFAs) according to the procedure by Kaiser et al. (2010) and Hu et al. (2018).
2.3. NaOH extractable protein
Proteins in soils are stabilized by sorption to humic substances, clay minerals and Fe-/Al- hydroxides (Schulten and Schnitzer, 1997; Lehmann and Kleber, 2015). NaOH extracts free and loosely bound proteins but not proteins stabilized in metal-organic complexes (Wattel-Koekkoek et al., 2001). We assumed that the more labile fraction extractable by NaOH therefore represents the protein fraction available for enzymatic cleavage. Aliquots of 2 g field-moist soil were extracted with 0.5 M NaOH for 2 h in an ultrasonic bath (160 W, Sonorex RK510, Germany) and subsequently for 16 h on a rotary shaker (1:10 (w:v)). Extracts were centrifuged for 15 min at 10.000×g at room temperature and total free amino acid (FAA) and ammonium concentrations measured fluorimetrically and colorimetrically in the supernatant as mentioned above. In a second aliquot of the supernatant, proteins were hydrolyzed with 6 M HCl at 90 °C overnight. The hydrolyzed extracts were dried under a gentle N2 stream, dissolved in ultra-pure water, and dried again in a vacuum concentrator (SC 110 SpeedVac, Savant, USA) to remove residual HCl. After re-dissolving in ultra-pure water the amino acid and ammonium concentrations were re-measured as above. The difference between the FAA concentrations in the hydrolyzed extracts (total bound amino acids) and in the non-hydrolyzed extracts (total free amino acids) was used as a measure for NaOH-extractable protein.
2.4. Incubation experiments
To assess short-term temperature and moisture/O2 sensitivities of organic N cycling processes and enzyme activities, we conducted two incubation experiments. One week prior to starting the temperature experiment and isotope pool dilution measurements, soils were adjusted to 60% WHC and stored at 15 °C in polyethylene bags. Soils were aerated every second day and water content was adjusted when necessary. For the temperature sensitivity measurements aliquots of 4 g pre-incubated soil were transferred to 50 mL polypropylene centrifuge tubes and soils were allowed to adjust to the respective temperature (5, 15 and 25 °C) overnight. The moisture/O2 sensitivities of organic N cycling processes were studied in a second incubation experiment, run four weeks after the first experiment. Aliquots of soil (stored at 15 °C and 60% WHC before) were first adjusted to the respective soil moisture contents (30, 60 and 90% WHC), weighed into polypropylene tubes and pre-incubated for 1 week at the respective WHC and O2 contents at 20 °C. Because water logging in nature comes with suboxic conditions causing microbial oxygen limitation, the 90% WHC treatment aliquots were incubated in a suboxic chamber at 1% O2. All other moisture treatments were run under normal air (21% O2). To avoid changes in O2 concentration during the pre-incubation phase, incubation vials were opened every second day for a few minutes. The isotope pool dilution experiments were started immediately after pre-incubation.
2.5. 15N isotope pool dilution (IPD) experiments
Gross protein depolymerization and amino acid uptake rates were assessed by an isotope pool dilution (IPD) assay as described by Wanek et al. (2010) and Noll et al. (2019) using 15N labeled amino acids (U-15N-98 at% 15N amino acid mixture from crude algal protein, Cambridge Isotope Laboratories, Radeberg, Germany). Pool sizes of FAA were determined immediately before starting the IPD experiments. The added 15N-label amounts were adjusted to account for ∼20% of the FAA pool size, avoiding a strong fertilization effect by tracer addition. Preliminary experiments showed that the added 15N-FAAs are rapidly immobilized in soils. Therefore, we used highly enriched 15N labeled amino acids (98 at %) as tracers added to the soils. The tracer was dissolved and diluted with ultra-pure water to the respective concentrations and 400 μL of label solution were added drop wise to 4 g soil with a pipette and vigorous shaking to guarantee good mixing. Soil samples were incubated at the respective temperatures (5, 15 and 25 or 20 °C) for 15 and 45 min. Preliminary experiments showed that the recovery of the amino acid label decreases at constant rates between 15 and 60 min after tracer addition (Noll et al., 2019). The incubation was stopped by adding 20 mL of 1 M KCl solution at room temperature. Hu et al. (2017) showed that enzyme activity is effectively quenched with KCl at 4° and 15 °C in the absence of formaldehyde. Soils were extracted for 1 h on a rotary shaker and then filtered through ash-free cellulose filters. The charged nature of amino acids renders them to bind to positively (anion-exchange mechanisms) or negatively charged sites in soils (cation-exchange mechanisms) and therefore to be unavailable for extraction by water. 1 M KCl has been shown to achieve higher extraction efficiency than 0.01 M CaSO4 or water (Noll et al., 2019). Moreover, Dippold and Kuzyakov (2013) showed in experiments with life soils and soils inhibited with HgCl2 and NaN3, that only 3–6% of added alanine was sorbed to the soil matrix after 36 h and 83–90% of the sorbed alanine could be extracted with 0.5 M CaCl2. Extracts were stored at −20 °C until further analyses. Isotope analyses were performed based on a novel isotope ratio mass spectrometry (IRMS) protocol adopted from Zhang and Altabet (2008) as modified by Noll et al. (2019). Prior to measuring free amino acid concentrations and isotope ratios by IRMS NH4+ was removed from the extracts by microdiffusion as NH4+ would also be oxidized to nitrite simultaneously as α-amino N (Zhang and Altabet, 2008). Acid traps were prepared from small cellulose filter discs soaked with 4 μL 2.5 M KHSO4 and sealed between two pieces of PTFE-tape (Lachouani et al., 2010). Aliquots of extracts (10 mL) were transferred to 20 mL scintillation vials pre-filled with ∼100 mg of MgO and acid traps were added. Micro-diffusions were run on a shaker for 48 h, and afterwards acid traps were removed and NH4+-free extracts were further processed. Concentrations and 15N enrichments of FAA were measured after conversion to N2O by purge-and-trap (PT)-IRMS (Lachouani et al., 2010). In the first step α-amino groups of FAA were oxidized to NO2− by the Strecker reaction (Zhang and Altabet, 2008). Therefore aliquots (2 mL) of soil extracts were transferred to 12 mL exetainer glass vials with PTFE septa. Thereafter, 50 μL 60 mM KBr and 20 μL 10 M NaOH were added and exetainers were closed tightly. Finally, 50 μL NaClO (30 mM) were added with a syringe and samples were incubated for 30 min at 50 °C. To deactivate excess NaClO, 100 μL 0.4 M Na-metaarsenite was added after incubation. Subsequently NO2− was reduced by NaN3 to N2O by adding 140 μL NaN3 (20 mM; Sigma) in 50% acetic acid (Merck) with a gas tight syringe and samples were incubated for 30 min on an orbital shaker at room temperature (Lachouani et al., 2010). Finally, NaN3 was deactivated by addition of 80 μL 6 M NaOH. Free amino acid concentrations and 15N enrichments in the produced N2O were measured by PT-IRMS, and concentrations (equimolar 20 FAA mix) and isotope enrichments (natural abundance to 20 at% 15N) calibrated by respective standards (Lachouani et al., 2010). PT-IRMS analyses were performed on a purge-and-trap isotope ratio mass spectrometer (Finnigan Delta V Advantage IRMS, Thermo Fisher, Germany) consisting of a Gasbench II headspace analyzer (Thermo Fisher, Germany) with a cryo-focusing unit.
2.6. Potential extracellular peptidase activity
Leucine-amino peptidase activities (EC 3.4.11.1) were measured in 2 mL soil slurries using a fluorometric microtiter plate assay with 50 μL L-leucine-7-amido-4-methyl coumarin (AMC-leucine, 1 mM) as a substrate in Na-acetate buffer (100 mM, pH 5.5, Kaiser et al., 2010). The samples were run in triplicates and were incubated for 2 h at the respective temperatures (5, 15 and 25 °C for the temperature experiment, and 20 °C for the moisture experiment) and measured every 30 min. Fluorescence was measured with a TECAN InfiniteR M200 (Austria) at an excitation wavelength of 365 nm and an emission wavelength of 450 nm, and was corrected for sample blank and quenching prior to calculations of AMC concentration.
2.7. Data analysis
Gross protein depolymerization rates (GP) and gross microbial amino acid uptake rates (GI) were calculated following the equations by Kirkham and Bartholomew (1954) and Wanek et al. (2010):
| (1) |
| (2) |
where Nt1 and Nt2 are the concentrations of FAA-N at the time points t1 (15 min) and t2 (60 min). 15N enrichments in amino acids at the time points of termination are expressed as at%15Nt1 and at%15Nt2, and at %15Nb is the background 15N abundance (0.366 at%15N) in non-labeled samples.
Temperature sensitivities of gross protein depolymerization rates, amino acid uptake rates and peptidase activity between 5 and 25 °C were calculated by a log linear Q10 function (Janssens and Pilegaard, 2003):
where R is the measured rate or enzyme activity, Q10 is the temperature sensitivity, T is the incubation temperature and b a fitted coefficient. Q10 and b where determined by non-linear least square estimates.
2.8. Statistics
Statistics and estimations of Q10 values were performed with R version 3.1.3 R core (R Development Core Team, 2008). Data were log transformed or rank normalized if not normally distributed or hetero-scedastic. Correlations were done using Pearson's product moment correlation. In addition, we performed generalized linear mixed models (GLMM) of the influence of soil parent material on the relationship of selected soil parameters and gross protein depolymerization rates using the “lmer” function included in the “lme4” package (Bates et al., 2014). Soil parent material was specified as random factor. Regressions between gross protein depolymerization rates and NaOH-extractable protein were done using nonlinear least-squares estimates of the parameters in a nonlinear model. Differences in gross rates and peptidase activities between soil parent material, land use or the respective temperature and moisture/O2 treatments were addressed by three-way analysis of variance (ANOVA). If effects were significant (p < 0.05) differences between groups were tested with Tukey's HSD. Significant differences in Q10 values between parent material and land use were addressed by two-way ANOVA followed by Tukey's HSD. We used soil parameters which correlated significantly with gross protein depolymerization rates or free amino acid contents to develop a base multiple regression model. The most parsimonious model was determined by step wise backward/forward multiple regression analysis using Akaike's Information Criterion (AIC). R-squared values were obtained by regressing model predictions against observed values. Community composition based on PLFA analyses was visualized by nonmetric multidimensional scaling (NMDS) ordinations of individual fatty acid methyl esters. Protein depolymerization and soil parameters were fitted onto the ordination using the ‘envfit’ function in the R package ‘vegan’ (Oksanen et al., 2016). Significance of correlations was tested with 999 permutations.
3. Results
3.1. Basic soil parameters and organic N pool sizes
The studied soils showed significant differences in physicochemical soil parameters and N pool sizes depending on parent material and land use (Table 1). Clay and silt contents were higher in calcareous than in silicate soils, while silicate soils had significantly higher sand contents than calcareous soils (p < 0.001, ANOVA). Soil pH, cation-exchange capacity (CECeff), base saturation (BS) and exchangeable Ca2+ were higher in calcareous soils than in silicate soils (p < 0.001, ANOVA). Amorphous Al (Alo) contents were significantly higher in silicate soils than in calcareous soils (p < 0.001, ANOVA), while calcareous soils had significantly higher crystalline Fe (Fed-o) contents (p < 0.01, ANOVA). We also found a strong land use effect (p < 0.001, ANOVA) with increasing soil pH in the order forest < pasture < cropland. NaOH-extractable protein-N was significantly higher in calcareous soils than in silicate soils (F = 16.2, p < 0.001), and ranged between 11 and 66% of total soil N (Table 1). We also observed a significant effect of land use (F = 15.9, p < 0.001) represented by increasing NaOH-extractable protein-N contents in the order cropland < forest < pasture. Total free amino acids (FAA) were higher in silicate soils than in calcareous soils (p < 0.001) and accounted for 27 ± 10% of the dissolved organic N pool in silicate soils and for 13 ± 5% in calcareous soils, but were unaffected by land use. Significant effects of soil parent material and land use were found for total, extractable and microbial C and N pools (Table S1). PLFA contents were highest in calcareous soils, but did not vary with land use, and there was no significant parent material or land use effect on fungi:bacteria ratios (Table S1).
Table 1.
Initial soil physicochemical and biological properties (mean ± s.d., n = 4). Effects of soil parent material (P), land use (L) and their interaction (PxL) were tested by two-way ANOVA. Asterisks represent significance levels.
| Calcareous | Silicate | p value | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Arable | Pasture | Forest | Arable | Pasture | Forest | P | L | PxL | |
| Sand (%) | 29 ± 3 | 48 ± 13 | 23 ± 3 | 44 ± 1 | 61 ± 5 | 47 ± 2 | ** | ** | n.s. |
| Silt (%) | 56 ± 1 | 4 ± 11 | 63 ± 4 | 47 ± 1 | 33 ± 5 | 35 ± 2 | *** | * | n.s. |
| Clay (%) | 15 ± 2 | 8 ± 2 | 15 ± 3 | 9 ± 1 | 6 ± 1 | 17 ± 1 | *** | *** | *** |
| pH | 7.2 ± 0.1 | 5.9 ± 0.4 | 5.6 ± 0.1 | 5.4 ± 0.7 | 4.5 ± 0.1 | 3.5 ± 0.1 | *** | *** | *** |
| CECeff (cmolc kg−1) | 33.6 ± 1.0 | 22.7 ± 6.0 | 22.8 ± 1.0 | 8.4 ± 4.0 | 5.4 ± 1.0 | 9.7 ± 1.0 | *** | ** | ** |
| BS (%) | 99.9 ± 0.03 | 99.6 ± 0.3 | 99.3 ± 0.2 | 93.4 ± 7.0 | 82.7 ± 6.0 | 5.0 ± 0.4 | *** | *** | *** |
| exch. Ca2+ (cmolc kg−1) | 31.9 ± 0.5 | 19.6 ± 6.5 | 21.0 ± 0.9 | 6.8 ± 3.8 | 4.0 ± 0.6 | 0.2 ± 0.1 | *** | *** | * |
| exch. Al3+ (mmolc kg−1) | 0 ± 0 | 0.03 ± 0.05 | 0.15 ± 0.06 | 3.03 ± 0.3 | 7.9 ± 2.2 | 87.3 ± 12.8 | *** | *** | *** |
| Alo (mg g−1) | 1.6 ± 0.1 | 1.66 ± 0.9 | 2.1 ± 0.3 | 3.7 ± 0.3 | 2.9 ± 0.4 | 3.2 ± 0.1 | *** | n.s. | * |
| Ald (mg g−1) | 2.7 ± 0.9 | 3.2 ± 1.5 | 3.5 ± 0.4 | 5.5 ± 0.5 | 4.1 ± 1.0 | 4.3 ± 0.5 | *** | n.s. | * |
| Ald-o (mg g−1) | 1.0 ± 0.2 | 1.6 ± 1.1 | 1.3 ± 0.5 | 1.8 ± 0.6 | 1.2 ± 0.7 | 1.1 ± 0.4 | n.s. | n.s. | n.s. |
| Feo (mg g−1) | 4.8 ± 0.4 | 7.9 ± 0.7 | 4.9 ± 0.1 | 8.5 ± 0.7 | 7.1 ± 0.9 | 11.2 ± 2.0 | *** | n.s. | ** |
| Fed (mg g−1) | 24.9 ± 2.2 | 39.7 ± 20 | 26.3 ± 3.4 | 24.2 ± 2.7 | 18.8 ± 3.6 | 26.1 ± 3.6 | ** | n.s. | *** |
| Fed-o (mg g−1) | 20.1 ± 2.3 | 31.8 ± 17.7 | 21.4 ± 3.3 | 15.7 ± 2.8 | 11.8 ± 3.6 | 14.9 ± 2.4 | ** | n.s. | n.s. |
| SOC (mg g−1) | 47.0 ± 1.7 | 43.9 ± 10.7 | 36.8 ± 4.7 | 21.8 ± 2.2 | 26.7 ± 1.8 | 49.9 ± 15.2 | ** | n.s. | *** |
| Total N (mg g−1) | 4.8 ± 0.2 | 4.6 ± 1.2 | 3.3 ± 0.2 | 2.2 ± 0.2 | 2.8 ± 0.2 | 2.5 ± 0.8 | *** | * | * |
| DOC (μg g−1) | 53 ± 4 | 54 ± 25 | 53 ± 14 | 65 ± 7 | 86 ± 9 | 161 ± 26 | *** | *** | *** |
| DON (μg g−1) | 16.9 ± 2.0 | 16 ± 3.0 | 15.2 ± 4.0 | 15.7 ± 2.0 | 32.4 ± 5.0 | 13.6 ± 4.0 | ** | *** | *** |
| NH4+ (μg N g−1) | 1.8 ± 1.6 | 0.7 ± 0.5 | 1.2 ± 1.2 | 1.3 ± 1.3 | 0.7 ± 0.1 | 6.3 ± 1.5 | ** | *** | *** |
| NO3− (μg N g−1) | 20.0 ± 1.6 | 18.9 ± 4.1 | 15.7 ± 3.2 | 10.0 ± 3.5 | 2.1 ± 2.2 | 4.5 ± 1.9 | *** | ** | * |
| FAA (μg N g−1) | 1.5 ± 0.3 | 2.2 ± 1.0 | 2.4 ± 1.0 | 4.1 ± 2.0 | 7.0 ± 1.0 | 4.1 ± 1.0 | *** | n.s. | n.s. |
| ProteinNaOH (mg N g−1) | 0.8 ± 0.3 | 3.0 ± 2.1 | 1.6 ± 0.8 | 0.2 ± 0.2 | 1.9 ± 0.9 | 0.3 ± 0.1 | *** | *** | n.s. |
| % ProteinNaOH from total N | 18.8 ± 7.0 | 62.9 ± 43.0 | 46.8 ± 22.0 | 10.9 ± 7.0 | 66.3 ± 31.0 | 13.3 ± 2.0 | n.s. | *** | n.s. |
| Cmic (mg g−1) | 1.40 ± 0.11 | 1.81 ± 0.67 | 0.89 ± 0.03 | 0.28 ± 0.04 | 0.65 ± 0.09 | 0.58 ± 0.16 | *** | ** | * |
| Nmic (mg g−1) | 0.32 ± 0.22 | 0.36 ± 0.08 | 0.20 ± 0.01 | 0.07 ± 0.01 | 0.16 ± 0.02 | 0.10 ± 0.03 | *** | *** | ** |
| PLFA Gram positive (mg g−1) | 0.60 ± 0.28 | 0.80 ± 0.22 | 0.44 ± 0.16 | 0.18 ± 0.14 | 0.29 ± 0.19 | 0.24 ± 0.18 | *** | n.s. | n.s. |
| PLFA Gram negative (mg g−1) | 0.64 ± 0.28 | 0.86 ± 0.26 | 0.38 ± 0.16 | 0.17 ± 0.10 | 0.46 ± 0.43 | 0.40 ± 0.06 | * | n.s. | n.s. |
| PLFA Bacteria (mg g−1) | 1.51 ± 0.35 | 1.66 ± 0.33 | 1.04 ± 0.19 | 0.43 ± 0.15 | 0.76 ± 0.08 | 0.63 ± 0.28 | *** | n.s. | n.s. |
| PLFA Fungi (mg g−1) | 0.98 ± 0.40 | 1.39 ± 0.36 | 0.84 ± 0.49 | 0.22 ± 0.11 | 0.38 ± 0.19 | 0.62 ± 0.32 | *** | n.s. | n.s. |
| PLFA Fungi:Bacteria | 0.72 ± 0.17 | 0.47 ± 0.15 | 1.54 ± 0.29 | 0.69 ± 0.25 | 0.87 ± 0.33 | 0.66 ± 0.26 | n.s. | n.s. | n.s. |
Significance levels: n.s.: not significant, *: p < 0.05, **: p < 0.01, ***: p < 0.001.
3.2. Effects of parent material and land use on soil processes
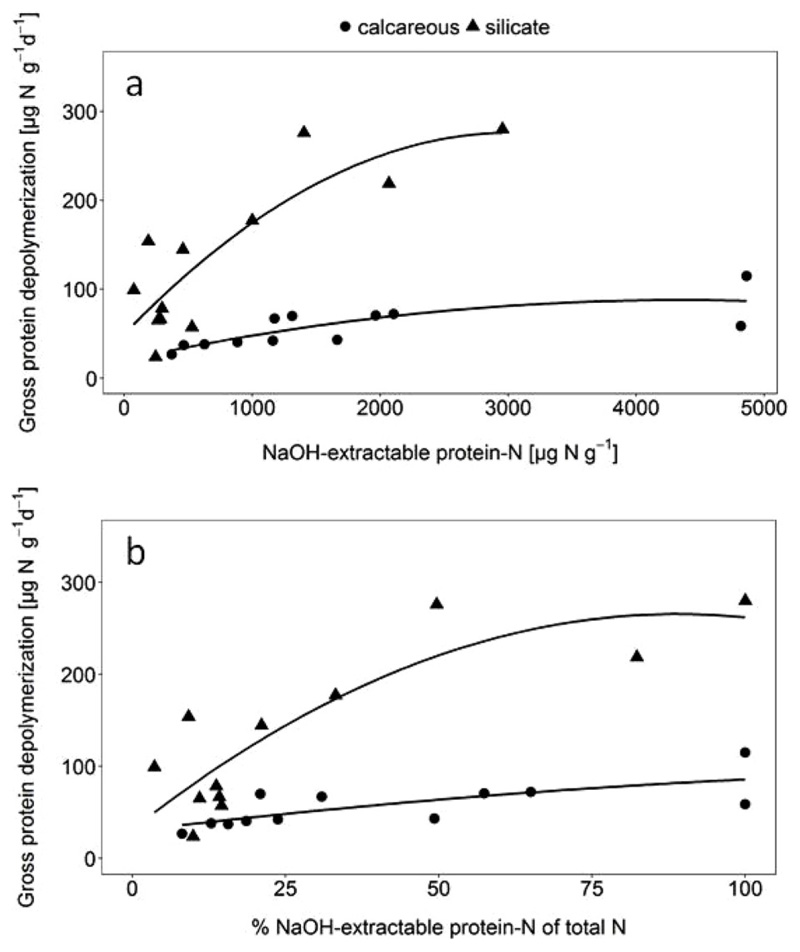
Gross protein depolymerization rates ranged between 24 and 280 μg N g d. w.−1 d−1 and showed significant parent material (F = 18.21, p < 0.001) and land use effects (F = 10.45, p < 0.001, Table S2). In detail, gross protein depolymerization rates were significantly higher in silicate soils than in calcareous soils, and in pasture soils compared to cropland and forest soils. Highly similar patterns were observed for gross rates of microbial amino acid uptake. To examine the effects of soil physicochemical parameters on gross rates of protein depolymerization and microbial amino acid uptake, we performed linear regression analyses (Table 2). Gross protein depolymerization and amino acid uptake rates were significantly negatively correlated with soil properties such as clay and silt content, exchangeable Ca2+ and CECeff and strongly positively correlated with sand content, DON and FAA-N, but not with microbial biomass C and N or PLFA contents (Table 2). Considering the strong parent material effects, we used generalized linear mixed models of gross protein depolymerization with soil parent material as random factor. We obtained significant results for the mentioned soil parameters, including the significant positive effect of NaOH-extractable protein and negative ones of CECeff and clay content (Table S3). The relation of NaOH-extractable protein-N and gross protein depolymerization was fitted to a quadratic polynomial model using a generalized non-linear least squares fit, which explained 70% of the variation in silicate soils and 60% of the variation in calcareous soils (Fig. 2). However, across and within all soil types no significant correlations were found between potential peptidase activity and gross protein depolymerization rates. Following linear regression analyses, multiple regression models were tested for gross protein depolymerization, and the final most parsimonious model included soil Ntot (positive effect), NaOH-extractable protein (positive effect), CECeff (negative effect) and crystalline Fe (negative effect, Table S4). The model explained about 86% of the variance in gross protein depolymerization (r2 = 0.86, F = 28.11, p < 0.001). Gross microbial amino acid uptake rates across different parent materials and land use types (Table 2) showed similar correlations as described above for gross protein depolymerization rates. Significant effects on amino acid up-take rates were observed for parent material (F = 19.3, p < 0.001) and land use (F = 13.5, p < 0.001) and their interaction (F = 8.6, p = 0.001). Furthermore, amino acid uptake rates were strongly correlated with protein depolymerization rates across all sites (Table 2). Potential leucine-amino peptidase activity was positively correlated to PLFA contents, microbial C and N, NaOH-extractable protein and soil pH (Table 2) while negative correlations were found with DOC, soil C:N ratio, Feo, Alo, Ald and exchangeable Al3+. Soil NaOH-extractable protein was positively correlated to peptidase activity, microbial C and N as well as gram negative PLFA markers and fungal PLFAs while clay content and Alo negatively affected NaOH-extractable protein (Table 2). Microbial community composition obtained from PLFA analyses correlated with soil pH and leucine-amino peptidase activity, CECeff, DOP, total N and organic C but not with protein depolymerization rates (NMDS, Fig. S1).
Table 2.
Correlation analyses of gross protein depolymerization rates (Depoly), microbial amino acid uptake rates (Uptake), peptidase activities (Peptidase), NaOH-extractable protein contents (Protein) and total free amino acid (FAA) contents (Pearson's R) with selected soil physicochemical and biological parameters (n = 24).
| Depoly | Uptake | Peptidase | Protein | FAA | |
|---|---|---|---|---|---|
| Depoly | 1 | 0.98*** | n.s. | n.s. | 0.89*** |
| Uptake | 0.98*** | 1 | n.s. | n.s. | 0.87*** |
| Peptidase | n.s. | n.s. | 1 | 0.51** | n.s. |
| Protein | n.s. | n.s. | 0.51** | 1 | n.s. |
| FAA | 0.89*** | 0.87*** | n.s. | n.s. | 1 |
| Sand | 0.63*** | 0.64*** | n.s. | n.s. | 0.70*** |
| Silt | −0.52** | −0.53** | n.s. | n.s. | −0.72*** |
| Clay | −0.65*** | −0.68*** | n.s. | −0.52** | n.s. |
| CECeff | −0.66*** | −0.63*** | 0.51* | n.s. | −0.79*** |
| BS | n.s. | n.s. | 0.57** | n.s. | −0.48* |
| pH | n.s. | n.s. | 0.64*** | n.s. | −0.74*** |
| exch. Ca2+ | −0.53** | −0.50* | 0.58** | n.s. | −0.78*** |
| exch. Al3+ | n.s. | n.s. | −0.53** | n.s. | n.s. |
| Alo | n.s. | n.s. | −0.78*** | −0.55** | 0.48* |
| Ald | n.s. | n.s. | −0.59** | n.s. | n.s. |
| Ald-o | n.s. | n.s. | n.s. | n.s. | n.s. |
| Feo | n.s. | n.s. | −0.62** | n.s. | n.s. |
| Fed | n.s. | n.s. | n.s. | n.s. | −0.42* |
| Fed-o | n.s. | n.s. | n.s. | n.s. | −0.51** |
| Corg | −0.50* | −0.51* | n.s. | n.s. | n.s. |
| Ntot | n.s. | n.s. | 0.58** | n.s. | n.s. |
| DOC | n.s. | n.s. | −0.43* | n.s. | 0.62** |
| DON | 0.83*** | 0.87*** | n.s. | n.s. | 0.84** |
| NH4+ | n.s. | n.s. | n.s. | −0.42* | n.s. |
| NO3− | −0.66*** | −0.59** | 0.51* | n.s. | −0.81*** |
| Cmic | n.s. | n.s. | 0.62** | 0.44* | −0.41* |
| Nmic | n.s. | n.s. | 0.67*** | 0.45* | −0.48* |
| soil C:N | n.s. | n.s. | −0.44* | n.s. | n.s. |
| soil C:P | n.s. | n.s. | n.s. | n.s. | n.s. |
| soil N:P | −0.47* | −0.46* | n.s. | n.s. | n.s. |
| PLFA Gram+ | n.s. | n.s. | 0.60** | n.s. | −0.41* |
| PLFA Gram- | n.s. | n.s. | 0.51* | 0.58** | n.s. |
| PLFA Bacteria | n.s. | n.s. | 0.69*** | n.s. | −0.41* |
| PLFA Fungi | n.s. | n.s. | 0.66*** | 0.53** | n.s. |
Significance levels: n.s. Not significant, *p < 0.05, **p < 0.01, ***p < 0.001
Fig. 2.
a) Relationship between gross protein depolymerization (at 15 °C) and NaOH-extractable protein contents in calcareous and silicate soils. Solid lines represent 2nd polynomial regression model for silicate (r2 = 0.721, p < 0.001, n = 12) and calcareous soils (r2 = 0.598, p < 0.010, n = 12). Regression models were significantly different between silicate and calcareous soils (p < 0.001). b) Relationship between gross protein depolymerization (at 15 °C) and the contribution of NaOH-extractable protein-N to total soil N contents in calcareous and silicate soils. Solid lines represent 2nd polynomial regression models for silicate (r2 = 0.672, p < 0.01, n = 12) and calcareous soils (r2 = 0.504, p < 0.05, n = 12). Regression models were significantly different between silicate and calcareous soils (p < 0.001, ANOVA).
3.3. Soil temperature effects
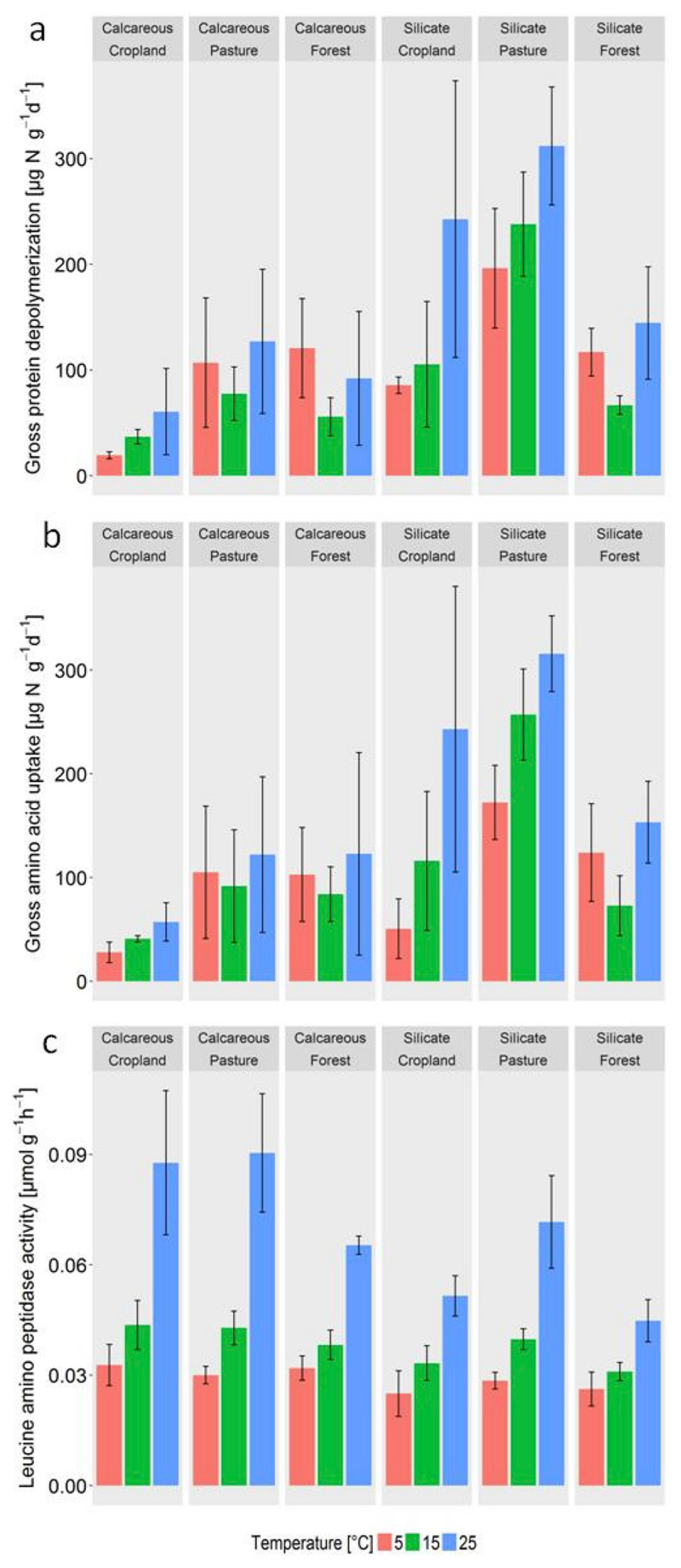
Incubation temperature (5, 15 25 °C) had a weak positive effect on gross protein depolymerization rates compared to strong effects of parent material and land use according to three-way ANOVA (Table 3, Fig. 3.). We also found a weak interaction effect of land use x temperature as cropland soils showed an increase of gross rates with temperature, while forest soils showed a minimum at 15 °C (Fig. 3). Interaction effects of soil parent material x temperature were not significant. Tukey's HSD did not reveal significant differences between single treatments across soils. Within and across all temperature treatments, peptidase activity was not related to depolymerization rates. However, we found a strong positive temperature effect on peptidase activity as well as weaker parent material and land use effects (Table 3, Fig. 3). Peptidase activities increased in the order forest < arable = pasture and were higher in calcareous soils than in silicate soils (Fig. 3). Gross amino acid uptake rates showed a similar pattern as depolymerization rates across the temperature treatments (Fig. 3), with a weak but significant positive temperature effect, and strong effects of parent material and land use (Table 3).
Table 3.
Three-way ANOVA of gross protein depolymerization rates, gross microbial amino acid uptake rates and peptidase activities for 6 soils differing in parent material and land use, exposed to three soil temperature levels, and their respective interactions (n = 4).
| Gross depolymerization rate | Gross amino acid uptake rate | Leucine-amino peptidase activity | ||||
|---|---|---|---|---|---|---|
| F | p | F | p | F | p | |
| Parent material | 57.5 | < 0.001 | 30.5 | < 0.001 | 52.5 | < 0.001 |
| Land use | 21.3 | < 0.001 | 16.1 | < 0.001 | 11 | < 0.001 |
| Temperature | 7.8 | < 0.01 | 7.6 | < 0.01 | 206.3 | < 0.001 |
| Parent material x Land use | 7 | < 0.01 | 4.4 | 0.017 | 4.1 | 0.022 |
| Parent material x Temperature | 0.6 | 0.53 | 1.3 | 0.281 | 3.6 | 0.033 |
| Land use x Temperature | 3.4 | 0.015 | 2.5 | 0.053 | 3.4 | 0.015 |
| Parent material x Land use x Temperature | 0.8 | 0.504 | 0.5 | 0.759 | 0.1 | 0.995 |
Fig. 3.
(a) Gross protein depolymerization, (b) microbial amino acid uptake and (c) peptidase activities at 5, 15 and 25 °C (mean ± s.d., n = 4) in 6 soils differing in parent material (calcareous, silicate) and land use (croplands, pastures, forests). Statistical analyses are provided in Table 3.
Results from a two-way ANOVA showed significant land use effects on Q10 values of gross protein depolymerization (F = 6.5, p = 0.007) but no effect of soil parent material. Q10 values of gross protein depolymerization (and amino acid uptake) showed an increase in the order forest (0.8–1.1) < pasture (1.2–1.4) < crop land (1.6–2.2; Table 4). Q10 values of amino acid uptake rates (F = 7.6, p = 0.004) and peptidase activity (F = 9.7, p < 0.005) were also affected by land use and significant effects of parent material were observed for the Q10 values of peptidase activity (F = 7.9, p < 0.05). Mean Q10 values of peptidase activity ranged between 1.3 and 1.7 and were slightly higher in calcareous soils than in silicate soils (p = 0.01, Table 4). Q10 values of gross protein depolymerization (1.6–2.2) therefore matched (or even exceeded) those of amino peptidase activity in cropland soils, were slightly lower in pasture soils (1.2–1.4 for protein depolymerization compared to 1.6–1.7 for peptidase activity) and no temperature sensitivity was apparent for forest soils (0.8–1.1) though Q10 values of peptidase activity in forests ranged between 1.3 and 1.4 (Table 4).
Table 4.
Q10 of gross depolymerization, amino acid uptake and peptidase activity (mean ± s.d., n = 4) and effects of parent material, land use types as well as their interaction (p-values) obtained from two-way ANOVA.
| Parent material | Land use | Q10 gross depolymerization rate | Q10 gross amino acid uptake rate | Q10 Leucine-amino peptidase activity |
|---|---|---|---|---|
| silicate | Crop land | 1.6 ± 0.4 | 2.2 ± 0.5 | 1.5 ± 0.2 |
| Pasture | 1.3 ± 0.1 | 1.4 ± 0.1 | 1.6 ± 0.1 | |
| Forest | 1.1 ± 0.3 | 1.1 ± 0.2 | 1.3 ± 0.1 | |
| calcareous | Crop land | 1.7 ± 0.7 | 1.5 ± 0.3 | 1.6 ± 0.2 |
| Pasture | 1.2 ± 0.4 | 1.3 ± 0.4 | 1.7 ± 0.1 | |
| Forest | 0.8 ± 0.2 | 1 ± 0.4 | 1.4 ± 0.1 | |
| Parent material | n.s. | n.s. | * | |
| Land use | ** | ** | ** | |
| Parent material x land use | n.s. | n.s. | n.s. | |
Significance levels: n.s. Not significant, *p < 0.05, **p < 0.01, ***p < 0.001.
3.4. Soil moisture effects
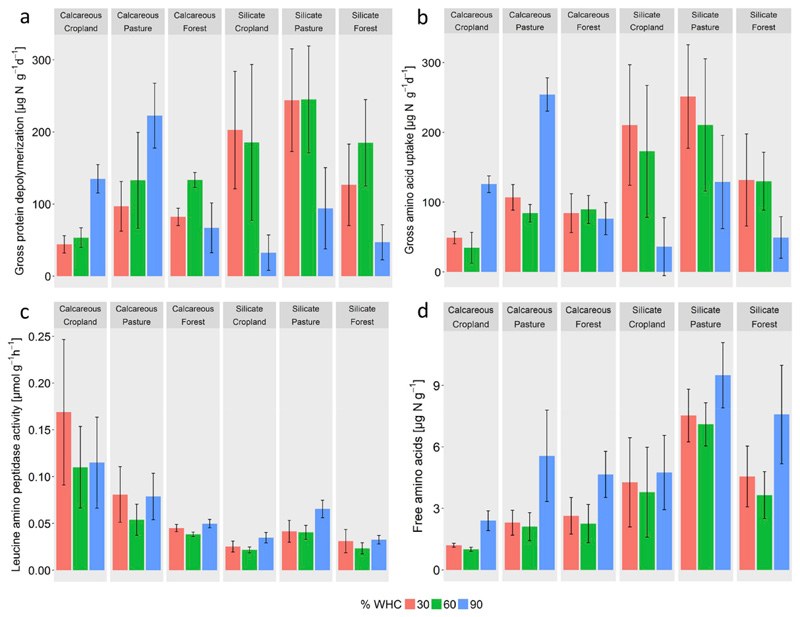
Prior to IPD experiments soils were pre-incubated at the respective soil moisture (30, 60, 90% WHC) and O2 concentration (21% O2 at 30 and 60% WHC, 1% O2 at 90% WHC) for 7 days to allow equilibration of biochemical processes. After pre-incubation free amino acid concentrations (measured 1 day before starting the IPD experiment) were significantly higher in suboxic treatments (90% WHC, 1% O2) than in oxic treatments (30 and 60% WHC, 21% O2, Fig. 4d), whereas nitrate was almost entirely depleted in all suboxic treatments (Fig. S2). Three-way ANOVA of gross protein depolymerization with the factors parent material, land use and soil moisture/O2 showed significant main effects of parent material, land use and soil moisture/O2, though interaction effects of parent material x soil moisture/O2 had the highest explanatory power (Table 5). More specifically, gross depolymerization rates did not differ significantly between 30 and 60% WHC across all parent material and land use types (Fig. 4a). In oxic treatments (30 and 60% WHC) we found similar patterns as described for the temperature treatments including lower gross depolymerization rates in calcareous soils and a parent material dependent land use effect (Fig. 4a). This pattern changed for the suboxic treatment at 90% WHC. In silicate soils at 90% WHC/1% O2 gross protein depolymerization rates decreased significantly across all land use types (p < 0.05, Tukey HSD), and the average decreases accounted for 54–74% of the rates at 60% WHC. In calcareous soils at 90% WHC/1% O2 gross protein depolymerization rates increased more than two-fold in cropland soils (p < 0.05, Tukey HSD) compared to the 60% WHC treatment and in pasture soils (p < 0.01, Tukey HSD) compared to the 30% WHC treatment, while no significant changes were observed for the calcareous forest soil.
Fig. 4.
(a) Gross protein depolymerization, (b) microbial amino acid uptake, (c) peptidase activities and (d) free amino acid contents at different moisture/O2 levels (30% WHC/21% O2, 60% WHC/21% O2 and 90% WHC/1% O2) at 20 °C in 6 soils differing in parent material (calcareous, silicate) and land use (croplands, pastures, forests; mean ± s.d.), n = 4). Statistical analyses are provided in Table 5.
Table 5.
3-Way ANOVA of gross protein depolymerization rates, gross microbial amino acid uptake rates and peptidase activities for 6 soils differing in parent material and land use, exposed to three soil moisture/O2 treatments (30% WHC/21% O2, 60% WHC/21% O2, 90% WHC/1% O2, n = 4), and their respective interactions.
| Gross depolymerization rate | Gross amino acid uptake rate | Leucine-amino peptidase activity | Total free amino acids | |||||
|---|---|---|---|---|---|---|---|---|
| F | p | F | p | F | p | F | p | |
| Parent material | 11.2 | < 0.001 | 12.30 | < 0.001 | 124 | < 0.001 | 90 | < 0.001 |
| Land use | 16 | < 0.001 | 20.20 | < 0.001 | 20 | < 0.001 | 23 | < 0.001 |
| WHC | 10 | < 0.001 | 1.60 | 0.2 | 8 | < 0.001 | 20 | < 0.001 |
| Parent material x Land use | 1 | 0.29 | 1.10 | 0.34 | 28 | < 0.001 | 6 | < 0.01 |
| Parent material x WHC | 40.4 | < 0.001 | 29.70 | < 0.001 | 3 | 0.08 | 0.1 | 0.89 |
| Land use x WHC | 4 | < 0.05 | 2.10 | 0.09 | 0 | 0.8 | 1.5 | 0.23 |
| Parent material x Land use x WHC | 4.6 | 0.003 | 2.90 | 0.031 | 0.9 | 0.48 | 0.5 | 0.71 |
We found similar effects of parent material and land use on gross amino acid uptake rates as we reported for gross depolymerization rates but no significant moisture effect/O2 (Table 5). On average gross amino acid uptake rates in the 90% WHC/1%O2 WHC treatment decreased in silicate soils by 46–68% and increased by 2- to 3-fold in calcareous cropland (p < 0.05, Tukey HSD) and pasture soils (p < 0.01, Tukey HSD, Fig. 4b, Table 5) compared to the 60% WHC treatments. In contrast to gross depolymerization and amino acid uptake rates, parent material had the strongest explanatory effect on peptidase activity, followed by land use and soil moisture/O2 (Table 5). Differences in peptidase activities between soil moisture/O2 treatments were significant and showed a slight increase between 60 and 90% WHC (p < 0.001, Tukey HSD). Again peptidase activities were not related to gross protein depolymerization rates within or across soil moisture treatments.
4. Discussion
4.1. Parent material and land use effects on soil organic N cycling
Our results demonstrate that gross protein depolymerization as catalyzed by extracellular enzymes varied considerably with soil parent material and land use. Soil parent material explained 29% of the variability in protein depolymerization and 23% of the variability in microbial amino acid uptake (Table S2). In detail, gross depolymerization rates responded negatively to clay and silt content, exchangeable Ca2+ and CECeff, resulting in lower gross rates in calcareous than in silicate soils (Fig. 3a). The dampened response of gross protein depolymerization rates to increasing NaOH extractable protein contents (representing a large proportion of the total soil protein pool) in calcareous compared to silicate soils (Table 1) suggests that proteins are less available for enzymatic cleavage, likely due to stronger sorption to mineral surfaces in calcareous soils (Rillig et al., 2007). This is supported by the negative effects of CECeff and crystalline Fe on gross protein depolymerization rates (Table S4). The studied calcareous soils had higher clay and silt contents than the silicate soils (Table 1 and Table S1), indicating a higher soil surface area and a higher proportion of more strongly sorbed or occluded protein. Moreover, in calcareous soils exchangeable Ca2+ contents were higher, allowing enhanced Ca2+ ion bridging between negatively charged proteins and negatively charged mineral surfaces to promote protein sorption (Lützow et al., 2006). These findings and the missing relation between gross depolymerization rates and potential peptidase activity support the hypothesis that gross rates of organic N cycling are substrate limited rather than enzyme limited, triggered by varying mechanisms of protein sorption and occlusion. In line with substrate limitation, Prommer et al. (2014) showed that protein depolymerization decreased after addition of biochar likely through protein sorption in micropores rendering them non-accessible to microbiota and extracellular enzymes. Soil mineralogy affects organic N cycling by stabilization of proteins, but also by abiotic fragmentation of proteins on Mn-oxides. Recent studies showed that proteins can be abiotically degraded in the presence of birnessite, a pedogenic Mn-oxide produced under alternating oxic and suboxic conditions (Russo et al., 2009; Reardon et al., 2016). Soils developed on limestone often contain considerable amounts of Mn-oxides (Dixon and White, 2002), however exchangeable Mn concentrations were not significantly different between the studied calcareous (0.07 ± 0.07 cmolc kg−1) and silicate soils (0.06 ± 0.04 cmolc kg−1). Though intriguing, it is unclear if abiotic degradation of proteins might enhance organic N availability due to liberation of small peptides and amino acids, or even decreases organic N availability due to deactivation of extracellular proteolytic enzymes (Reardon et al., 2016).
Moreover, the studied soils covered three land use types (forest, pasture, cropland) which differ in organic matter quality, quantity and type of N input, cycling of N and other essential nutrients such as P and K, and in microbial community structure (Tables 1 and S1). Land use explained about 30% of the variability of gross protein depolymerization and microbial amino acid uptake (Table S2), with highest NaOH-extractable protein contents and gross protein depolymerization rates in pasture soils, likely induced by high organic N inputs by cattle or sheep in the form of slurry or feces. Protein availability is also influenced by plant-microbe-mineral interactions. Root exudates, like organic acids, weaken organo-mineral interactions and thereby release mineral-associated proteins. Moreover, excretion of labile root exudates might enhance microbial N mining due to stimulated microbial activity (Jilling et al., 2018). Since pasture soils are often characterized by high root densities, root exudates might hamper protein stabilization in pasture soils compared to cropland soils. Low protein depolymerization and amino acid uptake rates in cropland soils corresponded to lowest NaOH-extractable protein contents; however in cropland soils Q10 values of gross depolymerization were similar to those of peptidase activity, indicating enzyme limitation rather than substrate limitation (Table 4). The lower depolymerization rates in forest soils compared to pasture soils might be caused by complexation of proteins with organic substances such as tannins rendering them less accessible for soil proteases (Joanisse et al., 2007). At low soil pH < 4.5, as in the silicate forest soils, formation of organo-metal-complexes also contributes to the stabilization of SOM and of proteins (Nierop et al., 2002). This might also explain the negative effect of organic C on gross depolymerization rates observed in our model (Table S4). Although microbial community composition was driven by soil pH and had a significant effect on peptidase activity, we found no effect on protein depolymerization rates, supporting the crucial role of soil physicochemical properties and substrate availability on organic N transformation rates in situ.
4.2. Temperature effects on soil organic N cycling
Microbial and enzyme activities are generally assumed to be accelerated by higher temperatures, causing faster decomposition of soil organic matter (Kirschbaum, 1995). In our study the short-term experiment temperature changes explained only 7% of the variability of gross protein depolymerization rates and 11% of the variability of microbial amino acid uptake rates (Table 3). In contrast, potential peptidase activity increased significantly with temperature for all soils, with an overall Q10 value of 1.53 (range 1.3–1.7, Table 4) and temperature explained 72% of the variability (Table 3). Similar Q10 values of potential peptidase activity around 1.6 have also been reported for alpine organic and mineral forest soils (Koch et al., 2007; Schindlbacher et al., 2015). Low temperature sensitivity of N cycling processes are in accordance with results from soil warming experiments, reporting also no significant response of organic N mineralization rates to instantaneous and long-term increased soil temperatures (Schindlbacher et al., 2015).
Moreover, the temperature sensitivity of protein depolymerization of fungal and bacterial cell wall material (e.g. chitin and peptidoglycan) measured on the same soils was lower than that of exochitinase (Hu et al., 2018). However, in the here studied forest soils increasing potential peptidase activity with temperature did not lead to faster protein depolymerization in the short term, indicating that protein stabilization/occlusion was the main control, likely fostered by protein sorption and by complexation of SOM and of proteins with tannins or metal ions (Nierop et al., 2002; Joanisse et al., 2007; Hu et al., 2018).
4.3. Soil moisture effects on soil organic N cycling
Soil moisture had no distinct effect on gross protein depolymerization and microbial amino acid uptake rates (Fig. 4a and b). Non-significant changes in gross protein depolymerization rates between 30 and 60% WHC indicate that diffusion of proteins and enzymes was not the limiting factor within the studied soil moisture range (30–60% WHC) which most likely also does not translate into a stimulation of depolymerization at higher soil moisture i.e. 90% WHC. Most notable, we observed strong inverse responses of protein depolymerization rates to suboxic conditions in calcareous and silicate soils, i.e. increases in calcareous soils and decreases in silicate soils. To explain these adverse suboxic effects on protein depolymerization the following mechanisms might be relevant: (i) energy limitation induced by lower energetic efficiency of anaerobic compared to aerobic respiration and subsequent reduction of microbial N mining (Kögel-Knabner et al., 2010), (ii) soil pH increases due to redox reactions with alternate electron acceptors such as NO3−, Mn4+, Fe3+, SO42−, and CO2 (Thompson et al., 2006; Grybos et al., 2009), (iii) release of adsorbed organic matter including proteins due to reductive dissolution of Fe and Mn oxyhydroxides (Herndon et al., 2017), and (iv) release of Al3+ with negative effects on protein availability and enzyme conformation (Garcidueñas Piña and Cervantes, 1996; Nierop et al., 2002).
-
(i)
Energy limitation: The first possible relevant mechanism relies on the lower energy efficiency of anaerobic metabolism and decomposition processes, eventually causing energy limitation of the microbial communities. This might be followed by decreases in growth and respiration, in extracellular enzyme production for N mining and inhibition of protein depolymerization, while utilization of FAA as C source might increase due to energy limitation. Though intriguing, and important for long-term responses, we found little support for the generality of this mechanism to explain changes in protein depolymerization in the short-term here. The increase of TFAA concentrations in all soils during pre-incubation at suboxic conditions (90% WHC, Fig. 4d) indicates an imbalance between production rates of amino acids by protein depolymerization and microbial amino acid utilization, and provides direct evidence that amino acids were not increasingly used as a C source. Moreover, in soils from both parent materials, microbial respiration and microbial biomass remained unaltered under suboxic conditions while microbial growth and microbial C use efficiency as reported by Zheng et al. (2019) in associated experiments declined strongly.
-
(ii)
Increasing pH: The second mechanism is based on the increase of soil pH by 0.3–2 units due to denitrification (Grybos et al., 2009) and reduction of Fe(III)- and Mn(IV)-oxyhydroxides (Thompson et al., 2006), both effectively consuming H+. We found an almost complete depletion of the soil NO3− pool by dissimilatory nitrate reduction after 7 days at 1% O2, indicating increases in soil pH in all soils. Increasing pH negatively affects the sorption capacity of organic matter to Fe-oxides (Gu et al., 1994; Kaiser and Zech, 1999) and this is assumed to be a further control on organic matter desorption under anoxic conditions besides reductive dissolution of Fe oxyhydroxides (Grybos et al., 2009). Optimal pH for sorption of proteins on clay minerals depends on the isoelectric point (IEP) of the adsorbed protein i.e. the pH where they have zero net charge. In laboratory experiments adsorption of albumin showed an optimum close to the IEP, decreasing above that due to repulsion by clays of more negatively charged proteins (Quiquampoix and Ratcliffe, 1992; Staunton and Quiquampoix, 1994). Since Fe-oxyhydroxides are the main sorption sites of organic matter in soils (Kaiser and Guggenberger, 2003), desorption of proteins due to rising pH might explain the observed increase of protein depolymerization in calcareous soils, but not the patterns found in silicate soils.
-
(iii)
Fe-Mn oxide reduction: In the studied soils the NO3− pool was exhausted after pre-incubation at 1% O2 (Fig. S2), forcing heterotrophic microbes from dissimilatory nitrate reduction (e.g. denitrification) to anoxic Mn(IV) and Fe(III) respiration. As Fe(III) is reduced to Fe(II), Fe-oxyhydroxides are dissolved, and thereby strongly sorbed organic matter is released and becomes bioavailable (Herndon et al., 2017), which can enhance the pH-dependent solubilization process of proteins under suboxic conditions. A combination of increasing pH and Fe(III) reduction therefore might explain the stimulation of protein depolymerization in calcareous soils under O2 limitation. Considering the different parent materials of the studied soils, the specific mineral composition might influence the response of gross depolymerization to anoxia. The studied silicate soils developed on sericite-quartz and phyllite which have a higher content in easily decomposable Al-silicates such as muscovite and chlorites (Keil and Neubauer, 2011). In contrast, the studied calcareous soils developed on limestone and dolomite deposits originating from the northern calcareous alps, with enhanced contents of kaolinite and Fe-(hydr)oxides such as goethite and hematite (Egli et al., 2008). The differences in pedogenesis resulted in higher dithionite-extractable and crystalline Fe in the studied calcareous soils compared to silicate soils and higher oxalate and dithionite extractable Al in silicate soils (Table 1). The silicate soils showed higher depolymerization rates at the same level of NaOH-extractable protein compared to calcareous soils (Fig. 2), indicating that proteins are more strongly sorbed in calcareous than in silicate soils. This might have also contributed to greater effects of increasing pH and Fe(III) respiration on calcareous soils where protein depolymerization is more strongly constrained by protein sorption than in silicate soils.
-
(iv)
Al3+ release: In contrast to Fe-oxyhydroxides, poorly crystalline Al phases, the most reactive sites for DOC complexation (Davis and Gloor, 1981; Kaiser, 2003; Kothawala et al., 2009), are not directly affected by redox changes. However, due to reductive dissolution of Fe-oxyhydroxides Al3+ and at pH > 4.5, Al(OH)3 are released from binding sites and might inhibit proteolytic enzymes through conformational changes as well as decreased protein availability by complexation and precipitation of Al-humus complexes (Nierop et al., 2002). Protein depolymerization rates may also decrease due to inhibition of soil microorganisms by elevated Al3+ under acidic conditions (Garcidueñas Piña and Cervantes, 1996; Scheel et al., 2007), such as found in the silicate soils studied here. In the studied soils this negative Al-effect most likely has overridden the stimulating effect of anoxic conditions on protein availability and depolymerization as demonstrated in calcareous cropland and pasture soils. This is strongly supported by the negative correlations of peptidase activity with exchangeable Al3− and amorphous Al contents (Table 1).
5. Conclusions
The presented study aimed at deciphering major controls of gross protein depolymerization and microbial amino acid uptake rates in soils considering temperature, soil moisture/O2 concentration, parent material and land use. Our results showed that cleavage of soil proteins and microbial amino acid uptake are tightly co-regulated and are mainly controlled by substrate availability, resulting in low sensitivity to short-term temperature and moisture changes compared to e.g. N mineralization. The prominent role of substrate availability is also underlined by the relatively small effects of potential peptidase activities on protein depolymerization rates. The different response of protein depolymerization rates to suboxic conditions in calcareous and silicate soils implies that in calcareous soils Fe-oxyhydroxides are the main sorption sites for soil proteins while on the contrary protein stabilization in silicate soils is rather driven by organo-metal complexes. Our findings imply that soil physicochemical properties, such as pH, mineral composition and clay content caused by different soil parent materials are the key controls of organic N cycling over biotic controls such as microbial activity, microbial community structure or potential enzyme activity.
Supplementary Material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.soilbio.2019.02.016.
Acknowledgements
We thank Theresa Böckle for help with the phosphorus measurements. Annika Retzmann, Johanna Irrgeher and Thomas Prohaska are acknowledged for ICP-SFMS measurements. This study was funded by the Austrian Science Fund (FWF; project P-28037-B22).
References
- Adamczyk B, Godlewski M, Zimny J, Zimn A. Wheat (Triticum aestivum) seedlings secrete proteases from the roots and, after protein addition, grow well on medium without inorganic nitrogen. Plant Biology. 2008;10:718–724. doi: 10.1111/j.1438-8677.2008.00079.x. [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. Journal of Statistical Software. 2014;67:1–48. [Google Scholar]
- Brookes P, Landman A, Pruden G, Jenkinson D. Chloroform fumigation and the release of soil nitrogen: a rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biology and Biochemistry. 1985;17:837–842. [Google Scholar]
- Brzostek ER, Finzi AC. Substrate supply, fine roots, and temperature control proteolytic enzyme activity in temperate forest soils. Ecology. 2011;92:892–902. doi: 10.1890/10-1803.1. [DOI] [PubMed] [Google Scholar]
- Brzostek ER, Finzi AC. Seasonal variation in the temperature sensitivity of proteolytic enzyme activity in temperate forest soils. Journal of Geophysical Research: Biogeosciences. 2012;117:G1. [Google Scholar]
- Conant RT, Ryan MG, Agren GI, Birge HE, Davidson EA, Eliasson PE, Evans SE, Frey SD, Giardina CP, Hopkins FM, Hyvonen R, et al. Temperature and soil organic matter decomposition rates - synthesis of current knowledge and a way forward. Global Change Biology. 2011;17:3392–3404. [Google Scholar]
- Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiological Reviews. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrouzet-Nardi A, Ladd MP, Weintraub MN. Fluorescent microplate analysis of amino acids and other primary amines in soils. Soil Biology and Biochemistry. 2013;57:78–82. [Google Scholar]
- Davis JA, Gloor R. Adsorption of dissolved organics in lake water by aluminum oxide. Effect of molecular weight. Environmental Science and Technology. 1981;15:1223–1229. doi: 10.1021/es00092a012. [DOI] [PubMed] [Google Scholar]
- Dippold MA, Kuzyakov Y. Biogeochemical transformations of amino acids in soil assessed by position-specific labelling. Plant and Soil. 2013;373:385–401. [Google Scholar]
- Dixon JB, White GN. Manganese oxides. In: Dixon JB, Schulze DG, editors. Soil Mineralogy with Environmental Applications. Soil Science Society of America; Madison, WI: 2002. pp. 367–388. 2002. [Google Scholar]
- Egli M, Merkli C, Sartori G, Mirabella A, Plötze M. Weathering, mineralogical evolution and soil organic matter along a Holocene soil toposequence developed on carbonate-rich materials. Geomorphology. 2008;97:675–696. [Google Scholar]
- Fanin N, Bertrand I. Aboveground litter quality is a better predictor than belowground microbial communities when estimating carbon mineralization along a land-use gradient. Soil Biology and Biochemistry. 2016;94:48–60. [Google Scholar]
- Farrell M, Hill PW, Farrar J, DeLuca TH, Roberts P, Kielland K, Dahlgren R, Murphy DV, Hobbs PJ, Bardgett RD, Jones DL. Oligopeptides represent a preferred source of organic N uptake: a global phenomenon? Ecosystems. 2012;16:133–145. [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuka MM, Engel M, Gattinger A, Bausenwein U, Sommer M, Munch JC, Schloter M. Factors influencing the variability of proteolytic genes and activities in arable soils. Soil Biology and Biochemistry. 2008;40:1646–1653. [Google Scholar]
- Gallet-Budynek A, Brzostek E, Rodgers VL, Talbot JM, Hyzy S, Finzi AC. Intact amino acid uptake by northern hardwood and conifer trees. Oecologia. 2009;160:129–138. doi: 10.1007/s00442-009-1284-2. [DOI] [PubMed] [Google Scholar]
- Garcidueñas Piña R, Cervantes C. Microbial interactions with aluminum. Biometals. 1996;9:311–316. doi: 10.1007/BF00817932. [DOI] [PubMed] [Google Scholar]
- Geisseler D, Horwath WR. Regulation of extracellular protease activity in soil in response to different sources and concentrations of nitrogen and carbon. Soil Biology and Biochemistry. 2008;40:3040–3048. [Google Scholar]
- Grybos M, Davranche M, Gruau G, Petitjean P, Pédrot M. Increasing pH drives organic matter solubilization from wetland soils under reducing conditions. Geoderma. 2009;154:13–19. [Google Scholar]
- Gu B, Schmitt J, Chen Z, Liang L, McCarthy JF. Adsorption and desorption of natural organic matter on iron oxide: mechanisms and models. Environmental Science and Technology. 1994;28:38–46. doi: 10.1021/es00050a007. [DOI] [PubMed] [Google Scholar]
- Herndon E, Al Bashaireh A, Singer D, Roy Chowdhury T, Gu B, Graham D. Influence of iron redox cycling on organo-mineral associations in Arctic tundra soil. Geochimica et Cosmochimica Acta. 2017;207:210–231. [Google Scholar]
- Hill PW, Quilliam RS, DeLuca TH, Farrar J, Farrell M, Roberts P, Newsham KK, Hopkins DW, Bardgett RD, Jones DL. Acquisition and assimilation of nitrogen as peptide-bound and D-enantiomers of amino acids by wheat. PLoS One. 2011;6:e19220. doi: 10.1371/journal.pone.0019220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood-Nowotny R, Umana NH-N, Inselbacher E, Oswald-Lachouani P, Wanek W. Alternative methods for measuring inorganic, organic, and total dissolved nitrogen in soil. Soil Science Society of America Journal. 2010;74:1018–1027. [Google Scholar]
- Hu YT, Zheng Q, Wanek W. Flux analysis of free amino sugars and amino acids in soils by isotope tracing with a novel liquid chromatography/high resolution mass spectrometry platform. Analytical Chemistry. 2017;89:9192–9200. doi: 10.1021/acs.analchem.7b01938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zheng Q, Zhang S, Noll L, Wanek W. Significant release and microbial utilization of amino sugars and D-amino acid enantiomers from microbial cell wall decomposition in soils. Soil Biology and Biochemistry. 2018;123:115–125. doi: 10.1016/j.soilbio.2018.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan MT, Roberts P, Tonheim SK, Jones DL. Protein breakdown represents a major bottleneck in nitrogen cycling in grassland soils. Soil Biology and Biochemistry. 2009;41:2272–2282. [Google Scholar]
- Jangid K, Williams MA, Franzluebbers AJ, Sanderlin JS, Reeves JH, Jenkins MB, Endale DM, Coleman DC, Whitman WB. Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biology and Biochemistry. 2008;40:2843–2853. [Google Scholar]
- Janssens IA, Pilegaard KIM. Large seasonal changes in Q10 of soil respiration in a beech forest. Global Change Biology. 2003;9:911–918. [Google Scholar]
- Jilling A, Keiluweit M, Contosta AR, Frey S, Schimel J, Schnecker J, Smith RG, Tiemann L, Grandy AS. Minerals in the rhizosphere: overlooked mediators of soil nitrogen availability to plants and microbes. Biogeochemistry. 2018;139:103–122. [Google Scholar]
- Joanisse GD, Bradley RL, Preston CM, Munson AD. Soil enzyme inhibition by condensed litter tannins may drive ecosystem structure and processes: the case of Kalmia angustifolia. New Phytologist. 2007;175:535–546. doi: 10.1111/j.1469-8137.2007.02113.x. [DOI] [PubMed] [Google Scholar]
- Jones DL, Owen AG, Farrar JF. Simple method to enable the high resolution determination of total free amino acids in soil solutions and soil extracts. Soil Biology and Biochemistry. 2002;34:1893–1902. [Google Scholar]
- Jones D, Kielland K, Sinclair F, Dahlgren R, Newsham K, Farrar J, Murphy D. Soil organic nitrogen mineralization across a global latitudinal gradient. Global Biogeochemical Cycles. 2009;23:GB1016. [Google Scholar]
- Kaiser K. Sorption of natural organic matter fractions to goethite (α-FeOOH): effect of chemical composition as revealed by liquid-state13C-NMR and wet-chemical analysis. Organic Geochemistry. 2003;34:1569–1579. [Google Scholar]
- Kaiser K, Guggenberger G. Mineral surfaces and soil organic matter. European Journal of Soil Science. 2003;54:219–236. [Google Scholar]
- Kaiser K, Zech W. Release of natural organic matter sorbed to oxides and a subsoil. Soil Science Society of America Journal. 1999;63:1157–1166. [Google Scholar]
- Kaiser C, Koranda M, Kitzler B, Fuchslueger L, Schnecker J, Schweiger P, Rasche F, Zechmeister-Boltenstern S, Sessitsch A, Richter A. Belowground carbon allocation by trees drives seasonal patterns of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytologist. 2010;187:843–858. doi: 10.1111/j.1469-8137.2010.03321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallenbach CM, Frey SD, Grandy AS. Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nature Communications. 2016;7:1–10. doi: 10.1038/ncomms13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil M, Neubauer F. The Miocene Enns valley basin (Austria) and the North Enns valley fault. Austrian Journal of Earth Sciences. 2011;104:49–65. [Google Scholar]
- Kirkham D, Bartholomew W. Equations for following nutrient transformations in soil, utilizing tracer data. Soil Science Society of America Journal. 1954;18:33–34. [Google Scholar]
- Kirschbaum MU. The temperature dependence of soil organic matter decomposition, and the effect of global warming on soil organic C storage. Soil Biology and Biochemistry. 1995;27:753–760. [Google Scholar]
- Koch O, Tscherko D, Kandeler E. Temperature sensitivity of microbial respiration, nitrogen mineralization, and potential soil enzyme activities in organic alpine soils. Global Biogeochemical Cycles. 2007;21:GB4017. [Google Scholar]
- Kögel-Knabner I, Amelung W, Cao Z, Fiedler S, Frenzel P, Jahn R, Kalbitz K, Kölbl A, Schloter M. Biogeochemistry of paddy soils. Geoderma. 2010;157:1–14. [Google Scholar]
- Kögel-Knabner I, Guggenberger G, Kleber M, Kandeler E, Kalbitz K, Scheu S, Eusterhues K, Leinweber P. Organo-mineral associations in temperate soils: integrating biology, mineralogy, and organic matter chemistry. Journal of Plant Nutrition and Soil Science. 2008;171:61–82. [Google Scholar]
- Kothawala D, Moore T, Hendershot W. Soil properties controlling the adsorption of dissolved organic carbon to mineral soils. Soil Science Society of America Journal. 2009;73:1831–1842. [Google Scholar]
- Kuo S. Phosphorus. In: Sparks DL, editor. Methods of Soil Analysis Part 3 Chemical Methods. Soil Sience Society of America Inc., American Society of Agronomy Inc.; Madison, WI: 1996. pp. 869–919. [Google Scholar]
- Lachouani P, Frank AH, Wanek W. A suite of sensitive chemical methods to determine the d15N of ammonium, nitrate and total dissolved N in soil extracts. Rapid Communications in Mass Spectrometry. 2010;24:3615–3623. doi: 10.1002/rcm.4798. [DOI] [PubMed] [Google Scholar]
- Lajtha K, Driscoll C, Jarrell W, Elliott E. Standard Soil Methods for Long-Term Ecological Research. Oxford University Press; New York: 1999. Soil phosphorus: characterization and total element analysis; pp. 115–142. [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. Soil pH as a predictor of soil bacterial community structure at the continental scale: a pyrosequencing-based assessment. Applied and Environmental Microbiology. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann J, Kleber M. The contentious nature of soil organic matter. Nature. 2015;528:60–68. doi: 10.1038/nature16069. [DOI] [PubMed] [Google Scholar]
- Lützow Mv, Kögel-Knabner I, Ekschmitt K, Matzner E, Guggenberger G, Marschner B, Flessa H. Stabilization of organic matter in temperate soils: mechanisms and their relevance under different soil conditions–a review. European Journal of Soil Science. 2006;57:426–445. [Google Scholar]
- Martens DA, Loeffelmann KL. Soil amino acid composition quantified by acid hydrolysis and anion chromatography-pulsed amperometry. Journal of Agricultural and Food Chemistry. 2003;51:6521–6529. doi: 10.1021/jf034422e. [DOI] [PubMed] [Google Scholar]
- Moldrup P, Olesen T, Komatsu T, Schjønning P, Rolston D. Tortuosity, diffusivity, and permeability in the soil liquid and gaseous phases. Soil Science Society of America Journal. 2001;65:613–623. [Google Scholar]
- Mooshammer M, Wanek W, Hämmerle I, Fuchslueger L, Hofhansl F, Knoltsch A, Schnecker J, Takriti M, Watzka M, Wild B. Adjustment of microbial nitrogen use efficiency to carbon: nitrogen imbalances regulates soil nitrogen cycling. Nature Communications. 2014;5 doi: 10.1038/ncomms4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Mueller R, Myrold D. Diversity and Phylogenetic Distribution of Extracellular Microbial Peptidases EGU 2017. Geophysical Research Abstracts; Vienna, Austria: 2017. p. 2674. [Google Scholar]
- Nierop KG, Jansen B, Verstraten JM. Dissolved organic matter, aluminium and iron interactions: precipitation induced by metal/carbon ratio, pH and competition. The Science of the Total Environment. 2002;300:201–211. doi: 10.1016/s0048-9697(02)00254-1. [DOI] [PubMed] [Google Scholar]
- Noll L, Zhang S, Wanek W. Novel high-throughput approach to determine key processes of soil organic nitrogen cycling: gross protein depolymerization and microbial amino acid uptake. Soil Biology and Biochemistry. 2019 doi: 10.1016/j.soilbio.2018.12.005. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin P, O'Hara R, Simpson G, Solymos P. Vegan: Community Ecology Package. R Package Version 2.3-5. R Foundation; Vienna, Austria: 2016. [Google Scholar]
- Picek T, Šimek M, Šantrůčková H. Microbial responses to fluctuation of soil aeration status and redox conditions. Biology and Fertility of Soils. 2000;31:315–322. [Google Scholar]
- Ponnamperuma FN. The chemistry of submerged soils. Advances in Agronomy. 1972;24:29–96. [Google Scholar]
- Prommer J, Wanek W, Hofhansl F, Trojan D, Offre P, Urich T, Schleper C, Sassmann S, Kitzler B, Soja G, Hood-Nowotny RC. Biochar decelerates soil organic nitrogen cycling but stimulates soil nitrification in a temperate arable field trial. PLoS One. 2014;9:e86388. doi: 10.1371/journal.pone.0086388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiquampoix H. Mechanisms of protein adsorption on surfaces and consequences for extracellular enzyme activity in soil. In: Bollag J-M, Stotzky G, editors. Soil Biochemistry. Marcel Dekker, Inc.; New York: 2000. pp. 171–206. [Google Scholar]
- Quiquampoix H, Ratcliffe RG. A31P NMR study of the adsorption of bovine serum albumin on montmorillonite using phosphate and the paramagnetic cation Mn2+: modification of conformation with pH. Journal of Colloid and Interface Science. 1992;148:343–352. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]
- Reardon PN, Chacon SS, Walter ED, Bowden ME, Washton NM, Kleber M. Abiotic protein fragmentation by manganese oxide: implications for a mechanism to supply soil biota with oligopeptides. Environmental Science and Technology. 2016;50:3486–3493. doi: 10.1021/acs.est.5b04622. [DOI] [PubMed] [Google Scholar]
- Rillig MC, Caldwell BA, Wösten HAB, Sollins P. Role of proteins in soil carbon and nitrogen storage: controls on persistence. Biogeochemistry. 2007;85:25–44. [Google Scholar]
- Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporaso J, GKnight R, Fierer N. Soil bacterial and fungal communities across a pH gradient in an arable soil. The ISME Journal. 2010;4:1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- Russo F, Johnson C, Johnson C, McKenzie D, Aiken J, Pedersen J. Pathogenic prion protein is degraded by a manganese oxide mineral found in soils. Journal of General Virology. 2009;90:275–280. doi: 10.1099/vir.0.003251-0. [DOI] [PubMed] [Google Scholar]
- Schädel C, Bader MKF, Schuur EA, Biasi C, Bracho R, Čapek P, De Baets S, Diáková K, Ernakovich J, Estop-Aragones C, Graham DE, et al. Potential carbon emissions dominated by carbon dioxide from thawed permafrost soils. Nature Climate Change. 2016;6:950–953. [Google Scholar]
- Scheel T, Dörfler C, Kalbitz K. Precipitation of dissolved organic matter by aluminum stabilizes carbon in acidic forest soils. Soil Science Society of America Journal. 2007;71:64–74. [Google Scholar]
- Schimel JP, Bennett J. Nitrogen mineralization: challenges of a changing paradigm. Ecology. 2004;85:591–602. [Google Scholar]
- Schimel J, Balser TC, Wallenstein M. Microbial stress-response physiology and its implications for ecosystem function. Ecology. 2007;88:1386–1394. doi: 10.1890/06-0219. [DOI] [PubMed] [Google Scholar]
- Schindlbacher A, Schnecker J, Takriti M, Borken W, Wanek W. Microbial physiology and soil CO2 efflux after 9 years of soil warming in a temperate forest - no indications for thermal adaptations. Global Change Biology. 2015;21:4265–4277. doi: 10.1111/gcb.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulten H-R, Schnitzer M. The Chemistry of Soil Organic Nitrogen: A Review. Biology and Fertility of Soils. 1997;26:1–15. [Google Scholar]
- Six J, Jastrow JD. Organic matter turnover. In: Lal R, editor. Encyclopedia of Soil Science. Marcel Dekker; Boca Raton, FL: 2002. p. 936. 9420. [Google Scholar]
- Spohn M, Widdig M. Turnover of carbon and phosphorus in the microbial biomass depending on phosphorus availability. Soil Biology and Biochemistry. 2017;113:53–59. [Google Scholar]
- Spohn M, Klaus K, Wanek W, Richter A. Microbial carbon use efficiency and biomass turnover times depending on soil depth – implications for carbon cycling. Soil Biology and Biochemistry. 2016;96:74–81. [Google Scholar]
- Starz W, Steinwidder A, Pfister R, Rohrer H. Vergleich zwischen kurzrasenweide und schnittnutzung unter ostalpinen klimabedingungen. Beiträge zur 11. Wissenschaftstagung Ökologischer Landbau; Justus-Liebig-Universität Gießen: 2011. pp. 93–96. [Google Scholar]
- Staunton S, Quiquampoix H. Adsorption and conformation of bovine serum albumin on montmorillonite: modification of the balance between hydrophobic and electrostatic interactions by protein methylation and pH variation. Journal of Colloid and Interface Science. 1994;166:89–94. [Google Scholar]
- Steinweg JM, Dukes JS, Wallenstein MD. Modeling the effects of temperature and moisture on soil enzyme activity: linking laboratory assays to continuous field data. Soil Biology and Biochemistry. 2012;55:85–92. [Google Scholar]
- Steinweg JM, Kostka JE, Hanson PJ, Schadt CW. Temperature sensitivity of extracellular enzymes differs with peat depth but not with season in an ombrotrophic bog. Soil Biology and Biochemistry. 2018;125:244–250. [Google Scholar]
- Thompson A, Chadwick OA, Rancourt DG, Chorover J. Iron-oxide crystallinity increases during soil redox oscillations. Geochimica et Cosmochimica Acta. 2006;70:1710–1727. [Google Scholar]
- Vranova V, Rejsek K, Formanek P. Proteolytic activity in soil: a review. Applied Soil Ecology. 2013;70:23–32. [Google Scholar]
- Wallenstein MD, Weintraub MN. Emerging tools for measuring and modeling the in situ activity of soil extracellular enzymes. Soil Biology and Biochemistry. 2008;40:2098–2106. [Google Scholar]
- Wanek W, Mooshammer M, Blöchl A, Hanreich A, Richter A. Determination of gross rates of amino acid production and immobilization in decomposing leaf litter by a novel15N isotope pool dilution technique. Soil Biology and Biochemistry. 2010;42:1293–1302. [Google Scholar]
- Wattel-Koekkoek EJW, van Genuchten PPL, Buurman P, van Lagen B. Amount and composition of clay-associated soil organic matter in a range of kaolinitic and smectitic soils. Geoderma. 2001;99:27–49. [Google Scholar]
- Wickings K, Grandy AS, Reed SC, Cleveland CC. The origin of litter chemical complexity during decomposition. Ecology Letters. 2012;15:1180–1188. doi: 10.1111/j.1461-0248.2012.01837.x. [DOI] [PubMed] [Google Scholar]
- Wild B, Schnecker J, Knoltsch A, Takriti M, Mooshammer M, Gentsch N, Mikutta R, Alves RJE, Gittel A, Lashchinskiy N. Microbial nitrogen dynamics in organic and mineral soil horizons along a latitudinal transect in western Siberia. Global Biogeochemical Cycles. 2015;29:567–582. doi: 10.1002/2015GB005084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witteveen CF, Visser J. Polyol pools in Aspergillus Niger. FEMS Microbiology Letters. 1995;134:57–62. doi: 10.1111/j.1574-6968.1995.tb07914.x. [DOI] [PubMed] [Google Scholar]
- WRB IWG. International Soil Classification System for Naming Soils and Creating Legends for Soil Maps. World Soil Resources Reports. World Soil Resources Reports. FAO; Rome: World Reference Base for Soil Resources 2014, Update 2015. [Google Scholar]
- Zhang L, Altabet MA. Amino-group-specific natural abundance nitrogen isotope ratio analysis in amino acids. Rapid Communications in Mass Spectrometry. 2008;22:559–566. doi: 10.1002/rcm.3393. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Hu Y, Zhang S, Noll L, Böckle T, Richter A, Wanek W. Growth explains microbial carbon use efficiency across soils differing in land use and geology. Soil Biology and Biochemistry. 2019;128:45–55. doi: 10.1016/j.soilbio.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.