Abstract
Background:
A significant fraction of prostate cancer (PC) patients experience post-radical prostatectomy (RP) biochemical recurrence (BCR). New predictive markers are needed for optimizing post-operative PC management. STAT5 is an oncogene in PC which undergoes amplification in 30% of PCs during progression.
Methods:
We evaluated the significance of a positive status for nuclear STAT5 protein expression vs. STAT5 locus amplification vs. combined positive status for both in predicting BCR after RP in 300 patients.
Results:
Combined positive STAT5 status was associated with a 45% disadvantage in BCR in Kaplan-Meier survival analysis in all Gleason grade patients. Patients with Gleason grade groups (GG) 2 and 3 PCs and combined positive status for STAT5 had a more pronounced disadvantage of 55–60% at 7 years after RP in univariate analysis. In multivariate analysis including the CAPRA-S nomogram variables, combined positive STAT5 status was independently associated with a shorter BCR-free survival in all Gleason GG patients (HR=2.34, p=0.014) and in intermediate Gleason GG 2 or 3 patients (HR=3.62, p=0.021). The combined positive STAT5 status improved the predictive value of the CAPRA-S nomogram in both receiver-operating-characteristic area-under-the-curve analysis and in decision-curve-analysis for BCR.
Conclusions:
Combined positive status for STAT5 was independently associated with shorter disease-free survival in univariate analysis and was an independent predictor for BCR in multivariate analysis using the CAPRA-S variables in PC.
Impact:
Our results highlight potential for a novel precision medicine concept based on a pivotal role of STAT5 status in improving selection of PC patients who are candidates for early adjuvant interventions to reduce the risk of recurrence.
Keywords: STAT5, gene rearrangement, prostate cancer, recurrence, radical prostatectomy, intermediate risk group, CAPRA-S, risk-stratification, Gleason grade group
Introduction
Prostate cancer (PC) is among the three most common cancers in men worldwide, and the second leading cause of cancer-related deaths of men in North America(1). The standard treatment for organ-confined prostate cancer is surgery or radiation therapy(1). After radical prostatectomy (RP), approximately 30–60% of patients with intermediate risk PC experience biochemical disease recurrence (BCR) as evidenced by rising post-operative serum PSA levels(2). BCR is a surrogate marker of disease progression and triggers follow-up treatments aiming to prevent clinical PC recurrence. While post-prostatectomy PSA is a specific indicator of disease status, by the time it becomes detectable, it is likely that significant regrowth of PC has already occurred.
Risk stratification of PC patients at the time of prostatectomy aims to identify patients at high risk of PC mortality who are likely to experience BCR and would require early multimodal therapy vs. those who are at a relatively low risk and might be spared from the potential impact of aggressive treatment on quality of life. A number of clinical variables are predictive for recurrence risk. The histological grade of PC itself provides prognostic information at the extremes of the scale where Gleason Grade Group (GG) 1 PCs are at a low risk and Gleason GGs 4 and 5 PCs are at a high risk(3). However, the prognostic information of Gleason GGs 2 and 3 is limited and, consequently, there is a great need for potent predictive biomarkers for identification of patients undergoing RP whose intermediate risk organ-confined PC is likely to progress early. A number of models for predicting post-prostatectomy BCR have been published including the D’Amico risk stratification scheme(4), the Waltz nomogram(5), Stephenson nomogram(6), Suardi nomogram(7) and the Cancer of the Prostate Risk Assessment Postsurgical nomogram (CAPRA-S)(8–10). CAPRA-S score has been shown to outperform Stephenson nomogram and D’Amico scheme in terms of calibration and decision curve analysis (DCA)(11,12). Key predictive variables of the CAPRA-S nomogram for post-prostatectomy BCR include preoperative PSA, pathological Gleason score, extraprostatic extension and seminal vesicle invasion(13). CAPRA-S score has been demonstrated to predict rapid PSA doubling time(14) as well as metastatic progression and disease-specific mortality in PC patients(15). However, improvement of the prediction models for better identification of patients at increased risk for BCR would allow initiation of adjuvant treatment strategies in association with minimal residual disease volume in order to achieve greater curative benefit to avoid development of metastatic disease and death due to PC.
We have shown that active STAT5 protein is a critical driver of PC progression by sustaining cancer cell viability in vitro and by inducing PC growth in vivo when grown as xenograft tumors in mice(16–19). In addition, active STAT5 triggers epithelial-mesenchymal transition in PC cells and induces metastatic dissemination of PC in preclinical models(20). STAT5 is comprised of two highly homologous isoforms STAT5a and STAT5b, which act both as signaling proteins and nuclear transcription factors and are encoded by separate juxtaposed genes on chromosome 17q21(21,22). Our previous studies demonstrated that nuclear STAT5 levels are high in PC tissues but not in normal prostate epithelium(21). We further showed that STAT5 gene locus undergoes amplification during PC progression to metastatic castrate-resistant PC, and that STAT5 locus amplification results in increased STAT5 protein levels in PC(21). Based on analyses of three independent cohorts, we demonstrated that elevated levels of active nuclear STAT5 protein in PC at intent-to-cure RP predicted disease recurrence and early PC-specific death(23,24). These findings corroborate the role of STAT5 in growth and metastatic behavior of PC shown in preclinical models.
In the present study, we evaluated the predictive value of a positive status for STAT5 locus amplification vs. nuclear STAT5 protein expression vs. combined positive status for both STAT5 gene locus amplification and nuclear STAT5 protein expression for BCR after RP in univariate and multivariate analyses. Patients with combined positive status for STAT5 gene amplification and nuclear protein overexpression suffered a 45% disadvantage in BCR in Kaplan-Meier survival analysis compared to patients with negative status for STAT5. Importantly, patients with Gleason grade group (GG) 2 and 3 PCs and a combined positive status for STAT5 had even a more pronounce disadvantage of 55–60% at 7 years after RP in univariate analysis. We further evaluated the predictive value of combined positive STAT5 status as an additional variable in the CAPRA-S nomogram(8) which is currently used for risk assessment for BCR following RP. Our results show that when combined positive STAT5 status was added to the CAPRA-S variables in multivariate analysis, a positive status for STAT5 was a significant independent predictor for BCR. Moreover, combined positive status for STAT5 improved the receiver operating characteristic-area under the curve (ROC-AUC) analysis and the decision curve analysis (DCA)(25) in disease-free survival in all Gleason GG patients and in Gleason GG 2 and 3 patients. This work introduces a novel concept that patients with combined positive STAT5 status for both locus amplification and nuclear protein expression at RP may be at an increased risk of PC recurrence post-RP, which needs to be evaluated in larger cohorts. Incorporation of STAT5 status into the evaluation of surgically removed prostate cancer may significantly improve the selection of patients with the potential to benefit from early adjuvant therapy following RP.
Materials and Methods
Patient Characteristics
Our patient cohort included 532 patients having undergone RP at the Turku University Hospital, Finland, during years 2000–2005 and analyzed as a subcohort in our previous study for nuclear STAT5 protein expression status (24). The study protocol was approved by the Ethics Committee of the University of Helsinki (Helsinki, Finland), and the National Data Protection Ombudsman (Helsinki, Finland) was notified about the collection of the information. After excluding patients who received neoadjuvant treatments (the exclusion criteria included any neoadjuvant treatments prior to RP), complete relapse-free survival-related clinical data and follow-up information, RP tissue materials for re-review and tissue microarray construction were available for 457 patients (Table 1). The cohort for BCR did not include any patients who received salvage radiation therapy. The cohort was not controlled for other post-RP adjuvant therapies. Complete CAPRA-S variables were available for 396 patients, STAT5 protein expression status for 417 patients and STAT5 gene locus amplification status for 376 patients. Complete information for CAPRA-S and STAT5 variables were available for 300 patients (Table 1). The 300 patients were comparable in clinico-pathologic variables to the remaining 157 patients from the original study group as demonstrated in Table 1 and compared by Student’s t-test and Fisher’s exact test. Approximately 24% of patients in both the entire cohort and in the subcohort of 300 patients with complete CAPRA-S and STAT5 information experienced biochemical recurrence (BCR). Univariate Cox regression analyses of this subcohort of 300 patients demonstrate that PSA above 6, positive surgical margins, seminal vesicle involvement, Gleason GG above 1, extracapsular extension and lymph node involvement, each predicted shorter BCR-free survival (Suppl. Table 1), as expected, thus validating the use of the subcohort.
Table 1.
Demographics of the study cohort
| Total Cohort (n = 457) | Patients without Complete Data (n = 157) | Patients with Complete Data (n = 300) | P-value | |
|---|---|---|---|---|
| Age at RP, years (mean, SD) (n = 457) | 62.0 (5.8) | 61.62 (6.0) | 61.65 (5.6) | 0.953a |
| Preoperative PSA, ng/ml (n, %) (n = 457) | ||||
| ≤10.0 | 294 (69.2) | 80 (70.4) | 206 (68.7) | 0.443b |
| 10.1–20.0 | 96 (22.6) | 30 (24.0) | 66 (22.0) | |
| >20.0 | 35 (8.2) | 7 (5.6) | 28 (9.3) | |
| Grade group at RP (n, %) (n = 457) | ||||
| 1 | 168 (36.8) | 65 (41.4) | 103 (34.3) | 0.294b |
| 2 | 134 (29.3) | 48 (30.6) | 86 (28.7) | |
| 3 | 63 (13.8) | 20 (12.7) | 43 (14.3) | |
| 4 | 70 (15.3) | 17 (10.8) | 53 (17.7) | |
| 5 | 22 (4.8) | 7 (4.5) | 15 (5.0) | |
| pT (n, %) (n = 440) | ||||
| 2 | 233 (53.0) | 72 (51.4) | 161 (53.7) | 0.785b |
| 3a | 153 (34.8) | 52 (37.1) | 101 (33.7) | |
| 3b | 54 (12.2) | 16 (11.5) | 38 (12.6) | |
| Lymph node status (n, %) (n = 454) | ||||
| Negative | 434 (95.6) | 150 (97.4) | 284 (94.7) | 0.230b |
| Positive | 20 (4.4) | 4 (2.6) | 16 (5.3) | |
| Follow-up time after RP, yr (median, range) (n = 457) | 9.5 (0.2–14.0) | 10.17 (0.6–13.99) | 9.52 (0.73–13.97) | 0.006a |
| Biochemical recurrence (n, %) (n = 457) | 110 (24.1) | 37 (23.7) | 73 (24.3) | 0.909b |
| Death from any cause (n, %) (n = 457) | 73 (16.0) | 21 (13.4) | 52 (17.3) | 0.286b |
| Death from prostate cancer (n, %) (n = 457) | 19 (4.2) | 8 (5.1) | 11 (3.7) | 0.468b |
| Patients receiving secondary therapy after RP (n, %) (n = 438) | 136 (31.1) | 42 (28.0) | 94 (32.6) | 0.330b |
| STAT5 IHC (n, %) (n = 417) | ||||
| Negative | 254 (60.9) | 74 (63.2) | 180 (60.0) | 0.576b |
| Positive | 163 (39.1) | 43 (36.8) | 120 (40.0) | |
| STAT5 FISH (n, %) (n = 376) | ||||
| Negative | 282 (75.0) | 61 (80.3) | 221 (73.7) | 0.300b |
| Positive | 94 (25.0) | 15 (19.7) | 79 (26.3) |
RP, radical prostatectomy; PSA, prostate specific antigen; IHC, immunohistochemistry; FISH, fluorescence in situ hybridization;
Student’s t-test,
Fisher’s exact test.
Tissue Microarray (TMA) Construction
Slides of the whole-mounted RP specimens were re-reviewed by T.M. and M.K. according to the current Gleason grading and WHO/ISUP Grade grouping criteria(26). TMAs were constructed as described previously(24). Briefly, each patient’s RP H&E slides were evaluated and a minimum of three cores (on average 9–10 cores), 1mm in diameter each, from the dominant cancer focus were transferred to the recipient TMA block. In addition, at least one core representing benign prostate epithelium remote from the cancer site was also selected for each patient. The representativeness of each core was confirmed by H&E staining in a sequential section following the STAT5 IHC and STAT5 FISH analyses.
STAT5 Immunohistochemistry and Scoring of the TMAs
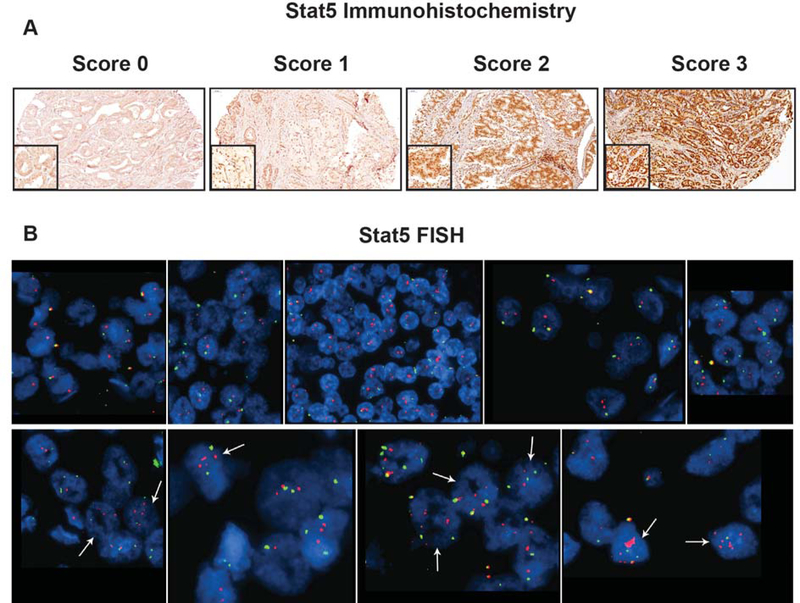
Immunohistochemistry (IHC) was conducted as described previously(16,23,24,27). In brief, for evaluation of nuclear STAT5 protein levels, each TMA core was given a score based on both intensity and proportion of immunostained cells where 0 represented negative, 1 weak, 2 moderate and 3 strong immunostaining for STAT5. The final score for each patient was the highest score of the individual scores. For example, if a subject had three cores scored 0, 1 and 3, the final STAT5 score for that subject would be 3(24). Representative pictures of IHC scores are shown in Figure 1A. Scores 0 and 1 were considered negative while scores 2 and 3 represented positive status for nuclear STAT5 protein. These cutoff-points for nuclear STAT5 protein levels as a function of survival were defined in three previously analyzed cohorts(23,24). In the current study, we evaluated the predictive value of the STAT5 locus amplification status alone or in combination with the Stat5 protein status.
Figure 1.
STAT5 protein levels and STAT5 locus amplification in prostate cancer tissue microarrays. A: Individual prostate cancer (PC) tissue microarray (TMA) cores were scored for nuclear STAT5a/b levels, detected by immunohistochemistry (IHC), on a scale from 0 to 3, where 0 represented negative, 1 weak, 2 moderate and 3 strong immunostaining (top panel). B: STAT5A/B FISH analysis of PC TMA cores (bottom panel). The top row shows representative sections from 5 cases showing no STAT5 locus amplification. Most cells show 2 copies of STAT5A/B (red signal). A chromosome 17 centromeric probe was used (green signal) as a control. The bottom row shows representative sections from 4 cases with STAT5 locus amplification. Arrows point to cells with STAT5 locus amplification.
Fluorescence in situ Hybridization (FISH)
Evaluation of STAT5 locus amplification in TMA tissue cores using FISH analysis was performed as described earlier(21). Briefly, we designed a STAT5A/B probe consisting of a contig of four overlapping bacterial artificial chromosome (BAC) clones containing sequences of the STAT5A/B gene: RP11–60B4, RP11–1151C17, RP11–1151G10, and RP11–365D24 (BACPAC Resources, Oakland, CA). As a control probe, we used a chromosome 17 centromeric BAC clone: RP11–299G20 (BACPAC Resources, Oakland, CA). FISH analysis of the TMAs was performed using a standard protocol(21). The STAT5A/B probe was detected using red fluorescence and chromosome 17 centromeric control probe using green fluorescence. Scoring of cells and digital image acquisition were performed as described earlier(21). STAT5 locus amplification was defined as a signal ratio of gene probe to control probe ≥2 or five or more copies of the gene signal in ≥10% of the tumor nuclei(21). STAT5 locus amplification status was scored either positive or negative if any of the cores from a given patient was positive for STAT5 locus amplification. Thus, STAT5 locus amplification evaluation did not require a cutoff analysis in a training cohort.
Statistical Analyses
Biochemical recurrence (BCR) was defined as a minimum of two consecutive post-RP PSA measurements of 0.2 ng/ml or higher. Mortality data was registered as death due to PC or death due to any other cause for disease-specific survival (DSS) and overall survival (OS) analyses, respectively. Kaplan-Meier, ROC-AUC, and Decision Curve Analyses (DCA) were run in R version 3.3.2 using the survival, survminer, pROC, and MASS packages.
Nuclear STAT5 levels were determined by IHC (Figure 1A) and STAT5 gene copy number status by FISH (Figure 1B), as described previously(21,23,24). The analysis of a cohort of 457 PCs, treated exclusively by RP, yielded complete information for all CAPRA-S and STAT5 variables for 300 patients (Table 1). Since the FISH and IHC were not conducted simultaneously, these two analyses were performed on serial sections of the TMAs and do not represent the FISH/IHC status of the same cells. The maximal patient-wise marker status (either STAT5 protein or STAT5 locus amplification) was used to assess the final patient marker status. Concordance of STAT5 protein status and STAT5 locus amplification was high at patient level (88.9%) and is shown in Supplementary Table 2 (all spots disconcordant).
Results
Kaplan-Meier Analysis of Disease-Free Survival of Prostate Cancer Patients with Combined Positive STAT5 Status
The levels of nuclear STAT5a/b expression in PCs were examined by IHC, as described previously(23,27,28). Representative images of PCs with no detectable nuclear STAT5a/b (score 0), weak (score 1), moderate (score 2) or strong (score 3) nuclear STAT5a/b expression are shown in Figure 1A. To identify PCs with the STAT5 gene locus amplification, we used FISH analysis of paraffin embedded (FFPE) tissue cores of localized PCs removed by RP, as described previously(21). Figure 1B shows examples of PC cases with STAT5a/b diploid pattern (top panel) vs. PCs with STAT5a/b gene locus amplification (bottom panel). Optimal cutoff-points for nuclear STAT5 protein levels as a function of disease-free survival were defined in three cohorts that we have described previously(23). Scores 0 and 1 represented negative status for STAT5 protein, while scores 2–3 were considered positive for nuclear STAT5. The purpose of the current study was to evaluate the predictive value of STAT5 gene locus amplification alone or in combination with positive nuclear STAT5 protein status for biochemical recurrence of PC after RP.
The KM analysis showed, for the first time, that combined positive status for both nuclear STAT5 protein expression and STAT5 gene locus amplification (FISH+/IHC+) was associated with 55–60% disadvantage in disease-free survival at 7 years after RP in patients with Gleason GG 2 or 3 PCs (Figure 2). At the same time, combined positive status for STAT5 (FISH+/IHC+) was associated with approximately 45% disadvantage in disease-free survival in patients with PCs of all histological grades (Figure 2 A, B, C). Positive status for STAT5 locus amplification alone (STAT5 FISH+) was associated with less significant disadvantage of 20% in disease-free survival at 7 years after RP in patients with all Gleason GG PCs and in patients with Gleason GG 2 or 3 PCs (Figure 2 D, E, F). In addition, positive STAT5 locus amplification status predicted shorter disease-specific survival (DSS) and overall survival (OS) in Gleason GG 2 or 3 patients (Suppl. Figures 1 and 2). Consistent with our previous results(23,24), nuclear STAT5 protein expression alone (STAT5 IHC+) in Kaplan-Meier (KM) survival analysis predicted shorter BCR-free survival in patients with PCs of all Gleason GG (Figure 2 G, H, I). Collectively, our results suggest that positive status for STAT5 gene locus amplification, when combined with positive status for nuclear STAT5 protein expression, provides a more potent predictor of BCR than positive status for either STAT5 locus amplification or nuclear protein expression alone for PC after RP and, especially, for patients with Gleason GGs 2 or 3 PCs. These findings are important because only a limited number of predictive markers of PC recurrence after RP are currently available for patients with Gleason GG 2 or 3 PCs.
Figure 2.
Univariate analysis of the combined status for both STAT5 gene locus amplification status and protein expression and progression-free survival with biochemical recurrence (BCR) as the endpoint in prostate cancers of all Gleason GG (A) vs. prostate cancers of Gleason GG 2 and 3 (B) vs. GG 4 and 5 (C). Univariate analysis of the STAT5 gene locus amplification status and progression-free survival in prostate cancers of all Gleason Grade Groups (GG) (D) vs prostate cancers of Gleason GG 2 and 3 (E) vs GG 4 and 5 (F). Univariate analysis of the STAT5 protein status and progression-free survival in prostate cancers of all Gleason Grade Groups (GG) (G) vs. prostate cancers of Gleason GG 2 and 3 (H) vs. GG 4 and 5 (I). Kaplan Meier curves, with global p-values calculated by Mantel-Haenszel tests. The STAT5 protein status detected by immunohistochemistry (IHC) and STAT5 locus amplification by fluorescence in situ hybridization (FISH). Global Mantel-Haenszel log-rank p-values were calculated comparing the difference in survival between all patient groups.
The Predictive Value of Combined Positive STAT5 Status in Prostate Cancer in Relation to CAPRA-S Variables
To further investigate the independent predictive value of combined positive status for both nuclear STAT5 protein expression and gene locus amplification, we performed multivariate Cox regression analyses that included the CAPRA-S variables(8) (Tables 2a–c). Table 2a demonstrates multivariate analysis of the predictive value of the CAPRA-S variables alone without the STAT5 status for BCR in this cohort of 300 patients with complete STAT5 and CAPRA-S information showing the expected risk stratification by the CAPRA-S variables in this cohort. When both the STAT5 status and the CAPRA-S variables were included in the multivariate analysis, combined positive status for STAT5 was associated with a 2.34-fold increased risk of BCR compared to patients with combined negative status for STAT5 (FISH-/IHC-) in PC (p = 0.014, 95% CI = 1.184 – 4.625) (Table 2b). At the same time, patients having positive status for nuclear STAT5 protein expression alone in PC were at a 1.88-fold increased risk of BCR compared to patients negative for STAT5 protein (p=0.049) (Table 2b). When patients with Gleason GG 2 or 3 PC were analyzed separately, combined positive status for STAT5 was associated with the highest risk of BCR (a 3.62-fold increased risk) in multivariate analysis (p=0.021, 95% CI 1.22 – 10.78) (Table 2c). Importantly, in Gleason GG 2 and 3 PCs combined positive STAT5 status was a stronger predictor of BCR than any of the clinical parameters of the CAPRA-S nomogram alone (Table 2c). These data indicate that combined positive STAT5 status of PC, even when accounting for clinical variables in the CAPRA-S nomogram, was independently associated with increased risk of BCR after RP.
Table 2a.
Multivariate Cox regression analysis of the risk of biochemical recurrence by CAPRA-S variables in all PC patients with complete data (n = 300)
| Variable | Level | HR | 95% CI | P-Value |
|---|---|---|---|---|
| PSA | 0–6 | Reference | - | |
| 6.01–10 | 2.673 | (1.161 – 6.153) | 0.021 | |
| 10.01–20 | 2.996 | (1.261 – 7.12) | 0.013 | |
| > 20 | 8.059 | (3.14 – 20.689) | < 0.001 | |
| SM | Negative | Reference | - | |
| Positive | 0.563 | (0.32 – 0.99) | 0.046 | |
| SVI | No | Reference | - | |
| Yes | 0.921 | (0.487 – 1.743) | 0.802 | |
| Grade Group | 1 | Reference | - | |
| 2–3 | 1.697 | (0.826 – 3.485) | 0.15 | |
| 4–5 | 3.283 | (1.577 – 6.835) | 0.001 | |
| EPE | No | Reference | - | |
| Yes | 3.071 | (1.714 – 5.5) | < 0.001 | |
| LNI | No | Reference | - | |
| Yes | 1.495 | (0.691 – 3.234) | 0.307 |
PSA, prostate specific antigen; SM, surgical margin; SVI, seminal vesicle involvement; EPE, extraprostatic extension; LNI, lymph node involvement.
Table 2c.
Multivariate Cox regression analysis of the risk of biochemical recurrence (BCR) by CAPRA-S variables and STAT5 status in patients with Gleason GG 2 or 3 PC (n = 129)
| Variable | Level | HR | 95% CI | P-Value |
|---|---|---|---|---|
| PSA | 0–6 | Reference | - | - |
| 6.01–10 | 1.461 | (0.41 – 5.24) | 0.560 | |
| 10.01–20 | 2.442 | (0.71 – 8.42) | 0.157 | |
| > 20 | 10.422 | (2.69 – 40.43) | 0.001 | |
| SM | Negative | Reference | - | - |
| Positive | 0.418 | (0.15 – 1.17) | 0.095 | |
| SVI | No | Reference | - | - |
| Yes | 0.800 | (0.27 – 2.41) | 0.692 | |
| EPE | No | Reference | - | - |
| Yes | 2.571 | (0.97 – 6.82) | 0.058 | |
| LNI | No | Reference | - | - |
| Yes | 1.189 | (0.23 – 6.05) | 0.834 | |
| STAT5 | STAT5 FISH−/IHC− | Reference | - | - |
| STAT5 FISH+/IHC− | 2.442 | (0.79 – 7.52) | 0.120 | |
| STAT5 FISH−/IHC+ | 2.543 | (0.91 – 7.07) | 0.074 | |
| STAT5 FISH+/IHC+ | 3.623 | (1.22 – 10.78) | 0.021 |
PSA, prostate specific antigen; SM, surgical margin; SVI, seminal vesicle involvement; EPE, extraprostatic extension; LNI, lymph node involvement.
Table 2b.
Multivariate Cox regression analysis of the risk of biochemical recurrence (BCR) by CAPRA-S variables and STAT5 status in all PC patients with complete data (n = 300)
| Variable | Level | HR | 95% CI | P-Value |
|---|---|---|---|---|
| PSA | 0–6 | Reference | - | |
| 6.01–10 | 2.559 | (1.099 – 5.960) | 0.029 | |
| 10.01–20 | 2.776 | (1.165 – 6.614) | 0.021 | |
| > 20 | 7.256 | (2.776 – 18.964) | < 0.001 | |
| SM | Negative | Reference | - | |
| Positive | 0.544 | (0.304 – 0.972) | 0.04 | |
| SVI | No | Reference | - | |
| Yes | 0.98 | (0.522 – 1.840) | 0.95 | |
| Grade Group | 1 | Reference | - | |
| 2–3 | 1.812 | (0.878 – 3.740) | 0.108 | |
| 4–5 | 2.905 | (1.381 – 6.111) | 0.005 | |
| EPE | No | Reference | - | |
| Yes | 2.934 | (1.624 – 5.301) | < 0.001 | |
| LNI | No | Reference | - | |
| Yes | 1.471 | (0.677 – 3.196) | 0.33 | |
| STAT5 | STAT5 FISH−/IHC− | Reference | - | |
| STAT5 FISH+/IHC− | 1.813 | (0.875 – 3.754) | 0.109 | |
| STAT5 FISH−/IHC+ | 1.876 | (1.003 – 3.508) | 0.049 | |
| STAT5 FISH+/IHC+ | 2.340 | (1.184 – 4.625) | 0.014 |
PSA, prostate specific antigen; SM, surgical margin; SVI, seminal vesicle involvement; EPE, extraprostatic extension; LNI, lymph node involvement.
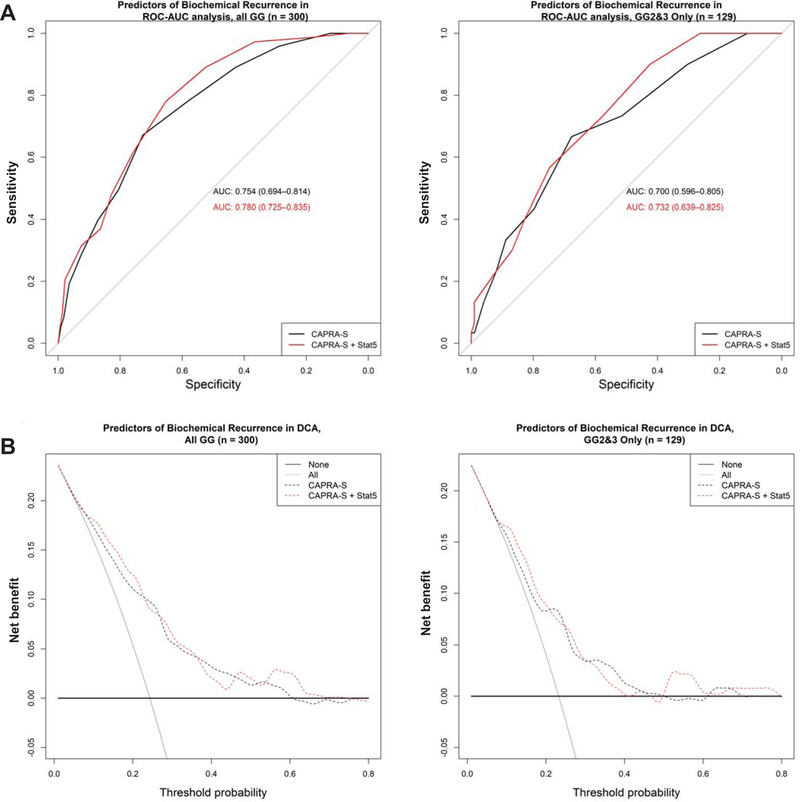
Given that data from both univariate and multivariate survival analyses supported a new concept that combined positive STAT5 status identifies a group of PC patients with the highest risk of recurrence, we next sought to assess whether the combined positive STAT5 status adds predictive value to the CAPRA-S nomogram in ROC-AUC analyses (Figure 3A, Suppl. Figure 3, Table 3). In ROC-AUC analyses of all PC patients (Table 3a), positive status for either STAT5 IHC or STAT5 FISH, each, added predictive value alone (both by 0.013; 0.767 minus 0.754). Combined positive status for both STAT5 IHC and FISH added the most predictive value for BCR by 0.026 (0.780 minus 0.754), when compared to CAPRA-S alone (Table 3a, Figure 3A). When exclusively Gleason GG 2 and 3 PC patients were evaluated, combined positive STAT5 status added predictive value to CAPRA-S by 0.032 (0.732 minus 0.700) in ROC-AUC analysis (Table 3b, Figure 3A). To assess potential added value of STAT5 status to treatment decisions of patients, we conducted decision curve-analysis (DCA) (Figure 3B, Suppl. Figure 4) where models are compared against the percentage likelihood of an outcome event to occur (25). In DCA of both the whole cohort and sub-analysis of Gleason GG 2 and 3 patients exclusively, combined Stat5 status with CAPRA-S (as compared to CAPRA-S alone) added clinical benefit across most probabilities of BCR occurring. Specifically, for the whole cohort (Figure 3B), DCA added benefit in approximately the 10–25%, 30–40%, and 50–70% ranges. In Gleason GG 2 or 3 PCs, DCA added benefit in the 10–25%, 30–35%, and 50%−80% ranges (Figure 3B). While the decision curves intersect at a number of probabilities, this result suggests that for patients with a low-risk of BCR (<25% risk) assessing combined Stat5 status adds clinical benefit. Taken together, these results suggest that combined positive status for STAT5 protein and gene locus amplification may add value to the CAPRA-S nomogram to predict PC recurrence after RP, a finding that requires follow-up studies in other cohorts.
Figure 3.
Receiver-operating characteristic-area under the curve (ROC-AUC) and decision curve analyses (DCA). A: ROC-AUC and (B) DCA of CAPRA-S with and without the combined positive STAT5 status in the entire cohort, and a separate sub-analysis of Gleason GG 2 and 3 PC patients exclusively.
Table 3a.
ROC-AUC values for CAPRA-S and STAT5 status in all PC patients (n = 300)
| Model | BCR | 95% CI | OS | 95% CI | DSS | 95% CI |
|---|---|---|---|---|---|---|
| CAPRA-S | 0.754 | (0.694 – 0.814) | 0.608 | (0.522 – 0.694) | 0.818 | (0.685 – 0.952) |
| CAPRA-S + STAT5 IHC | 0.767 | (0.710 – 0.824) | 0.605 | (0.519 – 0.692) | 0.825 | (0.786 – 0.948) |
| CAPRA-S + STAT5 FISH | 0.767 | (0.709 – 0.826) | 0.621 | (0.536 – 0.705) | 0.841 | (0.724 – 0.957) |
| CAPRA-S + STAT5 FISH+IHC | 0.780 | (0.725 – 0.835) | 0.619 | (0.534 – 0.704) | 0.849 | (0.743 – 0.956) |
CAPRA-S, cancer of the prostate postsurgical assessment; IHC, immunohistochemistry; FISH, fluorescence in situ hybridization.
Table 3b.
ROC-AUC values for CAPRA-S and STAT5 status in patients with Gleason GG 2 or 3 PC (n = 129)
| Model | BCR | 95% CI | OS | 95% CI | DSS | 95% CI |
|---|---|---|---|---|---|---|
| CAPRA-S | 0.700 | (0.596 – 0.805) | 0.503 | (0.375 – 0.631) | 0.617 | (0.365 – 0.869) |
| CAPRA-S + STAT5 IHC | 0.718 | (0.622 – 0.814) | 0.494 | (0.366 – 0.622) | 0.635 | (0.635 – 0.861) |
| CAPRA-S + STAT5 FISH | 0.715 | (0.614 – 0.816) | 0.517 | (0.391 – 0.643) | 0.674 | (0.449 – 0.900) |
| CAPRA-S + STAT5 FISH+IHC | 0.732 | (0.639 – 0.825) | 0.517 | (0.390 – 0.645) | 0.698 | (0.498 – 0.898) |
CAPRA-S, cancer of the prostate postsurgical assessment; IHC, immunohistochemistry; FISH, fluorescence in situ hybridization.
Discussion
Radical prostatectomy is the mainstay of the treatment of localized PC. Numerous prediction models based on preoperative and/or postoperative status have been developed to identify patients who are likely to experience post-RP BCR(10) and eventually metastatic progression and PC-specific death(10,13). Here, we evaluated the predictive value of combined positive status for both STAT5 locus amplification and nuclear protein expression in PCs at the time of RP in relation to CAPRA-S nomogram for BCR in the cohort of 300 PCs. Our data show that combined positive status for STAT5 in this cohort was independently associated with shorter disease-free survival in univariate analysis and was an independent predictor for BCR in multivariate analysis using the CAPRA-S variables in PC. In Gleason GG 2 and 3 PCs, combined positive STAT5 status was a stronger predictor of BCR than any other clinical variables of the CAPRA-S nomogram. In addition, combined positive STAT5 status added predictive value to the CAPRA-S nomogram in ROC-AUC and DCA analyses in the cohort analyzed here.
A key finding of this study is that combined positive STAT5 status added predictive value to the CAPRA-S nomogram for BCR in this cohort of PCs treated by RP. Future studies are needed in independent larger cohorts to validate the finding of the present study. Moreover, it will be crucial to evaluate whether combined positive status for STAT5 in PC at RP predicts development of metastatic PC and PC-specific mortality in patients experiencing BCR in relation to the CAPRA-S nomogram. This will require a larger patient cohort with enough events occurring within the follow-up period. Also, the risk of BCR in patient groups with conflicting CAPRA-S risk vs. STAT5 status will need to be specifically interrogated.
Another clinical implication of the findings of the present study is that the patients with combined positive status for STAT5 in PC at RP may benefit from post-RP adjuvant therapy in the form of chemotherapy, radiation or anti-androgens. In addition, identification of the JAK2-STAT5 signaling pathway as an independent predictive variable for disease recurrence/progression provides a novel target for innovative adjuvant treatment strategies. Numerous Jak2-inhibitors are currently in phase II trials in myeloproliferative disorders(29) and therefore readily available for evaluation of their efficacy for PC. Evaluation of nuclear STAT5 protein status by immunohistochemistry and gene locus amplification by FISH in paraffin-embedded tissue sections of surgically resected PC are both simple and highly feasible tests that can be easily standardized for clinical pathology laboratories.
Combined positive status for STAT5 gene locus amplification and nuclear STAT5 protein expression in the cohort evaluated here was a more powerful predictor of PC recurrence than either STAT5 locus amplification or nuclear protein expression alone. This finding suggests that somatic amplifications of the STAT5 locus may not always result in an intact reading frame and expression of a fully functional STAT5 protein in PC. However, it is important to note that the FISH and IHC were not conducted simultaneously on the same TMA section, but these two analyses were performed on serial sections of the TMAs and, therefore, the data do not represent the FISH/IHC status of the same cells. Furthermore, the STAT5 IHC was analyzed in a 4-tier grouping system where scores 0 and 1 were considered negative while scores 2 and 3 were considered positive for nuclear STAT5 protein status. Accordingly, the maximal patient-wise marker status (either STAT5 protein or STAT5 locus amplification) was used to assess the final patient marker status. STAT5 has been shown to be a potent inducer of PC proliferation, epithelial-mesenchymal transition and castrate-resistant PC growth(16–20) and, therefore, the finding of combined positive status for STAT5 locus amplification and protein expression as a predictor of PC recurrence in patients is consistent with the data on the role of STAT5 in promoting PC growth and progression in preclinical PC model systems. Future studies need to evaluate the STAT5 protein and locus amplification status simultaneously in the same sections.
In conclusion, the results of this study introduce a new concept that combined positive status for both STAT5 protein expression and gene amplification in PC at the time of RP is a predictor of BCR in multivariate analysis that includes the variable of the CSAPRA-S nomogram. These findings will need to be investigated further in larger cohorts in the future studies. Evaluation of STAT5 protein status by IHC and gene locus amplification by FISH in paraffin-embedded tissue sections of surgically resected PC are both simple and highly feasible tests that can be easily standardized for clinical pathology laboratories. By identifying patients at increased risk for treatment failure the incorporation of STAT5 status in the evaluation of surgically removed PC at RP would allow for improved patient counseling and identification of patients likely to benefit from early adjuvant therapy following surgery.
Supplementary Material
Acknowledgments
Funding
This work was supported in part by grants from the National Institutes of Health / National Cancer Institute to M.T.N., B. R. H. and J. D. R (7R01CA113580–10, 5R21CA178755–02, AHW 5520368), from Finnish Medical Foundation (T.M.), Academy of Finland (T.M. and A.E.) and Finnish Cancer Society (T.M.), P.S.L (R01CA218144 and R21CA231892).
Footnotes
Conflict of Interest: None
References
- 1.Attard G, Parker C, Eeles RA, Schroder F, Tomlins SA, Tannock I, et al. Prostate cancer. Lancet 2016;387(10013):70–82 doi 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 2.Grossfeld GD, Latini DM, Lubeck DP, Mehta SS, Carroll PR. Predicting recurrence after radical prostatectomy for patients with high risk prostate cancer. J Urol 2003;169(1):157–63 doi 10.1097/01.ju.0000036470.57520.a0. [DOI] [PubMed] [Google Scholar]
- 3.Epstein JI, Zelefsky MJ, Sjoberg DD, Nelson JB, Egevad L, Magi-Galluzzi C, et al. A Contemporary Prostate Cancer Grading System: A Validated Alternative to the Gleason Score. Eur Urol 2016;69(3):428–35 doi 10.1016/j.eururo.2015.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Amico AV, Whittington R, Malkowicz SB, Fondurulia J, Chen MH, Tomaszewski JE, et al. The combination of preoperative prostate specific antigen and postoperative pathological findings to predict prostate specific antigen outcome in clinically localized prostate cancer. J Urol 1998;160(6 Pt 1):2096–101. [DOI] [PubMed] [Google Scholar]
- 5.Walz J, Chun FK, Klein EA, Reuther A, Saad F, Graefen M, et al. Nomogram predicting the probability of early recurrence after radical prostatectomy for prostate cancer. J Urol 2009;181(2):601–7; discussion 7–8 doi 10.1016/j.juro.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 6.Stephenson AJ, Scardino PT, Eastham JA, Bianco FJ Jr., Dotan ZA, DiBlasio CJ, et al. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Clin Oncol 2005;23(28):7005–12 doi 10.1200/JCO.2005.01.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suardi N, Porter CR, Reuther AM, Walz J, Kodama K, Gibbons RP, et al. A nomogram predicting long-term biochemical recurrence after radical prostatectomy. Cancer 2008;112(6):1254–63 doi 10.1002/cncr.23293. [DOI] [PubMed] [Google Scholar]
- 8.Cooperberg MR, Hilton JF, Carroll PR. The CAPRA-S score: A straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer 2011;117(22):5039–46 doi 10.1002/cncr.26169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooperberg MR, Pasta DJ, Elkin EP, Litwin MS, Latini DM, Du Chane J, et al. The University of California, San Francisco Cancer of the Prostate Risk Assessment score: a straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J Urol 2005;173(6):1938–42 doi 10.1097/01.ju.0000158155.33890.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lughezzani G, Briganti A, Karakiewicz PI, Kattan MW, Montorsi F, Shariat SF, et al. Predictive and prognostic models in radical prostatectomy candidates: a critical analysis of the literature. Eur Urol 2010;58(5):687–700 doi 10.1016/j.eururo.2010.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vickers A Prediction models in urology: are they any good, and how would we know anyway? Eur Urol 2010;57(4):571–3; discussion 4 doi 10.1016/j.eururo.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lughezzani G, Budaus L, Isbarn H, Sun M, Perrotte P, Haese A, et al. Head-to-head comparison of the three most commonly used preoperative models for prediction of biochemical recurrence after radical prostatectomy. Eur Urol 2010;57(4):562–8 doi 10.1016/j.eururo.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Brockman JA, Alanee S, Vickers AJ, Scardino PT, Wood DP, Kibel AS, et al. Nomogram Predicting Prostate Cancer-specific Mortality for Men with Biochemical Recurrence After Radical Prostatectomy. Eur Urol 2015;67(6):1160–7 doi 10.1016/j.eururo.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroeck FR, Aronson WJ, Presti JC Jr., Terris MK, Kane CJ, Amling CL, et al. Do nomograms predict aggressive recurrence after radical prostatectomy more accurately than biochemical recurrence alone? BJU Int 2009;103(5):603–8 doi 10.1111/j.1464-410X.2008.08118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooperberg MR, Broering JM, Carroll PR. Risk assessment for prostate cancer metastasis and mortality at the time of diagnosis. J Natl Cancer Inst 2009;101(12):878–87 doi 10.1093/jnci/djp122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahonen TJ, Xie J, LeBaron MJ, Zhu J, Nurmi M, Alanen K, et al. Inhibition of transcription factor Stat5 induces cell death of human prostate cancer cells. J Biol Chem 2003;278(29):27287–92. [DOI] [PubMed] [Google Scholar]
- 17.Dagvadorj A, Kirken RA, Leiby B, Karras J, Nevalainen MT. Transcription factor signal transducer and activator of transcription 5 promotes growth of human prostate cancer cells in vivo. Clin Cancer Res 2008;14(5):1317–24. [DOI] [PubMed] [Google Scholar]
- 18.Gu L, Liao Z, Hoang DT, Dagvadorj A, Gupta S, Blackmon S, et al. Pharmacologic Inhibition of Jak2-Stat5 Signaling By Jak2 Inhibitor AZD1480 Potently Suppresses Growth of Both Primary and Castrate-Resistant Prostate Cancer. Clin Cancer Res 2013;19(20):5658–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan SH, Dagvadorj A, Shen F, Gu L, Liao Z, Abdulghani J, et al. Transcription factor Stat5 synergizes with androgen receptor in prostate cancer cells. Cancer Res 2008;68(1):236–48. [DOI] [PubMed] [Google Scholar]
- 20.Talati PG, Gu L, Ellsworth EM, Girondo MA, Trerotola M, Hoang DT, et al. Jak2-Stat5a/b Signaling Induces Epithelial-to-Mesenchymal Transition and Stem-Like Cell Properties in Prostate Cancer. Am J Pathol 2015;185(9):2505–22 doi 10.1016/j.ajpath.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haddad BR, Gu L, Mirtti T, Dagvadorj A, Vogiatzi P, Hoang DT, et al. STAT5A/B gene locus undergoes amplification during human prostate cancer progression. Am J Pathol 2013;182(6):2264–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy DE, Darnell JE Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol 2002;3(9):651–62. [DOI] [PubMed] [Google Scholar]
- 23.Li H, Zhang Y, Glass A, Zellweger T, Gehan E, Bubendorf L, et al. Activation of signal transducer and activator of transcription-5 in prostate cancer predicts early recurrence. Clin Cancer Res 2005;11(16):5863–8. [DOI] [PubMed] [Google Scholar]
- 24.Mirtti T, Leiby BE, Abdulghani J, Aaltonen E, Pavela M, Mamtani A, et al. Nuclear Stat5a/b predicts early recurrence and prostate cancer-specific death in patients treated by radical prostatectomy. Hum Pathol 2013;44(3):310–9 doi 10.1016/j.humpath.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making 2006;26(6):565–74 doi 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA, et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am J Surg Pathol 2016;40(2):244–52 doi 10.1097/PAS.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 27.Nevalainen MT, Xie J, Bubendorf L, Wagner KU, Rui H. Basal activation of transcription factor signal transducer and activator of transcription (Stat5) in nonpregnant mouse and human breast epithelium. Mol Endocrinol 2002;16(5):1108–24. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Ahonen TJ, Alanen K, Xie J, LeBaron MJ, Pretlow TG, et al. Activation of signal transducer and activator of transcription 5 in human prostate cancer is associated with high histological grade. Cancer Res 2004;64(14):4774–82. [DOI] [PubMed] [Google Scholar]
- 29.Komrokji RS, Seymour JF, Roberts AW, Wadleigh M, To LB, Scherber R, et al. Results of a phase 2 study of pacritinib (SB1518), a JAK2/JAK2(V617F) inhibitor, in patients with myelofibrosis. Blood 2015;125(17):2649–55 doi 10.1182/blood-2013-02-484832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.