Abstract
Purpose:
Metabolomics is a discovery tool for novel associations of metabolites with disease. Here, we interrogated the metabolome of human breast tumors to describe metabolites whose accumulation affects tumor biology.
Experimental Design:
We applied large-scale metabolomics followed by absolute quantification and machine learning-based feature selection using LASSO to identify metabolites that show a robust association with tumor biology and disease outcome. Key observations were validated with the analysis of an independent dataset and cell culture experiments.
Results:
LASSO-based feature selection revealed an association of tumor glycochenodeoxycholate levels with improved breast cancer survival, which was confirmed using a Cox proportional hazards model. Absolute quantification of four bile acids, including glycochenodeoxycholate and microbiome-derived deoxycholate, corroborated the accumulation of bile acids in breast tumors. Levels of glycochenodeoxycholate and other bile acids showed an inverse association with the proliferation score in tumors and the expression of cell cycle and G2/M checkpoint genes, which was corroborated with cell culture experiments. Moreover, tumor levels of these bile acids markedly correlated with metabolites in the steroid metabolism pathway and increased expression of key genes in this pathway, suggesting that bile acids may interfere with hormonal pathways in the breast. Lastly, a proteome analysis identified the Complement & Coagulation Cascade as being up-regulated in glycochenodeoxycholate-high tumors.
Conclusions:
We describe the unexpected accumulation of liver- and microbiome-derived bile acids in breast tumors. Tumors with increased bile acids show decreased proliferation, thus fall into a good prognosis category, and exhibit significant changes in steroid metabolism.
Keywords: Breast cancer, metabolism, bile acid, proliferation, steroid synthesis, survival
INTRODUCTION
Breast cancer is a heterogeneous disease and its biology is closely related to the expression of hormone receptors. Studies of the transcriptome discovered that breast tumors can be classified into subtypes with distinct biology (1,2). Of all subtypes, basal-like and HER2-positive tumors tend to produce the most aggressive disease (3,4). Estrogen receptor-positive tumors mainly fall into the luminal A subtype category while tumors lacking the expression of the estrogen, progesterone, and HER2 receptors have been described as triple-negative tumors and largely overlap with the basal-like subtype. Factors besides molecular subtypes that associate with the prognosis of breast cancer patients include disease stage, the tumor proliferation index, the differentiation status and vascularization of a tumor, and a patient’s body mass (5-8).
The relationship of tumor metabolism with breast cancer subtypes and disease prognosis remains incompletely understood (9). Large differences in metabolite abundance have been observed between tumor and adjacent non-cancerous tissues and by estrogen receptor status (10,11). It has also been shown that TP53 and Myc signaling influence breast cancer metabolism (9,11,12). Preclinical and clinical studies have linked tumor metabolism to therapeutic response and patient survival (9,13). Thus, current research suggests that metabolic profiling of tumors may provide new opportunities to improve outcomes in breast cancer. Here, we interrogated the metabolome of human breast tumors to describe metabolites whose accumulation affects tumor biology and disease prognosis. We applied large-scale metabolomics and machine learning-based feature selection to identify metabolites that show a robust association with tumor biology and disease outcome. Key observations were corroborated using absolute quantification of selected metabolites and analysis of an independent dataset, and cell culture experiments. Our approach led to the identification of bile acids as metabolites that accumulate in breast tumors. Bile acids are important signaling molecules (14). Accordingly, tumors with increased bile acid content exhibited a distinct disease biology and an association with patient survival.
MATERIALS AND METHODS
Tissue collection.
Breast cancer patients were recruited between 1993 and 2003, as described previously (11,15,16). Patients completed a questionnaire and provided biospecimens. Samples of fresh-frozen tumor tissue and adjacent non-cancerous tissue were processed by a pathologist immediately after surgery at the Department of Pathology, University of Maryland. Clinical and pathological information was obtained from medical records and pathology reports. The collection of biospecimens and the clinical and pathological information was approved by the University of Maryland Institutional Review Board for the participating institutions (UMD protocol #0298229). IRB approval of this protocol was then obtained at all institutions (Veterans Affairs Medical Center, Union Memorial Hospital, Mercy Medical Center, and Sinai Hospital, Baltimore, MD). The research was also reviewed and approved by the NIH Office of Human Subjects Research Protections (OHSRP #2248). Informed written consent was obtained from all patients and the research followed the ethical guidelines set by the Declaration of Helsinki.
Cell lines and bile acids.
Human breast cancer cell lines (MCF7, MDA-MB-175-VII, T47D, ZR-75–30) were acquired from the American Tissue Culture Collection (ATCC, Manassas, VA) and were cultured in RPMI (Life Technologies, Carlsbad, CA) with 10% FBS (Life Technologies). Cells were regularly authenticated using either a short tandem repeat analysis with GenePrint10 or STR profile creation (17 loci plus Amelogenin) from ATCC and tested for mycoplasma contamination. Four bile acids, deoxycholate (≥ 99%), chenodeoxycholate (≥ 97%), glycodeoxycholate (≥ 97%) and glycochenodeoxycholate (≥ 97%), were purchased from Sigma-Aldrich (St. Louis, MO) and resolved in DMSO to a stock concentration of 100 mM and then added to culture medium at indicated concentrations. The TGR5 and OATP1B1/3 antagonists, NF449 and rifampicin, were also obtained from Sigma-Aldrich.
Bromodeoxyuridine (BrdU)-based proliferation assay.
Cells were pre-seeded in 96-well cell culture plates (Corning Life Sciences, Tewksbury, MA) overnight and then cultured with added bile acids. Untreated and bile acid-treated cells were cultured for 24 hours and then assessed for cell proliferation using the BrdU colorimetric ELISA assay (Roche Applied Science, Indianapolis, IN) according to the provided protocol. Six to 12 biological replicates were analyzed per treatment condition and reported as mean ± standard error of the mean (S.E.M.). To study the ability of TGR5 and OATP1B1/3 antagonists to inhibit the anti-proliferative effects of deoxycholate, T47D were pretreated with 10 μM of antagonist for 24 hours before 20 μM deoxycholate was added for an additional 48 hours.
Metabolome analysis.
Metabolomic profiling of human breast tissues was performed using both an untargeted discovery approach and a targeted approach for validation and absolute quantification. Untargeted metabolic profiling of known and unknown metabolites in the discovery set included 67 human breast tumors and 65 tumor-adjacent noncancerous tissues and was performed by Metabolon Inc, as described previously (11,17). Additional metabolic profiling with absolute quantification of the four bile acids, deoxycholate, chenodeoxycholate, glycodeoxycholate, and glycochenodeoxycholate was performed at the Alkek Center for Molecular Discovery of Baylor College of Medicine using selective reaction monitoring and the Agilent 6490 Triple Quadrupole Mass Spectrometer system. For absolute quantification of bile acids in tissue extracts from 20 tumor-adjacent noncancerous tissue pairs, a random subset of the 67 tumors described above, we prepared serial dilutions of bile acid standards (all Sigma-Aldrich) to generate the calibration curve.
Gene expression analysis.
Gene expression data existed for 61 breast tumors with metabolome data in the discovery set, as described (11). We performed RNA sequencing to obtain additional gene expression data for bile acid-treated cell lines (T47D, MDA-MB-175-VII). Here, cells were treated with deoxycholate for 24 hours prior to RNA isolation. RNA sequencing was performed using 3–4 biological replicates. The data were deposited in the NCBI database (http://www.ncbi.nlm.nih.gov). The NCBI BioProject ID is PRJNA544091. See Supplementary Methods for more details about the gene expression analysis.
Pathway analysis.
For pathway enrichment analysis, genes were ranked by t‐statistic and imported into the Gene Set Enrichment Analysis Preranked module (https://software.broadinstitute.org/gsea/index.jsp) (18). Hallmark and KEGG gene sets were selected within MSigDB as references for pathway analysis. Visualization of particular pathways was performed using pathview, a tool set for pathway-based data integration and visualization in Bioconductor.
Proteomics data.
Large-scale proteome and metabolome data existed for 58 breast tumors. The mass spectrometry (MS)-based analysis of the proteome was performed as previously described (19). More details are provided in Supplementary Methods.
Tissue proliferation score.
We selected the array-based gene expression profiles of 11 cell cycle genes (BIRC5, CCNB1, CDC20, CEP55, MKI67, NDC80, NUF2, PTTG1, RRM2, TYMS, UBE2C) and summed them into a meta-gene score as a marker for tissue proliferation, as described previously (19-21). This proliferation signature also contains MKI67, the transcript which encodes Ki67, a commonly used proliferation marker using immunohistochemistry.
LASSO feature selection and concordance index.
We built predictive models for patient survival from a training set using Cox Proportional-Hazards Regression modeling with L1 penalized log partial likelihood estimation (22). The predictive power of metabolomics data - when integrated with clinical variables - was then assessed by the concordance index (C-index) (23). The C-index is a nonparametric value to quantify the power of a predictive model, wherein a C-index of 1 indicates perfect prediction accuracy while a C-index of 0.5 is indicates a model not better than random chance. More details are provided in Supplementary Methods.
Statistical analysis.
Analyses were conducted using R version 3.5 (R Foundation for Statistical Computing; http://www.r-project.org/). Within R, the following packages were used: the following packages were used: pheatmap_1.0.12, pathview_1.24.0, org.Hs.eg.db_3.8.2, beeswarm_0.2.3, survival_2.44–1.1, ggplot2_3.1.1, DESeq2_1.24.0, Biobase_2.44.0, GenomicRanges_1.36.0, S4Vectors_0.22.0. All statistical tests were two-sided. P < 0.05 was considered statistically significant. The non-parametric Wilcoxon rank test was used for group comparisons with continuous data. Box and whisker plots were used to display data in graphs. We used the Spearman rank correlation test and calculated rho to assess correlations between tumor markers. Survival analysis was performed for the 67 breast cancer patients in the discovery set with existing metabolome data. These patients had long-term follow up for breast cancer-specific survival. The Cox Proportional-Hazards Regression model was applied to estimate hazard ratios and a Wald test was used to evaluate the significance of outcome differences between risk groups. In the analysis, tumor metabolite levels were generally median-dichotomized to define high-abundance and low-abundance groups, except for glycochenodeoxycholate abundance measurements in the Metabolon dataset where the cutoff was set at the detection limit (above vs. at/below) because this metabolite was at/below the detection limit in more than 50% of the tumors in this dataset. Ptrend for the Cox regression analysis was calculated using continuous data for tumor metabolite levels.
RESULTS
Glycochenodeoxycholate, a primary bile acid, is increased in a subset of human breast tumors and improves survival prediction in breast cancer.
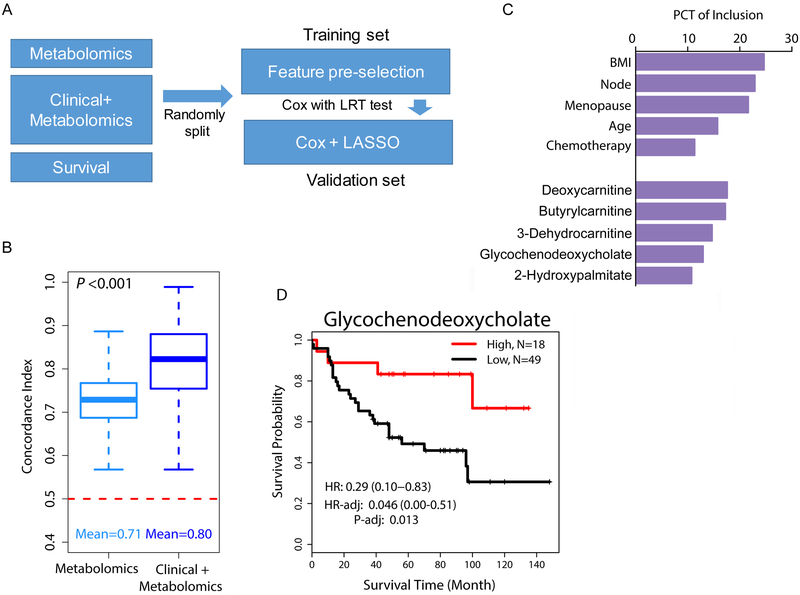
In a previous study of 67 human breast tumors and 65 tumor-adjacent noncancerous tissues, our group described relative abundance levels of 536 metabolites in these tissues (11). Here, we further investigated the association of metabolites with patient survival and disease biology using this cohort. The characteristics of the 67 breast cancer patients are described in Supplementary Table 1. We restricted our analysis to those metabolites that were measurable in at least 25% of the tumors (n = 398) and explored their association with breast cancer survival in the context of other patient data using a cross-validation approach and LASSO-based feature selection for Cox regression modeling (Figure 1A). The predictive power of metabolomics data - when integrated with clinical variables – was assessed by the C-index, as described under Methods. Using only metabolomics data, top models achieved substantial predictive power with C-indexes being significantly higher than 0.5 (mean C-index = 0.71) (Figure 1B), indicating the usefulness of metabolome data for survival prediction. When we applied an integrated model of metabolomics and clinical variables, we significantly improved the predictive power (mean C-index = 0.80, Wilcoxon signed rank test P < 0.001 comparing top metabolomics-based models vs. integrated models of metabolomics and clinical variables). The features that were most commonly selected by LASSO for survival prediction in the integrated models included the variables patient’s age, body mass index (BMI), menopausal and node status, and receipt of chemotherapy, as well as the five metabolites, deoxycarnitine, butyrylcarnitine, 3-dehydrocarnitine, glycochenodeoxycholate (GCDC) and 2-hydroxypalmitate (Figure 1C). Among these metabolites, GCDC is a liver metabolism-derived primary bile acid with an unexpected accumulation in a subset of breast tumors and adjacent non-cancerous tissue (Supplementary Figure 1). The other four metabolites are surrogates for changes in fatty acid metabolism in mitochondria and peroxisomes. The inclusion of these 5 metabolites into the integrated model for breast cancer survival prediction was substantiated with Kaplan-Meier plots and Cox proportional hazards analyses for GCDC (Figure 1D), deoxycarnitine, butyrylcarnitine, 3-dehydrocarnitine, and 2-hydroxypalmitate (Supplementary Figure 2). Of these metabolites, only the increase in tumor GCDC levels associated with improved patient survival (HR = 0.29, 95% CI: 0.1 to 0.83; Ptrend = 0.032 when continuous data were used in the Cox regression model). GCDC was associated with survival independent of patient’s age, BMI, menopausal and node status, and receipt of chemotherapy, or when the multivariable Cox regression analysis was additionally adjusted for tumor subtypes (HR = 0.08, 95% CI: 0.02 to 0.65). Noteworthy, GCDC levels in breast tumors did not associate with patients’ BMI, arguing against an obesity effect that led to GCDC accumulation.
Figure 1. Metabolomics improves survival prediction for breast cancer.
A. Computational approach to build a survival prediction model using LASSO feature selection for Cox proportional hazards modeling. B. Concordance indexes for two models, either including only metabolomics data (398 metabolites) or a combination of clinical and metabolomics data (clinical + metabolomics). The dashed red line (C-index = 0.5) would represent models with no prognostic power whereas a C-index of 1 indicates a perfect prediction accuracy. Shown are the medians and confidence intervals not crossing the red line. C. Top ten features selected from the training set using LASSO feature selection for survival prediction in the validation set. PCT, percentage. D. Association of tumor glycochenodeoxycholate content with breast cancer-specific survival. Kaplan-Meier plot and hazard ratio (HR) estimates in the unadjusted and adjusted Cox regression analysis. High: elevated glycochenodeoxycholate levels (n=18). Low: detection limit or below (n=49). Adjusted analysis with patient’s age, BMI, menopausal and node status, and receipt of chemotherapy as covariables.
Absolute quantification of primary and secondary bile acids in human breast tissues shows their presence in tumor and adjacent non-cancerous tissues.
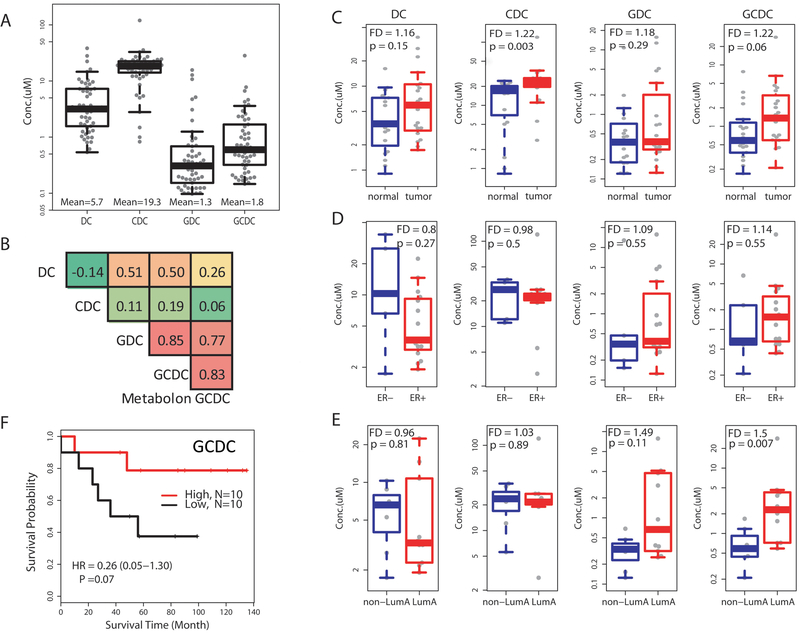
To further examine the occurrence of GCDC and other bile acids in breast tumors, we selected four bile acids, deoxycholate (DC), chenodeoxycholate (CDC), glycodeoxycholate (GDC) and GCDC, for the analysis. DC is a gut microbiome-derived bile acid, while the other three originate from liver metabolism. Forty breast tissues (20 tumor-adjacent normal pairs) were analyzed using selective reaction monitoring and the Agilent 6490 Triple Quadrupole Mass Spectrometer system for absolute measurements. With this approach, we could detect these bile acids in all 40 tissues at mean concentrations of 5.7 μM for DC, 19.3 μM for CDC, 1.3 μM for GDC and 1.8 μM for GCDC (Figure 2A). A robust correlation (r = 0.83) was observed when we compared between GCDC measurements using the Metabolon platform and our absolute quantification method (Figure 2B). GDC, GCDC and DC levels showed high correlations with each other (r > 0.5) in the breast tissues, whereas CDC followed a dissimilar accumulation pattern. The bile acids tended to be slightly elevated in tumor tissue when compared to adjacent non-cancerous tissues (Figure 2C), but no clear difference emerged between estrogen receptor (ER)-positive and ER-negative tumors (Figure 2D). GCDC levels were higher in luminal A tumors than other tumors (Figure 2E), which we confirmed with the analysis of the only publicly available dataset (“Duke cohort”) (Supplementary Figure 3A & B) that contains bile acid measurements for 25 breast tumors (12). Lastly, in an exploratory survival analysis using the absolute measurements of GCDC in the 20 breast tumors, we could corroborate our previous finding that increased GCDC in breast tumors is associated with improved patient survival (Figure 2F).
Figure 2. Absolute quantification of bile acids in 20 breast tumor-adjacent non-cancerous tissue pairs.
A. Shown is the concentration of four bile acids across the 20 tissue pairs (n = 40) measured by mass spectrometry. Average concentration of the bile acids is 5.7 μM for deoxycholate (DC), 19.3 μM for chenodeoxycholate (CDC), 1.3 μM for glycodeoxycholate (GDC), and 1.8 μM for glycochenodeoxycholate (GCDC). Box plots with median values (horizontal line). B. Pearson’s correlation coefficients matrix that includes measurements for the four bile acids and the relative abundance measurement of GCDC by Metabolon in these 40 tissues. In-house absolute measurements of GCDC and the relative abundance measurements by Metabolon show a high correlation (r = 0.83). Shades of red indicate a high correlation while shades of green indicate a low correlation. C-E. Bile acid concentrations after stratification into normal vs. tumor with fold difference (FD) (C), ER-negative (ER−) versus ER-positive (ER+) tumors (D), and luminal A tumors vs. other tumor subtypes (E). Fold difference was calculated from the mean value for each group and the Wilcoxon rank test was used for significance analysis. F. Kaplan-Meier survival plot after stratifying tumors into low and high GCDC with the median as cutoff.
Transcriptome and proteome profiles of breast tumors indicate that increased bile acid levels inhibit tumor cell proliferation.
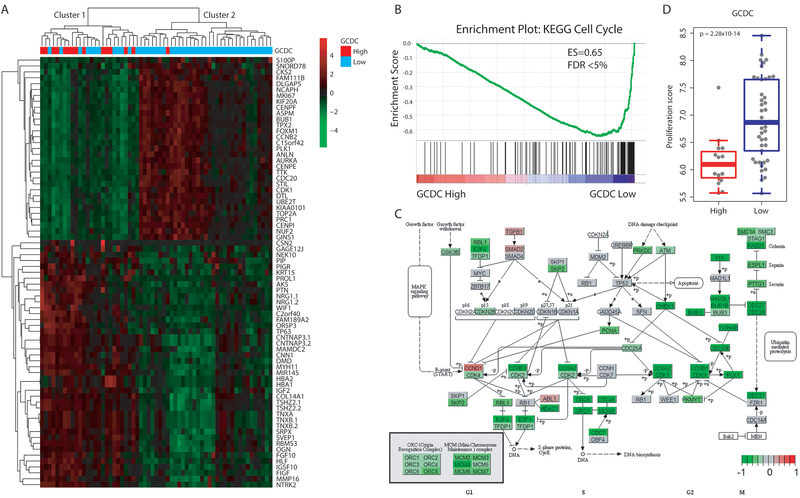
To further characterize the tumor biology associated with increased GCDC, we examined relationships of GCDC with the transcriptome and proteome. Having existing gene expression and proteome data from 61 and 58 breast tumors, respectively, in our NCI-Maryland breast cancer cohort (11,19), we initially analyzed the transcriptome differences between the 15 GCDC-high versus the 46 GCDC-low tumors. The investigation identified 73 transcripts that were significantly differentially expressed using a false discovery rate (FDR) < 5% and fold-difference > 2 as stringent cutoffs (Supplementary Table 2). A list of 15 genes including BUB1, NUF2, AURKA, CDC20, CENPE/F/I, CCNB2, CDK1, FOXM1, PLK1 and TOP2A, all key regulators of the cell cycle and cell proliferation, showed significantly decreased expression in GCDC-high tumors (all were decreased between 2.1 to 2.5-fold), while genes such as PROL1, PIP, WIF1, HBA1/2, IGF2 and COL14A1 that have a relationship with extracellular matrix and exosome secretion were up-regulated in these tumors (Figure 3A). A gene set enrichment analysis (GSEA) including all differently expressed genes further corroborated these findings. The Cell Cycle KEGG pathway was the top down-regulated pathway (Enrichment score 0.65, FDR = 0) in GCDC-high tumors, with the down-regulation of G2/M checkpoint genes being the most significant change (Figure 3B-C). We also assessed the proliferation score of each tumor, as described under Methods, and then correlated this score with GCDC levels in tumors. This analysis showed that GCDC-high tumors tend to have a low proliferation score, while the score is prominently higher in the GCDC-low tumors (Figure 3D). Our finding was validated with an analysis of the “Duke cohort” (12), the independent dataset with both metabolome and TCGA transcriptome data, showing that increased GCDC levels are associated with a significantly decreased proliferation rate in breast tumors (Supplementary Figure 3C). Similar inverse associations between tumor bile acid levels and the proliferation score were observed for GCDC, DC and GDC based on the analysis of their absolute measurements in 20 breast tumors (Supplementary Figure 4). However, we did not find that GCDC levels affected the tissue proliferation score in tumor-adjacent non-cancerous tissues (Supplementary Figure 5), indicating a distinct effect of GCDC on tumor biology.
Figure 3. Transcriptome profile of GCDC-high tumors indicates that GCDC may target cell cycle pathways and inhibit tumor cell proliferation.
A. Hierarchical clustering based on differentially expressed genes between tumors with high (n=15) or low (n=46) abundance of GCDC. Heatmap represents the most differentially expressed transcripts associated with GCDC abundance with ∣fold change∣ > 2 and FDR < 0.05 (n = 73). Low GCDC: GCDC is at the detection limit in these tumors. Red indicates up-regulated genes and green indicates down-regulated genes. Cluster 1 is enriched for GCDC-high tumors. B. Gene set enrichment analysis identifies the KEGG Cell Cycle pathway as the top down-regulated pathway in GCDC-high breast tumors (Enrichment score = −0.65 and FDR < 0.05). C. Genes were mapped to the KEGG Cell Cycle pathway and labeled as red and green when up- or down-regulated in GCDC-high tumors, respectively, indicating common down-regulation of G1, S, and G2/M phase genes. Dark green indicates down-regulation of many G2/M phase genes. D. Tumor proliferation score is higher in GCDC-low than GCDC-high tumors. A Wilcoxon rank test was applied for significance testing to compare proliferation scores between GCDC-high (n = 15) and low tumors (n = 46).
As proteome data existed for 58 breast tumors in our study, we analyzed the relationship of tumor GCDC levels with proteome-defined tumor characteristics by applying GSEA with proteome-annotated genes. We made two key observations. GSEA identified the Complement & Coagulation Cascade gene set as one of the top up-regulated proteome-defined gene sets in GCDC-high tumors and corroborated the down-regulation of G2/M checkpoint genes in these tumors (Supplementary Figures 6A-C & 7). The proteome data also suggested a down-regulation of fatty acid metabolism genes in GCDC-high tumors.
To gain an understanding of how bile acids are taken up into breast tumors, we examined the relationship of tumor bile acid content with the following known bile acid transporters: ASBT (SLC10A2), NTCP (SLC10A1), and OATP1B1 & B3 (SLCO1B1 and SLCO1B3). We used the tumor content data for GCDC and DC from the absolute measurements in 20 tumors (Figure 2A) for this analysis. We found that GCDC tumor content significantly correlated with the transcript expression of SLCO1B1 (rho = 0.56, P = 0.02) whereas DC correlated with the expression of both SLCO1B1 (rho = 0.37, P = 0.03) and SLCO1B3 (rho = 0.34, P = 0.04). To further assess the contribution of the organic anion transporter OATP1B1 to bile acid uptake into breast cancer cells, we pre-treated T47D cells, which express SLCO1B1 but not SLCO1B3, with an antagonist of the OATP1B1/3 transport system, rifampicin (24), and then challenged the cells with DC (Supplementary Figure 8). This experiment showed rifampicin attenuates the growth inhibitory effect of DC, indicating the importance of OATP1B1 for bile acid uptake into T47D cells.
Association of GCDC with metabolome profiles of breast tumors denotes activation of the sterol/steroid metabolism in GCDC-high tumors.
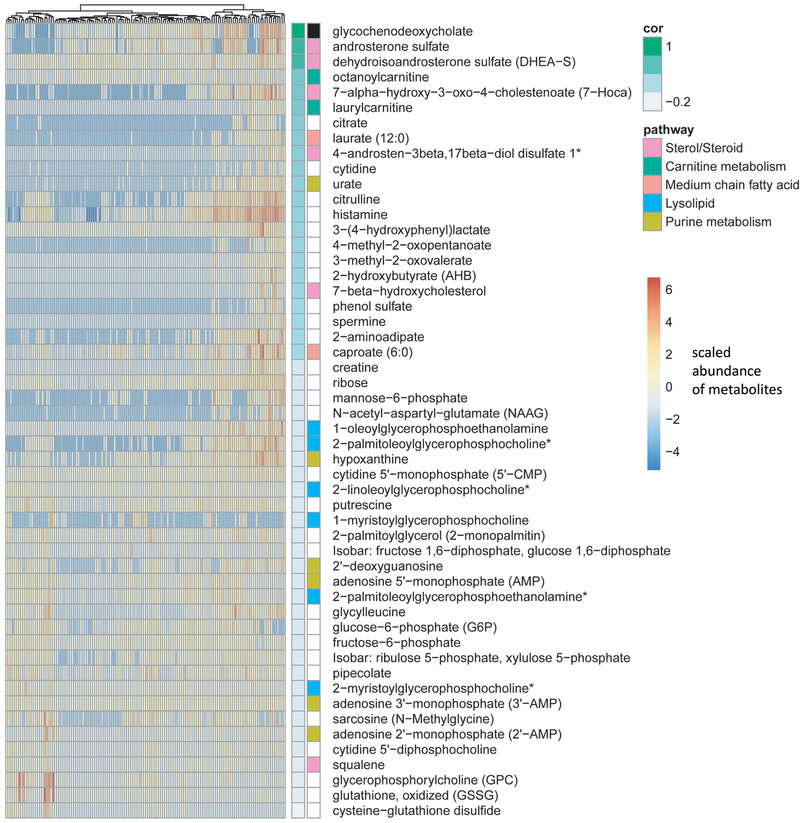
To further gain an understanding of metabolites that are correlated with GCDC tissue levels, we conducted a correlation analysis between GCDC and the other 397 metabolites across all 132 tissue samples in the study. The analysis identified 51 metabolites that significantly correlated with GCDC (Figure 4). Of those, 21 were positively and 30 were negatively correlated with GCDC (all P < 0.05). Further analysis showed that these metabolites were enriched for members in the Sterol/Steroid Metabolism sub-pathway. Enrichment in this pathway positively correlated with GCDC levels (hypergeometric test P =2.1×10−4). Androsterone sulfate and dehydroepiandrosterone sulfate (DHEA-S) were among these metabolites and had the highest correlation of all metabolites with GCDC (r = 0.72 and r = 0.69, respectively; adj. P < 0.001 each, adjusted for multiple comparison analysis). When we restricted the analysis to tumor samples only, this positive correlation with metabolites in the sterol/steroid pathway remained (r = 0.63 each; adj. P < 0.001). We then validated this finding with the analysis of our independent dataset (“Duke cohort”), verifying the positive correlation between tumor GCDC levels and various sterol/steroid pathway metabolites, including androsterone sulfate, DHEA-S, epiandrosterone sulfate, and 4-androsten-3beta,17beta-diol disulfate 1 (r = 0.38 to 0.61, all P < 0.05).
Figure 4. GCDC abundance correlates with metabolites in the Sterol/Steroid pathway.
Shown is the correlation between abundance of GCDC and other metabolites across 132 breast tissues (67 breast tumors and 65 adjacent noncancerous tissues). Fifty-one metabolites were identified as significantly correlated with GCDC using Pearson’s correlation. The Sterol/Steroid pathway is the most significantly enriched metabolic pathway and positively correlated with GCDC levels (hypergeometric test: P =2.1×10−4). Androsterone sulfate and DHEA-S are among the most significantly correlated metabolites, having correlation coefficients of r = 0.72 and 0.69, respectively, with GCDC tissue content.
Bile acids inhibit proliferation of human breast cancer cells.
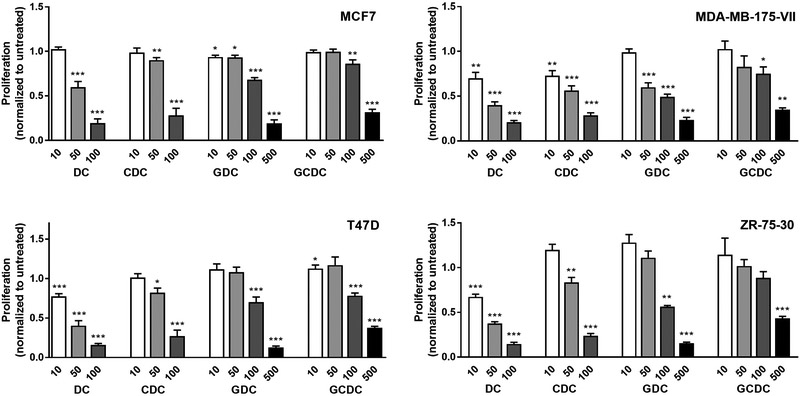
Multiple studies previously examined effects of bile acids on cancer cell growth and reported both pro- and anti-proliferative effects of these bile acids in different cancer cell models including breast cancer (25-29). To re-visit this question and to obtain further clarification whether bile acids are growth-inhibitory in breast cancer cells, four luminal A breast cancer cell lines, MCF7, MDA-MB-175-VII, T47D, and ZR-75–30 (30), were treated with either 10 to 100 μM of DC or CDC, or with 10 to 500 μM of GDC or GCDC, and proliferation was assessed with a BrdU incorporation assay (Figure 5). Although all bile acids had a growth inhibitory effect in the luminal A breast cancer cells, both DC and CDC were more potent in inhibiting cell growth than their glycine conjugates, GDC and GCDC, which is likely explained by the limited uptake of GDC and GCDC into cultured cells. DC showed significant growth inhibitory effects in cell culture for three cell lines, T47D, MDA-MB-175-VII and ZR-75–30, at concentrations that are found in breast tumors (see Figure 2A) while GCDC did not.
Figure 5. Bile acids inhibit proliferation of breast cancer cells.
Proliferation was measured by BrdU incorporation and normalized to untreated cells. Shown are relative ratios (BrdU incorporation of untreated = 1) with mean ± S.E.M. Cells were seeded into 96 well plates and incubated with 10, 50, 100, or 500 μM of bile acid (DC, CDC, GDC and GCDC) for 24 hours. * P < 0.05, ** < 0.01, *** < 0.001, for significantly altered BrdU incorporation. Significance testing was performed with ANOVA and a posthoc t-test.
RNA sequencing shows that DC inhibits cell cycle progression and increases steroid biosynthesis in breast cancer cells.
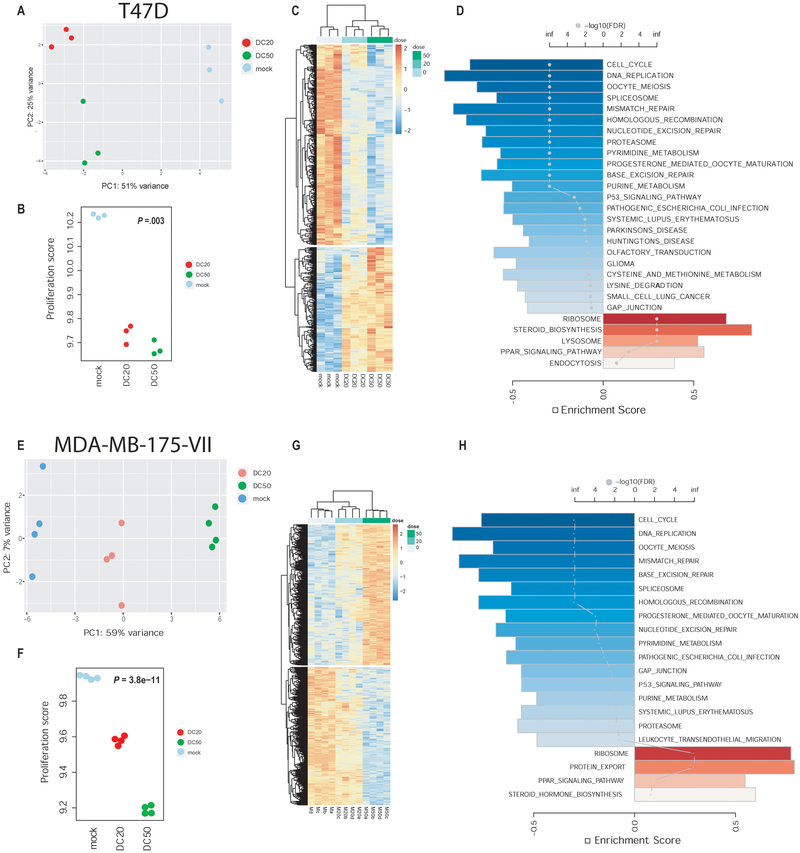
To further characterize the changes that are induced by bile acids in breast cancer cells, T47D and MDA-MB-175-VII cells were treated with 20 and 50 μM DC, which are physiologically relevant concentrations and induce growth arrest in these cells according to our BrdU assay. Following treatment for 24 hours, we performed RNA sequencing to examine the gene expression changes induced by DC. A principle component analysis pointed to significant differences in gene expression between treated and untreated cells (Figure 6A,E), accompanied by significantly decreased cell proliferation scores in treated cells (Figure 6B,F), consistent with our data from human breast tumors. The DC-induced expression changes were dose-dependent (Figure 6C,G) and affected a core set of pathways, as shown by GSEA (Figure 6D,H). Notable, cell cycle- and DNA replication-related genes were down-regulated, and so were many DNA repair-related genes. In contrast, DC treatment led to the induction of steroid biosynthesis genes (Supplementary Figure 9 and Supplementary Table 3), consistent with human breast tumor data that pointed to an increase in sterol/steroid metabolism-related metabolites in GCDC- and DC-high breast tumors. DC also altered expression of genes in the Steroid Hormone Biosynthesis pathway (Supplementary Figure 10 and Supplementary Table 4). We followed up on this observation and examined the expression of key enzymes in the estrogen metabolism pathway and their correlation with tumor GCDC content. Within the group of luminal A tumors that tend to have the highest GCDC content and express estrogen receptor α, we found a positive correlation with GCDC for catechol-O-methyltransferase (COMT; rho = 0.45, P = 0.03), 17β-hydroxysteroid dehydrogenase 2 (HSD17B2; rho = 0.49, P = 0.02), and sulfotransferase 1E1 (SULT1E1; rho = 0.37, P = 0.08), with an average increase in expression of these genes in GCDC-high tumors by 15–30%, based on transcript levels. Notable, all three enzyme activities are predicted to decrease estradiol availability in the tumor.
Figure 6. Pathway alterations in human breast cancer cells treated with deoxycholic acid (DC).
Analysis of RNA sequencing data from T47D (A-D) and MDA-MB-175-VII (E-H) cells. A,E. Principle component analysis of gene expression data from untreated cells (mock), or cells treated with either 20 μM (DC20) or 50 μM DC (DC50). Untreated and treated groups are clearly separated. B,F. DC treatment decreases the cell proliferation score. Regression analysis with P value calculated using the Wald test. C,G. Changes in gene expression are related to the dose of DC. Heatmaps represent the most differentially expressed genes (FDR < 0.05). D,H. Gene set enrichment analysis. A bar plot shows the top-ranked pathways with the directional enrichment score (FDR < 0.05 for inclusion). Blue bars represent an inhibition and red bars an activation with treatment. Color scale corresponds with statistical significance while the dots represent the enrichment scores.
DISCUSSION
Here, we report a distinctive tumor biology and less aggressive disease in breast cancer patients that accumulate bile acids in their tumors. Most notable, these tumors exhibit a low proliferation index based on gene expression data, which is consistent with our cell culture data showing that bile acids tend to inhibit cell cycle progression. Increased tumor proliferation is one of the most important adverse markers for clinical outcome in breast cancer (7), specifically in luminal A tumors (8). Since luminal A tumors may contain the highest bile acid concentrations, as our data for GCDC indicate, GCDC and other bile acids may determine the tumor biology of this subtype more so than of other breast cancer subtypes. The reason of why bile acids may accumulate in these tumors remains to be determined, but our data imply that the process is not related to body mass or obesity.
Bile acids are produced by the liver and are commonly modified by the gut microbiome after their secretion into the bile and the lumen of the small intestine (31). It remains unclear whether they are modified by the microbiome of organs other than the gut. Recent publications indicate the presence of bacteria in breast tumors (32,33). Similarly, bacteria can be found in normal human breast tissue and breast milk (34-36). Thus, it is possible that this microflora has an influence on the composition of bile acids in human breast tissue. Few previous studies investigated the relationships of bile acids with breast disease. It was shown that DC, CDC, lithocholate, and cholate can be detected in breast cyst fluid from women with fibrocystic disease (37). In a follow up study, the same authors showed that deuterated CDC quickly appears in cyst fluid after being ingested by volunteers (38). Thus, there is uptake of bile acids into breast tissue. Others reported slightly elevated plasma bile acid concentrations in postmenopausal women with breast cancer comparing 20 patients with matched controls (39). We observed that the bile acid contents of tumor and adjacent non-cancerous tissues are not significantly different, indicating that bile acids are not selectively taken up into tumors. However, we observed an inverse relationship between GCDC tissue level and the proliferation score only in tumor tissue. The data argue for a distinct effect of bile acids on tumor biology.
Several studies have investigated the effects of bile acids in breast cancer cell lines. The findings are rather heterogenous suggesting pro- and anti-tumor effects. Two studies reported growth stimulatory effects of bile acids at up to 100 μM concentrations in MCF-7 cells (25,40). Others reported that DC promotes survival of MDA-MB-231 and 4T1 breast cancer cells (41,42). In contrast, anti-tumor effects, including inhibition of growth, were observed by different investigators (27-29). One of these studies reported decreased long-term survival of CDC- and DC-treated MCF-7 cells and destabilization of HIF1α (28). In our investigations, DC but not GCDC led to an inhibition of proliferation in human luminal A breast cancer cell lines at concentrations that we observed in breast tumors. Bile acids may directly interfere with the cell cycle and synergize in their effects on tumor biology because GCDC, GDC and DC levels show high correlations with each other in breast tissues, as shown by data in this study. Yet, the inverse relationship between their tumor levels and the tissue proliferation rate may involve other mechanisms, such as the interference of bile acids with the hypoxia response, as postulated by Phelan et al. (28). Additional mechanisms may include an increase to drug sensitivity in presence of bile acids, as was reported recently (43), or an effect on steroid hormone biosynthesis towards a decreased estrogen availability in tumors, as suggested from our gene expression data in GCDC-high luminal A breast tumors. Additionally, bile acids may also accumulate in tumor-associated fibroblasts and adipocytes and interfere with estrogen metabolism in these cells.
Bile acids signal through the farnesoid X receptor (FXR) and G-protein-coupled bile acid receptor 1 (TGR5) (14). Our assessment of available gene expression data for these two receptors in breast tumors and breast cancer cell lines indicated a rather low expression. Nevertheless, treatment of T47D cells with NF449, a TRG5 antagonist, attenuated the antiproliferative effects of DC (Supplementary Figure 8). Other studies have shown FXR protein expression in these cell lines and tumors using Western blot analysis and immunohistochemistry (27,44-46). As a general finding, FXR signaling in these cell lines led to oncosuppressive effects (27,44,45). Consistent with these observations, high FXR expression in human breast tumors was associated with favorable patient survival in one analysis (46). These data further substantiate our findings that the tumor bile acid content associates with a less aggressive disease and improved patient survival in breast cancer. Moreover, the upregulation of genes in the sterol/steroid metabolism in GCDC-high breast tumors is consistent with FXR and TGR5 activation and signaling, although most of our understanding about these receptors comes from studies of the liver and gut (47,48). Lastly, our observation that the Complement & Coagulation Cascade is upregulated in GCDC-high tumors based on proteome data for these tumors could be related to the presence of FXR and TGR5 in immune cells and adipose tissue, or an activation of FXR signaling in the liver (49).
Bile acids interfere with glucose, fatty acid and lipid metabolism through FXR signaling (14). Our proteome data suggest a down-regulation of fatty acid metabolism genes in GCDC-high tumors. In addition, bile acids can inhibit carnitine acetyltransferase activity, as shown previously (50). Carnitines facilitate the transport of long-chain fatty acids into mitochondria for β-oxidation. Interestingly, the three carnitines that were found to be associated with reduced breast cancer survival in our study, butyrylcarnitine, 3-dehydrocarnitine, and deoxycarnitine, all show an inverse association in tissue levels with GCDC (rho = −0.39, P < 0.001; rho = −0.35, P = 0.003; rho = −0.43, P < 0.001, respectively). Thus, the accumulation of bile acids in breast tumors may also lead to a disturbance in carnitine metabolism and inhibition of fatty acid metabolism, which should be investigated in future studies.
In summary, we report the first evidence that liver- and microbiome-derived bile acids accumulate in a subset of human breast tumors and inhibit their growth and improve patient survival. Future research is needed to gain more insight into the bile acid-induced signaling cascade in breast tumors and the involvement of cancer-associated fibroblasts and adipocytes in this biology.
Supplementary Material
TRANSLATIONAL RELEVANCE.
The intrinsic potential for a breast tumor to progress is often difficult to evaluate. Here, we analyzed large-scale metabolome data and their relationship to the transcriptome and proteome in human breast tumors and found that liver- and microbiome-derived bile acids accumulate in a subset of the tumors. These tumors have distinct disease characteristics including a low proliferation score and alterations in the steroid metabolism pathway. Our findings were corroborated in in vitro experiments with bile acid-treated human breast cancer cells. Together, our findings suggest that bile acids may cause a reprogramming in breast cancer biology towards a less aggressive disease and thereby improve breast cancer outcomes.
ACKNOWLEDGEMENTS
We would like to thank personnel at the University of Maryland and the Baltimore Veterans Administration Hospital for their contributions with the recruitment of subjects. The research was supported by the Intramural Research Program of the Center for Cancer Research (ZIA BC 010887 to S.A.); American Cancer Society grant 127430-RSG-15–105-01-CNE and National Institutes of Health grants R01CA220297 and R01CA216426 (all to N.P), and support grants for Metabolomics Shared Resources (P30 CA125123) and CPRIT Proteomics and Metabolomics Core Facility at Dan L. Duncan Cancer Center (RP170005).
Abbreviations:
- CDC
chenodeoxycholate
- DC
deoxycholate
- GDC
glycodeoxycholate
- GCDC
glycochenodeoxycholate
- GSEA
gene set enrichment analysis
- BrdU
bromodeoxyuridine
- LASSO
least absolute shrinkage and selection operator
Footnotes
Conflict of interest: The authors declare no potential conflicts of interest.
REFERENCES
- 1.Perou CM, Sorlie T, Eisen MB, Van De RM, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature 2000;406:747–52 [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001;98:10869–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 2004;10:5367–74 [DOI] [PubMed] [Google Scholar]
- 4.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 2006;295:2492–502 [DOI] [PubMed] [Google Scholar]
- 5.Taneja P, Maglic D, Kai F, Zhu S, Kendig RD, Fry EA, et al. Classical and Novel Prognostic Markers for Breast Cancer and their Clinical Significance. Clin Med Insights Oncol 2010;4:15–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan DS, Vieira AR, Aune D, Bandera EV, Greenwood DC, McTiernan A, et al. Body mass index and survival in women with breast cancer-systematic literature review and meta-analysis of 82 follow-up studies. Ann Oncol 2014;25:1901–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desmedt C, Sotiriou C. Proliferation: the most prominent predictor of clinical outcome in breast cancer. Cell Cycle 2006;5:2198–202 [DOI] [PubMed] [Google Scholar]
- 8.Desmedt C, Haibe-Kains B, Wirapati P, Buyse M, Larsimont D, Bontempi G, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res 2008;14:5158–65 [DOI] [PubMed] [Google Scholar]
- 9.Mishra P, Ambs S. Metabolic Signatures of Human Breast Cancer. Mol Cell Oncol 2015;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budczies J, Brockmoller SF, Muller BM, Barupal DK, Richter-Ehrenstein C, Kleine-Tebbe A, et al. Comparative metabolomics of estrogen receptor positive and estrogen receptor negative breast cancer: alterations in glutamine and beta-alanine metabolism. J Proteomics 2013;94:279–88 [DOI] [PubMed] [Google Scholar]
- 11.Terunuma A, Putluri N, Mishra P, Mathe EA, Dorsey TH, Yi M, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest 2014;124:398–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang X, Lin CC, Spasojevic I, Iversen ES, Chi JT, Marks JR. A joint analysis of metabolomics and genetics of breast cancer. Breast Cancer Res 2014;16:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan LB, Gui DY, Vander Heiden MG. Altered metabolite levels in cancer: implications for tumour biology and cancer therapy. Nat Rev Cancer 2016;16:680–93 [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez FJ, Jiang C, Xie C, Patterson AD. Intestinal Farnesoid X Receptor Signaling Modulates Metabolic Disease. Dig Dis 2017;35:178–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boersma BJ, Howe TM, Goodman JE, Yfantis HG, Lee DH, Chanock SJ, et al. Association of breast cancer outcome with status of p53 and MDM2 SNP309. J Natl Cancer Inst 2006;98:911–9 [DOI] [PubMed] [Google Scholar]
- 16.Prueitt RL, Boersma BJ, Howe TM, Goodman JE, Thomas DD, Ying L, et al. Inflammation and IGF-I activate the Akt pathway in breast cancer. Int J Cancer 2007;120:796–805 [DOI] [PubMed] [Google Scholar]
- 17.Mishra P, Tang W, Putluri V, Dorsey TH, Jin F, Wang F, et al. ADHFE1 is a breast cancer oncogene and induces metabolic reprogramming. J Clin Invest 2018;128:323–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang W, Zhou M, Dorsey TH, Prieto DA, Wang XW, Ruppin E, et al. Integrated proteotranscriptomics of breast cancer reveals globally increased protein-mRNA concordance associated with subtypes and survival. Genome Med 2018;10:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin M, Prat A, Rodriguez-Lescure A, Caballero R, Ebbert MT, Munarriz B, et al. PAM50 proliferation score as a predictor of weekly paclitaxel benefit in breast cancer. Breast Cancer Res Treat 2013;138:457–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goeman JJ. L1 penalized estimation in the Cox proportional hazards model. Biom J 2010;52:70–84 [DOI] [PubMed] [Google Scholar]
- 23.Mayr A, Schmid M. Boosting the concordance index for survival data--a unified framework to derive and evaluate biomarker combinations. PLoS One 2014;9:e84483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Bruyn T, van Westen GJ, Ijzerman AP, Stieger B, de Witte P, Augustijns PF, et al. Structure-based identification of OATP1B1/3 inhibitors. Mol Pharmacol 2013;83:1257–67 [DOI] [PubMed] [Google Scholar]
- 25.Baker PR, Wilton JC, Jones CE, Stenzel DJ, Watson N, Smith GJ. Bile acids influence the growth, oestrogen receptor and oestrogen-regulated proteins of MCF-7 human breast cancer cells. Br J Cancer 1992;65:566–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Payne CM, Crowley-Skillicorn C, Holubec H, Dvorak K, Bernstein C, Moyer MP, et al. Deoxycholate, an endogenous cytotoxin/genotoxin, induces the autophagic stress-survival pathway: implications for colon carcinogenesis. J Toxicol 2009;2009:785907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alasmael N, Mohan R, Meira LB, Swales KE, Plant NJ. Activation of the Farnesoid X-receptor in breast cancer cell lines results in cytotoxicity but not increased migration potential. Cancer Lett 2016;370:250–9 [DOI] [PubMed] [Google Scholar]
- 28.Phelan JP, Reen FJ, Dunphy N, O’Connor R, O’Gara F. Bile acids destabilise HIF-1alpha and promote anti-tumour phenotypes in cancer cell models. BMC Cancer 2016;16:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miko E, Vida A, Kovacs T, Ujlaki G, Trencsenyi G, Marton J, et al. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim Biophys Acta Bioenerg 2018;1859:958–74 [DOI] [PubMed] [Google Scholar]
- 30.Jiang G, Zhang S, Yazdanparast A, Li M, Pawar AV, Liu Y, et al. Comprehensive comparison of molecular portraits between cell lines and tumors in breast cancer. BMC Genomics 2016;17 Suppl 7:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem 2003;72:137–74 [DOI] [PubMed] [Google Scholar]
- 32.Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci Rep 2016;6:30751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Altemus J, Niazi F, Green H, Calhoun BC, Sturgis C, et al. Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget 2017;8:88122–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urbaniak C, Cummins J, Brackstone M, Macklaim JM, Gloor GB, Baban CK, et al. Microbiota of human breast tissue. Appl Environ Microbiol 2014;80:3007–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khalkhali S, Mojgani N. Characterization of Candidate probionts isolated from human breast milk. Cell Mol Biol (Noisy-le-grand) 2017;63:82–8 [DOI] [PubMed] [Google Scholar]
- 36.Fernandez L, Langa S, Martin V, Maldonado A, Jimenez E, Martin R, et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res 2013;69:1–10 [DOI] [PubMed] [Google Scholar]
- 37.Raju U, Levitz M, Javitt NB. Bile acids in human breast cyst fluid: the identification of lithocholic acid. J Clin Endocrinol Metab 1990;70:1030–4 [DOI] [PubMed] [Google Scholar]
- 38.Javitt NB, Budai K, Miller DG, Cahan AC, Raju U, Levitz M. Breast-gut connection: origin of chenodeoxycholic acid in breast cyst fluid. Lancet 1994;343:633–5 [DOI] [PubMed] [Google Scholar]
- 39.Costarelli V, Sanders TA. Plasma deoxycholic acid concentration is elevated in postmenopausal women with newly diagnosed breast cancer. Eur J Clin Nutr 2002;56:925–7 [DOI] [PubMed] [Google Scholar]
- 40.Raju U, Katz J, Levitz M. Effect of bile acids and estradiol on thymidine incorporation into DNA in MCF-7 and MCF-10A breast cell lines. Steroids 1997;62:643–6 [DOI] [PubMed] [Google Scholar]
- 41.Silva J, Dasgupta S, Wang G, Krishnamurthy K, Ritter E, Bieberich E. Lipids isolated from bone induce the migration of human breast cancer cells. J Lipid Res 2006;47:724–33 [DOI] [PubMed] [Google Scholar]
- 42.Krishnamurthy K, Wang G, Rokhfeld D, Bieberich E. Deoxycholate promotes survival of breast cancer cells by reducing the level of pro-apoptotic ceramide. Breast Cancer Res 2008;10:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chewchuk S, Boorman T, Edwardson D, Parissenti AM. Bile Acids Increase Doxorubicin Sensitivity in ABCC1-expressing Tumour Cells. Sci Rep 2018;8:5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giordano C, Catalano S, Panza S, Vizza D, Barone I, Bonofiglio D, et al. Farnesoid X receptor inhibits tamoxifen-resistant MCF-7 breast cancer cell growth through downregulation of HER2 expression. Oncogene 2011;30:4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barone I, Vircillo V, Giordano C, Gelsomino L, Gyorffy B, Tarallo R, et al. Activation of Farnesoid X Receptor impairs the tumor-promoting function of breast cancer-associated fibroblasts. Cancer Lett 2018;437:89–99 [DOI] [PubMed] [Google Scholar]
- 46.Giaginis C, Karandrea D, Alexandrou P, Giannopoulou I, Tsourouflis G, Troungos C, et al. High Farnesoid X Receptor (FXR) expression is a strong and independent prognosticator in invasive breast carcinoma. Neoplasma 2017;64:633–9 [DOI] [PubMed] [Google Scholar]
- 47.Kuipers F, Bloks VW, Groen AK. Beyond intestinal soap--bile acids in metabolic control. Nat Rev Endocrinol 2014;10:488–98 [DOI] [PubMed] [Google Scholar]
- 48.de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab 2013;17:657–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Pircher PC, Schulman IG, Westin SK. Regulation of complement C3 expression by the bile acid receptor FXR. J Biol Chem 2005;280:7427–34 [DOI] [PubMed] [Google Scholar]
- 50.Sekas G, Paul HS. Inhibition of carnitine acetyltransferase by bile acids: implications for carnitine analysis. Anal Biochem 1989;179:262–7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.