Abstract
Folate-dependent one-carbon (C1) metabolism is compartmentalized into the mitochondria and cytosol and supports cell growth through nucleotide and amino acid biosynthesis. Mitochondrial C1 metabolism, including serine hydroxymethyltransferase (SHMT) 2, provides glycine, NAD(P)H, ATP, and C1 units for cytosolic biosynthetic reactions, and is implicated in the oncogenic phenotype across a wide range of cancers. Whereas multi-targeted inhibitors of cytosolic C1 metabolism such as pemetrexed are used clinically, there are currently no anticancer drugs that specifically target mitochondrial C1 metabolism. We used molecular modeling to design novel small-molecule pyrrolo[3,2-d]pyrimidine inhibitors targeting mitochondrial C1 metabolism at SHMT2. In vitro antitumor efficacy was established with the lead compounds (AGF291, AGF320, AGF347) toward lung, colon, and pancreatic cancer cells. Intracellular targets were identified by metabolic rescue with glycine and nucleosides, and by targeted metabolomics using a stable isotope tracer, with confirmation by in vitro assays with purified enzymes. In addition to targeting SHMT2, inhibition of the cytosolic purine biosynthetic enzymes, β-glycinamide ribonucleotide formyltransferase and/or 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase, and SHMT1 was also established. AGF347 generated significant in vivo antitumor efficacy with potential for complete responses against both early stage and upstage MIA PaCa-2 pancreatic tumor xenografts, providing compelling proof-of-concept for therapeutic targeting of SHMT2 and cytosolic C1 enzymes by this series. Our results establish structure-activity relationships and identify exciting new drug prototypes for further development as multi-targeted antitumor agents.
Keywords: SHMT2, purine, folate, one-carbon, antifolates
Introduction
Metabolic reprogramming to support tumor progression has emerged as a hallmark of cancer (1). Of the many altered metabolic pathways associated with the malignant phenotype, one-carbon (C1) metabolism is particularly notable (2–4). C1 metabolism depends on an adequate supply of tetrahydrofolate (THF) metabolites and generates critical purine, thymidylate, and glycine metabolites essential for cell proliferation and tumor progression (3–5). Purine nucleotides are synthesized de novo in the cytosol whereas thymidylate is synthesized in both the cytosol and nucleus (3–6). C1 enzymes such as thymidylate synthase (TS) and the purine biosynthetic enzymes β-glycinamide ribonucleotide (GAR) formyltransferase (GARFTase) and 5-aminoimidazole-4-carboxamide (AICA) ribonucleotide (AICAR) formyltransferase (AICARFTase) (respectively, the third and ninth steps in de novo purine biosynthesis) are important therapeutic targets for cancer (7–9). Serine biosynthesis from glycine in the cytosol involves serine hydroxymethyltransferase (SHMT) 1 and uses C1 units from 5,10-methylene-THF (5,10-me-THF) (2–4). SHMT1, like TS, is localized to both the cytosol and nucleus (5,6); however, nuclear SHMT1 does not appear to be an important source of C1 units for TS (6,10).
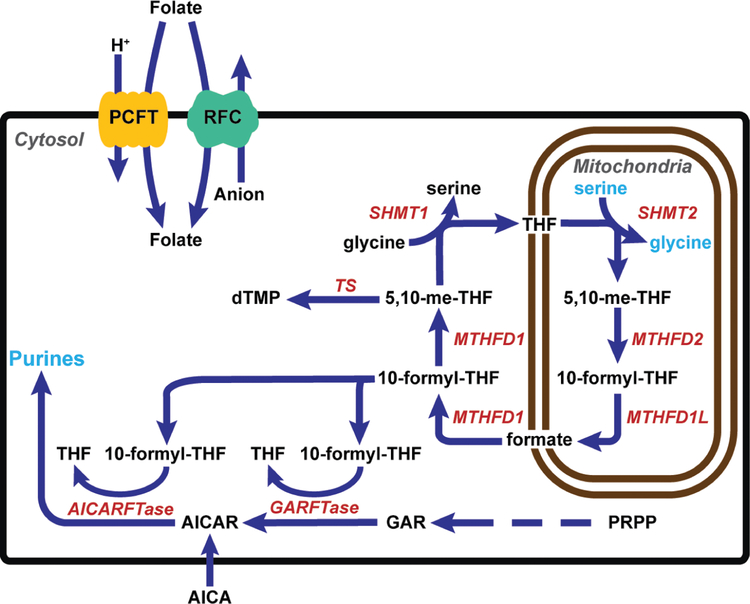
Cytosolic and mitochondrial C1 metabolic pathways are interconnected by exchange of serine, glycine and formate (Figure 1). Extracellular folates are transported into cells by the reduced folate carrier (RFC), proton-coupled folate transporter (PCFT) and folate receptors (FRs) (11,12). Whereas cytosolic folates are transported into mitochondria via the mitochondrial folate transporter (SLC25A32) (13,14), mitochondrial folates do not exchange with those in the cytosol (15). In cancer cells, the 3-carbon of serine is the major source of C1 units, and in mitochondria, serine catabolic enzymes including SHMT2, 5,10-me-THF dehydrogenase 2 (MTHFD2) and 10-formyl-THF synthetase (reverse) (MTHFD1L) generate glycine and C1 units (i.e. formate) to sustain C1-dependent nucleotide and amino acid biosynthesis in the cytosol (Figure 1). 10-Formyl-THF is synthesized from formate in the cytosol by the trifunctional enzyme MTHFD1. 10-Formyl-THF is utilized for purine nucleotide biosynthesis and can be further converted by MTHFD1 to 5,10-me-THF for TS and SHMT1.
Figure 1. C1 metabolism is compartmentalized in the cytosol and mitochondria.
Folates enter the cell through the plasma membrane facilitated folate transporters, PCFT and RFC, and enter the mitochondria via SLC25A32. Serine catabolism in the mitochondria beginning with SHMT2 generates glycine and formate, the latter of which is required for downstream cytosolic de novo purine biosynthesis (by GARFTase and AICARFTase) as 10-formyl-THF, and thymidylate biosynthesis, following conversion to 5,10-me-THF by MTHFD1. SHMT1 catalyzes conversion of glycine to serine in the cytosol. AICA is metabolized to AICAR (ZMP), the AICARFTase substrate which circumvents the GARFTase step. The arrows denote the net flux of C1 metabolism in proliferating cells, although most reactions are reversible. Both SHMT1 and TS are also localized to the nucleus; however, nuclear SHMT1 is not an important source of C1 units for TS (6,10) .
Several studies have implicated mitochondrial C1 metabolism as critical to the malignant phenotype (16–19). A study (18) of messenger RNA profiles for 1451 metabolic enzymes spanning 1981 tumors across nineteen different cancer types and 951 matched normal tissues identified SHMT2 and MTHFD2 among the top five most differentially expressed genes, highlighting an oncogenic role of mitochondrial C1 metabolism. Metabolomics analyses of 219 extracellular metabolites from the NCI-60 cancer cell lines showed that glycine consumption and the glycine biosynthetic pathway correlated with cell proliferation (16). These findings, combined with evidence of functional shortages of amino acids (e.g. glycine) in tumors (20), suggested a therapeutic opportunity for SHMT2 targeting in cancer.
SHMT2 is induced by hypoxic stress in Myc-transformed cells (21) and is critical to tumor cell survival in the hypoxic, nutrient-poor tumor microenvironment (17,21). SHMT2 (or MTHFD2) knockout (KO) cells are viable and tumorigenic (albeit with decreased growth rates) in nutrient-rich conditions, as reversal of cytosolic SHMT1 (serine-to-glycine) provides sufficient C1 units to sustain some level of de novo nucleotide biosynthesis (22). However, SHMT1 only restores a small fraction of the C1 pools in wild-type (WT) cells (22). Further, SHMT1 does not generate sufficient glycine for protein, nucleotide and glutathione biosynthesis, rendering both SHMT2 and MTHFD2 KO cells as glycine auxotrophs (4). This was the impetus for studies of pyrazolopyran compounds (e.g., SHIN1) with dual SHMT1/SHMT2 inhibition (23). While structurally unrelated to folates, these compounds bound to the folate binding site in SHMT2 and showed in vitro antitumor efficacy (particularly with B-cell cancers).
In this report, we describe a novel series of 5-substituted pyrrolo[3,2-d]pyrimidine compounds that inhibit SHMT2, and also SHMT1, GARFTase and/or AICARFTase. Our lead compounds (AGF291, AGF320, and AGF347) show broad-spectrum in vitro antitumor efficacies against H460 non-small cell lung cancer (NSCLC), HCT116 colon cancer, and MIA PaCa-2 pancreatic cancer cells. For AGF347, in vitro findings were extended in vivo to MIA PaCa-2 tumor xenografts, providing compelling proof-of-concept of the therapeutic potential of our multi-targeted SHMT2 therapeutics for cancer.
Materials and Methods
Chemicals.
[2,3,3-2H]L-Serine (98%) was purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA). Leucovorin (6R,S) 5-formyl-THF) was provided by the National Cancer Institute (Bethesda, MD). Pemetrexed (Alimta) (PMX) was purchased from LC Laboratories (Woburn, MA). Gemcitabine (Gemzar; GEM) was purchased from Fresenius Kabi USA, LLC (Lake Zurich, IL). Serine-, glycine- and folate-free RPMI 1640 was custom-ordered from ThermoFisher (Waltham, MA) and supplemented with tissue-culture grade glycine (ThermoFisher) or serine (Sigma-Aldrich), as needed. Other chemicals were obtained from commercial sources in the highest available purities. Chemical synthesis of the pyrrolopyrimidine compounds is provided in the Supplementary Methods.
Molecular modeling and computational studies.
Molecular modeling was performed for all analogs with the human SHMT2 crystal structure (PDB: 5V7I) (23) using the induced fit docking protocol of Maestro (2,3) (see Supplemenary Methods). The compounds were also docked into rabbit SHMT1 (PDB: 1LS3) (24) binding sites. The docking scores of the analogs are reported in Supplementary Table S1.
Cell culture and proliferation/protection assays.
The HCT116 cell lines including the SHMT1, SHMT2, and MTHFD2 KO cells were previously described (22,23). The wild-type HCT116 and H460 human tumor cell lines were obtained from the American Type Culture Collection (Manassas, VA), whereas MIA PaCa-2 cells were provided by Dr. Yubin Ge (Karmanos Cancer Institute). Cell lines were frozen in aliquots in liquid nitrogen upon receipt with aliquots from the original freezes thawed and used for experiments. As warranted, cell lines were re-verified by STR analysis by Genetica DNA Laboratories (Burlington, NC) (H460, November 2015; MIA PaCa-2 (April 2019)).
MTXRIIOuaR2–4 (i.e. R2) Chinese hamster ovary (CHO) cells were provided by Dr. Wayne Flintoff (University of Western Ontario) (25). From this parental R2 cell line, human RFC and PCFT were individually transfected to generate the isogenic CHO cell lines designated PC43–10 (RFC) and R2/PCFT4 (PCFT) (26–28). Other than to confirm phenotypic expression of individual RFC or PCFT expression levels and/or inhibitor sensitivities, CHO cells were not further authenticated.
Cell lines were tested for Mycoplasma by PCR using a Mycloplasma testing kit (Venor™ GeM Mycoplasma Detection Kit, Sigma) (Feb 2018). As warranted, cells were treated (Plasmocin, Invivogen) and re-tested for Mycoplasma. To avoid cell line “drift”, fresh cultures were reconstituted no less than every month from the original freezer stocks (above).
Human tumor cell lines were cultured in folate-free RPMI 1640 supplemented with 10% dialyzed fetal bovine serum (Sigma-Aldrich), 1% penicillin/streptomycin solution, 2 mM L-glutamine, and 25 nM leucovorin in a humidified atmosphere at 37°C in the presence of 5% CO2 and 95% air. The CHO cell lines were cultured in alpha-minimal essential medium (alpha-MEM) supplemented with 10% bovine calf serum, 1% penicillin/streptomycin solution, and 2 mM L-glutamine. The transfected CHO cell lines (i.e. R2/PCFT4 and PC43–10) were cultured in the presence of 1 mg/ml of G418.
Proliferation assays in CHO cells were performed as described (29), except glycine-free folate-free RPMI 1640 was used. For the HCT116, H460, and MIA PaCa-2 tumor cell lines, an identical procedure was followed except that the maximal drug concentration was increased to 10 μM from 1 μM. For the HCT116 KO cells, glycine (130 μM) was included.
Glycine/nucleoside rescue experiments in CHO and tumor cell lines were performed in folate- and glycine-free RPMI 1640/10% dialyzed fetal bovine serum, supplemented with 25 nM leucovorin without additions, or in the presence of adenosine (60 μM), thymidine (10 μM), glycine (130 μM) and/or AICA (320 μM). Growth of metabolite-treated cells was normalized to controls treated with metabolites and vehicle (i.e., DMSO), both singly and in combination.
Generation of H460 SHMT2 knockdown cell line.
H460 SHMT2 knockdown (KD) cells were generated by transduction with MISSION lentivirus particles (Sigma-Aldrich) containing shRNA targeting SHMT2 (TRCN0000034805) or non-targeted control (NTC) lentivirus as previously described (29). Single clones were isolated and cultures expanded. The extent of SHMT2 KD was confirmed by Western blotting (below).
Real-time PCR.
Cells were harvested from 60 mm dishes or T25 flasks at ~80% confluence and RNAs extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). cDNAs were synthesized with random hexamers, MuLV reverse transcriptase, and RNase inhibitor (Applied Biosystems, Waltham, MA), and purified with a QIAquick PCR Purification Kit (QIAGEN, Valencia, CA). Quantitative real-time RT-PCR was performed using a Roche LightCycler 480 (Roche Diagnostics, Indianapolis, IN) with gene-specific primers to the major folate transporters and Universal Probe Library probes (SHMT2-#83, RFC-#32, PCFT-#89, FRα-#65 (Roche Diagnostics)). Transcript levels were normalized to β-actin and GAPDH. Primer sequences are available upon request.
Gel electrophoresis and Western blots.
H460 WT, H460 NTC, H460 SHMT2 KD, HCT116 WT, and HCT116 SHMT2 KO cell lines were plated (1 × 106 cells/dish) in 60 mm dishes and harvested when the cells were ~80% confluent. Cells were disrupted by sonication and cell debris removed by centrifugation (1800 rpm, 5 min, 4°C). The soluble cell fraction was assayed for protein (30). Equal amounts of protein (37 μg) from each sample were electrophoresed on 10% polyacrylamide gels with SDS (31) and transferred to polyvinylidene difluoride membranes (ThermoFisher) (32). To detect SHMT2, membranes were incubated for 72 h with rabbit anti-SHMT2 primary antibody (#12762 (22); Cell Signaling Technology, Danvers, MA). The blots were developed by incubating in IRDye800CW-conjugated goat anti-rabbit IgG secondary antibody (LICOR Biosciences, Omaha, NE) for 90 min and scanning with an Odyssey infrared imaging system (LICOR Biosciences). Protein loading was normalized to β-actin using anti-β-actin mouse antibody (Sigma-Aldrich).
In vitro targeted metabolomics by liquid chromatography-mass spectrometry (LC-MS).
Cells were seeded in triplicate 60 mm dishes in folate-free RPMI 1640 (contains glycine and unlabeled serine) supplemented with 10% dialyzed fetal bovine serum, 1% penicillin/ streptomycin, 2 mM L-glutamine, and 25 nM leucovorin. The cells were allowed to adhere for 24 h. The media was aspirated and replaced with culture media (contains 25 nM leucovorin, glycine, and unlabeled serine) and 10 μM AGF291, AGF320 or AGF347, or vehicle (DMSO) (with or without 1 mM formate). After 16 h, the cells were washed with phosphate-buffered saline (3x), and the media was replaced with folate- and serine-free media (containing glycine) supplemented with 10% dialyzed fetal bovine serine, 25 nM leucovorin, and [2,3,3-2H]serine (250 μM), including 10 μM drug or vehicle. The cells were incubated for 24 h. Metabolites were extracted and analyzed (22,33) with normalization to total protein from the post-extraction pellet (solubilized with 0.5 N NaOH) by the Folin-phenol method (30). Samples were run on a ThermoElectron Corporation Exactive mass spectrometer operating in negative ion mode (34). Separation used reverse phase ion-pairing chromatography on a 100 mm/2.5 μm Synergi Hydro-RP C18 column (Phenomenex). Individual metabolites were identified from their exact masses and comparison of retention times with standard compounds using the MAVEN software suite (35). Values below the limit of detection were assigned a value of 100 for normalization.
Enzyme expression and purification.
N-terminal His-tagged GARFTase (formyltransferase domain; residues 100–302) was expressed and purified (36). Full-length human AICARFTase/IMP cyclohydrolase (ATIC), SHMT1 (residues 7–478, Uniprot ID P34896), SHMT2 (residues 42–504, Uniprot ID 34897), and MTHFD2 (residues 36–333, Uniprot ID P13995) with N-terminal, cleavable hexahistidine tags were expressed and purified by essentially the same protocol. Samples were further purified on ÄKTA FPLC (GE Healthcare) via Superdex 200 16/60 (GE Healthcare) column. Further details in the Supplementary Methods.
In vitro enzymatic assays and Ki determinations.
AICARFTase and GARFTase activities were measured by monitoring formation of THF spectrophotometrically from 10-formyl-THF in the presence of various concentrations of inhibitor (37,38). SHMT1/2 activities were assayed by a coupled reaction with His-MTHFD2 in 200-fold molar excess with NADH production monitored by fluorescence. Kis were calculated from the IC50 values [Ki = IC50/([S]/KM+1)], using the Km and substrate concentrations for 10-formyl-THF. The calculated Km values for 10-formyl-THF with His-ATIC and His-GARFTase were 100 μM and 84.8 μM respectively. The calculated Km values for THF with His-SHMT1 and His-SHMT2 were 62.8 μM and 108 μM, respectively. To confirm that MTHFD2 was not inhibited by our inhibitors, MTHFD2 activity was evaluated with an NAD(P)H-Glo™ Detection System Kit (Promega, Ref G9061). Further details in the Supplementary Methods.
In vivo efficacy trial with MIA PaCa-2 pancreatic cancer xenografts.
Methods for in vivo maintenance of MIA PaCa-2 tumor xenografts and drug efficacy evaluations are analogous to those previously described (39–44). The mouse study in this report was approved by the Wayne State University Institutional Animal Care and Use Committee (IACUC). Female NCr severe compromised immunodeficient (SCID) mice were purchased from Charles River Labs. For the early stage trial, chemotherapy was initiated one day post-tumor implantation with AGF347; for the upstage trial, chemotherapy was initiated seven days post-tumor implantation (when tumors had grown to 100–150 mg) with AGF347. For both designs, dosing for AGF347 was 15 mg/kg/injection every 2 days × 8 (total dose of 120 mg/kg); for GEM, dosing was 120 mg/kg/injection every 4 days × 4 (total dose of 480 mg/kg). Both drugs were administered intravenously (0.2 ml/injection). The tumor masses from both flanks of each mouse were added together, and the total mass per mouse was used for calculations of anti-tumor activity. Quantitative end-points include: (i) tumor growth delay [T-C, where T is the median time in days required for the treatment group tumors to reach a predetermined size (e.g., 1000 mg), and C is the median time in days for the control group tumors to reach the same size; tumor-free survivors are excluded from these calculations]; and (ii) gross log10 cell kill (LCK), determined by the formula LCK = (T-C; tumor growth delay in days)/3.32 × Td (tumor doubling time in days determined by growth plot). Qualitative analysis included determination of T/C values (in percent) on all days of tumor measurement using the median total tumor burden for treatment (T) and control (C) groups. The end point %T/C value for this study corresponds to the first measurement taken post last treatment (day 16 for early stage or day 21 for late stage), when control tumors were still in exponential growth phase (i.e., 500–1250 mg). Further details are described in the Supplementary Methods.
From the upstage in vivo study, a separate AGF347 treatment arm (3 mice) was designated for metabolomics analysis in comparison with the untreated control (3 mice). Six hours after the 6th dose of AGF347, mice were sacrificed, and tumors removed and immediately frozen in liquid nitrogen. Frozen isolated tumors were weighed; ~50 mg pieces were disrupted with a cryomill (Retsch) in 1 mL ice-cold acetonitrile:methanol: water (40:40:20). Solids were collected by centrifugation then re-extracted with 1 mL lysis solution (above). Combined supernatants were dried down, then resuspended in water to a concentration of 50 mg/mL (original tissue mass) for LC-MS analysis, as described above.
Statistics.
All data shown reflect at least three biological replicates unless noted otherwise. Statistical analyses were performed by the Karmanos Cancer Institute Biostatistics Core. The expression levels were assessed for the normality assumption. The log2 transformation was used when all values were positive and the square root transformation was used when values included zero. Statistical tests were carried out using an unpaired t-test. P-values were not adjusted for multiple comparisons.
Results
Rationale for design of 5-substituted pyrrolo[3,2-d]pyrimidine compounds targeting mitochondrial C1 metabolism at SHMT2.
Given the association of mitochondrial C1 metabolism with malignancy (16–19) , including reports of SHMT2 as a potential “onco-driver” (17,45), it was of interest to develop inhibitors of SHMT2 as antitumor agents. We initially looked for potential SHMT2 inhibitors among prototypes of different classes of GARFTase and AICARFTase inhibitors from our published studies (26,27,40,41,43,46,47). We tested these compounds (Supplementary Figure S1) for inhibition of proliferation of CHO cells engineered from the transporter-null R2 CHO subline (25) to express human PCFT (R2/PCFT4) (27), as we reasoned that tumor-selective uptake of a putative SHMT2 inhibitor by PCFT (48) would be desirable. Since inhibition of mitochondrial C1 metabolism (at SHMT2) should induce glycine auxotrophy (22), and mitochondrial C1 metabolism generates glycine and C1 units (i.e., formate) critical for nucleotide biosynthesis, these experiments were performed in glycine- and nucleoside-free media, and the protective effects of added glycine (130 μM) and/or adenosine (60 μM) were determined. With the exception of AGF147, these compounds inhibited cell proliferation and purine biosynthesis in R2/PCFT4 cells (reflected in adenosine rescue); however, glycine was not protective with or without adenosine addition, indicating a lack of mitochondrial C1 targeting. Thus, it became imperative to design new analogs.
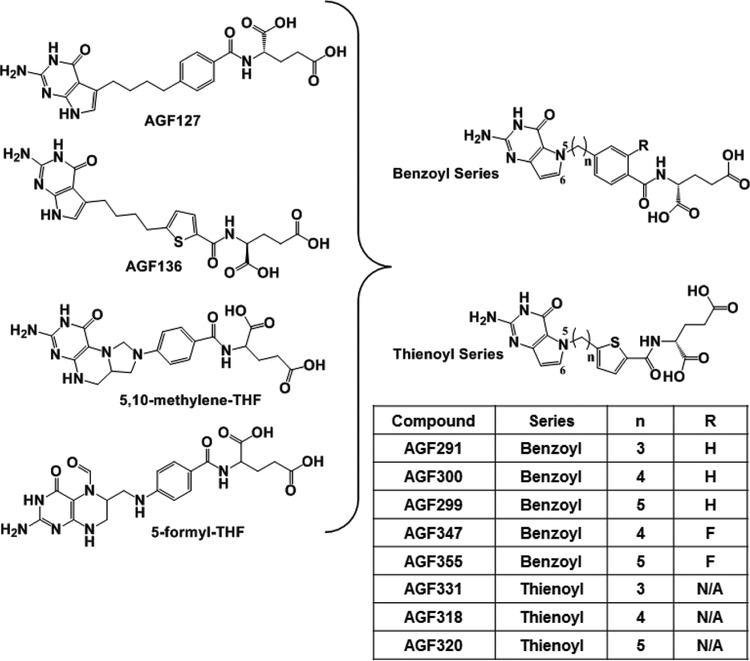
To engineer SHMT2 activity into our compound series, we merged structural features of our previous 5-substituted pyrrolo[2,3-d]pyrimidine benzoyl and thienoyl compounds (46,47) with those of 5,10-me-THF (SHMT2 product) and 5-formyl-THF (SHMT2 inhibitor) (24) (Figure 2). The resulting 5-substituted pyrrolo[3,2-d]pyrimidine analogs included 3–5 bridge carbons linked to benzoyl (i.e., AGF291, AGF300, and AGF299) or thienoyl (i.e., AGF331, AGF318, and AGF320) moieties. Based on the reported impact of 2’ fluorine substitutions in increasing inhibitory potencies of pyrrolo[2,3-d]pyrimidine compounds (41), we designed 2’-fluorinated analogs of AGF300 and AGF299, as well (AGF347 and AGF355, respectively) (Figure 2). Compounds were docked into human SHMT2 (Supplementary Figure S2) with docking scores (Supplementary Table S1) better than for the pyrazolopyran inhibitor SHIN1 (23) (−5.58 kcal/mol) upon re-docking.
Figure 2. Rational design of novel 5-substituted pyrrolo[3,2-d]pyrimidine benzoyl and thienoyl analogs.
Structural features of the 5-substituted pyrrolo[2,3-d]pyrimidines AGF127 and AGF136 (46,47) were merged with the SHMT2 substrate 5,10-me-THF and inhibitor 5-formyl-THF to generate the novel analogs in this report. Key structural features for the various compounds are summarized including the bridge lengths (3 or 4 carbons), thienoyl or benzoyl side-chains, and for the latter, the 2’ ring substituent (F or H). NA, not applicable.
Discovery of 5-substituted pyrrolo[3,2-d]pyrimidine compounds that target mitochondrial C1 metabolism.
We synthesized the novel analogs and screened these for inhibition of cell proliferation. To encompass the major modes of facilitative transport, we initially assessed inhibition by these compounds (from 0 to 1000 nM) toward PCFT-expressing R2/PCFT4 CHO cells and an isogenic CHO subline engineered to express human RFC (PC43–10) (26). Results were compared to those for folate transporter-null R2 CHO cells (25). IC50 values for “active” compounds are shown in Table 1 for the analogs, along with those for AGF94 (a “pure” GARFTase inhibitor) (43) and PMX. Active compounds toward R2/PCFT4 and PC43–10 cells included AGF291, AGF320, AGF331 and AGF347. These analogs were further tested with human tumor cell lines, including H460 NSCLC, HCT116 colon cancer, and MIA PaCa-2 pancreatic cancer cells, characterized by expression of PCFT and RFC but not FRα (49,50) (Supplementary Figure S2). IC50 values for growth inhibition are in Table 1. Although there were differences in inhibitor sensitivities among the assorted tumor models, AGF291, AGF320 and AGF347 were consistently the most active of the series.
Table 1.
IC50 values for anti-proliferative activities toward engineered CHO and human tumor cell lines, and cell-free enzyme inhibition by 5-substituted pyrrolo[3,2-d]pyrimidine inhibitors compared to AGF94 and PMX.
| Compound | Proliferation Inhibition [IC50 (nM)] | In vitro enzyme assays [Ki (μM)] | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 (null) |
PC43–10 (RFC) | R2/PCFT4 (PCFT) | HCT116 | HCT116 SHMT2 KO | H460 | MIA PaCa-2 | GARFTase | AICARFTase | SHMT2 | MTHFD2 | SHMT1 | |
| AGF94 | 136 (3) | 110 (15) | 3 (1) | 191 (62) | 295 (59)* | 166 (59) | 1924 (539) | 0.88 (0.71) | NI | 188.0 (91.8) | NI | 112 (57) |
| PMX | 395 (83) | 124 (39) | 24 (5) | 293 (81) | 333 (40) | 165 (59) | 281 (23) | 5.19 (1.63) | 0.88 (0.56) | NI | NI | NI |
| AGF291 | NI | 454 (87) | 282 (22) | 2266 (450) | 108 (66)** | 461 (163) | 3664 (721) | NI | 13.31 (3.16) | 0.63 (0.29) | NI | 0.90 (0.06) |
| AGF347 | NI | 224 (17) | 479 (63) | 437 (180) | 42(19)*** | 214 (88) | 1381 (182) | 3.13 (0.66) | 3.72 (1.61) | 2.19 (0.23) | NI | 2.91 (0.59) |
| AGF331 | NI | NI | 661 (62) | 2199 (817) | 969 (254) | 2367 (711) | 6681 (604) | ND | ND | ND | ND | ND |
| AGF320 | NI | NI | 694 (56) | 737 (195) | 117 (60)* | 573 (144) | 2703 (400) | 0.33 (0.22) | 2.36 (1.99) | 0.056 (0.020) | NI | 0.126 (0.030) |
Proliferation inhibition assays were performed using the engineered CHO cell lines R2 (folate transporter-null), PC43–10 (expresses RFC only), and R2/PCFT4 (expresses PCFT only), and human tumor cell lines, including HCT116 (colon cancer), H460 (lung cancer), and MIA PaCa-2 (pancreatic cancer). For HCT116, both wild-type (WT) and CRISPR/Cas9 SHMT2 KO cells (22,23) were tested. For SHMT2 KO cells, glycine was added to the media to circumvent the impact of loss of SHMT2, permitting cell growth. Results are shown as mean IC50 values (± standard deviations), corresponding to the concentrations that inhibit growth by 50% relative to vehicle-treated control cells, from at least four biological replicates. p < 0.10;
p < 0.05;
p < 0.01;
p < 0.001 in HCT116 SHMT2 KO relative to WT cells. For the in vitro enzyme assays, Ki values are presented as mean values (+/− standard deviations) from at least 3 replicate experiments. “ND” denotes assay not done. “NI” denotes “no inhibition” at 1000 nM inhibitor for the CHO cell line proliferation experiments and up to 200 μM inhibitor for the in vitro enzyme assays.
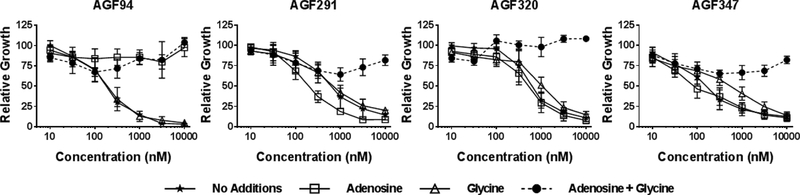
We again used glycine and nucleoside rescue studies (above) in H460, HCT116, and MIA PaCa-2 cells treated with AGF291, AGF320, or AGF347 to identify the targeted pathways. Results were compared to those for AGF94 and are shown in Figure 3 for H460 cells. Adenosine (60 μM) was completely protective up to 10 μM AGF94, whereas glycine (130 μM) had no effect. We tested the protective effects of AICA (320 μM) which is metabolized to AICAR (ZMP) (AICARFTase substrate), thus circumventing GARFTase in de novo purine biosynthesis (43) (Figure 1). As AICA was completely protective (Supplementary Figure S4), GARFTase must be the intracellular target for AGF94 (43). For AGF291, AGF347, and AGF320, combined adenosine and glycine was substantially protective for all compounds (Figure 3). These results strongly suggest that these compounds target both mitochondrial C1 metabolism and cytosolic de novo purine biosynthesis. Thymidine provided no protection from any of the compounds and did not increase the extent of rescue by glycine and adenosine. For some analogs, notably AGF320, growth inhibition was modestly (and incompletely) reversed by AICA (with glycine), suggesting a secondary target (below). Analogous results were obtained with HCT116 and MIA PaCa-2 tumor cells (Supplementary Figure S4). These results establish that AGF291, AGF320, and AGF347 likely target both mitochondrial and cytosolic C1 metabolism.
Figure 3. In vitro antitumor efficacy and identification of targeted pathways and enzymes by novel pyrrolo[3,2-d]pyrimidine analogs in H460 tumor cells by proliferation assays.
Dose-response growth inhibition curves are shown for AGF94, an established GARFTase inhibitor (43), and for AGF291, AGF320, and AGF347 without additions, or in the presence of adenosine (60 μM) and/or glycine (130 μM). The results are mean values +/−standard deviations for three biological replicates.
Identification of the mitochondrial enzyme target for AGF291, AGF320 and AGF347 by targeted metabolomics.
To confirm the intracellular enzyme targets of the lead compounds AGF291, AGF320, and AGF347, we performed targeted metabolomics with LC-MS using a [2,3,3-2H]serine tracer in HCT116, H460 and MIA PaCa-2 tumor cells. The cells were processed for LC-MS analysis of total serine, and of isotopically labeled M+3, M+2, M+1, and M+0 serine (M+n represents species with n deuterium atoms). Results for inhibitor-treated cells were compared to those for untreated cells. For WT HCT116 cells, the results were compared to those for SHMT1 KO, MTHFD2 KO, and SHMT2 KO cells (22). For H460 cells, controls included NTC and SHMT2 shRNA KD cells.
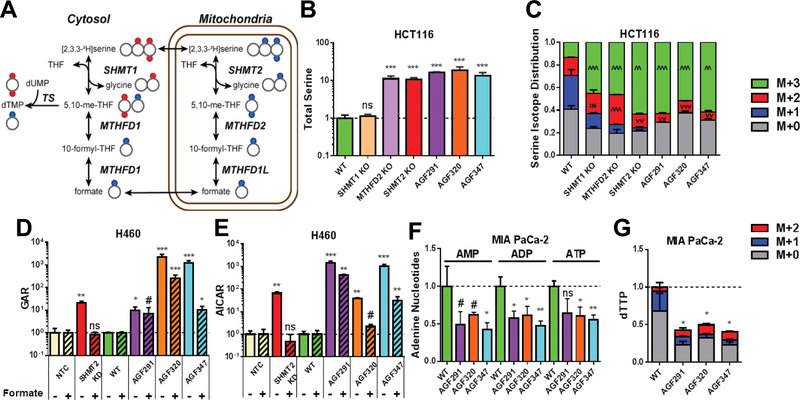
KO of cytosolic SHMT1 (SHMT1 KO) in HCT116 cells had no impact on total serine pools. Serine pools were elevated ~10-fold in the HCT116 sublines with mitochondrial C1 KOs (SHMT2 KO and MTHFD2 KO) relative to WT controls (Figure 4B). This further supports the notion that SHMT2 rather than SHMT1 is the primary catabolic enzyme for serine in HCT116 cells (22). In AGF291-, AGF320- and AGF347-treated HCT-116 cells, serine also increased ~10-fold, indicating a profound loss of serine catabolism. Analogous results were seen with the H460 and MIA PaCa-2 cell lines, including the H460 SHMT2 KD cells (Supplementary Figures S5A and B).
Figure 4. Targeted metabolomics analysis to identify intracellular enzyme targets of AGF291, AGF320, and AGF347.
A: A schematic of serine isotope scrambling and dTTP isotope analysis is shown. Heavy (2H) atoms in serine (red circles) enter the C1 flux in the cytosol through reversal of the SHMT1 reaction and TS leading to dTMP and dTTP (as M+2). In the mitochondria, 2H atoms in serine (blue circles) are metabolized via SHMT2, MTHFD2, and MTHFD1 to formate which is converted to 10-formyl-THF and 5,10-me-THF, leading to dTMP and dTTP (as M+1). Most steps are reversible as noted. Adapted from (22). B and C: Total serine pools (B) and the corresponding serine isotope distributions (C) for WT and KO HCT116 sublines, and for inhibitor-treated WT cells, are shown. D and E: Total GAR (D) and AICAR (E) pools are shown for WT and SHMT2 KD H460 sublines, including inhibitor-treated WT cells, with and without 1 mM formate. F and G: Relative adenine nucleotide pools (AMP, ADP, and ATP) and dTTP pools with isotope distributions are shown for untreated and inhibitor-treated MIA PaCa-2 cells. For D and E, results for inhibitor-treated and SHMT2 KD cells were normalized to vehicle-treated WT ± formate or NTC ± formate samples, as appropriate. Data are mean values +/− standard deviations for three technical replicates. #, p < 0.10; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ^ or v are used in place of * to specify a significant increase or decrease, respectively. Statistical comparisons were with vehicle-treated WT ± formate or NTC ± formate samples, as appropriate. ns = not significant.
The flux of [2H]metabolites originating from [2,3,3-2H]serine tracer is depicted in Figure 4A. In proliferating tumor cells, C1 metabolism flows in a clockwise manner, with serine catabolized in mitochondria (starting with SHMT2) and regenerated in the cytosol (via SHMT1) (22). The reactions are reversible (e.g., serine is resynthesized from formate in the mitochondria (22)). The compartmental metabolism of serine can be revealed by analyzing the isotope tracing (“scrambling”) patterns from [2,3,3-2H]serine in HCT116 cells (22,23). These patterns include both unmetabolized (i.e., M+3) and metabolized (M+2 and M+1) forms of serine. The latter species are derived from the recombination of unlabeled glycine with doubly labeled (M+2) 5,10-me-THF and the synthesis of serine from an unlabeled glycine and a singly labeled 5,10-me-THF derived from a formate/10-formyl-THF precursor (M+1), respectively (Figure 4A).
In WT HCT116 cells fed [2,3,3-2H]serine, most of the serine was catabolized (only 10% M+3 serine remained) (Figure 4C). A large fraction of serine was incompletely labeled (scrambled; i.e., M+1 or M+2), reflecting resynthesized serine from M+1 or M+2 precursors. SHMT1 KO led to a drop (~60%) in the M+1 fraction without a change in the M+2 serine fraction, consistent with SHMT1 in the cytosol being responsible for bulk synthesis of serine from M+1 5,10-me-THF and unlabeled (media) glycine (22). In SHMT2 KO cells, the M+3 fraction was >60% of the total serine (Figure 4C), consistent with a profound loss of serine catabolism. Reflecting this and the depletion of formate downstream of SHMT2, there was a ~90% decrease in M+1 serine (22). M+2 serine also decreased (~25%) compared to WT cells. Although similar results were seen for the M+1 and M+3 serine fractions with MTHFD2 KO cells (decreased ~75% and increased 3.5-fold, respectively, from WT levels), M+2 serine was increased (~1.7-fold) rather than decreased (Figure 4C). This reflects accumulation of M+2 5,10-me-THF when MTHFD2 is lost which drives synthesis of M+2 serine from the largely unlabeled glycine pool. Thus, changes in serine isotope labeling from [2,3,3-2H]serine (particularly in the M+2 fraction) are diagnostic for specific perturbations in folate metabolism, and also inform upon the particular enzymatic step that is inhibited.
Treatment of WT HCT116 cells with AGF291, AGF320 or AGF347 closely recapitulated the effects of the SHMT2 KO, including a substantial increase in M+3 serine fraction (~55–60% of total serine) and decreased M+2 serine (~2–3-fold) compared to WT cells, accompanied by nearly complete loss of M+1 serine (Figure 4C). Similar results were obtained with SHMT2 KD H460 cells and inhibitor-treated WT H460 and MIA PaCa-2 cells (Supplementary Figure S5C and D, respectively). These results identify SHMT2 as the likely mitochondrial target for AGF291, AGF320 and AGF347.
Identification of the de novo purine biosynthetic pathway as a direct target for AGF291, AGF320 and AGF347.
Both GARFTase and AICARFTase require 10-formyl-THF derived from formate, most of which is generated via mitochondrial C1 metabolism from serine (22) (Figures 1 and 4A). Consistent with this, loss of SHMT2 in H460 SHMT2 KD cells induced significant increases in purine intermediates which are dependent on C1 pools (i.e., 10-formyl-THF), including GAR (GARFTase substrate; 21-fold) (Figure 4D) and AICAR (AICARFTase substrate; 65-fold) (Figure 4E). Likewise, treatment of H460 cells with AGF291, AGF320, and AGF347 (10 μM) increased GAR (10–2300-fold) and AICAR (40–1500-fold) relative to untreated controls (Figures 4D and E). Similar increases in GAR and AICAR pools resulted in inhibitor-treated HCT116 and MIA PaCa-2 cells (Supplementary Figure S5E-H). For the HCT116 sublines, the increases in GAR and AICAR upon inhibitor treatments generally exceeded those resulting from the SHMT2 KO.
To assess the possibility that AGF291, AGF320 and AGF347 directly inhibit cytosolic enzyme targets in de novo purine biosynthesis (i.e., GARFTase and/or AICARFTase), we treated the H460 cells with 1 mM formate, to replenish the cytosolic C1 pool while circumventing the mitochondrial C1 pathway. We reasoned that formate treatment of SHMT2 KD cells should restore levels of GAR and AICAR to those seen in NTC (WT) cells. However, if the cytosolic enzymes were directly inhibited, formate should not effectively reverse GAR and/or AICAR accumulation. In H460 SHMT2 KD cells, treatment with formate completely reversed elevated GAR (Figure 4D) and AICAR (Figure 4E) to NTC levels. However, for inhibitor-treated H460 cells, reversal by formate was incomplete, albeit to different extents for different compounds. These results implicate direct targeting of GARFTase and/or AICARFTase by the novel inhibitors, in addition to SHMT2.
We measured the effects of the inhibitors on de novo purine biosynthesis, as reflected in total cellular pools of AMP, ADP and ATP. In Mia PaCa-2 cells, all the adenine nucleotides were comparably suppressed (~50%) with inhibitor treatments (Figure 4F), whereas in HCT116 cells (Supplementary Figure S5I), AMP and ADP pools were impacted more (50% suppression) than ATP (30% suppression), although SHMT2 KO had no effect. In H460 cells (Supplementary Figure S5J), SHMT2 KD decreased adenine nucleotide pools (20–50%), yet in response to the inhibitors, AMP increased (~50%), ADP decreased (up to ~60%) and ATP was essentially unaffected. Thus, in spite of the requirement for adenosine (with glycine) for complete rescue of all three cell lines from the inhibitory effect of the pyrrolopyrimidine compounds (Figure 3, Supplementary Figure S4), the extent of adenine nucleotide depletion varied.
Identification of SHMT1 as a target for pyrrolo[3,2-d]pyrimidine analogs.
As SHMT2 (UniProtKB: P34897) maintains 66% sequence identity (51) to SHMT1 (UniProtKB: 34896), we considered the possibility that AGF291, AGF320 and AGF347 could also target cytosolic SHMT1. By molecular modeling (Supplementary Figure S2), these analogs bound to rabbit SHMT1 (UniProtKB: 07511, 93% homology with human SHMT1(52)) with docking scores from −8.9 to −11.14 kcal/mol (Supplementary Table S1), raising the possibility that they may directly inhibit this enzyme, as well.
To gauge potential cytosolic SHMT1 inhibition by our compounds, we traced [2,3,3-2H]serine into dTTP (dTMP) via C1 transfer from 5,10-me-THF to dUMP by TS (22) (Figure 4A). In all three cell lines, WT cells incubated with [2,3,3-2H]serine generated M+1 dTTP with little-to-no M+2 dTTP (Figure 4G, and Supplementary Figures S5K and S5L). This confirms preferential [2,3,3-2H]serine metabolism through the mitochondrial C1 pathway to [2H]formate and into dTTP (M+1) (22). KD of SHMT2 in H460 cells or KO of SHMT2 in HCT116 cells induced a robust M+2 dTTP signal, reflecting serine-to-glycine flux through SHMT1 in the cytosol (22). Consistent with SHMT2 targeting, treatment with AGF291, AGF320, or AGF347 (10 μM) resulted in decreased M+1 and total dTTP in the cell lines (Figure 4G, and Supplementary Figures S5K and S5L). However, only in MIA PaCa-2 cells were these changes accompanied by increased M+2 dTTP (reflecting a SHMT1 compensation response (22)); the other two cell lines showed no measurable M+2 dTTP. These results likely reflect (i) sufficient residual flux through SHMT2 to preserve the primary M+1 labeling of dTTP, and/or (ii) direct targeting of SHMT1 by our compounds, along with cell line-specific differences in SHMT1 activity and inhibition by our compounds. In spite of decreased dTTP pools, thymidine did not rescue cells from effects of any of our inhibitors (Figure S4), a marked difference from TS-targeted inhibitors such as PMX (53). Thus, decreased dTTP likely reflects a secondary impact of targeting SHMT2/SHMT1 rather than directly targeting TS.
Further evidence for direct SHMT1 targeting by our inhibitors involves the substantially increased anti-proliferative effects of AGF291, AGF320 and AGF347 toward HCT116 SHMT2 KO cells compared with WT cells (21-fold for AGF291, 6-fold for AGF320, and 10-fold for AGF347; Table 1). Conversely, sensitivities to AGF94 and PMX were similar between WT and SHMT2 KO cells. These results mirror the increased potency toward SHMT2 KO cells relative to WT HCT116 cells seen with SHIN1 (23) and further suggest that our pyrrolo[3,2-d]pyrimidine compounds inhibit SHMT1 in addition to SHMT2.
In vitro confirmation of enzyme targets.
To confirm the enzyme targets (SHMT2, GARFTase, AICARFTase, and SHMT1) identified from our metabolomics experiments, we performed in vitro assays using purified recombinant enzymes. Our results demonstrate direct inhibition of SHMT2 by AGF291, AGF320 and AGF347 (KIs of 0.63, 0.056 and 2.19 μM, respectively; Table 1). While the compounds inhibited AICARFTase within a 4–5-fold range, AGF320 was the most potent GARFTase inhibitor, corroborating the metabolomics results (Figure 4D and E). For SHMT1, inhibitions paralleled those for SHMT2. None of the compounds inhibited MTHFD2 at concentrations of up to 200 μM of inhibitor (Table 1). These results confirm that SHMT2, the purine biosynthetic enzymes GARFTase and AICARFTase, and SHMT1 are direct targets of our lead pyrrolo[3,2-d]pyrimidine compounds. As a control, AGF94 was tested up to 200 μM inhibitor. AGF94 inhibited GARFTase (0.88 μM), however, there was no inhibition for AICARFTase or MTHFD2 and only weak inhibition of SHMT2 and SHMT1 (Kis of 188 and 112 μM, respectively). Notably, for PMX there was no significant inhibition of MTHFD2, SHMT1 or SHMT2 up to 200 μM; however, substantial inhibition was seen for GARFTase and AICARFTase (Table 1).
In vivo antitumor efficacy study of AGF347 with MIA PaCa-2 tumor xenografts.
We performed an in vivo efficacy trial with AGF347 in a head-to-head comparison with gemcitabine (GEM) toward MIA PaCa-2, an aggressive tumor model that reaches a euthanasia endpoint within 2 weeks of implant. We initially used early stage disease as a primary test of drug efficacy. Fourteen days before SC tumor implant, the mice were fed a folate-deficient diet to ensure that serum folate levels approximated those of humans (40,42–44,54). The mice were unselectively distributed to each group’s treatment and control arms (5 mice per group), and dosed with AGF347 (15 mg/kg/injection every 2 days × 8; total dose of 120 mg/kg) or GEM (120 mg/kg/injection every 4 days × 4; total dose of 480 mg/kg) just below their respective maximum tolerated doses. GEM was efficacious, with a median tumor burden on day 16 of 420 mg (range 284–552 mg) compared to 1189 mg (range 601–1711 mg) for the control cohort; AGF347 was distinctly superior, yielding a median tumor burden of 0 mg (range 0–276 mg). Median T/C values (see Materials and Methods for definition of quantitative endpoints) on day 16 were 35% for GEM and 0% for AGF347 (Figure 5A). Tumor growth delays (median T-C to reach 1000 mg) were 6 days for GEM, compared to 39 days for 4 of 5 mice treated with AGF347 and one tumor-free survivor (TFS) on day 136 (121 days post last treatment). To confirm whether immune factors contributed to this “curative” response, this mouse was “re-challenged” with MIA PaCa-2 tumor (bilateral SC implant). The tumor reached 1000 mg 16 days post-implant, with comparable growth and doubling time to the control group (median, 15.5 days; range, 13–20 days) (Figure 5A). Both drugs were well tolerated, with modest weight loss as the only adverse symptom (for AGF347, 1% (−0.2 g) median nadir on day 17; for GEM, 6.9%(−1.4 g) median nadir on day 3). After cessation of therapy, host recovery time was rapid for AGF347 (1 day) compared to GEM (14 days). Thus, at equitoxic dose levels, AGF347 showed superior antitumor efficacy over GEM (>5 logs of cell kill compared to 1.1 logs for GEM), with a 4-fold decreased dose requirement, no acute or long-term toxicities other than reversible weight loss, rapid host recovery time, and a tumor growth delay of >38 days, with 1 of 5 TFS (Supplementary Table S2).
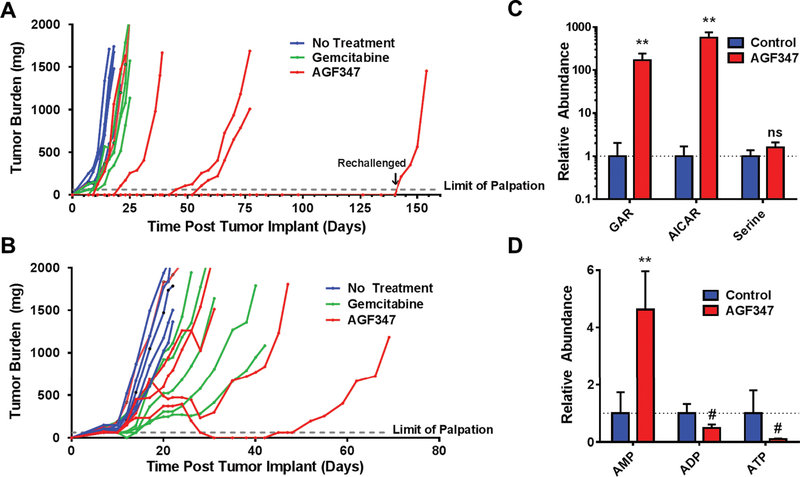
Figure 5. In vivo efficacy in early stage and upstage trials and pharmacodynamics of AGF347 toward MIA PaCa-2 PaC xenografts.
A: Female NCr SCID mice (11 weeks old 20.2 g average body weight) were implanted bilaterally with human MIA PaCa-2 pancreatic cancer xenograft fragments. Treatment was initiated on day 1 following SC tumor implantation. The mice were dosed as follows: AGF347, Q2dx8 at 15 mg/kg/injection, total dose 120 mg/kg; and GEM, Q4dx4 at 120 mg/kg/IV injection, total dose 480 mg/kg. Results are shown for individual mice. B: Female ICR SCID mice (10 weeks old; 19 g average body weight) were implanted bilaterally with human MIA PaCa-2 PaC tumors. On day 7 following SC implantation, the mice were dosed as for A. C,D: Targeted metabolomics on tumor samples from upstage trial mice. GAR, AICAR, and serine (C) and adenine nucleotides (AMP, ADP, ATP) (D) were quantified in control and AGF347-treated mice. Results for metabolomics (mean ± standard deviation) are normalized to control values and represent one tumor each taken from three different control and AGF347-treated mice. The changes in GAR and AICAR were statistically significant (**, p < 0.01) as were the changes in adenine nucleotides (**, p<0.01, #, p < 0.1) while the changes in serine did not reach statistical significance (“ns”= not significant), likely due to circulating serine pools.
Studies were extended to upstage MIA PaCa-2 xenografts. For these experiments, after implantation, MIA PaCa-2 tumors were allowed to grow for 7 days (100–150 mg), unselectively distributed (5 mice/group), then treated as for the early stage trial (Figure 5B). AGF347 and GEM were both efficacious, with median tumor burdens on day 21 of 733 mg (range 373–1835 mg) and 524 mg (range 247–1045 mg), respectively, compared to 1736 mg (range 1117–2046 mg) for the control cohort. AGF347 effected two partial remissions (PRs), including one complete response (CR); no PRs or CRs were seen with GEM. The PRs involved substantial decreases in tumor burden (688 to 220 mg, and 400 to 0 mg) and occurred on day 31 (10 days post last dose). Overall tumor growth delays (median T-C to reach 1000 mg) of 11 days (GEM) and 7 days (AGF347) were recorded. For the PRs, AGF347 induced tumor growth delays of 26 and 52 days, as compared to 16 and 25 days for the best GEM-responding mice. Both drugs were well tolerated, with modest weight loss during treatment (AGF347, 1% (−0.2g) median nadir on day 21 with full recovery on day 24; GEM, 5.1% (−1.0 g) median nadir on day 20 with full recovery on day 24).
For matched cohorts of mice maintained on a standard diet (results in ~10-fold increased serum folate), antitumor activity of AGF347 was ablated (Supplementary Figure S7). Thus, excessive levels of circulating folates largely abolish the antitumor activity of AGF347.
To confirm that the enzyme targets of AGF347 identified in vitro (GARFTase, AICARFTase, and SHMT1/2 ) were inhibited in vivo, tumor samples from a separate cohort of AGF347-treated and control mice (n=3 each) were harvested six hours after the 6th injection of AGF347 (tumors > 1000 mg). The tumors were homogenized and metabolites analyzed by targeted metabolomics (Figures 5C and 5D). In vivo administration of AGF347 induced substantially increased GAR (200-fold) and AICAR (500-fold), consistent with direct inhibition of GARFTase and AICARFTase as seen in vitro (Figures 4D and 4E); total serine trended up but did not differ significantly, likely due to exchange with circulating serine pools. We measured changes in purine nucleotide pools in vivo, including 5-fold increased AMP, ~2-fold decreased ADP and ~10-fold decreased ATP. This severe perturbation of energy charge resulted in a compensatory upregulation of cytochrome c oxidase activity (55) (Supplementary Figure S8), consistent with cytotoxic drug effects over the chronic course of in vivo treatment.
Discussion
The clinical utility of standard chemotherapy drugs is often limited by toxicities toward normal tissues (reflecting a lack of tumor selectivity) and/or drug resistance. Discovery of new and potent inhibitors of tumor-selective pathways remains a formidable challenge.
In this report, we identified novel inhibitors of C1 metabolism in mitochondria and cytosol with in vivo antitumor activity. Serine catabolism by mitochondrial C1 metabolism is the principal source of C1 units for cytosolic de novo purine and thymidylate biosynthesis, and a source of reducing equivalents and ATP (2–5,21). We targeted SHMT2, the first enzyme in the mitochondrial C1 pathway, with 5-substituted pyrrolo[3,2-d]pyrimidine analogs, rationally designed by molecular modeling from the SHMT2 crystal structure (23) and based on structural similarities to our previously reported 5-substituted pyrrolo[2,3-d]pyrimidine compounds (46,47) and N-substituted THF metabolites. We identified lead compounds AGF291, AGF320 and AGF347 that inhibited in vitro proliferation of a broad spectrum of tumor subtypes including lung (H460), colon (HCT116), and pancreatic (MIA PaCa-2) cancers.
We assessed critical enzyme targets for our compounds through glycine and nucleoside rescue, and by targeted metabolomics with a [2,3,3-2H]serine tracer, and identified SHMT2 in mitochondria as an intracellular target, along with SHMT1, GARFTase and AICARFTase in the cytosol. Inhibition of all these targets was confirmed by in vitro assays with purified recombinant enzymes. While the in vitro assays used “monoglutamyl” inhibitors, greater inhibition should result following metabolism to polyglutamates (7,9). Inhibition of SHMT1 by our analogs prevents metabolic “compensation” by reversal of SHMT1 catalysis and synthesis of glycine in response to loss of SHMT2 activity (22,23).
A number of pharmacodynamic factors could contribute to the in vitro antitumor effects of the novel analogs described herein. These include facilitated transport across the plasma membrane by PCFT and/or RFC and into mitochondria (potentially by SLC25A32 (13,14)), and metabolism to drug polyglutamates (7,9). Variations in these parameters likely account for differences in relative anti-proliferative activities toward the assorted tumor models. As AGF347 showed the most potent in vitro activity of the series, this compound was tested in vivo with both early stage and upstage MIA PaCa-2 xenograft models in SCID mice. AGF347 exhibited potent in vivo efficacy superior to that of GEM, even at a fold decreased total dose, resulting in sustained tumor growth delay and a 20% complete response rate. For the upstage trial, for the PRs, substantial decreases in tumor burden occurred 10 days after the last dose, likely reflecting a long term impact of AGF347 polyglutamates, associated with target inhibition.
Primary inhibition of SHMT2 in mitochondria results in decreased glycine and C1 units for cytosolic biosynthesis (4,23), and complete ablation through genetic means leads to defective mitochondrial respiration due to impaired synthesis of respiratory chain proteins (56,57). Other effects include decreased NADPH and glutathione with impacts on mitochondrial redox balance and reactive oxygen species (21), and decreased ATP synthesis due to impaired oxidative phosphorylation (56,57) and C1-flux through MTHFD1L (58). These are exacerbated by direct inhibitions of the de novo purine biosynthetic enzymes GARFTase and AICARFTase by our analogs. Although adenosine (with glycine) was essential for in vitro rescue of the tumor cell lines from our lead compounds, their effects on purine nucleotide pools were variable in both magnitude and direction. This suggests that depletion of critical subcellular purine nucleotide pools may not be reflected in measurements of total cellular purine nucleotides. The sustained treatment for AGF347 in vivo resulted in tumor killing with net energy depletion resulting in increased AMP/ATP ratio. This was accompanied by activation of cytochrome c oxidase, possibly reflecting posttranslational modification (dephosphorylation) (55).
Our results with active 5-substituted pyrrolo[3,2-d]pyrimidine compounds expand upon earlier findings with non-folate pyrazolopyran inhibitors of human SHMT2 (23) and, to our knowledge, this series represents the first bona fide inhibitors of this intracellular target with demonstrated in vivo antitumor efficacy. Thus, inhibition of SHMT2, coupled with direct inhibition of multiple C1-dependent targets including de novo purine biosynthesis and SHMT1, affords a valuable and exciting new platform for future drug development.
Supplementary Material
Acknowledgements
This work was supported in part by R01 CA53535 (LHM and ZH), R01 CA152316 (LHM and AG), K99 CA215307 (GSD), DP1DK113643 (JDR), R01 CA163591 (JDR), and R01 CA166711 (AG, LHM and CED) from the National Institutes of Health, a pilot Small Molecule Drug Discovery Grant from the Karmanos Cancer Institute (ZH and LHM), the Eunice and Milton Ring Endowed Chair for Cancer Research (LHM), and the Duquesne University Adrian Van Kaam Chair in Scholarly Excellence (AG). Mr. Dekhne was supported by T32 CA009531 (LHM) and F30 CA228221. The Animal Model and Therapeutics Evaluation Core (LP, SD, KW, JK), the Pharmacology Core (XB, JL) and Biostatistics Core (SK) were supported, in part, by NIH Center grant P30 CA022453 to the Karmanos Cancer Institute and the Wayne State University.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab 2016;23:27–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman AC, Maddocks ODK. One-carbon metabolism in cancer. British Journal of Cancer 2017;116:1499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang M, Vousden KH. Serine and one-carbon metabolism in cancer. Nat Rev Cancer 2016;16:650–62 [DOI] [PubMed] [Google Scholar]
- 4.Ducker GS, Rabinowitz JD. One-Carbon Metabolism in Health and Disease. Cell Metabolism 2017;25:27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tibbetts AS, Appling DR. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr 2010;30:57–81 [DOI] [PubMed] [Google Scholar]
- 6.Field MS, Kamynina E, Chon J, Stover PJ. Nuclear folate metabolism. Annu Rev Nutr 2018;38:219–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattopadhyay S, Moran RG, Goldman ID. Pemetrexed: biochemical and cellular pharmacology, mechanisms, and clinical applications. Molecular cancer therapeutics 2007;6:404–17 [DOI] [PubMed] [Google Scholar]
- 8.Matherly LH, Wilson MR, Hou Z. The Major Facilitative Folate Transporters Solute Carrier 19A1 and Solute Carrier 46A1: Biology and Role in Antifolate Chemotherapy of Cancer. Drug metabolism and disposition: the biological fate of chemicals 2014;42:632–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visentin M, Zhao R, Goldman ID. The Antifolates. Hematology/oncology clinics of North America 2012;26:629–ix [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer AM, Kamynina E, Field MS, Stover PJ. Folate rescues vitamin B12 depletion-induced inhibition of nuclear thymidylate biosynthesis and genome instability. Proc Natl Acad Sci U S A 2017;114:E4095–E102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matherly LH, Hou Z, Deng Y. Human reduced folate carrier: translation of basic biology to cancer etiology and therapy. Cancer metastasis reviews 2007;26:111–28 [DOI] [PubMed] [Google Scholar]
- 12.Zhao RD-B, Ndeye; Visentin, Michele; Goldman, David I. Mechanisms of Membrane Transport of Folates into Cells and Across Epithelia. Annual Review of Nutrition 2011;31:177–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy EA, Titus SA, Taylor SM, Jackson-Cook C, Moran RG. A mutation inactivating the mitochondrial inner membrane folate transporter creates a glycine requirement for survival of chinese hamster cells. J Biol Chem 2004;279:33829–36 [DOI] [PubMed] [Google Scholar]
- 14.Lawrence SA, Hackett JC, Moran RG. Tetrahydrofolate Recognition by the Mitochondrial Folate Transporter. J Biol Chem 2011;286:31480–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence SA, Titus SA, Ferguson J, Heineman AL, Taylor SM, Moran RG. Mammalian mitochondrial and cytosolic folylpolyglutamate synthetase maintain the subcellular compartmentalization of folates. J Biol Chem 2014;289:29386–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science (New York, NY) 2012;336:1040–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D, Fiske BP, Birsoy K, Freinkman E, Kami K, Possemato R, et al. SHMT2 drives glioma cell survival in the tumor microenvironment but imposes a dependence on glycine clearance. Nature 2015;520:363–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun 2014;5:3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Chen Z, Xue D, Zhang Q, Liu X, Luh F, et al. Prognostic and therapeutic value of mitochondrial serine hydroxyl-methyltransferase 2 as a breast cancer biomarker. Oncology reports 2016;36:2489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loayza-Puch F, Rooijers K, Buil LC, Zijlstra J, Oude Vrielink JF, Lopes R, et al. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature 2016;530:490–4 [DOI] [PubMed] [Google Scholar]
- 21.Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer discovery 2014;4:1406–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, et al. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab 2016;23:1140–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ducker GS, Ghergurovich JM, Mainolfi N, Suri V, Jeong SK, Hsin-Jung Li S, et al. Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A 2017;114:11404–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu TF, Scarsdale JN, Kazanina G, Schirch V, Wright HT. Location of the pteroylpolyglutamate-binding site on rabbit cytosolic serine hydroxymethyltransferase. J Biol Chem 2003;278:2645–53 [DOI] [PubMed] [Google Scholar]
- 25.Flintoff WF, Davidson SV, Siminovitch L. Isolation and partial characterization of three methotrexate-resistant phenotypes from Chinese hamster ovary cells. Somat Cell Mol Genet 1976;2:245–61 [DOI] [PubMed] [Google Scholar]
- 26.Deng Y, Wang Y, Cherian C, Hou Z, Buck SA, Matherly LH, et al. Synthesis and discovery of high affinity folate receptor-specific glycinamide ribonucleotide formyltransferase inhibitors with antitumor activity. J Med Chem 2008;51:5052–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng Y, Zhou X, Desmoulin SK, Wu J, Cherian C, Hou Z, et al. Synthesis and biological activity of a novel series of 6-substituted thieno[2,3-d]pyrimidine antifolate inhibitors of purine biosynthesis with selectivity for high affinity folate receptors over the reduced folate carrier and proton-coupled folate transporter for cellular entry. J Med Chem 2009;52:2940–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong SC, Proefke SA, Bhushan A, Matherly LH. Isolation of human cDNAs that restore methotrexate sensitivity and reduced folate carrier activity in methotrexate transport-defective Chinese hamster ovary cells. J Biol Chem 1995;270:17468–75 [DOI] [PubMed] [Google Scholar]
- 29.Hou Z, Gattoc L, O’Connor C, Yang S, Wallace-Povirk A, George C, et al. Dual targeting of epithelial ovarian cancer via folate receptor alpha and the proton-coupled folate transporter with 6-substituted pyrrolo[2,3-d]pyrimidine antifolates. Molecular cancer therapeutics 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem 1951;193:265–75 [PubMed] [Google Scholar]
- 31.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680–5 [DOI] [PubMed] [Google Scholar]
- 32.Matsudaira P Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem 1987;262:10035–8 [PubMed] [Google Scholar]
- 33.Ducker GS, Ghergurovich JM, Mainolfi N, Suri V, Jeong SK, Hsin-Jung Li S, et al. Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proceedings of the National Academy of Sciences 2017;114:11404–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Analytical chemistry 2010;82:3212–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clasquin MF, Melamud E, Rabinowitz JD. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Current protocols in bioinformatics 2012;Chapter 14:Unit14.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Golani LK, Wallace-Povirk A, Deis SM, Wong JE, Ke J, Gu X, et al. Tumor Targeting with Novel 6-Substituted Pyrrolo [2,3-d] Pyrimidine Antifolates with Heteroatom Bridge Substitutions Via Cellular Uptake by Folate Receptor α and the Proton-coupled Folate Transporter and Inhibition of De Novo Purine Nucleotide Biosynthesis. J Med Chem 2016;59:7856–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shih C, Chen VJ, Gossett LS, Gates SB, MacKellar WC, Habeck LL, et al. LY231514, a pyrrolo[2,3-d]pyrimidine-based antifolate that inhibits multiple folate-requiring enzymes. Cancer Res 1997;57:1116–23 [PubMed] [Google Scholar]
- 38.Wang L, Wallace A, Raghavan S, Deis SM, Wilson MR, Yang S, et al. 6-Substituted Pyrrolo[2,3-d]pyrimidine Thienoyl Regioisomers as Targeted Antifolates for Folate Receptor alpha and the Proton-Coupled Folate Transporter in Human Tumors. J Med Chem 2015;58:6938–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cherian C, Kugel Desmoulin S, Wang L, Polin L, White K, Kushner J, et al. Therapeutic targeting malignant mesothelioma with a novel 6-substituted pyrrolo[2,3-d]pyrimidine thienoyl antifolate via its selective uptake by the proton-coupled folate transporter. Cancer Chemother Pharmacol 2013;71:999–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Golani LK, Wallace-Povirk A, Deis SM, Wong JE, Ke J, Gu X, et al. Tumor targeting with novel 6-substituted pyrrolo[2,3-d]pyrimidine antifolates with heteroatom bridge substitutions via cellular uptake by folate receptor α and the proton-coupled folate transporter and inhibition of de novo purine nucleotide biosynthesis. J Med Chem 2016;59:7856–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravindra MP, Wilson MR, Tong N, O’Connor C, Karim MA, Polin L, et al. Fluorinated substituted pyrrolo[2,3-d]pyrimidine antifolates with tumor-targeting via cellular uptake by folate receptor α and the proton-coupled folate transporter and inhibition of de novo purine nucleotide biosynthesis. J Med Chem 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Cherian C, Desmoulin SK, Polin L, Deng Y, Wu J, et al. Synthesis and antitumor activity of a novel series of 6-substituted pyrrolo[2,3-d]pyrimidine thienoyl antifolate inhibitors of purine biosynthesis with selectivity for high affinity folate receptors and the proton-coupled folate transporter over the reduced folate carrier for cellular entry. J Med Chem 2010;53:1306–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Desmoulin SK, Cherian C, Polin L, White K, Kushner J, et al. Synthesis, biological and antitumor activity of a highly potent 6-substituted pyrrolo[2,3-d]pyrimidine thienoyl antifolate inhibitor with proton-coupled folate transporter and folate receptor selectivity over the reduced folate carrier that inhibits β-glycinamide ribonucleotide formyltransferase. J Med Chem 2011;54:7150–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L, Wallace A, Raghavan S, Deis SM, Wilson MR, Yang S, et al. 6-Substituted Pyrrolo[2,3-d]pyrimidine Thienoyl Regioisomers as Targeted Antifolates for Folate Receptor α and the Proton-Coupled Folate Transporter in Human Tumors. J Med Chem 2015;58:6938–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee GY, Haverty PM, Li L, Kljavin NM, Bourgon R, Lee J, et al. Comparative oncogenomics identifies PSMB4 and SHMT2 as potential cancer driver genes. Cancer research 2014;74:3114–26 [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Cherian C, Orr S, Mitchell-Ryan S, Hou Z, Raghavan S, et al. Tumor-Targeting with Novel Non-Benzoyl 6-Substituted Straight Chain Pyrrolo[2,3-d]pyrimidine Antifolates via Cellular Uptake by Folate Receptor α and Inhibition of de novo Purine Nucleotide Biosynthesis. Journal of medicinal chemistry 2013;56:10.1021/jm401139z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Mitchell-Ryan S, Raghavan S, George C, Orr S, Hou Z, et al. Novel 5-substituted pyrrolo[2,3-d]pyrimidines as dual inhibitors of glycinamide ribonucleotide formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase and as potential antitumor agents. J Med Chem 2015;58:1479–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desmoulin SK, Hou Z, Gangjee A, Matherly LH. The human proton-coupled folate transporter: Biology and therapeutic applications to cancer. Cancer Biology & Therapy 2012;13:1355–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson MR, Hou Z, Yang S, Polin L, Kushner J, White K, et al. Targeting Nonsquamous Nonsmall Cell Lung Cancer via the Proton-Coupled Folate Transporter with 6-Substituted Pyrrolo[2,3-d]Pyrimidine Thienoyl Antifolates. Mol Pharmacol 2016;89:425–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kugel Desmoulin S, Wang L, Hales E, Polin L, White K, Kushner J, et al. Therapeutic targeting of a novel 6-substituted pyrrolo [2,3-d]pyrimidine thienoyl antifolate to human solid tumors based on selective uptake by the proton-coupled folate transporter. Mol Pharmacol 2011;80:1096–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, et al. Protein database searches using compositionally adjusted substitution matrices. The FEBS journal 2005;272:5101–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology 1990;215:403–10 [DOI] [PubMed] [Google Scholar]
- 53.Rong S, Zou L, Zhang Y, Zhang G, Li X, Li M, et al. Determination of purine contents in different parts of pork and beef by high performance liquid chromatography. Food chemistry 2015;170:303–7 [DOI] [PubMed] [Google Scholar]
- 54.Varela-Moreiras G, Selhub J. Long-term folate deficiency alters folate content and distribution differentially in rat tissues. J Nutr 1992;122:986–91 [DOI] [PubMed] [Google Scholar]
- 55.Huttemann M, Lee I, Grossman LI, Doan JW, Sanderson TH. Phosphorylation of mammalian cytochrome c and cytochrome c oxidase in the regulation of cell destiny: respiration, apoptosis, and human disease. Advances in experimental medicine and biology 2012;748:237–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Minton DR, Nam M, McLaughlin DJ, Shin J, Bayraktar EC, Alvarez SW, et al. Serine Catabolism by SHMT2 Is Required for Proper Mitochondrial Translation Initiation and Maintenance of Formylmethionyl-tRNAs. Mol Cell 2018;69:610–21 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morscher RJ, Ducker GS, Li SH, Mayer JA, Gitai Z, Sperl W, et al. Mitochondrial translation requires folate-dependent tRNA methylation. Nature 2018;554:128–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meiser J, Tumanov S, Maddocks O, Labuschagne CF, Athineos D, Van Den Broek N, et al. Serine one-carbon catabolism with formate overflow. Sci Adv 2016;2:e1601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.