Abstract
Chromosomal passenger complex (CPC) has been demonstrated to be a potential target of cancer therapy by inhibiting Aurora B or Survivin in different types of cancer including neuroblastoma (NB). However, chemical inhibition of either Aurora B or Survivin doesn’t target CPC specifically due to off-target effects or CPC independent activities of these two components. In a previous chromatin-focused siRNA screen, we found that NB cells were particularly vulnerable to loss of INCENP, a gene encoding a key scaffolding component of the CPC. In this study, INCENP was highly expressed by NB cells and its expression decreased following retinoic acid-induced NB differentiation. Elevated levels of INCENP were significantly associated with poor prognosis in primary tumors of NB patients with high-risk disease. Genetic silencing of INCENP reduced the growth of both MYCN-wild-type (MYCN-WT) and MYCN-amplified (MYCN-amp) NB cell lines in vitro and decreased the growth of NB xenografts in vivo with significant increases in murine survival. Mechanistically, INCENP depletion suppressed NB cell growth by inducing polyploidization, apoptosis and senescence. In most NB cell lines tested in vitro, apoptosis was the primary cell fate after INCENP silencing due to induction of DNA damage response and activation of the p53-p21 axis. These results confirm that CPC is a therapeutic target in NB and targeting INCENP is a novel way to disrupt the activity of CPC and inhibit tumor progression in NB.
Keywords: INCENP, CPC, Polyploidy, DNA damage, Apoptosis, Senescence
Introduction
Neuroblastoma (NB) is the most common extracranial solid tumor in early childhood. It accounts for more than 7% of pediatric malignancies in patients under the age of 15 yet causes 15% of all pediatric cancer deaths (1,2). NB is a neural crest-derived malignancy and is presumed to arise from a failure of sympathoadrenal progenitor differentiation during the normal development of sympathetic nervous system (3). During last few decades, there has been significant progress in delineating the genetic underpinnings involved in NB tumorigenesis leading to better risk stratification. This and the addition of immunotherapy has contributed to the increased survival rate of NB patients (4–6). However, overall survival of high-risk NB patients is still <50% despite intensive multimodal treatment (2,6). Therefore, a better understanding of the functional consequences of the genetic alterations in high-risk NB patients and a function-based target selection strategy should lead to novel therapeutics.
In a chromatin-focused siRNA screen we identified 53 targets whose loss of function led to decreased NB cell growth and/or increased NB differentiation. INCENP was identified as one of those candidate genes whose silencing significantly inhibits NB cell proliferation (7,8). INCENP encodes the Inner centromere protein (INCENP), which is the structural and regulatory component of the chromosomal passenger complex (CPC) comprised of INCENP, Survivin, Borealin, and the Aurora B kinase (9). CPC is responsible for proper chromosomal alignment, segregation and cytokinesis during the mitosis (9). In the CPC, INCENP plays two critical roles: firstly, it functions as a scaffold protein coordinating assembly of this complex by interacting with all the other three components and secondarily, the interaction between INCENP and Aurora B is necessary for activation of the Aurora B kinase, the catalytic subunit of this complex (9). Thus, disruption of INCENP expression leads to dissociation of the whole complex and limits Aurora B kinase activity (10,11). Targeting the CPC has been led by strategies aimed at targeting Survivin or Aurora B kinase in NB since inhibition of either of them significantly blocks NB tumor cell growth in vitro and xenograft growth in vivo (12–15). Recent Genome-Wide Meta-Analyses (GWAS) studies have identified SNPs in INCENP that are linked to increased susceptibility and risk in Breast, Ovarian, and Prostate Cancer (16,17). In addition, INCENP has been found to be overexpressed in high grade non-Hodgkin B-cell lymphomas and non-small-cell lung cancer, and proposed to be a biomarker for poor prognosis in these types of cancer (18,19). However, the function and therapeutic potential of targeting INCENP in NB remains unclear.
In this study we investigated how genetic targeting of INCENP affected NB cell growth. We find that decreased INCENP levels are associated with increases in polyploidization, apoptosis and senescence in vitro and decreased NB tumor growth in vivo.
Methods and Reagents
Cell culture
All the human NB cell lines used in this study were obtained from the cell line bank of the Pediatric Oncology Branch of the National Cancer Institute and have been genetically verified by short tandem repeat (STR) analysis and were routinely tested to be free of mycoplasma. NB cells were routinely cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 2mM L-glutamine, 100 units/mL penicillin and 100 μg/mL streptomycin (termed complete RPMI-1640). Stable shRNA NB cell lines were maintained in complete RPMI-1640 containing 0.5 or 1μg/ml puromycin. Tet21N cells were maintained in complete RPMI-1640 supplemented with 1μg/ml Tetracycline (Tet). HeLa, 293T cells and ARPE-19 cells were cultured in DMEM with 10% FBS, 2mM L-glutamine, 100 units/mL penicillin and 100 μg/mL streptomycin. For retinoic acid (RA) treatment experiments, NB cells were treated with 5 μM of all trans-RA dissolved in 95% ethanol and fresh RA containing medium was changed every 48 hrs.
Antibodies and Reagents
Primary antibodies against human INCENP (sc-376514), GAPDH (sc-25778), p53 (DO-I, sc-126), MYCN (sc-53993) and Borealin (sc-376635) as well as HRP-labeled secondary antibodies including Goat anti-mouse IgG-HRP (sc-2005) and Goat anti-Rabbit IgG-HRP(sc-2004) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The anti-MYC (5605S), anti-PARP (9532s), anti-Caspase 3 (9662s), anti-p21 (2947s), anti-Survivin (2808s), anti-pH2A.X (2577s), anti-pCHK2 (2197s), anti-Vinculin (13901s), anti-Tubulin (3873s) and anti-Aurora B (3094s) antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). Doxycycline (D9891) were purchased from Sigma-Aldrich. DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) was purchased from Thermo Fisher Scientific.
INCENP silencing and rescue experiment
Transient INCENP knockdown was conducted by nucleofection or Lipofectamine® transfection of different NB (except KCNR and Kelly cells) or control cell lines with two different siRNAs targeting human INCENP (siINCENP-#2: 5’- GGATGGATCTGAATA GCGA −3’; siINCENP-#4:5’- CCACGAUGCUGACUAAGAA-3’) from Dharmacon, GE. For INCENP knockdown in KCNR and Kelly cells, ON-TARGETplus pool siRNAs targeting INCENP (Dharmacon, GE) were used. For MYCN knockdown, siMYCN (5’- CAGTATT AGACTGGAAGTTCA-3’, QIAGEN) was used. Nucleofection was performed in an Amaxa Nucleofector II with Cell Line Nucleofector™ Kit V (VVCA-1003, Amaxa Biosystems, Gaithersburg, MD, USA) according to the manufacturer’s instructions. A non-targeting siRNA (siCTRL: 5’-UGGUUUACAUGU CGACUAA-3’, Dharmacon) was used as a negative control for gene knockdown experiments. In order to establish an inducible INCENP knockdown system in NB cell lines, we packaged a pTRIPZ inducible lentiviral shRNA construct targeting INCENP (RHS4740-EG3619, clone ID V2THS_364633, 5’-AACTGTGACAGATGATGCG −3’, Dharmacon, GE) and a non-targeting pTRIPZ control shRNA (RHS4743) into lentiviral particles for efficient transduction and gene silencing. Different NB cell lines were transduced with the shCTRL lentiviruses and the shINCENP lentiviruses and then the stable shRNA transduced NB cell lines were selected with puromycin (0.5 μg/mL for SY5Y and NGP or 1 μg/mL for BE2C). Gene silencing could be efficiently induced when these cells were cultured in complete RPMI-1640 containing 1 μg/mL doxycycline (dox). For the rescue experiment, we first generated a lentiviral construct (pLVX-tetOne-INCENP mutant) in which siINCENP-#2 resistant form of INCENP ( the target sequence of siINCENP-2# was mutated from GG-ATG-GAT-CTG-AAT-AGC-GA to GG-ATG-GAC-TTA-AAC-TCA-GA ) could be induced upon dox treatment. After lentivirus packaging and transduction, we generated a stable BE2C cell line with doxycycline inducible expression of siINCENP-#2 resistant form of INCENP that still encoded the same protein. To perform the rescue experiement, the expression of siRNA resistant form of INCENP was induced immediately by adding 0.5 μg/mL dox after siRNAs were transfected into this cell line.
Cell growth and clonogenicity assays
To assess the effect of INCENP knockdown on NB cell growth, NB cells transfected with siCTRL and siINCENP were plated in 24-well plates and the growth kinetics were monitored in IncuCyte ZOOM (Essen BioScience) using the integrated confluence algorithm as a surrogate for cell number. MTS assay (Promega) was performed according to manufacturer’s instructions in 96-well plate at day 3 after transfection. To investigate the effects of INCENP knockdown on clonogenic ability of NB cells, we performed both colony formation assay (adherent colony formation) and soft agar assay (anchorage-independent colony formation) using inducible shRNA NB cell lines. For colony formation assays, 103 NB cells were plated into each well of 6-well plates and cultured in complete RPMI-1640 medium containing puromycin (0.5 μg/mL for SY5Y and NGP or 1μg/mL for BE2C) and treated with or without 1ug/ml dox. For soft agar assay, 104 NB cells were cultured in 0.7% top agarose in media containing puromycin (± doxy) and plated over a bottom layer of 1.4% bottom agar/media. In both assays fresh medium was changed twice weekly and visible colonies were allowed to be formed in two to four weeks. The number of colonies was counted directly or after crystal violet staining.
Immunofluorescent cytochemical staining
Subconfluent cells were seeded on 8 well chamber slide after nucleofection with siRNAs.
The cells were fixed in 4% paraformaldehyde/PBS for 10 min at 48 hr post transfection. Immunostaining was performed as previously described [7]. Antibodies used for immunostaining are listed as follow: anti-pH2A.X (2577s) and anti-α-Tubulin (3873s). DAPI was used to stain the nuclei.
Cell cycle analysis
Cell cycle analysis was performed using FxCycle™ PI/RNase Staining Solution (Thermo Fisher Scientific) according to the manufacturer’s instructions. At day 2 post transfection, siRNA transfected NB cells were harvested, washed with 1× PBS twice and fixed with 70% ethanol for at least two hours. Before acquiring data on a flow cytometer, stained the samples with FxCycle™ PI/RNase Staining Solution for 30 minutes at room temperature and wrapped them in foil to protect from light. Samples were analyzed in a flow cytometer, using 532 nm excitation and emission was collected in a 585/42 bandpass or equivalent. The cell cycle results were analyzed using FlowJo software.
Clinical patient cohorts
Clinical patient data were analyzed using R2: Genomic Analysis and Visualization Platform (r2.amc.nl/). Kaplan-Meier plots and the graphs of gene expression were generated based on the expression of INCENP in tumors of NB patients in the TARGET dataset (249 patients) or Kocak dataset (649 patients in total but only 476 patients have survival data).
RNA and protein analyses
Total RNA was isolated with RNeasy Mini plus Kit (Qiagen Inc., Valencia, CA, USA). The reverse transcription reactions were conducted with High-Capacity RNA-to-cDNA™ Kit (Applied Biosystems, Poster City, CA, USA). Housekeeping gene GAPDH was used to normalize the gene expression. Primers used in this study were as follows: INCENP-Forward 5’-AGGCTCCTGAATGTTGAGGTGC-3’ and INCENP-reverse 5’-GTGTGCTGTTGGCAATCTCCGT-3’; GAPDH-forward 5’-AGAAGGCTGG GGCTCATTTG-3’, GAPDH-reverse 5’-AGGGGCCATCCACAGTCT TC-3’. Quantitative real-time PCR was performed on CFX384 Touch™ Real-Time PCR Detection System (Biorad) using SYBR Green SuperMix as described previously [7]. The gene expression levels were analyzed by 2−ΔΔCt method. Transcriptional changes in BE2C cells following 24 hr of transfection with INCENP siRNAs were analyzed by RNA-seq (HiSeq4000, Illumina). Total proteins were extracted from cultured cell lines or tumor tissues in RIPA buffer (50 mM Tris pH 8, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP40, 1 mM EDTA and a mix of protease/phosphatase inhibitors). Western blot was performed as previously described (7).
Caspase-Glo 3/7 assay
NB cells nucleofected with siRNAs were plated in triplicate in 96-well plates. At day 3 after transfection, Caspase 3/7 activity were detected using Caspase-Glo 3/7 Assay Systems (Promega) according to the manufacturer’s instruction.
Xenograft assay
All animal studies were performed at National Cancer Institute (NCI) and approved by the Animal Care and Use Committee of the NCI (PB-023). SY5Y shCTRL, SY5Y shINCENP, BE2C shCTRL and BE2C shINCENP cells were re-suspended in HBSS and Matrigel (1:1, Trevigen, Gaitherburg, MD, USA) respectively. Cell suspension (100ul) containing 2×106 cells was injected subcutaneously into the left flank of 5- to 6-week-old female athymic nude mice (Frederick Animal Facility, NCI) using a 28-gauge needle (Becton Dickinson, Franklin Lakes, NJ). When tumors reached approximately 100–200 mm3, mice were given chow containing doxycycline (Bioserv, Flemington, NJ, USA) or regular chow. The dimensions, length (L) and width (W) of the tumors were measured thrice weekly using a digital caliper, and the tumor volume (mm3) was calculated as (L × W2)/4. Mice were euthanized once the tumor diameter reached 2 cm.
Statistical analysis
Statistical analyses were performed by a standard two-tailed Student’s t-test using GraphPad Prism 7.0 software. The Kaplan-Meier survival analsyis was used to estimate murine survival in the animal studies. To evaluate the differences between the survival curves of control group and experiment group, we used a log-rank test method (Mantel-Cox). All p values of less than 0.05 are considered to be statistically significant and ns stands for not statistically significant..
Results
INCENP is overexpressed in NB cells and high INCENP level correlates with poor outcome in high-risk NB patients
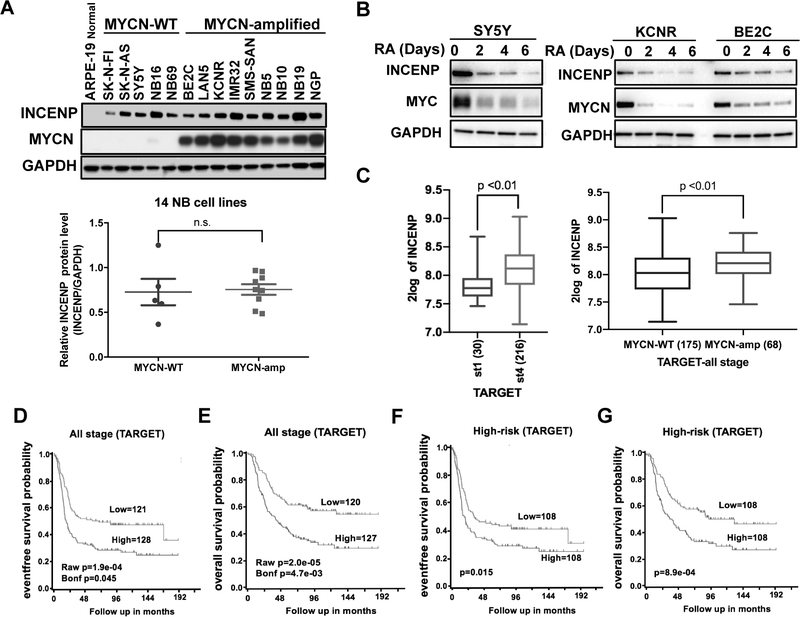
From our previous siRNA screen (7), we found silencing of INCENP using either a pool of siRNAs or any of four individual siRNAs targeting INCENP significantly decreased the cell number in SY5Y cells. To understand the nature of this dependency, we first evaluated the expression level of INCENP in 14 commonly used NB cell lines and one non-transformed cell line-ARPE-19 cells. Western blot showed that INCENP protein was highly expressed in both MYCN-WT and MYCN-amp NB cell lines (Fig.1 A). This pattern of INCENP expression is similar to levels of the other components of the CPC-Aurora B, Survivin and Borealin, which are also expressed in NB cells (Fig.S1A). In addition, we detected the expression pattern of all the components of CPC during retinoic acid (RA)-induced differentiation of NB cells and found that RA treatment decreased MYC level in SY5Y cells (Fig.1B, left panel) and MYCN level in both KCNR and BE2C cells as early as day 2 (Fig.1B, right panel). Meanwhile, decreased INCENP was observed in all selected NB cell lines (SY5Y, KCNR and BE2C) upon RA treatment (Fig.1B). Notably, different from further decrease of MYC or MYCN level in SY5Y and NGP cells, MYCN was decreased at day 2 but sustained at a stable level in BE2C cells despite of continuous RA treatment (Fig. 1B). The expression of other CPC components (Aurora B, Survivin and Borealin) decreased in RA-treated SY5Y and KCNR cells but their levels did not show obvious change in RA-treated BE2C cells (Fig. S1B). Next, we evaluated publicly available microarray or RNA-seq data of primary NB tumor tissues in R2 database (r2.amc.nl/) (20). We chose and analyzed two different datasets -TARGET with 249 patient samples (21) and Kocak with 649 patient samples (22) and found that there was higher expression of INCENP in Stage 4 tumors compared to Stage 1 tumors (Fig.1C, left panel and Fig.S1C, left panel). Additionally, we found significantly higher INCENP mRNA levels in MYCN-amp tumors compared to MYCN-WT tumors (Fig.1C, right panel and Fig.S1C, right panel). Consistent with a prevailing view that MYCN is an amplifier of transcriptionally active genes (23,24), MYCN ChIP-seq data [24] show that MYCN binds to the promoter regions of all the CPC component genes including INCENP in BE2C cells, indicating an involvement of MYCN in the transcriptional regulation of INCENP and other CPC components (Fig.S1D). Consistently, in Tet21N NB cell line with Tetracycline (Tet)-regulated expression of MYCN (Tet-off), Tet removal induced the expression of MYCN and upregulated expression of CPC components (Fig.S1E, left panel). In contrast, knockdown of MYCN led to a downregulation of the expression of CPC components in BE2C cells (Fig.S1E, right panel). These results indicate that in NB cells MYCN directly regulates the expression of CPC components and sustained CPC levels in RA-treated BE2C cells may be due to residual MYCN expression. A Kaplan–Meier analysis of all NB stages showed high INCENP expression was associated with worse prognosis, whereas low level of INCENP was associated with a better outcome (Fig. 1D, Fig. 1E, Fig.S1F and Fig. S1G). Moreover, in high-risk NB patients including stage 4 patients >18 months and patients whose tumors harbor MYCN amplification, high INCENP expression was associated with poor prognosis which was statistically significant in both TARGET dataset and Kocak dataset (Fig. 1F, Fig. 1G, Fig.S1H and Fig.S1I). Taken together, our results show INCENP is highly expressed in NB cells and patients whose tumors have elevated levels of INCENP have worse prognoses.
Figure 1.
INCENP is highly expressed in NB cells and high INCENP expression is associated with worse prognosis in NB tumors.
A. Upper panel: western blot analysis of INCENP protein levels in fourteen NB cell lines and one immortal but non-tumorigenic cell line-ARPE-19; lower panel: densitometric analysis of INCENP protein levels normalized to GAPDH in both MYCN AMP and MYCN WT NB cell lines was calculated using Imaging J software. Bars show the mean ± SEM. B. Western blot analysis of INCENP, and MYC or MYCN protein levels in SY5Y (left panel), KCNR and BE2C cells (right panel) treated with 5 μM Retinoic Acid (RA) for 0, 2, 4 and 6 days. C. Left panel: INCENP mRNA levels in stage 1 and stage 4 NB tumors (TARGET-249 dataset). Right panel: INCENP mRNA levels in MYCN-WT and MYCN-amp NB tumors (all stages, TARGET-249 dataset). D and E. Event-free (D) and overall (E) Kaplan-Meier plots based on the expression of INCENP in tumors from NB patients at all stages. All the data are from TARGET-249 dataset. F and G. Event-free (F) and overall (G) Kaplan-Meier plots based on the expression of INCENP in tumors from high-risk NB patients in TARGET-249 dataset.
INCENP depletion inhibits NB cell growth and tumorigenicity in vitro
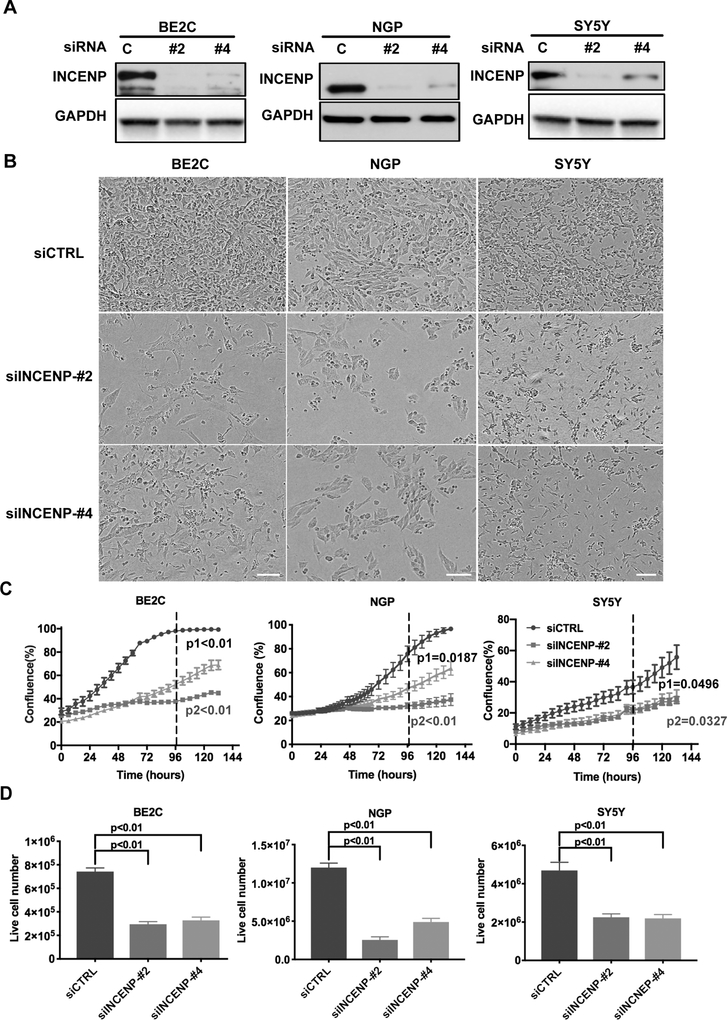
To evaluate the biological effects of loss of INCENP in NB cells, we utilized short interfering RNAs (siRNAs) to transiently knock down the expression of endogenous INCENP in 3 genetically distinct NB cell lines; BE2C (MYCN-amp and p53 mutant), NGP (MYCN-amp and p53-WT) and SY5Y (MYCN-WT and p53-WT). These NB cell lines were transfected with control siRNA (siCTRL) or INCENP siRNAs (siINCENP-#2 and siINCENP-#4) targeting two different regions of INCENP mRNA and cultured for three to five days. Western blot showed that INCENP protein levels were significantly downregulated in either of INCENP siRNAs transfected NB cells compared to control siRNA transfected NB cells (Fig.2A). Phenotypically, we observed INCENP silencing resulted in significant cell growth inhibition as both cell confluence and the number of attached live cells in the siINCENP transfected NB cells significantly decreased (Fig. 2B–D). A rescue experiment was performed to exclude the possibility that these siRNA-induced phenotypes were due to off-target effects. Briefly, we first generated a stable BE2C cell line with doxycycline inducible expression of siINCENP-#2 resistant form of INCENP that still encoded the same protein (Fig.S2A). We found that overexpression of siRNA resistant form of INCENP could rescue all of the siINCENP-mediated phenotypes including cell growth inhibition (Fig. S2B and S2C). This finding indicates that functional activities due to INCENP depletion could be rescued by INCENP overexpression. In addition, the inhibitory effects of INCENP silencing in NB cells have also been verified in the KCNR and Kelly NB cell lines using pool siRNAs (Fig.S2D and S2E). Consistent with above results, gene dependency analysis in multiple NB cell lines based on CRISPR-Cas9 screen data from Project Achilles (http://www.broadinstitute.org/achilles, Broad Institute) further demonstrates that NB cells are dependent on INCENP as well as other CPC components for their growth (Fig.S2F).
Figure 2.
INCENP depletion inhibits NB cell growth in vitro.
A. Western blot analysis of INCENP knockdown efficiency in BE2C, NGP and SY5Y cells transfected with two different siRNAs targeting INCENP (#2: siINCENP-#2; #4: siINCENP-#4) and control siRNA (C: siCTRL) for 72 hr. B. The bright field pictures of BE2C, NGP and SY5Y cells at 72 hr (BE2C and NGP) or 96 hr (SY5Y) after siRNA transfection. Scale bars, 100 μm. C. The confluence analysis of siCTRL, siINCENP-#2 and #4 transfected BE2C, NGP and SY5Y cells in the IncuCyte ZOOM. The values of cell confluence at 96 hr is used for statistical analysis by standard two-tailed student’s t test (p1: siCTRL vs. siINCENP-2#; p2: siCTRL vs. siINCENP-4#). Bars show the average of three replicates ± SD. D. Live cell number counting of siCTRL, siINCENP-#2 and #4 transfected BE2C, NGP and SY5Y cells at 72 hr (BE2C and NGP) or 96 hr (SY5Y) post-transfection. Data is represented as mean ± SD of triplicates.
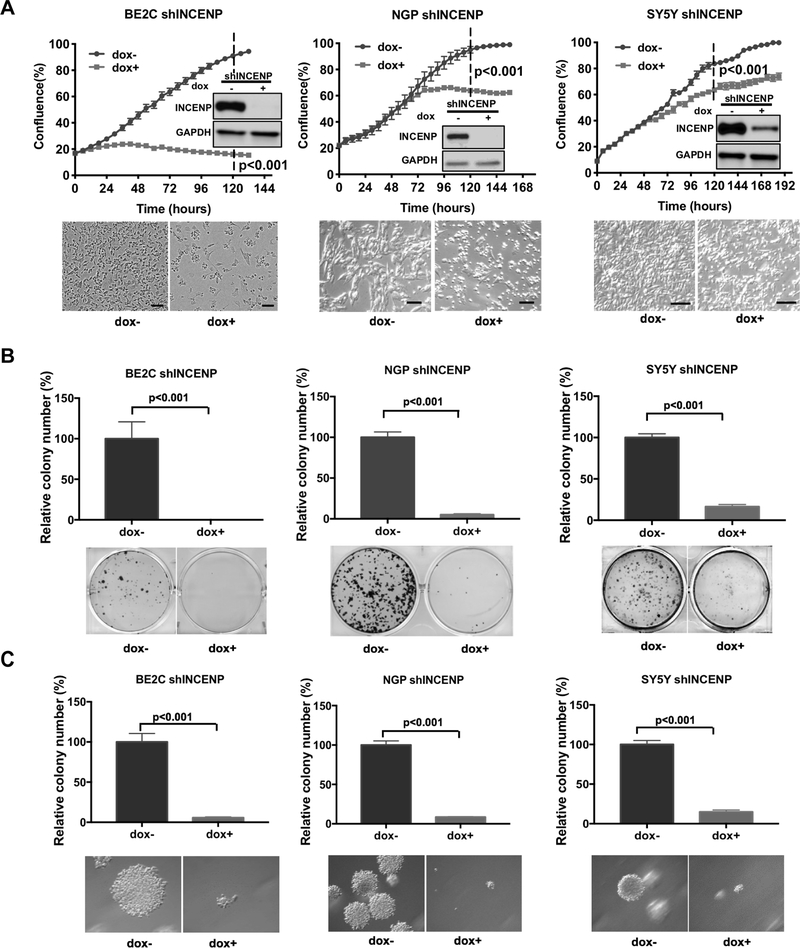
To investigate whether INCENP depletion could affect the clonogenicity and tumorigenicity of NB cells, we utilized doxycycline (dox)-inducible lentiviral shRNA knockdown system to generate stable NB cell lines expressing dox-regulated shRNAs targeting INCENP (hereinafter refer to as shINCENP). We found that dox-mediated INCENP silencing caused a significant decrease in INCENP levels (Fig. 3A) and this was accompanied by a significant inhibition of NB cell growth (Fig. 3A). In clonogenic assays, we found that INCENP knockdown significantly decreased colony numbers in all three NB cell lines tested (Fig. 3B). In soft-agar colony formation assays, both the size and the number of the colonies generated from INCENP depleted cells were reduced compared to the colonies from control treated cells (Fig.3C). However, control shRNA (hereinafter refer to as shCTRL) had no effects on cell growth and the anchorage-dependent (clonogenic assay) or -independent colony formation (soft agar assay) ability of NB cells (Fig.S3A–S3C). These results show that INCENP is important for NB cell clonogenicity and tumorigenicity in vitro.
Figure 3.
INCENP depletion suppresses the colony formation ability and tumorigenesis of NB cells in vitro.
A. The confluence analysis of BE2C, NGP and SY5Y inducible shINCENP cells treated with doxycycline (dox) or vehicle in the IncuCyte ZOOM (upper panel). The representative pictures for each cell type under different treatment are shown (lower panel). Western blot analysis of INCENP knockdown level after 72 hr dox treatment are also shown. The values of cell confluence at 120 hr is used for statistical analysis by standard two-tailed student’s t test. Bars show the average of three replicates ± SD. Scale bars, 100 μm. B. Clonogenicity of BE2C, NGP and SY5Y inducible shINCENP cells was determined by colony formation assays. The relative number of colonies (upper panel) and the representative pictures for each cell type after crystal violet staining are shown (lower panel). Bars show the mean ± SD of triplicates. C. The tumorigeneicity of BE2C, NGP and SY5Y inducible shINCENP cells was determined by soft agar assays in vitro. The relative colony numbers (upper panel) and the representative pictures of colonies grown in soft agar for each cell type upon different treatment are shown (lower panel). Data is represented as mean ± SD of three replicates.
INCENP depletion inhibits NB cell growth and tumorigenesis in vivo
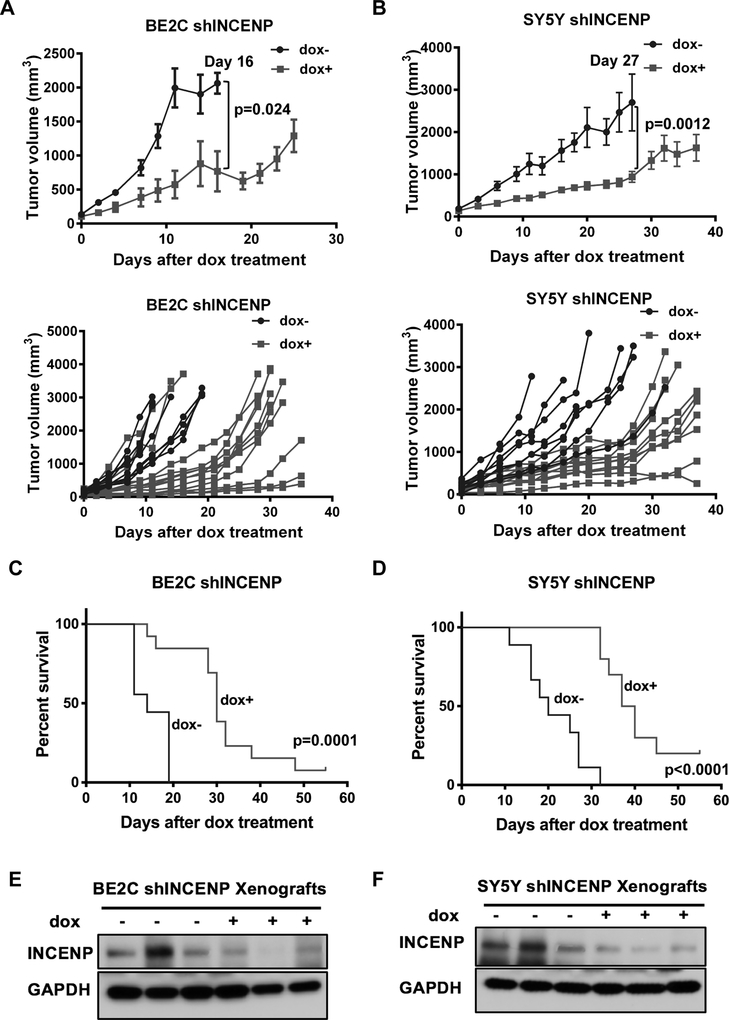
To investigate the effects of INCENP depletion in vivo, we implanted doxycycline inducible shCTRL or shINCENP BE2C cells (BE2C-dox-shCTRL or BE2C-dox-shINCENP) and SY5Y cells (SY5Y-dox-shCTRL or SY5Y-dox-shINCENP) into nude mice subcutaneously. When tumor volume reached 100–200 mm3, mice were stratified into two different groups that received either dox-chow or regular chow. The BE2C-dox-shCTRL and SY5Y-dox-shCTRL cell lines showed that doxycycline did not significantly affect tumor growth and murine survival in vivo (Fig.S4A–S4D). Mice receiving dox-chow had significantly reduced tumor size in both the BE2C-dox-shINCENP (Fig. 4A) and the SY5Y-dox-shINCENP cell lines (Fig. 4B) and this was associated with increased murine survival compared to mice receiving normal chow (Fig. 4C and 4D). The knockdown level of INCENP in the tumor samples was confirmed by western blot (Fig. 4E and 4F). These studies demonstrate that targeting INCENP expression in NB cells significantly inhibits in vivo tumor xenograft growth and prolongs survival of mice.
Figure 4.
Genetic silencing of INCENP inhibits the growth of NB tumor xenografts in vivo.
A. BE2C-shINCENP xenograft tumor growth in mice treated with doxycycline (dox)-containing (n=9) or control chow (n=13). Upper panel: xenograft growth curve by average tumor volume of each group; Lower panel: xenograft growth curve by tumor volume of individual mouse in each group. Bars show the tumor size average of the mice/group ± SEM. B. SY5Y-shINCENP xenograft tumor growth in mice treated with doxycycline (dox)-containing (n=8) or control chow (n=10). Upper panel: xenograft growth curve by average tumor volume of each group; Lower panel: xenograft growth curve by tumor volume of individual mouse in each group. Bars show the tumor size average of the mice/group ± SEM. C and D. Kaplan-Meier graphs showing the murine survival of BE2C-shINCENP (C) and SY5Y-shINCENP (D) tumor-bearing mice upon INCENP silencing. The statistical significance between two treatment groups was evaluated using a log rank test. E and F. Western blot analysis of INCENP knockdown levels in BE2C-shINCENP xenografts and SY5Y-shINCENP xenografts two weeks after dox treatment.
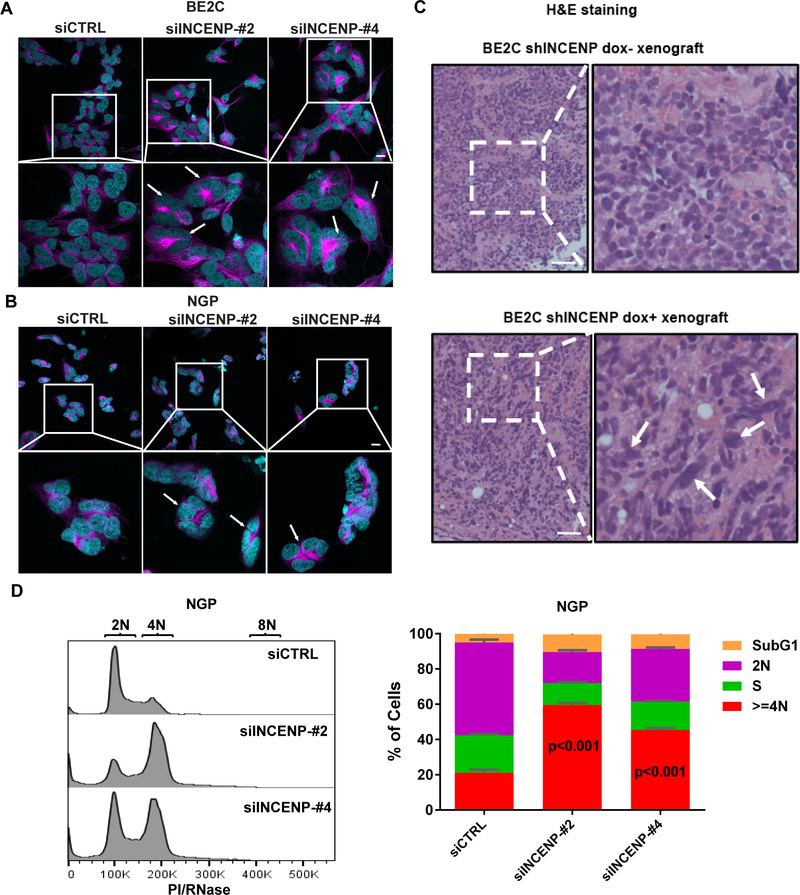
INCENP depletion in NB cells leads to multinucleation and polyploidy
Next we investigated the mechanisms of how INCENP depletion affected NB growth and tumorigenesis. As a critical scaffold protein and regulatory component of the Chromosomal Passenger Complex (CPC), INCENP silencing would disrupt the normal function of CPC in mitosis and cytokinesis by causing complex dissociation and mis-localization and inactivation of Aurora B kinase which could lead to multinucleation and generation of polyploid cells (9–11). As a result, we hypothesized that INCENP knockdown-induced polyploidy contributes to cell growth inhibition in NB cells. To examine this, we co-stained siRNA transfected NB cells with the nuclear dye DAPI and cytoskeleton marker α-Tubulin. After transfection with siINCENP, we observed an increase in multinulceated and polyploid cells compared to siCTRL treated BE2C cells (Fig. 5A) and NGP cells (Fig.5B). Consistently, H&E staining of tumor xenografts showed cells with irregular nuclear shape from dox-chow fed mice bearing BE2C shINCENP xenografts compared to controls (Fig. 5C). Cell cycle analysis of NGP cells transfected with siINCENP-#2 or -#4 showed significant increases in the percentage of cells at the G2/M phase and increases in the population of cells with DNA content≥4N (Fig.5D). In INCENP silenced SY5Y cells, we also observed cells with polyploidy or abnormal nuclei (Fig.S5A,S5B and S5C). As expected, multi-nucleated cells have also been found in HeLa and two non-tumorigenic cell lines-293T and ARPE19 cells after transfection of siRNAs targeting INCENP (Fig.S5D). Taken together, these data support our hypothesis that INCENP depletion increases the percentage of G2/M population and induces the generation of multinucleated and polyploid NB cells, which contributes to decreased cell growth.
Figure 5.
Genetic Inhibition of INCENP in NB cells leads to multinucleation and polyploidy.
A. The results of immunostaining (DAPI: Cyan; a-Tubulin: Magenta) showing the irregular nuclear shape of INCENP depleted BE2C cells at 48 hr post transfection. White arrows indicate multinucleated cells with enlarged and abnormal nuclei. Scale bars, 20 μm. B. The results of immunostaining (DAPI: Cyan; a-Tubulin: Magenta) showing the irregular nuclear shape of INCENP depleted NGP cells at 48 hr post transfection. White arrows indicate multinucleated cells with enlarged and abnormal nuclei. Scale bars, 20 μm. C. H&E staining results of BE2C-shINCENP xenografts without (upper panel) or with (lower panel) dox treatment. Scale bars, 50 μm. D. Cell cycle analysis of NGP cells transfected with siCTRL, siINCENP-#2 and siINCENP-#4 for 48 hr. Data represent mean ± SD of three replicates.
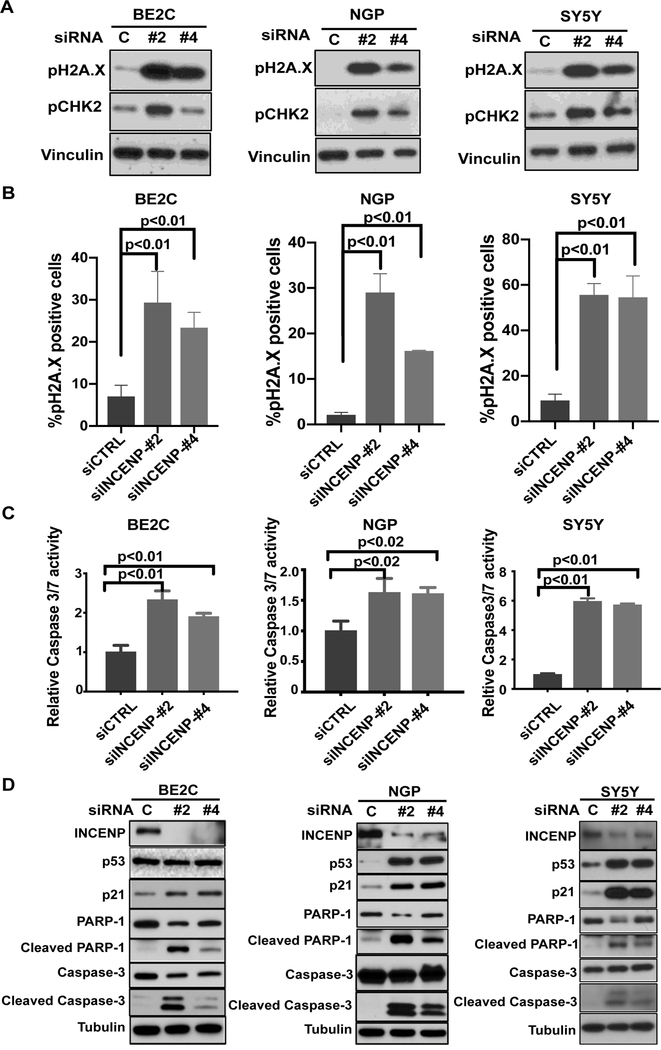
INCENP silencing induces DNA damage and apoptosis in NB cells
To further identify mechanisms involved in the growth inhibition after INCENP silencing, we performed RNA-seq analysis after INCENP knockdown for 24 hr. Gene ontology (GO) analysis of differentially expressed genes showed enrichment of a series of genes associated with cell growth and differentiation in INCENP silenced BE2C cells (Table S1, Fig.S6A and S6B). Among the differentially expressed genes, GADD45B was upregulated significantly upon INCENP knockdown. Previous studies show that the upregulation of GADD45B frequently occurs in response to different cell stressors and implicates DNA damage as a prelude to an apoptotic cellular response (25–27). We found that INCENP knockdown increased the expression of DNA damage markers (pH2A.X and pCHK2) in all three NB cell lines (Fig.6A and Fig.S6C) with some 30–50% of the cells exhibiting activation of DNA damage signaling as evidenced by pH2A.X immunostaining (Fig. 6B). Next we determined the apoptosis level in these control and INCENP silenced cells and found there was a significant increase in Caspase-3/7 activity when INCENP expression was silenced (Fig. 6C and Fig.S6D). Western blot analysis further confirmed that INCENP depletion was accompanied by increases in p53-p21 signaling in p53-WT NB cell lines (NGP, SY5Y) and upregulation of apoptosis markers including cleaved Caspase-3 and cleaved PARP-1 in all three NB cell lines (Fig. 6D). Notably, p53 and p21 were significantly increased in INCENP depleted NGP cells and SY5Y cells, both of which had wild type p53. In p53 mutant BE2C cells, p53 levels did not change but the levels of p21 slightly increased upon INCENP knockdown (Fig. 6D left panel and Fig.S6E). To verify whether p21 upregulation is p53 dependent in BE2C cells, we performed INCENP and p53 double knockdown experiment. We found that knockdown of p53 didn’t block p21 upregulation in INCENP silenced BE2C cells but resulted in a slight increase of the protein levels of p21, which is consistent with mutant p53 playing a dominant negative role in BE2C cells. (Fig.S6F). This result suggests that induction of p21 in INCENP depleted BE2C cells is p53 independent. Thus, our results indicate that loss of INCENP activates p21 in both p53 WT NB cells and p53 mutant cells through different mechanisms. Moreover, treatment of INCENP silenced BE2C cells with the pan-caspase inhibitor Z-VAD-fmk (Z-VAD) partially rescued the BE2C cell numbers and this finding further demonstrates that knockdown INCENP induces cell apoptosis (Fig. S6G, left panel). Western blot results also showed Z-VAD treatment inhibited the upregulation of pH2A.X and cleaved Caspase-3 upon INCENP silencing (Fig. S6G, right panel). Interestingly, different from our observation in NB cells, we found that INCENP knockdown could only induce polypoidy but no obvious apoptosis was detected in 293T, ARPE19 and even HeLa cells whereas massive apopotosis has been observed in siINCENP silenced NB cells (Fig.S6H). This observation indicates that NB cells are more sensitive to INCENP knockdown. Collectively, these data show that INCENP silencing increases the DNA damage signaling and induces apoptosis in NB cells.
Figure 6.
Genetic Inhibition of INCENP induces DNA damage response and apoptosis in NB cells.
A. Western blot analysis of DNA damage markers-pH2A.X and pCHK2 in BE2C, NGP and SY5Y cells transfected with two different siRNAs targeting INCENP (#2: siINCENP-#2; #4: siINCENP-#4) and control siRNA (C: siCTRL) for 48 hr. B. Quantification of the percentage of pH2A.X positive cells in each cell type based on the results of immunocytochemistry for DNA damage marker pH2A.X in BE2C, NGP and SY5Y cells transfected with siCTRL, siINCENP-#2 and siINCENP-#4 for 48 hr. Bars show the mean ± SD of triplicates. C. Caspase-3/7 activity assay was conducted to determine the activation of apoptosis in INCENP depleted BE2C (left), NGP (middle) and SY5Y (right) cells at 72 hr post transfection. Data represent mean ± SD of three replicates. D. Western blot analysis of INCENP knockdown efficiency, the activation of p53-p21 pathway and the upregulation of apoptosis markers including cleaved caspase-3/7 and cleaved PARP1 in BE2C (left), NGP (middle) and SY5Y (right) cells transfected with siRNAs targeting INCENP.
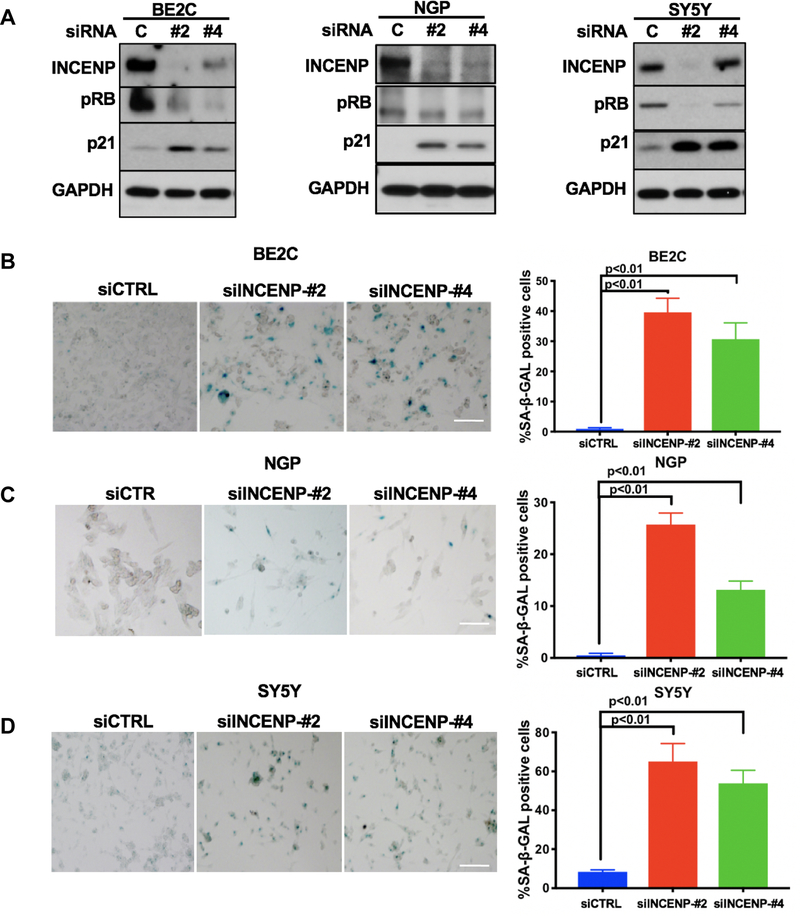
INCENP depletion induces a cellular senescence phenotype in NB cells
Since several studies have reported that inhibition of Aurora kinase activity using specific chemical inhibitors or genetic silencing induced significant cellular senescence in different types of cancer cell lines (28–32), we tested whether INCENP knockdown would also induce cellular senescence in NB cell lines. Western blot results showed dynamic regulation of two markers of cell cycle exit 5 days after INCENP silencing with increased levels of p21 while levels of phosphorylated forms of RB (pRB) decreased (Fig.7A). Treatment of BE2C cells with Barasertib, a specific Aurora B kinase inhibitor, for 7 days significantly increased SA-β-gal activity, a marker of senescent cells (Fig.S7A). Similarly, INCENP knockdown in BE2C, NGP and SY5Y cells also induced a 30–50% increase in the fraction of cells with SA-β-gal activity (Fig.7B, 7C and 7D, Fig.S7B and S7C).
Figure 7.
INCENP depletion induces cellular senescence phenotype in NB cells.
A. Western blot analysis of senescence markers-downregulation of pRB and upregulation of p21 in BE2C, NGP and SY5Y cells transfected with two different siRNAs targeting INCENP (#2: siINCENP-#2; #4: siINCENP-#4) and control siRNA (C: siCTRL) for 5 days. B, C and D. β-gal staining of senescent cells in siCTRL, siINCENP-#2 and #4 transfected BE2C (B), NGP (C) and SY5Y (D) at day 7 post transfection to determine the cellular senescence in each cell population. Bright field pictures (left panel) and quantification data (right panel) are shown. Bars show the mean ± SD of triplicates. Scale bars, 50 μm.
Discussion:
In this study, we find that high expression of INCENP mRNA levels is associated with poor prognosis of NB patients. Our genetic inhibition studies show that INCENP is indispensable for the growth of both MYCN-WT and MYCN-amp NB cell lines in vitro and in vivo. The functional response of NB cells to INCENP silencing is heterogenous with the evidence of induction of multinucleated cells, increased apoptosis and cellular senescence which all contribute to decreased cell numbers. Our findings suggest that strategies aimed at decreasing INCENP levels selectively target the CPC and inhibit NB tumor growth.
High INCENP expression has been reported in some cancer types. In high grade non-Hodgkin B-cell lymphomas (18), there is significantly higher nuclear immunohistochemical signal of INCENP compared to low grade ones. High expression of three components of CPC (Aurora B, INCENP, and Survivin) except Borealin, is associated with poor survival rate in patients with NSCLC (19). Recent evidence shows both Aurora B and Survivin are highly expressed in NB cells and high levels of either Aurora B or Survivin are associated with poor prognosis (12–15). We find that INCENP is highly expressed and required for cell growth in both MYCN-AMP and MYCN-WT NB cell lines although MYCN amp-NB patient tumors have higher INCENP mRNA levels (Fig.1C right panel and Fig.S1C right panel). High INCENP level is associated with poor prognosis even in high-risk NB patients (Fig.1F, Fig.1G, Fig.S1H and Fig.S1I). The results of MYCN knockdown and overexpression experiments as well as analysis of publically available ChIP-seq data support a model in which MYCN directly activates INCENP expression and thus contribute to its elevated levels in NB cells expressing high levels of MYCN. However, the mechanisms leading to dysregulation of INCENP in the high-risk non-MYCN amplified tumors have not been delineated. It is known that some MYCN-WT NB tumors have an elevated MYC level or MYCN/MYC transcriptome signature (33,34) which might contribute to elevated INCENP level in these non-MYCN amplified tumors. Another intriguing possibility is that INCENP is located at chromosomal location at 11q12.3 adjacent to the commonly deleted region on chromosome 11q which is a characteristic of the high-risk MYCN subgroup and may impact INCENP regulation (35,36). Genome-wide association studies (GWAS) identified several single nucleotide polymorphisms (SNPs) in INCENP which contributed to the susceptibility of Breast, Ovarian, and Prostate Cancer (16,17) but GWAS analyses in NB have not reveal any susceptibility loci in INCENP’s chromosomal region (37,38).
Similar to the functions of the other two components of CPC-Aurora B and Survivin in NB, we find INCENP is also indispensable for NB growth in vitro and in vivo.
Previously, Aurora B and Survivin have been shown to be necessary for NB growth (12–15). Survivin was firstly described as a member of the inhibitor of apoptosis protein (IAP) family (39) and the subsequent finding that survivin was expressed in a cell cycle-dependent manner led to its involvement in the CPC (40–42). A combined siRNA and drug screen identified Aurora Kinase B as a potent and selective target in NB cells (15). Targeting either Survivin or Aurora B with small molecule inhibitors or siRNAs inhibits NB cell growth by induction of multinucleation and apoptosis (12–15). In the present study, we observed similar phenotypes in INCENP depleted NB cells. Consistent with the important function of INCENP and CPC, DAPI staining and cell cycle analysis showed that INCENP knockdown induced multinucleation and generated more cells with DNA content≥4N. In addition, in INCENP silenced NB cells there was massive apoptosis marked by activation of p53-p21 pathway (only in p53 WT NB cells), upregulation of Caspase-3/7 activity and increases in cleaved Caspase-3 and cleaved PARP-1 (Fig. 6C, Fig. 6D and Fig.S6D) and increases in the sub-G1 fraction of NB cells in the cell cycle analysis (Fig. 5D and Fig. S5C). In contrast, there was no obvious cell death in INCENP silenced non-tumorigeneic 293T and ARPE-19 cells or tumorigenic HeLa cells (Fig.S6H). These findings suggest that NB cells are more sensitive to INCENP silencing than non-tumorigenic cells and some other types of cancer cells. One possible explanation why NB cells respond so dramatically to INCENP silencing is that NB cells have a dysregulated postmitotic checkpoint due to disruption of the p53-p21 axis (43,44) or loss of pRb or amplification/overexpression of MYC family genes (44–46). The significant increase in DNA damage signaling in INCENP knockdown cells supports a model in which polyploid NB cells arising after INCENP depletion are able to re-enter the next cell cycle and undergo S-phase and subsequent mitosis and these events eventually lead to endoreduplication, increased genomic instability and apoptosis (47).
In this study, we report that both Aurora B inhibiton and INCENP knockdown significantly increase the number of senescent cells in NB, as judged by cell morphology, SA-β-gal expression, induction of p53-p21cip1 signaling pathway, hypo-phosphorylated RB and reduction of cell proliferation. Aurora B kinase inhibition has been associated with an increase in sensescent cells in several different types of cancer cells and normal cells (28–32) which is variably reported to be dependent on p53 (29) (31). Here, we find INCENP knockdown induces senescence in both p53 wild-type NGP and p53 mutant BE2C cells and this indicates that senescence induced by INCENP depletion in NB is not p53 dependent.
CPC has been proved as a very potential therapeutic target for cancer by inhibiting either Aurora B or Suvivin. However, targeting these two components couldn’t always specifically inhibit CPC function for following reasons. Firstly, to date, many Aurora kinase inhibitors have been developed to interfere with the activation of these kinases by binding ATP-binding pocket competitively, including Barasertib (AZD1152) (48) used in this study. As many kinases share similar ATP-binding pocket, these chemical inhibitors have non-specific activities against many other kinases (49). Furthermore, it is relatively straightforward to generate mutant cells resistant to these Aurora inhibitors after long term treatment, and the utility of these inhibitors in the cancer therapy may therefore be limited (49,50). Secondly, Survivn also plays important roles in regulating apoptosis independent on CPC (39). Therefore, inhibition of Survivin could suppress tumor cell growth through both CPC dependent and CPC independent mechanism. Targeting INCENP or the interaction between INCENP and Aurora B kinase may provide a novel and alternative strategy to inhibit the CPC function and block NB cell growth (51,52). However, alternative strategies to limit the CPC function by targeting INCENP need to be further tested. The identification of peptides specifically interacting with INCENP may enable a PROTAC strategy (53–55) to be developed that selectively degrades INCENP and destabilize the CPC complex. Additionally an antisense oligo mediated gene silencing approach has been shown to effectively target nuclear protein-STAT3 in lymphomas and lung cancer (56) as well as NB (57) and such an approach could also provide another feasible strategy to selectively target CPC by inhibiting INCENP.
In conclusion, our study demonstrated the importance of INCENP for NB cell growth and the association of high INCENP expression with poor prognosis for NB patients. We further showed INCENP silencing inhibited NB tumor growth in NB xenograft model. Finally, we demonstrated INCENP silencing inhibited NB growth by induction of polyploidization, DNA damage response, massive cell apoptosis and cellular senescence.
Supplementary Material
Acknowledgements
We would like to thank Dr. Karen M. Wolcott and Norris Lam for their technical assistance with cell cycle analysis. We are grateful to Dr. Alexander E. Kelly for his helpful suggestions about this work. We thank all members of the Cell and Molecular Biology Section (CMBS), National Cancer Institute (NCI) for helpful discussions and comments during the course of all the experiments and preparation of manuscript. This research was supported by the Center for Cancer Research, National Cancer Institute, NIH Intramural Research program.
Footnotes
Conflict of interest
The authors declare no potential conflicts of interest.
References
- 1.Maris JM. Recent advances in neuroblastoma. N Engl J Med 2010;362:2202–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet 2007;369:2106–20 [DOI] [PubMed] [Google Scholar]
- 3.Schulte JH, Eggert A. Neuroblastoma. Crit Rev Oncog 2015;20:245–70 [DOI] [PubMed] [Google Scholar]
- 4.Bosse KR, Maris JM. Advances in the translational genomics of neuroblastoma: From improving risk stratification and revealing novel biology to identifying actionable genomic alterations. Cancer 2016;122:20–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luksch R, Castellani MR, Collini P, De Bernardi B, Conte M, Gambini C, et al. Neuroblastoma (Peripheral neuroblastic tumours). Crit Rev Oncol Hematol 2016;107:163–81 [DOI] [PubMed] [Google Scholar]
- 6.Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, et al. Advances in Risk Classification and Treatment Strategies for Neuroblastoma. J Clin Oncol 2015;33:3008–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veschi V, Liu Z, Voss TC, Ozbun L, Gryder B, Yan C, et al. Epigenetic siRNA and Chemical Screens Identify SETD8 Inhibition as a Therapeutic Strategy for p53 Activation in High-Risk Neuroblastoma. Cancer Cell 2017;31:50–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veschi V, Thiele CJ. High-SETD8 inactivates p53 in neuroblastoma. Oncoscience 2017;4:21–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol 2012;13:789–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honda R, Korner R, Nigg EA. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol Biol Cell 2003;14:3325–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein UR, Nigg EA, Gruneberg U. Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of Borealin, Survivin, and the N-terminal domain of INCENP. Mol Biol Cell 2006;17:2547–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azuhata T, Scott D, Takamizawa S, Wen J, Davidoff A, Fukuzawa M, et al. The inhibitor of apoptosis protein survivin is associated with high-risk behavior of neuroblastoma. J Pediatr Surg 2001;36:1785–91 [DOI] [PubMed] [Google Scholar]
- 13.Lamers F, Schild L, Koster J, Versteeg R, Caron HN, Molenaar JJ. Targeted BIRC5 silencing using YM155 causes cell death in neuroblastoma cells with low ABCB1 expression. Eur J Cancer 2012;48:763–71 [DOI] [PubMed] [Google Scholar]
- 14.Lamers F, van der Ploeg I, Schild L, Ebus ME, Koster J, Hansen BR, et al. Knockdown of survivin (BIRC5) causes apoptosis in neuroblastoma via mitotic catastrophe. Endocr Relat Cancer 2011;18:657–68 [DOI] [PubMed] [Google Scholar]
- 15.Bogen D, Wei JS, Azorsa DO, Ormanoglu P, Buehler E, Guha R, et al. Aurora B kinase is a potent and selective target in MYCN-driven neuroblastoma. Oncotarget 2015;6:35247–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabisch M, Lorenzo Bermejo J, Dunnebier T, Ying S, Michailidou K, Bolla MK, et al. Inherited variants in the inner centromere protein (INCENP) gene of the chromosomal passenger complex contribute to the susceptibility of ER-negative breast cancer. Carcinogenesis 2015;36:256–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kar SP, Beesley J, Amin Al Olama A, Michailidou K, Tyrer J, Kote-Jarai Z, et al. Genome-Wide Meta-Analyses of Breast, Ovarian, and Prostate Cancer Association Studies Identify Multiple New Susceptibility Loci Shared by at Least Two Cancer Types. Cancer Discov 2016;6:1052–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbanis S, Ioannou M, Kouvaras E, Karasavvidou F, Nakou M, Papamichali R, et al. INCENP (inner centromere protein) is overexpressed in high grade non-Hodgkin B-cell lymphomas. Pathol Oncol Res 2009;15:11–7 [DOI] [PubMed] [Google Scholar]
- 19.Xia R, Chen S, Chen Y, Zhang W, Zhu R, Deng A. A chromosomal passenger complex protein signature model predicts poor prognosis for non-small-cell lung cancer. Onco Targets Ther 2015;8:721–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koster J, Molenaar JJ, Versteeg R. R2: Accessible web-based genomics analysis and visualization platform for biomedical researchers. Cancer Research 2015;75 [Google Scholar]
- 21.Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet 2013;45:279–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kocak H, Ackermann S, Hero B, Kahlert Y, Oberthuer A, Juraeva D, et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis 2013;4:e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 2014;159:1126–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeid R, Lawlor MA, Poon E, Reyes JM, Fulciniti M, Lopez MA, et al. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat Genet 2018;50:515–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaelis KA, Knox AJ, Xu M, Kiseljak-Vassiliades K, Edwards MG, Geraci M, et al. Identification of growth arrest and DNA-damage-inducible gene beta (GADD45beta) as a novel tumor suppressor in pituitary gonadotrope tumors. Endocrinology 2011;152:3603–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nettersheim D, Jostes S, Fabry M, Honecker F, Schumacher V, Kirfel J, et al. A signaling cascade including ARID1A, GADD45B and DUSP1 induces apoptosis and affects the cell cycle of germ cell cancers after romidepsin treatment. Oncotarget 2016;7:74931–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ou DL, Shen YC, Yu SL, Chen KF, Yeh PY, Fan HH, et al. Induction of DNA damage-inducible gene GADD45beta contributes to sorafenib-induced apoptosis in hepatocellular carcinoma cells. Cancer Res 2010;70:9309–18 [DOI] [PubMed] [Google Scholar]
- 28.Bonet C, Giuliano S, Ohanna M, Bille K, Allegra M, Lacour JP, et al. Aurora B is regulated by the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signaling pathway and is a valuable potential target in melanoma cells. J Biol Chem 2012;287:29887–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadaie M, Dillon C, Narita M, Young AR, Cairney CJ, Godwin LS, et al. Cell-based screen for altered nuclear phenotypes reveals senescence progression in polyploid cells after Aurora kinase B inhibition. Mol Biol Cell 2015;26:2971–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuda Y, Iimori M, Nakashima Y, Nakanishi R, Ando K, Ohgaki K, et al. Mitotic slippage and the subsequent cell fates after inhibition of Aurora B during tubulin-binding agent-induced mitotic arrest. Sci Rep 2017;7:16762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Leite de Oliveira R, Wang C, Fernandes Neto JM, Mainardi S, Evers B, et al. High-Throughput Functional Genetic and Compound Screens Identify Targets for Senescence Induction in Cancer. Cell Rep 2017;21:773–83 [DOI] [PubMed] [Google Scholar]
- 32.Wang LX, Wang JD, Chen JJ, Long B, Liu LL, Tu XX, et al. Aurora A Kinase Inhibitor AKI603 Induces Cellular Senescence in Chronic Myeloid Leukemia Cells Harboring T315I Mutation. Sci Rep-Uk 2016;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valentijn LJ, Koster J, Haneveld F, Aissa RA, van Sluis P, Broekmans ME, et al. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc Natl Acad Sci U S A 2012;109:19190–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimmerman MW, Liu Y, He S, Durbin AD, Abraham BJ, Easton J, et al. MYC Drives a Subset of High-Risk Pediatric Neuroblastomas and Is Activated through Mechanisms Including Enhancer Hijacking and Focal Enhancer Amplification. Cancer Discov 2018;8:320–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caren H, Kryh H, Nethander M, Sjoberg RM, Trager C, Nilsson S, et al. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc Natl Acad Sci U S A 2010;107:4323–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mlakar V, Jurkovic Mlakar S, Lopez G, Maris JM, Ansari M, Gumy-Pause F. 11q deletion in neuroblastoma: a review of biological and clinical implications. Mol Cancer 2017;16:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barr EK, Applebaum MA. Genetic Predisposition to Neuroblastoma. Children (Basel) 2018;5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hungate EA, Applebaum MA, Skol AD, Vaksman Z, Diamond M, McDaniel L, et al. Evaluation of Genetic Predisposition for MYCN-Amplified Neuroblastoma. J Natl Cancer Inst 2017;109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 1997;3:917–21 [DOI] [PubMed] [Google Scholar]
- 40.Uren AG, Beilharz T, O’Connell MJ, Bugg SJ, van Driel R, Vaux DL, et al. Role for yeast inhibitor of apoptosis (IAP)-like proteins in cell division. Proc Natl Acad Sci U S A 1999;96:10170–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL, et al. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol 2000;10:1319–28 [DOI] [PubMed] [Google Scholar]
- 42.Yue Z, Carvalho A, Xu Z, Yuan X, Cardinale S, Ribeiro S, et al. Deconstructing Survivin: comprehensive genetic analysis of Survivin function by conditional knockout in a vertebrate cell line. J Cell Biol 2008;183:279–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gizatullin F, Yao Y, Kung V, Harding MW, Loda M, Shapiro GI. The Aurora kinase inhibitor VX-680 induces endoreduplication and apoptosis preferentially in cells with compromised p53-dependent postmitotic checkpoint function. Cancer Res 2006;66:7668–77 [DOI] [PubMed] [Google Scholar]
- 44.Giet R, Petretti C, Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol 2005;15:241–50 [DOI] [PubMed] [Google Scholar]
- 45.Hook KE, Garza SJ, Lira ME, Ching KA, Lee NV, Cao J, et al. An integrated genomic approach to identify predictive biomarkers of response to the aurora kinase inhibitor PF-03814735. Mol Cancer Ther 2012;11:710–9 [DOI] [PubMed] [Google Scholar]
- 46.Yang D, Liu H, Goga A, Kim S, Yuneva M, Bishop JM. Therapeutic potential of a synthetic lethal interaction between the MYC proto-oncogene and inhibition of aurora-B kinase. Proc Natl Acad Sci U S A 2010;107:13836–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hilton JF, Shapiro GI. Aurora Kinase Inhibition As an Anticancer Strategy. J Clin Oncol 2014;32:57–9 [DOI] [PubMed] [Google Scholar]
- 48.Mortlock AA, Foote KM, Heron NM, Jung FH, Pasquet G, Lohmann JJM, et al. Discovery, synthesis, and in vivo activity of a new class of pyrazoloquinazolines as selective inhibitors of aurora B kinase. J Med Chem 2007;50:2213–24 [DOI] [PubMed] [Google Scholar]
- 49.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, Mclauchlan H, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J 2007;408:297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Failes TW, Mitic G, Abdel-Halim H, Po’uha ST, Liu M, Hibbs DE, et al. Evolution of Resistance to Aurora Kinase B Inhibitors in Leukaemia Cells. Plos One 2012;7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gohard FH, St-Cyr DJ, Tyers M, Earnshaw WC. Targeting the INCENP IN-box-Aurora B interaction to inhibit CPC activity in vivo. Open Biol 2014;4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Unsal E, Degirmenci B, Harmanda B, Erman B, Ozlu N. A small molecule identified through an in silico screen inhibits Aurora B-INCENP interaction. Chem Biol Drug Des 2016;88:783–94 [DOI] [PubMed] [Google Scholar]
- 53.Sakamoto KM. Protacs for treatment of cancer. Pediatr Res 2010;67:505–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neklesa TK, Winkler JD, Crews CM. Targeted protein degradation by PROTACs. Pharmacol Ther 2017;174:138–44 [DOI] [PubMed] [Google Scholar]
- 55.Zou Y, Ma D, Wang Y. The PROTAC technology in drug development. Cell Biochem Funct 2019;37:21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hong D, Kurzrock R, Kim Y, Woessner R, Younes A, Nemunaitis J, et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Transl Med 2015;7:314ra185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Odate S, Veschi V, Yan S, Lam N, Woessner R, Thiele CJ. Inhibition of STAT3 with the Generation 2.5 Antisense Oligonucleotide, AZD9150, Decreases Neuroblastoma Tumorigenicity and Increases Chemosensitivity. Clin Cancer Res 2017;23:1771–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.