Abstract
Dynein-2 assembles with polymeric intraflagellar transport (IFT) trains to form a transport machinery crucial for cilia biogenesis and signaling. Here we recombinantly expressed the ~1.4 MDa human dynein-2 complex and solved its cryo-EM structure to near-atomic resolution. The two identical copies of the dynein-2 heavy chain are contorted into different conformations by a WDR60-WDR34 heterodimer and a block of two RB and six LC8 light chains. One heavy chain is steered into a zig-zag, which matches the periodicity of the anterograde IFT-B train. Contacts between adjacent dyneins along the train indicate a cooperative mode of assembly. Removal of the WDR60-WDR34-light chain subcomplex renders dynein-2 monomeric and relieves auto-inhibition of its motility. Our results converge on a model in which an unusual stoichiometry of non-motor subunits control dynein-2 assembly, asymmetry, and activity, giving mechanistic insight into dynein-2’s interaction with IFT trains and the origin of diverse functions in the dynein family.
Introduction
Cilia and flagella are constructed by an intricate motor-protein-driven process of intraflagellar transport1 (IFT). Dynein-2, the ubiquitous motor for retrograde IFT, is crucial for cilia biogenesis2–5 and the essential roles of these organelles in cell propulsion, signaling, and sensing environmental stimuli6,7. Dynein-2-powered movement of cargo along the cilium’s microtubules enables dynamic remodeling of the ciliary proteome. For example, dynein-2 exports G-protein coupled receptors out of the cilium, enabling proper signal transduction8,9, and also has emerging roles10 including in organization of the ciliary transition zone11,12. Loss of dynein-2 is embryonic lethal in mammals13, and its mutation is associated with a group of developmental disorders arising from defects in ciliary signaling, encompassing short-rib thoracic dysplasias with or without polydactyly14–22.
Dynein-2 operates in the context of IFT “trains”: linear polymers of IFT-A and -B complexes which bind cargoes, either directly23 or via associating factors such as the BBSome8. Dynein-2 assembles with IFT trains at the ciliary base, moves to the tip in an inhibited state under the power of kinesin-II, then restructures to drive retrograde transport back to the cell body24,25–31. How the fundamental recognition event between dynein-2 and the IFT train at the base of the cilium is achieved is unclear. Dynein-2 is one of the three major classes of dynein. The other two major classes drive intracellular transport (cytoplasmic dynein-1) and ciliary beating (multiple axonemal isoforms)7,32. Biochemical studies indicate that dynein-2 is a >1 MDa complex with a distinctive composition, incorporating three types of light chain (LC) shared with other dyneins2,33,34 (DYNLRB, DYNLL, DYNLT; referred to here as RB, LC8, and TCTEX) and five components specific to dynein-2: the motor-domain-containing heavy chain35,3–5 (DYNC2H1; here “DHC2”), light-intermediate chain36,37 (DYNC2LI1; here “LIC3”), light chain19,22,34,38 (TCTEX1D2), and two different intermediate chains33,34,39 (WDR60 and WDR34) (Supplementary Table 1). In the absence of structural information, the roles of dynein-2’s multitude of subunits in regulation of motor activity and IFT train recognition have been unclear40.
Here, we recombinantly expressed the complete dynein-2 complex and determined its structure to near-atomic resolution using cryo-EM. We show how an unusual stoichiometry of non-motor subunits brings together two copies of the dynein-2 heavy chain, controls their activity, and contorts them into an architecture with extreme asymmetry that matches the periodicity of the IFT-B train. Together, these results give insight into the molecular mechanism of IFT and the origin of diverse functions in the dynein family.
Results
Dynein-2 cryo-EM structure determination
We purified the human dynein-2 complex by co-expressing all of its subunits from a single baculovirus in insect cells (Supplementary Fig. 1a-d) and determined its structure by cryo-EM (Fig. 1). Initial EM analysis revealed that dynein-2’s “tail” domain flexed by as much as 50° and 250 Å relative to its motor domains, displaying a continuum of positions with a favored kink to one side (Supplementary Fig. 1e,f; Supplementary Video 1). Despite this large-scale flexibility, we found that focused classifications on the tail excluding the motor domains yielded detailed averages (and vice versa) (Supplementary Fig. 2), suggesting that the tail and motor domains pivot largely as rigid bodies about a hinge point near their junction. We were therefore able to determine cryo-EM structures of the tail and motor domains at average resolutions of 4.5 Å and 3.9 Å respectively, with varying local resolution as depicted in Supplementary Fig. 2f,g. Further local refinements within the tail yielded maps at 4.4 Å. We built a model of the dynein-2 tail using Rosetta comparative modeling41–44 and de novo tracing (Table 1, Supplementary Fig. 3), refined the crystal structure of the motor domain as a monomer45 into the dimer conformation (Supplementary Fig. 3c), and thus revealed the architecture of the dynein-2 complex.
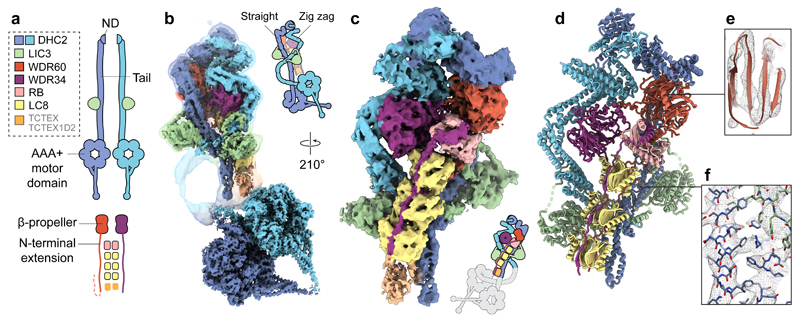
Figure 1. Cryo-EM structure of the dynein-2 complex.
(a) Overview of the human dynein-2 subunits and their stoichiometry in the structure, coloured according to the code in the upper left. The two copies of DHC2 are coloured in different shades of blue for distinction. ND; N-terminal domain. (b) Cryo-EM reconstructions of the dynein-2 tail domain and motor domains (solid). An unsharpened map showing the flexible connection between them is overlaid (transparent isosurface). Isosurfaces are coloured by subunit. (c) Enlarged view of the tail domain. (d) Ribbon representation of the tail domain. (e) Example density within the dynein-2 tail at the WDR60 region (mesh representation), showing β-strand separation. Model is shown in ribbon representation. (f) Enlargement showing density for bulky amino acid side chains in the DHC2/LIC3 region. Model is shown in stick representation.
Table 1. Cryo-EM data collection, refinement and validation statistics.
| Motor domain (EMD-4917, PDB 6RLA) |
Tail (EMD-4918, PDB 6RLB) |
|
|---|---|---|
| Data collection and processing | ||
| Magnification | 105,000 | 105,000 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e–/Å2) | 49.6a, 46.8b, 45.4c | 49.6a, 46.8b, 45.4c |
| Defocus range (μm) | –1 to –3.5 | –1 to –3.5 |
| Pixel size (Å) | 1.39 | 1.39 |
| Symmetry imposed | C2 | C1 |
| Initial particle images (no.) | 229,969 | 461,684 |
| Final particle images (no.) | 57,265 | 68,623 |
| Map resolution (Å) | 3.9 | 4.5 |
| FSC threshold | 0.143 | 0.143 |
| Map resolution range (Å) | 3.7–5.5 | 4.3–8.0 |
| Refinement | ||
| Initial model used (PDB code) | 4RH7 | 5OWO, 6F1T, 6F1Y, 6F1U, 6F1Z, 2PG1, 2XQQ & de novo |
| Model resolution (Å) | 3.8 | 4.5 |
| FSC threshold | 0.143 | 0.143 |
| Model resolution range (Å) | 3.7–5.5 | 4.3–8.0 |
| Map sharpening B factor (Å2) | –100 | –125 |
| Model composition | ||
| Nonhydrogen atoms | 43,034 | 20,408 |
| Protein residues | 5,500 | 4,122 |
| Ligands | ADP: 6, ATP: 2, Mg: 2 | 0 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.007 |
| Bond angles (°) | 1.141 | 0.951 |
| Validation | ||
| MolProbity score | 1.65 | backbone model |
| Clashscore | 3.81 | 0.41 |
| Poor rotamers (%) | 0.35 | backbone model |
| Ramachandran plot | ||
| Favored (%) | 92.07 | 95.91 |
| Allowed (%) | 7.93 | 4.04 |
| Disallowed (%) | 0 | 0.05 |
Dataset 1a, 2b, 3c
Dynein-2 has subunit copy numbers ranging from one to six
The most striking features of dynein-2 are its stoichiometry and asymmetry (Fig. 1, Supplementary Video 2). The subunit copy number in the complex ranges from one to six, contrasting with dynein-1, which is built from homodimers. The two copies of DHC2 span the complex, each comprising a compact N-terminal domain, an elongated tail region, and a C-terminal AAA+ motor domain. Whereas one copy of DHC2 is straight in the tail region, the other is in a zig-zag. Both copies of DHC2 bind a LIC3 subunit. The two intermediate chains, WDR60 and WDR34, form a heterodimer. Their C-terminal β-propeller domains bind DHC2, whereas their N-proximal regions are held together by an array of light chains consisting of one RB dimer and three LC8 dimers. We attribute a flexible, weaker density (Supplementary Fig. 4) to the TCTEX-TCTEX1D2 dimer identified in biochemical studies38. The presence of three LC8 dimers bridging WDR60 and WDR34 is unexpected, as only binding motifs for one dimer had been predicted using the most sensitive computational searches46. Indeed, the additional motifs deviate from the consensus sequence (Supplementary Fig. 4a), suggesting that LC8 binding to these sites is individually weak and enhanced by avidity in the complex42,47. While the stoichiometry of dynein-2 is unexpected, its structural organization is in agreement with previous studies highlighting DHC2-LIC3 and WDR60-WDR34-RB-LC8-TCTEX-TCTEX1D2 as separable interacting sub-complexes 38,48,49 (Fig. 1).
Dynein-2’s two identical heavy chains adopt different conformations
The two copies of DHC2 display a contrasting mixture of symmetric and asymmetric regions (Fig. 2a). The motor domains pack against each other with 180° rotational (C2) symmetry, forming the auto-inhibited conformation first visualized in 2D25 and at ~40 Å resolution in situ29, suggesting that the dynein-2 complex intrinsically exists in a regulated state ready for anterograde transport. The compact N-terminal domains also have C2 symmetry. In contrast, the intervening tail region (DHC2TAIL) is highly asymmetric. Each copy, which we designate as DHC2TAIL-A and DHC2TAIL-B, follows a different path after emerging from the motor such that the N-proximal segments are related by ~90° (Fig. 2a). Both copies of DHC2TAIL are composed of a series of α-helical bundles with a β-sheet bridging bundles 6 and 7 (Fig. 2b,c). The large-scale differences in the architecture of DHC2TAIL-A (straight) and -B (zig-zag) arise from hinge-like movements at the junctions between bundles 2-3 and 3-4 (Fig. 2c).
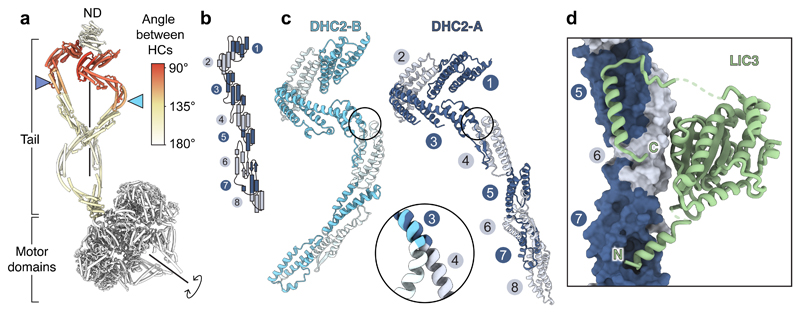
Figure 2. DHC2 asymmetry and LIC3 binding.
(a) Cylinder representation of the two copies of the DHC2, coloured by the rotation angle relating them. Other subunits omitted for clarity. Blue arrowheads indicate how equivalent helical bundles in DHC2TAIL are offset along the rotation axis. ND; N-terminal domain. (b) Topology of helical bundles (numbered) in the tail region. (c) Different conformations of the two copies of DHC2, which are shown after alignment on bundle 3. Enlargement shows major hinge site between bundles 3 and 4. (d) LIC3 binds to bundles 5-7 of the DHC2.
LIC3 stabilizes both heavy chains
LIC3 binds in a similar way to DHC2-A and -B, stabilizing the portion of the tail distal to the hinge sites (Fig. 2d). LIC3’s globular core has a Ras-like fold50, with a conserved nucleotide-binding pocket (Supplementary Note 1). This globular core associates with DHC2 bundle 6 and sends out two projections: an N-terminal α-helix that inserts into bundle 7, and a C-terminal region that extends toward bundle 4 before packing back as an α-helix against bundle 5. The C-terminal helix of LIC3 on DHC2-A forms a platform with which WDR60 and RB interact (Fig. 3c). Jeune Syndrome-associated mutation DHC2L871P (ref 14) locates near to the DHC2-LIC3 interface at bundle 7. This and other disease-associated mutations with the potential to disrupt subunit-subunit interfaces are highlighted in Supplementary Note 2. We conclude that the intimate association between LIC3 and DHC2 provides structural support to bundles 5-7 of DHC2, generates interaction surfaces for other subunits, and indicates why perturbation of LIC3 destabilizes DHC2 in vivo51,52.
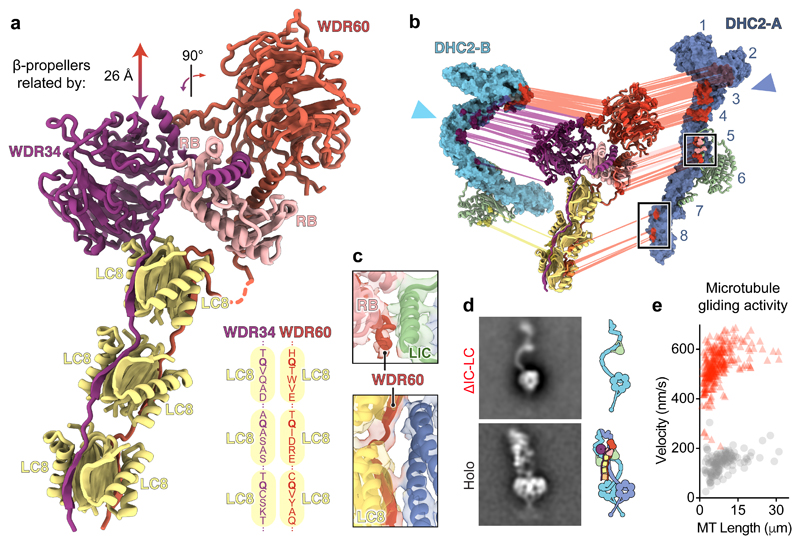
Figure 3. A block of intermediate and light chains controls dynein-2 asymmetry, oligomerization, and activity.
(a) Ribbon diagram of the intermediate chains WDR34 and WDR60, whose N-proximal extensions are held together by a block of RB and LC8 light chains. Other subunits are omitted for clarity. The translation and rotation relating the WDR34 and WDR60 β-propeller domains is indicated. Schematic (lower right) shows the LC8-binding motifs of WDR34 and WDR60. LC8 is known to be promiscuous in the peptides it can bind, but a glutamine (Q) – typically flanked by small hydrophobic or polar residues – near the C-terminus is important for binding47. All six of the LC8-binding motifs in WR60 and WDR34 contain this glutamine but are otherwise divergent in sequence. (b) Exploded view of asymmetric interactions between the IC-LC block and the two copies of DHC2. Interacting residues predicted by Rosetta modelling are connected by lines and coloured according to IC-LC subunit type. (c) Enlargements of the boxed regions in b highlighting interactions between DHC2-A and WDR60’s N-proximal region. The cryo-EM map and fitted structural model are shown. (d) Negative stain EM class averages of the dynein-2 holoenzyme (Holo) and a mutant lacking the intermediate chains and light chains (ΔIC-LC), with corresponding cartoons. (e) Plot of microtubule gliding velocity as a function of microtubule (MT) length for dynein-2 Holo (grey circles; n = 130 microtubule gliding events from three separate experiments) and ΔIC-LC (red triangles; n = 308 microtubule gliding events from three separate experiments).
Structural origins of dynein-2 asymmetry
We asked how and why the two copies of DHC2 are driven into different conformations. DHC2TAIL-A and -B, are bound by the β-propeller of WDR60 and WDR34 respectively, mainly via bundle 3 (Fig. 3a, b). In dynein-1, which has a homodimeric intermediate chain, the two β-propellers are related by a ~130° rotation as a result of the way in which their N-termini are tethered by RB53 (Supplementary Fig. 5). In contrast, in dynein-2, the β-propellers of WDR60 and WDR34 have a markedly different relationship (Fig. 3a). Owing to distinct interactions made by their surface loops, the WDR60 β-propeller binds to the top of RB, whereas the WDR34 β-propeller contacts the side of RB and the adjacent LC8. As a consequence, WDR60 and WDR34 hold the two copies of DHC2TAIL at right angles and stagger them such that DHC2TAIL-B is closer to the motor domains (Fig. 3b). As the motor domains are constrained by C2 symmetry, the two copies of DHC2TAIL are forced into different conformations to accommodate the symmetry mismatch (Fig. 2a).
Protruding from the β-propellers, the extended N-proximal regions of WDR60 and WDR34 template the block of dimeric light chains (Fig. 3a). Akin to a splint, this block of intermediate and light chains (“IC-LC block”) forms a structural element that stabilizes straightening of DHC2TAIL-A (Fig. 3b). For example, WDR60, RB, and LIC3 interact near DHC2-A bundle 5 (Fig. 3c, top). Furthermore, WDR60 peptides in LC8 contact DHC2-A directly at bundles 7 and 8 (Fig. 3c, bottom).
Intermediate and light chains control dynein-2 assembly, asymmetry, and activity
To assess the role of IC-LC subcomplex in dynein-2 complex formation, we generated a construct lacking these subunits (“ΔIC-LC”; Fig. 3d). DHC2, while maintaining a tight association with LIC3, did not homodimerize in the absence of the bridging IC-LC subunits and was monomeric (Fig. 3d). DHC2’s propensity to dissociate from dimer to monomer is compatible with the ~18 S and ~12 S forms of its ortholog found in Chlamydomonas reinhardtii extracts33,54, suggesting that weak dimerization is a conserved property of the dynein-2 heavy chain. DHC2TAIL was flexible in ΔIC-LC, adopting a range of different curvatures (Supplementary Note 3). Motor activity of ΔIC-LC was unregulated: whereas the holoenzyme bound microtubules weakly and exhibited slow (~140 nm/s) microtubule gliding, consistent with the majority of complexes being in an auto-inhibited state, ΔIC-LC drove rapid (~530 nm/s) microtubule movement, similar to the isolated motor domain25 and the velocity of retrograde IFT in mammalian cells8 (Fig. 3e, Supplementary Video 3). Together, these data indicate that the IC-LC block i) brings together two copies of DHC2; ii) stabilizes their auto-inhibited form; and iii) breaks their symmetry, sculpting their tails into straight and zig-zag conformations.
Dynein-2’s asymmetric structure matches the periodicity of the anterograde IFT-B train
How do dynein-2’s subunits enable it to assemble with IFT trains? To address this question, we docked our structures of the dynein-2 complex into a sub-tomogram average of the anterograde IFT-B train from C. reinhardtii at 37 Å resolution29. The distinctive shape of dynein-2’s tail and motor domains yielded an unambiguous fit (Supplementary Video 4). The angle between the dynein-2 tail and motor domains in the IFT-train-binding configuration lies within the range of angles observed in the isolated molecule (Supplementary Fig. 1). Docking successive dynein-2 complexes along the long-axis of the train reveals how they interdigitate and interact with one another (Fig. 4a). For example, there is a major contact between LIC3 of one dynein-2 complex and the motor domain of its neighbor (Fig. 4a; inset). Hence the presence of one dynein-2 complex on the train creates extra binding surface for the next. Each dynein-2 spans out over 7-8 IFT-B repeats (Fig. 4b). This expanded footprint suggests that dynein-2 affinity is selective for the assembled train over individual IFT-B proteins. Remarkably, the principal contacts between dynein-2 and the train all involve the heavy chain, DHC2, rather than the non-motor subunits. Moreover, they specifically involve the zig-zag heavy chain (DHC2-B), which matches the periodicity of the IFT-B polymer (Fig. 4b). Following bundle 1, DHC2-B arches up then down to interact via the bundle 3-4 junction, and then rises up and down again to form a contact via AAA5-AAA6 within the motor domain. The docking also places two of dynein-2’s flexible elements – the stalk of DHC2-A and the putatively disordered N-terminus of WDR60 – within potential contact distance to the IFT-B polymer. Together, these results reveal that the contorted architecture of the dynein-2 heavy chains is tailored to the polymeric structure of the anterograde IFT-B train.
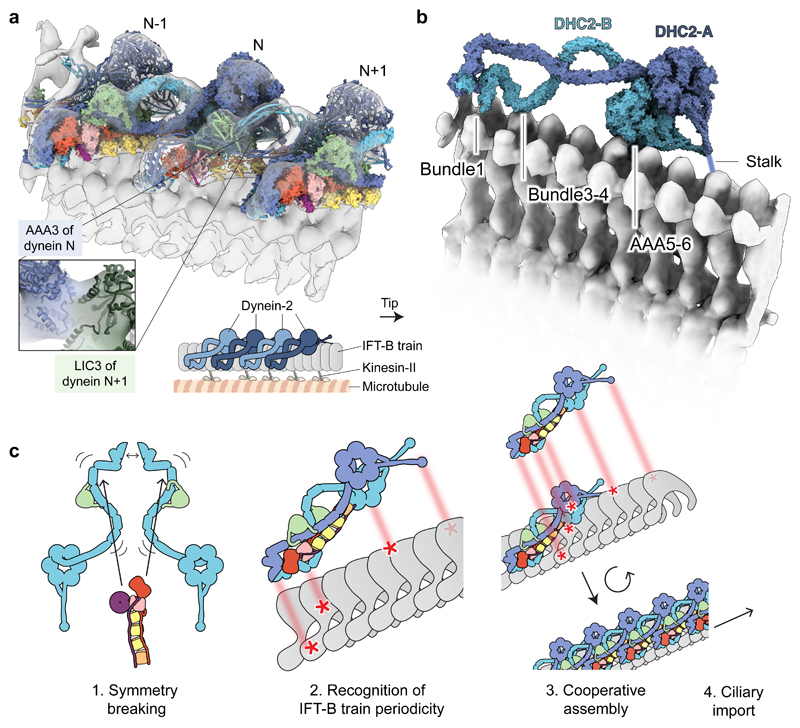
Figure 4. Dynein-2’s asymmetric structure matches the periodicity of the IFT train.
(a) Docking of dynein-2 complexes into a tomogram of an anterograde IFT-B train (EMDB-4303; transparent isosurface)29. Individual dynein-2 complexes are shown in alternating surface and cylinder representation for distinction. The enlargement shows contact between AAA3 of one dynein-2 complex (N) and LIC3 of the adjacent complex (N+1). The cartoon depicts the positions of dynein-2, IFT-B, kinesin-II and the microtubule doublet within the cilium. (b) View of the two copies of DHC2 in dynein-2 complex N. Sub-domains in proximity to IFT-B are indicated. Other subunits are omitted for clarity. (c) Model. See main text for details. An interesting variant of this model may apply in trypanosomatids, which feature two distinct dynein-2 heavy chains that exist in a heterodimer57 and may thus be intrinsically asymmetric.
Discussion
Our work shows how dynein-2 uses a mixed stoichiometry of non-motor subunits to generate a radically asymmetric structure that matches its cognate partner, the IFT train. We propose a model (Fig. 4c) in which the IC-LC block brings together two copies of DHC2-LIC3 and sculpts them into different conformations. The zig-zag heavy chain recognizes the periodicity of the assembling IFT train at the ciliary base. In mutants lacking intermediate or light chains, the heavy chain’s flexibility may enable it to reach its IFT-train-binding shape stochastically, albeit inefficiently, contributing to the impaired but non-null IFT phenotypes observed11,55. Binding of one dynein-2 complex to the train creates extra binding surface for the next, enabling cooperative loading of dynein-2 along the train, before the entire assembly is imported into the cilium by kinesin-II28. In this model, IFT train disassembly at the ciliary tip24 would destroy the multivalent binding site for inhibited dynein-2, allowing dynein-2 to reconfigure for retrograde motility25,29. The IC-LC block stabilizes the autoinhibited state of dynein-2. It must therefore either reconfigure dramatically or dissociate56,33 during dynein-2 activation at the ciliary tip. Our structure of the dynein-2 complex may help to dissect this event. The structural principles illuminated here – namely, modular use of non-motor subunits to sculpt the large, flexible dynein heavy chains into a configuration matching their function – may be used widely in the dynein family, as axonemal dyneins powering ciliary beating are also likely to contain heteromeric intermediate chains and light chain arrays7.
Methods
Expression of the dynein-2 complex
To generate a plasmid for expression of the human dynein-2 complex, the following genes were synthesised with codon-optimisation for insect cells (Epoch): the dynein-2 heavy chain (DYNC2H1), two intermediate chains (WDR60 and WDR34), light-intermediate chain (DYNC2LI1), light chains (DYNLRB1/2, DYNLL1/2, DYNLT1/3, and TCTEX1D2), and one component (LC8-like; LOC392067) that proved not to be part of dynein-2 and thus served as a control for the specificity of the complex. See Supplementary Table 1 for alternate subunit nomenclature. Using Gibson assembly, each subunit was inserted into one of a family of plasmids derived from pACEBac1 (Geneva Biotech). Each plasmid had an expression cassette containing a polH promoter and a SV40 terminator, followed by a SwaI site. Using flanking PmeI sites, expression cassettes were excised from the parent plasmid and inserted into the SwaI site of another plasmid in the family using Gibson assembly. Using this strategy in a pyramid fashion (Supplementary Fig. 1a), a plasmid containing all subunits was assembled. DHC2 has an N-terminal His tag, ZZ tag, TEV cleavage sites, and SNAPf tag and WDR34 has a C-terminal Strep tag. A construct lacking the intermediate and light chains (ΔIC-LC) was made by excising their expression cassettes using restriction enzymes. DNA sequences were verified by complete plasmid next-generation sequencing (Massachusetts General Hospital Center for Computational & Integrative Biology).
For baculovirus-based expression in insect cells, constructs were inserted into the Tn7 site of the EMBacY bacmid58. DH10EMBacY E. coli (Geneva Biotech) were transformed with plasmids via electroporation, followed by selection on antibiotic plates and blue/white screening. Successful transposants (white colonies) were used to inoculate 3 ml cultures for overnight growth, followed by purification of bacmid DNA using alkaline lysis and isopropanol precipitation as described59.
For baculovirus production and protein expression, Sf9 insect cells were grown in Insect-XPRESS Medium + L-glutamine (Lonza) at 27 °C. Non-adherent cultures were maintained at 1 – 2 x 106 cells/ml in flasks with shaking at 100 rpm. Baculoviruses were prepared as described25. Medium (100 μl), bacmid DNA (2 μg), and FuGene HD transfection reagent (3 μl) were mixed and incubated for 15 min, then added drop-wise to 1 x 106 Sf9 cells growing adherently in a 6-well plate with 2 ml of medium. After 3 days, the efficiency of transfection was assessed by monitoring YFP expression from EMBacY using a Countess II FL cell counter with an EVOS light cube (Thermo Fisher Scientific). The supernatant (V0 virus) was added to a 50 ml Sf9 culture and incubated for 3 days. The resulting supernatant (V1 virus) was stored at 4 °C, and used to infect 0.25 – 4 L cultures at a ratio of 1% (v/v) for protein production. Three-days after infection, cell pellets were harvested by centrifugation, washed in 1x PBS, flash frozen in liquid nitrogen, and stored at -80 °C.
Purification of the dynein-2 complex
All steps were performed at 4 °C. In a typical preparation, frozen cell pellets from 4 L of Sf9 culture were resuspended in purification buffer (50 mM HEPES [pH 7.5], 100 mM KCl, 150 mM K-acetate, 2 mM Mg-acetate, 1 mM EGTA, 10% [v/v] glycerol, 1 mM DTT, 0.2 mM Mg-ATP, 1 mM PMSF, Roche cOmplete™ EDTA-free Protease Inhibitor Cocktail) to a total volume of 200 ml. Cells were lysed in batches using a Dounce homogenizer with 10 – 20 strokes with a tight clearance pestle. Lysates were clarified by ultracentrifugation in a Type 70 Ti rotor at 360,562 g for 45 min. The supernatant was incubated for 1 h on a roller with 10 ml IgG Sepharose 6 resin (GE Healthcare) that had been pre-washed in purification buffer. The resin was then transferred to a glass column, washed with 2 x 100 ml volumes of purification buffer and 1x 25 ml volume of TEV buffer (as purification buffer, lacking KCl), and transferred to two 15 ml plastic conical tubes. Each tube was supplemented with 400 μg TEV protease, filled with TEV buffer, capped and and incubated overnight on a roller. TEV-cleaved proteins were separated from the resin using an empty column, concentrated to ~2 mg/ml using Amicon Ultra centrifugal filter devices (100 kDa cutoff). Aliquots of 50 – 100 μl were flash frozen in liquid nitrogen and stored at -80 °C. Finally, before use, aliquots were thawed and loaded onto a Superose 6 Increase 3.2/300 column using an ÄKTAmicro system (GE Healthcare) pre-equilibrated with gel filtration buffer (50 mM Tris [pH 7.5], 150 mM K-acetate, 2 mM Mg-acetate, 1 mM EGTA, 1 mM DTT, 0.1 mM MgATP). Fractions (50 μl) were collected and analyzed by SDS-PAGE (4 – 12% Tris-Bis gels with Sypro Red staining (Thermo Frisher Scientific)). Subunits (except LC8-like) were detected in the peak fraction by mass spectrometry (BSRC Mass Spectrometry and Proteomics Facility, University of St Andrews).
Negative stain electron microscopy
Immediately following gel filtration, samples were diluted to 5 – 130 nM and applied to EM grids using the rapid staining method. By pipetting ~4 μl of 2% uranyl acetate, a small air gap and then ~4 μl of sample into a single tip, the contents were rapidly deposited onto a glow-discharged continuous carbon grid (Electron Microscopy Sciences). Grids were then stained in three sequential drops of 75 μl 2% uranyl acetate, blotted and air dried.
Micrographs were collected using a Tecnai T12 microscope (Thermo Fisher Scientific) operating at 120 keV with a tungsten electron source and a 4k x 4k CCD camera (US4000, Thermo Fisher Scientific). Nominal magnification was 67,000X, giving a 1.67 Å/pixel sampling at the object level. Images were collected with a dose of 40 − 50 e-/Å2 and a nominal defocus of -1 μm. In total 458 and 415 micrographs were collected for the dynein-2 holoenzyme and ΔIC-LC complexes respectively.
Dynein-2 holoenzyme particles were manually picked and subjected to initial 2D classification using Relion v2.160. Flexibility between the tail and motor domains was further analysed using multivariate statistical analysis in Imagic61. A total of 4,465 particles were aligned and classified with a mask encompassing the entire molecule. By inspection of the 2D class averages, a subset of particles corresponding to the major tail view was selected and re-aligned based on the motor domain region. The aligned particles were then subjected to further classification using a mask that encompassed all observed tail positions but excluded the motor domains. To quantify the tail positions in the resulting 27 class averages (1,261 particles), the angle between the C2 symmetry axis in the motor domains and the dimerization domain in the tail was measured using ImageJ. A polar plot histogram of tail angles was generated using MatLab R2017b.
Particles of the ΔIC-LC sample were picked manually and subjected to 2D classification using Relion. To analyze flexibility in the tail, Imagic was used to align and classify 2,827 ΔIC-LC particles based on the tail region. By inspection of the 2D class averages, a subset of particles corresponding to the major tail view was selected and subjected to further classification to analyse tail flexibility (25 class averages, 1,116 particles).
Cryo-electron microscopy
Immediately following gel filtration, the dynein-2 complex was diluted to 60 – 70 nM and vitrified using a Vitrobot Mark IV system (Thermo Fisher Scientific) set to 4 °C and 95% humidity. Sample (3 μl) was applied to glow-discharged EM grids containing a lacey carbon support film covered with a 3 nm layer of continuous carbon (Agar Scientific). Following 10 s incubation, the grid was manually blotted within the Vitrobot chamber, and a second 3 μl aliquot of sample was then added and incubated for 20 s. The grid was blotted for 5 – 6 s using a force setting of 0 before vitrification in liquid ethane and storage under liquid nitrogen.
Data collection
An initial cryo-EM dataset of dynein-2 holoenzyme was collected on a Tecnai G2 Polara instrument (Thermo Fisher Scientific) operating at 300 keV, equipped with a K2 Summit direct electron detector and GIF Quantum energy filter (slit width 20 eV). Using SerialEM software, micrographs were collected in counting mode with a nominal magnification of 125,000 X (1.39 Å/pixel sampling), 18 s exposures, and 60 frame movies. The total dose per movie was 47.5 e-/Å2. Three further datasets were collected on a Titan Krios instrument equipped with a K2 Summit direct electron detector and GIF Quantum energy filter (slit width 20 eV) at the eBIC imaging facility (Diamond Light Source, Didcot). Data collection parameters are shown in Table 1.
Image pre-processing
Movies were aligned, dose-weighted and summed using MotionCor262. Micrographs were then visually screened and mis-targeted images containing thick carbon support in more than ~50% of the field of view and empty areas were discarded. Gctf63 was used to determine CTF parameters.
Tail reconstruction
Negative stain classification of the dynein-2 holoenzyme complex revealed extreme flexibility between the tail and motor domains (Supplementary Video 1). These two portions of the molecule were processed separately. An initial reconstruction of the tail was generated from the Polara dataset. A total of 3,236 particles were picked manually using Boxer64 and subjected to 2D classification using Relion (v2.0 or v2.1, used for all subsequent image processing steps unless stated). Five of the resulting 2D averages were used as templates for automated particle picking using Gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/). The resulting 21,191 picks were subjected to 2D classification and 11,095 particles from well-defined averages were subjected to 3D classification, using a 60 Å low-pass-filtered dynein-1 tail map53 (EMDB-3707) as a starting model, and refinement to a 10 Å resolution initial map.
Projections of this 10 Å tail map were generated using SPIDER65 and eight views were used as templates for automated particle picking of Krios Dataset 1 using Gautomatch. Class averages from Dataset 1 were used as templates for automated particle picking of Datasets 2 and 3. Permissive parameters were used to ‘over pick’ the micrographs, yielding 757,402 initial picks, from which 461,684 putative particles were isolated. Following extensive rounds of 2D and 3D classification, 68,623 particles were refined to yield a 4.5 Å tail reconstruction, which was sharpened using a B-factor of -125 Å.
Further focused refinements were used to determine maps of three tail sub-regions with improved local density (Supplementary Fig. 2h–j). The first, encompassing DHC2 TAIL-A, LIC3-A, WDR60, WDR34, RB and LC8, refined to 4.4 Å. The second, encompassing DHC2 N-terminal domain (ND), DHC2 TAIL-A (bundles 1–5), DHC2 TAIL-B (bundles 1–3), WDR60, WDR34 and RB subunits, also refined to 4.4 Å. The third, encompassing DHC2 TAIL-A (bundles 5–8), DHC2 TAIL-B (bundles 3–8), LIC3-A and –B, and LC8 subunits, was subjected to 3D classification, revealing flexibility in the position of TCTEX/TCTEX1D2 density (Supplementary Fig. 4c). Particles from class 1 containing the best defined TCTEX/TCTEX1D2 and DHC2 TAIL-B densities were refined to 7.5 Å. Half maps, refined maps, sharpened and masked maps, and mask used have been deposited to the EMDB.
Motor domain reconstruction
A model of the autoinhibited state of dynein-2 motor domains25, derived from PDB 4RH745, was low-pass filtered and projected in SPIDER. Eight distinct views were used as templates for automated particle picking of the initial Polara dataset. The resulting picks were subjected to 2D classification using Relion and 8,313 particles from well-defined averages were selected for 3D classification. Particles from the best-defined 3D class were refined to give an 11 Å initial map of the motor domains from 5,133 particles.
Six 2D averages from the Polara dataset were used as templates for automated particle picking of Krios Datasets 1 – 3 (Table 1), from which 229,969 putative particles were isolated. Following extensive rounds of 2D and 3D classification, 57,265 particles were refined with C2 symmetry to yield a 3.9 Å motor domain reconstruction, which was sharpened using a B-factor of -100 Å. Half maps, refined map, sharpened and masked map, and mask used have been deposited to the EMDB.
Local resolutions in the tail and motor domain maps were calculated using Relion. Map visualization was carried out in UCSF Chimera66 and UCSF Chimera X67.
Model building
Tail domain
RosettaCM44, COOT68, iMOD fit69, SWISS-MODEL70, RaptorX Contact71, and PSI-PRED72 software were used to build an atomic model of the tail as detailed in Supplementary Table 2. RosettaCM was used for density-guided rebuilding, completion, and refinement of homology models. Template-target alignments were generated using HHpred73 and templates were aligned to the cryo-EM density using Chimera’s ‘Fit in Map’ command. A total of 100-300 Rosetta models were generated and the conformation with the lowest energy (including fit-to-density energy) was used. Coordinates for all subunits were combined in the final tail model and subjected to rounds of relaxation into the density using Rosetta, real-space refinement using Phenix (phenix.real_space_refine, default parameters)74, and manual adjustments. For deposition to wwPDB, the tail model was truncated to poly-alanine (phenix.pdbtools) to denote that the majority of side chains positions are not experimentally determined. Register shifts cannot be excluded in the peripheral lower-resolution regions of the cryo-EM map (Supplementary Fig. 2f) and none of our conclusions rest on the sequence register in these regions.
Motor domains
The crystal structure of the dynein-2 motor domain as a monomer (PDB 4RH7)45 was separated into rigid bodies and docked into the motor domain EM density using UCSF Chimera’s ‘Fit in Map’ command, followed by manual adjustment using COOT. A second copy of the motor domain was then generated and fit into the second motor domain EM density using ‘Fit in Map’. Real-space refinement was performed using Phenix (phenix.real_space_refine, default parameters)74.
Holoenzyme
The dynein-2 tail and motor domain structures were fitted into the sub-tomogram average of anterograde IFT-B trains from C. reinhardtii cilia29 using Chimera’s ‘Fit in Map’ command. The two DHC2 bundles connecting the tail and motor domain were modelled using SWISS-MODEL [template PDB 3VKG, Chain A75] and RaptorX Contact (deposition as UNK).
Microtubule gliding assays
Dynein-2 holoenzyme and ΔIC-LC samples were biotinylated for microtubule gliding assays via their N-terminal SNAPf tag as described25. Flow chambers were made between glass slides, biotin-PEG coverslips, and double-sided tape. Fluorescently-labelled microtubules were visualized on an Eclipse Ti-E inverted microscope with a CFI Apo TIRF 1.49 N.A. oil objective, Perfect Focus System, H-TIRF module, LU-N4 laser unit (Nikon) and a quad band filter set (Chroma). Images were recorded with 100 ms exposures on an iXon DU888 Ultra EMCCD camera (Andor), controlled with NIS-Elements AR Software (Nikon). Temperature of the flow chamber was maintained at 25ºC by an objective heater (Okolab). Chambers were sequentially incubated with 1) blocking solution (0.75% Pluronic F-127, 5 mg/ml casein) for 5 min, followed by two washes with gel filtration buffer containing taxol, SEC-T (50 mM Tris [pH 7.5], 150 mM K-acetate, 2 mM Mg-acetate, 1 mM EGTA, 1 mM DTT, 20 μM taxol); 2) 0.5 mg/ml neutravidin for 2 min, followed by two washes with SEC-T supplemented with 1 mg/ml casein; 3) biotinylated motor protein (~40 μg/ml) for 2 min, followed by two washes with SEC-T supplemented with 1 mg/ml casein; 4) 0.1 μM Alexa-488 microtubules in assay solution (SEC-T supplemented with 1 mg/ml casein, 1 mM Mg-ATP, 71 mM β-mercaptoethanol, 20 mM glucose, 300 μg/ml glucose oxidase, 60 μg/ml catalase). Microtubule lengths and gliding velocities were calculated from kymographs generated in FIJI76,77. Graphing was performed in Prism5 (GraphPad).
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Cryo-EM maps are available from the EMDB under accession codes EMD-4918 (dynein-2 tail domain) and EMD-4917 (dynein-2 motor domains). Coordinates are available from the RCSB Protein Data Bank under accession codes 6RLB (dynein-2 tail domain), 6RLA (dynein-2 motor domains), and 6SC2 (dynein-2, docked into sub-tomogram average of the anterograde IFT-B train29 [EMDB-4303]). All other data supporting the conclusions of this manuscript are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank H. Mitchison, C. Moores, S. Webb, and G. Zanetti for comments on the manuscript; Diamond Light Source for cryo-EM facilities at the UK national electron bio-imaging centre (eBIC) supported by the Wellcome Trust, MRC and BBSRC; N. Lukoyanova, J. van Rooyen, A. Sielbert and D. Clare for help with cryo-EM data collection; and D. Houldershaw for computational support. This work was funded by Wellcome Trust and Royal Society (104196/Z/14/Z), BBSRC (BB/P008348/1), Royal Society (RG170260) grants to A.J.R; Wellcome Trust (WT100387) and MRC grants (MC_UP_A025_1011) to A.P.C; and Wellcome Trust (079605/Z/06/Z) and BBSRC (BB/L014211/1) grants supporting cryo-EM equipment at Birkbeck.
Footnotes
Author contributions: K.T: investigation, methodology, visualization, writing – original draft. R.Z: investigation, methodology. M.M: investigation, methodology. A.G.M: investigation, writing – review & editing. A.P.C: investigation, methodology, funding acquisition, supervision, writing – review & editing. A.J.R: conceptualization, investigation, methodology, funding acquisition, supervision, visualization, writing – original draft.
Competing interests: The authors declare no competing interests.
References
- 1.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 2.Pazour GJ, Wilkerson CG, Witman GB. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porter ME, Bower R, Knott JA, Byrd P, Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol Biol Cell. 1999;10:693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pazour GJ, Dickert BL, Witman GB. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Signor D, et al. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J Cell Biol. 1999;147:519–530. doi: 10.1083/jcb.147.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nachury MV, Mick DU. Establishing and regulating the composition of cilia for signal transduction. Nat Rev Mol Cell Biol. 2019 doi: 10.1038/s41580-019-0116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King SM. Axonemal Dynein Arms. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye F, Nager AR, Nachury MV. BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J Cell Biol. 2018;217:1847–1868. doi: 10.1083/jcb.201709041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lechtreck K-F, et al. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol. 2009;187:1117–1132. doi: 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao M, et al. Uni-directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. Elife. 2015;4 doi: 10.7554/eLife.05242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vuolo L, Stevenson NL, Heesom KJ, Stephens DJ. Dynein-2 intermediate chains play crucial but distinct roles in primary cilia formation and function. Elife. 2018;7 doi: 10.7554/eLife.39655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen VL, et al. Role for intraflagellar transport in building a functional transition zone. EMBO Rep. 2018;19 doi: 10.15252/embr.201845862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidts M, et al. Mutations in the gene encoding IFT dynein complex component WDR34 cause Jeune asphyxiating thoracic dystrophy. Am J Hum Genet. 2013;93:932–944. doi: 10.1016/j.ajhg.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dagoneau N, et al. DYNC2H1 mutations cause asphyxiating thoracic dystrophy and short rib-polydactyly syndrome, type III. Am J Hum Genet. 2009;84:706–711. doi: 10.1016/j.ajhg.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merrill AE, et al. Ciliary abnormalities due to defects in the retrograde transport protein DYNC2H1 in short-rib polydactyly syndrome. Am J Hum Genet. 2009;84:542–549. doi: 10.1016/j.ajhg.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huber C, et al. WDR34 mutations that cause short-rib polydactyly syndrome type III/severe asphyxiating thoracic dysplasia reveal a role for the NF-κB pathway in cilia. Am J Hum Genet. 2013;93:926–931. doi: 10.1016/j.ajhg.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McInerney-Leo AM, et al. Short-rib polydactyly and Jeune syndromes are caused by mutations in WDR60. Am J Hum Genet. 2013;93:515–523. doi: 10.1016/j.ajhg.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidts M, et al. TCTEX1D2 mutations underlie Jeune asphyxiating thoracic dystrophy with impaired retrograde intraflagellar transport. Nat Commun. 2015;6:7074. doi: 10.1038/ncomms8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor SP, et al. Mutations in DYNC2LI1 disrupt cilia function and cause short rib polydactyly syndrome. Nat Commun. 2015;6:7092. doi: 10.1038/ncomms8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessler K, et al. DYNC2LI1 mutations broaden the clinical spectrum of dynein-2 defects. Sci Rep. 2015;5 doi: 10.1038/srep11649. 11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gholkar AA, et al. Tctex1d2 associates with short-rib polydactyly syndrome proteins and is required for ciliogenesis. Cell Cycle. 2015;14:1116–1125. doi: 10.4161/15384101.2014.985066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taschner M, Lorentzen E. The intraflagellar transport machinery. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chien A, et al. Dynamics of the IFT machinery at the ciliary tip. Elife. 2017;6 doi: 10.7554/eLife.28606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toropova K, Mladenov M, Roberts AJ. Intraflagellar transport dynein is autoinhibited by trapping of its mechanical and track-binding elements. Nat Struct Mol Biol. 2017;24:461–468. doi: 10.1038/nsmb.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi P, Li W-J, Dong M-Q, Ou G. Dynein-Driven Retrograde Intraflagellar Transport Is Triphasic in C. elegans Sensory Cilia. Curr Biol. 2017;27:1448–1461.e7. doi: 10.1016/j.cub.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Mijalkovic J, Prevo B, Oswald F, Mangeol P, Peterman EJG. Ensemble and single-molecule dynamics of IFT dynein in Caenorhabditis elegans cilia. Nat Commun. 2017;8 doi: 10.1038/ncomms14591. 14591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wingfield JL, et al. IFT trains in different stages of assembly queue at the ciliary base for consecutive release into the cilium. Elife. 2017;6 doi: 10.7554/eLife.26609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan MA, Diener DR, Stepanek L, Pigino G. The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat Cell Biol. 2018;20:1250–1255. doi: 10.1038/s41556-018-0213-1. [DOI] [PubMed] [Google Scholar]
- 30.Mijalkovic J, van Krugten J, Oswald F, Acar S, Peterman EJG. Single-Molecule Turnarounds of Intraflagellar Transport at the C. elegans Ciliary Tip. Cell Rep. 2018;25:1701–1707.e2. doi: 10.1016/j.celrep.2018.10.050. [DOI] [PubMed] [Google Scholar]
- 31.Pedersen LB, Geimer S, Rosenbaum JL. Dissecting the molecular mechanisms of intraflagellar transport in chlamydomonas. Curr Biol. 2006;16:450–459. doi: 10.1016/j.cub.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 32.Reck-Peterson SL, Redwine WB, Vale RD, Carter AP. The cytoplasmic dynein transport machinery and its many cargoes. Nat Rev Mol Cell Biol. 2018;19:382–398. doi: 10.1038/s41580-018-0004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rompolas P, Pedersen LB, Patel-King RS, King SM. Chlamydomonas FAP133 is a dynein intermediate chain associated with the retrograde intraflagellar transport motor. J Cell Sci. 2007;120:3653–3665. doi: 10.1242/jcs.012773. [DOI] [PubMed] [Google Scholar]
- 34.Asante D, Stevenson NL, Stephens DJ. Subunit composition of the human cytoplasmic dynein-2 complex. J Cell Sci. 2014;127:4774–4787. doi: 10.1242/jcs.159038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbons BH, Asai DJ, Tang WJ, Hays TS, Gibbons IR. Phylogeny and expression of axonemal and cytoplasmic dynein genes in sea urchins. Mol Biol Cell. 1994;5:57–70. doi: 10.1091/mbc.5.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikami A, et al. Molecular structure of cytoplasmic dynein 2 and its distribution in neuronal and ciliated cells. J Cell Sci. 2002;115:4801–4808. doi: 10.1242/jcs.00168. [DOI] [PubMed] [Google Scholar]
- 37.Grissom PM, Vaisberg EA, McIntosh JR. Identification of a novel light intermediate chain (D2LIC) for mammalian cytoplasmic dynein 2. Mol Biol Cell. 2002;13:817–829. doi: 10.1091/mbc.01-08-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamada Y, Tsurumi Y, Nozaki S, Katoh Y, Nakayama K. Interaction of WDR60 intermediate chain with TCTEX1D2 light chain of the dynein-2 complex is crucial for ciliary protein trafficking. Mol Biol Cell. 2018;29:1628–1639. doi: 10.1091/mbc.E18-03-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel-King RS, Gilberti RM, Hom EFY, King SM. WD60/FAP163 is a dynein intermediate chain required for retrograde intraflagellar transport in cilia. Mol Biol Cell. 2013;24:2668–2677. doi: 10.1091/mbc.E13-05-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts AJ. Emerging mechanisms of dynein transport in the cytoplasm versus the cilium. Biochem Soc Trans. 2018;46:967–982. doi: 10.1042/BST20170568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urnavicius L, et al. Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature. 2018;554:202–206. doi: 10.1038/nature25462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams JC, et al. Structural and thermodynamic characterization of a cytoplasmic dynein light chain-intermediate chain complex. Proc Natl Acad Sci USA. 2007;104:10028–10033. doi: 10.1073/pnas.0703614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rapali P, et al. Directed evolution reveals the binding motif preference of the LC8/DYNLL hub protein and predicts large numbers of novel binders in the human proteome. PLoS One. 2011;6:e18818. doi: 10.1371/journal.pone.0018818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DiMaio F, et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt H, Zalyte R, Urnavicius L, Carter AP. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature. 2015;518:435–438. doi: 10.1038/nature14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erdős G, et al. Novel linear motif filtering protocol reveals the role of the LC8 dynein light chain in the Hippo pathway. PLoS Comput Biol. 2017;13:e1005885. doi: 10.1371/journal.pcbi.1005885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark SA, Jespersen N, Woodward C, Barbar E. Multivalent IDP assemblies: Unique properties of LC8-associated, IDP duplex scaffolds. FEBS Lett. 2015;589:2543–2551. doi: 10.1016/j.febslet.2015.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou Y, Witman GB. Dynein and intraflagellar transport. Exp Cell Res. 2015;334:26–34. doi: 10.1016/j.yexcr.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ichikawa M, Watanabe Y, Murayama T, Toyoshima YY. Recombinant human cytoplasmic dynein heavy chain 1 and 2: observation of dynein-2 motor activity in vitro. FEBS Lett. 2011;585:2419–2423. doi: 10.1016/j.febslet.2011.06.026. [DOI] [PubMed] [Google Scholar]
- 50.Schroeder CM, Ostrem JML, Hertz NT, Vale RD. A Ras-like domain in the light intermediate chain bridges the dynein motor to a cargo-binding region. Elife. 2014;3:e03351. doi: 10.7554/eLife.03351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hou Y, Pazour GJ, Witman GB. A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol Biol Cell. 2004;15:4382–4394. doi: 10.1091/mbc.E04-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reck J, et al. The role of the dynein light intermediate chain in retrograde IFT and flagellar function in Chlamydomonas. Mol Biol Cell. 2016;27:2404–2422. doi: 10.1091/mbc.E16-03-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang K, et al. Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-inhibited and Activated. Cell. 2017;169:1303–1314.e18. doi: 10.1016/j.cell.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perrone CA, et al. A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in chlamydomonas and Mammalian cells. Mol Biol Cell. 2003;14:2041–2056. doi: 10.1091/mbc.E02-10-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsurumi Y, Hamada Y, Katoh Y, Nakayama K. Interactions of the dynein-2 intermediate chain WDR34 with the light chains are required for ciliary retrograde protein trafficking. Mol Biol Cell. 2019;30:658–670. doi: 10.1091/mbc.E18-10-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li W, Yi P, Ou G. Somatic CRISPR-Cas9-induced mutations reveal roles of embryonically essential dynein chains in Caenorhabditis elegans cilia. J Cell Biol. 2015;208:683–692. doi: 10.1083/jcb.201411041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blisnick T, et al. The intraflagellar transport dynein complex of trypanosomes is made of a heterodimer of dynein heavy chains and of light and intermediate chains of distinct functions. Mol Biol Cell. 2014;25:2620–2633. doi: 10.1091/mbc.E14-05-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vijayachandran LS, et al. Gene gymnastics: Synthetic biology for baculovirus expression vector system engineering. Bioengineered. 2013;4:279–287. doi: 10.4161/bioe.22966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 2014;33:1855–1868. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 62.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 65.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 66.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 67.Goddard TD, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D, Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopéz-Blanco JR, Chacón P. iMODFIT: efficient and robust flexible fitting based on vibrational analysis in internal coordinates. J Struct Biol. 2013;184:261–270. doi: 10.1016/j.jsb.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 70.Waterhouse A, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang S, Sun S, Li Z, Zhang R, Xu J. Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model. PLoS Comput Biol. 2017;13:e1005324. doi: 10.1371/journal.pcbi.1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 73.Zimmermann L, et al. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 74.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D, Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kon T, et al. The 2.8 Å crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- 76.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps are available from the EMDB under accession codes EMD-4918 (dynein-2 tail domain) and EMD-4917 (dynein-2 motor domains). Coordinates are available from the RCSB Protein Data Bank under accession codes 6RLB (dynein-2 tail domain), 6RLA (dynein-2 motor domains), and 6SC2 (dynein-2, docked into sub-tomogram average of the anterograde IFT-B train29 [EMDB-4303]). All other data supporting the conclusions of this manuscript are available from the corresponding author upon reasonable request.