Abstract
The humoral immune response demands that B cells undergo a sudden anabolic shift and high cellular nutrient levels which are required to sustain the subsequent proliferative burst. Follicular lymphoma (FL) originates from B cells that have participated in the humoral response, and 15% of FL samples harbor point, activating mutations in RRAGC, an essential activator of mTORC1 downstream of the sensing of cellular nutrients. The impact of recurrent RRAGC mutations in B cell function and lymphoma is unexplored. RRAGC mutations, targeted to the endogenous locus in mice, confer a partial insensitivity to nutrient deprivation, but strongly exacerbate B cell responses and accelerate lymphomagenesis, while creating a selective vulnerability to pharmacological inhibition of mTORC1. This moderate increase in nutrient signaling synergizes with paracrine cues from the supportive T cell microenvironment that activates B cells via the PI3K-Akt-mTORC1 axis. Hence, Rragc mutations sustain induced germinal centers and murine and human FL in the presence of decreased T cell help. Our results support a model in which activating mutations in the nutrient signaling pathway foster lymphomagenesis by corrupting a nutrient-dependent control over paracrine signals from the T cell microenvironment.
Keywords: B cell lymphoma, germinal center, B lymphocytes, mTOR, rapamycin, RRAGC, nutrient signaling, cell growth, apoptosis, T follicular helper
Follicular lymphoma (FL) is the second most frequent form of non-Hodgkin lymphoma and, in spite of its indolent nature, remains mostly incurable 1. This lymphoma derives from B lymphocytes that have participated in germinal centers (GCs), transient anatomical structures formed primarily by B and T cells during the humoral response to infection and immunization 2. The GC enables the production of high affinity antibodies by iterative rounds of a two-step process consisting of affinity-based selection of antibody-producing B cells, which requires paracrine signals from T follicular helper (Tfh) cells, followed by proliferation and programmed mutation of the immunoglobulin locus in selected B cells 3–6. Importantly, FL maintains a follicular architecture reminiscent of the GC, and non-B lymphocyte cellular components of the GC, including Tfh, are also present in the FL 7. Ninety percent of FL harbor a t(14;18) chromosome translocation that leads to the overexpression of the anti-apoptotic gene BCL2 by placing it under the control of the IGH heavy chain enhancer 8. Additional genetic alterations include mutations in the epigenetic regulators KMT2D, CREBBP, EZH2 9–13, and genes involved in the interaction with the microenvironment of both FL and aggressive lymphoma, such as TNFRSF14, B2M and CD58 9,14–18.
We and others have recently identified recurrent mutations in components of the nutrient sensing pathway 17,19–21 that activates the mechanistic target of rapamycin complex 1 (mTORC1), a serine/threonine kinase driver of cellular anabolism 22. A large fraction of these missense mutations cluster within the nucleotide binding domain of the gene encoding the RagC GTPase (also known as RRAGC), which forms a heterodimeric complex with RagA responsible for activating mTORC1 upon cellular nutrient sufficiency 23. In the presence of cellular nutrients, such as certain amino acids, the Rag heterodimer acquires a specific nucleotide configuration that enables its interaction with mTORC1, leading to its recruitment to the outer lysosomal surface 24,25. Importantly, ectopic expression of these mutant variants in cancer cells lines leads to increased recruitment and activation of mTORC1 19,20,26.
Independent to the cellular nutrient signals, growth factors and cytokines, including paracrine signals from Tfh to B cells, activate mTORC1 via the PI3K-Akt signaling cascade 27,28. Mutations in the Rag GTPases are rare in other lymphoid and solid malignancies, arguing for a particular effect of such genetic alterations on the physiology and pathology of B lymphocytes. Although point mutations in either RRAGC or RRAGA can functionally activate the mTORC1 pathway 25,29, the reason for a selective genetic activation of RRAGC, sparing RagA, remains unclear.
We have engineered mouse strains to endogenously express RagC mutants in order to address their oncogenic potential and to understand the physiological impact that nutrient signaling may have on normal and pathological B cell responses. These genetic tools accelerated experimental lymphomagenesis, while creating a vulnerability to mTORC1 inhibitors. Mutations in the nutrient signaling pathway enhanced B cell autonomous activation, corrupting a nutrient-dependent control of paracrine positive signals from the T cell microenvironment.
Results
Novel RagC knock-in mice show partial insensitivity to nutrient withdrawal
To determine the impact of mutations in the nutrient sensing pathway on B cell function and FL development in a physiological setting, we used CRISPR/Cas9 genome engineering 30 to generate two independent murine Rragc knock-in models carrying point mutations recurrently observed in human FL samples: S74C and T89N (Supplementary Figure 1a), corresponding to S75C and T90N, respectively, in human RAGC protein 17,19–21. T90 was the most frequently observed variant 20, and S75 was mutated to at least three different amino acids (S75C, S75A, S75F); both mutants are likely to have functional consequences 26,29. In addition to the amino acid change, we introduced additional silent mutations for diagnostic and genotyping purposes and in the protospacer adjacent motive (PAM) sequence to prevent re-targeting (Supplementary Figure 1b).
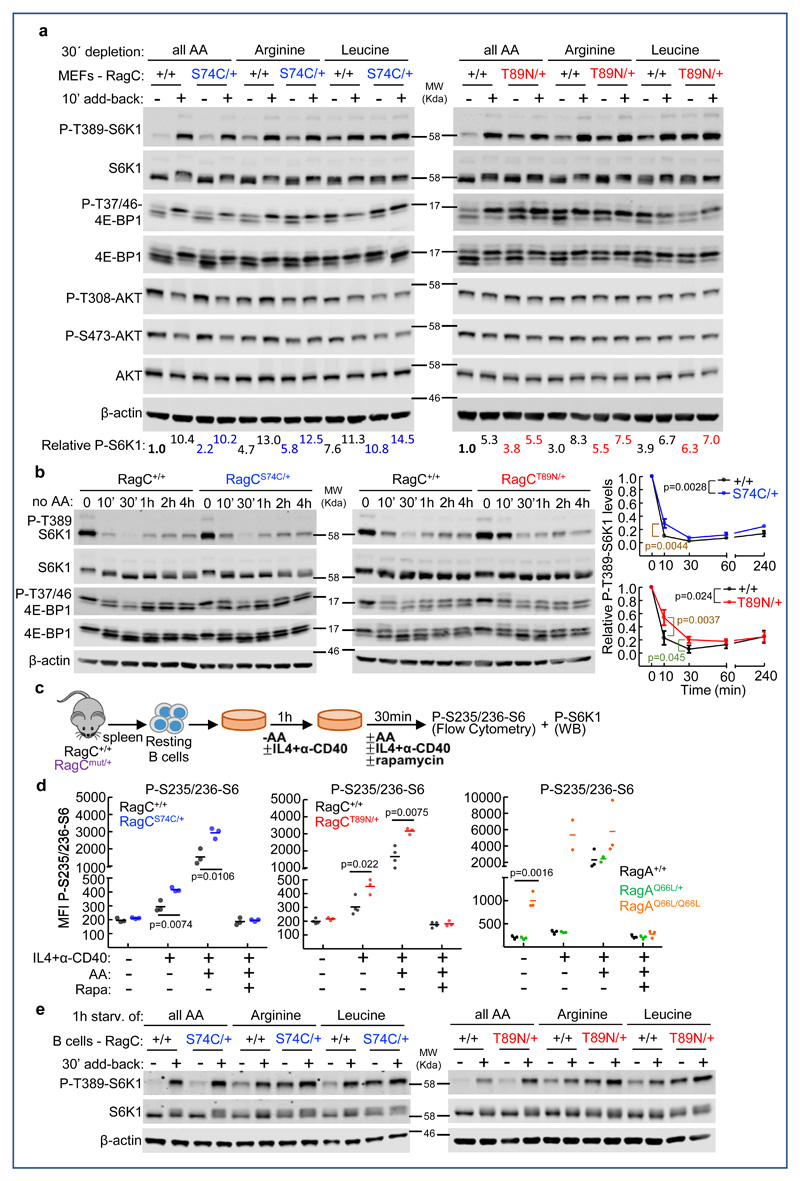
RagCS74C/+ and RagCT89N/+ mice were obtained with sub-Mendelian ratios (Supplementary Figure 1c and 1d), suggesting that partially penetrant lethality occurs before weaning. Moreover, crossing heterozygous RagCS74C/+ or RagCT89N/+ yielded no viable homozygous E19.5 neonates. These findings were not surprising, as fully-penetrant neonatal lethality was seen in mice endogenously expressing a constitutively-active form of RagA (RagAQ66L or GTP) 31. Surviving young heterozygous RagC mutant mice showed no obvious phenotypic alterations. We tested whether the expression of RagC mutants in heterozygosity conferred insensitivity to cellular nutrient withdrawal in cultured mouse embryonic fibroblasts (MEFs). MTORC1 activity resulted only partially resistant to withdrawal of all amino acids in both RagCS74C/+ and RagCT89N/+ cultures, as revealed by phosphorylation of the mTORC1 targets T389-S6K and T37/46-4EBP1 (Figure 1a, P-S6K1 quantified in Supplementary Figure 1e, with additional quantification of independent experiments in Supplementary Figure 1f). Importantly, compared to the maximal activity observed in wild-type cells, no supra-physiological increase in mTORC1 activity was seen in MEFs that endogenously expressed RagC mutant variants. Partial resistance to nutrient deprivation was more evident, albeit still incomplete, upon withdrawal of either leucine or arginine (Figure 1a), two key amino acids involved in Rag GTPase-mediated activation of mTORC132,33. As expected, phosphorylation of Akt at serine 473 and threonine 308, which occurs independently of the activation of the nutrient signaling cascade but depends on growth factor signaling, is unaffected in RagC mutant cells (Figure 1a). In time-lapse experiments as in 26, we observed that RagC mutants delayed the deactivation of mTORC1 by amino acid withdrawal (Figure 1b). Partial reactivation of the pathway by extended amino acid withdrawal was likely a consequence of autophagic degradation of internal cellular storages that occurs upon mTORC1 inhibition, as addition of chloroquine to starved cells prevented the partial reactivation of mTORC1 (Supplementary Figure 1g).
Figure 1. RagC mutant cells are partially resistant to amino acid withdrawal.
(A) Primary E13.5 mouse embryonic fibroblasts (MEFs) of RagC+/+, RagCS74C/+ and RagCT89N/+ genotypes were deprived of all amino acids, arginine or leucine in RPMI supplemented with dialyzed FBS for 30 min and re-stimulated for 10 min. Whole-cell protein lysates were immunoblotted for the indicated proteins. Quantification of P-S6K1 relative to the levels in RagC+/+ cells without amino acids is shown. (B) Same as in A, but MEFs were deprived of all amino acids for the indicated times. Quantification of P-S6K1 is shown for n=3 independent MEFs per genotype. Statistically significant increase in mTORC1 signaling was found for 10 and 30 min in RagCT89N/+ MEFs and for 10 min in RagCS74C/+ MEFs. The p values next to the genotypes state the statistical significance of the analysis of area under the curve calculated during the first 30 min of starvation. Error bars indicate standard deviation. Statistical significance was calculated by two-tailed Student´s t-test. (C) Experimental setup for purification and acute in vitro stimulation of B cells. CD43- naïve B cells were harvested from spleens of indicated genotypes and stimulated in culture with anti-CD40 and IL-4 in the presence or absence of all amino acids. WB, western blot. (D) mTORC1 activity revealed by intracellular phospho-S6 staining after stimulation and quantification by flow cytometry of the indicated RagC and RagA genotypes. (n=3 for RagC+/+, S74C and T89N/+; RagA+/+ and Q66L/+; n=2 for RagAQ66L/Q66L). (E) Primary B cells were treated as in D and whole-cell protein lysates were immunoblotted for the indicated proteins.
We next evaluated whether naïve B lymphocytes purified from RagC mutant mice (Figure 1c) also showed nutrient-independent mTORC1 activity. In the absence of a growth factor-like stimulus and amino acids in the medium, mTORC1 activity was very low and comparable to the level of rapamycin-treated samples, as revealed by the intracellular immunostaining for phosphorylated serines 235/236 of S6 (Figure 1d). Without amino acids in the medium, stimulation with ligands that mimic paracrine activating signals from T cells that stimulate mTORC1 via PI3K-Akt (IL-4 and an anti-CD40 activating antibody), resulted in a greater stimulation of mTORC1 activity in RagCS74C/+ and RagCT89N/+ B cells compared to RagC+/+ cells (Figure 1d). Simultaneous addition of amino acids and cytokines further activated mTORC1 activity in all cells, indicating that, as in MEFs, presence of amino acids was only partially dispensable for mTORC1 activation in the mutant B cells. As a comparison, we performed these signaling experiments side-by-side with B cells expressing the tool RagAGTP (Q66L) mutation in heterozygosity and homozygosity. We have previously shown 31 that mTORC1 in MEFs and neonatal tissues from RagAGTP/GTP mice was completely insensitive to nutrient starvation; this was also evident in RagAGTP/GTP B lymphocytes (Figure 1d). Heterozygous RagAGTP/+ cells, in contrast to RagCS74C/+ and RagCT89N/+ cells, showed no detectable signaling perturbation (Figure 1d and 31). The partial resistance to amino acid, leucine and arginine withdrawal was also observed in whole protein extracts from stimulated B cells (Figure 1e). Collectively, these results show that RagC mutations, when expressed endogenously and in heterozygosity, have a modest but significant activating effect on the regulation of mTORC1 by nutrients, while the presence of an activating mutation in RagA in heterozygosity is inconsequential. These data, generated without relying on the limitations of heterologous expression systems, provide cellular evidence for a biochemical asymmetry of the Rag heterodimeric components toward the control of mTORC1 activation, and are consistent with previous in vitro studies 26. In addition, they also provide a potential explanation for the exclusive occurrence of RRAGC heterozygous mutations, and the absence of mutations in RRAGA in human FL.
Because the expression of the point-mutant RagC variants is not restricted to B lymphocytes, we next quantified the proportions of different populations of the hematopoietic lineage, to exclude potential consequences of the expression of these variants in non-B cell populations. Lymphoid (B cells, CD4+ and CD8+ T cells) and myeloid (granulocytes, macrophages, myeloid cells and myeloid-derived suppressor cells (MDSCs)) populations were present in RagC mutant mice in frequencies similar to those of wild-type counterparts, in both spleen and bone marrow (BM) (Supplementary Figure 1h). Hence, while these quantifications are not a thorough functional examination of all populations in the hematopoietic lineage, it is apparent that the expression of RagC mutations endogenously has no obvious effect on lymphoid and myeloid lineages that could complicate further functional studies on B cells. Further support for this conclusion is provided below.
RagC mutations accelerate FL that are sensitive to rapamycin
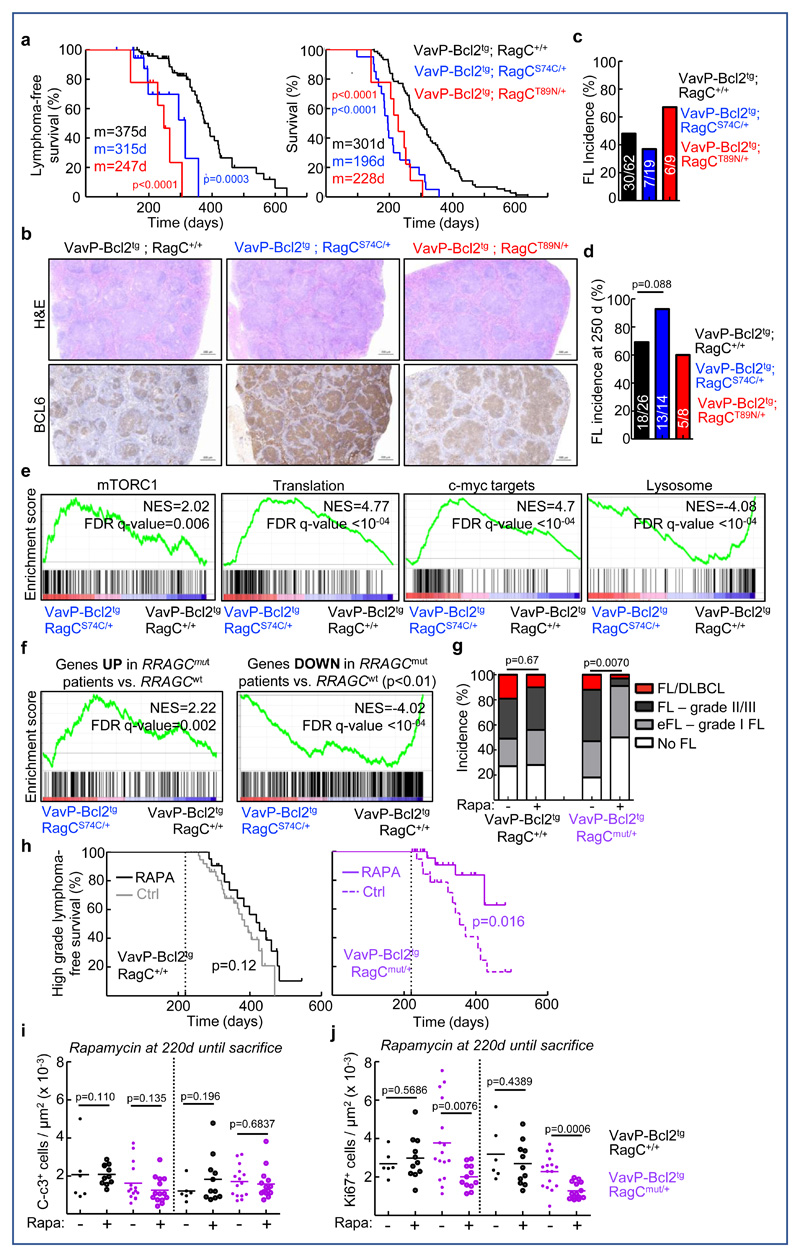
We next bred RagC mutant S74C and T89N strains with the transgenic mouse strain VavP-Bcl2, which expresses BCL2 under the lymphoid lineage-specific promoter VavP (VavP-Bcl2tg), the standard genetic tool to generate autochthonous FL in mice 34. The partial insensitivity to amino acid withdrawal observed in RagC mutant B cells was also observed in the VavP-Bcl2 background (Supplementary Figure 2a) Latencies to lymphoma development of VavP-Bcl2tg; RagCS74C/+ and VavP-Bcl2tg; RagCT89N/+ mice were significantly accelerated (Figure 2a), which was also true for BM chimeras (Supplementary Figure 2b). Murine FL were diagnosed by pathognomonic histological features of FL, such as expression of Bcl6 in spleen and lymph nodes, indicating that these tumors were of GC B cell origin (Figure 2b). Histological analysis of lymphoid organs from FL-bearing mice of all three genotypes revealed similar incidence (Figure 2c) and spread (Supplementary Figure 2c and 2d). Lymphoma incidence and spread were also similar in all three genotypes when complete cohorts of mice were sacrificed at 250 days (Figure 2d and Supplementary Figure 2e).
Figure 2. Accelerated lymphomagenesis by heterozygous expression of mutant RagC in mice, with selective sensitivity to rapamycin.
(A) Kaplan-Meier lymphoma-free survival (left) and survival (right) curves of VavP-Bcl2tg; RagC+/+ (n=74), VavP-Bcl2tg; RagCS74C/+ (n=20) and VavP-Bcl2tg; RagCT89N/+ (n=9) mice. In Lymphoma-free survival curves, mice that developed FL were scored as 1. Statistical significance was calculated with the logrank test; m: median survival. (B) Representative H&E and anti-Bcl6 IHC from spleen sections from FL-bearing mice (more than 90 tumors analyzed). Scale bar: 500μm. (C) Incidence of FL in cohorts shown in A. (D) Incidence of FL in cohorts euthanized at 250d. Statistical significance was calculated by Chi-Square test. (E) Enrichment of gene signatures (GSEA) related to mTORC1 signaling, genes involved in translation, c-myc targets and lysosomal genes in VavP-Bcl2tg; RagCS74C/+ FL sorted cells (n=4 mice) compared to VavP-Bcl2tg; RagC+/+ FL sorted cells (n=7 mice). (F) GSEA in VavP-Bcl2tg; RagCS74C/+ compared to VavP-Bcl2tg; RagC+/+ FL sorted cells, on the list of significantly upregulated and downregulated genes in RRAGC mutant human FL samples. NES: normalized enrichment score; FDR: false discovery rate. (G) Incidence and histological grades of tumors in mice treated with rapamycin and controls. (all mice were VavP-Bcl2tg: RagC+/+ -Rapa n=41, RagC+/+ +Rapa n=32; RagCmut -Rapa n=17; RagCmut +Rapa n=32). Mice that developed tumors before 220d were excluded. Statistical significance was calculated with Chi-Square test. (H) Kaplan-Meier lymphoma-free survival curves (FL-grade II, FL-grade III and diffused tumors were scored as 1, mice bearing no FL, early FL and FL-grade I: 0) of mice untreated or treated with rapamycin starting at 220d (vertical dotted line). All mice were VavP-Bcl2tg: RagC+/+ -Rapa n=43, RagC+/+ +Rapa n=32; RagCmut -Rapa n=19; RagCmut +Rapa n=34). Statistical significance was calculated with the log-rank test (I) Quantification of C-c3 in spleens and lymph nodes (LN) from rapa-treated (n=11 VavP-Bcl2tg; RagC+/+ and n=14 VavP-Bcl2tg; RagCmut) and untreated mice (n=6 VavP-Bcl2tg; RagC+/+ and n=15 VavP-Bcl2tg; RagCmut). (J) Quantification of Ki67 in VavP-Bcl2tg; RagC+/+ and VavP-Bcl2tg; RagCmut in spleens and LN from rapa-treated (n=11 VavP-Bcl2tg; RagC+/+ and n=14 VavP-Bcl2tg; RagCmut) and untreated mice (n=6 VavP-Bcl2tg; RagC+/+ and n=15 VavP-Bcl2tg; RagCmut). In i) and j) statistical significance was calculated by two-tailed Student´s t-test and lines indicate mean.
Of note, VavP-Bcl2 mice were also prone to develop autoimmunity 34, so their limited overall survival was due to occurrence of FL and/or autoimmune disease. To our surprise, in addition to the acceleration of FL development, we observed that VavP-Bcl2tg; RagCS74C/+ and VavP-Bcl2tg; RagCT89N/+ mice and BM chimeras also exhibited shortened latency to autoimmunity (Supplementary Figure 2f). Consistent with the acceleration of both prevalent pathologies in VavP-Bcl2 mice, overall survival (including lymphoma-free and autoimmunity-free) was significantly shorter in VavP-Bcl2tg; RagCmut (RagCS74C/+ and RagCT89N/+) mice (Figure 2a and Supplementary Figure 2g). These results suggest that deregulation of the nutrient sensing pathway deeply affects B cell functions to promote not only FL but also other pathological outcomes of aberrant B cell activation, such as autoimmune disease.
Considering that the expression of the mutations is neither conditional nor restricted to B cells, the striking similarities of latencies and diagnoses of RagCmut mice and RagCmut BM chimeras support a negligible impact of systemic expression the mutations in B cell biology and lymphomagenesis.
To investigate the mechanisms underlying the oncogenicity of RagCmut, we sorted B220+ cells from VavP-Bcl2tg; RagCmut and RagCwt FLs and performed transcriptional profiling analysis. The agnostic gene set enrichment analysis (GSEA) 35 showed the expected mTORC1 signaling signature enrichment in RagC mutant FL cells, together with other signatures consistent with mTORC1 activation, such as translation, c-myc targets, and suppression of lysosomal biogenesis (Figure 2e, see details of the signatures on Supplementary Table 1). We next performed a side-by-side comparison of transcriptional profiles obtained from murine and human FL samples of RagCwt and RagCmut genotypes 20 and observed that mRNAs selectively upregulated in murine RagC mutant FL samples were consistently enriched in RRAGC mutant human FL (and vice versa for downregulated genes (Figure 2f), supporting the translational relevance of analyzing RagC mutant mice for understanding human FL.
We reasoned that the mTOR inhibitor rapamycin would be particularly efficacious against FL arising in RagC mutant mice, and we orally administered rapamycin starting at 220 days to a cohort of VavP-Bcl2tg; RagCmut and VavP-Bcl2tg; RagCwt mice and monitored their survival thereafter. Rapamycin treatment strongly suppressed the phosphorylation of S240/244 of S6, a readout of mTORC1 signaling, in all lymphomas (Supplementary Figure 2h). Importantly, the incidence and grade of lymphomas were lower in the rapamycin-treated RagCmut mice, compared to untreated controls (Figure 2g), with no statistical change in the incidence or the grade of lymphomas arising in RagCwt mice. Pharmacological inhibition of mTORC1 did not extend the survival of mice with lymphomas of all grades (Supplementary Figure 2i), but interestingly, when we limited the analysis of lymphoma-free survival to mice with lymphomas of higher grade (grade II/III FL and FL/DLBCL, Figure 2g), rapamycin selectively extended the survival of VavP-Bcl2tg; RagCmut mice, but not that of VavP-Bcl2tg; RagCwt mice (Figure 2h). A short oral rapamycin treatment (for 7 days starting at 220 days) induced an acute shrinking of spleens and a strong cell-cycle arrest as measured by Ki67 staining, both in VavP-Bcl2tg; RagCwt and VavP-Bcl2tg; RagCmut mice (Supplementary Figures 2j and k). Importantly, and supporting a selective antitumoral effect of rapamycin for RagCmut mice, only the spleens of treated RagCmut mice were significantly smaller after long-term rapamycin treatment (Supplementary Figure 2j). Finally, and without a detectable pro-apoptotic effect (Figure 2i) long-term rapamycin treatment inhibited proliferation selectively in RagCmut FL (Figure 2j), selectivity also seen in the quantification of tumor burden (Supplementary Figure 2l). Altogether, these data strongly supports a selective long-term sensitivity to mTORC1 inhibition in tumors with activating mutations in the nutrient signaling pathway, and that this sensitivity may be more profound in FL of higher grade.
As a whole, the results on Figure 2 provide bona fide support for a critical oncogenic role for the RRAGC mutations in human FL. Moreover, our results show features of selective sensitivity of Rragc mutant FL to the pharmacological inhibition of mTORC1 in vivo. Hence, our data nominate mutations in the nutrient signaling pathway as potential markers for patients who would benefit from treatment with mTOR inhibitors.
B cell functions are exacerbated in RagC mutant mice
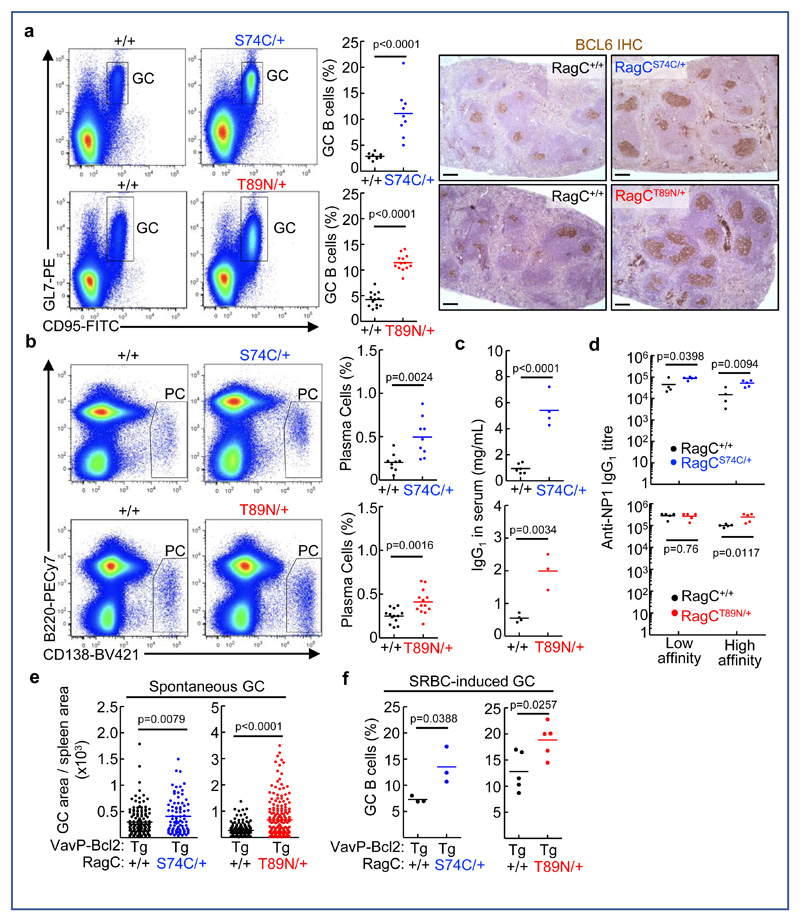
To determine whether RagC mutations would drive aberrant B cell function in the context of physiological humoral responses, we analyzed the primary response of RagC mutant B cells to intraperitoneal immunization with sheep red blood cells (SRBC) to induce GC formation (Supplementary Figure 3a). We observed a ˜3-fold increase in abundance of GC B cells in the spleens of RagCS74C/+ and RagCT89N/+ compared to RagC+/+ mice at ten days after immunization, close to the peak of the GC reaction (Figure 3a). Histological analysis and immunohistochemical staining of spleen sections for GC marker Bcl6 showed that GCs were both larger and more abundant in RagC mutant mice (Figure 3a and Supplementary Figure 3b). In contrast, activating mutations in RagA, which is not mutated in human FL, either in heterozygosity or homozygosity, led to no measurable GC enlargement under similar conditions (Supplementary Figure 3c), indicating an exquisite sensitivity of B cells to different degrees of activation of the nutrient signaling pathway. GC formation leads eventually to differentiation of GC B cells into antibody-secreting plasma cells (PC). Accordingly, RagC mutant mice showed a ˜3-fold increase in the abundance of splenic PC (Figure 3b). Consistently with increased PC production in RagCS74C/+ and RagCT89N/+ mice, the titer of class-switched, IgG1 antibodies in serum quantified 10 days after immunization was markedly increased compared to that of RagC+/+ mice (Figure 3c). The exacerbated humoral response to SRBC in RagC mutant mice was sensitive to rapamycin treatment, when administered either for the entire humoral response period assessed (10 days) and when restricted to the last 2 days, after GC entry (Supplementary Figure 3d). Collectively, these results show that RagC mutations enhance the humoral response to SRBC, but do not block differentiation of GC B cells to PC.
Figure 3. Exacerbated humoral response in RagC mutant mice.
(A) Representative flow cytometry plots (B220+ gated, GL7+ and CD95+), its quantification and IHC staining anti-Bcl6, in spleens harvested from RagC+/+ (n=9) and RagCS74C/+ (n=9), RagC+/+ (n=13) and RagCT89N/+ (n=13) chimeras 10d after immunization with SRBC. Scale bar: 100μm. (B) Representative flow cytometry plots (B220low and CD138+) and quantification of plasma cells in spleens from RagC+/+ (n=9) and RagCS74C/+ (n=9), RagC+/+ (n=13) RagCT89N/+(n=13) chimeras 10d after immunization with SRBC. (C) IgG1 quantification from sera from RagC+/+ (n=6 and n=4), RagCS74C/+ (n=4); and RagCT89N/+ (n=3) chimeras 10d after immunization with SRBC, measured by ELISA. Horizontal lines indicate the mean. (D) Anti-NP-IgG1 low and high affinity serum titers were determined by ELISA in RagC+/+ (n=4 and n=5), RagCS74C/+ (n=5); and RagCT89N/+ (n=5) chimeras. Serum was collected 28d after NP-KLH immunization. (E) Quantification of spontaneous GC formation, as determined by Bcl6 IHC, from the spleen sections of young VavP-Bcl2tg; RagCS74C/+ mice (n=133 GCs analyzed from 3 VavP-Bcl2tg ; RagC+/+ mice and 83 GCs analyzed from 3 VavP-Bcl2tg RagCS74C/+ mice; 6-13w old) and VavP-Bcl2tg; RagCT89N/+ mice (n=169 GCs analyzed from 5 VavP-Bcl2tg ; RagC+/+ mice, n=193 GCs analyzed from 6 VavP-Bcl2tg ; RagCT89N/+ mice, 6-11w old). Dots represent Bcl6 area of each GC normalized by total area of the spleen section. (F) Quantification of GC B cells (B220+ gated, GL7+ and CD95+) in spleens harvested from VavP-Bcl2tg; RagCS74C/+ and VavP-Bcl2tg; RagCT89N/+ chimeras 10d after immunization with SRBC (n=3 VavP-Bcl2tg; RagC+/+, n=3 VavP-Bcl2tg; RagCS74C/+, n=5 VavP-Bcl2tg; RagC+/+ and n=5 VavP-Bcl2tg; RagCT89N). In all panels, horizontal lines indicate the mean; statistical significance was calculated by two-tailed Student´s t-test.
Constitutive activation of the mTORC1 pathway by expression of RagAGTP or by deletion of Tsc1 in B cells is detrimental for affinity maturation 36, outcome of the GC reaction together with the production of memory B cells. This impairment occurs presumably because constitutive mTORC1 activity in all GC B cells, instead of its selective dynamic increase in cells with comparatively higher affinity antibodies, precludes affinity-based positive selection 36. To determine whether affinity maturation was also impaired in mice with mutations in RagC, we immunized mice intraperitoneally with the hapten-carrier complex 4-hydroxy-3-nitro-phenylacetyl-keyhole limpet hemocyanin (NP-KLH) in Alum, and quantified the increase in high-affinity NP antibodies in serum over a 28 days period by affinity-dependent ELISA (Supplementary Figure 3e). Surprisingly, and in contrast to decreased high-affinity titer of antibodies of RagAGTP and Tsc1-KO models of deregulated mTORC1 activity 36, RagCS74C/+ and RagCT89N/+ mice showed increased production of high-affinity antibodies to NP (Figure 3d).
Next we determined whether exacerbated B cell functions were also at work in mice expressing RagC mutations in the context of overexpression of BCL2, but before the onset of FL. We first quantified spontaneous GC formation in unimmunized mice, readily observed in VavP-Bcl2tg; RagCwt controls 34 and observed a significant expansion of GC area in VavP-Bcl2tg; RagCmut, (Figure 3e). Moreover, 10 days after SRBC immunization we observed a similar ˜2-3 fold expansion in GC B cells in VavP-Bcl2tg; RagCmut (Figure 3f). These results indicate that exacerbated B cell activation caused by endogenous expression of RagC mutations also occur in the cellular context of Bcl2 overexpression that will result in follicular lymphomagenesis.
Altogether, the results obtained with SRBC and NP-KLH immunizations show that RagC mutant mice have an exacerbated humoral response, with increased GC size and cellularity and PC production, but without impaired selection of high-affinity B cells nor compromised production of high-affinity antibodies. Hence, a mild, partial gain of function in the nutrient signaling arm of mTORC1 favors expansion of activated B cells, even in the context of Bcl2 overexpression. This aberrant behavior is consistent with the oncogenic potential of RagC mutations, and contrasts to the cell-intrinsic deleterious effects of more profound perturbations in mTORC1 activity as elicited by RagAGTP or loss of Tsc1 36.
Enhanced B cell activation in RagC mutants is cell intrinsic
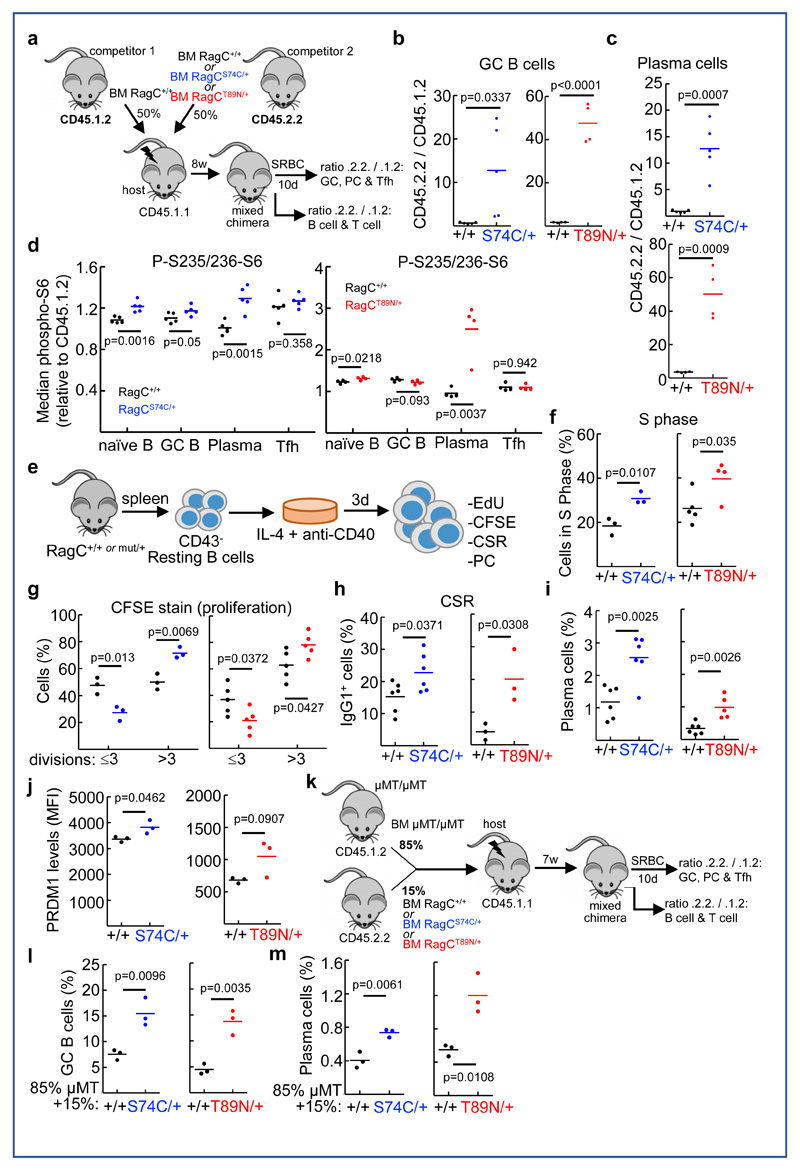
To ascertain whether RagC mutant cells had a competitive advantage relative to RagC wild-type cells, as suggested by the results of immunization (Figure 3), we performed competitive reconstitution in lethally-irradiated hosts by co-injecting RagC+/+ and either RagCS74C/+ or RagCT89N/+ BM cells in a 1:1 ratio, tractable by the differential expression of CD45.1 and CD45.2 (Figure 4a). The behavior and abundance of RagC mutant cells during competitive hematopoiesis, B cell lineage development and immunization and GC formation were then monitored. We first determined the contribution to lymphoid populations (mature B and T lymphocytes) in the spleen 2 months after reconstitution by cell surface immunostaining of the alleles CD45.1 and CD45.2. We observed that cells of the RagC mutant genotype marginally but significantly outnumbered wild-type cells in the B220+ naïve B cell compartment (Supplementary Figure 4a), while showing no competitive advantage in generating T cells (Supplementary Figure 4a). More importantly, when we immunized the mixed chimeras with SRBC, we observed a greater enrichment of RagC mutant cells in the GC B cell population (˜18-fold for RagCS74C/+; and ˜27-fold for RagCT89N/+). A similar enrichment was found in the PC population (˜14-fold for RagCS74C/+; and ˜14-fold for RagCT89N/+) (Figure 4b and 4c and Supplementary Figure 4b and 4c). Monitoring of mTORC1 activity in different B cell populations (naïve, GC and PC) showed increased phospho-S6 in RagCmut cells compared to RagCwt cells cohabiting the same spleen (Figure 4d). In contrast to the competitive advantage of B cells, RagC mutant GC-resident T lymphocytes (T follicular helper cells, Tfh) were only marginally, and not significantly enriched over wild-type competitors (Supplementary Figure 4d), with no significant differences in mTORC1 activity (Figure 4d). Together, our results show that RagC mutations confer a modest but significant increase in mTORC1 activity in B cell populations, which is associated with increased competitiveness of mutant B cells during B cell lineage development, and, markedly, during the GC response. The increased fitness of RagC mutant GC B cells is prone to play a role at enriching the population of RagC mutant cells during the multiple cellular competition events occurring during the protracted course of the development of FL.
Figure 4. B cell-intrinsic activation and increased fitness by expression of RagC mutations.
(A) Experimental setup of in vivo B lymphocyte competition experiments with mixed BM chimeras. BM reconstitution of lethally irradiated CD45.1.1 hosts, by co-injecting RagC+/+ (CD45.1.2) and either RagC+/+, S74C/+ or T89N/+ (CD45.2.2) BM cells in a 1:1 ratio. After 8w, reconstituted mice were immunized with SRBC. (B) Quantification of CD45.2.2/CD45.1.2 ratio in GC B cells (B220+CD95+GL7+gated) in splenocytes harvested from mixed chimeras 10d after immunization (n=5 for RagC+/+ - RagC+/+ and n=5 for RagC+/+ - RagCS74C/+; n=5 for RagC+/+ - RagC+/+ and n=4 for RagC+/+ - RagCT89N/+). (C) Quantification of CD45.2.2/CD45.1.2 ratio in Plasma Cells (PC) (B220lowCD138+gated) in splenocytes harvested from mixed chimeras 10d after immunization (N for each genotype as in b)). (D) Quantification of S235/236-phosphorylation of S6 in naïve (B220+CD95-GL7-), GC B (B220+CD95+GL7+), PC (B220lowCD138+) and Tfh (CD4+CXCR5+PD1+) populations from spleens harvested from mixed chimeras 10d after immunization (n=5 for RagC+/+ - RagC+/+ and n=5 for RagC+/+ - RagCS74C/+; n=4 for RagC+/+ - RagC+/+ and n=4 for RagC+/+ - RagCT89N/+). (E) Experimental setup of in vitro B cell activation. Naïve B cells were isolated from spleens of RagC+/+, S74C/+ and T89N/+ mice using CD43 magnetic beads. CD43- cells were stimulated with anti-CD40 and IL-4. Three days after activation, the following parameters were quantified: (F) S phase determined by EdU incorporation in CD19+ cells (n=3 for RagC+/+ and RagCS74C/+; n=5 for RagC+/+ and n=4 for RagCT89N/+). (G) proliferation by CFSE staining in B220+ cells (n=3 for RagC+/+ and RagCS74C/+; n=5 for RagC+/+ and RagCT89N/+). (H) Quantification of class-switched B220+IgG1+ cells (n=6 for RagC+/+ and RagCS74C/+; n=3 for RagC+/+ and RagCT89N/+). (I) PC production by expression of cell surface marker CD138 in B220low cells (n=6 for RagC+/+ and RagCS74C/+; n=6 for RagC+/+ and n=5 for RagCT89N/+); and (J) by intracellular immunostaining of PRDM1 in B220low CD138+ cells quantified by flow cytometry. n=3 mice per genotype. MFI: median fluorescence intensity. (K) Experimental setup for determining cell-intrinsic effects of RagC mutations using mixed BM chimeras with the IghmμMT genetic system (Kitamura et al. 1991). BM reconstitution of lethally irradiated CD45.1.1 hosts, by co-injecting IghmμMT/μMT (CD45.1.2) and either RagC+/+, S74C/+ or T89N/+ (CD45.2.2) BM cells in a 9:1 ratio. After 8w, mice were immunized with SRBC. Quantification of GC B cells (B220+CD95+GL7+) (L), and PC (B220lowCD138+) (M), in spleens harvested from IghmμMT/μMT : RagC+/+, S74C/+ or T89N/+ 10d after immunization (n=3 mice per genotype). In all panels, lines indicate the mean and statistical significance was calculated by two-tailed Student´s t-test.
In contrast to the strong effects on B cells, RagC mutations had minimal impact on T cell development or Tfh differentiation, suggesting that B cell phenotypes are strongly cell-autonomous. In addition, these results suggested that RagC mutant B cells may be less dependent on T cell help for activation. To test this, we mimicked T cell help to B cells in vitro by incubation with anti-CD40 (Figure 4e). This treatment elicited several features of enhanced dose-dependent activation of RagC mutant B cells, such as increased proliferation, measured by EdU incorporation and by CFSE label decay, increased class-switch recombination to IgG1, and increased PC differentiation, as measured by expression of PC surface marker CD138 and intracellular Prdm1 (Figure 4f-j and Supplementary Figure 4e-i). As seen in vivo (Supplementary Figure 3d) and as expected, in vitro activation of B cells and PC differentiation were sensitive to acute rapamycin treatment (Supplementary Figure 4j).
To confirm the B cell-intrinsic causality of exacerbated B cell activation by RagC mutations in vivo, we undertook a genetic approach consisting of BM reconstitution with a majority (85-90%) of IghmμMT/μMT hematopoietic stem cells (HSCs), which are unable to produce a B cell lineage 37, mixed with 10-15% of RagC+/+, RagCS74C/+ or RagCT89N/+ HSC (Figure 4k). In this setting, T lymphocytes and all myeloid cells derive almost exclusively from (RagC wild-type) IghmμMT/μMT HSCs, but the B cell lineage descends entirely from RagC mutant (or wild-type in control mice) progenitors (Supplementary Figure 4k). When these mixed BM chimeras were immunized with SRBC as before, we observed that expression of RagCS74C or RagCT89N mutations restricted to B cells sufficed to drive the same enhancement in GC formation and size (Figure 4l) and PC generation (Figure 4m) as that observed in single-population RagC mutant BM chimeras and in full-body RagC mutant mice. Hence, exacerbated B cell activation occurs by the expression of RagC mutants in a cell-intrinsic manner.
RagC mutants show decreased supportive T cell signals
To gain insight into the cellular events underlying the exacerbated B cell activation, we next compared the transcriptional profiles of sorted GC B cells from RagC wt and RagC mutant mice. The transcriptome of sorted GC cells arising from RagC mutant and wild-type mice showed a striking overlap with signatures identified in FL cells (Supplementary Figure 5a). This overlap supports the idea that the analysis of the GC reaction is a meaningful mirror of cellular states and processes occurring in FL. C-myc target gene signatures, indicative of a state of enhanced B cell activation 38,39, and synergistic with mTORC1 activity in GC cells 36, were also observed in RagC mutant GC and FL samples (Figure 5a, Figure 2e, and Supplementary Figure 5b). Furthermore, GSEA analysis also showed upregulation of gene signatures related to the activation of mTORC1 in RagC mutant GC, including ribosome biogenesis genes, and genes downregulated in lysosomal biogenesis (Figure 5a and Supplementary Figure 5b), and downregulated in a human B cell lymphoma line (BJAB) treated with rapamycin or deprived of leucine 40 (Supplementary Figure 5c).
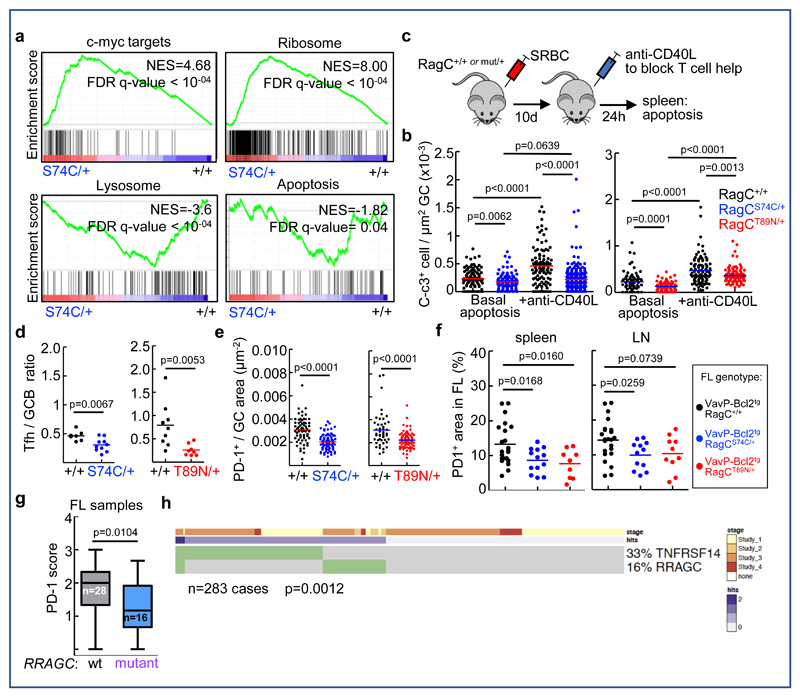
Figure 5. Impact of RagC mutations in Tfh-mediated B cell activation and apoptosis in FL.
(A) Enrichment of gene signatures (GSEA) related to c-myc targets and ribosomal genes in RagCS74C/+ (n=4 mice) versus RagC+/+ GC B cells (n=4 mice); and enrichment of gene signatures (GSEA) related to lysosomal genes and apoptotic genes in RagC+/+ versus RagCS74C/+ GC B cells. NES: normalized enrichment score; FDR: false discovery rate. (B) Quantification of C-c3+ cells within the GC area (Bcl6+ cells) in RagC+/+, S74C/+ and T89N/+ mice 10d after immunization with SRBC and following a 24h injection of 200μg of anti-CD40L. Dots represent C-c3+ cells per GC area (basal apoptosis: n=101 GCs analyzed in 5 RagC+/+ mice, n=118 GCs from 5 RagCS74C/+ mice, n=77 GCs from 5 RagC+/+ mice, n=80 GCs analyzed in 5 RagCT89N/+ mice, anti-CD40L induced apoptosis: n=105 GCs from 3 RagC+/+ mice, n=273 GCs from 4 RagCS74C/+ mice; n=102 GCs from 4 RagC+/+ mice, n=105 GCs from 4 RagCT89N/+ mice. (C) Experimental setup for induction of apoptosis by blocking Tfh-mediated B cell activation in mice immunized with SRBC. (D) Quantification of Tfh / GC B cell ratio in spleens harvested from RagC+/+, S74C/+ and T89N/+. Percentage of GC B cells was calculated from CD19+CD95+CD38- cells. Percentage of Tfh cells were calculated from CD4+CXCR5+PD1+ cells (n=7 RagC+/+, n=10 RagCS74C/+, n=10 RagC+/+, n=8 RagCT89N/+. (E) Quantification of PD-1+ cells within the GC area in RagC+/+, S74C/+ and T89N/+ mice 10d after immunization with SRBC (n= 67 GCs from 3 RagC+/+ mice and n=87 GCs from 3 RagCS74C/+ mice; n=47 GCs from 3 RagC+/+ mice and n=70 GCs from 4 RagCT89N/+ mice) (F) Quantification of PD1+ area in RagC+/+ (n=22), RagCS74C/+ (n=13) and RagCT89N/+ (n=9) (all VavP-Bcl2tg) mice, in spleen and lymph node (LN) areas affected by FL. In b), d), e) and f), lines indicate the mean; statistical significance was calculated by two-tailed Student´s t-test. (G) Quantification of PD1+ cells in untreated human FL samples relative to RRAGC mutational status. The box-whiskers plot indicate the mean (horizontal line), 25-75% range (box), and whiskers indicate maximum and minimum values. Statistical significance was calculated with one-tailed Mann-Whitney test. (H) Oncoprint of 283 cases with TNFRSF14 and RRAGC mutations showing a mutually-exclusive pattern. Study 1 52: 109 cases, Study 2 19: 12 cases; Study 3 20: 141 cases, Study 4 17: 21 cases. Total: 283 cases. Statistical significance was calculated with one-sided Fisher’s exact test.
GSEA also revealed a signature of suppressed apoptosis in the RagC mutant GCs (Figure 5a and Supplementary Figure 5b), a process not generally perceived as regulated by mTORC1 41–44. However, apoptosis has particular relevance in GC B cells, as programmed cell death is a frequent fail-safe response triggered upon poor affinity or lack of T cell help, unresolved DNA damage, or self-reactivity 2. Indeed, the increased GC size and exacerbated B cell response were consistent with increased proliferation, suppressed apoptosis, or both. Steady-state proliferation measured within the increased GC B cell population from RagC mutant mice was similar to that of wild-type mice, (Supplementary Figure 5d), suggesting that decreased death, rather than increased division, could partially explain the increase in the GC B cell population, and provide an explanation for the oncogenicity of RagC mutations. We validated the suppression of programmed cell death signature in RagC mutant GC B cells by immunostaining against cleaved-caspase 3 (c-c3) 10 days after induction of GC by SRBC injection (Figure 5b and Supplementary Figure 5e). In an attempt to mechanistically connect decreased apoptosis in RagC mutant B cells to an indifference to T cell support, we induced a ‘death-by-neglect’ fate by injecting a blocking antibody to CD40 ligand (anti-CD40L), the key driver of Tfh help to GC B cells 36,45,46. During the B-T synapse, ligation of the CD40 receptor in B cells activates several signaling cascades, including PI3K-Akt. Injection of anti-CD40L antibody into mice previously immunized with SRBC resulted in increased apoptosis, but significantly less so in RagC mutant versus wild-type chimeras, as determined by quantification of Bcl6/c-c3 double-positive GC B cells (Figure 5b). Importantly, this reduction in apoptosis was not a reflection of a general resistance to cell death, as gamma-irradiation with 5Gy induced equally large amounts of Bcl6/c-c3 positivity cells in wild-type and RagC mutant GC B cells (Supplementary Figure 5f). This result indicates that RagC mutant GC B cells are intrinsically resistant to apoptosis upon withdrawal of T cell help, and is consistent with a reduced need for microenvironmental activating Tfh signals in RagC mutant B cells. Indeed, while the overall abundance of Tfh gated on its parental CD4+ population is not decreased in RagC mutant mice, the relative abundance of supportive Tfh cells in GC is markedly decreased because of the massive expansion of the GC, as evidenced by a higher ratio of GC B cells to Tfh cells determined by flow cytometry (Figure 5d), and by lower Tfh abundance per GC area by immunostaining with the Tfh marker PD-1 (Figure 5e), supporting again that a relative reduction in T follicular help is well-tolerated in RagC mutant B cells. We did not find evidence for transcriptional changes in RagCmut GC cells in genes known to be involved in B-Tfh cross-talk in the GC synapse (Supplementary Table 2), supporting that B cells are less dependent on Tfh signals without paracrinally repelling Tfh or affecting their function47–50.
Increased independence of GC B cells from Tfh cell signals conferred by RagC mutations may or may not be meaningful in the context of FL induced by VavP-Bcl2tg, particularly if this independence is related to a suppression of apoptosis. To test this possibility, we first quantified steady-state apoptosis in GC induced upon immunization in VavP-Bcl2tg; RagCwt or mut mice. While apoptosis was suppressed by the expression of VavP-Bcl2, regardless of the RagC mutational status, we observed that VavP-Bcl2tg; RagCmut mice had significantly lower C-c3, compared to their VavP-Bcl2tg; RagCwt counterparts (Supplementary Figure 5g). Next, we quantified Tfh cells in murine FL samples. RagC mutant FL showed a similar decrease in the abundance of Tfh cells by IHC (Figure 5f) and by flow cytometry (CXCR5+ PD1+ Foxp3-) (Supplementary Figure 5h), again, without transcriptional changes in B-to-T signals (Supplementary Table 2)
We then analyzed collections of untreated human FL biopsies harboring mutations in RRAGC, and observed a similar decrease in the abundance of PD-1 positive T cells in the follicular areas of these tumors (RagC-wt: 28 cases, RagC-mutant: 16 cases; Figure 5g and Supplementary Figure 5i). The decrease in Tfh cell abundance was even stronger when samples harboring co-occurring mutations in RRAGC and the lysosomal v-ATPase, both involved in the control of mTORC1 activation 51, were grouped (Supplementary Figure 5j).
Finally, loss of TNFRSF14 (HVEM) function (by genetic loss or mutation) is a frequent lesion in human FL because it induces a tumor-supportive microenvironment that includes increased Tfh cellularity 14. If our hypothesis of RagC mutations conferring some degree of independence toward T cell help is true, then loss of TNFRSF14 would be functionally redundant with mutations in RRAGC, and hence these two genetic events would be unlikely to be co-selected in the same patient sample. Hence, we performed a meta-analysis of all 4 reports of human FL samples for which the information regarding the genomic mutational landscape, including the status of RRAGC and TNFRSF14, was available 17,19,20,52 and found that these two genetic lesions were mutually-exclusive (Figure 5h; p=0.0012). Because TNFRSF14 can be inactivated by mutation or deletion, we performed an additional validation with a cohort of FL samples for which we obtained deletion plus mutation information for TNFRSF14, plus mutational data on RRAGC. In this cohort of 109 cases we validated the existence of mutual exclusivity between RRAGC mutations and TNFRSF14 mutations or deletions (Fisher’s exact test: p=0.0367; Supplementary Figure 5k). Taken together, the lower abundance of Tfh cells from the tumor microenvironment and the mutually exclusive genetic alterations in RRAGC and HVEM suggest that FL may rely alternative avenues to support its growth: attracting Tfh cells by loss of HVEM, or autonomously activating intracellular pathways that synergize with, and thus decrease its need for, Tfh cell-derived signals by mutations in RRAGC (Figure 6).
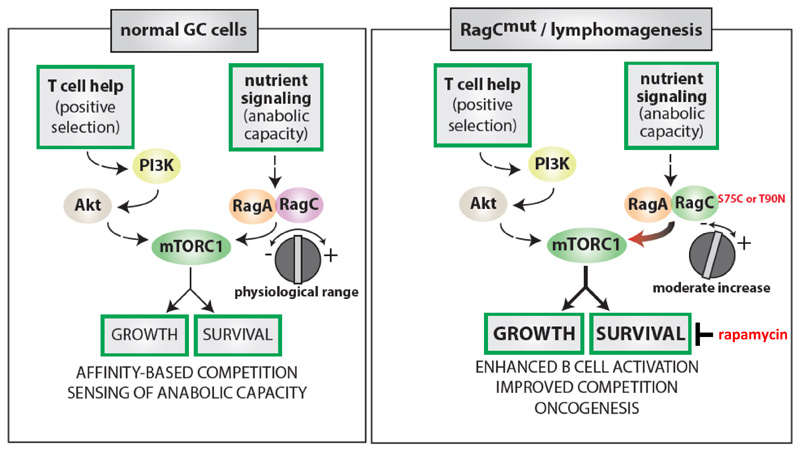
Figure 6. Consequences of RagC mutations for GC B cell and lymphomagenesis.
The nutrient signaling pathway imposes an ‘anabolic capacity’ barrier over B cell activation by T cell help and BCR signaling, so that growth only occurs upon nutrient sufficiency. RagC mutations weaken this barrier to allow B cell activation, while still retaining the ability to suppress activation upon profound reduction in nutrient levels. During lymphomagenesis, these mutations enhance B cell growth and survival but impose a selective vulnerability to pharmacological inhibition of mTORC1.
Discussion
Using novel genetically engineered mice, we provide the first bona fide evidence for an oncogenic role of the Rag GTPase pathway. The exclusiveness of recurrent mutations in RRAGC in human B cell lymphomas (FL, and a smaller percentage of DLBCL) suggests that B lymphocytes are exquisitely sensitive to perturbations in the nutrient sensing pathway. Our previous work in mice with genetic deletion 53 and constitutive activation 36 of RagA showed striking phenotypic aberrations in the B cell lineage and severe impairment of B cell competitiveness in the GC and consequently of the entire antibody response. This was paradoxical, given that strong physiological activation of mTORC1 was a requirement for positive selection of GC B cells. Our present data clarifies this apparent paradox, by showing that more modest activation of the mTORC1 pathway afforded by patient-derived mutations in RagC provides the correct “dose” of mTORC1 hyperactivation required to increase rather than decrease GC B cell fitness, and suggests an explanation for why gain-of-function mutations in RagA do not arise in primary lymphomas 31. Thus, the use of genetically-engineered mice expressing RagC mutations under endogenous control and physiological levels revealed an exquisite sensitivity of B cells to different degrees of activation in the nutrient signaling pathway. The sudden burst of growth and proliferation that occurs upon B cell activation during the GC reaction, driven by c-myc, PI3K-Akt, and NF-κB 36,38,39,45,54,55, among other pathways, depends on an adequate nutrient and energetic supply to sustain the associated anabolic needs. Hence, proper sensing of nutrient sufficiency is imperative to match the extent of the burst to nutrient availability and anabolic capacity, while mismatches caused by excessive activation of the mTORC1 pathway would lead to loss of competitiveness. In this regard, a complete inability to integrate nutrient level cues in the control of mTORC1 could result in an energetic catastrophe during anabolism and proliferation, whereas a mild increase in nutrient signaling may offer a competitive advantage if still retaining the ability to respond to dramatic fluctuations in nutrient levels, as oncogenic mutations in RRAGC do.
It was recently shown that GC B cells are intrinsically resistant to activation of mTORC1 in response to T cell help via the PI3K-Akt axis, at least when compared to naïve B cells 45. The two independent arms upstream of mTORC1, PI3K-Akt downstream of BCR and Tfh cell help and activating mutations in the nutrient-sensing pathway, significantly synergize, but do not substitute each other (Figure 6). As predicted given the decreased requirement for T-cell mediated signals for B cell activation, RagC mutations correlated with decreased presence of Tfh cells (PD1+ T lymphocytes) in both RagC mutant FL samples and human RRAGC mutant FL samples (Figure 5).
We found little evidence of a direct control of mTORC1 over proliferation of GC B cells in vivo, consistent with previous results from us and others 36,56,57, albeit more proliferating GC cells were present in RagC mutant mice at all times due to the extraordinary enlargement of the GC B cell population. Transcriptional profiling and functional experiments in GC B cells support a role for RagC mutations in the suppression of apoptosis, even when the anti-apoptotic protein BCL2 was overexpressed (Figure 5). We postulate that increased mTORC1 activity reinforces B cell activation by T cell help, and suppression of apoptosis is one of the outcomes of B cell activation, likely a critical one in the long process of follicular lymphomagenesis, but not the only one. Indeed, when FL-bearing mice were treated with rapamycin, we observed a selective extension of survival that occurred with a decrease in proliferation of RagC mutant FL cells (Figure 2), and without a significant increase in C-c3 positive cells. This could be indicative of increased resistance to programmed cell death in established lymphomas when compared to GC B cells, including those GC B cells that over-express Bcl2. It is also consistent with an early induction of apoptosis with rapid turnover upon pharmacological inhibition of mTORC1, followed by a halt in proliferation among remaining viable cells, in line with the well-established cytostatic effects of mTORC1 inhibitors.
Loss-of-function mutation or deletion of TNFRSF14 in FL induces a supportive microenvironment that includes abundant Tfh 14, which would be partially redundant with RRAGC mutations if they confer some degree of independence from Tfh signals. Indeed, mutual exclusiveness of RRAGC and TNFRSF14 mutations was precisely what we observed when we performed a meta-analysis of available mutational data on RRAGC and TNFRSF14 (Figure 5). It will be worthwhile to evaluate the interplay between the supportive FL microenvironment, as an inheritance of the requirements of GC B cells, and the autochthonous immune response against FL.
Altogether, we demonstrate that a mild increase in nutrient signaling upstream of mTORC1 has dramatic consequences for FL development and sensitizes toward mTORC1 inhibitors. Our work suggests that pharmacological inhibition of mTORC1 with rapalogs or ATP-competitive inhibitors may be particularly efficacious for the 15% FL harboring activating mutations in the nutrient signaling pathway. This percentage may be an underestimation, if other genetic lesions or epigenetic means of activating the nutrient signaling pathway are at work in FL patients without RRAGC mutations. Clinical trials with rapalogs as monotherapy have been performed recently in FL 58–60, with a fraction of patients showing impressive responses. These intriguing clinical observations, viewed in the context of our work and the existence of oncogenic mutations in RRAGC, trigger speculative thoughts, such as whether the biologic underpinnings of the mTOR dependencies and drug response in FL may be determined, at least in part, precisely by the mutational status of genes involved in this nutrient sensing.
It remains to be determined whether non-genetic perturbations in nutrient signaling (as in the case of chronically increased or decreased nutrient levels), would significantly impact the biology of GC and FL cells. Epidemiology-based connection between elevated nutrient intake, body mass index (BMI) and B cell lymphoma is far from clear, with reports showing opposite conclusions 61–64. The humoral response is affected by BMI in humans and rodents 65,66. Hence, further research is needed to examine if therapeutic manipulation of nutrient levels may serve as a means of modulating the humoral response and indeed if this provides a novel strategy against FL tumors.
Methods
Generation of RagC Crispr-edited KI mice
To introduce the patient-derived RRAGC mutations, C57BL/6 mouse blastocysts were injected with the Cas9 mRNA, a single-guide RNA (sgRNA) targeting the sequence of interest (mouse Rragc), and a single-stranded DNA oligo (ssDNA) containing the desired mutation flanked by 40–60 bases homologous to the sequence adjoining the DNA double-strand break (DSB). Following clone selection, genotyping was performed by specific PCR followed by restriction fragment length polymorphisms (RFLP) or Sanger sequencing.
Sequences used:
For RagC-S74C: TCC->AAC
ssDNA:
CGGCCCGGGCGGCGCTGACAGCTCCAAGCCGAGGATCCTGCTTATGGGGCTCCGGCGCAGCGGCAAAAACAGTATCCAAAAAgtgagcgccccgcagccgccgcctgcccttggttttgcccggcccacggcgctcaggtggcgggt
gRNA: CGGCAAATCCTCCATCCAGA
For RagC-T89N: ACT->AAT
ssDNA
cttgtatagttaaatagttacaaagttgtctctgtttctcctcacagGTGGTGTTTCATAAGATGTCACCCAATGAGAATCTGTTCCTAGAAAGTACCAACAAGATTTATAAAGATGACATTTCCAACAGCTCCTTTGTGAACTTCCA
gRNA: ACCCAATGAGACTCTCTTTT
Mice
All animal procedures carried out at the CNIO were performed according to protocols approved by the CNIO-ISCIII Ethics Committee for Research and Animal Welfare (CEIyBA). IghmμMT mice 37 were obtained from Jackson Laboratories (JAX stock #002288). Mice were housed under specific pathogen-free (SPF) conditions, at 22°C, and with 12-hr dark/light cycles (light cycle from 8:00am to 8:00pm). Mice were fed with a standard chow diet (Harlan Teklad 2018). All mice were observed weekly by trained personnel. Upon signs of morbidity, mice were closely inspected daily until application of Humane End Point (HEP) criteria (http://dels.nas.edu/global/ilar/Guide) in consultation with veterinary staff. From our experience, the humane end point is applied when the life expectancy of the mice is on average shorter than one week. No maximal tumor volume criteria was used because health decline caused by FL always precedes the growth of measurable bulky tumors. Rapamycin was supplied encapsulated in chow diet at 42 ppm (Rapamycin Holdings and Purina Lab Diet).
Immunizations and mouse experimentation
For the generation of GC response, 8-10 week old mice were immunized by intraperitoneal (i.p.) injection of (1x109) Sheep Red Blood Cells (SRBC, Oxoid, #SR0053B) in PBS or 50µg of 4-hydroxy-3-nitrophenylacetyl conjugated to Keyhole Limpet Hemocyanin (NP-KLH, BioSearch, #N-5060-5) in Alum Adjuvant (Thermo Scientific, #77161) (2:1) and analyzed at day 10.
To generate larger amounts of GC B cells (i.e. for gene expression profiling), we did two sequential i.p. SRBC injections (day 0, 5x108; day 5, 1x109) and collected cells at day 12. This protocol yielded about three-four fold more GC B cells (~12-15% of the B cell fraction) than with a single immunization.
For the memory response assays, mice were immunized by intraperitoneal injection of 50μg of NP-KLH in complete Alum Adjuvant and blood was collected at day 0, 7, 14 and 28.
To measure proliferation in GC B cells, mice were immunized by intraperitoneal injection of (1x109) SRBC, and after 10d, 2 hours prior to sacrifice, mice were intraperitoneal injected with 0.5 mg of EdU. Analysis of EdU incorporation was performed using the Click-iT® Plus EdU Alexa Fluor® 647 Flow Cytometry Assay Kit (Life Technologies, #C10634) following manufacturer instructions
For anti-CD40L induced apoptosis, mice were immunized by intraperitoneal injection of (1x109) SRBC, and after 10d, mice were injected intravenously with 200μg of anti-CD40L agonist antibody (BioXCell, clone FGK4.5).
To study the effect of rapamycin in GC formation, 10-12 week old chimeric mice were immunized by intraperitoneal injection of (1x109) SRBC and simultaneously treated with encapsulated rapamycin chow diet for 10 days (see above) or administered i.p. at 2mg/kg at day 8, prior to assessment of GC formation by flow cytometry at day 10 (see Supplementary Figure 3d)
Diagnose of murine lymphomas and autoimmunity
Lymphoproliferative diseases developing in the VavP-Bcl2tg; RagCmut cohort were diagnosed based on morphology according to the following criteria, in analogy to the classification of human lymphoma: (i) early stages of FL (FL in situ), defined by the presence of oversized follicles with partial or absent mantle zone and loss of confinement in the context of a yet preserved tissue architecture; (ii) overt FL of various grades, characterized by the effacement of the nodal and/or splenic architecture by a proliferation of follicle center B cells with a follicular growth pattern occupying the medullary and/or paracortical areas; (iii) DLBCL, defined by the effacement of the lymphoid organ architecture due to the expansion of large cells, with occasional infiltration beyond the capsule into surrounding soft tissues 67. FL was classified into the following three histological grades: grade I, 0-5 centroblasts in 40x field; grade II, 6-15 centroblasts in 40x field; grade III, >15 centroblasts in 40x field. Lymph nodes, spleen, bone marrow and other tissues with abnormal mononuclear infiltrate were evaluated. Mice showed numerous grossly enlarged and fused germinal centers replete with more than 5 germinal centers per nodule of white pulp in the spleen were considered lymphoma. Bcl6 stain was used in doubtful cases to highlight the follicular pattern. The genotype of the animal was not disclosed to the pathologist (A de Martino and E Caleiras). Autoimmunity was scored by perivascular inflammatory infiltrate, predominantly mononuclear of lymphoid aspect, with or without necrosis of the vessel wall, and was most prevalently seen in salivary glands and kidney. Autoimmune glomerulonephritis was defined by hypercellular glomeruli containing eosinophilic deposits, generally accompanied by Bowman epithelium hyperproliferation (a representative example is shown in Supplementary Figure 2d).
Signaling in Mouse Embryonic Fibroblasts
For amino acid deprivation experiments, sub-confluent cell cultures were rinsed twice and incubated in RPMI (US Biological #R8999-04A) and supplemented with 10% dialyzed FBS and all 20 amino acids during one hour. After that, cells were rinsed three times and were placed in RPMI without amino acids, Arginine or Leucine, supplemented with 10% dialyzed FBS, during the indicated time-points. Re-stimulation with amino acids was performed during 10 minutes.
mTORC1 activity in naïve B cells
To study mTORC1 activity in naïve B cells, we isolated CD43- resting B cells from 8-10 week-old non-immunized mice using magnetic beads, (Myltenyi, #130-049-801) following manufacturer instructions. 1x106 cells CD43- resting B cells were plated with B cell media (RPMI (US Biological #R8999-04A) +10% dialyzed FBS +55μM β-mercaptoethanol (Gibco, #31350-010) + 10mM HEPES (Lonza #BE17-737E) +100μg/mL penicillin/streptomycin (Gibco, #15070-063) containing no amino acids with or without cytokines (1μg/mL anti-CD40, (R&D Systems, #MAB440) and 25ng/mL mouse Interleukin-4 (R&D Systems, #404-ML)) during 1h. After this time, stimulation with amino acids was performed for 30min and cells where harvested and processed for flow cytometry.
Class-switch recombination and differentiation in vitro
To study the process of class-switch recombination (CSR) and plasma cell differentiation in activated mouse B cells, we isolated CD43- resting B cells from 8-10 week-old non-immunized mice using magnetic beads, (Myltenyi, #130-049-801) following manufacturer instructions. Cells were washed and resuspended in B cell medium (RPMI (Sigma #R8758) +10% FBS (Hyclone #SV30160.03) +55μM β-mercaptoethanol (Gibco, #31350-010) +10mM HEPES (Lonza #BE17-737E) +100ug/mL penicillin/streptomycin (Gibco, #15070-063) containing 1ug/mL a-CD40, (R&D Systems, #MAB440) and 25ng/mL mouse Interleukin-4 (R&D Systems, #404-ML). The culture medium was replenished every two days to avoid exhaustion. Four days later, cells were harvested and processed for flow cytometry analysis.
To study PRDM1 expression in activated B cells, after 4 days of differentiation, cells were harvested and processed for flow cytometry using Alexa Fluor-647 anti-mouse Blimp-1 detection kit (Biolegend #149902)
To study the effect of rapamycin in plasma cell production, B cells were treated with 1nM or 10nM 48h before collection and flow cytometry analysis (day 4).
B cell proliferation
To study cell proliferation in activated B cells, CD43- resting B cells were labeled with 5mM CFSE (Life Technologies #C34554) following manufacturer instructions. This reagent monitors distinct generations of proliferating cells by a fluorescent dye dilution. Data were acquired at days 2 to 4 on a FACSCanto II (BD Biosciences) flow cytometer with 488nm excitation and an emission filter in the 530/30nm. Alternatively, analysis of EdU incorporation in 3d-activated B cells was performed using the Click-iT® Plus EdU Alexa Fluor® 647 Flow Cytometry Assay Kit (Life Technologies, #C10634) following manufacturer instructions. To study the effect of rapamycin in B cell proliferation, B cells were treated with 1nM or 10nM 24h before the addition of Edu (day 3).
Staining and Flow Cytometry Analysis
Mononuclear cell pools were isolated from mouse spleens at the indicated times postimmunization. Cells were separated by crushing the spleens through a 70 micron mesh (Corning) in ice-cold PBS +0.1% BSA +3mM EDTA, and red blood cells were lysed using Erythrocyte Lysis Buffer (Qiagen, #79217). Cell staining was performed on ice in PBS +0.1% BSA + 3mM EDTA. We included a prior step of incubation with Fc-block Reagent (Anti-CD16/CD32, Pharmigen, #553142). GC B cells were identified within the B cell fraction (B220+) as GL7+ CD95+. Alternatively, GC B cells were identified within the CD19+ cell fraction, as CD95+CD38-. Plasma cells were identified by gating on CD138+, B220int cells. Granulocytes were identified as B220-/CD4-/CD8-/CD11b-/Gr-1+. MDSC were identified as B220-/CD4-/CD8-/CD11b+/Gr-1+. Myeloid cells were identified as B220-/CD4-/CD8-/CD11b+. Macrophages were identified as B220-/CD4-/CD8-/CD11blow/F4/80+. T follicular helper cells (Tfh) were identified as B220-/CD4+/CXCR5+/PD-1+. For the detection of phosphorylated S235/236 of S6 with intracellular staining, cell suspensions were fixed and permeabilized using the Cytofix-Cytoperm and Cytoperm-Wash buffers (BD Biosciences #554714) and subsequently stained for 120min at RT. Complete information on antibodies used in flow cytometry is shown in Supplementary Table 3.
All flow cytometry analyses were done at the Flow Cytometry Facility (CNIO), using BD LSR-Fortessa or BD CantoB cell analyzers, running BD FACSDiva software (BD Biosciences). FlowJo software (v 9.8.1 and v.10; TreeStar) was used for data analyses and plot rendering. See details on the gating strategies in Supplementary Figure 6.
Immunoblotting
Cells were rinsed once with ice-cold PBS and lysed in ice-cold lysis buffer (50 mM HEPES [pH 7.4], 40mM NaCl, 2mM EDTA, 1.5mM sodium orthovanadate, 50mM NaF, 10mM pyrophosphate, 10mM glycerophosphate, and 1% Triton X-100, and one tablet of EDTA-free complete protease inhibitors [Roche] per 25ml). Cell lysates were cleared by centrifugation at 13,000rpm for 10 min. Proteins extracts were denatured by the addition of sample buffer, boiled for 5min, resolved by SDS-PAGE, and analyzed by immunoblotting. Western blot analyses were performed according to standard procedures. Antibodies from Cell Signaling were used for detection of P-T389-S6K1 (#9234), S6K1 (#2708), P-S235/236-S6 (#2211), S6 (#2217), P-T37/46-4EBP1 (#2855), 4EBP1 (#9644), P-T308-AKT (#2965), P-S743-AKT (#4060), AKT (#4691). For detection of β-actin we used an antibody from Sigma (#A5441).
Gene expression profiling of GC B cells and FL
GC B cells (B220+, CD95+, CD38-) were sorted from RagCmut and RagCwt splenocytes after 12 days of SRBC immunization in a BD FACSAria Ilu (Becton Dickinson) and InFlux (Cytopeia-Becton Dickinson) cell sorters. Total RNA from sorted GCB was extracted using TRIZOL (Invitrogen, #15596026) and PicoPure RNA Isolation kit (Arcturus, #12204-01) following manufacturer instructions. For gene expression profiling of murine lymphomas, B220+ were isolated from mouse lymphoma tumors by immunomagnetic enrichment with CD45R (B220) microbeads (Miltenyi Biotech # 130-049-501).
RagC S74C GCB samples
Samples of 500ng of total RNA were used. Average sample RNA Integrity Number was 9.75 (range 9.0 - 10) when assayed on an Agilent 2100 Bioanalyzer. PolyA+ fraction was extracted from 1µg of total RNA, randomly fragmented and converted to double stranded cDNA and processed through subsequent enzymatic treatments of end-repair, dA-tailing, and ligation to adapters as in Illumina's "TruSeq Stranded mRNA Sample Preparation Part # 15031047 Rev. D" kit (this kit incorporates dUTP during 2nd strand cDNA synthesis, which implies that only the cDNA strand generated during 1st strand synthesis is eventually sequenced). Adapter-ligated library was completed by PCR with Illumina PE primers. The resulting purified cDNA library was applied to an Illumina flow cell for cluster generation and sequenced on an Illumina instrument (Illumina HiSeq2500) by following manufacturer's protocols. Image analysis, per-cycle basecalling and quality score assignment was performed with Illumina Real Time Analysis software. Conversion of Illumina BCL files to bam format was performed with the Illumina2bam tool (Wellcome Trust Sanger Institute –New Pipeline Group (NPG); https://github.com/wtsi-npg/illumina2bam).
RagC T89N GCB and RagC S74C FL samples
Samples of 500ng of total RNA (but 100ng for YRT-236, and 200-300ng for YRT-224, -230 and -235) were used. Average sample RNA Integrity Number was 8.5 (range 7.9-10) when assayed on an Agilent 2100 Bioanalyzer. PolyA+ fraction was purified and randomly fragmented, converted to double stranded cDNA and processed through subsequent enzymatic treatments of end-repair, dA-tailing, and ligation to adapters as in "NEBNext Ultra II Directional RNA Library Prep Kit for Illumina" (NEB #E7760) [this kit incorporates dUTP during 2nd strand cDNA synthesis, which implies that only the cDNA strand generated during 1st strand synthesis is eventually sequenced]. Adapter-ligated library was completed by PCR with Illumina PE primers. The resulting purified cDNA library was applied to an Illumina flow cell for cluster generation and sequenced on an Illumina instrument (Illumina HiSeq2500) by following manufacturer's protocols. Image analysis, per-cycle base calling and quality score assignment was performed with Illumina Real Time Analysis software. Conversion of BCL files to FASTQ format was performed with the bcl2fastq Software (Illumina).
Sequencing reads were analyzed with the nextpresso pipeline (http://bioinfo.cnio.es/nextpresso/) as follows: sequencing quality was checked with FastQC v0.11.0 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were aligned to the mouse genome (NCBI37/mm9) with TopHat-2.0.10 68 using Bowtie 1.0.0 69 and SAMtools 0.1.19 70, allowing 2 mismatches and 20 multihits. Differential expression was tested with DESeq2 71, using the mouse NCBI37/mm9 transcript annotations from https://ccb.jhu.edu/software/tophat/igenomes.shtml. GSEAPreranked 35 was used to perform gene set enrichment analysis of the described gene signatures on a pre-ranked gene list, setting 1000 gene set permutations. GSEA Preranked calculates an enrichment score for each gene set using Kolmogorov-Smirnov test. The nominal p value estimates the statistical significance of the enrichment score for a single gene set and it was corrected for gene set size and multiple hypothesis testing” For GSEA analysis, non-expressed genes were removed from the ranking.
ELISA
Total IgG1 serum titers after 10d of SRBC immunization were measured by ELISA using Abcam #ab133045-IgG1 Mouse ELISA kit and following manufacturer instructions. High-affinity and total NP-specific antibodies were measured by ELISA using 10mg/ml of NP(2.5)-BSA or NP(25)-BSA as the coating reagent, respectively. Serum was assayed in 3-fold dilutions starting at 1/100. NP-specific IgG1 was detected using goat anti–mouse IgG1 Fc-specific antibody conjugated to horseradish peroxidase (Jackson Immunoresearch, # 115-035-071) and developed with tetramethylbenzidine (Sigma, T4319). OD450 was measured using a Synergy4 microplate reader (Izasa Scientific). Titers were calculated by logarithmic interpolation of the dilutions with readings immediately above and immediately below an OD450 of 0.2 (for example, if OD450 readings at dilution 2 (1/900) = 0.3 and at dilution 3 (1/2700) = 0.1, then Titer = dilution = 2.5 = 1/1558).
Histological and histochemical analysis of mouse tissues
Tissue samples were fixed in 10% neutral buffered formalin (4% formaldehyde in solution), paraffin-embedded and cut at 3μm, mounted in superfrost®plus slides and dried overnight. For different staining methods, slides were deparaffinized in xylene and re-hydrated through a series of graded ethanol until water. Consecutive sections were stained with hematoxylin and eosin (H&E), and several immunohistochemistry reactions were performed in an automated immunostaining platform (Ventana Discovery XT). Antigen retrieval was first performed with the appropriate pH buffer, (CC1m, Ventana, Roche) and endogenous peroxidase was blocked (peroxide hydrogen at 3%). Then, slides were incubated with the appropriate primary antibody as detailed: rabbit monoclonal anti-PD-1 (D7D5W, 1/50, Cell Signaling, #84651), mouse monoclonal anti-Bcl6 (191E/A8; 1/30; CNIO Monoclonal Antibodies Core Unit #AM(191E/A8)) and rabbit polyclonal anti-Cleaved Caspase 3 (1/300, Cell Signaling, #9661). After the primary antibody, slides were incubated with the visualization systems (Omni Map anti-Rabbit, Ventana, Roche) conjugated with horseradish peroxidase. Immunohistochemical reaction was developed using 3, 30-diaminobenzidine tetrahydrochloride (DAB) (Chromo Map DAB, Ventana, Roche; DAB Dako) and nuclei were counterstained with Carazzi’s hematoxylin. Finally, the slides were dehydrated, cleared and mounted with a permanent mounting medium for microscopic evaluation. Positive control sections known to be primary antibody positive were included for each staining run. For PD-1 quantification, whole slides were acquired with a slide scanner (AxioScan Z1, Zeiss). After ROI (Regions Of Interest) selection (lymph nodes and spleen), areas for quantification were selected and exported as subsets of images in TIFF format. All images were then checked and those with staining or cutting artifacts were eliminated. Different images from different slides were chosen for quantification program training (AxioVision 4.6 software package, Zeiss) and an appropriate script for each antibody was created for PD-1 quantification, positivity was evaluated in one phase (phase 1, positive cells) and compared with total tissue area (phase 2, total cells). After training and script optimization, the quantification program was run and results exported as excel files with scoring data for each tiff file. Data obtained was then compiled and appropriately assessed.
Histology and immunohistochemistry FL patient samples
Immunohistochemistry (IHC) staining was performed in an automated system following the manufacturer’s instructions (Leica Bond-Max, Epitope Retrieval Solution 2 AR9640 for 20 minutes and Bond Polymer Refine Detection DS9800; Leica Microsystems K.K., Tokyo, Japan). The primary antibody was a mouse monoclonal anti-PD1, clone NAT105C/E3 lot num. 0208, provided by Dr. Giovanna Roncador from the Monoclonal Antibodies Core Unit of the CNIO. Incubation time of the primary antibody was 15 minutes. The tissue microarray (TMA) had 3 punches per biopsy that were distributed discontinuously. The resulting cores had a diameter of 1-1.5 mm. The PD1 analysis was set up in 72,73 and was done as follows: first, observation was made using a brightfield Olympus BX63 microscope (Olympus K.K. Tokyo, Japan). Then, the slide was scanned at 400x magnification using a digital pathology system (Hamamatsu NanoZoomer C9600) and visualized with NDP.view2 software (Hamamatsu Photonics, Hamamatsu, Japan). PD1 was quantified based on the expression within the follicles except when the follicular lymphoma had a completely diffuse pattern. PD1 was measured as 0 (<1%), +1 (1-5%), +2 (5-20%) and +3 (>20%). The analysis of the cores was made without previous knowledge of the mutational status of RRAGC. The value of each core was recorded and the average of the three cores was calculated. Occasional broken cores with insufficient tissue or inadequate immunohistochemical staining were excluded from the analysis. Comparisons were made by non-parametric Mann-Whitney U test, one-tailed and with a priori significance level of 0.05 (www.socscistatistics.com).
Quantification, Statistical Analysis and Reproducibility
The n, indicating the total number of animals per group, as well as the definition of center, dispersion and precision measures are reported in each figure and figure legend. Unless otherwise stated, two-sided Student’s t-test or Chi square test were performed as depicted in the Figures. Survival in mouse experiments was represented with Kaplan–Meier curves, and significance was estimated with the log-rank test. Analyses were carried out using Prism software (GraphPad). Further information on statistical parameters, software, study design and reagents used can be found in the Reporting Summary.
Reproducibility: the experiments in Figure 1a were performed four independent times for RagCS74C/+ mice and three independent times for RagC T89N/+ mice showing similar results. The experiments quantified in Supplementary Figure 1f were performed seven independent times for RagCS74C/+ mice and four independent times for RagC T89N/+ mice showing similar results. The experiments in Figure 1b were performed three independent times per genotype showing similar results. The experiments in Figure 1d were performed 8 independent times in RagCS74C/+B cells, 2 independent times in RagCT89N/+ B cells and 1 time in RagA B cells showing similar results. The experiments in Figure 1e were performed two independent times per genotype showing similar results. The experiment in Supplementary Figure 1g was performed once for each genotype. The experiment in Supplementary Figure 1h was performed once. The experiment in Figure 2a, 2c, 2d, 2e, 2g, 2h, 2i, 2j was performed once. The experiment in Supplementary Figure 2a was performed two independent times for each genotype. The experiment in Supplementary Figure 2b, 2f, 2g, 2j, 2i, 2l was performed once. The experiment in Supplementary Figure 2k was performed two independent times showing similar results. The experiments in Figure 3a were performed 19 independent times in RagCS74C/+ mice and 14 independent times in RagCT89N/+ mice showing similar results. The experiments in Figure 3b were performed four independent times in RagCS74C/+ mice and three independent times in RagCT89N/+ showing similar results. The experiments in Figure 3c were performed two independent times in both strains showing similar results. The experiments in Figure 3d were performed one time in the RagCS74C/+ mice and two independent times in RagCT89N/+ mice showing similar results. The experiments in Figure 3e were performed once. The experiments in Figure 3f were performed two independent times in VavP-Bcl2tg; RagCS74C/+ and Bcl2tg; RagCT89N/+ mice showing similar results. The experiments in Supplementary Figure 3b were performed twice in each genotype showing similar results.
The experiment in Supplementary Figure 3c was performed twice showing similar results. The experiments in Supplementary Figure 3d were performed three times showing similar results. The experiments in Figure 4b, 4c, 4d, Supplementary Figure 4a, 4b and 4c were performed once. The experiments in Figure 4f, 4g and 4j were performed two independent times showing similar results. The experiments in Figure 4h, Supplementary Figure 4h, and Supplementary Figure 4i were performed three independent times showing similar results. The experiments in Figure 4l, 4m and Supplementary Figure 4d were performed two independent times for RagCS74C/+ mice and once for RagCT89N/+ mice showing similar results. The experiment in Supplementary Figure 4f were performed once. The experiments in Supplementary Figure 4j were performed two independent times for each genotype showing similar results. The experiments in Figure 5a, Supplementary Figure 5a, 5b and 5c were performed once. The experiments in Figure 5b and Supplementary Figure 5e were performed five independent times in RagCS74C/+ mice and two independent times in RagCT89N/+ mice showing similar results. The experiments in Figure 5d were performed three independent times for each genotype showing similar results. The experiments in Figure 5e, 5f and 5g were performed once. The results obtained in Figure 5h were validated on a different cohort of FL patients (Supplementary Figure 5k). The experiments in Supplementary Figure 5d were performed four independent times in RagCS74C/+ mice and five independent times in RagCT89N/+ mice showing similar results. The experiments in Supplementary Figure 5f, 5g and 5j were performed two independent times showing similar results.
Supplementary Material
Editor’s Summary.
Some follicular B cell lymphomas harbor activating mutations in RRAGC, activator of the nutrient sensor mTORC1. Here the authors show that these mutations confer insensitivity to nutrient deprivation and synergize with paracrine cues from the supportive T cell microenvironment to accelerate lymphomagenesis, but impose vulnerability to inhibition of mTORC1.
Acknowledgements
We are indebted to David M. Sabatini (R01 CA129105, R01 CA103866 and R37 AI047389) and thank R. Jaenisch, S. Markoulaki and the Whitehead Institute for Biomedical Research CRISPR facility for zygote injections. We thank Andrew Clear and Koorosh Korfi for generating TMAs from lymphoma patients, and Pekka Aleksi Katajisto for critical reading of the manuscript. Research was supported by the RETOS projects Programme of Spanish Ministry of Science, Innovation and Universities (MICIU), Spanish State Research Agency (AEI), co-funded by the European Regional Development Fund (ERDF) (grant SAF2015-67538-R), EU-H2020 Programme (ERC-2014-STG-638891), Excellence Network Grant from MICIU/AEI (SAF2016-81975-REDT), a Ramon y Cajal Award from MICIU/AEI (RYC-2013-13546), Spanish Association Against Cancer Research Scientific Foundation (AECC) Lab Grant, Beca de Investigación en Oncología Olivia Roddom, FERO Grant for Research in Oncology; Miguel Servet Fellowship and Grant Award (MS16/00112 and CP16/00112) and Project PI18/00816 within the Health Strategic Action from the ISCIII (to A.O.-M.), both co-funded by the European Regional Development Fund (ERDF), Marcos Fernandez Fellowship from the Spanish Leukemia and Lymphoma Foundation/ Vistare Foundation (to A.O.-M.) and L’Oreal For Women in Science Award (to A.O.-M.). J.F. is a recipient of a Cancer Research UK Programme Award (15968) and J.O. is a recipient of a Cancer Research UK Clinician Scientist Fellowship (22742). N.M.M. is a Ramon y Cajal Awardee MICIU/AEI (RYC-2016-20173). N.D.S., C.C.A., A.B.P.-G. and K.T. are recipients of Ayudas de contratos predoctorales para la formacion de doctores from MICIU/AEI (BES-2016-077410, BES-2015-073776, BES-2017-081381, BES-2016-078082).
Footnotes
Data Availability
Sequence data that support the findings of this study have been deposited in GEO, with the accession codes GSE125393 and GSE125394. The data that support the findings of this study are available from the corresponding author upon request. The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
Author Contributions: A.O.-M. performed most experiments, contributed to experimental design, data analysis, and writing of the manuscript. N.D.-S, A.S., C.L.F, C.M., C.C.A., A.V., L.M.-A, B.F.-R, A.B.P-G., and N.M.M. provided help with experimentation. K.T and E.P-Y performed bioinformatics analysis of RNA-seq and meta-analysis of mutually exclusive mutations. E.C. and A.D.M. performed and diagnosed the histology and pathology. J.C. and N.N. performed and analyzed immunohistochemistry studies on human samples. S.A., J.O. and J.F. performed the mutation analyses on the patient samples, provided the corresponding tissue microarrays and clinical information. J.O., J.F. and G.D.V. contributed critical intellectual input in design and interpretation of data. A.E. conceived and supervised the study, analyzed the data, wrote the manuscript and secured funding. All authors read and commented on the manuscript and figures.
Competing Interests statement. No competing interests.
References
- 1.Kahl BS, Yang DT. Follicular lymphoma: evolving therapeutic strategies. Blood. 2016 doi: 10.1182/blood-2015-11-624288. [DOI] [PubMed] [Google Scholar]
- 2.Victora GD, Nussenzweig MC. Germinal Centers. Annu Rev Immunol. 2012 doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 3.Shlomchik MJ, Weisel F. Germinal center selection and the development of memory B and plasma cells. Immunol Rev. 2012 doi: 10.1111/j.1600-065X.2012.01124.x. [DOI] [PubMed] [Google Scholar]
- 4.Mesin L, Ersching J, Victora GD. Germinal Center B Cell Dynamics. Immunity. 2016 doi: 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannard O, Cyster JG. Germinal centers: programmed for affinity maturation and antibody diversification. Current Opinion in Immunology. 2017 doi: 10.1016/j.coi.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Tas J, et al. Visualizing antibody affinity maturation in germinal centers. Science (80-. ) 2016 doi: 10.1126/science.aad3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Jong D, Fest T. The microenvironment in follicular lymphoma. Best Pr Res Clin Haematol. 2011 doi: 10.1016/j.beha.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Huet S, Sujobert P, Salles G. From genetics to the clinic: A translational perspective on follicular lymphoma. Nature Reviews Cancer. 2018 doi: 10.1038/nrc.2017.127. [DOI] [PubMed] [Google Scholar]
- 9.Pasqualucci L, et al. Genetics of Follicular Lymphoma Transformation. Cell Rep. 2014 doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasqualucci L, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011 doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010 doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okosun J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46:176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boice M, et al. Loss of the HVEM Tumor Suppressor in Lymphoma and Restoration by Modified CAR-T Cells. Cell. 2016 doi: 10.1016/j.cell.2016.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Challa-Malladi M, et al. Combined Genetic Inactivation of β2-Microglobulin and CD58 Reveals Frequent Escape from Immune Recognition in Diffuse Large B Cell Lymphoma. Cancer Cell. 2011 doi: 10.1016/j.ccr.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung KJJ, et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res. 2010 doi: 10.1158/0008-5472.CAN-10-2460. [DOI] [PubMed] [Google Scholar]
- 17.Kridel R, et al. Histological Transformation and Progression in Follicular Lymphoma: A Clonal Evolution Study. PLoS Med. 2016 doi: 10.1371/journal.pmed.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Launay E, et al. High rate of TNFRSF14 gene alterations related to 1p36 region in de novo follicular lymphoma and impact on prognosis. Leukemia. 2012 doi: 10.1038/leu.2011.266. [DOI] [PubMed] [Google Scholar]
- 19.Ying ZX, et al. Recurrent mutations in the MTOR regulator RRAGC in follicular lymphoma. Clinical Cancer Research. 2016 doi: 10.1158/1078-0432.CCR-16-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okosun J, et al. Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma. Nat Genet. 2015 doi: 10.1038/ng.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green MR, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci U S A. 2015;112:E1116–25. doi: 10.1073/pnas.1501199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-Sahra I, Manning BD. mTORC1 signaling and the metabolic control of cell growth. Current Opinion in Cell Biology. 2017 doi: 10.1016/j.ceb.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–310. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science (80-. ) 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen K, Choe A, Sabatini DM. Intersubunit Crosstalk in the Rag GTPase Heterodimer Enables mTORC1 to Respond Rapidly to Amino Acid Availability. Molecular Cell. 2017 doi: 10.1016/j.molcel.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15:155–162. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- 28.Valvezan AJ, Manning BD. Molecular logic of mTORC1 signalling as a metabolic rheostat. Nat Metab. 2019;1:321–333. doi: 10.1038/s42255-019-0038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsun ZY, et al. The Folliculin Tumor Suppressor Is a GAP for the RagC/D GTPases That Signal Amino Acid Levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/cas-mediated genome engineering. Cell. 2013 doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Efeyan A, et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–83. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hara K, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 33.Wang S, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science (80-. ) 2015;347:188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egle A, Harris AW, Bath ML, O’Reilly L, Cory S. VavP-Bcl2 transgenic mice develop follicular lymphoma preceded by germinal center hyperplasia. Blood. 2004;103:2276–2283. doi: 10.1182/blood-2003-07-2469. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL. Gene set enrichment analysis : A knowledge-based approach for interpreting genome-wide. Proc Natl Acad Sci U S A. 2005 doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ersching J, et al. Germinal Center Selection and Affinity Maturation Require Dynamic Regulation of mTORC1 Kinase. Immunity. 2017 doi: 10.1016/j.immuni.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 38.Calado DP, et al. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol. 2012 doi: 10.1038/ni.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominguez-Sola D, et al. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol. 2012 doi: 10.1038/ni.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng T, Golub TR, Sabatini DM. The Immunosuppressant Rapamycin Mimics a Starvation-Like Signal Distinct from Amino Acid and Glucose Deprivation. Mol Cell Biol. 2002 doi: 10.1128/MCB.22.15.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami M, et al. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol. 2004;24:6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zoncu R, Efeyan A, Sabatini DM. MTOR: From growth signal integration to cancer, diabetes and ageing. Nature Reviews Molecular Cell Biology. 2011 doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo W, Weisel F, Shlomchik MJ. B Cell Receptor and CD40 Signaling Are Rewired for Synergistic Induction of the c-Myc Transcription Factor in Germinal Center B Cells. Immunity. 2018 doi: 10.1016/j.immuni.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han S, et al. Cellular interaction in germinal centers. Roles of CD40 ligand and B7-2 in established germinal centers. J Immunol (Baltimore, Md 1950) 1995 [PubMed] [Google Scholar]
- 47.Papa I, Vinuesa CG. Synaptic interactions in germinal centers. Frontiers in Immunology. 2018 doi: 10.3389/fimmu.2018.01858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vinuesa CG, Cyster JG. How T cells earn the follicular rite of passage. Immunity. 2011 doi: 10.1016/j.immuni.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 49.Vinuesa CG, Linterman MA, Yu D, MacLennan ICM. Follicular Helper T Cells. Annu Rev Immunol. 2016 doi: 10.1146/annurev-immunol-041015-055605. [DOI] [PubMed] [Google Scholar]
- 50.Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 2008 doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zoncu R, et al. mTORC1 senses lysosomal aminoacids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science (80-. ) 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krysiak K, et al. Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma. Blood. 2017 doi: 10.1182/blood-2016-07-729954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Efeyan A, et al. RagA, but Not RagB, Is Essential for Embryonic Development and Adult Mice. Dev Cell. 2014;29:321–329. doi: 10.1016/j.devcel.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boothby M, Rickert RC. Metabolic Regulation of the Immune Humoral Response. Immunity. 2017 doi: 10.1016/j.immuni.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heise N, et al. Germinal center B cell maintenance and differentiation are controlled by distinct NF-κB transcription factor subunits. J Exp Med. 2014 doi: 10.1084/jem.20132613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dowling RJO, et al. mTORC1-Mediated Cell Proliferation, But Not Cell Growth, Controlled by the 4E-BPs. Science (80-. ) 2010 doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barbet NC, et al. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996 doi: 10.1091/mbc.E12-03-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith SM, et al. Temsirolimus has activity in non-mantle cell non-Hodgkin’s lymphoma subtypes: The University of Chicago phase II consortium. J Clin Oncol. 2010;28:4740–4746. doi: 10.1200/JCO.2010.29.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bennani NN, et al. Efficacy of the oral mTORC1 inhibitor everolimus in relapsed or refractory indolent lymphoma. Am J Hematol. 2017 doi: 10.1002/ajh.24671. [DOI] [PubMed] [Google Scholar]
- 60.Witzig TE, et al. A phase II trial of the oral mTOR inhibitor everolimus in relapsed aggressive lymphoma. Leukemia. 2011 doi: 10.1038/leu.2010.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willett EV, et al. Non-Hodgkin lymphoma and obesity: A pooled analysis from the InterLymph consortium. Int J Cancer. 2008 doi: 10.1002/ijc.23344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet. 2008;371:569–578. doi: 10.1016/S0140-6736(08)60269-X. [DOI] [PubMed] [Google Scholar]
- 63.Bhaskaran K, et al. Body-mass index and risk of 22 specific cancers: a population-based cohort study of 5.24 million UK adults. Lancet. 2014;384:755–765. doi: 10.1016/S0140-6736(14)60892-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Body-mass index and all-cause mortality: individual-participant-data meta-analysis of 239 prospective studies in four continents. Lancet. 2016 doi: 10.1016/S0140-6736(16)30175-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kosaraju R, et al. B Cell Activity Is Impaired in Human and Mouse Obesity and Is Responsive to an Essential Fatty Acid upon Murine Influenza Infection. J Immunol. 2017 doi: 10.4049/jimmunol.1601031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheridan Pa, et al. Obesity is associated with impaired immune response to influenza vaccination in humans. Int J Obes (Lond) 2012 doi: 10.1038/ijo.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang J, et al. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov. 2017 doi: 10.1158/2159-8290.CD-16-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012 doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009 doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carreras J, et al. High numbers of tumor-infiltrating programmed cell death 1-positive regulatory lymphocytes are associated with improved overall survival in follicular lymphoma. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.18.0513. [DOI] [PubMed] [Google Scholar]
- 73.Carreras J, et al. Genomic Profile and Pathologic Features of Diffuse Large B-Cell Lymphoma Subtype of Methotrexate-associated Lymphoproliferative Disorder in Rheumatoid Arthritis Patients. Am J Surg Pathol. 2018 doi: 10.1097/PAS.0000000000001071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.