Abstract
Background
Both quantitative and qualitative aspects of plasma cell‐free DNA (plasma cfDNA, pcfDNA) have been well‐studied as potential biomarkers in non‐small cell lung cancer (NSCLC). Accumulating evidence has proven that saliva also has the potential for the detection and analysis of circulating free DNA (saliva cfDNA, scfDNA).
Methods
In the current study, we aimed to explore the potential application of scfDNA in NSCLC diagnostics and consistency of epidermal growth factor receptor (EGFR) mutation detection in paired pcfDNA and scfDNA using droplet digital PCR (ddPCR) and analyze the relationship between EGFR mutations and clinical treatment response.
Results
In the quantitative cohort study, scfDNA concentration in NSCLC patients was no different from that in healthy donors, or in benign patients. ScfDNA concentration was significantly lower than pcfDNA concentration, yet they were not statistically significant in relevance (Spearman's rank correlation r = −0.123, P = 0.269). In the qualitative cohort study, the overall concordance rate of EGFR mutations between pcfDNA and scfDNA was 83.78% (31 of 37; k = 0.602; P < 0.001). EGFR mutation detection in paired pcfDNA and scfDNA was significantly correlated with the clinical treatment response (Spearman's rank correlation r = 0.664, P = 0.002).
Conclusions
Our results demonstrated that saliva might not be the idea material for a cfDNA quantitative test, and scfDNA concentration is not applicable for NSCLC diagnostics. Conversely, scfDNA was capable of acting as the supplement for EGFR mutations due to the coincidence rate of EGFR mutation detection between scfDNA and pcfDNA.
Keywords: ddPCR, EGFR mutation detection, NSCLC, scfDNA
Introduction
Non‐small cell lung cancer (NSCLC) comprises 80% of all lung cancer cases and is the leading cause of cancer‐associated mortality worldwide.1 Most patients have local or distant metastasis at the time of diagnosis, whereas earlier tumor detection is associated with excellent survival.2 In recent years, circulating‐free DNA (cfDNA), released by both healthy and cancer cells into the bloodstream during apoptosis or necrosis, or by active secretion,3 has been proven to greatly impact molecular diagnostics of NSCLC patients due to simple, noninvasive access to genetic material detectable in plasma.4
The utility of plasma cell‐free DNA (pcfDNA) is not only to overcome the disadvantage of sampling limitations and heterogeneity in tissue biopsy, but also to reflect the status of genetic variation in real‐time.5 PcfDNA concentration has been shown to act as the biomarker in the diagnosis of NSCLC due to its ability to discriminate healthy subjects and NSCLC patients,6, 7, 8 as well as the predictor for disease progression since it is significantly related with treatment response.9, 10 Nowadays, the use of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) has become the standard therapeutic approach for NSCLC patients harboring sensitizing EGFR mutations, among which L858R mutation and 19 exon deletions (E19‐Dels) together account for approximately 90% of EGFR‐mutant tumors in the clinic.11 However, the majority of patients with EGFR mutations are found to be resistant (primary resistance) or gradually develop resistance (acquired resistance) after EGFR‐TKIs therapy, and T790M mutation is found in approximately 50%–60% of these cases.12 In mid‐2016, pcfDNA was approved by the U.S. Food and Drug Administration (FDA) for the identification of EGFR sensitizing mutations in basal setting (patients naive to any treatment) when tissue was not available or inadequate and in the progression setting for the identification of the EGFR T790M,13 suggesting the mutation detection in pcfDNA appears to be a promising and minimally invasive alternative to tumor biopsy for NSCLC patients.
Accumulating evidence has shown that saliva also has the potential for the detection and analysis of cfDNA. Saliva provides good‐quality genomic DNA which is comparable to blood as a template for genotyping.14, 15, 16 Saliva is produced by acinar cells in the salivary glands, which are highly permeable and surrounded by abundant capillaries, allowing molecules in the blood to exchange freely with those in adjacent salivary cells,17 thereby, most analytes detected in the blood are also found in saliva, indicating saliva should be considered as an ideal even better diagnostic fluid. It has been reported that the combined use of N‐α‐acetyltransferase 10 protein (Naa10p) and carcinoembryonic antigen (CEA) as tumor markers for oral squamous cell carcinoma (OSCC) in saliva were more sensitive than that in serum18; MiR‐21 in saliva was increased in colorectal cancer patients with a sensitivity of 97% and a specificity of 91%.19 Importantly, saliva can also be used for cfDNA (scfDNA) detection and analysis. EGFR mutations can be detected in the saliva of NSCLC patients using a novel core technology, called electric field‐induced release and measurement,20 processing the excellent detection efficiency with AUC (area under curve, after ROC analysis) of 0.96 and 0.94 for L858R and E19‐Dels, respectively. Despite these favorable attributes, the use of saliva as a diagnostic fluid seems to not yet have become a mainstream idea, mainly because the levels of most analytes in saliva which are quite different from those in blood are substantially diminished.21
In the current study, we aimed to explore the potential application of scfDNA at quantitative and qualitative levels in NSCLC. We studied the differences of scfDNA concentration between the case and control groups, then explored the consistency of EGFR mutation detection in paired pcfDNA and scfDNA and analyzed the relationship between EGFR mutations and clinical treatment response, providing novel insights of using saliva as a supplement to fluid biopsy materials.
Methods
Patients and healthy donors
The study included NSCLC patients admitted to the Department of Shandong Cancer Hospital Affiliated to Shandong University from June 2015 to August 2018. For the quantitative cohort study, 78 basal NSCLC patients naive to any anti‐tumor treatment, 15 patients with pulmonary benign disease and 26 healthy donors were recruited. For the qualitative cohort study, 40 NSCLC patients diagnosed with known EGFR mutations clinically using tumor tissue samples and six healthy donors were enrolled. No surgery was performed until collection of paired blood and saliva. All patients and healthy donors gave their informed consent for specimen collection and clinical information collection.
Samples collection and cfDNA extraction
Peripheral blood samples were collected into EDTA tubes and centrifuged at 1900 g for 10 minutes at 4°C to separate the peripheral blood cells. The plasma was then further centrifuged at 16 000 g for 10 minutes at 4°C to pellet any remaining cells and was immediately stored at −80°C until DNA extraction.
Saliva was collected as reported previously.22 Briefly, all subjects were asked to refrain from eating, drinking, or oral hygiene for at least one hour prior to collection. They rinsed their mouths with water and used their tongues against the upper jaws to allow saliva to flow into an aseptic container. Participants were instructed not to cough or strongly expectorate in order to collect unstimulated saliva samples. Saliva was then centrifuged for 20 minutes at 300 × g to remove cells and another 20 minutes at 10 000 × g to remove cellular debris within one hour of collection, and was then stored at −80°C until DNA isolation.
PcfDNA and scfDNA were extracted using QIAamp Circulating Nucleic Acid Kit (Qiagen, Dusseldorf, Germany) according to the manufacturer's protocol. They were eluted into RNase free water, and the eluate reapplied onto the column for re‐elution. The final eluate was collected and stored at −20°C. Samples from paired plasma and saliva were always extracted together to avoid batch effects.
Quantification of cfDNA by qPCR
Quantification of cfDNA was performed by SYBR Green based qPCR as described previously.23 Human‐specific primers were used for detection of human Long Interspersed Nuclear Element 1 (LINE1) retrotransposon, the primer sequences as follows: Forward, 5'‐GAAGTCAGTGTGGCGATTCC‐3′; and Reverse, 5'‐GGTTCCAAGTCTTTGCTATTGTG −3′. Serial dilutions from human leukocyte genomic DNA were used as calibrators for cfDNA quantification. For every independent experiment we made standard curves based on RNase free water to calculate cfDNA concentration of plasma and saliva, respectively.
Cell line, gDNA extraction, droplet digital PCR
Droplet digital PCR (ddPCR) was established and analyzed using genomic DNA (gDNA) derived from the cell lines including human lung cancer‐derived cell lines HCC827 harboring EGFR E19‐Dels mutation, H1975 harboring EGFR T790M and L858R mutations and A549 harboring wild‐type EGFR, which were purchased from American Type Culture Collection (Manassas, VA, USA) and China Center for Type Culture Collection (Wuhan, China). All these cells were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco, Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco, Invitrogen) and antibiotics (penicillin/streptomycin, 100 U/mL) at 37°C in 5% CO2.
Cellular gDNA was extracted using the genomic DNA extraction kit (Tiangen, Beijing, China) according to the manufacturer's protocol, and the gDNA concentration was determined by Nanodrop (TheromoFisher, Waltham, MA, USA) to calculate the amount of the sample.
DdPCR was performed on Bio‐Rad QX200 Droplet Digital PCR (Bio‐Rad Laboratories, Hercules, CA, USA) platform according to the manufacturer's protocol. Analysis of the ddPCR data was performed with QuantaSoft analysis software (version 1.7.4; Bio‐Rad Laboratories, Hercules, CA, USA) which accompanied the droplet reader. Sequences of primers and probes were purchased from Life Technologies (ThermoFisher) and are listed in Table 1.
Table 1.
Sequence information of the primers and probes for the ddPCR assays
| Mutation | Primer/probe ID | Sequence |
|---|---|---|
| E19‐Dels | E19‐F | 5′‐ GTGAGAAAGTTAAAATTCCCGTC ‐ 3′ |
| E19‐R | 5′ – TGGGCCTGAGGTTCAGA ‐ 3′ | |
| E19‐Ref probe | 5′ – FAM ‐ TGAGTTTCTGCTTTGCTGTGT‐MGB ‐ 3′ | |
| E19‐Tar probe | 5′ – VIC ‐ AGGAATTAAGAGAAGCAACAT – MGB ‐ 3′ | |
| T790M | E20‐F | 5′ – GCCTGCTGGGCATCTGC ‐ 3′ |
| E20‐R | 5′ – TCTTTGTGTTCCCGGACATAGTC ‐ 3′ | |
| E20‐MUT probe | 5′ – FAM – TCATCATGCAGCTCAT – MGB ‐ 3′ | |
| E20‐WT probe | 5′ – VIC – TCATCACGCAGCTCAT – MGB ‐ 3' | |
| L858R | E21‐F | 5′ ‐ CCGCAGCATGTCAAGATCAC ‐ 3' |
| E21‐R | 5′ – CCTCCTTCTGCATGGTATTCTTTCT ‐ 3' | |
| E21‐MUT probe | 5′ – FAM – AGTTTGGCCCGCCCAA ‐ MGB ‐ 3' | |
| E21‐WT probe | 5′ – VIC – AGTTTGGCCAGCCCAA – MGB ‐ 3' |
ddPCR, droplet digital PCR; E19‐Dels, exon 19 deletions; F, forward primer; R, reverse primer; MUT, mutant allele; WT, wild‐type allele.
The design principle of the ddPCR assays is shown in Fig 1. Briefly, for the L858R and T790M assays, two probes targeting a mutated region were labeled with FAM and VIC to detect the mutant and wild‐type EGFR allele with one nucleotide difference, respectively11; For E19‐Dels assay, as described previously,24 the Ref probe was designed to target the nonmutated region, and the Tar probe was designed to target the mutation region. WT molecules were double positive (VIC+/FAM+), and MUT molecules has low VIC signal (VIClow/FAM+). E19‐Dels assay was capable of detecting eight kinds of common exon19 deletions, as listed in Table 2.
Figure 1.
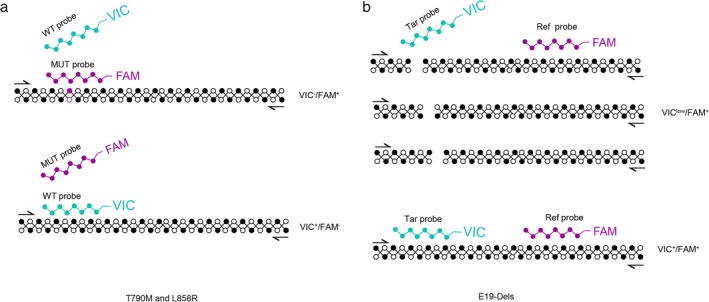
Design of the two assays for detection of EGFR E19‐Dels, T790M and L858R. For T790M and L858R assay (a), FAM− and VIC− labeled probes were designed to target the mutant and wild‐type EGFR alleles, respectively. For E19‐Del assay (b), The Ref probe was designed to target the nonmutated region, and the Tar probe was designed to target the mutation region. The Ref probe and Tar probe were labeled with FAM and VIC to detect the WT (VIC+/FAM+) and MUT (VIClow/FAM+). Our assay used a single probe covering the mutated region to detect all the mutations contained in hotspot regions. E19‐Dels, exon 19 deletions; MUT, mutant allele; WT, wild‐type allele.
Table 2.
Eight kinds of common exon19 deletions detected by E19‐Dels assay
| Mutation | Type | AA mutation | CDS mutation | Genomic coordinates | Mutation ID |
|---|---|---|---|---|---|
| E746_A750del | Deletion‐In frame | p.E746_A750delELREA | c.2235_2249del15 | 7:55174772… 55 174 786 |
COSM6223 |
| E746_A750del | Deletion‐In frame | p.E746_A750delELREA | c.2236_2250del15 | 7:55174773… 55 174 787 |
COSM6225 |
| L747_P753del | Complex‐deletion inframe | p.L747_P753 >S |
c.2240_2257del18 | 7:55174777… 55 174 794 |
COSM12370 |
| L747_T751del | Deletion‐In frame | p.L747_T751 delLREAT |
c.2240_2254del15 | 7:55174777… 55 174 791 |
COSM12369 |
| L747_A750del | Complex‐deletion inframe | p.L747_A750 > P | c.2239_2248TTAAGAGAAG > C | 7:55174775… 55 174 785 |
COSM12422 |
| E746_T751del | Complex‐deletion inframe | p.E746_T751 >A |
c.2237_2251del | 7:55174774… 55 174 788 |
COSM12678 |
| L747_S752del | Deletion‐In frame | p.L747_S752 delLREATS |
c.2239_2256del18 | 7:55174776… 55 174 793 |
COSM6255 |
| L747_T751del | Deletion‐In frame | p.L747_T751 delLREAT |
c.2238_2252del15 | 7:55174775… 55 174 789 |
COSM23571 |
Statistical analysis
Statistical analysis was performed using SPSS 22.0 statistical software (SPSS, Chicago, IL, USA) and GraphPad Prism 6.0 (GraphPad Software, San Diego, CA, USA). For comparisons of non‐normal continuous variables, Wilcoxon test was used for two related samples and the Mann‐Whitney U‐test and the Kruskal‐Wallis H tests for two or more independent samples, respectively. Spearman correlation was used to compare the correlation between two variables. The EGFR status consistency between plasma and saliva was assessed by Kappa test. Data are presented as the median ± interquartile range (IQR) (range, minimum‐maximum). Significance was established at P < 0.05.
Results
Quantitative study cohort
Study population
For the quantitative cohort study, a total of 78 basal NSCLC patients were recruited, three patients without definite diagnosis and seven pulmonary metastases from other cancers were ruled out, thereby, 68 NSCLC patients and 41 nontumor subjects including 15 patients with pulmonary benign disease and 26 healthy donors were subjected for the next research. Demographic information is listed in Table 3: age, gender, behavioral factors, pathological type, disease stage and tumor long diameter. Patients with pulmonary benign disease and healthy subjects matched for age, gender and risk factors were recruited as controls.
Table 3.
Numbers and characteristics of cancer and noncancer donors
| Characteristics | No. (%) of cancer and noncancer persons |
|---|---|
| NSCLC | 68 |
| Male | 44 (64.7) |
| Age, median (min, max) | 59 (26, 77) |
| Behavioral factors | |
| Smoking | 38 (55.9) |
| Non‐smoking | 30 (44.1) |
| Drinking | 33 (48.5) |
| Nondrinking | 35 (51.5) |
| Pathological type | |
| AC | 39 (57.4) |
| SCC | 28 (41.2) |
| Other | 1 (1.5) |
| Disease stage | |
| I | 37 (54.4) |
| II | 6 (8.8) |
| III | 16 (23.5) |
| IV | 4 (5.9) |
| Unknown | 5 (7.4) |
| T category | |
| T1 | 13 (19.1) |
| T2 | 38 (55.9) |
| T3 | 8 (11.8) |
| T4 | 3 (4.4) |
| Unknown | 6 (8.8) |
| Extracapsular spread (for N1–N3) | |
| No | 42 (61.8) |
| Yes | 20 (29.4) |
| Unknown | 6 (8.8) |
| Distant metastasis | |
| M0 | 59 (86.8) |
| M1 | 3 (4.4) |
| Unknown | 6 (8.8) |
| Tumor long diameter (cm) | |
| ≥2.7 | 23 (33.8) |
| <2.7 | 24 (35.3) |
| Unknown | 21 (30.9) |
| Patients with pulmonary benign diseases | 15 |
| Male | 11 (73.3) |
| Age, median (min, max) | 58 (48,66) |
| Pathological type | |
| Pneumonia | 3 (20) |
| Nodule | 1 (6.7) |
| Phthisis | 2 (13.3) |
| Benign tumor | 4 (26.7) |
| Cyst | 1 (6.7) |
| Lung space | 3 (20) |
| Right lower lobe isolation | 1 (6.7) |
| Behavioral factors | |
| Smoking | 7 (46.7) |
| Non‐smoking | 8 (53.3) |
| Drinking | 6 (40) |
| Nondrinking | 9 (60) |
| Healthy donors | 26 |
| Male | 12 (46.2) |
| Age, median (min, max) | 30 (24,62) |
| Behavioral factors | |
| Smoking | 3 (11.5) |
| Non‐smoking | 23 (88.5) |
| Drinking | 3 (11.5) |
| Nondrinking | 23 (88.5) |
AC, adenocarcinoma; NSCLC, non‐small cell lung cancer; SCC, squamous cell carcinoma.
ScfDNA concentration is not applicable for NSCLC diagnostics
First, we determined whether scfDNA concentration was capable of acting as a biomarker for NSCLC diagnostics as outlined in the above mentioned cohort. The median concentration of scfDNA in healthy individuals was 1.11 (range, 0.01–67.63) ng/mL; in patients with pulmonary benign disease 3.26 (range, 0.029–26.30) ng/mL and in NSCLC patients 0.531 (range, 0.018–285.420) ng/mL, respectively. Unexpectedly, as shown in Fig 2a, scfDNA concentration in NSCLC patients was not different from that in healthy donors, or in benign patients. We also analyzed the relationship between scfDNA level and clinicopathological characteristics as listed in Table 4, and concluded that the age of the patient, their smoking or drinking history, pathological type, tumor size and disease stage was irrelevant to the results. Taken together, our data supported our conclusion that scfDNA concentration is not applicable in NSCLC diagnostics.
Figure 2.
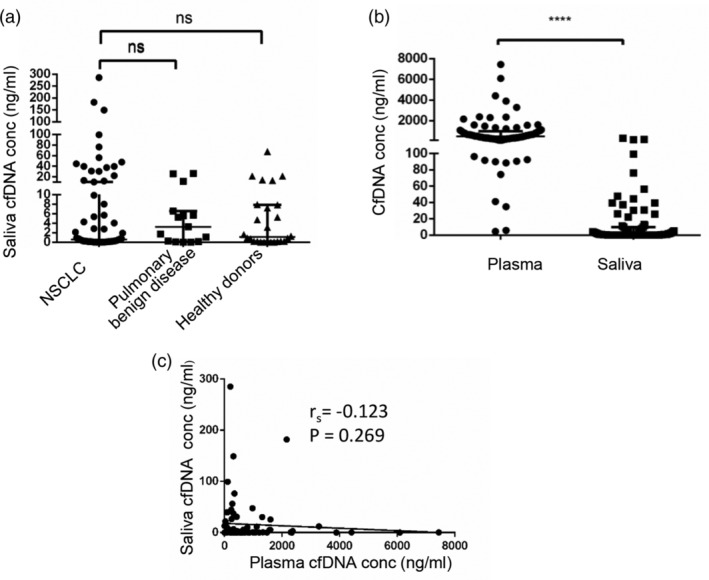
Quantification assays of scfDNA. (a) The differences of scfDNA level among NSCLC patients (n = 68), pulmonary benign disease patients (n = 15) and healthy donors (n = 26). (b) The differences of scfDNA and pcfDNA concentration in patients (n = 55 + 12). (c) The correlation of scfDNA and pcfDNA level in patients (n = 55 + 12). Spearman's correlation coefficient represented the degree of correlation. (Spearman's rank correlation, r = −0.123, P = 0.269). Solid lines represent median values. ****, P < 0.0001; NS, no significance; cfDNA, cell‐free DNA; scfDNA, saliva cfDNA; pcfDNA, plasma cfDNA; NSCLC, non‐small cell lung cancer; conc, concentration.
Table 4.
Relationships between scfDNA concentration and characteristics
| Characteristics | Numbers | P‐value |
|---|---|---|
| Age | 0.7722 | |
| ≤59 | 35 | |
| >59 | 33 | |
| Gender | 0.9518 | |
| Male | 44 | |
| Female | 24 | |
| Smoking history | 0.7427 | |
| Smoker | 38 | |
| Non‐smoker | 30 | |
| Drinking history | 0.7397 | |
| Drinker | 33 | |
| Nondrinker | 35 | |
| T category | 0.101* | |
| T1 | 13 | |
| T2 | 38 | |
| T3 | 8 | |
| T4 | 3 | |
| Nodal status | 0.0699 | |
| N0 | 42 | |
| N1 | 20 | |
| Disease stage | 0.1004 | |
| I | 37 | |
| IIa‐IV | 26 | |
| Pathological type | 0.7403 | |
| AC | 39 | |
| SCC | 28 | |
| Tumor long diameter (cm) | 0.6016 | |
| ≥2.7 | 23 | |
| <2.7 | 24 |
Kruskal‐Wallis H test was used.
AC, adenocarcinoma; SCC, squamous cell carcinoma; scfDNA, saliva cfDNA.
Next, we analyzed the difference and correlation between scfDNA and pcfDNA concentration. As shown in Fig 2b, cfDNA concentration in saliva was much lower than that in plasma. Notably, the correlation between paired scfDNA and pcfDNA concentration was not prominent (Spearman's rank correlation, r = −0.123, P = 0.269; Fig 2c), and further supporting scfDNA concentration was not applicable for NSCLC diagnostics.
Qualitative study cohort
Determination of the specificity and sensitivity of ddPCR assay
The ddPCR method was established as shown in “Methods” and followed the standard in Figure 3. To determine the specificity, the ddPCR assay were performed using related gDNA (cell lines HCC827 harboring EGFR E19‐Dels mutation, H1975 harboring EGFR T790M and L858R mutations and A549 harboring wild‐type EGFR) with different copies from 1000 to 50 000. As shown in Fig 4a, distinct separation between the positive and negative droplets demonstrated the excellent specificity of the established methods even gDNA amount reached 50 000 copies in the 1D Amplitude chart. Moreover, the sensitivity of ddPCR assay was also analyzed through testing serial dilutions of EGFR mutants by mixing DNA derived from the positive cell lines with wild‐type, and their sensitivity was 0.3%, 0.05%, 0.2%, respectively (Fig 4b).
Figure 3.
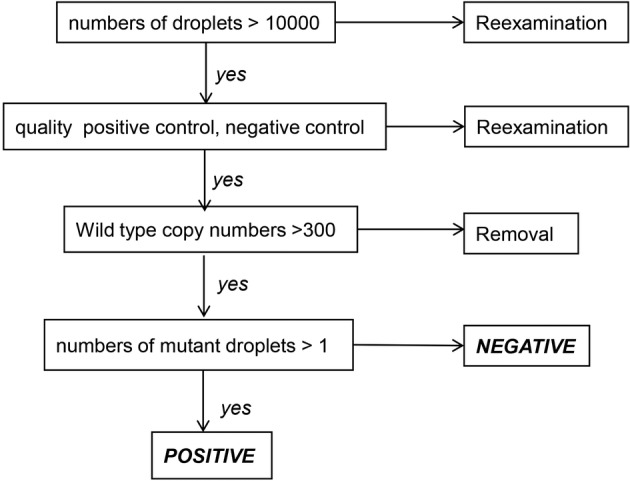
Establishment of EGFR mutations standard detection process. When ddPCR was performing, total numbers of droplets >10000, quality positive control and negative control, and wild type copy numbers >300 were required, otherwise, the samples were re‐examination or removal. It was defined as positive if numbers of mutant droplets >1, if not, it was defined as negative.
Figure 4.
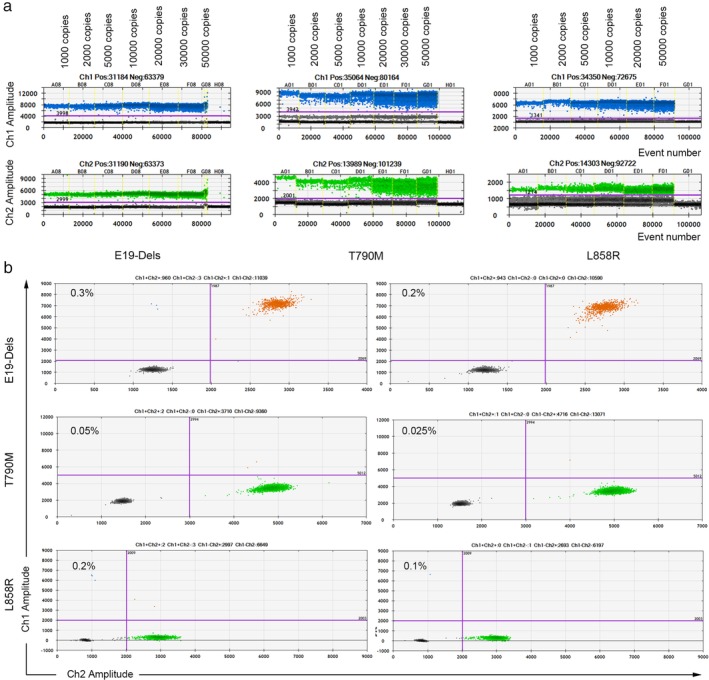
Specificity and sensitivity analysis for E19‐Dels, T790M and L858R methods. (a) For specificity analysis, the established ddPCRs were performed using related gDNA (HCC827 gDNA harboring EGFR E19‐Dels mutation, H1975 gDNA containing EGFR T790M and L858R double mutations) with different copies from 1000 to 50 000. (b) For sensitivity analysis, the established ddPCRs were performed using gradual diluted mutant gDNA with the wild‐type (A549 gDNA harboring wild‐type EGFR).
Study population
For the qualitative cohort study, paired scfDNA and pcfDNA samples were collected from 40 NSCLC patients diagnosed with indicated EGFR mutations clinically, as well as from six healthy donors,and subjected to ddPCR analysis. Representative two‐dimensional maps for EGFR mutations including E19‐Dels, T790M and L858R detection were displayed in Fig 5. At last, 13 paired samples were ruled out due to copy numbers of scfDNA <300, thereby 27 paired samples were enrolled in the research with full information (Table S1).
Figure 5.
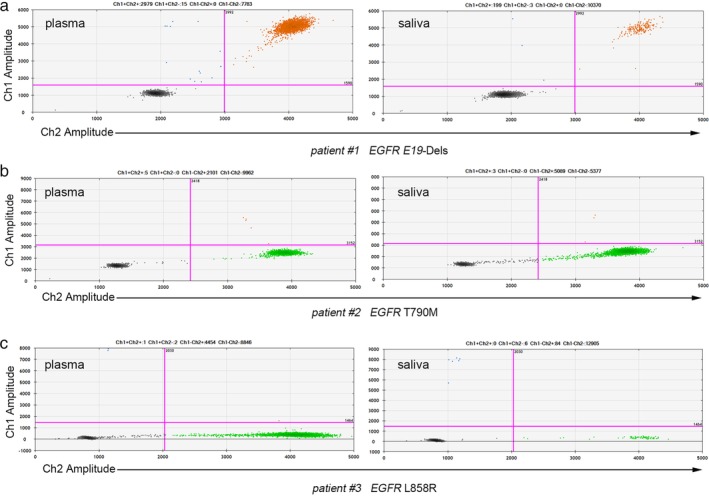
Two‐dimensional maps of EGFR mutation detection using ddPCR. DdPCR was performed via Bio‐Rad QX200TM Droplet Digital PCR system. Representative two‐dimensional maps for EGFR mutations including (a) E19‐Dels, (b) T790M and (c) L858R detection were displayed.
Consistency of EGFR mutation detection using scfDNA and pcfDNA
Next, we analyzed consistency of EGFR mutation detection in the above mentioned cohort using paired scfDNA and pcfDNA. As shown in Table 5, for the E19‐Dels ddPCR assay, 12 patients were detected, among which four patients were positive and five patients were negative in both plasma and saliva, whereas three patients were positive in plasma but undetectable in saliva. For the L858R ddPCR assay, 14 patients were detected, among which two patients were positive and nine patients were negative in both plasma and saliva, whereas three patients were positive in plasma, but undetectable in saliva. For the T790M ddPCR assay, only one patient was positive in the paired plasma and saliva samples. Healthy donors provided 10 paired blood and saliva samples, three of which were tested for L858R and T790M, respectively and four for E19‐Dels; the results were negative and coincident in paired plasma and saliva.
Table 5.
Consistency of EGFR mutation detection in paired plasma and saliva
| Saliva | |||
|---|---|---|---|
| + | − | Total | |
| NSCLC patients | |||
| Plasma | |||
| E19‐Dels | |||
| + | 4 | 3 | 7 |
| ‐ | 0 | 5 | 5 |
| Total | 4 | 8 | 12 |
| L858R | |||
| + | 2 | 3 | 5 |
| − | 0 | 9 | 9 |
| Total | 2 | 12 | 14 |
| T790M | |||
| + | 1 | 0 | 1 |
| − | 0 | 0 | 0 |
| Total | 1 | 0 | 1 |
| Healthy donors | |||
| Plasma | |||
| E19‐Dels | |||
| + | 0 | 0 | 0 |
| ‐ | 0 | 4 | 4 |
| Total | 0 | 4 | 4 |
| L858R | |||
| + | 0 | 0 | 0 |
| − | 0 | 3 | 3 |
| Total | 0 | 3 | 3 |
| T790M | |||
| + | 0 | 0 | 0 |
| − | 0 | 3 | 3 |
| Total | 0 | 3 | 3 |
Collectively, seven paired NSCLC samples were positive, 24 paired samples including 14 NSCLC and 10 healthy were negative for EGFR mutations (E19‐Dels, T790M, L858R) in the 37 qualified paired plasma and saliva samples. However, six samples positive in pcfDNA, were not detected with EGFR mutations in the paired scfDNA. Collectively, the overall concordance rate between pcfDNA and scfDNA was 83.78% (31 of 37; k = 0.602; P < 0.001) (Table 6). Our data suggested saliva‐based qualitative testing was applicable for EGFR mutation detection in NSCLC, indicating saliva as a supplement to blood‐ and tissue‐based biopsy.
Table 6.
Kappa analysis for consistency of EGFR mutation detection in paired plasma and saliva samples
| Saliva | ||||||
|---|---|---|---|---|---|---|
| + | − | Total | P‐value | Kappa value | ||
| Plasma | + | 7 | 6 | 13 | <0.001 | 0.602 |
| − | 0 | 24 | 24 | |||
| Total | 7 | 30 | 37 | |||
Application of EGFR mutation detection in pcfDNA and scfDNA for clinical treatment response assessment
Finally, we analyzed the relationship between EGFR mutation status detected in pcfDNA and scfDNA and clinical treatment response in the 27 NSCLC patients whose tumors harbored common EGFR mutations which were confirmed by tissue biopsy. They underwent a series of clinical anticancer treatments, and 19 had clear efficacy evaluation (Table S1) according to RECIST guidelines.25 For eight patients with stable disease (SD), two were positive (DP) and five were negative (DN) for EGFR mutations in blood and saliva, whereas one was positive in plasma but undetectable in saliva (SP). For seven patients with progressive disease (PD), two were DP and five were SP. Four patients with partial response (PR) were all DN. In total, EGFR mutation detection in paired pcfDNA and scfDNA was significantly correlated with the clinical treatment response (Spearman's rank correlation, r = 0.664, P = 0.002; Fig S1).
Discussion
In the current study, we explored the potential application of scfDNA in NSCLC diagnostics and consistency of EGFR mutation detection in paired plasma and saliva samples using ddPCR. The results demonstrated that saliva cfDNA is applicable for EGFR mutation detection but not for quantitation analysis in NSCLC.
We studied the relationship between scfDNA concentration and clinicopathological features of NSCLC patients, in which no significant differences were detected including age, gender, and pathological type, coincident with the studies on pcfDNA.26, 27 However, although previous studies had reported pcfDNA level acted as the biomarker in the diagnosis of NSCLC due to its ability to discriminate healthy subjects and NSCLC patients,6, 7, 8 our data demonstrated that scfDNA concentration was not applicable for NSCLC diagnostics. In fact, scfDNA originated from pcfDNA which originated from tumor tissue,28, 29 resulting in its low concentration, thus preventing scfDNA diagnostics in clinical practice.30 In addition, saliva viscosity might alter within a person and between individuals.31 Since saliva secretion is affected by various uncontrollable factors such as emotional and mental influences, even some participants are often unwilling or unable to actively participate in saliva expectoration,32 causing a large variation in scfDNA concentration, even in the same individual at different collection times. Another challenge is that saliva collecting, processing and testing methods are needed to standardize to eliminate the scientific validations.32, 33 In the current study, 68 NSCLC patients, 15 pulmonary benign disease patients and 26 healthy donors were enrolled, a tendency that scfDNA was elevated in NSCLC patient was observed, despite being of no statistical significance, thereby, we also would not deny the applicability of scfDNA for NSCLC diagnostics in an expanded cohort.
Owing to the development of hypersensitive techniques, the low concentration of analytes in saliva is no longer a limit.30 As an emerging platform, ddPCR distributed PCR reaction into discrete droplets, enabling the accurate detection and quantification of molecular targets, single molecule analysis by this manner is accurate, cost‐effective, and readily performed.34, 35 In our qualitative study, we performed ddPCR to detect and compare EGFR mutations in scfDNA and pcfDNA. The overall concordance rate between them was 83.78% (31 of 37; k = 0.602; P < 0.001) (Table 6), suggesting scfDNA would become a supplement for EGFR mutations beside plasma and tissue. Nevertheless, six patients whose EGFR mutations were positive in the pcfDNA, were not detected with EGFR mutations in the paired scfDNA. This might be attributed to the low scfDNA concentration and low mutations frequency as discussed above, since higher DNA input amounts could achieve associated increase in sensitivity and higher detection rate.36 Besides, 14 patients were still negative in the paired plasma and saliva samples, although 27 patients were previously diagnosed with EGFR mutations. This was because patients underwent TKIs targeted, radiation or other related treatments after pathological diagnosis and before collection of plasma and saliva samples, which reduced EGFR mutation frequency in NSCLC patients.37, 38
More importantly, our data revealed a potential correlation of EGFR mutation detection to response to clinical treatment, but conclusion was limited due to the small sample (only 19 patients involved with clear efficacy evaluation) and single arm study design. However, our data demonstrated a clear value to predict therapeutic effect of EGFR mutation detection in paired pcfDNA and scfDNA in NSCLC. Patients with EGFR mutations positive both in paired pcfDNA and scfDNA had worse clinical treatment response, which strengthened the potential clinical implication in monitoring treatment effect.
Nevertheless, although scfDNA concentration appears not to be applicable for NSCLC diagnostics because of its extremely low content, salivary circulating free miRNAs have recently become an emerging field for diagnosing or monitoring cancer.39 Due to its short length and resistance to RNases degradation, salivary miRNA could be exchanged more freely between plasma and saliva, thereby composing over 50% of total salivary RNA. It has been reported salivary miRNAs acted as biomarkers for oral cancer head and neck squamous cell carcinomas, esophageal cancer and even gastric cancer,40, 41, 42, 43 implying its potential role in the early diagnosis for NSCLC patients.
In conclusion, saliva might not be the ideal material for a cfDNA quantitative test, and scfDNA concentration not applicable for NSCLC diagnostics. However, scfDNA was capable of acting as the supplement for EGFR mutations beside plasma and tissue due to the coincidence rate of EGFR mutation detection between scfDNA and pcfDNA.
Disclosure
No authors report any conflict of interest.
Supporting information
Figure S1 The correlation between clinical response and EGFR mutations detection in paired pcfDNA and scfDNA. Three‐wire table (a) and Bar plot (b) illustrated EGFR mutations detection results in patients with different clinical response. DP represented patients were positive for EGFR mutations in both plasma and saliva; DN showed that patients were negative for EGFR mutations in both plasma and saliva; SP indicated that patients were positive in plasma but not in saliva. Spearman's correlation coefficient represented the degree of correlation (Spearman's rank correlation r = 0.664, P = 0.002). SD, stable disease; PD, progressive disease; PR, partial response; DP, double positive; DN, double negative; SP, single positive.
Table S1 Summary of patient demographics and mutations detection results by ddPCR.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81773237, 81672104), the Shandong Provincial Key Research and Development Program (2017GSF18183 and 2017CXGC1207), the Shandong Provincial Natural Science Foundation (ZR201808150069 and ZR2017BH074).
References
- 1. Nie K, Jia Y, Zhang X. Cell‐free circulating tumor DNA in plasma/serum of non‐small cell lung cancer. Tumour Biol 2015; 36 (1): 7–19. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Siegel R, Ward E et al Cancer statistics, 2006. CA Cancer J Clin 2006; 56 (2): 106–30. [DOI] [PubMed] [Google Scholar]
- 3. Tanic M, Beck S. Cell‐free DNA: Treasure trove for cancer medicine. Nat Mater 2017; 16 (11): 1056–7. [DOI] [PubMed] [Google Scholar]
- 4. Szpechcinski A, Chorostowska‐Wynimko J, Struniawski R et al Cell‐free DNA levels in plasma of patients with non‐small‐cell lung cancer and inflammatory lung disease. Br J Cancer 2015; 113 (3): 476–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zeng Q, Xie L, Zhou N, Liu M, Song X. Detection of PIK3CA mutations in plasma DNA of colorectal cancer patients by an ultra‐sensitive PNA‐mediated PCR. Mol Diagn Ther 2017; 21 (4): 443–51. [DOI] [PubMed] [Google Scholar]
- 6. Catarino R, Coelho A, Araujo A et al Circulating DNA: Diagnostic tool and predictive marker for overall survival of NSCLC patients. PLOS One 2012; 7 (6): e38559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ulivi P, Mercatali L, Casoni GL et al Multiple marker detection in peripheral blood for NSCLC diagnosis. PLoS One 2013; 8 (2): e57401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Szpechcinski A, Chorostowska‐Wynimko J, Kupis W et al Quantitative analysis of free‐circulating DNA in plasma of patients with resectable NSCLC. Expert Opin Biol Ther 2012; 12 (Suppl 1): S3–9. [DOI] [PubMed] [Google Scholar]
- 9. Sozzi G, Conte D, Mariani L et al Analysis of circulating tumor DNA in plasma at diagnosis and during follow‐up of lung cancer patients. Cancer Res 2001; 61 (12): 4675–8. [PubMed] [Google Scholar]
- 10. Lee YJ, Yoon KA, Han JY et al Circulating cell‐free DNA in plasma of never smokers with advanced lung adenocarcinoma receiving gefitinib or standard chemotherapy as first‐line therapy. Clin Cancer Res 2011; 17 (15): 5179–87. [DOI] [PubMed] [Google Scholar]
- 11. Zhu G, Ye X, Dong Z e a. Highly sensitive droplet digital PCR method for detection of EGFR‐activating mutations in plasma cell‐free DNA from patients with advanced non‐small cell lung cancer. J Mol Diagn 2015; 17 (3): 265–72. [DOI] [PubMed] [Google Scholar]
- 12. Stewart EL, Tan SZ, Liu G, Tsao MS. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations‐a review. Transl Lung Cancer Res 2015; 4 (1): 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Song X, Xie L, Wang X et al Temozolomide‐perillyl alcohol conjugate induced reactive oxygen species accumulation contributes to its cytotoxicity against non‐small cell lung cancer. Sci Rep 2016; 6: 22762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bahlo M, Stankovich J, Danoy P et al Saliva‐derived DNA performs well in large‐scale, high‐density single‐nucleotide polymorphism microarray studies. Cancer Epidemiol Biomarkers Prev 2010; 19 (3): 794–8. [DOI] [PubMed] [Google Scholar]
- 15. Hu Y, Ehli EA, Nelson K et al Genotyping performance between saliva and blood‐derived genomic DNAs on the DMET array: A comparison. PLOS One 2012; 7 (3): e33968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ponti G, Manfredini M, Tomasi A. Non‐blood sources of cell‐free DNA for cancer molecular profiling in clinical pathology and oncology. Crit Rev Oncol Hematol 2019; 141: 36–42. [DOI] [PubMed] [Google Scholar]
- 17. Yoshizawa JM, Schafer CA, Schafer JJ, Farrell JJ, Paster BJ, Wong DT. Salivary biomarkers: Toward future clinical and diagnostic utilities. Clin Microbiol Rev 2013; 26 (4): 781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zheng J, Sun L, Yuan W et al Clinical value of Naa10p and CEA levels in saliva and serum for diagnosis of oral squamous cell carcinoma. J Oral Pathol Med 2018; 47 (9): 830–5. [DOI] [PubMed] [Google Scholar]
- 19. Sazanov AA, Kiselyova EV, Zakharenko AA, Romanov MN, Zaraysky MI. Plasma and saliva miR‐21 expression in colorectal cancer patients. J Appl Genet 2017; 58 (2): 231–7. [DOI] [PubMed] [Google Scholar]
- 20. Wei F, Lin CC, Joon A et al Noninvasive saliva‐based EGFR gene mutation detection in patients with lung cancer. Am J Respir Crit Care Med 2014; 190 (10): 1117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller SM. Saliva testing‐‐a nontraditional diagnostic tool. Clin Lab Sci 1994; 7 (1): 39–44. [PubMed] [Google Scholar]
- 22. Gai C, Camussi F, Broccoletti R et al Salivary extracellular vesicle‐associated miRNAs as potential biomarkers in oral squamous cell carcinoma. BMC Cancer 2018; 18 (1): 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wei L, Xie L, Wang X et al Circulating tumor DNA measurement provides reliable mutation detection in mice with human lung cancer xenografts. Lab Invest 2018; 98 (7): 935–46. [DOI] [PubMed] [Google Scholar]
- 24. Decraene C, Silveira AB, Bidard F‐C et al Multiple hotspot mutations scanning by single droplet digital PCR. Clin Chem 2018; 64 (2): 317–28. [DOI] [PubMed] [Google Scholar]
- 25. Watanabe H, Okada M, Kaji Y et al New response evaluation criteria in solid tumours‐revised RECIST guideline (version 1.1). Gan to Kagaku Ryoho 2009; 36 (13): 2495–501 (In Japanese.). [PubMed] [Google Scholar]
- 26. Hyun MH, Sung JS, Kang EJ et al Quantification of circulating cell‐free DNA to predict patient survival in non‐small‐cell lung cancer. Oncotarget 2017; 8 (55): 94417–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sirera R, Bremnes RM, Cabrera A et al Circulating DNA is a useful prognostic factor in patients with advanced non‐small cell lung cancer. J Thorac Oncol 2011; 6 (2): 286–90. [DOI] [PubMed] [Google Scholar]
- 28. Pu D, Liang H, Wei F et al Evaluation of a novel saliva‐based epidermal growth factor receptor mutation detection for lung cancer: A pilot study. Thorac Cancer 2016; 7 (4): 428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Madhavan D, Wallwiener M, Bents K et al Plasma DNA integrity as a biomarker for primary and metastatic breast cancer and potential marker for early diagnosis. Breast Cancer Res Treat 2014; 146 (1): 163–74. [DOI] [PubMed] [Google Scholar]
- 30. Lee YH, Wong DT. Saliva: An emerging biofluid for early detection of diseases. Am J Dent 2009; 22 (4): 241–8. [PMC free article] [PubMed] [Google Scholar]
- 31. Aro K, Wei F, Wong DT, Tu M. Saliva liquid biopsy for point‐of‐care applications. Front Public Health 2017; 5: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Donzella B, Talge NM, Smith TL, Gunnar MR. To spear or not to spear: Comparison of saliva collection methods. Dev Psychobiol 2008; 50 (7): 714–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matthews AM, Kaur H, Dodd M et al Saliva collection methods for DNA biomarker analysis in oral cancer patients. Br J Oral Maxillofac Surg 2013; 51 (5): 394–8. [DOI] [PubMed] [Google Scholar]
- 34. Goh SK, Wong BKL, Muralidharan V, Christophi C, Do H, Dobrovic A. Adapting an established clinical chemistry quality control measure for droplet generation performance in digital PCR. Clin Chem 2018; 64 (8): 1255–7. [DOI] [PubMed] [Google Scholar]
- 35. Miotke L, Lau BT, Rumma RT, Ji HP. High sensitivity detection and quantitation of DNA copy number and single nucleotide variants with single color droplet digital PCR. Anal Chem 2014; 86 (5): 2618–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pietrasz D, Pecuchet N, Garlan F et al Plasma circulating tumor DNA in pancreatic cancer patients is a prognostic marker. Clin Cancer Res 2017; 23 (1): 116–23. [DOI] [PubMed] [Google Scholar]
- 37. Bai H, Wang Z, Chen K et al Influence of chemotherapy on EGFR mutation status among patients with non‐small‐cell lung cancer. J Clin Oncol 2012; 30 (25): 3077–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dowler Nygaard A, Spindler KL, Pallisgaard N, Andersen RF, Jakobsen A. Levels of cell‐free DNA and plasma KRAS during treatment of advanced NSCLC. Oncol Rep 2014; 31 (2): 969–74. [DOI] [PubMed] [Google Scholar]
- 39. Rapado‐Gonzalez O, Majem B, Muinelo‐Romay L et al Human salivary microRNAs in cancer. J Cancer 2018; 9 (4): 638–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Park NJ, Zhou H, Elashoff D et al Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res 2009; 15 (17): 5473–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wan Y, Vagenas D, Salazar C et al Salivary miRNA panel to detect HPV‐positive and HPV‐negative head and neck cancer patients. Oncotarget 2017; 8 (59): 99990–100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xie ZJ, Chen G, Zhang XC, Li DF, Huang J, Li ZJ. Saliva supernatant miR‐21: A novel potential biomarker for esophageal cancer detection. Asian Pac J Cancer Prev 2012; 13 (12): 6145–9. [DOI] [PubMed] [Google Scholar]
- 43. Li F, Yoshizawa JM, Kim KM et al Discovery and validation of salivary extracellular RNA biomarkers for noninvasive detection of gastric cancer. Clin Chem 2018; 64 (10): 1513–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The correlation between clinical response and EGFR mutations detection in paired pcfDNA and scfDNA. Three‐wire table (a) and Bar plot (b) illustrated EGFR mutations detection results in patients with different clinical response. DP represented patients were positive for EGFR mutations in both plasma and saliva; DN showed that patients were negative for EGFR mutations in both plasma and saliva; SP indicated that patients were positive in plasma but not in saliva. Spearman's correlation coefficient represented the degree of correlation (Spearman's rank correlation r = 0.664, P = 0.002). SD, stable disease; PD, progressive disease; PR, partial response; DP, double positive; DN, double negative; SP, single positive.
Table S1 Summary of patient demographics and mutations detection results by ddPCR.


