Abstract
Background
Tumor mutation burden (TMB) is an important determinant and biomarker for response of targeted therapy and prognosis in patients with lung cancer. The present study aimed to determine whether radiomics signature could non‐invasively predict the TMB status and driver mutations in patients with resectable early stage lung adenocarcinoma (LUAD).
Methods
A total of 61pulmonary nodules (PNs) from 51 patients post‐operatively diagnosed LUAD were enrolled for analysis. Two datasets were divided according to two‐thirds of patients from different commercial Comprehensive Genomic Profiling (CGP) panels: a training cohort including 41 PNs and a testing cohort including rest 20PNs. We sequenced all tumor specimens and paired blood cells using next generation sequencing (NGS), so as to detect TMB status and somatic mutations. We collected 718 quantitative 3D radiomics features extracted from segmented volumes of each PNs and 78 clinical and pathological features retrieved from medical records as well. Support vector machine methods were performed to establish the predictive model.
Results
We established an efficient fusion‐positive tumor prediction model that predicts TMB status and EGFR/TP53 mutations of early stage LUAD. The radiomics signature yielded a median AUC value of 0.606, 0.604, and 0.586 respectively. Combining radiomics with the clinical information can further improve the prediction performance, which the median AUC values are 0.671 for TMB, 0.697 and 0.656 for EGFR/TP53 respectively.
Conclusion
It is feasible and effective to facilitate TMB and somatic driver mutations prediction by using the radiomics signature and NGS data in early stage LUAD.
Keywords: Early stage lung cancer, next‐generation sequencing, radiomics, somatic mutations, tumor mutation burden
Introduction
Lung cancer is a common and prevalent malignant cancer worldwide, and it continues to be the main cause of cancer‐related death.1, 2 As a major subtype of non‐small cell lung cancer (NSCLC), the morbidity of lung adenocarcinomas (LUAD) has dramatically increased during the past decade, especially in China. Recent data showed that early stage LUAD increased from 6.25% in 1999 to 30.54% in 2012.3 More and more malignant pulmonary nodules (PNs) characterized as ground‐glass opacities (GGOs) have been detected by computed tomography (CT) scan screening.4, 5
Currently, we have stepped into the new era of next‐generation sequencing (NGS), which enables clinicians to conveniently harbor abundant somatic variations, as well as intergenic tumor mutation burden (TMB).6 TMB has gained widespread attention since it is an important biomarker predicting the response to PD‐1 blockade immunotherapy7, 8 and EGFR tyrosine kinase inhibitors (TKIs) therapy in NSCLC.9 For early stage lung cancer, previous studies demonstrate that high TMB predicts a better prognosis for resectable NSCLC.10 Under the guidance of TMB, a recent randomized clinical trial demonstrated neoadjuvant PD‐1 blockade immunotherapy was effective for early stage NSCLC.11 In addition, some driver mutations, such as TP53, EGFR, and KRAS, have previously shown specifically emerged in the malignant adenocarcinoma in situ (AIS) and minimally invasive adenocarcinoma (MIA) stages, but not the premalignant atypical adenomatous hyperplasia (AAH) stage.12 Based on the above findings, noninvasively harvesting the genomic information of lung lesions may facilitate clinicians to not only identify benign and malignant tumors,13 but also to formulate a better therapeutic plan.14
High‐dimensional radiomics based on highly dimensional features extracted from radiological images have shown promise for predicting diagnosis, prognosis, and optimal therapy in lung cancer.15 A recent study utilized a novel computer‐aided diagnosis (CAD) approach to predict the diagnosis of small PNs, and the positively predictive value from radiologists’ 0.70 to radiomics’ 0.86 was successfully improved.16 The radiomics signature could also be an independent biomarker estimating the disease‐free survival in patients with early‐stage NSCLC.17 Coroller et al. recently demonstrated that a combination of radiomics features of primary tumor and lymph nodes effectively predicts the pathological response to neoadjuvant chemotherapy in NSCLC patients.18 With respect to the driver mutations, such as EGFR and TP53 of NSCLC, radiomics also showed potential predictive values, which generated a new concept of “Radiogenomics.”15 Several groups have disclosed the association of CT radiomics features and somatic mutations.19, 20
So far, the majority of previous radiogenomics studies investigated were based on the single site mutation detection techniques, such as sanger sequencing or the amplification refractory mutation system, etc. Researchers are now focusing on the high throughput somatic variants data following the development of NGS.21 However, it is unknown whether the radiomics features can predict the TMB status, as well as somatic mutations based on NGS platform in NSCLC. Therefore, the aim of our study was to develop a radiomics signature to predict the TMB status and some driver mutations in patients with surgical‐resected early stage lung cancer.
Methods
Study population
A total of 51 patients with 61 PNs were enrolled for analysis. This research was performed according to the International Ethical Guidelines for Biomedical Research Involving Human Subjects (CIOMS). Written informed consent was obtained from all patients. This study was approved by the Ethics Committee of the Nanjing Medical University Affiliated Cancer Hospital and was performed in accordance with the provisions of the Ethics Committee of Nanjing Medical University Affiliated Cancer Hospital. All patients had pathologically‐confirmed lung adenocarcinoma. NGS tests and preoperative thin‐section CT images were available from November 2016 to July 2018.
Clinical data collected for analysis included age at diagnosis, gender, smoking status, histologic subtypes, pathologic stages based on the eighth edition of AJCC TNM staging system,22 and laboratory examinations. Smoking status was categorized into two groups; never smokers and smokers which included former or current smokers (detailed data available in Table S1).
Image acquisition
All patients underwent contrast‐enhanced chest CT using Discovery CT750 HD scanner (GE Medical Systems, Milwaukee, WI, USA). The entire thorax was scanned with the patient in a supine position and with suspended full inspiration. Technical parameters for CT were as follows: tube voltage, 120 kVp; tube current, 150–200 mA; pitch, 0.969; section thickness and reconstruction interval, 1.25 mm; reconstruction kernel, standard. CT scan was performed after 25 seconds delay following intravenous injection of 85 mL Iopromide (Uitravist‐300; Bayer Schering Pharma, Berlin Germany) at a rate of 3 mL/second for enhancement. All CT images were retrieved from a picture archiving and communication system (PACS; CAREstream Medical Ltd.) for image segmentation and analysis. Only images reconstructed in the transverse section were used in this study.
Tumor segmentation and radiomics feature extraction
As shown in Figure 1a, CT images were imported into the 3D‐Slicer 4.7.0 software and the tumors were then contoured manually by three independent observers using the built‐in paint tool. The delineation was performed in lung window setting (mean, −500 HU; width, 1 500 HU) on the transverse CT plane. Consensus was reached by discussion if the interobserver variability was apparent. Next, feature extraction was performed using a Radiomics plugin for 3D‐Slicer.23 For normalization, all CT voxels were resampled to 1 mm3 using a cubic interpolation. The intensities in the original image were discretized using a bin width of 25 Hounsfield units in order to increase sensitivity relative to the original image, reduce image noise and normalize the intensities across all the patients. Next, feature extraction was performed using a Radiomics plugin for 3D‐Slicer.
Figure 1.
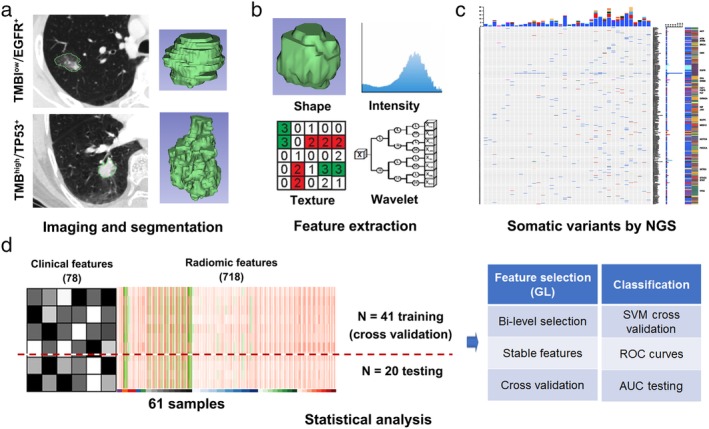
Radiogenomic data acquisition and analysis workflow. (a) CT imaging and tumor segmentation by 3DSlicer of two examples of lung adenocarcinomas with tumor mutation burden (TMB) and EGFR/TP53 mutation status. (b) Radiomics feature extraction and quantification of the tumor phenotype, including shape, intensity, texture, and wavelet features. (c) Schematic diagram of somatic variants by next generation sequencing (NGS). (d) Statistical analysis process of model construction and radiomics features selection.
A set of 718 radiomics features were extracted and categorized as follows (Fig 1b, 2a): (i) Shape features; (ii) Intensity features (first order); (iii) Texture features and (iv) Wavelet features. Group II and III features were also extracted after a wavelet transform of the CT images. In brief, “Coiflet 1” wavelet was applied on the original CT images, a same mother wavelet function was used as described in previous studies.24 Most features defined in this package are in compliance with feature definitions as described by the Imaging Biomarker Standardization Initiative (IBSI), which are available in a separate document by Zwanenburg et al.25
Figure 2.
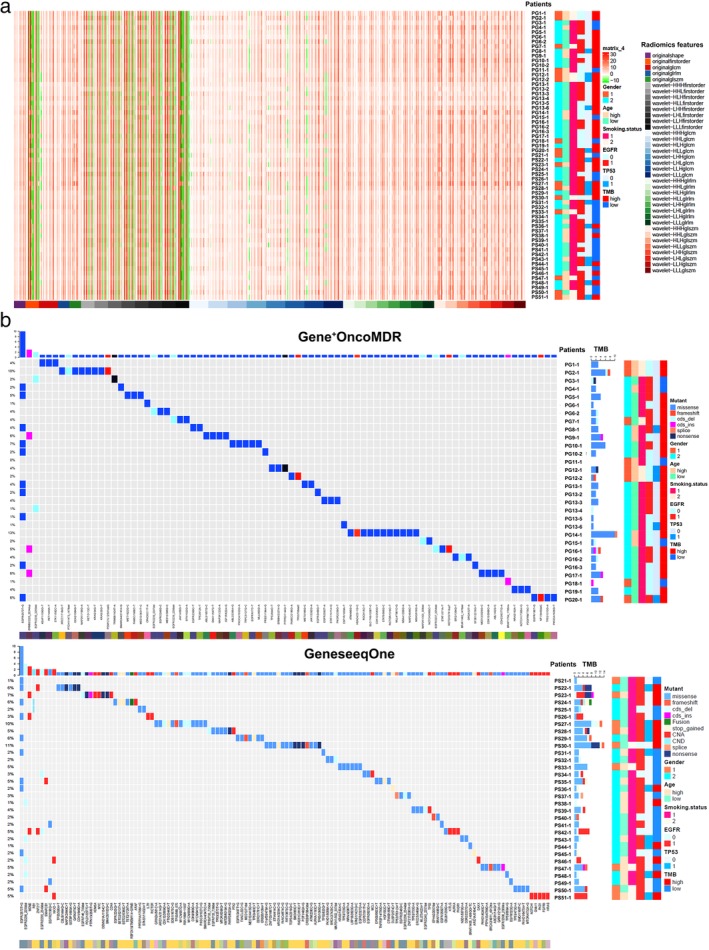
Radiomics and Somatic variants heatmap (Oncoprint) of 61 malignant pulmonary nodules. (a) Radiomics features and patients’ clinicopathological data with mutation status heatmap. (b) Oncoprint heatmap of somatic variants by next generation sequencing. Cohort 1: samples detected on Gene+OncoMDR platform. Cohort 2: samples detected on GeneseeqOne platform. Gender 1: male, Gender 2: female; Age high: ≥ 60 years, Age low: <60 years; Smoking status 1: Never smokers, Smoking status 2: Smokers; EGFR 0: EGFR wide‐type, EGFR 1: EGFR mutant; TP53 0: TP53 wide‐type, TP53 1: TP53 mutant; TMB: tumor mutation burden.
Targeted NGS and data processing
A NGS approach was performed on genomic DNA isolated from formalin‐fixed paraffin‐embedded (FFPE) surgically resected tumor samples (Fig 1c). Two commercial targeted pan‐cancer NGS panels were conducted by Gene+OncoMDR (1021 cancer related genes) and GeneseeqOne (416 cancer‐relevant genes) respectively, both of which were performed on the Hiseq NGS platforms (Illumina Inc., San Diego, CA, USA). TMB is defined as the rate of peptide‐changing single nucleotide variations (SNVs) per Mb. To estimate TMB of all tumors, SNVs with a mutation allelic fraction (MAF) of at least 0.1 after standard filtering and with high or moderate putative impact were retained. In a recent study, comprehensive analysis of more than 100 000 cases pan‐cancer genomes revealed that the median TMB in all cancers is 3.6.26 We therefore defined TMB > 4 as relatively high (TMBhigh) and TMB ≤ 4 as low (TMBlow).
Statistical analysis
We aimed to investigate the prediction performance of the radiomics features, clinical features and their combinations to the mutational status, including EGFR, TP53, and TMB. We collected 78 clinical features potentially related to gene mutation, and used the first 11 PCs which explains 70.3% of total variance, to reduce their dimension. From the 3D‐Slicer, we obtained 718 radiomics features within nine different groups. As previously mentioned, 41 individuals which were the randomly selected two‐thirds of patients from Gene+OncoMDR cohort and GeneseeqOne cohort were chosen as the training data set, and the rest of the 20 individuals were from the testing data set (Fig 1d). The purpose of the training data was to construct the model wrapping the feature selection and classification. Because the radiomics data consisted of nine different groups within a large correlation in each group, the group lasso was used to feature selection process. Considering the group correlation and different effect of individual covariates in the same group, the bilevel selection was used. The coefficient of different variables unveils their importance, which is the one with large absolute values of high importance. Then, a support vector machine (SVM) with Gaussian kernel function was tailored to high dimensional data. We performed cross validation to train the gamma of kernel function and regularization term in the Lagrange formulation of SVM, including best variable selection. In order to ensure the stability of cross validation results, five‐, four‐ and three‐fold cross validation were all performed 20 times for each, respectively. Finally, the performance of the model was evaluated by the testing data. The ability to predict the mutational status of the radiomics features was assessed by the area under the curve (AUC) of the receiver operator characteristic (ROC).
Statistical analysis was implemented by R program (Version 3.5.2). The group lasso used the grpreg package (Version 3.1‐3) and the bilevel selection was accomplished by “gel.” The SVM was implemented by the e1072 package (Version 1.6‐8) and the gamma and regularization term both range from 2e−7 to 2e7. The AUC and its testing were accomplished by pROC package (Version 1.11.0).
Results
Clinical characteristics and radiomics features
The demographic and clinical data of patients are presented in Tables 1 and S1. The Gene+OncoMDR group consisted of seven males and 13 females (mean age, 59.25 years; age range, 45–76 years), while the GeneseeqOne group comprised nine males and 22 females (mean age, 57.65 years; age range, 33–75 years). Five patients were diagnosed with multiple PNs, and 46 patients with solitary PNs. Within the multiple cases, three cases had two PNs, one (PG16) had three PNs and the other one (PG13) had six PNs. All PNs were less than 3 cm in diameter. More than 80% patients (35/51) were diagnosed as stage IA–IB NSCLC, except one case (PG19) who had premalignant atypical adenomatous hyperplasia (AAH). The other six patients (11.76%, 6/31) were classified as stage IIB (T1a‐cN1M0) because of the occasional metastasis of lymph nodes during operation. The heatmap of 718 radiomics features of 61 PNs were shown in Fig 2a. No statistical significance could be detected from the clinical information, gene mutation status, and radiomics characteristics when comparing the training to the testing set (Table 1).
Table 1.
The patients clinicopathological information and genomic mutation status
| All | Cohort 1 | Cohort 2 | EGFR+ | EGFR− | P‐value | TP53+ | TP53 − | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | (n = 51) | (n = 20) | (n = 31) | P‐value | (n = 39) | (n = 22) | (n = 15) | (n = 46) | p‐ value | ||
| Age | <60 | 28 | 11(54.05%) | 17(45.95%) | 0.3445 | 23(62.16%) | 14(37.84%) | 0.7204 | 7(18.92%) | 30(81.08%) | 0.2015 |
| ≥60 | 23 | 9(41.67%) | 14(58.33%) | 16(66.67%) | 8(33.33%) | 8(33.33%) | 16(66.67%) | ||||
| Gender | Male | 16 | 7(47.06%) | 9(52.94%) | 0.8368 | 10(58.82%) | 7(41.18%) | 0.6054 | 6(35.29%) | 11(64.71%) | 0.2275 |
| Female | 35 | 13(50.00%) | 22(50.00%) | 29(65.91%) | 15(34.09%) | 9(20.45%) | 35(79.55%) | ||||
| Smoking status | Smokers | 9 | 4(50.00%) | 5(50.00%) | 0.9548 | 5(50.00%) | 5(50.00%) | 0.3156 | 3(30.00%) | 7(70.00%) | 0.6639 |
| Never smokers | 42 | 16(49.02%) | 26(50.98%) | 34(66.67%) | 17(33.33%) | 12(23.53%) | 39(76.47%) | ||||
| Pathologic types | AAH | 1 | 1(100.00%) | 0(0.00%) | 0.2048 | 0(0.00%) | 1(100.00%) | 0.0651 | 0(0.00%) | 1(100.00%) | 0.5282 |
| AIS | 1 | 1(100.00%) | 0(0.00%) | 0(0.00%) | 1(100.00%) | 0(0.00%) | 1(100.00%) | ||||
| MIA | 10 | 7(70.00%) | 3(30.00%) | 4(40.00%) | 6(60.00%) | 1(10.00%) | 9(90.00%) | ||||
| IAD | 49 | 21(42.86%) | 28(57.14%) | 35(71.43%) | 14(28.57%) | 14(28.57%) | 35(71.43%) | ||||
| Stage | 0 | 2 | 2(100.00%) | 0(0.00%) | 0.347 | 0(0.00%) | 2(100.00%) | 0.357 | 0(0.00%) | 2(100.00%) | 0.1531 |
| Ia1 | 21 | 8(58.06%) | 13(41.94%) | 19(61.29%) | 12(38.71%) | 4(12.90%) | 27(87.10%) | ||||
| Ia2 | 14 | 5(35.71%) | 9(64.29%) | 11(78.57%) | 3(21.43%) | 7(50.00%) | 7(50.00%) | ||||
| Ia3 | 4 | 2(50.00%) | 2(50.00%) | 2(50.00%) | 2(50.00%) | 1(25.00%) | 3(75.00%) | ||||
| Ib | 4 | 1(25.00%) | 3(75.00%) | 3(75.00%) | 1(25.00%) | 1(25.00%) | 3(75.00%) | ||||
| IIb | 6 | 2(33.33%) | 4(66.67%) | 4(66.67%) | 2(33.33%) | 2(33.33%) | 4(66.67%) | ||||
AAH, atypical adenomatous hyperplasia; AIS, adenocarcinoma in situ; cohort 1, Gene+OncoMD panel; cohort 2, GeneseeqOne panel; IAD, invasive adenocarcinoma; MIA, minimally invasive adenocarcinoma.
Characteristics of somatic variants by NGS
The Oncoprint heatmaps of somatic mutations and copy number variation of two data sets are shown in Fig 2b. The gender, age, and smoke history are annotated in the right edge of the Oncoprint, suggesting that there was no relationship between the specific gene mutations and clinical features. All samples contained at least one detectable variant, except patient PG11. With regard to the mutation in EGFR/TP53, detected PNs for gene alterations were: 39 (63.93%) samples out of the full cohort of 61 PNs were identified as EGFR mutant and 22 as EGFR wild‐type; 15 (24.59%) samples out of the full cohort of 61 PNs were identified as TP53 mutant and 46 as TP53 wild‐type. Twelve samples were identified as both EGFR mutant and TP53 mutant. Finally, when defined TMB > 4 as high, there were 38 samples (62.3%) showed high TMB status; however, the other 23 samples (37.7%) were low.
Predictive radiomics signature of EGFR/TP53 mutation and TMB status
Since the frequency of most SNVs are lower than 10% except for EGFR and TP53 due to our limited samples, we therefore chose only EGFR and TP53, as well as TMB for further radiogenomic association analysis and predictive radiomics signature modeling. A total of eight radiomics features showed prognostic ability in the prediction of TMB status. For the prediction of TP53 mutant, six radiomics features demonstrated the significant correlation between feature values and mutation status. Besides, a total of 13 radiomics features have been proposed to identify EGFR mutant status (Fig 3).
Figure 3.
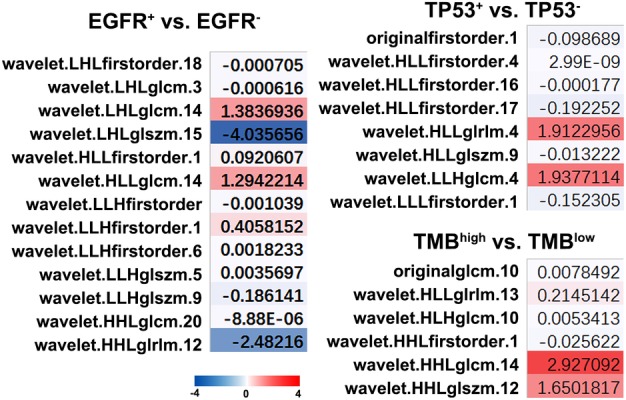
Radiomics signature to predict TMB status and EGFR/TP53 mutations. Somatic genotype‐imaging phenotype associations by comparing radiomics feature distributions between mutation subtypes. Heatmap shows the normalized mean difference of radiomics features feature distributions. TMB, tumor mutation burden.
In the ROC analysis for training cohort, when combining the radiomics and clinical signatures, we achieved an improved accuracy to predict the TMB status and EGFR/TP53 mutation when compared to clinical or radiomic features only (Fig 4a). Using five‐fold cross validation, the median AUC values of rad_clin group were 0.775, 0.764, and 0.842 for the prediction of TMB status and EGFR/TP53 mutations, respectively. Further, in the ROC analysis for testing cohort, we finally assessed the AUC combined the radiomics and clinical signatures, and the results demonstrated an improved accuracy (AUC = 0.671 for TMB, 0.697 for EGFR mutation, and 0.656 for TP53 mutation, respectively, Fig 4b). We achieved smilar median AUC values in training and testing cohorts under the methods of both four‐ and three‐folds‐ crossing validation (Fig S1) which ensured the stability of our cross validation results. Together, the above data showed an enhancement in combined signatures compared to respective signatures.
Figure 4.
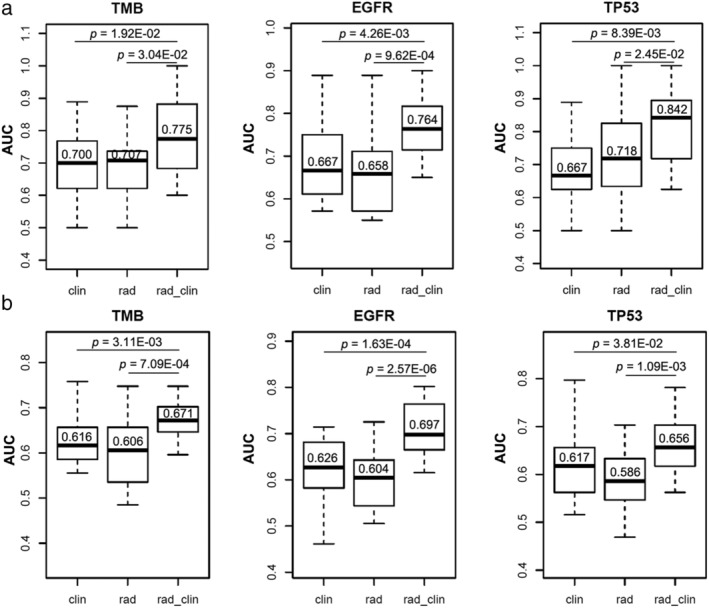
Predictive accuracy of radiomics signatures on TMB status and EGFR/TP53 mutations under the method of five‐fold cross validation in training (a) and testing (b) cohorts respectively. The median values of average area under the curve (AUC) were achieved for clinical factors alone (clin), radiomics features alone (rad), and a model that combined clinical factors and radiomics features (rad_clin), respectively.
It is worth noting that when comparing the patients with the same pathologic types, we found significant differences in either genetic mutations or TMB status. However, with the performance of our model, we could decode tumor phenotypes for TMB status and EGFR/TP53 mutations using a radiomics approach (Fig 5).
Figure 5.
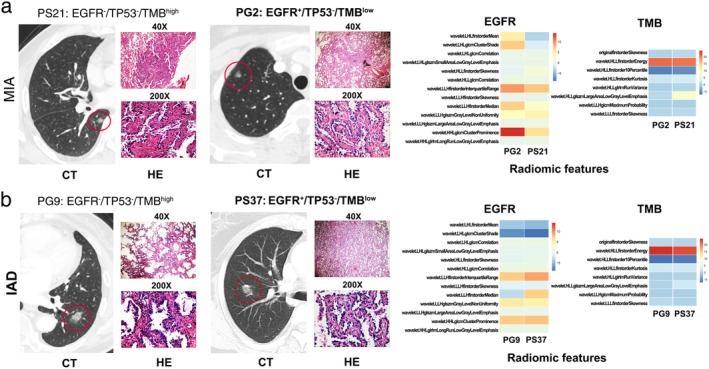
The radiomics features could predict different mutation and TMB status in the patients with same histological subtype (HE) and naked‐eye CT imaging. The representative CT images, pathological sections, and radiomics features of two patients (PG2 and PS21) with minimally invasive adenocarcinoma (MIA) were shown in (a) and another two patients (PG9 and PS37) with invasive adenocarcinoma (IAD) shown in (b).
Discussion
In the era of precision medicine, physicians are undoubtedly seeking to fully understand the phenotypic, pathologic, as well as the genomic characteristics of lung cancer as much as possible so a better treatment plan can be offered.26, 27 For early stage lung cancer, due to the difficulty of tissue biopsy, there is still a lack of sensitive biomarkers to predict TMB and oncogenic driver mutations before surgery. With the development of liquid biopsy techniques, such as circulating tumor DNA (ctDNA) or circulating tumor cells (CTCs), it is possible to obtain the genomic mutation status from a blood sample; however both the sensitivity and cost are prohibitive28, 29 Our study demonstrated that selected radiomics signatures using machine learning technique are feasible and effective to predict TMB and some driver mutations (EGFR and TP53) in patients with early stage LUAD.
A common shortcoming of radiomics studies is the inherent variability in CT acquisition and reconstruction parameters. In order to overcome this problem, the imaging data collected in our study were confined within a relatively short period of one year. On the other hand, the standard of care for both CT acquisition and routine clinical reconstruction was followed for most of the enrolled patients. These aspects can reduce the effect of heterogeneity in CT protocols. Further optimization and standardization of imaging data is still required for the introduction of imaging‐based biomarkers. Additionally, the results are provided by a single center, which makes it hard to assess the generalizability of outcomes to other institutions. Hence, future work needs to be carried out in independent validation sets from other institutions to evaluate the translational aspect of our models that generalized across institutions.
Recent studies have attempted to explore the relationship between mutations and radiomics features due to the development of targeted therapy in lung cancer. However, these studies are still relatively rare and their results inconsistent. Most focus on the predictive value of radiomics on EGFR mutation. One pilot study reported by Aerts et al. showed that radiomic‐feature Laws‐Energy derived from CT images could predict EGFR‐mutation status (AUC = 0.67, P = 0.03) reflected by the therapeutic responses of EGFR TKIs.30 A radiogenomic study predicting lung cancer somatic mutation in a real sense has also been recently reported and the CT imaging data from a total of 763 patients at four medical centers collected. The predictive accuracy combining radiomics feature with clinical phenotype has reached to 0.75 for EGFR mutation and 0.86 for KRAS mutation, respectively. Besides the CT images, Yip et al. also engaged metabolic imaging such as 18F‐FDG PET radiomics to predict mutation status of lung cancer. They enrolled 348 lung cancer patients and successfully predicted EGFR mutation status with an AUC = 0.67; however, the study failed to predict KRAS mutation.31 In our study, limited by a smaller sample size, we only tested the predictive effects for EGFR (39/61, 63.93%) and TP53 (15/61, 24.59%) due to lower frequency of other mutations. However, we have harbored relatively acceptable accuracy (0.697 for EGFR and 0.653 for TP53) based on only 61 PNs. We therefore have reason to believe that we will be able to gain higher accuracy if larger cohorts are collected in the future.
Another important aspect of our study is that we successfully harbored TMB prediction in early stage LUAD (AUC = 0.671), which has not been previously reported. We selected four as the cutoff value of TMB due to the recent pan‐cancer median TMB (3.6)26; an accurate cutoff value of TMB for NSCLC is still under investigation. Our data displayed the feasibility of TMB prediction using our approach and we may update our predictive signature once the cutoff value of TMB for NSCLC has been confirmed. In addition, a standard TMB value should be derived from the Whole Exon Sequencing (WES) of tumor tissues; however, our data were calculated from CGP panels. Latest data have confirmed that CGP assay was highly correlated with WES (r2 = 0.98),26 which also confirmed this opinion due to the consistent results derived from two different commercial CGP panels of our training and testing cohort. Last but not least, TMB is much more valuable in advanced NSCLC patients receiving PD‐1/PDL‐1 blockade immunotherapy,32 and this radiogenomic study which aims to predict TMB should be further investigated in patients with advanced NSCLC.
Finally, with respect to the statistical analysis, feature selection and classification methods should tail to different datasets. In our study, bilevel group lasso is possibly consistent with the characteristics of the data for the following two reasons. First, the features of radiomics have an intra‐group correlation. Second, the bilevel strategy analyzes the information of both groups and individuals at the same time, which facilitates us to further screen features already selected by group lasso. However, considering the sample size and the stability, cross validation is used to identify important feature in the screening and classification process.
In summary, this study demonstrates that it is feasible and effective to predict TMB and driver mutations by using the radiomics features and CGP sequencing data on NGS platform in early stage lung cancer. Prospective studies with other hypermutations are required to further validate larger and independent cohorts in the future.
Disclosure
The authors declare that there are no conflicts of interest.
Supporting information
Figure S1 Predictive accuracy of radiomics signatures on TMB status and EGFR/TP53 mutations under the method of four folds (training cohort in A, testing cohort in B) and three folds (training cohort in C, testing cohort in D) cross validation. The median values of average area under the curve (AUC) were achieved for clinical factors alone (clin), radiomics features alone (rad), and a model that combined clinical factors and radiomics features (rad_clin), respectively.
Table S1 Clinicopathological characteristics of patients.
Acknowledgments
None.
Contributor Information
Xiaoxiao Wang, Email: wxx1201@hotmail.com.
Rong Yin, Email: rong_yin@njmu.edu.cn.
Feng Chen, Email: fengchen@njmu.edu.cn.
References
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- 2. Zeng H, Chen W, Zheng R et al Changing cancer survival in China during 2003‐15: A pooled analysis of 17 population‐based cancer registries. Lancet Glob Health 2018; 6: e555–555e567. [DOI] [PubMed] [Google Scholar]
- 3. Zhang L, Li M, Wu N, Chen Y. Time trends in epidemiologic characteristics and imaging features of lung adenocarcinoma: A population study of 21,113 cases in China. PLOS One 2015; 10: e0136727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McWilliams A, Tammemagi MC, Mayo JR et al Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med 2013; 369: 910–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sun F, Xi J, Zhan C et al Ground glass opacities: Imaging, pathology and gene mutations. J Thorac Cardiovasc Surg 2018; 156: 808–13. [DOI] [PubMed] [Google Scholar]
- 6. Rizvi H, Sanchez‐Vega F, La K et al Molecular determinants of response to anti‐programmed cell death (PD)‐1 and anti‐programmed death‐ligand 1 (PD‐L1) blockade in patients with non‐small‐cell lung cancer profiled with targeted next‐generation sequencing. J Clin Oncol 2018; 36: 633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. High TMB. Predicts immunotherapy benefit. Cancer Discov 2018; 8: 668. [DOI] [PubMed] [Google Scholar]
- 8. Goodman AM, Piccioni D, Kato S et al Prevalence of PDL1 amplification and preliminary response to immune checkpoint blockade in solid tumors. JAMA Oncol 2018; 4: 1237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Offin M, Rizvi H, Tenet M et al Tumor mutation burden and efficacy of EGFR‐tyrosine kinase inhibitors in patients with EGFR‐mutant lung cancers. Clin Cancer Res 2018; 25: 1063–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Devarakonda S, Rotolo F, Tsao MS et al Tumor mutation burden as a biomarker in resected non‐small‐cell lung cancer. J Clin Oncol 2018; 36: 2995–3006.JCO2018781963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Forde PM, Chaft JE, Smith KN et al Neoadjuvant PD‐1 blockade in resectable lung cancer. N Engl J Med 2018; 378: 1976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Izumchenko E, Chang X, Brait M et al Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA. Nat Commun 2015; 6: 8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye M, Li S, Huang W et al Comprehensive targeted super‐deep next generation sequencing enhances differential diagnosis of solitary pulmonary nodules. J Thorac Dis 2018; 10: S820–820S829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hawkins S, Wang H, Liu Y et al Predicting malignant nodules from screening CT scans. J Thorac Oncol 2016; 11: 2120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thawani R, McLane M, Beig N et al Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer 2018; 115: 34–41. [DOI] [PubMed] [Google Scholar]
- 16. Huang P, Park S, Yan R et al Added value of computer‐aided CT image features for early lung cancer diagnosis with small pulmonary nodules: A matched case‐control study. Radiology 2018; 286: 286–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang Y, Liu Z, He L et al Radiomics signature: A potential biomarker for the prediction of disease‐free survival in early‐stage (I or II) non‐small cell lung cancer. Radiology 2016; 281 (3): 947–57. [DOI] [PubMed] [Google Scholar]
- 18. Coroller TP, Agrawal V, Huynh E et al Radiomic‐based pathological response prediction from primary tumors and lymph nodes in NSCLC. J Thorac Oncol 2017; 12: 467–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamamoto S, Korn RL, Oklu R et al ALK molecular phenotype in non‐small cell lung cancer: CT radiogenomic characterization. Radiology 2014; 272: 568–76. [DOI] [PubMed] [Google Scholar]
- 20. Rizzo S, Petrella F, Buscarino V et al CT radiogenomic characterization of EGFR, K‐RAS, and ALK mutations in non‐small cell lung cancer. Eur Radiol 2016; 26: 32–42. [DOI] [PubMed] [Google Scholar]
- 21. Incoronato M, Aiello M, Infante T et al Radiogenomic analysis of oncological data: A technical survey. Int J Mol Sci 2017; 18: 805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nicholson AG, Chansky K, Crowley J et al The International Association for the Study of Lung Cancer Lung Cancer Staging Project: Proposals for the revision of the clinical and pathologic staging of small cell lung cancer in the forthcoming eighth edition of the TNM classification for lung cancer. J Thorac Oncol 2016; 11: 300–11. [DOI] [PubMed] [Google Scholar]
- 23. van Griethuysen JJM, Fedorov A, Parmar C et al Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017; 77: e104–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aerts HJ, Velazquez ER, Leijenaar RT et al Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014; 5: 4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zwanenburg A, Leger S, Vallières M, Löck S. Initiative for the IBS. Image Biomarker Standardisation Initiative 2016. [Cited 16 May 2019]. Available from URL: http://arxiv.org/abs/1612.07003.
- 26. Chalmers ZR, Connelly CF, Fabrizio De a. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017; 9: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu CK, Kao SJ, Lai HC. Targeted therapy and immunotherapy lead to rapid regression of advanced non‐small cell lung cancer with multiple driver mutations. J Thorac Oncol 2018; 13: e103–103e105. [DOI] [PubMed] [Google Scholar]
- 28. Abbosh C, Birkbak NJ, Wilson GA et al Phylogenetic ctDNA analysis depicts early‐stage lung cancer evolution. Nature 2017; 545: 446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murlidhar V, Reddy RM, Fouladdel S et al Poor prognosis indicated by venous circulating tumor cell clusters in early‐stage lung cancers. Cancer Res 2017; 77: 5194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aerts HJ, Grossmann P, Tan Y et al Defining a radiomic response phenotype: A pilot study using targeted therapy in NSCLC. Sci Rep 2016; 6: 33860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yip SS, Kim J, Coroller TP et al Associations between somatic mutations and metabolic imaging phenotypes in non‐small cell lung cancer. J Nucl Med 2017; 58: 569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hellmann MD, Nathanson T, Rizvi H et al Genomic features of response to combination immunotherapy in patients with advanced non‐small‐cell lung cancer. Cancer Cell 2018; 33: 843–52.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Predictive accuracy of radiomics signatures on TMB status and EGFR/TP53 mutations under the method of four folds (training cohort in A, testing cohort in B) and three folds (training cohort in C, testing cohort in D) cross validation. The median values of average area under the curve (AUC) were achieved for clinical factors alone (clin), radiomics features alone (rad), and a model that combined clinical factors and radiomics features (rad_clin), respectively.
Table S1 Clinicopathological characteristics of patients.


