Abstract
Stryphnodendron adstringens is a medicinal plant belonging to the Leguminosae family, and it is commonly found in the southeastern savannas, endemic to the Cerrado biome. The goal of this study was to assemble and annotate the chloroplast genome of S. adstringens and to compare it with previously known genomes of the mimosoid clade within Leguminosae. The chloroplast genome was reconstructed using de novo and referenced-based assembly of paired-end reads generated by shotgun sequencing of total genomic DNA. The size of the S. adstringens chloroplast genome was 162,169 bp. This genome included a large single-copy (LSC) region of 91,045 bp, a small single-copy (SSC) region of 19,014 bp and a pair of inverted repeats (IRa and IRb) of 26,055 bp each. The S. adstringens chloroplast genome contains a total of 111 functional genes, including 77 protein-coding genes, 30 transfer RNA genes, and 4 ribosomal RNA genes. A total of 137 SSRs and 42 repeat structures were identified in S. adstringens chloroplast genome, with the highest proportion in the LSC region. A comparison of the S. adstringens chloroplast genome with those from other mimosoid species indicated that gene content and synteny are highly conserved in the clade. The phylogenetic reconstruction using 73 conserved coding-protein genes from 19 Leguminosae species was supported to be paraphyletic. Furthermore, the noncoding and coding regions with high nucleotide diversity may supply valuable markers for molecular evolutionary and phylogenetic studies at different taxonomic levels in this group.
Subject terms: Comparative genomics, Plant genetics
Introduction
The chloroplast, which is considered to have originated from free-living cyanobacteria through endosymbiosis, plays an essential role in photosynthesis and in many processes in plant cells1–3. In this evolutionary context, the chloroplast genome of angiosperms exhibit a highly conserved organization with a quadripartite structure, comprising two copies of inverted repeats (IRs), separated by large (LSC) and small (SSC) single-copy regions4.
The size of the circular chloroplast genome range between 120 and 160 kb in length5, but varies considerably both within and among plant families. For example, in the Geraniaceae, the size of the chloroplast genome ranges from 116,935 bp in Erodium carvifolium6 to 242,575 bp in Pelargonium transvaalense (Accession: NC_031206.1 unpublished). For Leguminosae, the size ranges from 120,289 bp in Lathyrus odoratus (Accession: NC_027150.1 unpublished) up to 178,887 in Pithecellobium flexicaule7. The variations in size can be attributed mostly to the expansion, contraction or loss of IRs, as well as variation in length of intergenic spacers5,8.
Most angiosperm chloroplast genome usually contain 100–130 distinct genes, comprising of 80–90 protein coding genes and approximately 30 transfer RNA (tRNA) genes and four ribosomal RNA (rRNA) genes9,10. The IR region comprises a duplicated set of tRNA and rRNA genes, whereas the single copy regions mostly consists of protein-coding genes involved in cell functions, which include components of the photosynthetic machinery (such as photosystem I (PSI), photosystem II (PSII), the cytochrome b6/f complex, and the ATP synthase), transcription, and translation. The two IRs are identical in their nucleotide sequence, so that every gene contained within them is present in two copies per genome which only differ in their relative orientation9–11.
The gene order and content of chloroplast genomes are generally highly conserved along plant evolution and the substitution rates are much lower than that of the nuclear genome12. This fact, coupled with the non-recombinant nature and maternal inheritance in most angiosperms, makes plant chloroplasts genomes valuable sources of genetic markers for analyzing evolutionary relationships at multiple scales, ranging from short-term phylogeographic patterns up to phylogenetic relationships among large clades13,14.
The first complete chloroplast genomes were determined over 30 years ago, for Nicotiana tabacum15 and Marchantia polymorpha16. However, because the time and cost associated with the conventional Sanger sequencing, the reconstruction of complete chloroplast genome was impractical for non-model species. More recently, with the advent of next-generation sequencing technology, whole genome sequencing has increased dramatically17. This offers an alternative way to obtain chloroplast genome based on downstream bioinformatics pipelines that allows distinguishing plastid reads from nuclear and mitochondrial reads18. Currently, approximately 1,654 eudicotyledons chloroplast genomes have been sequenced and deposited in the NCBI Organelle Genome, out of which 114 belong to the legume family (Leguminosae) and 19 to Caesalpinioideae subfamily.
The Leguminosae is the third-largest angiosperm family, with approximately 751 genera and ca. 19,500 species19,20. The Leguminosae was divided into three sub-families, the Caesalpinioideae, Mimosoid and Papilionoideae20. However, a new classification of the legumes has been proposed by The Legume Phylogeny Working Group21. They used a matK gene-based phylogeny and a wide Leguminosae sample (~90% of genera) to propose a new family organization consisting in six sub-families: Caesalpinioideae, Cercidoideae, Detarioideae, Dialioideae, Duparquetioideae and Papilionoideae21. The traditional subfamily Mimosoid is now recognized as a distinct ‘mimosoid clade’ nested in the reassigned Caesalpinioideae21. Within the ‘mimosoid clade’, the genus Stryphnodendron Mart. includes approximately 21 species and two subspecies, mainly found in the South-American neotropical savannas22. Recently phylogenetic analysis demonstrated that the genus Stryphnodendron are not monophyletic23, clustering with the monospecific genus Microlobius inside the Piptadenia group23.
The S. adstringens, popularly known as “barbatimão”, is a common tree in the Brazilian Savanna. It’s a small, hermaphroditic, deciduous tree with a rough, light-colored, thick, tortuous trunk. It can reach 4–5 meters tall and the trunk can be 20–30 cm in diameter. The leaves alternate between composed and binary. Flowering occurs in September24,25 and the fruits are sessile, thick and fleshy, linear, oblong, light brown in color, 10 cm long, producing many brown seeds. The stem bark of this plant is used, popularly, in the treatment of several diseases because of it´s anti-inflammatory, antimicrobial and antiulcerogenic properties26–29. These effects are directly correlated to the presence of high concentrations of tannin into the barks30.
The goal of this study was to assemble the chloroplast genome of S. adstringens from whole genome sequence data, reporting the annotation and its structural characterization providing new genomic resources for this species. We also used a phylogenetic analysis to evaluate the sequence divergence in chloroplast regions of S. adstringens when compared with other known species of the mimosoid clade.
Materials and Methods
DNA extraction and chloroplast genome sequencing
Fresh young leaves of S. adstringens were collected in Niquelândia, Goiás, Brazil (Sisgen Registration: A4EE2BE). Total genomic DNA was extracted using a CTAB protocol31. DNA quality was evaluated using horizontal electrophoresis with 1% agarose gel. In addition, DNA was quantified through fluorometry using Qubit 2.0 (Life Technologies). Genomic library preparation was performed using a Nextera DNA Sample Preparation Kit (Illumina, San Diego, USA). The resulted library was sequenced using the HiSeq2500 platform and V4 SBS kit (Illumina) on a single lane in paired-end mode (2 × 100 bp) at the University of São Paulo (Escola Superior de Agricultura Luiz de Queiroz da Universidade de São Paulo) in Piracicaba, Brazil.
Chloroplast genome assembly and annotation
Paired-end Illumina raw reads were filtered and trimmed using Trimmomatic V.0.3632 using the ILLUMINACLIP: NexteraPE-PE.fa:2:30:10 for adapter trimming, a sliding window of 10 base pairs with a minimum average quality score of 20 (SLIDINGWINDOW:10:20), and a minimum length of 40 bp (MINLEN:40).
The chloroplast genome of S. adstringens was reconstructed using a combination of de novo and reference-guided assemblies. To obtain the de novo chloroplast genome assembly, the paired-end sequence reads were mapped to five Mimosoid plastomes using Bowtie2 v.2.3.4.133 to exclude reads of nuclear and mitochondrial origins (Adenanthera microsperma Teijsm. & Binn. [accession no. NC_034986], Dichrostachys cinerea (L.) Wight & Arn. [accession no. NC_035346], Leucaena trichandra (Zucc.) Urb. [accession no. NC_028733], Parkia javanica (Lam.) Merr. [accession no. NC_034989], Piptadenia communis Benth. [accession no. NC_034990]). The obtained putative chloroplast reads were then used for de novo assembly using SPAdes 3.6.1 with iterative K-mer sizes of 55, 69 and 8734. Reference guided assembly was performed with YASRA 2.3235 using Piptadenia communis Benth. as reference chloroplast genome. Contigs with coverage below than 10x were eliminated. The remaining de novo contigs were merged with reference-guided contigs in Sequencher 5.4.6 (Genecodes, Ann Arbor, Michigan, USA) based on at least 20 bps overlap and 98% similarity. Any discrepancies between de novo and reference-guided contigs were corrected by searching the high quality read pool using the UNIX ‘grep’ function. A “genome walking” technique, using the Unix “grep” function, was used to find reads that could fill any gaps between contigs that did not assemble in the initial set of analyses. Assembly curation was performed by aligning sequencing reads on the chloroplast using the Bowtie2 program. Sequencing depth was measured using the samtools platform (samtools.sourceforge.net/). Additionally, we also compared the position of the chloroplast genome regions of S. adstringens related species in circle alignment graphs made with the Circus program (http://circos.ca/).
Annotation of the chloroplast genome was performed using Verdant36 and Dual Organellar Genome Annotator-DOGMA37, coupled with manual correction of start and stop codons and intron/exon boundaries. Transfer RNA (tRNA) genes were identified with DOGMA and the tRNAscan-SE program ver. 2.038 in organellar search mode with default parameters. The circular chloroplast genome map was drawn using OrganellarGenomeDRAW (OGDRAW)39. The codon usage analysis was performed in the web server Bioinformatics (https://www.bioinformatics.org/sms2/codon_usage.html).
Characterization of repeat sequences
The sizes and locations of forward, reverse, palindromic and complementary repeats in the S. adstringens chloroplast genome were determined by REPuter40 with a minimal size of 30 bp, hamming distance of 3 and over 90% identity. Simple sequence repeats (SSRs) were detected using the microsatellite identification tool MISA (available online: http://pgrc.ipk-gatersleben.de/misa/misa.html). The minimum number of SSRs was set to ten repeat units for mononucleotide, five repeat units for dinucleotide, four repeat units for trinucleotide and three repeat units for tetra-, penta- and hexanucleotide.
Nucleotide Diversity and Synonymous (Ks) and non-synonymous (Ka) substitution rate analysis
The complete chloroplast genome sequence of S. adstringens was compared with the chloroplast genome sequences of five Mimosoid chloroplast genomes used for assembling. To assess the complete nucleotide diversity (Pi) among the complete chloroplast genome of the six species, the complete chloroplast genome sequences were aligned using MAFFT aligner tool41, and manually adjusted with Bioedit42. We then performed a sliding window analysis to calculate the nucleotide variability (Pi) values using DnaSP 643 with window lenght of 600 bp and step size of 200 bp. The 77 protein-coding genes were extracted and aligned separately using MAFFT to estimate the synonymous (Ks) and non-synonymous (Ka) substitution rates. The Ka/Ks for each gene were estimated in DnaSP 6.
Comparative analysis of genome structure
The mVISTA program was applied to compare the complete chloroplast genome of S. adstringens against the whole chloroplast genome of the five mimosoid species using the shuffle-LAGAN mode44. The annotated S. adstringens chloroplast genome was used as reference. The expansion and contraction of the IR regions at junction sites between the six mimosoid species were verified and plotted using IRscope45.
Phylogenetic analyses
Seventy-three protein-coding genes were recorded from 19 species within the Leguminoseae – Caesalpinioideae, as well as from two outgroups (Cucumis sativus L. and Fragaria vesca L.). All genes sequences were obtained from GenBank (see Supplementary Table S6 for accession numbers). The accD, ycf1, rps16, rps12 genes were not considered for phylogenetic analysis as they were not present in all chloroplast genomes among the species under analysis.
The nucleotide sequences were aligned using MAFFT41 with default parameters. The Akaike Information Criterion (AIC) in JModelTest v2.1.10 was used to determine the best-fitting model of molecular evolution for each gene46 (models selected can be seen in Supplementary Table S7). The alignments from the 73 protein-coding genes were concatenated and a Bayesian inference was performed using BEAST v1.10.147 at the XSEDE Teragrid of the CIPRES science gateway48 (available online: www.phylo.org). The Markov chain Monte Carlo (MCMC) was set to run 50.000.000 generations and sampled every 1.000 generations, under a strict clock approach using the Yule speciation tree prior with the evolutionary models. The Convergence of parameters during MCMC runs were assessed by their Effective Sample Size (ESS) > 200 using TRACER v1.7.149. The phylogenetic tree was annotated as a maximum clade credibility tree using TREEANNOTATOR v.2.5.0 (part of the BEAST package), with burn in of 20%. The final tree was produced using FigTree v.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Results and Discussion
Genome assembly and annotation
A total of 563,117,260 raw Illumina paired-end reads from the S. adstringens genome were generated and filtered against Mimosoid chloroplast genomes. After trimming adapters, low quality bases, and mapping the reads to the reference set, a total of 10,549,708 reads were used to assemble the chloroplast genome. The filtered reads were assembled into 57 contigs with at least 10x of coverage with a total length of 271,468 bp and an N50 of 10,881. The reference guided assembly produced 64 contigs with an N50 of 3,938 (half of the assembly contained at least 3,938 bp). The final assembly from both approaches resulted in a single chromosome and was benchmarked based on the distribution of sequencing coverage by base. After that, for validation, a set of 18,937,635 raw paired-end Illumina reads were well aligned in the chloroplast genome. The average sequencing depth was 11,470X , with a standard deviation of 3,720 (median: 12,009; mode: 12,448; minimum: 29 and maximum: 72,245) (Supplementary Fig. 1A,B). The pairwise alignments with species close related to S. adstringens showed a high conservation of the general structure of the chromosome and correct arrangement of the chloroplast regions validating the proposed genome (Supplementary Fig. 2A–D). The sequence of the chloroplast genome was deposited in GenBank (accession number: MN196294).
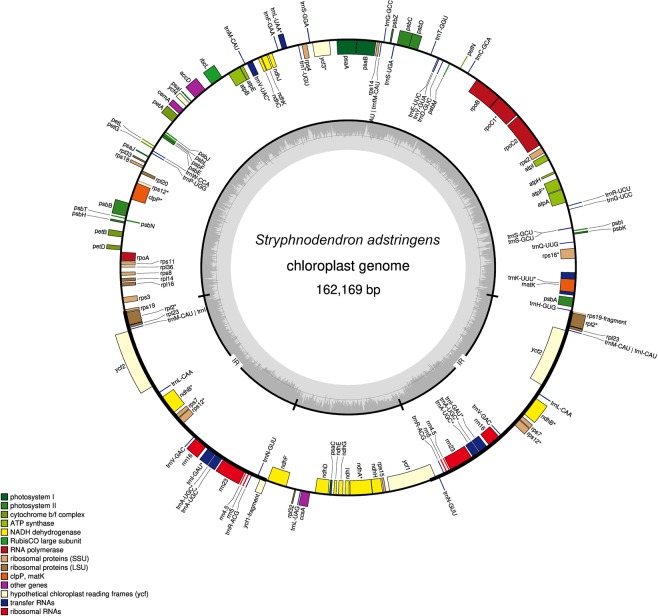
The complete chloroplast genome of S. adstringens was assembled with a total of 162,169 bp in size, divided in four regions, which included a large single-copy (LSC) region of 91,045 bp, a small single-copy (SSC) region of 19,014 bp, separated by two inverted repeat (IR) regions of 26,055 bp each (Fig. 1; Table 1). Analogous to most angiosperms, the S. adstringens chloroplast genome comprises a single circular molecule with a quadripartite structure4 and it is similar in size from other species from the Mimosoid clade (Leguminosae; Table 1; see also7).
Figure 1.
Gene map of the S. adstringens chloroplast genome. The genes drawn outside and inside of the circle are transcribed in clockwise and counterclockwise directions, respectively. Genes were colored based on their functional groups. The inner circle shows the quadripartite structure of the chloroplast: small single copy (SSC), large single copy (LSC) and a pair of inverted repeats (IRa and IRb). The gray ring marks the GC content with the inner circle marking a 50% threshold. Asterisks mark genes that have introns.
Table 1.
Chloroplast genome information from sampled Mimosoid species and the newly assembled S. adstringens.
| Species | Size (bp) | LSC (bp) | SSC (bp) | IR (bp) | GC (%) | Protein | RNA |
|---|---|---|---|---|---|---|---|
| Adenanthera microsperma | 159,389 | 88,577 | 18,756 | 26,028 | 36.5% | 77 | 34 |
| Dichrostachys cinerea | 161,240 | 90,430 | 18,526 | 26,142 | 35.9% | 77 | 34 |
| Leucaena trichandra | 164,692 | 93,690 | 18,890 | 26,056 | 35.6% | 77 | 34 |
| Parkia javanica | 161,681 | 91,093 | 18,574 | 26,007 | 35.9% | 77 | 34 |
| Piptadenia communis | 162,552 | 91,517 | 18,941 | 26,047 | 35.9% | 77 | 34 |
| Stryphnodendron adstringens | 162,169 | 91,045 | 19,014 | 26,055 | 35,9% | 77 | 34 |
LSC Large Single Copy, SSC Small Single Copy, IR Inverted Repeat.
The GC content of the S. adstringens chloroplast genome was 35.9%, which is also consistent with other Mimosoid, whose plastomes ranged from 35.6 to 36.5% overall GC content (Table 1; see also7). Among the LSC, SSC and IR regions, the highest GC content was found in the IR regions (42.7%), while the GC content of LSC and SSC was 33.3% and 30.0%, respectively (Supplementary Table S1). The high GC content in the IR regions are mainly due to high GC contents of the four ribosomal RNA (rRNA) genes, rrn23, rrn16, rrn5, rrn4.5, with 55.3%, 56.4%, 52.9% and 50%, respectively, that are located in this region.
The assembled chloroplast genome contained 111 different genes, with 77 protein-coding genes, 30 transfer RNA (tRNA) and 4 ribosomal RNA genes (rRNA) (Fig. 1; Table 2). A total of nine protein-coding genes and 6 tRNAs genes contained a single intron, whereas three genes (rps12, clpP and ycf3) exhibit two introns each (Supplementary Table S2). The rps12 gene was predicted to be trans-spliced, with the 5′ end located in the LSC region and the duplicated 3′ end in the IR region. The trnK-UUU has the largest intron encompassing the matK gene, with 2,558 bp, whereas the intron of trnL-UAA is the smallest (513 bp).
Table 2.
List of genes in the chloroplast genome of S. adstringens.
| Category | Gene groups | Name of genes |
|---|---|---|
| Self-replication | Large subunit of ribosomal proteins | rpl21,2, rpl14, rpl161, rpl20, rpl232, rpl32, rpl33, rpl36 |
| Small subunit of ribosomal proteins | rps2, rps3, rps4, rps72, rps8, rps11, rps121,2, rps14, rps15, rps161, rps18, rps19 | |
| DNA-dependent RNA polymerase | rpoA, rpoB, rpoC11, rpoC2 | |
| Ribosomal RNA genes | rrn4.52, rrn52, rrn162, rrn232 | |
| Transfer RNA genes | trnA-UGC1,2, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-UCC1, trnG-UCC, trnH-GUG, trnI-CAU2, trnI-GAU1,2, trnK-UUU1, trnL-CAA2, trnL-UAA1, trnL-UAG, trnM-CAU, trnN-GUU2, trnP-UGG, trnQ-UUG, trnR-ACG2, trnR-UCU, trnS-GCU, trnS-UGA, trnS-GGA, trnT-UGU, trnT-GGU, trnV-UAC1, trnV-GAC2, trnW-CCA, trnY-GUA | |
| Photosynthesis | Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| NADH dehydrogenase | NADH dehydrogenase | ndhA1, ndhB1,2, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| Cytochrome b/f complex | petA, petB1, petD1, petG, petL, petN | |
| ATP synthase | atpA, atpB, atpE, atpF1, atpH, atpI | |
| RubisCo large subunit | rbcL | |
| Other genes | Maturase K | matK |
| Envelope membrane protein | cemA | |
| Subunit of acetyl-CoAcarboxylase | accD | |
| C-type cytochrome synthesis gene | ccsA | |
| Protease | clpP 1 | |
| Conserved hypothetical chloroplast open reading frames | ycf1, ycf22, ycf31, ycf4 |
1 – Gene with introns.
2 – Gene completely duplicated in the inverted repeat.
Codon usage analysis performed using 77 protein coding gene sequences identified a total of 20,986 codons in the S. adstringens chloroplast genome (Table 3). Most identified codons are coders for amino acid leucine (2,227 codons, ~ 10.6% of the total number of codons) and the most abundant codon was TTA (33% of codons encoding leucine). The codons encoding the amino acid cysteine were identified as the least abundant in the S. adstringens chloroplast genome (256 codons, ~1.2%). Moreover, only one codon was identified for the coding of methionine (ATG) and tryptophan (TGG) amino acids.
Table 3.
Codon usage for S. adstringens chloroplast genome.
| Amino Acid | Codon | Number | Fraction | Amino Acid | Codon | Number | Fraction |
|---|---|---|---|---|---|---|---|
| Ala | GCG | 138 | 0.12 | Leu | CTA | 302 | 0.14 |
| GCA | 321 | 0.27 | CTT | 462 | 0.21 | ||
| GCT | 571 | 0.48 | CTC | 135 | 0.06 | ||
| GCC | 168 | 0.14 | Lys | AAG | 245 | 0.24 | |
| Arg | AGG | 134 | 0.11 | AAA | 759 | 0.76 | |
| AGA | 365 | 0.30 | Met | ATG | 504 | 1 | |
| CGG | 92 | 0.07 | Phe | TTT | 803 | 0.67 | |
| CGA | 282 | 0.23 | TTC | 397 | 0.33 | ||
| CGT | 275 | 0.22 | Pro | CCG | 112 | 0.13 | |
| CGC | 87 | 0.07 | CCA | 249 | 0.29 | ||
| Asn | AAT | 737 | 0.78 | CCT | 341 | 0.39 | |
| AAC | 208 | 0.22 | CCC | 170 | 0.19 | ||
| Asp | GAT | 655 | 0.81 | Ser | AGT | 324 | 0.20 |
| GAC | 149 | 0.19 | AGC | 97 | 0.06 | ||
| Cys | TGT | 186 | 0.74 | TCG | 145 | 0.09 | |
| TGC | 67 | 0.26 | TCA | 312 | 0.19 | ||
| Gln | CAG | 164 | 0.23 | TCT | 458 | 0.29 | |
| CAA | 559 | 0.77 | TCC | 267 | 0.17 | ||
| Glu | GAG | 264 | 0.25 | Thr | ACG | 113 | 0.11 |
| GAA | 784 | 0.75 | ACA | 322 | 0.30 | ||
| Gly | GGG | 238 | 0.16 | ACT | 442 | 0.42 | |
| GGA | 593 | 0.39 | ACC | 188 | 0.18 | ||
| GGT | 531 | 0.35 | Trp | TGG | 370 | 1 | |
| GGC | 152 | 0.10 | Tyr | TAT | 622 | 0.80 | |
| His | CAT | 395 | 0.78 | TAC | 153 | 0.20 | |
| CAC | 112 | 0.22 | Val | GTG | 168 | 0.14 | |
| Ile | ATA | 564 | 0.31 | GTA | 460 | 0.38 | |
| ATT | 933 | 0.51 | GTT | 434 | 0.36 | ||
| ATC | 341 | 0.19 | GTC | 134 | 0.11 | ||
| Leu | TTG | 466 | 0.21 | End | TGA | 29 | 0.28 |
| TTA | 727 | 0.33 | TAG | 21 | 0.21 | ||
| CTG | 138 | 0.06 | TAA | 52 | 0.51 |
The duplicated IR of the S. adstringens chloroplast genome resulted in complete duplication of sixteen genes (including five protein-coding genes [rpl2, rpl23, rps7, rps12 and ndhB], seven tRNAs [trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC], and all four rRNAs [rrn23, rrn16, rrn5, rrn4.5], see Table 2; Fig. 1) and parts of the 5′ end of ycf1 and rps19. Of the remaining genes, the LSC region contained 59 protein-coding and 22 tRNA genes, while the SSC region contained 11 protein-coding and one tRNA gene. These results corroborate the findings of Wang et al. (2017), which all species belonging to tribe Mimoseae had canonical IRs, with a typical gene content and general organization7.
It is important to point out that the structure of the chloroplast genome in species from Leguminosae family are highly variable because of either expansion or contraction of the IR7,50,51. This is mainly caused by the gene transfer from single copy regions to inverted repeat and vice versa in the boundaries of IRs, during evolution51. With the exception of Acacia ligulata52, the legume plastomes of all species documented to date within the clade formed by Ingeae and Acacia species, have IRs ca. 13 kb larger, and a SSC correspondingly smaller, than other legumes7,50. In addition, the size variation of the plastid genome has been explained, at least in part, by the loss of one IR. However, the mechanisms that led to IR loss are still unknown. Examples of variation within the Leguminosae include the loss of the IR in the monophyletic group within the subfamily Papilionoideae (including Trifolium, Pisum, Cicer, Medicago, Glycyrrhiza and Vicia), known as the “inverted repeat lacking clade”53,54.
Repeat sequences analysis
A total of 42 repeat structures, with lengths ranging from 30 bp to 128 bp, were detected in the S. adstringens chloroplast genome. They included 22 (52.38%) forward repeats, 18 (42.86%) palindromic repeats, and two (4.76%) reverse repeats (Supplementary Table S3), whereas none complementary structure was identified. The forward repeats ranged from 30 bp to 128 bp. The palindromic repeats were 30 bp to 60 bp, whereas the two reverse repeats were 30 bp and 38 bp (Supplementary Table S3). In the majority of Mimosoid species, the most abundant dispersed repeat identified were forward, then palindromic and the least was reverse7.
Among the 42 repeats, 64.29% are located in the LSC region, 14.29% in the SSC region and 16.67% in the IR region. Two repeats, which were 39 bp and 52 bp, located in the intron region of ycf3 and rpl16, was found to repeat thrice (LSC/SSC/IR) and twice (LSC/SSC), respectively, as forward repeat. Most of the repeats (80.95%) were found in the intergenic spacer regions (IGS), whereas 19.05% were located in the introns (ndhA, ycf3 and rpl16) and only two were located in coding region (psaB and trnS-GCU) (Supplementary Table S3).
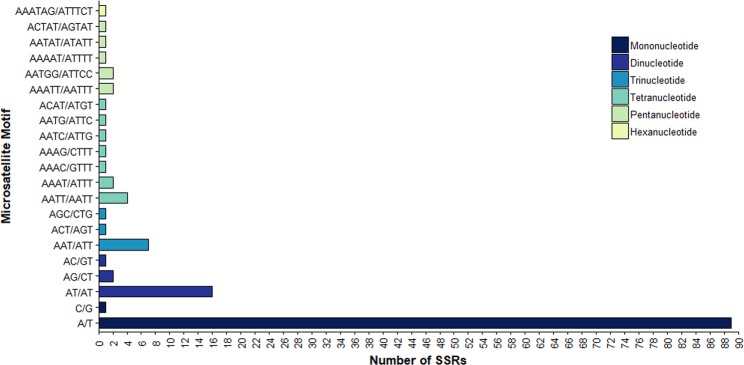
A total of 137 SSRs were detected in the S. adstringens chloroplast genome, which were composed by a length of at least 10 bp and repeated 3 to 21 times. Among them, 90 (65.69%) were mono-repeats, 19 (13.87%) were di-repeats, nine (6.57%) were tri-repeats, 11 (8.03%) were tetra-repeats, seven (5.11%) were penta-repeats and one (0.73%) was hexa-repeat. The majority (89 or 98.89%) of the mononucleotide repeats consisted of A/T motifs, while only one was composed of a G/C motif. Likewise, most of the dinucleotides and trinucleotides were AT-rich, being composed of AT/TA (84.21%) and AAT/TTA (77.78%) repeats, respectively (Fig. 2). These results showed that the SSRs exhibit a strong AT bias, which is consistent with the observed in other Leguminosae species, such as Vigna radiata55, Cajanus cajan10, Cajanus scarabaeoides10 and Arachis hypogaea56.
Figure 2.
Number and type of simple sequence repeats in S. adstringens chloroplast genome.
Concerning genomic localization, among the 137 SSRs, 114 (83.21%) were found in the LSC region, 17 (12.41%) in the SSC region and six (4.38%) in the IR region (Supplementary Table S4). Most of these SSRs were located in intergenic regions (102 or 74.45%), while 17 (12.41%) were in the introns and 18 (13.14%) were in the protein-coding genes (Supplementary Table S4). The ycf1 gene contained more SSRs than the other genes (Supplementary Table S4). In addition, the results found herein are in agreement with those from Cajanus cajan10, Cajanus scarabaeoides10, Vigna radiata55 and Glycine species57. The ycf1 encodes a protein of approximately 1,800 amino acids. The ycf1 located in the IR region is short and conserved, while the located in SSC region are extremely variable in seed plants58,59. Some studies reported that this region is the most variable locus for the design of primers, as well, as more variable than matK in many taxa, and thus suitable for molecular systematics at low taxonomic levels58,60.
Nucleotide diversity and Ka/Ks ratio
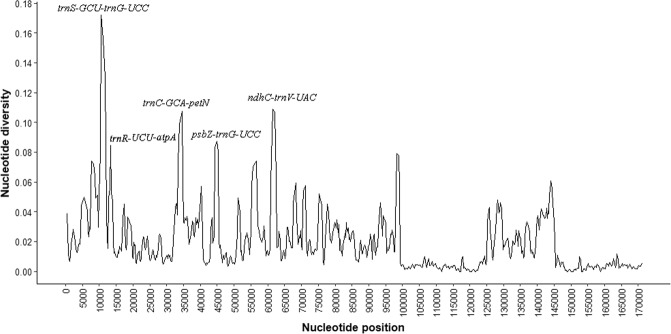
The average nucleotide variability (Pi) among the five chloroplast genomes of Mimosoid species was estimated to be 0.01771, ranging from 0 to 0.172. The nucleotide variability was higher in the SSC (Pi = 0.02712) and LSC (Pi = 0.02488), when compared to IR regions, which had a much lower nucleotide diversity (Pi = 0.00339). Similar results were obtained in the comparison of chloroplast genome sequences among Aconitum L. species, in which the average Pi in the IR region was 0.00146, whereas in the LSC and SSC regions the diversity estimates were 0.007140 and 0.008368, respectively61. Park et al. (2018) analyzed the nucleotide diversity in six Ipomoea L. chloroplast genome and also found that the IR regions were more conserved than the LSC and SSC regions, with average Pi values of 0.003 for IR and more than 0.006 for SSC and LSC regions62.
Five regions (trnS-GCU-trnG-UCC, trnR-UCU-atpA, trnC-GCA-petN, psbZ-trnG-UCC, ndhC-trnV-UAC) showed high levels of nucleotide diversity, with Pi values > 0.8 (Fig. 3). All of these highly variable regions are found in intergenic spacer from LSC region. Liu et al. (2018) analyzed the nucleotide diversity among seven species from caesalpinioid legumes and reported five regions including psbZ-trnG (trnT-trnL, rps3-rps19, rpl32 and ycf1) with higher Pi values, all with Pi > 0.1263. Highly variable regions among chloroplast genomes can be useful for phylogenetic reconstruction and may be used for further phylogenetic study of the genus Stryphnodendron.
Figure 3.
Sliding window analysis of Mimosoid chloroplast genomes.
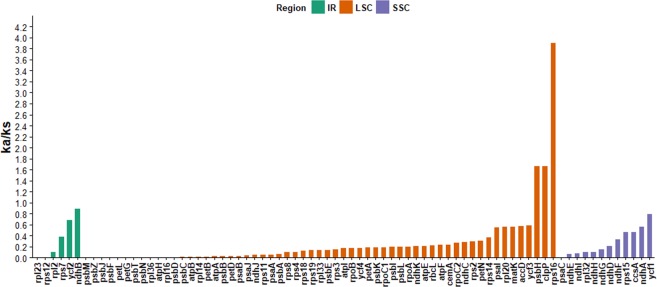
The non-synonymous (Ka) to synonymous (Ks) rate ratio (Ka/Ks) was calculated for the 77 protein-coding genes in common across all six chloroplast genomes (Fig. 4 and Supplementary Table S5). The synonymous substitution does not change the amino acid within a peptide chain, whereas nonsynonymous substitution does. The rpl32 gene associated with large subunit of ribosomal proteins had the highest synonymous rate, 0.09263, while the ycf1 gene with unknown functions, had the highest nonsynonymous rate, 0.03534.
Figure 4.
The Ka/Ks ratio of Mimosoid chloroplast genomes for individual genes.
The Ka/Ks ratio may indicate whether selective pressure is acting on a particular protein-coding gene. A Ka/Ks > 1 indicates that the gene is affected by positive selection, whereas Ka/Ks < 1 indicates that the gene is affected by negative selection or purifying selection. A value of 0, indicates the presence of neutral selection64. Concerning the different regions of chloroplast genomes, the Ka/Ks ratio were highest on average in the SSC region (0.2991) and lowest in the IR region (0.2856) and LSC region (0.2708). The lowest Ka/Ks ratio was observed for genes encoding subunits of ATP synthase, subunits of the cytochrome b/f complex, subunits of the large subunit of ribosomal proteins and subunits of photosystem II (Fig. 4; Supplementary Table S5).
Herein, the Ka/Ks ratio was calculated to be 0 for 13 genes, two inside the IR region (rpl23, rps12), ten in the LSC region (rpl36, psbM, psbZ, psbJ, psbF, psbT, psbN, petL, petG, atpH) and one in the SSC region (psaC) (Fig. 4). This occurred because the Ka or Ks is 0 or extremely low, thus Ka/Ks ratio could not be calculated65,66. Among the 77 protein-coding genes, Ka/Ks indicates purifying selection in 62 of them (Fig. 4; Supplementary Table S5). The Ka/Ks ration indicates positive selection for three genes analyzed, one of it is associated with small subunit of ribosomal proteins (rps16), the other is associated with Photosystem II (psbH) and the third with the clpP proteases (Fig. 4; Supplementary Table S5).
Liu et al. (2018) reported four genes with Ka/Ks ratio more than 1, indicating positive selection, ndhD, ycf1, infA and rpl23 in caesalpinioid legumes63, whereas, Park et al. (2018) observed positive selection in three genes, accD, cemA, and ycf2, among six Ipomoea species62. In addition, Tian et al. (2018) reported only one gene (rps12) with positive selection among nine Araceae species67.
In addition, it was demonstrated that Leguminosae chloroplast have regions with accelerated mutation rates, including genic regions such as the clpP in Mimosoids and rps16 in the IRLC clade50,52,68. The rps16 gene encodes the ribosomal protein S16 and it is present in the chloroplast genome of the majority of higher plants. Moreover, a multiple gene-loss event of rps16 was reported for various legumes lineages69,70. For instance, in the Leguminosae family, Cicer arietinum71, Caragana rosea72, Phaseolus vulgaris73,74, Lupinus species70 and Mucuna macrocarpa75 have lost this gene. As revealed in other studies, the multiple loss of the rps16 was assumed to be a consequence of the dual targeting of the nuclear rps16 copy to the plastid as well as the mitochondria, suggesting that the chloroplast-encoded rps16 has already been silenced and has become a pseudogene by the nuclear-encoded rps1669,70,76.
Comparative analysis of genome structure
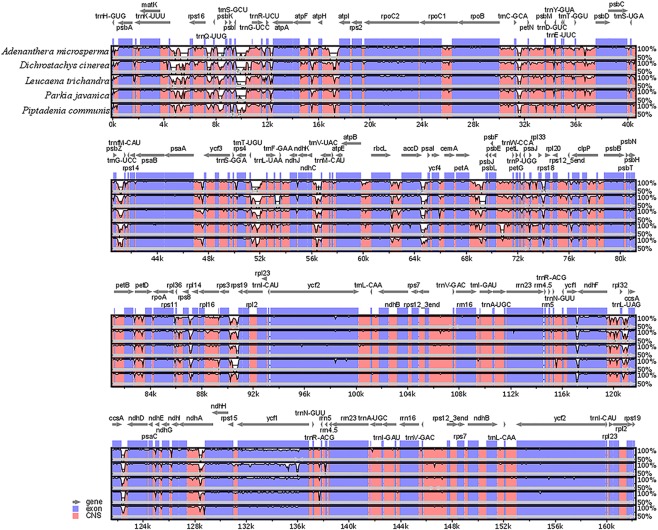
The structural characteristics in chloroplast genome among Mimosoid species revealed that gene coding regions were more conserved than the noncoding regions, and IRs were more conserved than LSC and SSC regions (Fig. 5). This result is consistent with the pattern revealed in other Leguminosae species, such as Glycine species77 and species from Caesalpinioideae subfamily7. Additionality, it was also observed that the intergenic spacers regions between several pairs of genes varied greatly, for example, between matK-rps16, rps16-psbK, trnS-GCU-trnG-UCC, atpH-atpI, trnC-GCA-petN, psbZ-trnG, trnT-UGU-trnL-UAA, ndhC-trnV-UAC and rps3-rps19. Some of these intergenic spacer regions had also the highest level of nucleotide diversity.
Figure 5.
Visualization of genome alignment of six Mimosoid chloroplast genomes using S. adstringens as reference. The vertical scale indicates the percent of identity, ranging from 50% to 100%. Coding regions are marked in purple, and non-coding as red. The horizontal axis indicates the coordinates within the chloroplast genome.
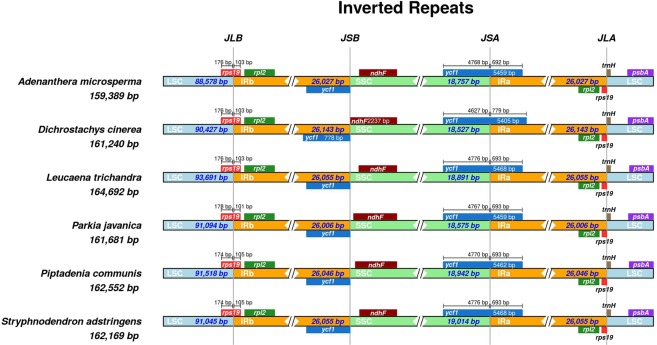
The expansion and contraction of the IR region and the single-copy boundary regions result in change in the position of the junction sites, which is considered as a primarily mechanism causing length variation of chloroplast genomes in higher plants78. The length of the IR regions was similar, ranging from 26,006 bp in Parkia javanica to 26,143 bp in Dichrostachys cinerea (Fig. 6). Wang et al. (2017) observed that species belonging to tribe Mimoseae had canonical IRs, whereas species belonging to tribes Ingae and Acacieae had much long IRs, as all of them experienced ca. 13 kb IR expansion into SSC previously reported by Dugas et al. (2015).
Figure 6.
Comparison of the junction sites between the Long Single Copy (LSC, light blue), Short Single Copy (SSC, light green) and Inverted Repeat (IRa and IRb, orange) regions among the six Mimosoid chloroplast genomes. JLB (IRb/LSC), JSB (IRb/SSC) JSA (SSC/IRa) and JLA (IRa/LSC) denote the junction sites between each corresponding regions on the genome.
The endpoint of the Mimoseae JLA (Junction between IRa and LSC) was located upstream of the rps19 and downstream of the trnH-GUG. Similar patterns were observed by Amiryousefi et al. (2018b) in Solanaceae chloroplast genomes79. The junction between IRb and SSC region (JSB) was located in the intergenic ycf1/ndhF, and the distance between the ndhF end to the junction of the IRb/SSC differs by 11 bp in Dichrostachys cinerea to 150 bp in Adenanthera microsperma. The junction between IRa and SSC (JSA) was located within the ycf1 gene, and the fragment located at the IRa region ranged from 692 bp to 779 bp (Fig. 6). The gene rps19 crossed the LSC/IRb region and the extent of the IR expansion to rps19 slightly varies among the Mimoseae species ranging from 101 bp to 105 bp.
Phylogenetic relationships
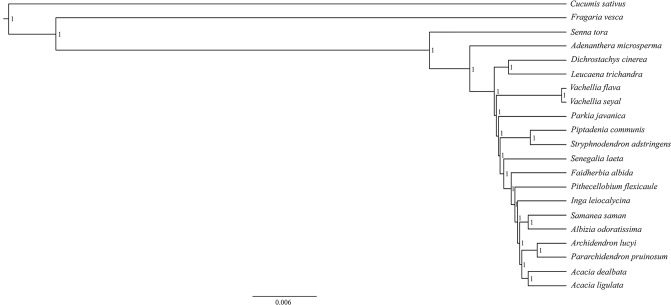
In this study, 19 species from Caesalpinioideae and two outgroups were analyzed based on 73 protein-coding genes of their chloroplast genomes. The total concatenated alignment length from the 73 protein-coding genes was 60,736 bp. The reconstructed phylogeny indicated that Caesalpinioideae was paraphyletic and that the species from tribe Mimoseae (Adenanthera microsperma, Dichrostachys cinerea, Leucaena trichandra, Parkia javanica, Piptadenia communis and S. adstringens) were deemed non-monophyletic (Fig. 7). All nodes were strongly supported, given the Bayesian posterior probability (Fig. 7). These results are consistent with those from Wang et al. (2017) and support the new classification system proposed for the Leguminosae21.
Figure 7.
Maximum credibility tree reconstructed based on 73 conserved coding-protein genes from twenty-one species. All nodes of the phylogenetic tree are supported by 1.00 Bayesian inference posterior probability.
Conclusion
In this work, we assemble the complete chloroplast genome of S. adstringens with 162,169 bp. Genome gene contents and orientation are similar to those found in the chloroplast genome of other Mimosoid (Leguminosae) species. This study also revealed the distribution and location of repeated structures and microsatellites along the chloroplast genome of S. adstringens. We also generated important genomic resources for Mimosoid group. Moreover, the Ka/Ks ratio was lower in the LSC region compared to SSC region. As expected, the comparison with other five Mimosoid species revealed that the coding regions are more conserved than non-coding regions, and IRs more conserved than LSC and SSC regions. Finally, the phylogenetic relationships built for 19 species of Caesalpinioideae, including the new data from S. adstringens and two outgroups, were fully resolved with high supports based on 73 conserved protein-coding genes. The maximum credibility tree revealed that the tribe Mimoseae is paraphyletic, consistent with the new classification proposed for the Leguminosae.
Supplementary information
Acknowledgements
Our research has been continuously supported by several grants and fellowships to the research network GENPAC (Geographical Genetics and Regional Planning for Natural Resources in Brazilian Cerrado) from CNPq/FAPEG (projects #563839/2010-4 and #201110267000125), by the FAPEG Universal (CH n. 07/2014) and by the CNPq/Universal project 402178/2016-5. Our study has also been developed in the context of the National Institutes for Science and Technology in Ecology, Evolution and Biodiversity Conservation (INCT_EECBio), supported by MCTIC/CNPq (process #465610/2014-5) and FAPEG, in addition to support from PPGS CAPES/FAPEG (Public Call #08/2014). U.J.B. Souza and R. Nunes were supported by Doctoral fellowships from CAPES. J.A.F. Diniz-Filho and M.P.C. Telles were supported by productivity fellowships from CNPq.
Author Contributions
U.J.B.S., M.P.C.T. and J.A.F.D.-F. conceived and designed research. M.P.C.T. and J.A.F.D.-F. provided financial resources to research. U.J.B.S. and C.P.T. performed the experiments. U.J.B.S. and R.N. did computational analyses. U.J.B.S. and R.N. analyzed data. U.J.B.S., R.N., C.P.T., M.P.C.T. and J.A.F.D.-F. wrote the paper. All authors reviewed the paper.
Data Availability
The complete chloroplast sequence generated and analyzed during the current study are available in GenBank, https://www.ncbi.nlm.nih.gov (accession numbers are described in the text).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50620-3.
References
- 1.Leister D. Chloroplast research in the genomic age. Trends in Genetics. 2003;19:47–56. doi: 10.1016/S0168-9525(02)00003-3. [DOI] [PubMed] [Google Scholar]
- 2.Neuhaus HE, Emes MJ. Nonphotosynthetic Metabolism in Plastids. Annual Review of Plant Physiology and Plant Molecular Biology. 2000;51:111–140. doi: 10.1146/annurev.arplant.51.1.111. [DOI] [PubMed] [Google Scholar]
- 3.Rodríguez-Ezpeleta N, et al. Monophyly of Primary Photosynthetic Eukaryotes: Green Plants, Red Algae, and Glaucophytes. Current Biology. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 4.Jansen Robert K., Ruhlman Tracey A. Advances in Photosynthesis and Respiration. Dordrecht: Springer Netherlands; 2012. Plastid Genomes of Seed Plants; pp. 103–126. [Google Scholar]
- 5.Palmer JD. Comparative Organization of Chloroplast Genomes. Annual Review of Genetics. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- 6.Blazier JC, Guisinger MM, Jansen RK. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae) Plant Molecular Biology. 2011;76:263–272. doi: 10.1007/s11103-011-9753-5. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y-H, Qu X-J, Chen S-Y, Li D-Z, Yi T-S. Plastomes of Mimosoideae: structural and size variation, sequence divergence, and phylogenetic implication. Tree Genetics & Genomes. 2017;13:41. doi: 10.1007/s11295-017-1124-1. [DOI] [Google Scholar]
- 8.Raubeson, L. A. & Jansen, R. K. Chloroplast genomes of plants. In Plant diversity and evolution: genotypic and phenotypic variation in higher plants. (ed. Henry, R.) 45–68 (Cambridge, MA:CABI, 2005).
- 9.Bock Ralph. Cell and Molecular Biology of Plastids. Berlin, Heidelberg: Springer Berlin Heidelberg; 2007. Structure, function, and inheritance of plastid genomes; pp. 29–63. [Google Scholar]
- 10.Kaila T, et al. Chloroplast Genome Sequence of Pigeonpea (Cajanus cajan (L.) Millspaugh) and Cajanus scarabaeoides (L.) Thouars: Genome Organization and Comparison with Other Legumes. Frontiers in Plant Science. 2016;7:1847. doi: 10.3389/fpls.2016.01847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li B, Zheng Y. Dynamic evolution and phylogenomic analysis of the chloroplast genome in Schisandraceae. Scientific Reports. 2018;8:9285. doi: 10.1038/s41598-018-27453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proceedings of the National Academy of Sciences. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Provan J, Powell W, Hollingsworth PM. Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends in Ecology & Evolution. 2001;16:142–147. doi: 10.1016/S0169-5347(00)02097-8. [DOI] [PubMed] [Google Scholar]
- 14.Ravi V, Khurana JP, Tyagi AK, Khurana P. An update on chloroplast genomes. Plant Systematics and Evolution. 2008;271:101–122. doi: 10.1007/s00606-007-0608-0. [DOI] [Google Scholar]
- 15.Shinozaki K, et al. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. The EMBO Journal. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohyama K, et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322:572–574. doi: 10.1038/322572a0. [DOI] [Google Scholar]
- 17.Nock CJ, et al. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnology Journal. 2011;9:328–333. doi: 10.1111/j.1467-7652.2010.00558.x. [DOI] [PubMed] [Google Scholar]
- 18.Twyford AD, Ness RW. Strategies for complete plastid genome sequencing. Molecular Ecology Resources. 2017;17:858–868. doi: 10.1111/1755-0998.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis, G. P., Schrire, B. D., Mackinder, B. & Lock, M. Legumes of the world. Royal Botanic Gardens, Kew, 577p (2005).
- 20.The Legume Phylogeny Working Group. Legume phylogeny and classification in the 21st century: Progress, prospects and lessons for other species-rich clades. Taxon. 2013;62:217–248. doi: 10.12705/622.8. [DOI] [Google Scholar]
- 21.Azani N, et al. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny – The Legume Phylogeny Working Group (LPWG) Taxon. 2017;66:44–77. doi: 10.12705/661.3. [DOI] [Google Scholar]
- 22.Souza, V. C. & Gibau, A. Stryphnodendron in Flora do Brasil 2020 em construção. Jardim Botânico do Rio de Janeiro (2018). Available at: http://reflora.jbrj.gov.br/reflora/floradobrasil/FB19133. (Accessed: 14th July 2018).
- 23.Simon MF, et al. Molecular phylogeny of Stryphnodendron (Mimosoideae, Leguminosae) and generic delimitations in the Piptadenia group. International Journal of Plant Sciences. 2016;177:44–59. doi: 10.1086/684077. [DOI] [Google Scholar]
- 24.Sanches ACC, et al. Estudo Morfológico Comparativo das Cascas e Folhas de Stryphnodendron adstringens, S. polyphyllum e S. obovatum-Leguminosae. Latin American Journal of Pharmacy. 2007;26:22–35. [Google Scholar]
- 25.Lorenzi, H. Árvores brasileiras: manual de identificação e cultivo de plantas arbóreas nativas do Brasil. Nova Odessa: Plantarum2, (1998).
- 26.Audi EA, et al. Gastric antiulcerogenic effects of Stryphnodendron adstringens in rats. Phytotherapy Research. 1999;13:264–266. doi: 10.1002/(SICI)1099-1573(199905)13:3<264::AID-PTR443>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 27.Ishida K, et al. Influence of tannins from Stryphnodendron adstringens on growth and virulence factors of Candida albicans. Journal of Antimicrobial Chemotherapy. 2006;58:942–949. doi: 10.1093/jac/dkl377. [DOI] [PubMed] [Google Scholar]
- 28.Lima JCS, Martins DTO, de Souza PT. Experimental evaluation of stem bark of Stryphnodendron adstringens (Mart.) Coville for antiinflammatory activity. Phytotherapy Research. 1998;12:218–220. doi: 10.1002/(SICI)1099-1573(199805)12:3<218::AID-PTR220>3.0.CO;2-4. [DOI] [Google Scholar]
- 29.Luiz RLF, et al. Proanthocyanidins polymeric tannin from Stryphnodendron adstringens are active against Candida albicans biofilms. BMC Complementary and Alternative Medicine. 2015;15:68. doi: 10.1186/s12906-015-0597-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos SC, et al. Tannin composition of barbatimão species. Fitoterapia. 2002;73:292–299. doi: 10.1016/S0367-326X(02)00081-3. [DOI] [PubMed] [Google Scholar]
- 31.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- 32.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bankevich A, et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. Journal of Computational Biology. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratan, A. Assembly algorithms for next-generation sequence data. (The Pennsylvania State University, University Park, PA, USA, 2009).
- 36.McKain MR, Hartsock RH, Wohl MM, Kellogg EA. Verdant: automated annotation, alignment and phylogenetic analysis of whole chloroplast genomes. Bioinformatics. 2017;33:130–132. doi: 10.1093/bioinformatics/btw583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 38.Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Research. 2016;44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current Genetics. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 40.Kurtz S. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall TH. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 1999;41:95–98. [Google Scholar]
- 43.Rozas J, et al. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Molecular Biology and Evolution. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 44.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Research. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amiryousefi A, Hyvönen J, Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- 46.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9:772–772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suchard MA, et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evolution. 2018;4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. in Gateway Computing Environments Workshop (GCE) 1–8 (2010).
- 49.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Systematic Biology. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dugas DV, et al. Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions, and accelerated rate of evolution in clpP. Scientific Reports. 2015;5:16958. doi: 10.1038/srep16958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu A, Guo W, Gupta S, Fan W, Mower JP. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytologist. 2016;209:1747–1756. doi: 10.1111/nph.13743. [DOI] [PubMed] [Google Scholar]
- 52.Williams AV, Boykin LM, Howell KA, Nevill PG, Small I. The Complete Sequence of the Acacia ligulata Chloroplast Genome Reveals a Highly Divergent clpP1 Gene. PLOS ONE. 2015;10:e0125768. doi: 10.1371/journal.pone.0125768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wojciechowski MF, Lavin M, Sanderson MJ. A phylogeny of legumes (Leguminosae) based on analysis of the plastid mat K gene resolves many well-supported subclades within the family. American Journal of Botany. 2004;91:1846–1862. doi: 10.3732/ajb.91.11.1846. [DOI] [PubMed] [Google Scholar]
- 54.Sabir J, et al. Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnology Journal. 2014;12:743–754. doi: 10.1111/pbi.12179. [DOI] [PubMed] [Google Scholar]
- 55.Tangphatsornruang S, et al. The Chloroplast Genome Sequence of Mungbean (Vigna radiata) Determined by High-throughput Pyrosequencing: Structural Organization and Phylogenetic Relationships. DNA Research. 2010;17:11–22. doi: 10.1093/dnares/dsp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yin D, et al. Development of chloroplast genome resources for peanut (Arachis hypogaea L.) and other species of Arachis. Scientific Reports. 2017;7:11649. doi: 10.1038/s41598-017-12026-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ozyigit II, Dogan I, Filiz E. In silico analysis of simple sequence repeats (SSRs) in chloroplast genomes of Glycine species. Plant Omics Journal. 2015;8:24–29. [Google Scholar]
- 58.Dong W, Liu J, Yu J, Wang L, Zhou S. Highly Variable Chloroplast Markers for Evaluating Plant Phylogeny at Low Taxonomic Levels and for DNA Barcoding. PLoS ONE. 2012;7:e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong W, et al. ycf1, the most promising plastid DNA barcode of land plants. Scientific Reports. 2015;5:8348. doi: 10.1038/srep08348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neubig KM, et al. Phylogenetic utility of ycf1 in orchids: a plastid gene more variable than matK. Plant Systematics and Evolution. 2009;277:75–84. doi: 10.1007/s00606-008-0105-0. [DOI] [Google Scholar]
- 61.Kong H, Liu W, Yao G, Gong W. A comparison of chloroplast genome sequences in Aconitum (Ranunculaceae): a traditional herbal medicinal genus. PeerJ. 2017;5:e4018. doi: 10.7717/peerj.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park I, et al. The Complete Chloroplast Genomes of Six Ipomoea Species and Indel Marker Development for the Discrimination of Authentic Pharbitidis Semen (Seeds of I. nil or I. purpurea) Frontiers in Plant Science. 2018;9:965. doi: 10.3389/fpls.2018.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu W, et al. Complete Chloroplast Genome of Cercis chuniana (Fabaceae) with Structural and Genetic Comparison to Six Species in Caesalpinioideae. International Journal of Molecular Sciences. 2018;19:1286. doi: 10.3390/ijms19051286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nei, M. & Kumar, S. Molecular evolution and phylogenetics. (Oxford university press, 2000).
- 65.Redwan RM, Saidin A, Kumar SV. Complete chloroplast genome sequence of MD-2 pineapple and its comparative analysis among nine other plants from the subclass Commelinidae. BMC Plant Biology. 2015;15:196. doi: 10.1186/s12870-015-0587-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang D, Liu F, Wang L, Huang S, Yu J. Nonsynonymous substitution rate (Ka) is a relatively consistent parameter for defining fast-evolving and slow-evolving protein-coding genes. Biology Direct. 2011;6:13. doi: 10.1186/1745-6150-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tian N, Han L, Chen C, Wang Z. The complete chloroplast genome sequence of Epipremnum aureum and its comparative analysis among eight Araceae species. PLOS ONE. 2018;13:e0192956. doi: 10.1371/journal.pone.0192956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Magee AM, et al. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Research. 2010;20:1700–1710. doi: 10.1101/gr.111955.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ueda M, et al. Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Molecular biology and evolution. 2008;25:1566–1575. doi: 10.1093/molbev/msn102. [DOI] [PubMed] [Google Scholar]
- 70.Keller J, et al. The evolutionary fate of the chloroplast and nuclear rps16 genes as revealed through the sequencing and comparative analyses of four novel legume chloroplast genomes from Lupinus. Dna Research. 2017;24:343–358. doi: 10.1093/dnares/dsx006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jansen RK, Wojciechowski MF, Sanniyasi E, Lee S-B, Daniell H. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae) Molecular phylogenetics and evolution. 2008;48:1204–1217. doi: 10.1016/j.ympev.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang M, et al. Sequencing, characterization, and comparative analyses of the plastome of Caragana rosea var. rosea. International journal of molecular sciences. 2018;19:1419. doi: 10.3390/ijms19051419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwarz EN, et al. Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. Journal of Systematics and Evolution. 2015;53:458–468. doi: 10.1111/jse.12179. [DOI] [Google Scholar]
- 74.Guo X, et al. Rapid evolutionary change of common bean (Phaseolus vulgaris L) plastome, and the genomic diversification of legume chloroplasts. BMC genomics. 2007;8:228. doi: 10.1186/1471-2164-8-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin D-P, Choi I-S, Choi B-H. Plastid genome evolution in tribe Desmodieae (Fabaceae: Papilionoideae) PloS one. 2019;14:e0218743. doi: 10.1371/journal.pone.0218743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roy S, Ueda M, Kadowaki K, Tsutsumi N. Different status of the gene for ribosomal protein S16 in the chloroplast genome during evolution of the genus Arabidopsis and closely related species. Genes & genetic systems. 2010;85:319–326. doi: 10.1266/ggs.85.319. [DOI] [PubMed] [Google Scholar]
- 77.Asaf S, et al. Comparative analysis of complete plastid genomes from wild soybean (Glycine soja) and nine other Glycine species. PLOS ONE. 2017;12:e0182281. doi: 10.1371/journal.pone.0182281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Niu Y-T, et al. Combining complete chloroplast genome sequences with target loci data and morphology to resolve species limits in Triplostegia (Caprifoliaceae) Molecular Phylogenetics and Evolution. 2018;129:15–26. doi: 10.1016/j.ympev.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 79.Amiryousefi A, Hyvönen J, Poczai P. The chloroplast genome sequence of bittersweet (Solanum dulcamara): Plastid genome structure evolution in Solanaceae. PLOS ONE. 2018;13:e0196069. doi: 10.1371/journal.pone.0196069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete chloroplast sequence generated and analyzed during the current study are available in GenBank, https://www.ncbi.nlm.nih.gov (accession numbers are described in the text).