Abstract
miRNAs can be found in serum and other body fluids and serve as biomarkers for disease. More importantly, secreted miRNAs, especially those in extracellular vesicles (EVs) such as exosomes, may mediate paracrine and endocrine communication between different tissues and thus modulate gene expression and the function of distal cells. When impaired, these processes can lead to tissue dysfunction, aging and disease. Adipose tissue is an especially important contributor to the pool of circulating exosomal miRNAs. As a result, alterations in adipose tissue mass or function, which occur in many metabolic conditions, can lead to changes in circulating miRNAs which then function systemically. Here we review the findings that led to these conclusions and discuss how this sets the stage for new lines of investigation in which extracellular miRNAs are recognized as important mediators of intercellular communication and potential candidates for therapy of disease.
Keywords: miRNA, exosome, extracellular vesicle, tissue communication, adipose tissue
eTOC Blurb
In this Review, Mori et al. discuss the emerging literature of extracellular miRNAs as mediators of tissue crosstalk. They outline the ability of these miRNAs to act as effective biomarkers, as well as a new class of hormones, and thus their potential as monitors of and therapeutic agents for disease.
Introduction
miRNAs are regulatory small non-coding RNAs of approximately 22-nt produced by virtually all cells in the body (Bartel, 2018; Ludwig et al., 2016). Many miRNAs are highly conserved across evolution, although their diversity and number correlate with organismal complexity (Berezikov, 2011; Deline et al., 2018; Tarver et al., 2015). Thus, the C. elegans repertoire contains 437 miRNAs, the mouse over 1,500 and humans produce between 2,000–3,000 miRNAs (data from the miRBase, Release 22). Many miRNAs are ubiquitously expressed, while others are tissue specific (Lagos-Quintana et al., 2002; Ludwig et al., 2016). This pattern of distribution is driven by both transcriptional and post-transcriptional regulation of miRNA precursors within the cell.
The synthesis of miRNAs is a multistep process [reviewed in (Bartel, 2018; Ha and Kim, 2014; Treiber et al., 2019)]. In the nucleus, primary miRNAs (primiRNA) are transcribed by RNA polymerase II then processed by the microprocessor complex, which contains the endoribonuclease DROSHA and its RNA binding partner DGCR8, or by components of the splicing machinery (Treiber et al., 2019). This results in pre-miRNAs of about 70-nt that are exported to the cytoplasm by XPO5 and Ran GTPase. Pre-miRNAs are further processed by type III endoribonuclease DICER in association with RNA binding proteins TRBP and PACT producing double-stranded miRNA duplexes. These duplexes are loaded into the RNA-induced silencing complex (RISC) where Argonaute-2 (AGO2) and chaperones like HSC70/HSP90 mediate the interaction of the guide strand of the miRNA with its target mRNA leading to inhibition of mRNA translation and/or increased mRNA degradation. In addition to this classical function, some miRNAs have been suggested to act through non-canonical pathways to produce the opposite effect, i.e., inducing transcription and upregulating protein expression (Vasudevan et al., 2007; Zhang et al., 2014). DICER-independent pathways of miRNA biogenesis have also been reported (Cheloufi et al., 2010), but their impact in miRNA synthesis is limited.
Similar to mRNAs, the miRNA expression profile can serve as a signature of cell identity. For instance, miR-122 is highly expressed in liver, representing up to 70% of the total miRNA expressed in this tissue (Bandiera et al., 2015; Jopling, 2012; Sekine et al., 2009). Similarly, muscle cells are enriched in miR-1, miR-133a, miR-133b, miR-206, miR-208a, miR-208b, miR-486 and miR-499, leading to the reference to these miRNAs as myomiRs (Horaka, 2016); miR-9 and miR-124 are expressed almost exclusively in brain, with the latter accounting for almost 50% of miRNA content in this tissue (Lagos-Quintana et al., 2002); and β-cells are unique for high abundance of miR-375 (Poy et al., 2004). On the other hand, some cells, like adipocytes and stem cells, express a wide range of miRNAs (Houbaviy et al., 2003; Kim et al., 2014; Kloting et al., 2009; Marson et al., 2008; Mori et al., 2012; Wang et al., 2008; Xie et al., 2009).
miRNAs Are More Mobile Than Expected
To help understand how miRNAs expressed in a given cell type contribute to development and homeostasis of that tissue, multiple cell type-specific DICER or DGCR8 knockout mice have been generated. These almost completely ablate production of mature miRNAs in the targeted cell. DICER knockout in the central nervous system, pancreas, or skeletal and cardiac muscles renders mice nonviable or with severe developmental defects (Chen et al., 2008; Kawase-Koga et al., 2009; Lynn et al., 2007; O’Rourke et al., 2007). This is not surprising given the important role of miRNAs in cell survival and differentiation. In contrast, liver-specific DICER knockout mice (LDicerKO) and adipocyte-specific DICER (ADicerKO) or DGCR8 (ADgcr8KO) knockout mice are indistinguishable from wildtype littermates until they reach adulthood, when they begin manifesting signs of metabolic dysfunction. These include hepatic steatosis and early-onset hepatocellular carcinoma in LDicerKO mice (Sekine et al., 2009) and partial lipodystrophy and insulin resistance in ADicerKO and ADgcr8KO mice (Kim et al., 2014; Mori et al., 2014; Reis et al., 2016).
While many aspects of these phenotypes are expected based on alterations in mRNA half-life and translation in the cell in which miRNA processing is disrupted, some phenotypes are secondary to changes in gene expression and function in other tissues suggesting cell non-autonomous effects of tissue-specific loss of miRNAs. For example, fat-specific DICER knockout (ADicerKO) mice exhibit changes in hepatic gene expression which are reversed when mice are transplanted with normal adipose tissue, suggesting that these changes are controlled by miRNAs secreted from the adipose tissue (Thomou et al., 2017). Further support for the possibility that adipose tissue-produced miRNAs contribute significantly to the pool of liver miRNAs is the fact that miRNAs that are highly enriched in fat are significantly decreased in liver of ADicerKO mice, and levels of these miRNAs in liver can be restored by adipose tissue transplantation without changes in hepatic levels of their corresponding pre-miRNAs (Thomou et al., 2017). This raises the hypothesis that the miRNA pool of each cell is the sum of endogenous miRNA production and uptake of exogenous miRNAs. This hypothesis may explain why following DICER knockout in hepatocytes, only 45 of the several hundred miRNAs detected in liver are downregulated (Sekine et al., 2009). While it is possible that a decrease in miRNAs in hepatocytes is masked by the presence of miRNAs in other cells in the liver, this seems unlikely, since hepatocytes are the predominant cell type in liver. It is also possible that changes in circulating miRNAs derived from one cell type impact on miRNA biogenesis in other cells. However, the relatively mild changes in miRNA expression in tissues following DICER knockout in vivo most likely reflects the mobile nature of miRNAs. Confirmation of this hypothesis will require development of robust techniques to track the origin, transport and fate of miRNAs in vivo.
Secretion and Transport of Extracellular miRNAs
The potential role of miRNAs in communication between cells and tissues is strongly supported by the fact that miRNAs can be exported and imported by cells through mechanisms involving vesicle trafficking and protein carriers. The notion was first described a little over a decade ago by Valadi et al., who identified a number of mRNAs and miRNAs within extracellular vesicles (EVs) secreted by different cell lines and found that these vesicles can be taken up by other cells, followed by release of their cargo into these target cells (Valadi et al., 2007). Concurrent studies demonstrated the presence of miRNAs in bodily fluids (Weber et al., 2010) and associated their levels to disease progression (see below). Since then, the mechanisms of extracellular miRNA transport have been explored in detail and are now known to occur via two main routes: a) active transport via EVs and b) transport as part of protein-miRNA complexes. In addition, there can be some leakage of miRNAs from broken or damaged cells. Below, we outline the details of these pathways.
a. Transport via Extracellular Vesicles
The classification and characterization of EVs has been a matter of debate, in part due to biological vagaries and in part due to methodological limitations [recently reviewed in (Mathieu et al., 2019; Shurtleff et al., 2018; van Niel et al., 2018)]. Indeed, there are many caveats in the methods for separation and isolation of the various types of EVs, both from each other and from protein complexes carrying miRNAs. As a result, in many studies, it is difficult to be certain of the exact carrier of an extracellular miRNA. Although there is no consensus regarding the nomenclature or methods to define different subtypes of EVs, the field is moving towards finding more accurate ways to classify EVs based on size, density, method of purification, surface markers, process of formation and release mechanism. This initiative includes a proposal by the International Society for Extracellular Vesicles (ISEV) with the aim to establish guidelines and recommendations on these issues (Mateescu et al., 2017; Thery et al., 2018) and multiple recent publications from the Extracellular RNA Communication Consortium characterizing the content of different extracellular particles (Das et al., 2019; Jeppesen et al., 2019; Murillo et al., 2019; Rozowsky et al., 2019; Srinivasan et al., 2019).
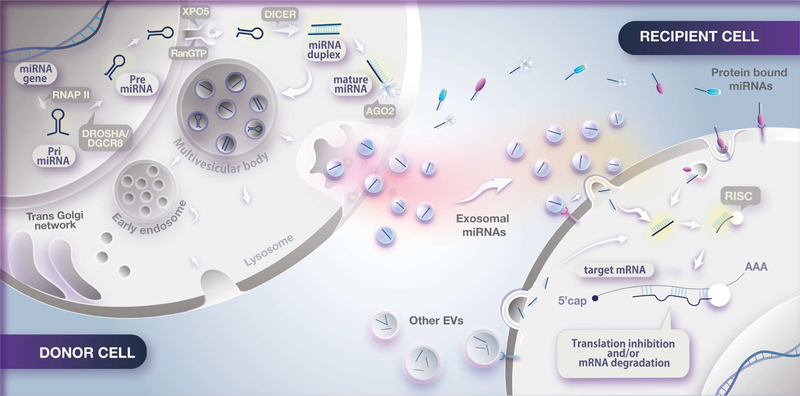
In general, smaller EVs (< 200 nm) generated by the fusion of multivesicular bodies (MVBs) and the plasma membrane are called exosomes (Figure 1), while larger EVs (> 200 nm) formed by direct outward budding and fission of the plasma membrane are called microvesicles. Direct budding can also produce small vesicles similar to exosomes which have been termed shed vesicles or ectosomes (Booth et al., 2006; Cocucci and Meldolesi, 2015). These EVs can also serve as mechanisms of miRNA release into the circulation or extracellular fluid, but “exosome” is the most frequently used term in studies analyzing forms of miRNA transport within EVs, even when the exact origin of these EVs is not demonstrated.
Figure 1. Intercellular communication via miRNAs.
RNAP II, RNA polymerase II. RISC, RNA-induced silencing complex. EVs, extracellular vesicles.
Exosomes are secreted after translocation of MVBs from the perinuclear cytoplasm to the plasma membrane, where they undergo fusion and release of their contents by the process of exocytosis. Several small GTPase proteins (e.g., Ral1, Rab27a and Rab27b) and proteins involved in ceramide biogenesis (e.g., nSMase2) participate in the control of intracellular formation, fusion and release of MVBs (Ostrowski et al., 2010; Trajkovic et al., 2008). The extent to which alterations in ceramide metabolism might be linked to alterations in exosome formation and release has not been studied, but altered ceramide metabolism is found in metabolic syndrome (Holland et al., 2011; Turpin et al., 2014).
What determines the miRNA content of EVs is a critical question and still poorly understood. There is growing evidence that the miRNA profile of EVs differs from that of the parent cell, indicating active loading or sorting of miRNAs into these vesicles (Mittelbrunn et al., 2011). Some studies have suggested a role of AGO2 and other RNA-binding proteins in the regulation of miRNA loading into exosomes. AGO2 has been found to be co-localized with the exosome protein CD63 in the cytoplasm during MVB formation, and AGO2 phosphorylation, which leads to uncoupling of miRNAs from the complex, alters miRNA sorting and loading, as well as exosome release (McKenzie et al., 2016). Other RNA binding proteins like hnRNPA2B (Villarroya-Beltri et al., 2013) and Y-box protein 1 (Shurtleff et al., 2016) may also confer specificity for miRNA loading into exosomes. This suggests an intricate mechanism coupling miRNA availability with exosome loading and is consistent with the fact that the composition of exosomes can differ considerably from tissue to tissue and vary according to the metabolic state of the cell (Vallabhajosyula et al., 2017).
Likewise, how exosomes and other EVs are taken up by potential target cells is not well understood. When injected intravenously, the half-life of EVs in the circulation varies from as little as 2 min to as much as 60 min, implying a large dynamic range in the circulating miRNA pool (Lai et al., 2014; Takahashi et al., 2013). The first step of EV uptake is attachment to the target cell [reviewed by (McKelvey et al., 2015; Mulcahy et al., 2014)], possibly by tetraspanins (e.g., CD9, CD53, CD63, CD81 and CD82), antigen recognition (e.g., MHC-I and MHC-II complexes) or other surface receptors. Bound EVs may then (a) activate intracellular signaling pathways, (b) release their content into the cell by membrane fusion, or (c) enter the cell via phagocytosis, macropinocytosis or receptor-mediated endocytosis. Following internalization, the various molecules carried in EVs may be released in the cytoplasm, delivered to lysosomes and be destroyed, or targeted to specific locations within the cell where they may elicit a functional response. Exactly how these mechanisms apply to different vesicles and host cells may vary; however, the process of EV uptake appears to show some selectively (Blanchard et al., 2002; Fitzner et al., 2011; Horibe et al., 2018).
b. miRNA Transport via Protein-miRNA complexes
In addition to EVs, miRNAs may be transported in the blood complexed with proteins. These complexes can also enter cells and deliver miRNAs to promote target mRNA inhibition. Both low density (LDL) and high density (HDL) lipoproteins can transport miRNAs in the circulation. In the case of HDL, the bound miRNAs can be taken up via class B type I scavenger receptors and released intracellularly where they may regulate gene expression in recipient cells (Vickers et al., 2011). In the case of miRNAs associated with lipoproteins, changes in nutritional and metabolic state, such as hypercholesterolemia, could alter the relative abundance of specific miRNAs complexes (Vickers et al., 2011). Importantly, the profile of miRNAs bound to HDL differs from that found in EVs (Vickers et al., 2011), indicating that the two are complementary and independent mechanisms of miRNA transport.
Despite being functionally important, EV-associated and lipoprotein-bound miRNAs are just a fraction of all the miRNAs found in circulation. In some studies, more than half of the miRNAs found in human serum may be bound to ribonucleoproteins, including argonaute (AGO2); however, only a fraction of these are carried exclusively by this means (Arroyo et al., 2011). The nucleolar protein nucleophosmin 1 (NPM1) has also been found to carry and protect extracellular miRNAs from degradation (Wang et al., 2010). It remains under debate whether either AGO2- or NPM1-bound miRNAs can act to alter function in distal cells or primarily are products of miRNA disposal and cell death. Prud’homme et al. have demonstrated internalization of AGO2-miRNA complexes into cells and found that, once inside the cell, these miRNAs may be able to regulate mRNA levels (Prud’homme et al., 2016). Another possibility is that RNA-binding proteins such as AGO2 and NPM1 carry miRNAs outside the cell and facilitate their loading onto lipoproteins. As discussed above, AGO2 has been implicated in exosome loading and may be bound to miRNAs within EVs (Gibbings et al., 2009; McKenzie et al., 2016; Melo et al., 2014; Squadrito et al., 2014), although a recent publication demonstrated that “classical” exosomes do not contain AGO2 (Jeppesen et al., 2019).
Regardless of their form, the presence of miRNAs in blood and other extracellular fluids has served as a stimulus to two important avenues of research. One is the potential that extracellular miRNAs may serve as biomarkers of disease, and the other, and even more exciting possibility, is the potential of extracellular miRNAs to serve as a novel mode of intercellular and intertissue communication, i.e., extracellular miRNAs act as hormones being produced in one cell or tissue and regulating gene expression in another. We will discuss both aspects below.
Extracellular miRNAs as Disease Biomarkers
A biomarker is a molecule that can be used for disease detection and/or prognosis prediction. Four of the most important features of a good biomarker are specificity, sensitivity and stability, but also that they can be obtained in a relatively non-invasive manner. Not surprisingly, most studies have focused on extracellular miRNAs as potential biomarkers, since they are stable (i.e., protected from ribonucleases) (Grasedieck et al., 2012; Mitchell et al., 2008) and can be detected in the blood, urine or other body fluids (Weber et al., 2010) using simple, sensitive and relatively cheap assays, even after periods of years of sample storage. Indeed, changes in levels of circulating miRNAs have been associated with a wide range of diseases including type 2 diabetes (T2D), obesity, cardiovascular disease, cancer, neurodegenerative disorders, and others. In addition, changes in levels of circulating miRNAs and EVs have been shown to correlate with differences in lifestyle activities, such as exercise (Flowers et al., 2015; Rome, 2015; Safdar et al., 2016; Whitham et al., 2018), and the composition of gut microbiota (Beatty et al., 2014), suggesting that extracellular miRNAs may serve as circulating indicators of the physiological status of the individual and a tool for targeted therapies in precision medicine.
a. Methods to Isolate and Detect Extracellular miRNAs
Although the concentration of miRNAs in body fluids is relatively low, i.e., in the femtomolar range (Williams et al., 2013), the fact that these molecules are stable and easily quantitated using RT-qPCR, microarrays or RNAseq allows their detection in relatively small samples. Each of these methods has its advantages and limitations. RT-qPCR is more sensitive and not limited by sequence-abundance bias (Dohm et al., 2008; Hansen et al., 2010; Zheng et al., 2011), but the technique requires optimization, proper standard curves, and ideally the inclusion of spiked-in exogenous miRNAs (Huggett et al., 2013; Mitchell et al., 2008; Zhuang et al., 2012a). In contrast, although more expensive, microarrays and RNAseq allow the simultaneous detection of a multiple miRNAs, opening the possibility to detect novel and combinatorial biomarkers.
When it comes to sensitivity and specificity in assessing extracellular miRNAs, standardizing the fluid and the fractionation method used in isolation are key variables. While whole serum or unfractionated biological fluids can be used for assay, most studies use isolated EVs to avoid products of cell lysis and artifacts; however, there is considerable variability as to the exact method used for isolation. In a database summarizing 1,226 publications, over 1,000 different protocols were used to retrieve EVs from biological fluids. These ranged from ultracentrifugation to the use of macromolecular crowding reagents (Consortium et al., 2017) resulting in a wide range of purity, different combinations of vesicle types, and “contamination” with miRNA-protein complexes [reviewed in (Shurtleff et al., 2018)]. Moreover, in most studies, the purity of the vesicle preparation was not determined. For these reasons, it is prudent to use the more general term “extracellular vesicle” or “EV” instead of “exosome” or “microvesicle” when referring to these various vesicle preparations. In addition to differences in isolation method, differences in methods of assessing the miRNAs and differences in the populations studied, as well as other factors, can lead to variability in findings. Thus, it is not surprising that there are examples of circulating miRNAs reported to be dysregulated in disease in one study but not in others. Despite this, there is a growing body of evidence associating changes in extracellular miRNAs with different disease states supporting their link to pathophysiology (reviewed below and Table 1).
Table 1:
Association of circulating miRNAs with metabolic and age-related diseases.
b. Disease-Associated Extracellular miRNAs
Obesity.
Several miRNAs have been found to be altered in the circulation of obese patients or obese animals, including miR-122, miR-142–3p, miR-192, miR-222, and miR-378a which are upregulated, and miR-138 and miR-221 which are downregulated (Can et al., 2016; Castano et al., 2018; Hsieh et al., 2015; Jones et al., 2017; Nunez Lopez et al., 2016; Ortega et al., 2013; Pescador et al., 2013; Villard et al., 2015; Wang et al., 2015; Willeit et al., 2017; Wu et al., 2015). These changes are accompanied by an increase in the levels of circulating EVs in obesity or type 2 diabetes (T2D) (Campello et al., 2016; Castano et al., 2018; Li et al., 2016; Stepanian et al., 2013), in part via an increase in EV secretion by adipocytes (Flaherty et al., 2019). Indeed, among tissues contributing to circulating miRNAs, adipose tissue is thought to play an important role (Thomou et al., 2017; Ying et al., 2017). The level of expression of multiple miRNAs and their target genes is altered in adipocytes in obesity and lipodystrophy (Arner et al., 2012; Arner and Kulyte, 2015; Heneghan et al., 2011; Meerson et al., 2013; Mori et al., 2012; Mori et al., 2014; Oliverio et al., 2016; Ortega et al., 2010; Torriani et al., 2016), and this may be reflected by changes in extracellular miRNAs. For example, miR-222 is a negative regulator of insulin sensitivity in adipocytes, reducing ERα expression and GLUT4-mediated glucose uptake (Shi et al., 2014). It also leads to insulin resistance in hepatocytes, where it targets insulin receptor substrate 1 (IRS1) (Ono et al., 2018). In obesity, miR-222 levels are increased in blood (Ortega et al., 2014; Villard et al., 2015), adipose tissue (Chartoumpekis et al., 2012; Xie et al., 2009) and liver (Ono et al., 2018). In blood, this miRNA is found both in EVs and associated with HDL (Santangelo et al., 2018; Sohn et al., 2015; Vickers et al., 2011).
Liver may also contribute to the pool of circulating miRNAs during the metabolic syndrome. miR-122 is a liver-enriched miRNA involved in regulation of fatty acid oxidation and cholesterol biosynthesis (Esau et al., 2006; Krutzfeldt et al., 2005). Circulating levels of miR-122 are positively associated with fatty liver disease, T2D, obesity and atherosclerosis (Jiang et al., 2014; Jones et al., 2017; Pirola et al., 2015; Wang et al., 2015; Willeit et al., 2017). miR-122 has been found bound to AGO2 in serum (Arroyo et al., 2011), as well as in circulating EVs (Castano et al., 2018). Interestingly, in mice with experimental hepatosteatosis the EV-associated fraction of miR-122 increases, while the AGO2-bound fraction is reduced (Povero et al., 2014), indicating that compartmentalization of these miRNAs may be altered by disease. Furthermore, lean mice injected repeatedly with EVs loaded with mimics of obesity-related miRNAs, such as miR-122, miR-192, miR-27a, and miR-27b, develop insulin resistance and dyslipidemia (Castano et al., 2018), suggesting that these miRNAs play a causal role in the pathophysiology of obesity.
Type 2 diabetes.
In addition to the circulating miRNAs associated with obesity which may be linked to insulin resistance, many extracellular miRNAs have been associated with T2D (Table 1), including miR-375, miR-223 and miR-155.
miR-375 is highly enriched in pancreatic islets where it suppresses glucose-stimulated insulin secretion by targeting myotrophin (MTPN), a protein involved in fusion of insulin-containing granules with the plasma membrane (Poy et al., 2004; Xia et al., 2011), and 3-phosphoinositide-dependent kinase 1 (PDK1), a key molecule in insulin’s metabolic signaling pathway (El Ouaamari et al., 2008). In addition, miR-375 participates in β-cell differentiation and islet development (Kloosterman et al., 2007; Lahmy et al., 2014; Nathan et al., 2015). miR-375 levels are increased in pancreatic islets from diabetic patients (Zhao et al., 2010), and several, but not all, studies have found elevated blood levels of miR-375 in T2D patients compared to non-diabetics (Al-Muhtaresh and Al-Kafaji, 2018; Higuchi et al., 2015; Karolina et al., 2012; Kong et al., 2011; Latreille et al., 2015; Sun et al., 2014; Zhu and Leung, 2015). While these reports have only analyzed whole plasma/serum, miR-375 is thought to be both carried in EVs (Bryant et al., 2012; Zaharie et al., 2015) and associated with HDL (Vickers et al., 2011), although the proportions in each fraction remain unknown.
miR-223 is reduced in blood from T2D patients compared to healthy controls and has been predicted in a meta-analysis to serve as a biomarker for T2D (Liang et al., 2018). Importantly, miR-223 is reduced in individuals up to 10 years prior to diagnosis of disease (Zampetaki et al., 2010) and is also reduced in obese patients (Wen et al., 2015). This reduction in miR-223 is believed to contribute to the increased adipose tissue inflammation observed in obesity, since miR-223 can inhibit inflammation by targeting NLRP3 (Nod-like receptor pyrin domain containing 3), a component of the inflammasome, and IL-1β production, a component of M1-like macrophage activation (Haneklaus et al., 2012). miR-223 downregulation also controls macrophage differentiation, primes the response of macrophages to proinflammatory stimuli (Li et al., 2010) and leads to M1-like macrophage polarization (Zhuang et al., 2012b). Accordingly, miR-223 knockout mice show severe insulin resistance and adipose tissue inflammation (Zhuang et al., 2012b). In the circulation miR-223 is found associated with EVs, HDL and AGO2 (Arroyo et al., 2011; Ismail et al., 2013; Vickers et al., 2011).
miR-155 is reduced in serum of prediabetic and T2D patients (Liang et al., 2018; Lin et al., 2016; Nunez Lopez et al., 2016), while it is upregulated in diabetic kidneys and retinal endothelial cells (Huang et al., 2014; Kovacs et al., 2011). This miRNA is sensitive to inflammation as adipose tissue macrophages from obese mice or LPS-induced macrophages or adipocytes treated with conditioned medium from pro-inflammatory macrophages have high miR-155 levels (Ortega et al., 2015; Ying et al., 2017). Indeed, miR-155 can be released in EVs from adipose tissue macrophages and transferred to other cell types such as adipocytes, myotubes or hepatocytes, where it worsens insulin resistance (Ying et al., 2017).
Cardiovascular Disease and Metabolic Syndrome.
Several miRNAs linked to T2D and obesity are also associated with cardiovascular disease (CVD), which is a major cause of morbidity and mortality in the elderly and in patients with metabolic syndrome. Thus, miR-92a, miR-126, and miR-222 are reduced, while miR-21, miR-122, miR-130a, miR-150, and miR-211 are elevated in the blood of patients with atherosclerosis (Jiang et al., 2014; Olivieri et al., 2012; Zhang et al., 2010). Another miRNA associated with cholesterol homeostasis is miR-223. In addition to its function in adipose tissue inflammation (see above), this miRNA is involved in the regulation of cholesterol synthesis and uptake (Vickers et al., 2014). miR-223-containing EVs have been shown to penetrate the vascular wall and decrease plaque size by inhibiting vascular smooth muscle cell proliferation and migration (Shan et al., 2015). Circulating miR-223 is elevated in mice and humans with atherosclerosis (Keller et al., 2017; Schulte et al., 2015; Shan et al., 2015), possibly as a compensatory mechanism to reduce cholesterol levels and plaque size. In combination with other miRNAs, miR-223 may predict the risk of cardiovascular events and mortality (Keller et al., 2017; Shan et al., 2015; Zampetaki et al., 2012). By contrast, circulating miR-30c is inversely correlated with cholesterol levels and plaque development in humans (Ceolotto et al., 2017). Mechanistically, miR-30c has been shown to attenuate inflammation (Ceolotto et al., 2017) and reduce hyperlipidemia in mice by inhibiting microsomal triglyceride transfer protein MTP (Soh et al., 2013).
A number of circulating miRNAs have also been associated with coronary artery disease. These include decreased levels of miR-126, miR-17, miR-92a, miR-145 and miR-155; and increased levels of miR-320a, miR-133a and miR-208a (Chen et al., 2015; Fichtlscherer et al., 2010). The compartmentalization of circulating miR-126 is important in its disease association, given that EVs containing miR-126, but not the circulating vesicle-free fraction of this miRNA, can predict the occurrence of cardiovascular events in patients with CVD (Jansen et al., 2014). miR-126 bound to AGO2 modulates smooth muscle gene expression and promotes a proatherogenic phenotype in smooth muscle cells (Zhou et al., 2013), whereas miR-126 in apoptotic bodies produced by endothelial cells in the course of atherosclerosis reduces atherosclerotic plaque formation in mice (Zernecke et al., 2009). miR-320a, on the other hand, is highly expressed in cardiomyocytes, endothelium and vascular smooth muscle cells, where it targets the IGF-1 signaling pathway and impairs angiogenesis and vascular remodeling (Chen et al., 2015; Goren et al., 2012; Ling et al., 2013; Wang et al., 2014; Wang et al., 2009).
Aging and Other Age-Related Diseases.
Aging is characterized by progressive dysfunction of multiple organs and tissues within an organism and is associated with increased risk of many diseases including T2D and metabolic syndrome (Kirkman et al., 2012). Dysregulated nutrient sensing and altered intercellular communication are hallmarks of aging (Lopez-Otin et al., 2013) and reflect dysfunction in whole-body metabolic homeostasis. In general, aging and senescence is characterized by downregulation of miRNA biogenesis. This occurs at the level of the whole organism (de Lencastre et al., 2010; Ibanez-Ventoso et al., 2006) and in several tissues, including adipose tissue, blood, kidney, brain vasculature and stem cells (Mori et al., 2012; Nidadavolu et al., 2013; Noren Hooten et al., 2010; Oliverio et al., 2016; Ungvari et al., 2013), although some studies reported overall upregulation of miRNAs in liver, brain and serum (Dhahbi et al., 2013; Li et al., 2011; Maes et al., 2008). Several of these differentially expressed miRNAs have been causally linked to lifespan extension in both invertebrates and vertebrates (Caravia et al., 2018; Du et al., 2014; Mori et al., 2012; Shen et al., 2012; Smith-Vikos and Slack, 2012).
Many miRNAs have been reported to differ in abundance in comparisons of serum, plasma and saliva samples from human donors of different ages (Ameling et al., 2015; Machida et al., 2015; Noren Hooten et al., 2013; Olivieri et al., 2014; Olivieri et al., 2012; Zhang et al., 2015), however these alterations have not been found consistently across studies reflecting possible environmental influences, methodological differences and/or the presence of undiagnosed diseases among the individuals studied. Despite the differences, increases in miR-126, miR-21 and miR-30c have been found in the circulation of elderly people in multiple studies (Ameling et al., 2015; Noren Hooten et al., 2013; Olivieri et al., 2014; Olivieri et al., 2012). The exact role of these changes in circulating miRNAs in the biology of aging remains unclear and needs further exploration.
Circulating miRNAs have been also proposed as biomarkers for detection and predicting prognosis of other age-related disorders linked to metabolism including neurodegenerative diseases, such as Alzheimer’s and Parkinson’s diseases, and cancer. While this topic is too extensive to review here, some of the miRNAs reproducibly linked to neurodegenerative diseases are listed in Table 1. The use of circulating miRNAs in detection and prognosis of cancer has also been the subject of many studies and been already reviewed (Anfossi et al., 2018; Fendler et al., 2016; He et al., 2015; Nedaeinia et al., 2017; Wong et al., 2018).
Extracellular miRNAs as Housekeepers of Metabolic Homeostasis
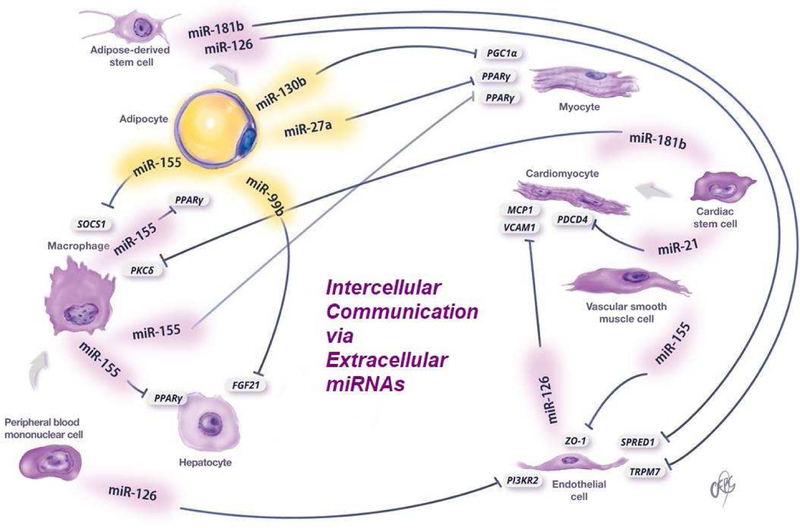
Adipose tissue, immune cells, pancreatic islets and many other cells linked to metabolic disease are known to secrete significant amounts of miRNAs both in EVs and in other forms. These miRNAs may, in turn, act in a paracrine or autocrine manner or reach the circulation to act in an endocrine manner. A list of extracellular miRNAs with their donor cells, recipient cells, target genes and disease association is presented in Table 2. The most relevant sites of systemic regulation via extracellular miRNAs are summarized below and in Figure 2.
Table 2. Intercellular crosstalk by extracellular miRNAs.
n.d., not determined. BCAA, branched chain amino acids. a, predicted.
| miRNA | Donor cell | Recipient cell | Target | Function | Reference |
|---|---|---|---|---|---|
|
miR-16 miR-27a miR-146b miR-222 |
Large adipocytes | Small adipocytes | n.d. | ↑ Lipid storage | (Müller et al., 2011) |
|
miR-16 let-7a |
Adipocytes | n.d. | Insulin signaling pathwaya | Associated with blood BCAA levels | (Hubal et al., 2017) |
| miR-21 | Cardiac stem cells | Cardiomyocytes | PDCD4 | ↓ Apoptosis | (Xiao et al., 2016) |
| miR-27a | Adipocytes | Skeletal muscle cells | PPARγ | ↑ Insulin resistance | (Yu et al., 2018) |
| miR-99b | Adipocytes | Hepatocytes | FGF21 | ↑ Glucose intolerance | (Thomou et al., 2017) |
| miR-124 | Microglia | Neurons | PDE4B | ↓ Inflammation ↑ Neurite outgrowth |
(Huang et al., 2018) |
| miR-126 | Endothelial cells | Cardiomyocytes | MCP1 VCAM1 | ↓ Cardiac hypertrophy | (Chen et al., 2017) |
| miR-126 | Adipose-derived stem cells | Endothelial cells | SPRED1 | ↑ Angiogenesis | (Togliatto et al., 2016) |
| miR-126 | Peripheral blood mononuclear cells | Human aortic endothelial cells | PI3KR2 | ↑ Angiogenesis | (Mocharla et al., 2012) |
| miR-130b | Adipocytes | Muscle cells | PGC1α | ↓ Oxidative metabolism | (Wang et al., 2013) |
| miR-150 | Monocytes | Vascular endothelial cells | c-Myb | ↑ Cell migration | (Zhang et al., 2010b) |
| miR-155 | Vascular smooth muscle cells | Vascular endothelial cells | ZO-1 | ↑ Endothelial permeability ↑ Atherosclerosis |
(Zheng et al., 2017) |
| miR-155 | Vascular endothelial cells | Macrophages | n.d. | ↑ M1 polarization ↑ Atherosclerosis |
(He et al., 2018) |
| miR-155 | Adipocytes | Macrophages | SOCS1 | ↑ M1 polarization | (Zhang et al., 2016) |
| ↑ Insulin resistance | |||||
| miR-155 | Macrophages | Adipocytes, Myocytes and Hepatocytes | PPARγ | ↑ Insulin resistance | (Ying et al., 2017) |
| miR-181b | Cardiac stem cells | Macrophages | PKCδ | ↑ Cardioprotection | (de Couto et al., 2017) |
| miR-181b | Adipose-derived stem cells | Brain microvascular endothelial cells | TRPM7 | ↑ Angiogenesis | (Yang et al., 2018) |
| miR-214 | Bone morrow stem cells | Cardiac stem cells | CaMKII | ↓ Oxidative stress ↓ Apoptosis |
(Wang et al., 2018) |
| miR-219 | n.d. | Oligodendrocyte precursor cells | NEUROD1 PDGFRα ELOVL7 | ↑ Myelination | (Pusic and Kraig, 2014) |
| miR-294 | Embryonic stem cells | Cardiac stem cells | n.d. | ↑ Survival ↑ Proliferation |
(Khan et al., 2015) |
Figure 2.
Examples of miRNAs that play a role in intercellular communication.
Adipose tissue.
In addition to its role in storing energy as triglycerides, adipose tissue is well known for its role in maintaining organismal homeostasis by secreting molecules that integrate whole-body metabolism. These molecules include adipose-derived hormones (known as adipokines) (Cao, 2014; Choi and Cohen, 2017; Stern et al., 2016), signaling lipids (Cao et al., 2008; Lynes et al., 2017; Stanford et al., 2018; Yore et al., 2014), inflammatory mediators (Brestoff and Artis, 2015; Glass and Olefsky, 2012; Mathis, 2013), and miRNA-containing EVs. The fact that adipose tissue contributes significantly to the pool of circulating miRNAs is evidenced by the finding that fat-specific knockout of the key miRNA processing enzyme Dicer (ADicerKO) results in significant decreases in about two-thirds of the circulating exosomal miRNAs (Thomou et al., 2017). Likewise, patients with various forms of lipodystrophy have major alterations in circulating exosomal miRNAs. Importantly, miRNAs secreted by adipose tissue have been shown to reach organs like liver and muscle, where they can modulate gene and protein expression (Thomou et al., 2017; Ying et al., 2017).
One example of an adipose tissue-derived circulating miRNA contributing to the control of metabolic homeostasis in an endocrine manner is the regulation of liver FGF21 by adipose tissue-derived miR-99b (Thomou et al., 2017). Thus, ADicerKO mice have reduced levels of miR-99b in circulating EVs and upregulation of Fgf21 mRNA and 3’UTR-reporter activity in liver, both of which can be significantly corrected by administering EVs loaded with miR-99b into the circulation. Consistent with this being part of a more generalized mechanism, AdicerKO mice exhibit a wide range of phenotypes reflecting dysfunction in other tissues, including muscle, b-cells and bone, as well as systemic insulin resistance (Mori et al., 2014; Reis et al., 2016). Which circulating exosomal miRNAs are potentially involved in these other phenotypes remains to be determined.
Other studies have shown that miRNAs in adipose-derived EVs may also serve a paracrine function. Thus, EVs released from large adipocytes containing miR-16, miR-27a, miR-146b and miR-222 can be transferred to small adipocytes to stimulate lipogenesis and adipocyte hypertrophy (Müller et al., 2011). Secretion of these miRNAs by adipocytes is induced by free fatty acids and H2O2, and is upregulated in the serum of old vs. young mice (Müller et al., 2011), suggesting that signals that promote lipid accumulation and insulin resistance may spread from insulin resistant adipocytes to newly formed adipocytes via adipocyte-secreted miRNAs.
Consistent with a potentially important role of adipose-secreted miRNAs in control of systemic metabolism is the finding that the circulating levels of miRNAs known to be secreted by adipose tissue have been associated with the pathophysiology of obesity- and age-related metabolic diseases (discussed above and listed in Table 1). Likewise, amelioration of metabolic dysfunction by weight loss may be explained, at least in part, due to changes in circulating miRNAs. Importantly, multiple adipose tissue-derived circulating miRNAs (identified by affinity purification of extracellular particles containing adipose-specific protein FABP4) are significantly changed in obese individuals one year after bariatric surgery (Hubal et al., 2017). These miRNAs are predicted to target components of the WNT/beta-catenin and insulin signaling pathways. Among the differentially expressed miRNAs after bariatric surgery, let-7a and miR-16 have targets involved in insulin receptor signaling, and the levels of these miRNAs correlate with the levels of branched-chain amino acids (BCAA), suggesting that they may be linked to systemic insulin resistance (Hubal et al., 2017). Modulation of WNT/beta-catenin and TGF-β signaling pathways is also a predicted mechanism of miRNAs differentially expressed in circulation of obese vs. lean subjects (Ferrante et al., 2015). For example, hepatoma cells treated with EVs isolated from adipocytes of obese donors exhibit changes in gene expression of components of the TGF-β signaling pathway (Koeck et al., 2014). Given previous reports relating TGF-β to fibrosis and late stages of non-alcoholic fatty liver disease (NAFLD) (Liu et al., 2016), these secreted miRNAs may serve as a mechanism of adipose tissue-liver crosstalk in response to metabolic dysregulation and contribute to NAFLD.
These adipose-derived miRNAs may be part of more complex regulatory loops. For example, TGF-β acts to induce miR-130b secretion from mature adipocytes, and this miRNA can then be transferred to muscle cells where it has been shown to reduce the expression of PGC-1α, thus controlling muscle oxidative metabolism (Wang et al., 2013). Skeletal muscle is also responsive to miR-20b - a miRNA that inhibits insulin-stimulated glycogen accumulation and is upregulated in serum EVs of T2D patients (Katayama et al., 2018). miR-27a, which is present in adipose-derived EVs and taken up by muscle cells, has been shown to induce insulin resistance via PPARγ repression (Yu et al., 2018). Serum levels of miR-27a are positively associated with obesity and insulin resistance in humans and mice indicating that miR-27a may be another modulator of obesity-associated insulin resistance (Yu et al., 2018).
Low-grade inflammation in adipose tissue and liver is a common feature of obesity and metabolic syndrome (Franceschi et al., 2018; Hotamisligil, 2017; Mori et al., 2010; Odegaard and Chawla, 2013). This is characterized by a polarization of adipose tissue macrophages towards the M1-like phenotype and increased proinflammatory cytokine production (Castoldi et al., 2015; Lackey and Olefsky, 2016; Shapouri-Moghaddam et al., 2018). Mice injected with EVs secreted from the adipose tissue of obese mice develop increased levels of the circulating proinflammatory cytokines IL-6 and TNF-α and insulin resistance (Deng et al., 2009). The proposed mechanism involves monocyte differentiation and macrophage activation through the TLR4 pathway (Deng et al., 2009). This appears to be controlled, in part, by miR-155 that can target SOCS1 in macrophages, thus promoting STAT1 and suppressing STAT6 signaling, thereby inducing M1-like macrophage polarization (Zhang et al., 2016). Similarly, when bone marrow-derived macrophages are pretreated with EVs secreted by adipocytes of obese mice, these macrophages secrete molecules that inhibit insulin signaling in myocytes or adipocytes (Deng et al., 2009; Zhang et al., 2016). Whether these molecules are cytokines or a second layer of EVs derived from the macrophages is unclear, but EVs obtained from adipose tissue macrophages of obese mice are able to induce insulin resistance when administrated to lean mice or incubated in vitro with adipocytes, myocytes or hepatocytes (Ying et al., 2017).
Among the miRNAs increased in EVs secreted by adipose tissue macrophages of obese mice is miR-155, a miRNA that downregulates PPARγ. Knockout of miR-155 in high fat diet-fed mice results in improved insulin sensitivity, and this is reversed by bone marrow transplantation from wild-type mice (Ying et al., 2017). On the other hand, another group found impaired glucose tolerance and insulin resistance in miR-155 knockout mice fed chow diet (Lin et al., 2016). Thus, the role of miR-155 may be more complex than believed. Also, no differences in glucose uptake or adipocyte differentiation were observed when adipocytes or preadipocytes were exposed to EVs derived from LPS-activated macrophages (De Silva et al., 2018). Likewise, miR-155 is not upregulated in EVs secreted by LPS-activated macrophages, suggesting that different pro-inflammatory stimuli (e.g., obesity vs. LPS) can lead to differential loading of EVs, which in turn may differentially affect their target tissues.
Along this line, EVs secreted by adipose-derived stem cells (ADSCs) from lean mice can lead to polarization of macrophages towards an anti-inflammatory M2-like phenotype (Zhao et al., 2017). Likewise, ADSC-released EVs obtained from lean individuals are enriched in miR-126 (Togliatto et al., 2016), and this is mechanistically associated with the pro-angiogenic potential of these vesicles in comparison to ADSC-released EVs isolated from obese individuals (Togliatto et al., 2016). Finally, EVs secreted by endothelial cells may also be involved in adipose tissue intercellular communication and macromolecule exchange, and this process can be regulated by feeding and fasting (Crewe et al., 2018).
Taken together, these data indicate that adipose tissue-secreted EVs containing miRNA may arise from different cells within the fat pad and be differentially regulated by various stimuli. Likewise, once they enter the blood or extravascular space, they have the ability to act on other cells within the fat pad or tissues at a distance, such as liver, muscle and hematopoietic cells, to coordinate metabolic homeostasis and energy balance. Thus, adipose tissue-derived miRNAs may therefore contribute broadly to the pathophysiology of metabolic diseases.
Pancreas.
Pancreatic islet cells may control metabolism not only via secretion of insulin and glucagon, but also by secretion of miRNAs. Primary islet cells and β-cell-derived MIN6 cells have been shown to release specific miRNAs within EVs in response to stimuli that induce insulin secretion (Zhang et al., 2018). For example, miR-223 is increased in serum, islets, liver and skeletal muscle of obese ob/ob mice in comparison to their lean controls. Interestingly, levels of its precursor, pri-miR-223, are increased only in islets, suggesting that islets are the likely source of the elevated levels of mature miR-223 in circulation and other tissues (Zhang et al., 2018). Studies on the potential function of miR-223 have yielded conflicting results, however, miR-223 has been shown to bind to the 3’UTR of Glut4 mRNA and downregulate GLUT4 in adipose tissue (Chuang et al., 2015) while promoting GLUT4 expression in cardiomyocytes (Lu et al., 2010).
Circulating miRNAs may also act on b-cells. For example, miR-155, miR-142–3p and miR-142–5p may be transferred from T-lymphocyte-derived EVs to β-cells, leading to activation of inflammatory pathways, apoptosis and development of insulin-deficient diabetes (Guay et al., 2019). Taken together, these studies suggest that islets could co-secrete miRNAs along with insulin, which could modulate insulin action in target tissues, while peripheral cells could send signals back to islets via EVs containing miRNAs.
Cardiovascular system.
EVs secreted by human monocytes after a proinflammatory challenge have high levels of miR-150 (Zhang et al., 2010). Incubation of microvascular endothelial cells with these EVs downregulates the miR-150 target gene c-Myb, a transcription factor involved in endothelial cell migration (Kopecki et al., 2007; Xiao et al., 2007; Zhang et al., 2010; Zhang et al., 2010b). Conversely, overexpression of miR-150 in vitro induces endothelial cell migration, and this effect can be mimicked by incubation with EVs from plasma of patients with atherosclerosis, which also have increased levels of miR-150 (Zhang et al., 2010). In addition to this crosstalk between monocytes and endothelial cells via miR-150, EVs from vascular smooth muscle cells have been shown to facilitate transfer of miR-155 to endothelial cells, affecting integrity of the endothelial barrier by decreasing levels of tight junction proteins (Zheng et al., 2017). EVs secreted by endothelial cells exposed to oxidized LDL have increased levels of miR-155, a miRNA that shifts macrophage polarization from the M2-like phenotype towards the pro-inflammatory M1-like phenotype (He et al., 2018; Zhang et al., 2016). In addition, changes in the levels of miR-126 in both serum and heart have been suggested to play a role in cardiac dysfunction by affecting the expression of MCP-1 and VCAM-1 (Chen et al., 2017). Together these processes could alter endothelial function and promote atherosclerosis.
Central nervous system.
There is growing evidence that circulating EVs may play a role in intertissue communication by crossing the ependymal layer and the blood brain barrier (BBB) to act on the central nervous system (Matsumoto et al., 2017). For instance, intranasal administration of serum-derived EVs containing miR-219 to aged rats results in increases in myelin content in the central nervous system (Pusic and Kraig, 2014). Neurodegenerative diseases which alter BBB permeability could facilitate the exchange of circulating miRNAs from the brain to the blood and vice-versa [reviewed in (Sweeney et al., 2018)]. Evidence has also been presented for transport of EVs across the BBB by a transcytosis mechanism (Chen et al., 2016).
Many extracellular miRNAs have been implicated in neurodegenerative diseases as disease biomarkers, although their role in pathophysiology of these disorders is uncertain. Zhang et al. demonstrated that aging impairs secretion of EV miRNAs by hypothalamic stem cells and that intracerebroventricular injection of EVs produced by these cells slows hypothalamic aging (Zhang et al., 2017b). EVs containing miRNAs have also been implicated in crosstalk between neurons, astrocytes, microglia and endothelial cells [reviewed by (Zagrean et al., 2018)]. For example, Huang et al. found increased levels of miR-124 in microglial EVs after brain injury and observed that this miRNA inhibits neuronal inflammation and promotes neurite outgrowth via transfer to neurons (Huang et al., 2018). Further studies will be required to fully define the role of extracellular miRNAs in brain function, development and degeneration, and to what extent peripherally produced EVs may act on the brain.
Challenges and Perspectives
a. Finding the source and mechanism of regulation of extracellular miRNAs
Although there is a growing body of evidence for a role of extracellular miRNAs in intercellular and intertissue communication, a major challenge in the field is determining the contribution of each tissue to the circulating miRNA pool in vivo, and how this might be modified in response to physiological changes or disease. It is also not clear why some miRNAs are preferentially loaded into EVs or onto carrier proteins for secretion, while others are retained in the cell; how circulating miRNAs in different forms differ in half-life, clearance or cellular uptake; and how all of these processes are regulated at a physiological level. In addition, our understanding of the degree to which these mechanisms contribute to normal physiology and disease pathophysiology remains far from complete.
For adipose tissue, a major contributor for the circulating miRNA pool, nutrient availability, β-adrenergic stimulation and cold exposure regulate miRNA biogenesis and secretion of EVs [our unpublished data and (Chen et al., 2016; Crewe et al., 2018; Mori et al., 2012; Mori et al., 2014; Oliverio et al., 2016; Reis et al., 2016; Torriani et al., 2016)]. Similarly, exercise can stimulate release of EVs containing miRNAs that can act on other tissues to enhance metabolic effects (Safdar and Tarnopolsky, 2018). To the extent that changes in miRNA biogenesis are coupled with EV secretion, these observations could reflect a mechanism to replenish the tissue with more miRNAs upon secretion or to control the abundance of miRNAs loaded in EVs. The clearest link that changes in miRNA biogenesis in adipose tissue significantly affect the levels of circulating miRNAs is found in individuals with HIV-associated lipodystrophy in whom reduced levels of DICER1 in adipose tissue lead to reduced serum levels of multiple exosomal miRNAs (Mori et al., 2014; Thomou et al., 2017; Torriani et al., 2016). While adipose tissue-derived miRNAs have been shown to suppress target mRNAs in distal tissues, such as the liver (Thomou et al., 2017), the full role of this form of intertissue communication in physiology and pathophysiology remains to be elucidated.
b. Defining targets and function of extracellular miRNAs
Current studies have focused on determining the target selectivity and mechanisms of action of extracellular miRNAs, i.e., if they have preferential target cells, specific mechanisms of delivery, etc. It is also of importance to determine how these miRNAs manage to avoid degradation in the recipient cell and become functional. Indeed, considering the relatively small amounts of miRNAs carried by EVs in circulation in comparison to the amounts found within the cells (Chevillet et al., 2014), it seems likely that there is some mechanism of selective delivery, such that miRNAs which enter their target cells are able to promote gene silencing.
As noted above, miRNAs can be carried in different forms in the circulation. Also, different types of extracellular particles may be taken up by cells differently (Horibe et al., 2018; McKelvey et al., 2015), and this could affect both effectiveness of entry and which miRNAs will remain active in the host cell. Proteins located on the EV membrane and the host cell membrane are thought to determine host cell selectivity and the mechanism of EV uptake, although details of this mechanism and how this varies from cell to cell is not clear (McKelvey et al., 2015). Whatever the mechanism(s), this could determine the fate of the EV content, whether released into the cytoplasm, loaded into intracellular vesicles or degraded in lysosomes. Hence, effects on intercellular communication may depend on what receptors mediate the internalization of the EVs or protein/lipoprotein/lipid-bound small RNAs.
c. Using extracellular miRNAs as biomarkers
While extracellular miRNAs have great potential as biomarkers of disease, important confounding factor in their utility are the wide variety of methods used to isolate miRNAs from body fluids, limitations of identifying the form in which they are carried and, most importantly, determining their source. About half of the publications in the field use ultracentrifugation as the primary method of EV isolation, but most of these studies differ in details of the protocol, which may be important in the ultimate nature of the fraction being isolated (Consortium et al., 2017). The second, and increasingly common, method to isolate EVs is through addition of macromolecular crowding reagents like polyethylene glycol. While both methods are subject to contamination by EVs of different kinds, as well as lipoproteins and high molecular weight protein complexes, contamination appears to be worse with the latter technique (Bobrie et al., 2012; Shurtleff et al., 2018). An alternative approach to minimize contaminants is to combine isolation methods with density gradient-based fractionation, size-exclusion chromatography, tangential flow filtration or magnetic beads targeting commonly found exosome markers, such as tetraspanins (Busatto et al., 2018; Haraszti et al., 2018; Jeppesen et al., 2019; Murillo et al., 2019). While these combinatorial methods may result in purer vesicle subtypes, they require larger volumes of blood, limit the amount of miRNA for analysis, are difficult to apply in large-scale studies, clinical trials or diagnostic assays, and may select only a subset of circulating miRNAs. Hopefully with more specific and standardized methods for isolation of miRNA containing EVs or protein complexes, and with techniques to identify EVs based on their tissue of origin and their surface protein patterns, circulating miRNAs in these different forms will provide more robust biomarkers of pathophysiological conditions and serve as accurate diagnosis tools.
d. Extracellular miRNAs in therapeutics
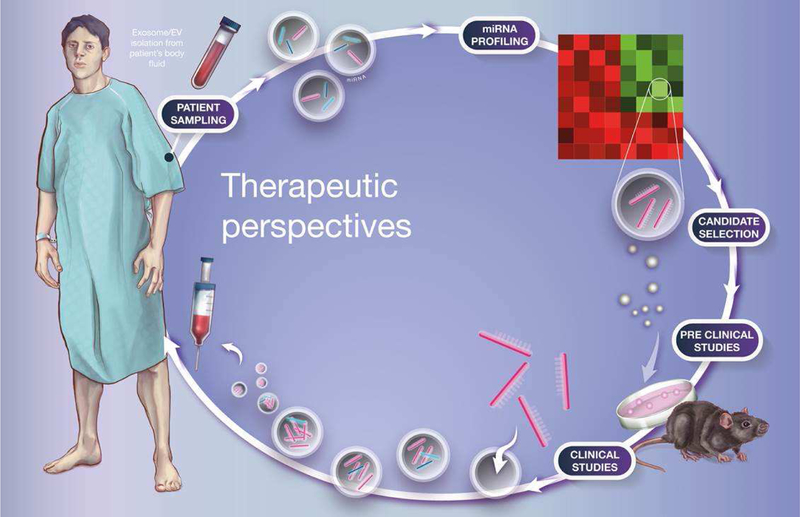
In the area of therapeutics, there are a number of potential benefits for considering the use of miRNA-containing complexes. Given the relatively easy access to these complexes, particularly circulating EVs, one could envision collecting EVs from healthy donors and injecting in patients to treat disease (Figure 3). The possibility of modifying the isolated EVs with exogenous miRNA mimics or anti-miRs, as well as targeting molecules on their surface, provides an opportunity to use EVs as carriers of specific miRNA molecules to specific tissues. In some cases, it is worth asking to what extent purified EVs might be sufficient to confer the positive effects of cell therapy. For example, adipose stem cell (ADSC)-derived EVs containing high levels of miR-181b are sufficient to induce angiogenesis (Yang et al., 2018) and may explain why ADSC transplantation is beneficial for amelioration of several cardiovascular complications (Suzuki et al., 2015). Likewise, miR-181b in cardiac stem cell-derived EVs (de Couto et al., 2017) and embryonic stem cell-derived EVs (Caspi et al., 2007; Chong et al., 2014; Khan et al., 2015) protect the heart after myocardial infarction. The latter appears to be mediated in part by miR-294 and is accompanied by increased cardiac neovascularization, improved cardiac function, and increased cardiac stem cell proliferation (Khan et al., 2015). Given the easy access to EV donors and straight forward techniques to manipulate their content and delivery, we envision EV-mediated therapy as a breakthrough in pharmacology.
Figure 3. Therapeutic perspectives for extracellular miRNAs.
Circulating EV-associated miRNAs are profiled in patients to identify differentially expressed molecules. These candidates are tested in pre-clinical studies and miRNAs that play a role in disease are selected for clinical trials. Clinically relevant miRNA mimics or antimiR molecules are loaded into the patient’s own EVs and reinjected in the blood stream to restore EV miRNA levels to normal.
One could also target easily accessible tissues, like adipose tissue, to produce therapeutic miRNAs which are secreted in EVs and act on tissues at a distance. Since adipose tissue is efficient in loading miRNAs into EVs, which are well taken up by liver (and perhaps other tissues), this could provide a novel way to manipulate gene expression in these target tissues without directly introducing genetic modifications in them. This would also have the advantage that the modified adipose tissue could always be removed if any adverse effect was observed from the gene therapy.
Conclusions
Even though the field is still young, the concept of extracellular miRNAs as a physiological mechanism of intercellular communication is exciting and gaining traction, as is the prospect of using extracellular miRNAs to better stage disease as well as for treatment. Developing proper tools and standardized methodology to assess miRNA transport and delivery are the bottlenecks in the field, but these are likely to be overcome in the next few years. Overcoming these hurdles will bring the field to another level – one in which specific extracellular miRNAs can be viewed as biomarkers of different physiological and pathophysiological conditions, and miRNAs within exosomes or other EVs can be used to treat diseases in a specific and efficient manner.
Acknowledgment and funding
We acknowledge Carolina Frandsen Pereira da Costa for the illustrations and artwork. This study has been supported by grants of Fundação de Amparo à Pesquisa do Estado de São Paulo (2017/01184–9, 2017/07975–8 and 2017/23920–9) to MAM and NIH grant R01DK082659 and the Mary K. Iacocca Professorship to CRK. RGM was funded by a Deutsche Forschungsgemeinschaft (DFG) fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Muhtaresh HA, and Al-Kafaji G (2018). Evaluation of Two-Diabetes Related microRNAs Suitability as Earlier Blood Biomarkers for Detecting Prediabetes and type 2 Diabetes Mellitus. Journal of clinical medicine 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameling S, Kacprowski T, Chilukoti RK, Malsch C, Liebscher V, Suhre K, Pietzner M, Friedrich N, Homuth G, Hammer E, et al. (2015). Associations of circulating plasma microRNAs with age, body mass index and sex in a population-based study. BMC medical genomics 8, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfossi S, Babayan A, Pantel K, and Calin GA (2018). Clinical utility of circulating non-coding RNAs - an update. Nature reviews. Clinical oncology 15, 541–563. [DOI] [PubMed] [Google Scholar]
- Arner E, Mejhert N, Kulyte A, Balwierz PJ, Pachkov M, Cormont M, Lorente-Cebrian S, Ehrlund A, Laurencikiene J, Heden P, et al. (2012). Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes 61, 1986–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner P, and Kulyte A (2015). MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol 11, 276–288. [DOI] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proceedings of the National Academy of Sciences of the United States of America 108, 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Tang Y, Yu M, Wu L, Liu F, Ni J, Wang Z, Wang J, Fei J, Wang W, et al. (2017). Downregulation of blood serum microRNA 29 family in patients with Parkinson’s disease. Scientific reports 7, 5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandiera S, Pfeffer S, Baumert TF, and Zeisel MB (2015). miR-122--a key factor and therapeutic target in liver disease. J Hepatol 62, 448–457. [DOI] [PubMed] [Google Scholar]
- Bartel DP (2018). Metazoan MicroRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty M, Guduric-Fuchs J, Brown E, Bridgett S, Chakravarthy U, Hogg RE, and Simpson DA (2014). Small RNAs from plants, bacteria and fungi within the order Hypocreales are ubiquitous in human plasma. BMC genomics 15, 933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E (2011). Evolution of microRNA diversity and regulation in animals. Nat Rev Genet 12, 846–860. [DOI] [PubMed] [Google Scholar]
- Blanchard N, Lankar D, Faure F, Regnault A, Dumont C, Raposo G, and Hivroz C (2002). TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. Journal of immunology 168, 3235–3241. [DOI] [PubMed] [Google Scholar]
- Bobrie A, Colombo M, Krumeich S, Raposo G, and Thery C (2012). Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. Journal of extracellular vesicles 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JE, and Gould SJ (2006). Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. The Journal of cell biology 172, 923–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta-Orfila T, Morato X, Compta Y, Lozano JJ, Falgas N, Valldeoriola F, Pont-Sunyer C, Vilas D, Mengual L, Fernandez M, et al. (2014). Identification of blood serum micro-RNAs associated with idiopathic and LRRK2 Parkinson’s disease. Journal of neuroscience research 92, 1071–1077. [DOI] [PubMed] [Google Scholar]
- Brestoff JR, and Artis D (2015). Immune regulation of metabolic homeostasis in health and disease. Cell 161, 146–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, Rhees B, Kuslich C, Visakorpi T, and Hamdy FC (2012). Changes in circulating microRNA levels associated with prostate cancer. British journal of cancer 106, 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busatto S, Vilanilam G, Ticer T, Lin WL, Dickson DW, Shapiro S, Bergese P, and Wolfram J (2018). Tangential Flow Filtration for Highly Efficient Concentration of Extracellular Vesicles from Large Volumes of Fluid. Cells 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campello E, Zabeo E, Radu CM, Spiezia L, Foletto M, Prevedello L, Gavasso S, Bulato C, Vettor R, and Simioni P (2016). Dynamics of circulating microparticles in obesity after weight loss. Internal and emergency medicine 11, 695–702. [DOI] [PubMed] [Google Scholar]
- Can U, Buyukinan M, and Yerlikaya FH (2016). The investigation of circulating microRNAs associated with lipid metabolism in childhood obesity. Pediatric obesity 11, 228–234. [DOI] [PubMed] [Google Scholar]
- Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, and Hotamisligil GS (2008). Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell 134, 933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H (2014). Adipocytokines in obesity and metabolic disease. J Endocrinol 220, T47–T59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caravia XM, Fanjul V, Oliver E, Roiz-Valle D, Moran-Alvarez A, Desdin-Mico G, Mittelbrunn M, Cabo R, Vega JA, Rodriguez F, et al. (2018). The microRNA-29/PGC1alpha regulatory axis is critical for metabolic control of cardiac function. PLoS biology 16, e2006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi O, Huber I, Kehat I, Habib M, Arbel G, Gepstein A, Yankelson L, Aronson D, Beyar R, and Gepstein L (2007). Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. Journal of the American College of Cardiology 50, 1884–1893. [DOI] [PubMed] [Google Scholar]
- Castano C, Kalko S, Novials A, and Parrizas M (2018). Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proceedings of the National Academy of Sciences of the United States of America 115, 12158–12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi A, Naffah de Souza C, Camara NO, and Moraes-Vieira PM (2015). The Macrophage Switch in Obesity Development. Frontiers in immunology 6, 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceolotto G, Giannella A, Albiero M, Kuppusamy M, Radu C, Simioni P, Garlaschelli K, Baragetti A, Catapano AL, Iori E, et al. (2017). miR-30c-5p regulates macrophage-mediated inflammation and pro-atherosclerosis pathways. Cardiovascular research 113, 1627–1638. [DOI] [PubMed] [Google Scholar]
- Chartoumpekis DV, Zaravinos A, Ziros PG, Iskrenova RP, Psyrogiannis AI, Kyriazopoulou VE, and Habeos IG (2012). Differential expression of microRNAs in adipose tissue after long-term high-fat diet-induced obesity in mice. PloS one 7, e34872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, and Hannon GJ (2010). A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465, 584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Wang Y, Yang S, Li H, Zhao G, Wang F, Yang L, and Wang DW (2015). MiR-320a contributes to atherogenesis by augmenting multiple risk factors and down-regulating SRF. Journal of cellular and molecular medicine 19, 970–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Liu L, Ma F, Wong CW, Guo XE, Chacko JV, Farhoodi HP, Zhang SX, Zimak J, Segaliny A, et al. (2016). Elucidation of Exosome Migration across the Blood-Brain Barrier Model In Vitro. Cellular and molecular bioengineering 9, 509–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Cui C, Yang X, Xu J, Venkat P, Zacharek A, Yu P, and Chopp M (2017). MiR-126 Affects Brain-Heart Interaction after Cerebral Ischemic Stroke. Transl Stroke Res 8, 374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, et al. (2008). Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proceedings of the National Academy of Sciences of the United States of America 105, 2111–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Doecke JD, Sharples RA, Villemagne VL, Fowler CJ, Rembach A, Martins RN, Rowe CC, Macaulay SL, Masters CL, et al. (2015). Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Molecular psychiatry 20, 1188–1196. [DOI] [PubMed] [Google Scholar]
- Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, et al. (2014). Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proceedings of the National Academy of Sciences of the United States of America 111, 14888–14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CHJ, and Cohen P (2017). Adipose crosstalk with other cell types in health and disease. Exp Cell Res 360, 6–11. [DOI] [PubMed] [Google Scholar]
- Chong JJ, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, et al. (2014). Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 510, 273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang TY, Wu HL, Chen CC, Gamboa GM, Layman LC, Diamond MP, Azziz R, and Chen YH (2015). MicroRNA-223 Expression is Upregulated in Insulin Resistant Human Adipose Tissue. J Diabetes Res 2015, 943659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocucci E, and Meldolesi J (2015). Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol 25, 364–372. [DOI] [PubMed] [Google Scholar]
- EV-TRACK Consortium, Van Deun J, Mestdagh P, Agostinis P, Akay O, Anand S, Anckaert J, Martinez ZA, Baetens T, Beghein E, et al. (2017). EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nature methods 14, 228–232. [DOI] [PubMed] [Google Scholar]
- Crewe C, Joffin N, Rutkowski JM, Kim M, Zhang F, Towler DA, Gordillo R, and Scherer PE (2018). An Endothelial-to-Adipocyte Extracellular Vesicle Axis Governed by Metabolic State. Cell 175, 695–708.e613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Ansel KM, Bitzer M, Breakefield XO, Charest A, Galas DJ, Gerstein MB, Gupta M, Milosavljevic A, McManus MT, et al. (2019). The Extracellular RNA Communication Consortium: Establishing Foundational Knowledge and Technologies for Extracellular RNA Research. Cell 177, 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Couto G, Gallet R, Cambier L, Jaghatspanyan E, Makkar N, Dawkins JF, Berman BP, and Marban E (2017). Exosomal MicroRNA Transfer Into Macrophages Mediates Cellular Postconditioning. Circulation 136, 200–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, and Slack FJ (2010). MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol 20, 2159–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva N, Samblas M, Martínez JA, Milagro FI (2018). Effects of exosomes from LPS-activated macrophages on adipocyte gene expression, differentiation, and insulin-dependent glucose uptake. J Physiol Biochem 74, 559–568. [DOI] [PubMed] [Google Scholar]
- Deline B, Greenwood JM, Clark JW, Puttick MN, Peterson KJ, and Donoghue PCJ (2018). Evolution of metazoan morphological disparity. Proceedings of the National Academy of Sciences of the United States of America 115, E8909–e8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng ZB, Poliakov A, Hardy RW, Clements R, Liu C, Liu Y, Wang J, Xiang X, Zhang S, Zhuang X, et al. (2009). Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes 58, 2498–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhahbi JM, Spindler SR, Atamna H, Yamakawa A, Guerrero N, Boffelli D, Mote P, and Martin DIK (2013). Deep sequencing identifies circulating mouse miRNAs that are functionally implicated in manifestations of aging and responsive to calorie restriction. Aging 5, 130–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Lottaz C, Borodina T, and Himmelbauer H (2008). Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res 36, e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Wang C, Lu S, Yu C, Huang L, Feng W, Xu H, Chen X, Zen K, Yan Q, et al. (2016). A panel of four decreased serum microRNAs as a novel biomarker for early Parkinson’s disease. Biomarkers : biochemical indicators of exposure, response, and susceptibility to chemicals 21, 129–137. [DOI] [PubMed] [Google Scholar]
- Du WW, Yang W, Fang L, Xuan J, Li H, Khorshidi A, Gupta S, Li X, and Yang BB (2014). miR-17 extends mouse lifespan by inhibiting senescence signaling mediated by MKP7. Cell death & disease 5, e1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, and van Obberghen E (2008). miR-375 targets 3’-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes 57, 2708–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. (2006). miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell metabolism 3, 87–98. [DOI] [PubMed] [Google Scholar]
- Fendler A, Stephan C, Yousef GM, Kristiansen G, and Jung K (2016). The translational potential of microRNAs as biofluid markers of urological tumours. Nature reviews. Urology 13, 734–752. [DOI] [PubMed] [Google Scholar]
- Ferrante SC, Nadler EP, Pillai DK, Hubal MJ, Wang Z, Wang JM, Gordish-Dressman H, Koeck E, Sevilla S, Wiles AA, et al. (2015). Adipocyte-derived exosomal miRNAs: a novel mechanism for obesity-related disease. 77, 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Roxe T, Muller-Ardogan M, et al. (2010). Circulating microRNAs in patients with coronary artery disease. Circulation research 107, 677–684. [DOI] [PubMed] [Google Scholar]
- Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, Regen T, Hanisch UK, and Simons M (2011). Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. Journal of cell science 124, 447–458. [DOI] [PubMed] [Google Scholar]
- Flaherty SE 3rd, Grijalva A, Xu X, Ables E, Nomani A, and Ferrante AW Jr. (2019) A lipase-independent pathway of lipid release and immune modulation by adipocytes. Science 363, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers E, Won GY, and Fukuoka Y (2015). MicroRNAs associated with exercise and diet: a systematic review. Physiological genomics 47, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Garagnani P, Parini P, Giuliani C, and Santoro A (2018). Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat Rev Endocrinol 14, 576–590. [DOI] [PubMed] [Google Scholar]
- Galimberti D, Villa C, Fenoglio C, Serpente M, Ghezzi L, Cioffi SM, Arighi A, Fumagalli G, and Scarpini E (2014). Circulating miRNAs as potential biomarkers in Alzheimer’s disease. Journal of Alzheimer’s disease : JAD 42, 1261–1267. [DOI] [PubMed] [Google Scholar]
- Gibbings DJ, Ciaudo C, Erhardt M, and Voinnet O (2009). Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 11, 1143–1149. [DOI] [PubMed] [Google Scholar]
- Glass CK, and Olefsky JM (2012). Inflammation and lipid signaling in the etiology of insulin resistance. Cell metabolism 15, 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren Y, Kushnir M, Zafrir B, Tabak S, Lewis BS, and Amir O (2012). Serum levels of microRNAs in patients with heart failure. European journal of heart failure 14, 147–154. [DOI] [PubMed] [Google Scholar]
- Grasedieck S, Scholer N, Bommer M, Niess JH, Tumani H, Rouhi A, Bloehdorn J, Liebisch P, Mertens D, Dohner H, et al. (2012). Impact of serum storage conditions on microRNA stability. Leukemia 26, 2414–2416. [DOI] [PubMed] [Google Scholar]
- Guay C, Kruit JK, Rome S, Menoud V, Mulder NL, Jurdzinski A, Mancarella F, Sebastiani G, Donda A, Gonzalez BJ, et al. (2019). Lymphocyte-Derived Exosomal MicroRNAs Promote Pancreatic beta Cell Death and May Contribute to Type 1 Diabetes Development. Cell metabolism 29, 348–361. [DOI] [PubMed] [Google Scholar]
- Ha M, and Kim VN (2014). Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15, 509–524. [DOI] [PubMed] [Google Scholar]
- Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB, Hammerschmidt W, O’Neill LA, and Masters SL (2012). Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. Journal of immunology 189, 3795–3799. [DOI] [PubMed] [Google Scholar]
- Hansen KD, Brenner SE, and Dudoit S (2010). Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res 38, e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraszti RA, Miller R, Stoppato M, Sere YY, Coles A, Didiot MC, Wollacott R, Sapp E, Dubuke ML, Li X, et al. (2018). Exosomes Produced from 3D Cultures of MSCs by Tangential Flow Filtration Show Higher Yield and Improved Activity. Mol Ther 26, 2838–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Wu C, Xiao J, Li D, Sun Z, and Li M (2018). Endothelial extracellular vesicles modulate the macrophage phenotype: Potential implications in atherosclerosis. Scand J Immunol 87, e12648. [DOI] [PubMed] [Google Scholar]
- He Y, Lin J, Kong D, Huang M, Xu C, Kim TK, Etheridge A, Luo Y, Ding Y, and Wang K (2015). Current State of Circulating MicroRNAs as Cancer Biomarkers. Clinical chemistry 61, 1138–1155. [DOI] [PubMed] [Google Scholar]
- Heneghan HM, Miller N, McAnena OJ, O’Brien T, and Kerin MJ (2011). Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. The Journal of clinical endocrinology and metabolism 96, E846–850. [DOI] [PubMed] [Google Scholar]
- Higuchi C, Nakatsuka A, Eguchi J, Teshigawara S, Kanzaki M, Katayama A, Yamaguchi S, Takahashi N, Murakami K, Ogawa D, et al. (2015). Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism: clinical and experimental 64, 489–497. [DOI] [PubMed] [Google Scholar]
- Holland WL, Miller RA, Wang ZV, Sun K, Barth BM, Bui HH, Davis KE, Bikman BT, Halberg N, Rutkowski JM, et al. (2011). Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nat Med. 17, 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Li Y, and Su B (2017). Identification of Circulating miR-125b as a Potential Biomarker of Alzheimer’s Disease in APP/PS1 Transgenic Mouse. Journal of Alzheimer’s disease : JAD 59, 1449–1458. [DOI] [PubMed] [Google Scholar]
- Horaka MN,J; Bienertova-Vaskuac J (2016). Muscle-specific microRNAs in skeletal muscle development. Developmental Biology 410, 1–13. [DOI] [PubMed] [Google Scholar]
- Horibe S, Tanahashi T, Kawauchi S, Murakami Y, and Rikitake Y (2018). Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer 18, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS (2017). Inflammation, metaflammation and immunometabolic disorders. Nature 542, 177–185. [DOI] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, and Sharp PA (2003). Embryonic stem cell-specific MicroRNAs. Dev Cell 5, 351–358. [DOI] [PubMed] [Google Scholar]
- Hsieh CH, Rau CS, Wu SC, Yang JC, Wu YC, Lu TH, Tzeng SL, Wu CJ, and Lin CW (2015). Weight-reduction through a low-fat diet causes differential expression of circulating microRNAs in obese C57BL/6 mice. BMC genomics 16, 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Ge X, Yu J, Han Z, Yin Z, Li Y, Chen F, Wang H, Zhang J, and Lei P (2018). Increased miR-124–3p in microglial exosomes following traumatic brain injury inhibits neuronal inflammation and contributes to neurite outgrowth via their transfer into neurons. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 32, 512–528. [DOI] [PubMed] [Google Scholar]
- Huang Y, Liu Y, Li L, Su B, Yang L, Fan W, Yin Q, Chen L, Cui T, Zhang J, et al. (2014). Involvement of inflammation-related miR-155 and miR-146a in diabetic nephropathy: implications for glomerular endothelial injury. BMC nephrology 15, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubal MJ, Nadler EP, Ferrante SC, Barberio MD, Suh JH, Wang J, Dohm GL, Pories WJ, Mietus-Snyder M, and Freishtat RJ (2017). Circulating adipocyte-derived exosomal MicroRNAs associated with decreased insulin resistance after gastric bypass. Obesity 25, 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, Hellemans J, Kubista M, Mueller RD, Nolan T, et al. (2013). The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clinical chemistry 59, 892–902. [DOI] [PubMed] [Google Scholar]
- Ibanez-Ventoso C, Yang M, Guo S, Robins H, Padgett RW, and Driscoll M (2006). Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging cell 5, 235–246. [DOI] [PubMed] [Google Scholar]
- Ismail N, Wang Y, Dakhlallah D, Moldovan L, Agarwal K, Batte K, Shah P, Wisler J, Eubank TD, Tridandapani S, et al. (2013). Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood 121, 984–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen F, Yang X, Proebsting S, Hoelscher M, Przybilla D, Baumann K, Schmitz T, Dolf A, Endl E, Franklin BS, et al. (2014). MicroRNA expression in circulating microvesicles predicts cardiovascular events in patients with coronary artery disease. Journal of the American Heart Association 3, e001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, et al. (2019) Reassessment of Exosome Composition. Cell 177, 428–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia LH, and Liu YN (2016). Downregulated serum miR-223 servers as biomarker in Alzheimer’s disease. Cell biochemistry and function 34, 233–237. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Wang HY, Li Y, Guo SH, Zhang L, and Cai JH (2014). Peripheral blood miRNAs as a biomarker for chronic cardiovascular diseases. Scientific reports 4, 5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A, Danielson KM, Benton MC, Ziegler O, Shah R, Stubbs RS, Das S, and Macartney-Coxson D (2017). miRNA Signatures of Insulin Resistance in Obesity. Obesity 25, 1734–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C (2012). Liver-specific microRNA-122: Biogenesis and function. RNA Biol 9, 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolina DS, Tavintharan S, Armugam A, Sepramaniam S, Pek SL, Wong MT, Lim SC, Sum CF, and Jeyaseelan K (2012). Circulating miRNA profiles in patients with metabolic syndrome. The Journal of clinical endocrinology and metabolism 97, E2271–2276. [DOI] [PubMed] [Google Scholar]
- Katayama M, Wiklander OPB, Fritz T, Caidahl K, Andaloussi SE, Zierath JR, and Krook A (2018). Circulating Exosomal miR-20b-5p is Elevated in Type 2 Diabetes and Could Impair Insulin Action in Human Skeletal Muscle. Diabetes 68, 515–526. [DOI] [PubMed] [Google Scholar]
- Kawase-Koga Y, Otaegi G, and Sun T (2009). Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn 238, 2800–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T, Boeckel JN, Gross S, Klotsche J, Palapies L, Leistner D, Pieper L, Stalla GK, Lehnert H, Silber S, et al. (2017). Improved risk stratification in prevention by use of a panel of selected circulating microRNAs. Scientific reports 7, 4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Nickoloff E, Abramova T, Johnson J, Verma SK, Krishnamurthy P, Mackie AR, Vaughan E, Garikipati VNS, Benedict C, et al. (2015). Embryonic Stem Cell-Derived Exosomes Promote Endogenous Repair Mechanisms and Enhance Cardiac Function Following Myocardial Infarction. Circulation research 117, 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Cho H, Alexander R, Patterson HC, Gu M, Lo KA, Xu D, Goh VJ, Nguyen LN, Chai X, et al. (2014). MicroRNAs are required for the feature maintenance and differentiation of brown adipocytes. Diabetes 63, 4045–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman MS, Briscoe VJ, Clark N, Florez H, Haas LB, Halter JB, Huang ES, Korytkowski MT, Munshi MN, Odegard PS, et al. (2012). Diabetes in older adults. Diabetes care 35, 2650–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, and Plasterk RH (2007). Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS biology 5, e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloting N, Berthold S, Kovacs P, Schon MR, Fasshauer M, Ruschke K, Stumvoll M, and Bluher M (2009). MicroRNA expression in human omental and subcutaneous adipose tissue. PloS one 4, e4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeck ES, Iordanskaia T, Sevilla S, Ferrante SC, Hubal MJ, Freishtat RJ, and Nadler EP (2014). Adipocyte exosomes induce transforming growth factor beta pathway dysregulation in hepatocytes: a novel paradigm for obesity-related liver disease. 192, 268–275. [DOI] [PubMed] [Google Scholar]
- Kong L, Zhu J, Han W, Jiang X, Xu M, Zhao Y, Dong Q, Pang Z, Guan Q, Gao L, et al. (2011). Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol 48, 61–69. [DOI] [PubMed] [Google Scholar]
- Kopecki Z, Luchetti MM, Adams DH, Strudwick X, Mantamadiotis T, Stoppacciaro A, Gabrielli A, Ramsay RG, and Cowin AJ (2007). Collagen loss and impaired wound healing is associated with c-Myb deficiency. J Pathol 211, 351–361. [DOI] [PubMed] [Google Scholar]
- Kovacs B, Lumayag S, Cowan C, and Xu S (2011). MicroRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Investigative ophthalmology & visual science 52, 4402–4409. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, and Stoffel M (2005). Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438, 685–689. [DOI] [PubMed] [Google Scholar]
- Kumar P, Dezso Z, MacKenzie C, Oestreicher J, Agoulnik S, Byrne M, Bernier F, Yanagimachi M, Aoshima K, and Oda Y (2013). Circulating miRNA biomarkers for Alzheimer’s disease. PloS one 8, e69807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey DE, and Olefsky JM (2016). Regulation of metabolism by the innate immune system. Nat Rev Endocrinol 12, 15–28. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, and Tuschl T (2002). Identification of tissue-specific microRNAs from mouse. Curr Biol 12, 735–739. [DOI] [PubMed] [Google Scholar]
- Lahmy R, Soleimani M, Sanati MH, Behmanesh M, Kouhkan F, and Mobarra N (2014). MiRNA-375 promotes beta pancreatic differentiation in human induced pluripotent stem (hiPS) cells. Molecular biology reports 41, 2055–2066. [DOI] [PubMed] [Google Scholar]
- Lai CP, Mardini O, Ericsson M, Prabhakar S, Maguire C, Chen JW, Tannous BA, and Breakefield XO (2014). Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano 8, 483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latreille M, Herrmanns K, Renwick N, Tuschl T, Malecki MT, McCarthy MI, Owen KR, Rulicke T, and Stoffel M (2015). miR-375 gene dosage in pancreatic beta-cells: implications for regulation of beta-cell mass and biomarker development. Journal of molecular medicine 93, 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidinger P, Backes C, Deutscher S, Schmitt K, Mueller SC, Frese K, Haas J, Ruprecht K, Paul F, Stahler C, et al. (2013). A blood based 12-miRNA signature of Alzheimer disease patients. Genome biology 14, R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Bates DJ, An J, Terry DA, and Wang E (2011). Up-regulation of key microRNAs, and inverse down-regulation of their predicted oxidative phosphorylation target genes, during aging in mouse brain. Neurobiology of aging 32, 944–955. [DOI] [PubMed] [Google Scholar]
- Li S, Wei J, Zhang C, Li X, Meng W, Mo X, Zhang Q, Liu Q, Ren K, Du R, et al. (2016). Cell-Derived Microparticles in Patients with Type 2 Diabetes Mellitus: a Systematic Review and Meta-Analysis. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology 39, 2439–2450. [DOI] [PubMed] [Google Scholar]
- Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, and Liu ZG (2010). MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nature immunology 11, 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YZ, Li JJ, Xiao HB, He Y, Zhang L, and Yan YX (2018). Identification of stress-related microRNA biomarkers in type 2 diabetes mellitus: A systematic review and meta-analysis. Journal of diabetes. [DOI] [PubMed] [Google Scholar]
- Lin X, Qin Y, Jia J, Lin T, Lin X, Chen L, Zeng H, Han Y, Wu L, Huang S, et al. (2016). MiR-155 Enhances Insulin Sensitivity by Coordinated Regulation of Multiple Genes in Mice. PLoS genetics 12, e1006308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S, Nanhwan M, Qian J, Kodakandla M, Castillo AC, Thomas B, Liu H, and Ye Y (2013). Modulation of microRNAs in hypertension-induced arterial remodeling through the beta1 and beta3-adrenoreceptor pathways. Journal of molecular and cellular cardiology 65, 127–136. [DOI] [PubMed] [Google Scholar]
- Liu W, Baker RD, Bhatia T, Zhu L, and Baker SS (2016). Pathogenesis of nonalcoholic steatohepatitis. Cellular and molecular life sciences : CMLS 73, 1969–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, and Kroemer G (2013). The hallmarks of aging. Cell 153, 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Buchan RJ, and Cook S.A.J.C.r. (2010). MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. 86, 410–420. [DOI] [PubMed] [Google Scholar]
- Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stahler C, Meese E, et al. (2016). Distribution of miRNA expression across human tissues. Nucleic Acids Res 44, 3865–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli G, Cohen AM, Bennett DA, Shah RC, Fields CJ, Hernandez AG, and Smalheiser NR (2015). Plasma Exosomal miRNAs in Persons with and without Alzheimer Disease: Altered Expression and Prospects for Biomarkers. PloS one 10, e0139233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynes MD, Leiria LO, Lundh M, Bartelt A, Shamsi F, Huang TL, Takahashi H, Hirshman MF, Schlein C, Lee A, et al. (2017). The cold-induced lipokine 12,13-diHOME promotes fatty acid transport into brown adipose tissue. Nature medicine 23, 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, and German MS (2007). MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes 56, 2938–2945. [DOI] [PubMed] [Google Scholar]
- Ma W, Li Y, Wang C, Xu F, Wang M, and Liu Y (2016). Serum miR-221 serves as a biomarker for Parkinson’s disease. Cell biochemistry and function 34, 511–515. [DOI] [PubMed] [Google Scholar]
- Machida T, Tomofuji T, Ekuni D, Maruyama T, Yoneda T, Kawabata Y, Mizuno H, Miyai H, Kunitomo M, and Morita M (2015). MicroRNAs in Salivary Exosome as Potential Biomarkers of Aging. International journal of molecular sciences 16, 21294–21309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes OC, An J, Sarojini H, and Wang E (2008). Murine microRNAs implicated in liver functions and aging process. Mechanisms of ageing and development 129, 534–541. [DOI] [PubMed] [Google Scholar]
- Margis R, Margis R, and Rieder CR (2011). Identification of blood microRNAs associated to Parkinsonis disease. J Biotechnol 152, 96–101. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. (2008). Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B, Kowal EJK, van Balkom BWM, Bartel S, Bhattacharyya SN, Buzás EI, Buck AH, de Candia P, Chow FWN, Das S, et al. (2017). Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. Journal of extracellular vesicles 6, 1286095–1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M, Martin-Jaular L, Lavieu G, and Thery C (2019) Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 21, 9–17. [DOI] [PubMed] [Google Scholar]
- Mathis D (2013). Immunological goings-on in visceral adipose tissue. Cell metabolism 17, 851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto J, Stewart T, Banks WA, and Zhang J (2017). The Transport Mechanism of Extracellular Vesicles at the Blood-Brain Barrier. Current pharmaceutical design 23, 6206–6214. [DOI] [PubMed] [Google Scholar]
- McKelvey KJ, Powell KL, Ashton AW, Morris JM, and McCracken SA (2015). Exosomes: Mechanisms of Uptake. Journal of circulating biomarkers 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie AJ, Hoshino D, Hong NH, Cha DJ, Franklin JL, Coffey RJ, Patton JG, and Weaver AM (2016). KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. Cell Rep 15, 978–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerson A, Traurig M, Ossowski V, Fleming JM, Mullins M, and Baier LJ (2013). Human adipose microRNA-221 is upregulated in obesity and affects fat metabolism downstream of leptin and TNF-alpha. Diabetologia 56, 1971–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, et al. (2014). Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America 105, 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, and Sanchez-Madrid F (2011). Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocharla P, Briand S, Giannotti G, Dörries C, Jakob P, Paneni F, Lüscher T, and Landmesser U (2013). AngiomiR-126 expression and secretion from circulating CD34+ and CD14+ PBMCs: role for pro-angiogenic effects and alterations in type-2 diabetics. Blood 121, 226–236. [DOI] [PubMed] [Google Scholar]
- Mori MA, Liu M, Bezy O, Almind K, Shapiro H, Kasif S, and Kahn CR (2010). A systems biology approach identifies inflammatory abnormalities between mouse strains prior to development of metabolic disease. Diabetes 59, 2960–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori MA, Raghavan P, Thomou T, Boucher J, Robida-Stubbs S, Macotela Y, Russell SJ, Kirkland JL, Blackwell TK, and Kahn CR (2012). Role of microRNA processing in adipose tissue in stress defense and longevity. Cell metabolism 16, 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori MA, Thomou T, Boucher J, Lee KY, Lallukka S, Kim JK, Torriani M, Yki J, xE, rvinen H, et al. (2014). Altered miRNA processing disrupts brown/white adipocyte determination and associates with lipodystrophy. The Journal of Clinical Investigation 124, 3339–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy LA, Pink RC, and Carter DR (2014). Routes and mechanisms of extracellular vesicle uptake. Journal of extracellular vesicles 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller G, Schneider M, Biemer-Daub G, and Wied S.J.C.s. (2011). Microvesicles released from rat adipocytes and harboring glycosylphosphatidylinositol-anchored proteins transfer RNA stimulating lipid synthesis. 23, 1207–1223. [DOI] [PubMed] [Google Scholar]
- Murillo OD, Thistlethwaite W, Rozowsky J, Subramanian SL, Lucero R, Shah N, Jackson AR, Srinivasan S, Chung A, Laurent CD, et al. (2019). exRNA Atlas Analysis Reveals Distinct Extracellular RNA Cargo Types and Their Carriers Present across Human Biofluids. Cell 177, 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan G, Kredo-Russo S, Geiger T, Lenz A, Kaspi H, Hornstein E, and Efrat S (2015). MiR-375 promotes redifferentiation of adult human beta cells expanded in vitro. PloS one 10, e0122108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedaeinia R, Manian M, Jazayeri MH, Ranjbar M, Salehi R, Sharifi M, Mohaghegh F, Goli M, Jahednia SH, Avan A, et al. (2017). Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer gene therapy 24, 48–56. [DOI] [PubMed] [Google Scholar]
- Nidadavolu LS, Niedernhofer LJ, and Khan SA (2013). Identification of microRNAs dysregulated in cellular senescence driven by endogenous genotoxic stress. Aging 5, 460–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, and Evans MK (2010). microRNA expression patterns reveal differential expression of target genes with age. PloS one 5, e10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren Hooten N, Fitzpatrick M, Wood WH 3rd, De S, Ejiogu N, Zhang Y, Mattison JA, Becker KG, Zonderman AB, and Evans MK (2013). Age-related changes in microRNA levels in serum. Aging 5, 725–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez Lopez YO, Garufi G, and Seyhan AA (2016). Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Molecular bioSystems 13, 106–121. [DOI] [PubMed] [Google Scholar]
- O’Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, Swanson MS, and Harfe BD (2007). Essential role for Dicer during skeletal muscle development. Dev Biol 311, 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, and Chawla A (2013). Pleiotropic actions of insulin resistance and inflammation in metabolic homeostasis. Science 339, 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliverio M, Schmidt E, Mauer J, Baitzel C, Hansmeier N, Khani S, Konieczka S, Pradas-Juni M, Brodesser S, Van TM, et al. (2016). Dicer1-miR-328-Bace1 signalling controls brown adipose tissue differentiation and function. Nat Cell Biol 18, 328–336. [DOI] [PubMed] [Google Scholar]
- Olivieri F, Bonafe M, Spazzafumo L, Gobbi M, Prattichizzo F, Recchioni R, Marcheselli F, La Sala L, Galeazzi R, Rippo MR, et al. (2014). Age- and glycemia-related miR-126–3p levels in plasma and endothelial cells. Aging 6, 771–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri F, Spazzafumo L, Santini G, Lazzarini R, Albertini MC, Rippo MR, Galeazzi R, Abbatecola AM, Marcheselli F, Monti D, et al. (2012). Age-related differences in the expression of circulating microRNAs: miR-21 as a new circulating marker of inflammaging. Mechanisms of ageing and development 133, 675–685. [DOI] [PubMed] [Google Scholar]
- Ono K, Igata M, Kondo T, Kitano S, Takaki Y, Hanatani S, Sakaguchi M, Goto R, Senokuchi T, Kawashima J, et al. (2018). Identification of microRNA that represses IRS-1 expression in liver. PloS one 13, e0191553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega FJ, Mercader JM, Catalan V, Moreno-Navarrete JM, Pueyo N, Sabater M, Gomez-Ambrosi J, Anglada R, Fernandez-Formoso JA, Ricart W, et al. (2013). Targeting the circulating microRNA signature of obesity. Clinical chemistry 59, 781–792. [DOI] [PubMed] [Google Scholar]
- Ortega FJ, Mercader JM, Moreno-Navarrete JM, Rovira O, Guerra E, Esteve E, Xifra G, Martinez C, Ricart W, Rieusset J, et al. (2014). Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes care 37, 1375–1383. [DOI] [PubMed] [Google Scholar]
- Ortega FJ, Moreno M, Mercader JM, Moreno-Navarrete JM, Fuentes-Batllevell N, Sabater M, Ricart W, and Fernandez-Real JM (2015). Inflammation triggers specific microRNA profiles in human adipocytes and macrophages and in their supernatants. Clinical epigenetics 7, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega FJ, Moreno-Navarrete JM, Pardo G, Sabater M, Hummel M, Ferrer A, Rodriguez-Hermosa JI, Ruiz B, Ricart W, Peral B, et al. (2010). MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PloS one 5, e9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, et al. (2010). Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 12, 19–30; sup pp 11–13. [DOI] [PubMed] [Google Scholar]
- Pescador N, Perez-Barba M, Ibarra JM, Corbaton A, Martinez-Larrad MT, and Serrano-Rios M (2013). Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PloS one 8, e77251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirola CJ, Fernandez Gianotti T, Castano GO, Mallardi P, San Martino J, Mora Gonzalez Lopez Ledesma, M., Flichman D, Mirshahi F, Sanyal AJ, and Sookoian S (2015). Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut 64, 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povero D, Eguchi A, Li H, Johnson CD, Papouchado BG, Wree A, Messer K, and Feldstein AE (2014). Circulating extracellular vesicles with specific proteome and liver microRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PloS one 9, e113651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. (2004). A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432, 226–230. [DOI] [PubMed] [Google Scholar]
- Prud’homme GJ, Glinka Y, Lichner Z, and Yousef GM (2016). Neuropilin-1 is a receptor for extracellular miRNA and AGO2/miRNA complexes and mediates the internalization of miRNAs that modulate cell function. Oncotarget 7, 68057–68071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusic AD, and Kraig RP (2014). Youth and environmental enrichment generate serum exosomes containing miR-219 that promote CNS myelination. Glia 62, 284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis FC, Branquinho JL, Brandão BB, Guerra BA, Silva ID, Frontini A, Thomou T, Sartini L, Cinti S, and Kahn CRJA (2016). Fat-specific Dicer deficiency accelerates aging and mitigates several effects of dietary restriction in mice. 8, 1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rome S (2015). Use of miRNAs in biofluids as biomarkers in dietary and lifestyle intervention studies. Genes & nutrition 10, 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y, Bao W, Shan Z, Liu J, Yu X, Xia S, Gao H, Wang X, Yao P, Hu FB, et al. (2013). Increased microRNA-146a levels in plasma of patients with newly diagnosed type 2 diabetes mellitus. PloS one 8, e73272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Hernandez H, Chigurupati S, Raymick J, Robinson B, Cuevas E, Hanig J, and Sarkar S (2018). Identification of altered microRNAs in serum of a mouse model of Parkinson’s disease. Neuroscience letters 687, 1–9. [DOI] [PubMed] [Google Scholar]
- Rozowsky J, Kitchen RR, Park JJ, Galeev TR, Diao J, Warrell J, Thistlethwaite W, Subramanian SL, Milosavljevic A, and Gerstein M (2019). exceRpt: A Comprehensive Analytic Platform for Extracellular RNA Profiling. Cell Syst 8, 352–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safdar A, Saleem A, and Tarnopolsky MA (2016). The potential of endurance exercise-derived exosomes to treat metabolic diseases. Nat Rev Endocrinol 12, 504–517. [DOI] [PubMed] [Google Scholar]
- Safdar A, and Tarnopolsky MA (2018). Exosomes as Mediators of the Systemic Adaptations to Endurance Exercise. Cold Spring Harb Perspect Med 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo A, Imbruce P, Gardenghi B, Belli L, Agushi R, Tamanini A, Munari S, Bossi AM, Scambi I, Benati D, et al. (2018). A microRNA signature from serum exosomes of patients with glioma as complementary diagnostic biomarker. Journal of neuro-oncology 136, 51–62. [DOI] [PubMed] [Google Scholar]
- Schulte C, Molz S, Appelbaum S, Karakas M, Ojeda F, Lau DM, Hartmann T, Lackner KJ, Westermann D, Schnabel RB, et al. (2015). miRNA-197 and miRNA-223 Predict Cardiovascular Death in a Cohort of Patients with Symptomatic Coronary Artery Disease. PloS one 10, e0145930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S, Ogawa R, Ito R, Hiraoka N, McManus MT, Kanai Y, and Hebrok M (2009). Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology 136, 2304–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Z, Qin S, Li W, Wu W, Yang J, Chu M, Li X, Huo Y, Schaer GL, Wang S, et al. (2015). An Endocrine Genetic Signal Between Blood Cells and Vascular Smooth Muscle Cells: Role of MicroRNA-223 in Smooth Muscle Function and Atherogenesis. Journal of the American College of Cardiology 65, 2526–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi A, Afshari JT, and Sahebkar A (2018). Macrophage plasticity, polarization, and function in health and disease. 233, 6425–6440. [DOI] [PubMed] [Google Scholar]
- Shen Y, Wollam J, Magner D, Karalay O, and Antebi A (2012). A steroid receptor-microRNA switch regulates life span in response to signals from the gonad. Science 338, 1472–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Zhao C, Guo X, Ding H, Cui Y, Shen R, and Liu J (2014). Differential expression of microRNAs in omental adipose tissue from gestational diabetes mellitus subjects reveals miR-222 as a regulator of ERalpha expression in estrogen-induced insulin resistance. Endocrinology 155, 1982–1990. [DOI] [PubMed] [Google Scholar]
- Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, and Schekman R (2016). Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff MJ, Temoche-Diaz MM, and Schekman R (2018). Extracellular Vesicles and Cancer: Caveat Lector. Annual Review of Cancer Biology 2, 395–411. [Google Scholar]
- Smith-Vikos T, and Slack FJ (2012). MicroRNAs and their roles in aging. Journal of cell science 125, 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, and Hussain MM (2013). MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nature medicine 19, 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, Shim SG, and Paik YH (2015). Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Experimental & molecular medicine 47, e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, Ibberson M, and De Palma M (2014). Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep 8, 1432–1446. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Yeri A, Cheah PS, Chung A, Danielson K, De Hoff P, Filant J, Laurent CD, Laurent LD, Magee R, et al. (2019). Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation. Cell 177, 446–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford KI, Lynes MD, Takahashi H, Baer LA, Arts PJ, May FJ, Lehnig AC, Middelbeek RJW, Richard JJ, So K, et al. (2018). 12,13-diHOME: An Exercise-Induced Lipokine that Increases Skeletal Muscle Fatty Acid Uptake. Cell metabolism 27, 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanian A, Bourguignat L, Hennou S, Coupaye M, Hajage D, Salomon L, Alessi MC, Msika S, and de Prost D (2013). Microparticle increase in severe obesity: not related to metabolic syndrome and unchanged after massive weight loss. Obesity 21, 2236–2243. [DOI] [PubMed] [Google Scholar]
- Stern JH, Rutkowski JM, and Scherer PE (2016). Adiponectin, Leptin, and Fatty Acids in the Maintenance of Metabolic Homeostasis through Adipose Tissue Crosstalk. Cell metabolism 23, 770–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Chang X, Yin L, Li J, Zhou T, Zhang C, and Chen X (2014). Expression and DNA methylation status of microRNA-375 in patients with type 2 diabetes mellitus. Molecular medicine reports 9, 967–972. [DOI] [PubMed] [Google Scholar]
- Suzuki E, Fujita D, Takahashi M, Oba S, and Nishimatsu H (2015). Adipose tissue-derived stem cells as a therapeutic tool for cardiovascular disease. World journal of cardiology 7, 454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MD, Sagare AP, and Zlokovic BV (2018). Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat Rev Neurol 14, 133–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Nishikawa M, Shinotsuka H, Matsui Y, Ohara S, Imai T, and Takakura Y (2013). Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J Biotechnol 165, 77–84. [DOI] [PubMed] [Google Scholar]
- Tan L, Yu JT, Tan MS, Liu QY, Wang HF, Zhang W, Jiang T, and Tan L (2014). Genome-wide serum microRNA expression profiling identifies serum biomarkers for Alzheimer’s disease. Journal of Alzheimer’s disease : JAD 40, 1017–1027. [DOI] [PubMed] [Google Scholar]
- Tarver JE, Cormier A, Pinzon N, Taylor RS, Carre W, Strittmatter M, Seitz H, Coelho SM, and Cock JM (2015). microRNAs and the evolution of complex multicellularity: identification of a large, diverse complement of microRNAs in the brown alga Ectocarpus. Nucleic Acids Res 43, 6384–6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, Rao TN, Winnay JN, Garcia-Martin R, Grinspoon SK, et al. (2017). Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542, 450–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togliatto G, Dentelli P, Gili M, Gallo S, Deregibus C, Biglieri E, Iavello A, Santini E, Rossi C, Solini A, et al. (2016). Obesity reduces the pro-angiogenic potential of adipose tissue stem cell-derived extracellular vesicles (EVs) by impairing miR-126 content: impact on clinical applications. Int J Obes (Lond). 40, 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani M, Srinivasa S, Fitch KV, Thomou T, Wong K, Petrow E, Kahn CR, Cypess AM, and Grinspoon SK (2016). Dysfunctional Subcutaneous Fat With Reduced Dicer and Brown Adipose Tissue Gene Expression in HIV-Infected Patients. The Journal of clinical endocrinology and metabolism 101, 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, and Simons M (2008). Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science (New York, N.Y.) 319, 1244–1247. [DOI] [PubMed] [Google Scholar]
- Treiber T, Treiber N, and Meister G (2019). Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol 20, 5–20. [DOI] [PubMed] [Google Scholar]
- Turpin SM, Nicholls HT, Willmes DM, Mourier A, Brodesser S, Wunderlich CM, Mauer J, Xu E, Hammerschmidt P, Bronneke HS, et al. (2014) Obesity-induced CerS6-dependent C16:0 ceramide production promotes weight gain and glucose intolerance. Cell metabolism 20, 678–686. [DOI] [PubMed] [Google Scholar]
- Ungvari Z, Tucsek Z, Sosnowska D, Toth P, Gautam T, Podlutsky A, Csiszar A, Losonczy G, Valcarcel-Ares MN, Sonntag WE, et al. (2013). Aging-induced dysregulation of dicer1-dependent microRNA expression impairs angiogenic capacity of rat cerebromicrovascular endothelial cells. The journals of gerontology. Series A, Biological sciences and medical sciences 68, 877–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, and Lotvall JO (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9, 654–659. [DOI] [PubMed] [Google Scholar]
- Vallabhajosyula P, Korutla L, Habertheuer A, Yu M, Rostami S, Yuan CX, Reddy S, Liu C, Korutla V, Koeberlein B, et al. (2017). Tissue-specific exosome biomarkers for noninvasively monitoring immunologic rejection of transplanted tissue. J Clin Invest 127, 1375–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G, D’Angelo G, and Raposo G (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology 19, 213–228. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, and Steitz JA (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934. [DOI] [PubMed] [Google Scholar]
- Vickers KC, Landstreet SR, Levin MG, Shoucri BM, Toth CL, Taylor RC, Palmisano BT, Tabet F, Cui HL, Rye KA, et al. (2014). MicroRNA-223 coordinates cholesterol homeostasis. Proceedings of the National Academy of Sciences of the United States of America 111, 14518–14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, and Remaley AT (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 13, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villard A, Marchand L, Thivolet C, and Rome S (2015). Diagnostic Value of Cell-free Circulating MicroRNAs for Obesity and Type 2 Diabetes: A Meta-analysis. Journal of molecular biomarkers & diagnosis 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, and Sanchez-Madrid F (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 4, 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang S, Weber J, Baxter D, and Galas DJ (2010). Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res 38, 7248–7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Hong J, Cao Y, Shi J, Gu W, Ning G, Zhang Y, and Wang W (2015). Elevated circulating microRNA-122 is associated with obesity and insulin resistance in young adults. European journal of endocrinology 172, 291–300. [DOI] [PubMed] [Google Scholar]
- Wang X, Huang W, Liu G, Cai W, Millard RW, Wang Y, Chang J, Peng T, and Fan GC (2014). Cardiomyocytes mediate anti-angiogenesis in type 2 diabetic rats through the exosomal transfer of miR-320 into endothelial cells. Journal of molecular and cellular cardiology 74, 139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Qian RZ, Zhang W, Chen SF, Jin HM, and Hu RM (2009). MicroRNA-320 expression in myocardial microvascular endothelial cells and its relationship with insulin-like growth factor-1 in type 2 diabetic rats. Clinical and experimental pharmacology & physiology 36, 181–188. [DOI] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, and Blelloch R (2008). Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 40, 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhao R, Liu D, Deng W, Xu G, Liu W, Rong J, Long X, Ge J, and Shi B (2018). Exosomes Derived from miR-214-Enriched Bone Marrow-Derived Mesenchymal Stem Cells Regulate Oxidative Damage in Cardiac Stem Cells by Targeting CaMKII. Oxid Med Cell Longev 2018, 4971261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Li Y, Wang XY, Zhang D, Zhang H, Wu Q, He YQ, Wang JY, Zhang L, and Xia HJD (2013). Circulating miR-130b mediates metabolic crosstalk between fat and muscle in overweight/obesity. 56, 2275–2285. [DOI] [PubMed] [Google Scholar]
- Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, and Wang K (2010). The microRNA spectrum in 12 body fluids. Clinical chemistry 56, 1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen D, Qiao P, and Wang L (2015). Circulating microRNA-223 as a potential biomarker for obesity. Obesity research & clinical practice 9, 398–404. [DOI] [PubMed] [Google Scholar]
- Whitham M, Parker BL, Friedrichsen M, Hingst JR, Hjorth M, Hughes WE, Egan CL, Cron L, Watt KI, Kuchel RP, et al. (2018). Extracellular Vesicles Provide a Means for Tissue Crosstalk during Exercise. Cell metabolism 27, 237–251. [DOI] [PubMed] [Google Scholar]
- Willeit P, Skroblin P, Moschen AR, Yin X, Kaudewitz D, Zampetaki A, Barwari T, Whitehead M, Ramirez CM, Goedeke L, et al. (2017). Circulating MicroRNA-122 Is Associated With the Risk of New-Onset Metabolic Syndrome and Type 2 Diabetes. Diabetes 66, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, and Tuschl T (2013). Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proceedings of the National Academy of Sciences of the United States of America 110, 4255–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CM, Tsang FH, and Ng IO (2018). Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nature reviews. Gastroenterology & hepatology 15, 137–151. [DOI] [PubMed] [Google Scholar]
- Wu L, Dai X, Zhan J, Zhang Y, Zhang H, Zhang H, Zeng S, and Xi W (2015). Profiling peripheral microRNAs in obesity and type 2 diabetes mellitus. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica 123, 580–585. [DOI] [PubMed] [Google Scholar]
- Xia HQ, Pan Y, Peng J, and Lu GX (2011). Over-expression of miR375 reduces glucose-induced insulin secretion in Nit-1 cells. Molecular biology reports 38, 3061–3065. [DOI] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, and Rajewsky K (2007). MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131, 146–159. [DOI] [PubMed] [Google Scholar]
- Xiao J, Pan Y, Li XH, Yang XY, Feng YL, Tan HH, Jiang L, Feng J, and Yu XY (2016). Cardiac progenitor cell-derived exosomes prevent cardiomyocytes apoptosis through exosomal miR-21 by targeting PDCD4. Cell death & disease 7, e2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Lim B, and Lodish HF (2009). MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 58, 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Cai Y, Zhang Y, Liu J, and Xu Z (2018). Exosomes Secreted by Adipose-Derived Stem Cells Contribute to Angiogenesis of Brain Microvascular Endothelial Cells Following Oxygen-Glucose Deprivation In Vitro Through MicroRNA-181b/TRPM7 Axis. J Mol Neurosci 65, 74–83. [DOI] [PubMed] [Google Scholar]
- Ying W, Riopel M, Bandyopadhyay G, Dong Y, Birmingham A, Seo JB, Ofrecio JM, Wollam J, Hernandez-Carretero A, Fu W, et al. (2017). Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 171, 372–384. [DOI] [PubMed] [Google Scholar]
- Yore MM, Syed I, Moraes-Vieira PM, Zhang T, Herman MA, Homan EA, Patel RT, Lee J, Chen S, Peroni OD, et al. (2014). Discovery of a Class of Endogenous Mammalian Lipids with Anti-Diabetic and Anti-inflammatory Effects. Cell 159, 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Du H, Wei S, Feng L, Li J, Yao F, Zhang M, Hatch GM, and Chen L (2018). Adipocyte-Derived Exosomal MiR-27a Induces Insulin Resistance in Skeletal Muscle Through Repression of PPARγ. Theranostics 8, 2171–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagrean AM, Hermann DM, Opris I, Zagrean L, and Popa-Wagner A (2018). Multicellular Crosstalk Between Exosomes and the Neurovascular Unit After Cerebral Ischemia. Therapeutic Implications. Front Neurosci 12, 811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharie F, Muresan MS, Petrushev B, Berce C, Gafencu GA, Selicean S, Jurj A, Cojocneanu-Petric R, Lisencu CI, Pop LA, et al. (2015). Exosome-Carried microRNA-375 Inhibits Cell Progression and Dissemination via Bcl-2 Blocking in Colon Cancer. Journal of gastrointestinal and liver diseases : JGLD 24, 435–443. [DOI] [PubMed] [Google Scholar]
- Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, et al. (2010). Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circulation research 107, 810–817. [DOI] [PubMed] [Google Scholar]
- Zampetaki A, Willeit P, Tilling L, Drozdov I, Prokopi M, Renard JM, Mayr A, Weger S, Schett G, Shah A, et al. (2012). Prospective study on circulating MicroRNAs and risk of myocardial infarction. Journal of the American College of Cardiology 60, 290–299. [DOI] [PubMed] [Google Scholar]
- Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Koppel T, Jahantigh MN, Lutgens E, et al. (2009). Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Science signaling 2, ra81. [DOI] [PubMed] [Google Scholar]
- Zhang A, Li D, Liu Y, Li J, Zhang Y, Zhang C-YJB, and communications, b.r. (2018). Islet β cell: An endocrine cell secreting miRNAs. 495, 1648–1654. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang H, Zhang C, Jing Y, Wang C, Liu C, Zhang R, Wang J, Zhang J, Zen K, et al. (2015). Investigation of microRNA expression in human serum during the aging process. The journals of gerontology. Series A, Biological sciences and medical sciences 70, 102–109. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yang R, Hu BL, Lu P, Zhou LL, He ZY, Wu HM, and Zhu JH (2017a). Reduced Circulating Levels of miR-433 and miR-133b Are Potential Biomarkers for Parkinson’s Disease. Frontiers in cellular neuroscience 11, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, et al. (2014). MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kim MS, Jia B, Yan J, Zuniga-Hertz JP, Han C, and Cai D (2017b). Hypothalamic stem cells control ageing speed partly through exosomal miRNAs. Nature 548, 52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, et al. (2010). Secreted monocytic miR-150 enhances targeted endothelial cell migration. Molecular cell 39, 133–144. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mei H, Chang X, Chen F, Zhu Y, and Han X (2016). Adipocyte-derived microvesicles from obese mice induce M1 macrophage phenotype through secreted miR-155. J Mol Cell Biol 8, 505–517. [DOI] [PubMed] [Google Scholar]
- Zhao H, Guan J, Lee HM, Sui Y, He L, Siu JJ, Tse PP, Tong PC, Lai FM, and Chan JC (2010). Up-regulated pancreatic tissue microRNA-375 associates with human type 2 diabetes through beta-cell deficit and islet amyloid deposition. Pancreas 39, 843–846. [DOI] [PubMed] [Google Scholar]
- Zhao H, Shang Q, Pan Z, Bai Y, Li Z, Zhang H, Zhang Q, Guo C, Zhang L, and Wang Q. (2018). Exosomes from adipose-derived stem cells attenuate adipose inflammation and obesity through polarizing M2 macrophages and beiging in white adipose tissues. Diabetes 67, 235–247. [DOI] [PubMed] [Google Scholar]
- Zhao N, Jin L, Fei G, Zheng Z, and Zhong C (2014). Serum microRNA-133b is associated with low ceruloplasmin levels in Parkinson’s disease. Parkinsonism & related disorders 20, 1177–1180. [DOI] [PubMed] [Google Scholar]
- Zheng B, Yin WN, Suzuki T, Zhang XH, Zhang Y, Song LL, Jin LS, Zhan H, Zhang H, Li JS, et al. (2017). Exosome-Mediated miR-155 Transfer from Smooth Muscle Cells to Endothelial Cells Induces Endothelial Injury and Promotes Atherosclerosis. Mol Ther 25, 1279–1294. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zheng W, Chung LM, and Zhao H (2011). Bias detection and correction in RNA-Sequencing data. BMC bioinformatics 12, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li YS, Nguyen P, Wang KC, Weiss A, Kuo YC, Chiu JJ, Shyy JY, and Chien S (2013). Regulation of vascular smooth muscle cell turnover by endothelial cell-secreted microRNA-126: role of shear stress. Circulation research 113, 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, and Leung SW (2015). Identification of microRNA biomarkers in type 2 diabetes: a meta-analysis of controlled profiling studies. Diabetologia 58, 900–911. [DOI] [PubMed] [Google Scholar]
- Zhuang F, Fuchs RT, and Robb GB (2012a). Small RNA expression profiling by high-throughput sequencing: implications of enzymatic manipulation. Journal of nucleic acids 2012, 360358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, Li H, Wang G, Evans AR, Safe S, et al. (2012b). A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation 125, 2892–2903. [DOI] [PubMed] [Google Scholar]