Abstract
The majority of colorectal cancers (CRCs) show hyperactivated WNT signaling due to inactivating mutations in the APC tumor suppressor. Genetically restoring Apc suppresses WNT and induces rapid and sustained tumor regression, implying that re-engaging this endogenous tumor suppressive mechanism may be an effective therapeutic strategy. Here, using new animal models, human cell lines, and ex vivo organoid cultures, we show that Tankyrase (TNKS) inhibition can control WNT hyperactivation and provide long-term tumor control in vivo, but that effective responses are critically dependent on how APC is disrupted. Mutant APC proteins truncated within the Mutation Cluster Region (MCR) region physically engage the destruction complex and suppress the WNT transcriptional program, while early APC truncations (i.e. ApcMin) show limited interaction with AXIN1 and β-catenin, and do not respond to TNKS blockade. Together, this work shows that TNKS inhibition, like APC restoration, can reestablish endogenous control of WNT/β-catenin signaling, but that APC genotype is a crucial determinant of this response.
Keywords: APC, CRC, Tankyrase, TNKS, Base editing, CRISPR
Introduction
Hyperactivation of the WNT signaling pathway is a hallmark of colorectal cancer (CRC), in most cases (~80%) due to inactivating mutations in the adenomatous polyposis coli (APC) tumor suppressor (1,2). APC is a scaffold protein that coordinates assembly of the destruction complex (DC), made up of core proteins APC, AXIN1, GSK3β, and CK1α. The DC regulates WNT signaling via sequential phosphorylation of the WNT master regulator β-catenin, marking it for degradation by the proteasome (3). Previously, using genetically engineered and transplantation-based mouse models, we showed that restoration of endogenous Apc expression drives WNT suppression and rapid regression of CRCs in both the colon and liver (4,5). This work provides in vivo evidence that re-engaging endogenous regulation of WNT signaling is a potent tumor suppressive strategy.
Pharmacologically, it is possible to re-engage DC activity via Tankyrase inhibition (6). Tankyrase 1 and 2 (TNKS and TNKS2) are functionally-redundant members of the poly-ADP ribose polymerase (PARP) family. Unlike other PARP proteins, they play no role in DNA repair, but rather potentiate WNT signaling by PARylating AXIN1/2, targeting it for degradation, and decreasing the stability and activity of the DC (7–9). Numerous small molecules that enable selective TNKS1/2 blockade and WNT pathway suppression in CRC cell lines (10–12) and normal mouse intestine (13,14) have been described, yet evidence for suppression of WNT-driven tumor growth in vivo is conflicting (15–17). Thus, it is unclear if, and in what context, re-engaging the tumor suppressive DC is an effective approach to control hyperactive WNT/β-catenin signaling. Given the well-defined and consistent tumor response seen following Apc restoration in shApc mice, we sought to exploit genetically defined in vivo and organoid models to determine how TNKS inhibition influences tumor cell behavior after Apc disruption.
Results
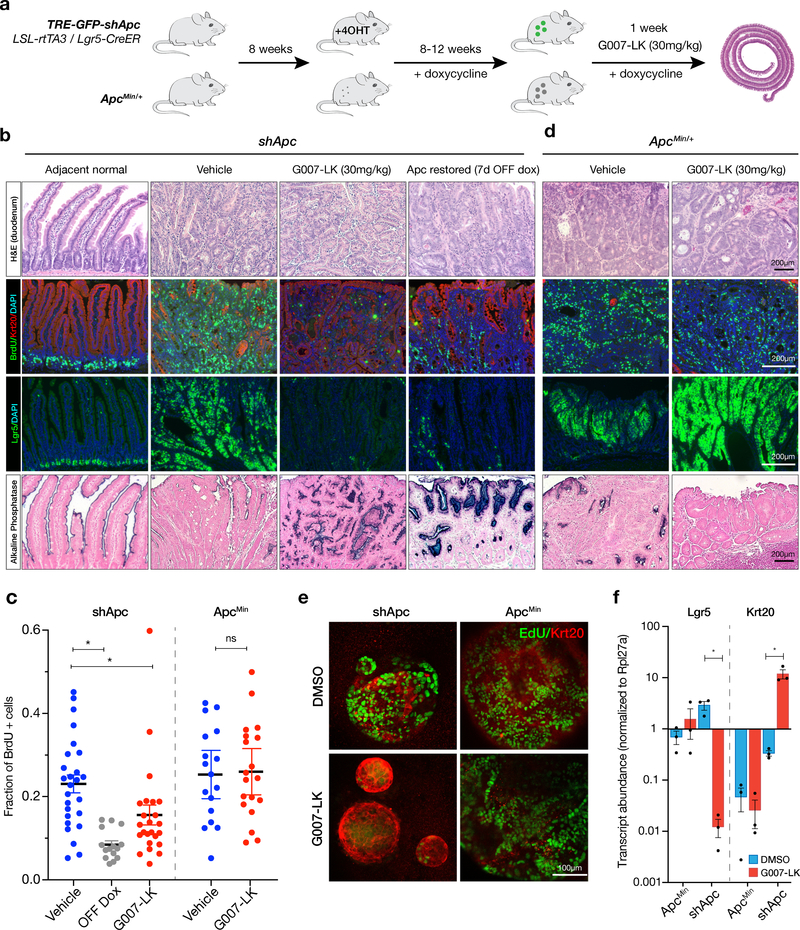
We previously showed that the acute response to Apc restoration in intestinal and colonic tumors is cell cycle arrest and differentiation (4,5). To determine whether TNKS1/2 inhibition could elicit a similar response in vivo, we established a cohort of tumor-bearing CAGs-LSL-rtTA3/TRE-GFP-shApc/Lgr5-CreER (hereafter TG-shApc) mice by treatment with 4-hydroxytamoxifen (4OHT; 25mg/kg, single i.p. injection) and doxycycline (dox; 200mg/kg in the chow, ad libitum) (Figure 1a, Supplementary Figure 1a). Eight - twelve weeks following tumor initiation, mice were randomized into two treatment arms: TNKS1/2 inhibitor G007-LK (30mg/kg, QD) (16) or vehicle control, for one week. Vehicle-treated mice showed highly proliferative lesions with elevated expression of the stem cell marker Lgr5, and limited and sporadic evidence of differentiation markers, Keratin 20 (Krt20) and Intestinal Alkaline Phosphatase (ALPi) (Figure 1b). In contrast, G007-LK-treated and Apc-restored tumors had reduced BrdU incorporation, loss of Lgr5 expression, and induction of both Krt20 and ALPi (Figure 1b-c). Thus, pharmacological TNKS1/2 inhibition phenocopies the acute, tumor-cell-intrinsic response to genetic restoration of Apc.
Figure 1. Tankyrase inhibition drives differentiation in Apc-silenced intestinal tumors.
a. Schematic depiction of the animal models and treatments to produce adenomas in vivo. For consistency, ApcMin mice were administered dox at 8 weeks (when tumors are first detectable), and continued for 8–12 weeks. shApc and ApcMin mice were treated with G007-LK (30mg/kg) or vehicle for 1 week, then sacrificed to assess tumor burden. b. Immunohistochemical, immunofluorescent, and in situ hybridization (Lgr5) stains of representative small intestinal adenomas from shApc mice as indicated. Adjacent normal crypt-villus architecture and shApc off dox (1 week) are shown for reference. c. Quantitation of the fraction of BrdU positive cells in individual tumors (n=3–6 tumors/mouse, n=3–6 mice/treatment). Bars show the mean and 95% CI. d. Immunohistochemical, immunofluorescent, and in situ hybridization (Lgr5) stains of representative small intestinal adenomas from ApcMin mice as indicated. e. Immunofluorescent images of shApc and ApcMin organoids cultured in DMSO or G007-LK (1μM) for 3 days. Organoids were pulsed with EdU for 4 hours to label proliferative cells. f. Quantitative RT-PCR analysis of gene expression of stem cell (Lgr5) and differentiation (Krt20) markers in shApc and ApcMin organoids cultured in DMSO or G007-LK (1μM) for 3 days (n=3, error bars = s.e.m, * p-value < 0.05, unpaired t-test with Welch’s correction).
We next sought to confirm our findings in an independent Apc-driven CRC model. ApcMin mice carry a heterozygous nonsense mutation at codon 850 of Apc and develop intestinal tumors following spontaneous loss of the wildtype allele (18). We treated tumor-bearing 15–20 week-old ApcMin mice for one week with G007-LK (30mg/kg, QD) and measured tumor response on histological cross sections, as described above. Surprisingly, and in contrast to what we observed in the TG-shApc model, ApcMin tumors were completely insensitive to G007-LK treatment. In fact, while adjacent normal intestine showed elevated Axin1 and decreased Lgr5 staining, confirming drug exposure (Supplementary Figure 1b-c), G007-LK-treated tumors maintained proliferation, Lgr5 expression, and showed minimal Krt20 staining or intestinal Alkaline Phosphatase activity (Figure 1c,d).
The difference in tumor response between these two Apc-driven models was striking, and in contrast to a recent report by Tanaka et. al., which suggested that early truncating mutations in APC sensitize cell lines to TNKS inhibition (19). To ensure this conflicting effect was not due to differences in in vivo tumor growth kinetics or timing of treatment, we generated shApc and ApcMin intestinal organoids and treated them ex vivo for 3 days with G007-LK (1μM). In support of our in vivo results, G007-LK-treated shApc organoids underwent rapid cell cycle arrest, while ApcMin organoids maintained a proliferative phenotype (Figure 1e). RNA expression by quantitative RT-PCR (qPCR) further confirmed that TNKS1/2 inhibition induced Krt20 and reduced Lgr5 in shApc organoids, but had no effect in ApcMin cultures (Figure 1f). Together, this data supports the notion that TNKS inhibition can effectively promote tumor cell arrest in some contexts.
Although BrdU incorporation was significantly decreased in G007-LK-treated shApc tumors, the response was less uniform than following genetic Apc restoration (Figure 1b), raising the possibility that variable drug dose or delivery may have impacted tumor response in some cases. Indeed, current TNKS inhibitors show limited oral bioavailability and require daily or twice daily intraperitoneal or intratumoral delivery to achieve effective TNKS blockade (16,20). In addition, in our hands, daily i.p. dosing also produced inflammatory adhesions on the liver spleen and peritoneum, precluding increased or prolonged treatment (Supplementary Figure 1d). To achieve potent, constitutive TNKS blockade without the confounding issue of drug delivery we produced two new transgenic strains (carrying independent tandem shRNAs) that enable dox-inducible silencing of Tnks1/2. To do this we first screened a series of Tnks and Tnks2 miRE-based shRNAs using a fluorescent sensor construct and identified the two most potent targeting each gene (Supplementary Figure 2a-c). Importantly, both tandem shTnks1/2 constructs induced Axin1 stabilization comparable to treatment with a small molecule TNKS inhibitor in vitro (Supplementary Figure 2d) and in vivo (Supplementary Figure 2e).
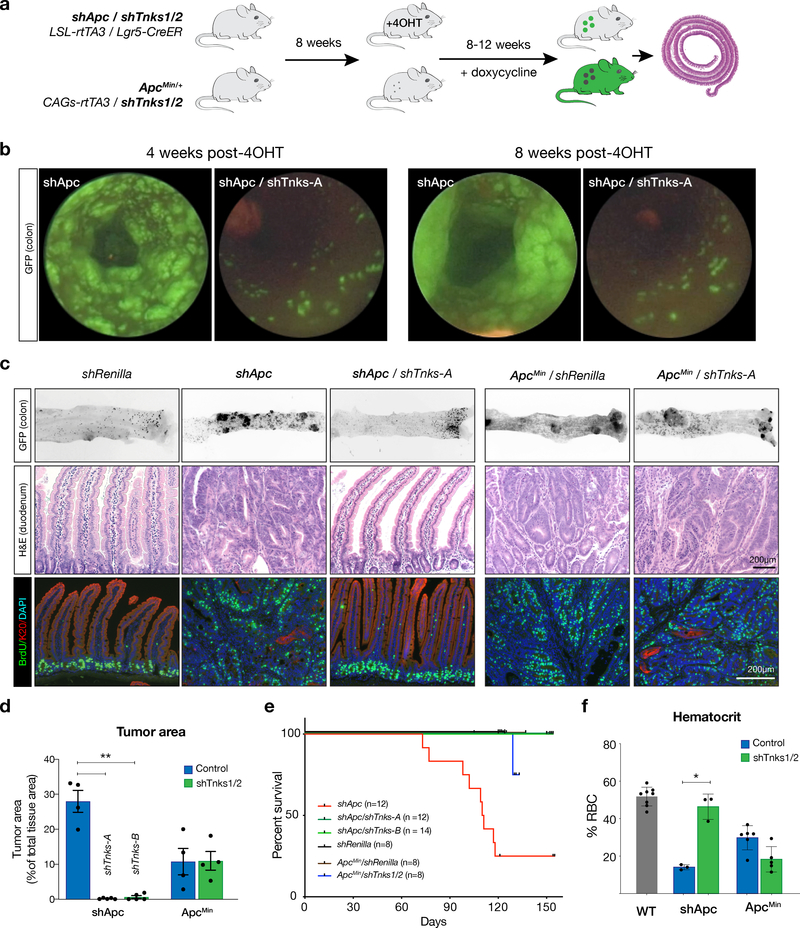
To assess the capacity of Tnks1/2 blockade to provide long-term tumor control, we crossed each TG-shTnks1/2 strain to TG-shApc mice and treated 6–8 week-old mice with 4-OHT and dox (Figure 2a). In shApc mice, numerous GFP+ colon lesions were detectable as early as 4 weeks (Figure 2b), and by 10–12 weeks (endpoint), tumor made up more than a quarter (28%) of the gut (Figure 2c-d). In contrast, shApc/shTnks mice showed only individual GFP+ crypts, similar to those seen in shRenilla (shRen) control mice (Figure 2b-c, Supplementary Figure 3a). No adenomas were observed in the colon of shApc/shTnks mice up to 150 days (endpoint), at which time most shApc mice had become moribund (median survival 111 days) (Figure 2e). Similarly, the small intestine of shApc/shTnks animals resembled wildtype tissue, with only a few animals displaying small hyperproliferative lesions that made up 0.1–0.5% of the total intestinal area (Figure 2d). Tumor suppression was not due to an off-target effect of the Tnks shRNAs, as we observed an identical effect using the second TG-shTnks1/2 strain (Supplementary Figure 3a-c). Consistent with the dramatically reduced tumor burden, shApc/shTnks mice showed no signs of anemia (reduced hematocrit) that is commonly seen in shApc mice with substantial intestinal tumor load (Figure 2f).
Figure 2. Sustained Tnks1/2 knockdown is sufficient to block shApc-mediated tumorigenesis.
a. Schematic of the experimental strategy to achieve Tnks1/2 knockdown in shApc and ApcMin adenomas. b. Colonic endoscopy images taken 4 and 8 weeks post 4-OHT/dox treatment in shApc or shApc//shTnks1/2 mice show a dramatic difference in adenoma formation. c. Epifluorescent images of whole mount colon from shRen, shApc, shApc//shTnks1/2, ApcMin//shRen, and ApcMin//shTnks1/2 mice sacrificed at the experimental endpoint; Black signal represents GFP fluorescence. Immunohistochemical (H&E) and immunofluorescent (BrdU/Keratin20) stains of representative small intestinal adenomas from mice sacrificed at experimental endpoint. d. Quantitation of tumor burden from H&E stained sections of each genotype, as indicated. Tumor area was calculated as a percentage of total tissue in representative cross-sections of the small intestine and colon (n=4–5, error bars = s.e.m., ** p-value < 0.005, unpaired t-test with Welch’s correction). e. Kaplan-Meier plot showing survival of each genotype, as indicated. Log-rank statistical tests are available in Table S5. f. Hematocrit from peripheral blood of each genotype at experimental endpoint (n=3–8, error bars = s.e.m., * p-value < 0.05, unpaired t-test with Welch’s correction).
Mirroring what we observed following G007-LK treatment of ApcMin mice, genetic Tnks1/2 suppression did not block ApcMin-driven tumor growth. Both ApcMin /shRen and ApcMin /shTnks mice developed highly proliferative colonic and small intestinal tumors with minimal evidence of differentiation (Figure 2c, Supplementary Figure 3b). Accordingly, ApcMin/shRen and ApcMin/shTnks mice showed no significant difference in relative tumor area, hematocrit, or survival (Figure 2d-f).
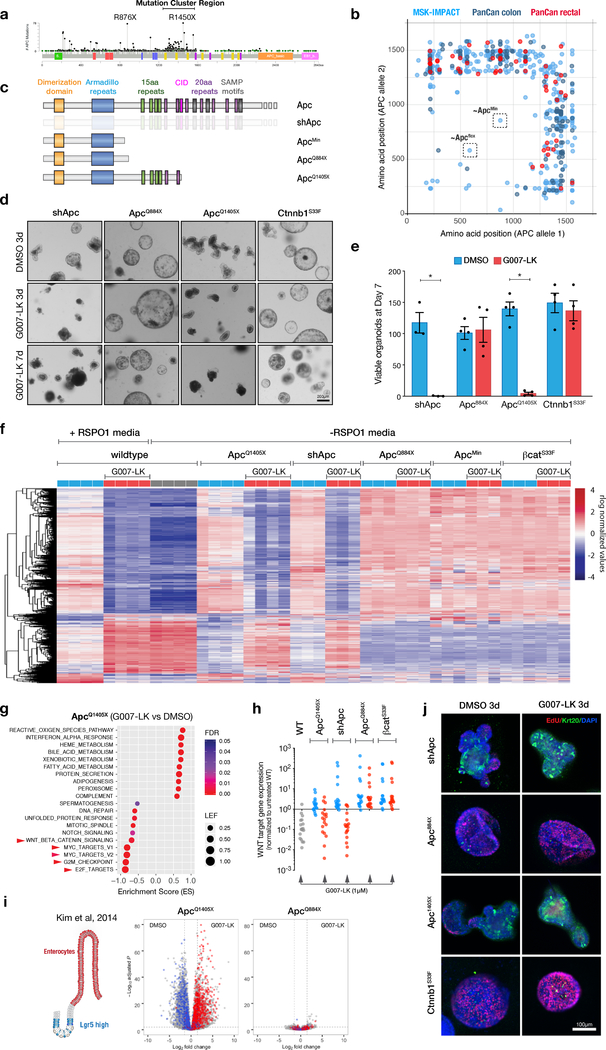
The in vivo data described above highlights that response to TNKS inhibition may be dependent on the genetic context of the tumor. We sought to reconcile which model better reflects the biology and therapeutic response of human CRC; therefore, we analyzed two independent clinical datasets (MSK-IMPACT colorectal and TCGA PanCan colorectal cancer) for the type and frequency of APC mutations. Consistent with previous analyses (1,2), APC mutations were present in 1231/1693 (73%) patient samples, with predominantly nonsense mutations throughout the N-terminal half of the gene (Figure 3a). Further examination of tumors with two identifiable APC mutations revealed a striking selection for cancers carrying at least one truncating mutation within the mutation cluster region (MCR) that spans amino acids ~1250–1580 (Figure 3b). Moreover, cases in which only one allele could be identified showed a further bias toward mutations in the MCR (Supplementary Figure 4a-b). These observations are consistent with previous analyses on an independent cohort (21), and strongly implies that accurate genetic models of human CRC should harbor at least one MCR mutation in APC, more C-terminal than either the ApcMin or ApcFlox alleles (Figure 3b).
Figure 3. An MCR truncation in Apc sensitizes organoids to Tnks1/2 blockade.
a. Lollipop plot of publicly available human CRC MSK-IMPACT and PanCan data showing mutation frequency across all codons in APC. b. Scatter plot of MSK-IMPACT and PanCan data from CRCs carrying two distinct mutations in APC, highlights the presence of late truncating mutations in most CRCs. c. Apc mutations generated in organoids. d. Brightfield images of Apc-mutant or βcatS33F organoids treated with DMSO or G007-LK (1μM) as indicated. e. Quantitation of viable organoids after 7 days of DMSO or G007-LK treatment (n=3–4, error bars = s.e.m., * p <0.05, unpaired t-test with Welch’s correction). All organoids were passaged once at day 3. f. Heatmap representing relative gene expression in wildtype, Apc and βcatS33F organoids cultured in DMSO or G007-LK for 3 days. Shown are 1472 genes with a log2FC >2 and adj. p-value <0.01 in G007-LK-treated WT organoids, compared to DMSO controls. g. Summary plot of GSEA results, including 10 significantly positively and negatively enriched gene sets in G007-LK treated ApcQ1405X organoids. Leading Edge Fraction (LEF) is the fraction of genes in the geneset included in the leading edge of the GSEA plot. h. Mean expression of 18 validated WNT target genes (Table S2) following 3 days treatment with G007-LK or DMSO, normalized to DMSO-treated WT organoids. i. Schematic depiction of the cell populations identified by gene signatures in Kim et al. 2014. Adjacent color-coded volcano plots of G007-LK treated ApcQ1405X and ApcQ884X organoids highlight genes included in these signatures. Genes upregulated following G007-LK treatment are shifted to the right. j. Immunofluorescent staining of Apc-mutant or βcatS33F mutant organoids cultured in DMSO or G007-LK (1μM) for 3 days. Organoids were pulsed with EdU for 4 hours to label proliferative cells.
To engineer defined nonsense mutations both preceding and within the MCR, we used recently described optimized base editing tools (22). We transfected wildtype C57Bl/6 intestinal organoids with the FNLS base editing enzyme (22), and sgRNAs targeting codons Q884 (ApcQ884X) and Q1405 (ApcQ1405X)(Figure 3c, Supplementary Figure 5a-b). The Q884X mutation closely mimics the R876X allele seen in human CRCs, which truncates APC after the armadillo repeats, but before the first set of 15 amino acid β-catenin binding repeats (Figure 3c). Truncation of Apc at Q1405, immediately after the second 20 amino acid repeat (20AAR), removes a conserved ‘catenin inhibitory domain’ (CID) (Figure 3c) that is thought to be critical for proper β-catenin regulation (23,24). Indeed, studies in Drosophila, cancer cell lines, and mice have shown this allele promotes high levels of WNT activation (23–26). In addition to engineered Apc mutations, we generated ApcMin and shApc organoids, as well as those with an activating missense mutation in serine 33 of Ctnnb1 (βcatS33F) (Supplementary Figure 5b); this mutant protein cannot be phosphorylated at S33 by GSK3β, and is therefore independent of the DC.
All mutant organoids could be maintained in the absence of WNT/RSPO ligands, confirming cell intrinsic constitutive activation of WNT signaling. To assess the response to TNKS inhibition, we treated each mutant with G007-LK (1μM) for 7 days, and quantified cell survival by counting viable organoids. As expected, βcatS33F showed no change in morphology or viability following G007-LK treatment. Similarly, and consistent with the lack of response in ApcMin cells, ApcQ884X organoids showed no response to G007-LK. In contrast, ApcQ1405X and shApc organoids showed reduced crypt branching after 3 days, and were unable to survive 7 days of G007-LK treatment (Figure 3d-e). Importantly, the lack of response to TNKS inhibition in ApcQ884X and ApcMin cultures was not specific to these similar truncating alleles, as organoids carrying an even earlier truncation, ApcΔ580 (generated from the Apcflox allele (27)), were also resistant to G007-LK treatment (Supplementary Figure 6a). Further, the effect was not specific to G007-LK, as we noted a similar genotype-dependent response with two independent TNKS inhibitors of different chemotypes: NVP656 (20) and XAV939 (6)(Supplementary Figure 6b).
To gain insight into the molecular events underlying the genotype-dependent response to G007-LK, we performed transcriptome profiling by RNAseq. We treated wildtype, Apc mutant (ApcMin, ApcQ884X, ApcQ1405X, and shApc), and Ctnnb1 mutant (βcatS33F) organoids with 1μM G007-LK (or DMSO vehicle) for three days and recovered cells for mRNA sequencing. Principle component analysis of the data revealed tight clustering of individual biological replicates of similar genotypes, but broad gene expression differences between organoid groups (Supplementary Figure 7a). More than 80% of the variance in gene expression between groups was accounted for by PC1, which included a number of canonical WNT target genes, including Sox17, Nkd1, Wnt6, and Prox1. In general, organoids with βcatS33F mutations and early Apc truncations clustered together and had higher WNT target gene expression, while ApcQ1405X and shApc organoids had lower WNT target induction, albeit markedly higher than wildtype organoids (Supplementary Figure 7b). Importantly, the transcriptome analysis showed that organoids derived from ApcMin mice (ApcL850X) and those with a similar base editing-induced mutation (ApcQ884X) had highly similar global gene expression patterns (Figure 3f, Supplementary Figure 7a), highlighting the fidelity of base editing for modeling specific genetic alleles in organoids.
Treatment with G007-LK did not dramatically alter global gene expression of βcatS33F, ApcMin, ApcQ884X cultures, but caused a striking shift in the transcriptome of shApc, ApcQ1405X, and wildtype organoids. Indeed, significant gene expression changes (log2FC >2, adj. p-value <0.01) observed in wildtype cultures following treatment with G007-LK (or RSPO1 withdrawal) were closely mirrored in shApc and ApcQ1405X organoids (Figure 3f). In contrast, few transcriptional changes were observed in G007-LK-treated ApcMin and ApcQ884X organoids (28 and 56 genes, respectively; log2FC >2, adj. p-value <0.01), and there were zero (0) significant changes in gene expression observed in βcatS33F cultures (Table S1). The profound lack of response in βcatS33F organoids strongly suggests that the primary effect of TNKS inhibition is modulating DC activity via Axin1/2.
Gene set enrichment analysis (GSEA) of G007-LK-treated ApcQ1405X organoid transcriptomes identified an upregulation of networks related to differentiated intestine function, such as protein and lipid metabolism, and a decrease in cell cycle-related MYC and E2F targets (Figure 3g). Conversely, MYC-related gene sets were actually enriched in G007-LK-treated ApcQ884X cells (Supplementary Figure 7c). As expected, G007-LK also induced a significant decrease in WNT/βcatenin signaling in ApcQ1405X organoids (Figure 3h, Table S2), while WNT signaling was not significantly altered in ApcQ884X, ApcMin, or βcatS33F G007-LK-treated organoids (Figure 3h). We next compared our data to two published, experimentally-derived intestinal gene signatures that distinguish crypt-based stem cells and differentiated enterocytes (28,29). In both cases, genes linked with terminal enterocyte differentiation were strongly upregulated following treatment of ApcQ1405X organoids with G007-LK, while genes associated with Lgr5-positive stem cells were downregulated (Figure 3i, Supplementary Figure 7d, Supplementary Figure 8). Consistent with transcriptional changes, G007-LK-treated shApc and ApcQ1405X organoids showed rapid cell cycle arrest and induction of differentiation markers (Krt20 and Alkaline Phosphatase) (Figure 3j, Supplementary Figure 9a-b). No apparent increase in differentiation or loss of stem cell identity were observed following treatment of ApcQ884X or βcatS33F organoids (Figure 3i-j, Supplementary Figures 7-9). The organoid response to G007-LK was not a transient cell cycle arrest, but likely reflects an enterocyte differentiation program, as organoid survival at 7 days was only slightly improved following washout of the drug after 3 days of treatment (Supplementary Figure 10a-b). Further, the intestinal differentiation response observed in organoids is consistent with what we observed following G007-LK treatment of shApc tumors in vivo (Figure 1) or genetic Apc restoration (4). In all, these data support the notion that Tankyrase inhibition re-engages endogenous WNT regulation and normal intestinal differentiation in cells carrying a late APC truncation, closely mimicking the effect Apc restoration (4).
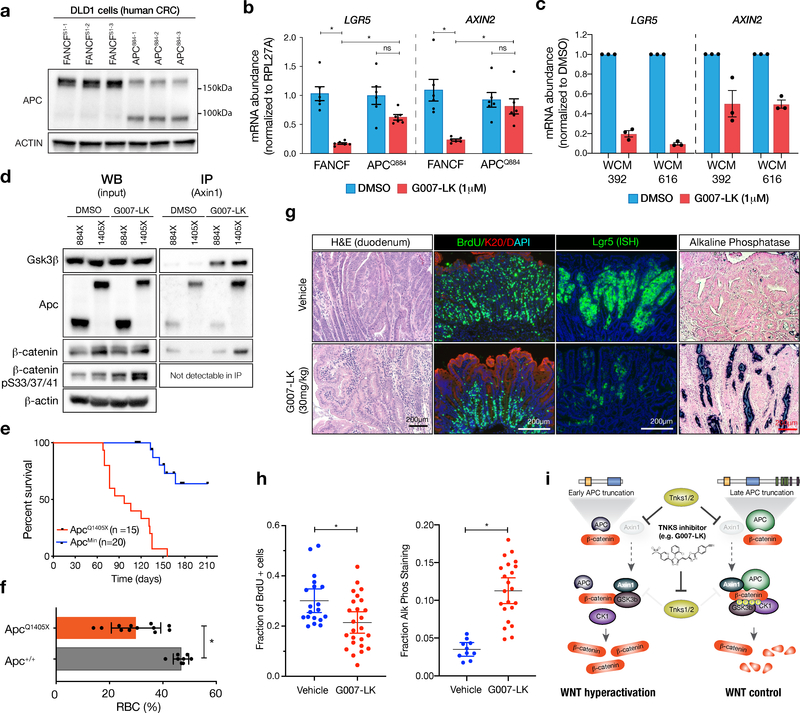
To determine whether similar a genotype-dependent response was apparent in human colorectal cancer cells, we used a well-characterized human CRC cell line (DLD1) that carries a late truncating allele of APC (1417X) and shows a marked WNT transcriptional response to TNKS inhibitors. Using Cas9 and an sgRNA analogous to the ApcQ884X guide used in the mouse studies (APCQ883), we created multiple independent DLD1 lines with shorter APC truncations (Figure 4a, Supplementary Figure 11a). Consistent with what we observed in mouse organoids, the WNT-suppressive response of G007-LK was abrogated in APCQ884 cells (Figure 4b). Similarly, patient-derived human CRC organoids (30) carrying late truncating mutations in APC showed decreased WNT target gene expression (LGR5 and AXIN2) after 3 days of G007-LK treatment (Figure 4c, Supplementary Figure 11b-c). Together these data show that TNKS inhibitors can suppresses WNT targets in human colorectal cancer cells carrying MCR APC truncations.
Figure 4. Human cells and ApcQ1405X mouse adenomas with MCR truncations respond to Tnks1/2 inhibition.
a. Western blot of DLD1 cells expressing Cas9, transduced with sgRNAs targeting either FANCFS1 (negative control) or APC (near codon Q884). Targeting with the APC sgRNA generated a shorter truncated protein. b. Expression of WNT target genes LGR5 and AXIN2 in FANCFS1 or APCQ884 transduced DLD1s following 72 hrs. treatment with G007-LK, as indicated. c. Expression of WNT target genes LGR5 and AXIN2 in patient-derived organoids following 72 hrs. treatment with G007-LK, as indicated. Raw values of individual replicate experiments are presented in Supplementary Figure 11. d. Immunoprecipitation of Axin1 in whole cell lysates from ApcQ1405X and ApcQ884X cells treated with G007-LK or DMSO for 24 hours. e. Kaplan-Meier plot showing survival of ApcQ1405X mice (red line) as compared to ApcMin mice (blue line). f. Bar graph showing hematocrit from ApcQ1405X/+ and Apc+/+ mice at 10–11 weeks. g. Immunohistochemical, immunofluorescent, and in situ hybridization (Lgr5) stains from the proximal small intestine of 10-week-old ApcQ1405X mice treated with vehicle or G007-LK (30mg/kg, QD) for one week. ApcQ1405X mice treated with G007-LK show a differentiation profile marked by decreased BrdU incorporation, decreased Lgr5 abundance, and increased alkaline phosphatase staining. h. Quantitation of the fraction of BrdU positive cells (left) and Alkaline phosphatase positive area (right) in individual tumors (n=3–6 tumors/mouse, n=3–6 mice/treatment). Bars represent mean and 95% CI. i. Model for data presented, indicating that late truncating mutations in Apc, but not early truncating mutations can engage the destruction complex in response to Tnks1/2 inhibition. Subsequent DC assembly re-establishes normal control of WNT signaling. DC assembly re-establishes normal control of WNT signaling.
Our cellular and transcriptome analyses demonstrate that ApcQ1405X organoids show a profound WNT pathway downregulation in response to TNKS1/2 inhibition. Indeed, ApcQ1405X organoids showed a marked decrease in non-phosphorylated (active) β-catenin following treatment with G007-LK (Supplementary Figure 12a). To directly test if this was due to increased activity of the DC, we measured physical association of key DC members by immunoprecipitation of Axin1. To produce the cell numbers required for effective immunoprecipitation, we generated 2D cell lines carrying KrasG12D and p53-null mutations and either ApcQ884X or ApcQ1405X truncations.
Baseline Axin1 levels were higher in ApcQ884X cells, but as expected, G007-LK treatment drove Axin1 stabilization in both contexts (Supplementary Figure 12b-c). The shorter ApcQ884X mutant protein was more abundant than ApcQ1405X in intestinal cells; however, Axin1 selectively associated with the longer truncated form (Figure 4d). G007-LK treatment further strengthened this binding and enabled increased association with β-catenin. Consistent with this, G007-LK-treated ApcQ1405X cells showed increased phosphorylation of β-catenin at S33/S37/T41 (Figure 4d); We were unable to detect phosphorylated β-catenin in Axin1 IPs, likely due to the rapid release of phospho-β-catenin from this complex. In contrast to ApcQ1405X, ApcQ884X mutant protein showed no increased binding to Axin1 in the presence of G007-LK, did not enable further β-catenin association, and produced only a minor increase in S33 phosphorylation (Figure 4d).
Our organoid data described above indicate that the position of Apc truncations dictates response to TNKS1/2 inhibition. To directly test this hypothesis in vivo, we generated a new ApcQ1405X mouse allele by direct base editing in C57Bl/6 zygotes (see Methods for detail). Sanger sequencing of DNA from viable founder mice confirmed the expected C>T transition in 14/30 (47%) animals, with the remaining mice showing a mixture of non-C>T changes, indels, and wildtype sequence (Table S3). As expected, no ApcQ1405X/Q1405X founder mice were obtained, due to early embryonic lethality of homozygous Apc mutants (31), while ApcQ1405X heterozygous mice became moribund by 12–14 weeks of age (median survival: 97 days) (Figure 4e). As commonly observed in other Apc-mutant models, early onset anemia correlated with multi-focal adenoma growth throughout the small intestine (most likely following LOH, as occurs in ApcMin (18)) (Figure 4f-g).
To determine whether established ApcQ1405X adenomas were sensitive to TNKS1/2 inhibition, we treated tumor-bearing 10-week-old ApcQ1405X/+ mice with G007-LK (30mg/kg, QD) or vehicle for 1 week. Histological sections of G007-LK-treated intestinal tumors showed decreased nuclear to cytoplasmic ratio, nuclear polarization, and a transition to differentiated villi-like structures (Figure 4g; H&E). Consistent with this, immunofluorescent staining showed reduced proliferation (Figure 4g-h; BrdU) and induction of differentiation markers (Figure 4g-h; Krt20 and ALPi). In contrast to treated ApcMin polyps, Lgr5 expression was dramatically reduced in ApcQ1405X lesions, confirming reduced WNT signaling within the adenoma (Figure 4g; Lgr5). Together, this data supports our findings in organoids that truncating mutations in the MCR of APC, render cells sensitive to WNT suppression via TNKS inhibition (Figure 4i).
Discussion
We previously reported that restoring Apc expression induces sustained tumor regression in CRC (4). Here, using genetically defined animal models, organoids and human cell lines, we show that TNKS inhibition can re-engage the same endogenous tumor suppressive mechanism, but this molecular switch is highly dependent on the specific Apc disruption. Tumor cells containing ‘early’ truncating mutations that eliminate all identified Axin1/2 and β-catenin motifs (e.g. ApcMin, ApcQ884X) do not respond to TNKS inhibition, yet those with an MCR mutation can regulate β-catenin and suppress oncogenic signaling in response to Axin1/2 stabilization. To our knowledge, this is the first in vivo demonstration that subtle changes in CRC initiating genetic events can lead to profound changes in response to targeted therapy and has implications for strategies that aim to modulate WNT activity through the DC.
Consistent with previously published data (21) we observed in three independent datasets that the majority of CRCs carry at least one nonsense mutation within the MCR (~1250–1580). This mutational pattern is hypothesized to be a selection for mutations that enable oncogenic transformation yet retain partial β-catenin regulation, providing a ‘just right’ level of WNT activation (32–34). Interestingly, APC structure-function studies in CRC cell lines and model organisms suggest that truncations at Q1405 that remove the ‘Catenin inhibitory domain’ (CID) immediately after the second 20AAR, disrupt β-catenin turnover, and thus drive high levels of WNT activation (23,24). Consistent with this, our ApcQ1405X organoids, carrying the same truncating allele, hyperactivate WNT signaling, and heterozygous ApcQ1405X/+ mice develop intestinal tumors with 100% penetrance. However, in response to TNKS inhibition, ApcQ1405X can engage the DC to suppress hyperactive WNT signaling; this response would not have been predicted from structure-function models. It is important to note that while most human CRCs harbor both an early and late truncation, and our organoids were engineered to carry two ApcQ1405X alleles, we observed that one of the biological replicates (replicate A) carried an unexpected heterozygous deletion in one allele of Apc (Supplementary Figure 13a-c). While these cells express only one truncated allele and show moderately higher baseline WNT activity, they have an identical response to TNKS inhibition (Supplementary Figure 13d), implying that the presence of one MCR truncated allele is sufficient to engage the DC.
Given the importance of Apc interaction with β-catenin and the DC, it is somewhat counter-intuitive that shApc organoids and tumors show such robust WNT-suppression and differentiation responses to G007-LK, despite extremely potent knockdown (Supplementary Figure 14). We propose that the reason underlying the response of shApc cells is the presence of a minimal residual amount of full-length Apc protein that can be detected in complex with Axin1, but not in whole cell lysates (Supplementary Figure 14). While the amount of Apc is dramatically reduced, the residual protein contains all β-catenin and Axin binding domains that allows it to effectively engage the DC, and following Axin stabilization, drive WNT suppression. The observation that only minimal full-length Apc protein is required for tumor suppression is consistent with our previously published work, which showed that intestinal adenomas have a phenotypic response to genetic Apc restoration even before Apc protein is detectable by western blot (4).
In all, these findings highlight that distinct mutations within the same gene can appear phenotypically similar (hyperactive WNT), but have profoundly different biochemical responses to therapies targeting their mechanism of action. This reinforces the need to create accurate and representative genetic models for understanding the molecular events of disease and therapy response.
Our results contrast with recent in vitro work, which concluded that cells with early APC truncations are uniquely sensitive to TNKS inhibition (19). In our experiments, WNT suppression correlated precisely with cell cycle arrest and differentiation, consistent with the role of WNT as a driver of self-renewal and tumorigenesis. In contrast, while almost all cell lines assessed by Tanaka et al. showed WNT suppression following TNKS inhibition, most were not dependent on WNT for survival, and thus classified as resistant (19). We believe it is likely that either the accumulation of specific oncogenic events, or prolonged in vitro culture relieved their WNT dependence, confounding the correlation of TNKSi response and APC/WNT alterations. Indeed, one recent study in lung adenocarcinoma demonstrated that WNT/RSPO-dependent 3D organoid cultures become WNT-independent when cultured in 2D (35). Further, Lord and colleagues recently proposed that activating Kras mutations may correlate with resistance to TNKS inhibitors (36). While the latter study did not ascribe a specific mechanism to this possible interaction, it is consistent with multiple observations that concurrent inhibition of classic oncogenic signaling pathways such as MAPK or AKT, enhances the response to TNKS inhibition (12,20,37–39).
While TNKS inhibition is unlikely to be used as a monotherapy for cancer treatment, it is perhaps worth (re)considering its utility in patients with Familial Adenomatous Polyposis (FAP), that most commonly carry heterozygous germline mutations in the MCR (40), and ultimately have few options other than surgical removal of the colon. Our data imply that FAP-associated adenomas may respond to TNKS inhibitors, though a number of issues of drug dose, bioavailability, and safety would need to be addressed before they could be considered a viable option.
Understanding how specific oncogenic insults drive cell transformation and impact therapy response is a key goal of precision oncology medicine. WNT pathway hyperactivation is a clear cancer driver in a number of malignancies, including CRC (1,4,41,42), and has been suggested as an immune-modulator that may impact response to checkpoint inhibitors (43–45). While WNT-targeted agents have yet to make significant progress in clinical medicine, approaches to block WNT hyperactivation are of keen interest for cancer treatment. Defining the genetic context/s in which they will be effective, as described here, is critical for the future development and application of WNT-targeted therapeutics.
Methods
Cloning
Individual Tnks and Tnks2 shRNAs were cloned into MLPE vector at XhoI and EcoRI sites as previously described (46). For tandem shRNA cloning, the destination vector containing shRNA #1 was digested with EcoRI for 4 hours at 37˚C and incubated with CIP for 30 min at 37˚C. shRNA #2 was PCR amplified using miRE-TX_For and miRE_EcoRI_Rev primers (Table S4). Positive clones were identified by restriction digest and validated by Sanger sequencing with mir30_seq primer (Table S4). Tandem shRNA cassettes were shuttled into the col1A1 targeting vector (cTGME) following PCR amplification with miRE_XhoI_For and miRE_EcoRI_Rev primers, and XhoI/EcoRI digest. For the shRNA sensor assay, shRNA target sequences against Tnks and Tnks2 were arrayed, produced as a synthetic gBlock (IDT), and cloned into the 3’UTR of a dTomato sensor vector (XhoI/ EcoRI). For Tnks or Tnks2 specific sensors, gBlocks with only Tnks or Tnks2 target sequences were designed and cloned as described above. sgRNA cloning into the vector LRT2B cloning was performed as previously described (22,47). The shApc.2235E retroviral vector used to transduce organoids was generated by Gibson cloning into the pMSCV-rtTA3-miRE vector.
Mutation detection by T7 assay
Cas9-induced mutations were detected using the T7 endonuclease I (NEB). Briefly, the target region surrounding the expected mutation site was PCR-amplified using Herculase II (600675, Agilent Technologies). PCR products were column purified (Qiagen) and subjected to a series of melt-anneal temperature cycles with annealing temperatures gradually lowered in each successive cycle. T7 endonuclease I was then added to selectively digest heteroduplex DNA. Digest products were visualized on a 2.5% agarose gel.
Cells
HEK293T (ATCC CRL-3216) and DLD1 cells (ATCC CCL-221) were purchased from ATCC and maintained in Dulbecco’s Modified Eagle’s Medium (Corning) supplemented with 10% (v/v) fetal bovine serum (FBS), at 37° with 5% CO2. NIH/3T3 (ATCC CRL-1658) were maintained in Dulbecco’s Modified Eagle’s Medium (Corning) supplemented with 10% (v/v) bovine calf serum. Cells were regularly tested for mycoplasma using the MycoAlert mycoplasma detection kit (Lonza, #LT07–418) and discarded if found positive.
Virus Production
HEK-293T cells cultured in DMEM + 10%FBS were co-transfected with retroviral vector, CMV-Gag-Pol, CMV-Eco envelope and pcSUPER-shPasha (to prevent miRNA processing of the nascent retroviral RNA transcript). Cells were washed 12–24hrs after transfection and viral supernatants were collected from 36–60hrs post-transfection. Detailed protocol available at: www.dowlab.org/Protocols. For LRT2B-sgRNA virus, no pcSUPER-shPasha was included.
shRNA Sensor Assay
The sensor assay was adapted (48) using NIH-3T3s were cultured in DMEM + 10% Calf Serum. Following transduction, the top 20% brightest dTomato-positive cells were collected by FACS. Sorted cells were subsequently transduced with shRNA virus at 1:10 to achieve approximately single MOI. Five days post infection, dTomato expression was measured by flow cytometry, and knockdown calculated as relative decrease from parental population after subtraction of background signal.
Human Colorectal Cancer Cell Lines and Patient-derived organoid
DLD1 cells were transduced with lentivirus expressing Cas9 (Addgene #110837), in the presence of polybrene (8μg/μl). Two days after transduction, cells were selected in Puromycin (2μg/ml). 1 × 104 DLD-Cas9 cells were plated in a 6 well plate, and transduced with the LRT2B vector (Addgene #110854) containing either hAPC.884 or FANCF.S1 control sgRNAs. Two days after transduction, cells were selected in Blasticidin S (5μg/ml) .3 × 104 LRT2B-transduced cells were plated for treatment with either DMSO or G007-LK 1μM for 3 days for RNA and protein collection. Deidentified, patient-derived human colorectal cancer organoids (WCM392 and WCM616; previously described (30)) were obtained from the Institute for Precision Medicine at Weill Cornell Medicine. Organoids were cultured in Basal Media + B27, and treated with either DMSO or G007-LK 1μM for 3 days.
ES cell targeting
Embryonic Stem (ES) cells were maintained on irradiated feeders, C57Bl/6 cells were cultured 2i media and KH2 cells were cultured in M15 media, both containing LIF as previously outlined (46). Cells were transfected with targeting vector and CAGs-FlpE vector using a Lonza X-unit nucleofector with P3 buffer kit (Lonza #V4XP-3032). Two days following transfections, cells were treated with media containing 150ug/ml hygromycin and individual surviving clones were picked after 9–10 days of selection. Two days after clones were picked hygromycin was removed from the media and cells were cultures in standard M15 thereafter. To confirm single copy integration at the col1a1 locus we first validated expected integration by multiplex col1a1 PCR 3, and second, confirmed the presence of a single GFP cassette using the Taqman copy number assay, according to the manufacturer’s instructions (Invitrogen).
Isolation and culture of intestinal organoids
Isolation, maintenance and staining of mouse intestinal organoids has been described previously (49,50). Briefly, for isolation, 15 cm of the proximal small intestine was removed and flushed with cold PBS. The intestine was then cut into 5 mm pieces, vigorously resuspended in 5mM EDTA-PBS using a 10ml pipette, and placed at 4ºC on a benchtop roller for 10 minutes. This was then repeated for a second time for 30 minutes. After repeated mechanical disruption by pipette, released crypts were mixed with 10ml DMEM Basal Media (Advanced DMEM F/12 containing Pen/Strep, Glutamine, 1mM N-Acetylcysteine (Sigma Aldrich A9165-SG)) containing 10 U/ml DNAse I (Roche, 04716728001), and filtered sequentially through 100μm and 70μm filters. 1ml FBS (final 5%) was added to the filtrate and spun at 1200 RPM for 4 minutes. The purified crypts were resuspended in basal media and mixed 1:10 with Growth Factor Reduced Matrigel (BD, 354230). 40μl of the resuspension was plated per well in a 48 well plate and placed in a 37ºC incubator to polymerize for 10 minutes. 250μl of small intestinal organoid growth media (Basal Media containing 50 ng/mL EGF (Invitrogen PMG8043), 100ng/ml Noggin (Peprotech 250–38), and 500 ng/mL R-spondin (R&D Systems, 3474-RS-050, or from conditioned media) was then laid on top of the Matrigel. Where indicated, dox was added to experiments at 500 ng/ml.
For sub-culture and maintenance, media was changed on organoids every two days and they were passaged 1:4 every 5–7 days. To passage, the growth media was removed and the Matrigel was resuspended in cold PBS and transferred to a 15ml falcon tube. The organoids were mechanically disassociated using a p1000 or a p200 pipette and pipetting 50–100 times. 7 ml of cold PBS was added to the tube and pipetted 20 times to fully wash the cells. The cells were then centrifuged at 1000 RPM for 5 minutes and the supernatant was aspirated. They were then resuspended in GFR Matrigel and replated as above. For freezing, after spinning the cells were resuspended in Basal Media containing 10% FBS and 10% DMSO and stored in liquid nitrogen indefinitely.
Organoid transfection
Murine small intestinal organoids were cultured in transfection medium containing CHIR99021 (5μM) and Y-27632 (10μM) for 2 days prior to transfection. Single cells suspensions were produced by dissociating organoids with TrypLE express (Invitrogen #12604) for 5 min at 37°C. After trypsinization, cell clusters in 300μl transfection medium were combined with 100μl DMEM/F12-Lipofectamine2000 (Invitrogen #11668)-DNA mixture (97ul-2ul-1ug), and transferred into a 48-well culture plate. The plate was centrifuged at 600g at 32°C for 60 min, followed by another 6h incubation at 37°C. The cell clusters were spun down and plated in Matrigel. For selecting organoids with Apc mutants, exogenous R-spondin1 and Noggin were withdrawn 2 days after transfection.
Organoid Drug Treatment and Counts
Organoids were plated in 100 μl Matrigel (2× 50 μl droplets) in one 12 well and cultured in Advanced DMEM/F12 + EGF media with either DMSO or G007-LK 1uM. Three days after plating 8 brightfield images were taken in a vertical line, approximately the length of one droplet. Images were then compiled in one document, and viable organoids were counted for each condition. Organoids were then passaged either 1:2 or 1:1 (based on confluency) and then cultured again in DMSO or G007-LK. Day 7 counts were calculated from brightfield images and multiplied by the passaging factor.
EdU Flow Cytometry
Organoid EdU flow cytometry was performed using the Click-iT™ Plus EdU Alexa Fluor™ 647 Flow Cytometry Assay Kit (Thermo Fisher, # C10634). Organoids were first incubated with 10 μM EdU for 4 hours at 37˚C. One well of a 12 well plate was broken up by pipetting vigorously 50 times in 1mL PBS, then diluted in 5 mL of PBS. Cells were pelleted at 1100rpm x 4 min at 4˚C, then resuspended in 50 μL TrypLE and incubated at 37˚C for 5 mins. 5 mL of PBS was then added to inactivate the TrypLE, and cells were pelleted. Cells were resuspended in 250 μL of 1% BSA in PBS, transferred to a 1.7 mL tube, and then pelleted at 3000rpm x 4 min. Cells were then resuspended in 100 μL Click-iT™ fixative, and processed as instructed in the Click-iT™ Plus EdU protocol (starting with Step 4.3). Wash and reaction volumes were 250 μL.
Animal studies
Production of mice and all treatments described were approved by the Institutional Animal Care and Use Committee (IACUC) at Weill Cornell Medicine (NY), under protocol number 2014–0038. ES cell-derived mice were produced by blastocyst injection and animals were either maintained on a mixed C57B6/129 background for experimental breeding or backcrossed to C57Bl/6N mice. Progeny of both sexes were used for experiments and were genotyped for specific alleles (Lgr5-GFP-IRES-CreER, CAGs-rtTA, Col1A1, TG-shApc.2235E, TG-Ren.713, TG-shTnks1/2–3341-1328, TG-shTnks1/2–1385-3004, ApcQ1405X, and ApcMin) using primers described in Supplementary Table 3 and protocols available at www.dowlab.org/Protocols. Production of mice and all treatments described were approved by the Institutional Animal Care and Use Committee (IACUC) at Weill Cornell Medicine (NY), under protocol number 2014–0038. For induction of the CreER transgene, animals were administered 4-hydroxytamoxifen (4-OHT; 25mg/kg in 90% v/v corn oil, 10% v/v Ethanol) via i.p. injection. Where required, doxycycline was administered via food pellets (200mg/kg) (Harlan Teklad) from 6–8 weeks of age. For G007-LK treatment studies, G007-LK (30mg/kg, MedChemExpress) was mixed with 20% CremaphorEL and 70% PBS, then delivered by daily i.p. injection. Mice were sacrificed after 7 or 14 days. Animal studies were not blinded during treatment, however, quantitation of tumor burden involved measurements by two parties, one blinded to the treatment groups.
Immunofluorescence and in situ hybridization (ISH)
Tissue, fixed in freshly prepared 4% paraformaldehyde for 24 hours, was embedded in paraffin and sectioned by IDEXX RADIL (Columbia, MO). Sections were rehydrated and unmasked (antigen retrieval) by heat treatment for 5 mins in a pressure cooker in 10mM Tris / 1mM EDTA buffer (pH 9) containing 0.05% Tween 20. Sections were blocked in TBS / 0.1% Triton X-100 containing 1% BSA. Organoids were stained as previously described (50), with the following exception: Prior to beginning the EdU/Keratin20 staining, organoids were washed once with 300 μl PBS, then incubated with 300 μl Cell Recovery Solution for 20 min on ice. Primary antibodies used were: rabbit anti-KRT20 (1:200, Cell Signaling Technologies, #13063), rat anti-BrdU (1:200, Abcam #ab6326). Secondary antibodies were applied in TBS for 1 hour at room temp in the dark, washed twice with TBS, counterstained for 5 mins with DAPI and mounted in ProLong Gold (Life Technologies, #P36930). Secondary antibodies used were: anti-rabbit 488 (1:500, Abcam, #ab150073) and anti-rat 594 (1:500, Abcam, #ab150156). Intestinal alkaline phosphatase staining was performed using the BCIP/NBT Alkaline Phosphatase (AP) Substrate Kit (Vector Laboratories, # SK-5400) [Note: 1 drop Lemavisole Solution/5mL was added prior to the reaction to inhibit endogenous alkaline phosphatase activity (Vector Labs, # SP-5000)] and counter-stained with Nuclear Fast Red (Sigma-Aldrich, #60700). For Lgr5 ISH, freshly cut 5 micron paraffin sections were stained using RNAscope 2.5 LS Red kit (ACD, cat#322150) and Bond Polymer Refine Red Detection kit (Leica, cat#DS9390) on Leica Bond RX instrument following routine manufacturer protocol ACD 2.5 Red. RNAscope 2.5 LS probes for Ms-LGR5 (ACD, cat# 312178). DapB-negative control (ACD, cat#312038) were used with hybridization at 42C for 2 hours. The sections were pre-treated with Leica Bond ER2 Buffer for 20 min at 95C and Protease III (ACD, cat#322102) for 20 min at 40C. After staining the sections were counterstained with Hematoxylin and 10ug/ml DAPI for 10 min and mounted with Mowiol mounting media.
Imaging
Images of fluorescent and IHC stained sections were acquired on a Zeiss Axioscope Imager (chromogenic stains), Nikon Eclipse T1 microscope (IF stains) or Zeiss LSM 880 Laser Scanning Confocal Microscope (organoid stains). Raw .tif files were processed using FIJI (Image J) and/or Photoshop CS (Adobe Systems Inc., San Jose, CA) to create stacks, adjust levels and/or apply false coloring.
Tumor Quantification
Tumor area in H&E stained slides were quantified using FIJI (Image J). Scanned slides were converted to 8-bit images, one intestinal roll was selected (four rolls per mouse), and an Over/Under threshold was set to capture only cells. The total intestinal area was measured using the ‘Measure’ feature, and then tumors were then identified, manually traced, and measured. All of the tumor areas were summed, then the ratio of total tumor area: total intestinal area was calculated to determine the Tumor Area for each mouse. Immunofluorescent images of BrdU/DAPI positive tumors were analyzed using FIJI. Images from each channel were exported as separate .tif files, then converted to 16-bit images. The tumor area was traced manually, and the nontumor regions of the image were cleared. A threshold was set to capture only BrdU/DAPI positive cells, then these cells were quantified using the ‘Analyze Particles’ feature. The ratio of BrdU/DAPI positive cells was then calculated to determine the Fraction of BrdU positive cells within a tumor. IHC images of Alkaline Phosphatase positive tumors were also analyzed using FIJI. Images were converted to 16 bit, and the entire tumor area was traced manually and nontumor regions were cleared from the image. A threshold was then set for the stained areas of the tumor, and the area was quantified using the ‘Measure’ feature. The total area of the tumor was also measured, and then ratio of Alkaline Phosphatase area: total tumor area was calculated to determine the Fraction of Alkaline Phosphatase Staining.
Protein analysis
Small intestine organoids were grown in 300μl of Matrigel in one well of a 6-well dish for 3 days post-passage. Organoids were then recovered from the Matrigel using Cell Recovery Solution(ref). Organoid pellets were lysed in 30 μl RIPA buffer. Antibodies used for Western blot were: anti-Apc (Millipore, #5535), anti-Axin1 (CST, #2087), anti-non-phosphorylated β-catenin (CST,), anti-phospho-β-catenin S33/S37/T41 (CST, #9561), total β-catenin (CST, #8480), anti-GSK3 (CST, #9832), anti-actin-HRP (Abcam, #ab49900), anti-Tnks1/2 (Santa Cruz, # sc-365897), anti-GFP (Abcam, #ab13970), Anti-α-Tubulin (Millipore Sigma, # CP06).
Immunoprecipitation
To generate 2D cultures, organoids were broken up by pipetting vigorously 50 times, then diluted in 5 mL of Basal Media. Cells were pelleted at 1100rpm x 4 min at 4˚C, then resuspended in 50 μl TrypLE and incubated at 37˚C for 5 mins. 5mL of PBS was then added to inactivate the TrypLE, and cells were pelleted. Organoids were resuspended in 2mL Basal Media and plated into one well of a 6 well plate, coated with Rat Tail Collagen I (Thermo Fisher, #A10483–01) at 3μg/ml in PBS for 30 min at 37˚C. When confluent (approximately 5 days), 2D cell lines were passaged with 500 μl Trypsin, neutralized in 1mL DMEM + 10% FBS, and pelleted. Cells were then resuspended in 2 mL Basal Media and plated on Collagen-coated plates as required. For Immunoprecipitation, cells were then expanded into 2 × 15 cm plates, and when near-confluent, cultured in DMSO or G007-LK (1uM) for 24 hours. Cells were washed once with PBS, then scraped into 1mL IP lysis buffer (10mM Tris 7.4, 150mM NaCl, 0.5mM EDTA, 0.5% NP40 + PPI/PI) and centrifuged at 13000rpm x 10 min at 4˚C. 200 μl of lysate was used for the immunoprecipitation using 25ul Protein-A Dynabeads (Thermo Fisher, #10001D) and incubated with Axin1 (1ug, CST, #2087) or IgG (1ug, CST, #2729). Beads were washed (200μl) and eluted (20μl) in IP lysis buffer. 5μl 5X SLB was added to the elution, and samples were denatured at 95˚C for 5 min.
RNA isolation, cDNA synthesis and qPCR
RNA was extracted using TRIzol (Thermo Fisher, #15596018) according to the manufacturer’s instructions and contaminating DNA was removed by DNase treatment for 10 mins and column purification (Qiagen RNeasy #74106). cDNA was prepared from 1 μg total RNA using qScript reverse transcription kit (Quantabio, #95047). Quantitative PCR detection was performed using SYBR green reagents (VWR, #101414–288) and specific primers listed in Table S4.
RNA sequencing
Total RNA was isolated using Trizol, DNAse treated and purified using the RNeasy mini kit (Qiagen, Hilden, Germany). Following RNA isolation, total RNA integrity was checked using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA concentrations were measured using the NanoDrop system (Thermo Fisher Scientific, Inc., Waltham, MA). Preparation of RNA sample library and RNA-seq were performed by the Genomics Core Laboratory at Weill Cornell Medicine. Messenger RNA was prepared using TruSeq Stranded mRNA Sample Library Preparation kit (Illumina, San Diego, CA), according to the manufacturer’s instructions. The normalized cDNA libraries were pooled and sequenced on Illumina NextSeq500 sequencer with single-end 75 cycles.
RNAseq analysis
The quality of raw FASTQ files were checked with FastQC and mapped to mouse reference GRCm38 using STAR two-pass alignment (v2.4.1d; default parameters) (51), and transcript abundance estimates were performed using Kallisto (52), aligned to the same (GRCm38) reference genome. Kallisto transcript count data for each sample was concatenated, and transcript per million (TPM) data was reported for each gene after mapping gene symbols to ensemble IDs using the following R packages (“tximport”, tximportData”, “ensembldb”, and “EnsDb.Mmusculus.v79”). Differential gene expression was estimated using DESeq2 (53). For data visualization and gene ranking, log fold changes were adjusted using the lfcShrink command in DESeq2, to minimize the effect size of poorly expressed genes. GSEA analysis (v3.0) was performed on pre-ranked gene sets from differential expression between DMSO and G007-LK-treated groups. We used R (v3.5.1) and R Studio (v1.1.383) to create all visualizations, perform hierarchical clustering and principal component analysis. Volcano plots, heatmaps and other visualizations were produced using the software packages:
Enhanced Volcano (https://bioconductor.org/packages/devel/bioc/html/EnhancedVolcano.html)
Pheatmap (https://cran.r-project.org/web/packages/pheatmap/index.html)
ggplot2 (https://cran.r-project.org/web/packages/ggplot2/index.html)
ggsashimi (https://hub.docker.com/r/guigolab/ggsashimi) (54)
Zygote Injections
mRNA was synthesized with mMESSAGE mMACHINE™ T7 ULTRA Transcription Kit (Thermo Fisher, #AM1345). 10 μg of DNA from the CMV-FNLS(RA) vector (Addgene, Plasmid #112671) was linearized with SalI, then DNA was ethanol precipitated, and used for the in vitro transcription reaction. For transgenesis, FNLS mRNA (100ng or 20ng) and 2’-O-methyl 3’phosphorothioate stabilized sgRNA (GTTCAGAGTGAGCCATGTAG; 100ng/ul; Synthego Corp, CA) were co-delivered by microinjection. Microinjections were performed by the Memorial Sloan Kettering Cancer Center (MSKCC) mouse transgenic core facility, using C57Bl/6J fertilized zygotes. Viable pups were genotyped by PCR and direct Sanger sequencing. Animals were maintained as heterozygotes by breeding to C57Bl/6N mice.
Statistical analyses
Statistical tests for all data presented here (other than RNAseq) were performed in GraphPad Prism. Detailed results of each test are provided in Table S5.
Data Availability
Raw sequence data is available at the NCBI Sequence Read Archive (SRA) under accession PRJNA524289
Supplementary Material
Statement of Significance.
This study reveals how subtle changes to the mutations in a critical colorectal tumor suppressor, APC, influence the cellular response to a targeted therapy. It underscores how investigating the specific genetic alterations that occur in human cancer can identify important biological mechanisms of drug response and resistance.
Acknowledgements
This work was supported by a project grant from the NIH/NCI (CA195787–01) and a Stand Up to Cancer Colorectal Cancer Dream Team Translational Research Grant (SU2C-AACR-DT22–17). Stand Up to Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. We thank Kevin Blighe for assistance with Enhanced Volcano R package. We thank Shiaoching Gong and the MSKCC Mouse Transgenic Core Facility who performed zygote microinjections, supported in part by a U54 grant from the NIH/NCI (U54OD020355). We thank Katia Manova and Mesruh Turkekul from the Molecular Cytology Core Facility who performed ISH experiments, supported in part by a core grant from the NIH (P30 CA0088748). EMS was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM07739 to the Weill Cornell / Rockefeller / Sloan-Kettering Tri-Institutional MD-PhD Program, and an F31 Award from the NCI/NIH under grant number 1 F31 CA224800–01. MPZ is supported in part by National Cancer Institute (NCI) Grant NIH T32 CA203702. LED was supported by a K22 Career Development Award from the NCI/NIH (CA 181280–01). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
COI statement
LED is a scientific advisory board member and stockholder in Mirimus Inc., who have licensed shRNA technology that is used in this study. All other authors declare no competing interested.
References
- 1.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487(7407):330–7 doi 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23(6):703–13 doi 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stamos JL, Weis WI. The beta-catenin destruction complex. Cold Spring Harb Perspect Biol 2013;5(1):a007898 doi 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dow LE, O’Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, et al. Apc Restoration Promotes Cellular Differentiation and Reestablishes Crypt Homeostasis in Colorectal Cancer. Cell 2015;161(7):1539–52 doi 10.1016/j.cell.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Rourke KP, Loizou E, Livshits G, Schatoff EM, Baslan T, Manchado E, et al. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat Biotechnol 2017;35(6):577–82 doi 10.1038/nbt.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 2009;461(7264):614–20 doi 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 7.Riffell JL, Lord CJ, Ashworth A. Tankyrase-targeted therapeutics: expanding opportunities in the PARP family. Nature reviews 2012;11(12):923–36 doi 10.1038/nrd3868. [DOI] [PubMed] [Google Scholar]
- 8.Lehtio L, Chi NW, Krauss S. Tankyrases as drug targets. Febs J 2013;280(15):3576–93 doi 10.1111/febs.12320. [DOI] [PubMed] [Google Scholar]
- 9.Haikarainen T, Krauss S, Lehtio L. Tankyrases: structure, function and therapeutic implications in cancer. Curr Pharm Des 2014;20(41):6472–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol 2009;5(2):100–7 doi 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao R, Christova T, Song S, Angers S, Yan X, Attisano L. Inhibition of tankyrases induces Axin stabilization and blocks Wnt signalling in breast cancer cells. PLoS ONE 2012;7(11):e48670 doi 10.1371/journal.pone.0048670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoumacher M, Hurov KE, Lehar J, Yan-Neale Y, Mishina Y, Sonkin D, et al. Inhibiting Tankyrases sensitizes KRAS-mutant cancer cells to MEK inhibitors via FGFR2 feedback signaling. Cancer Res 2014;74(12):3294–305 doi 10.1158/0008-5472.CAN-14-0138-T. [DOI] [PubMed] [Google Scholar]
- 13.Norum JH, Skarpen E, Brech A, Kuiper R, Waaler J, Krauss S, et al. The tankyrase inhibitor G007-LK inhibits small intestine LGR5(+) stem cell proliferation without altering tissue morphology. Biol Res 2018;51(1):3 doi 10.1186/s40659-017-0151-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye P, Chiang YJ, Qi Z, Li Y, Wang S, Liu Y, et al. Tankyrases maintain homeostasis of intestinal epithelium by preventing cell death. PLoS Genet 2018;14(9):e1007697 doi 10.1371/journal.pgen.1007697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong Y, Katavolos P, Nguyen T, Lau T, Boggs J, Sambrone A, et al. Tankyrase Inhibition Causes Reversible Intestinal Toxicity in Mice with a Therapeutic Index < 1. Toxicol Pathol 2016;44(2):267–78 doi 10.1177/0192623315621192. [DOI] [PubMed] [Google Scholar]
- 16.Lau T, Chan E, Callow M, Waaler J, Boggs J, Blake RA, et al. A novel tankyrase small-molecule inhibitor suppresses APC mutation-driven colorectal tumor growth. Cancer Res 2013;73(10):3132–44 doi 10.1158/0008-5472.CAN-12-4562. [DOI] [PubMed] [Google Scholar]
- 17.Waaler J, Machon O, Tumova L, Dinh H, Korinek V, Wilson SR, et al. A novel tankyrase inhibitor decreases canonical Wnt signaling in colon carcinoma cells and reduces tumor growth in conditional APC mutant mice. Cancer Res 2012;72(11):2822–32 doi 10.1158/0008-5472.CAN-11-3336. [DOI] [PubMed] [Google Scholar]
- 18.Levy DB, Smith KJ, Beazer-Barclay Y, Hamilton SR, Vogelstein B, Kinzler KW. Inactivation of both APC alleles in human and mouse tumors. Cancer Res 1994;54(22):5953–8. [PubMed] [Google Scholar]
- 19.Tanaka N, Mashima T, Mizutani A, Sato A, Aoyama A, Gong B, et al. APC Mutations as a Potential Biomarker for Sensitivity to Tankyrase Inhibitors in Colorectal Cancer. Mol Cancer Ther 2017;16(4):752–62 doi 10.1158/1535-7163.MCT-16-0578. [DOI] [PubMed] [Google Scholar]
- 20.Arques O, Chicote I, Puig I, Tenbaum SP, Argiles G, Dienstmann R, et al. Tankyrase Inhibition Blocks Wnt/beta-Catenin Pathway and Reverts Resistance to PI3K and AKT Inhibitors in the Treatment of Colorectal Cancer. Clin Cancer Res 2016;22(3):644–56 doi 10.1158/1078-0432.CCR-14-3081. [DOI] [PubMed] [Google Scholar]
- 21.Christie M, Jorissen RN, Mouradov D, Sakthianandeswaren A, Li S, Day F, et al. Different APC genotypes in proximal and distal sporadic colorectal cancers suggest distinct WNT/beta-catenin signalling thresholds for tumourigenesis. Oncogene 2013;32(39):4675–82 doi 10.1038/onc.2012.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zafra MP, Schatoff EM, Katti A, Foronda M, Breinig M, Schweitzer AY, et al. Optimized base editors enable efficient editing in cells, organoids and mice. Nat Biotechnol 2018;36(9):888–93 doi 10.1038/nbt.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohler EM, Chandra SH, Behrens J, Schneikert J. Beta-catenin degradation mediated by the CID domain of APC provides a model for the selection of APC mutations in colorectal, desmoid and duodenal tumours. Hum Mol Genet 2009;18(2):213–26 doi 10.1093/hmg/ddn338. [DOI] [PubMed] [Google Scholar]
- 24.Roberts DM, Pronobis MI, Poulton JS, Waldmann JD, Stephenson EM, Hanna S, et al. Deconstructing the sscatenin destruction complex: mechanistic roles for the tumor suppressor APC in regulating Wnt signaling. Molecular biology of the cell 2011. doi 10.1091/mbc.E10-11-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohler EM, Derungs A, Daum G, Behrens J, Schneikert J. Functional definition of the mutation cluster region of adenomatous polyposis coli in colorectal tumours. Hum Mol Genet 2008;17(13):1978–87 doi 10.1093/hmg/ddn095. [DOI] [PubMed] [Google Scholar]
- 26.Zeineldin M, Neufeld KL. Understanding phenotypic variation in rodent models with germline Apc mutations. Cancer Res 2013;73(8):2389–99 doi 10.1158/0008-5472.CAN-12-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuraguchi M, Wang X-P, Bronson RT, Rothenberg R, Ohene-Baah NY, Lund JJ, et al. Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS genetics 2006;2(9):e146 doi 10.1371/journal.pgen.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim TH, Li F, Ferreiro-Neira I, Ho LL, Luyten A, Nalapareddy K, et al. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature 2014;506(7489):511–5 doi 10.1038/nature12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moor AE, Harnik Y, Ben-Moshe S, Massasa EE, Rozenberg M, Eilam R, et al. Spatial Reconstruction of Single Enterocytes Uncovers Broad Zonation along the Intestinal Villus Axis. Cell 2018;175(4):1156–67 e15 doi 10.1016/j.cell.2018.08.063. [DOI] [PubMed] [Google Scholar]
- 30.Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov 2017;7(5):462–77 doi 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moser AR, Shoemaker AR, Connelly CS, Clipson L, Gould KA, Luongo C, et al. Homozygosity for the Min allele of Apc results in disruption of mouse development prior to gastrulation. Dev Dyn 1995;203(4):422–33. [DOI] [PubMed] [Google Scholar]
- 32.Albuquerque C, Breukel C, van der Luijt R, Fidalgo P, Lage P, Slors FJ, et al. The ‘just-right’ signaling model: APC somatic mutations are selected based on a specific level of activation of the beta-catenin signaling cascade. Hum Mol Genet 2002;11(13):1549–60. [DOI] [PubMed] [Google Scholar]
- 33.Lamlum H, Ilyas M, Rowan A, Clark S, Johnson V, Bell J, et al. The type of somatic mutation at APC in familial adenomatous polyposis is determined by the site of the germline mutation: a new facet to Knudson’s ‘two-hit’ hypothesis. Nat Med 1999;5(9):1071–5 doi 10.1038/12511. [DOI] [PubMed] [Google Scholar]
- 34.Crabtree M, Sieber OM, Lipton L, Hodgson SV, Lamlum H, Thomas HJ, et al. Refining the relation between ‘first hits’ and ‘second hits’ at the APC locus: the ‘loose fit’ model and evidence for differences in somatic mutation spectra among patients. Oncogene 2003;22(27):4257–65 doi 10.1038/sj.onc.1206471. [DOI] [PubMed] [Google Scholar]
- 35.Guimaraes PPG, Tan M, Tammela T, Wu K, Chung A, Oberli M, et al. Potent in vivo lung cancer Wnt signaling inhibition via cyclodextrin-LGK974 inclusion complexes. J Control Release 2018;290:75–87 doi 10.1016/j.jconrel.2018.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menon M, Elliott R, Bowers L, Balan N, Rafiq R, Costa-Cabral S, et al. A novel tankyrase inhibitor, MSC2504877, enhances the effects of clinical CDK4/6 inhibitors. Sci Rep 2019;9(1):201 doi 10.1038/s41598-018-36447-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Lu B, Castillo J, Zhang Y, Yang Z, McAllister G, et al. Tankyrase Inhibitor Sensitizes Lung Cancer Cells to Endothelial Growth Factor Receptor (EGFR) Inhibition via Stabilizing Angiomotins and Inhibiting YAP Signaling. J Biol Chem 2016;291(29):15256–66 doi 10.1074/jbc.M116.722967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X, Luo F, Li J, Zhong X, Liu K. Tankyrase 1 inhibitior XAV939 increases chemosensitivity in colon cancer cell lines via inhibition of the Wnt signaling pathway. Int J Oncol 2016;48(4):1333–40 doi 10.3892/ijo.2016.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solberg NT, Waaler J, Lund K, Mygland L, Olsen PA, Krauss S. TANKYRASE Inhibition Enhances the Antiproliferative Effect of PI3K and EGFR Inhibition, Mutually Affecting beta-CATENIN and AKT Signaling in Colorectal Cancer. Mol Cancer Res 2018;16(3):543–53 doi 10.1158/1541-7786.MCR-17-0362. [DOI] [PubMed] [Google Scholar]
- 40.Nieuwenhuis MH, Vasen HF. Correlations between mutation site in APC and phenotype of familial adenomatous polyposis (FAP): a review of the literature. Crit Rev Oncol Hematol 2007;61(2):153–61 doi 10.1016/j.critrevonc.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 41.Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017;169(6):985–99 doi 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 42.Kahn M Can we safely target the WNT pathway? Nature reviews 2014;13(7):513–32 doi 10.1038/nrd4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luke JJ, Bao R, Sweis RF, Spranger S, Gajewski TF. WNT/beta-catenin Pathway Activation Correlates with Immune Exclusion across Human Cancers. Clin Cancer Res 2019;25(10):3074–83 doi 10.1158/1078-0432.CCR-18-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grasso CS, Giannakis M, Wells DK, Hamada T, Mu XJ, Quist M, et al. Genetic Mechanisms of Immune Evasion in Colorectal Cancer. Cancer Discov 2018;8(6):730–49 doi 10.1158/2159-8290.CD-17-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 2015;523(7559):231–5 doi 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 46.Dow LE, Premsrirut PK, Zuber J, Fellmann C, McJunkin K, Miething C, et al. A pipeline for the generation of shRNA transgenic mice. Nat Protoc 2012;7(2):374–93 doi 10.1038/nprot.2011.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schatoff EM, Paz Zafra M, Dow LE. Base editing the mammalian genome. Methods 2019. doi 10.1016/j.ymeth.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol Cell 2011;41(6):733–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Rourke KP, Ackerman S, Dow LE, Lowe SW. Isolation, Culture, and Maintenance of Mouse Intestinal Stem Cells. Bio Protoc 2016;6(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Rourke KP, Dow LE, Lowe SW. Immunofluorescent Staining of Mouse Intestinal Stem Cells. Bio Protoc 2016;6(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21 doi 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34(5):525–7 doi 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 53.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550 doi 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garrido-Martin D, Palumbo E, Guigo R, Breschi A. ggsashimi: Sashimi plot revised for browser- and annotation-independent splicing visualization. PLoS Comput Biol 2018;14(8):e1006360 doi 10.1371/journal.pcbi.1006360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data is available at the NCBI Sequence Read Archive (SRA) under accession PRJNA524289