Abstract
Maize is an excellent nutritional source and is consumed as a staple food in different parts of the world, including India. Developing a maize genotype with a combination of higher lysine and tryptophan, along with β-carotene, can help alleviate the problem of protein-energy malnutrition (PEM) and vitamin A deficiency (VAD). This study is aimed at improving lysine and tryptophan content by transferring opaque-2 (o2) gene from donor HKI163 to β-carotene-rich inbred lines viz., UMI1200β+ and UMI1230β+. For this purpose, F1, BC1F1, BC2F1, BC2F2, and BC2F3 plants were developed using an o2 line HKI163 and two β-carotene-rich inbred lines, UMI1200β+ and UMI1230β+, as the parents. Foreground selection using the associated marker umc1066 for the o2 gene and the marker crtRB1 3′TE for the crtRB1 gene was used to select the target genes. A total of 236 simple sequence repeat (SSR) markers distributed evenly across the maize genome were employed for the background selection. To fix the crtRB1 allele in the BC1F1 stage, individual plants homozygous at the crtRB1 locus and heterozygous at the o2 locus were selected and used for backcrossing to produce BC2F1 plants. Furthermore, the selected heterozygous BC2F1 plants from both crosses were selfed to obtain the BC2F2 plants, which were then selected for the target gene and selfed to generate the BC2F3 lines. From each cross, five improved lines with homozygous marker alleles for the crtRB1 and o2 genes with a recurrent parent genome (RPG) recovery ranging from 86.75 to 91.21% in UMI1200β+×HKI163 and 80.00 to 90.08% in UMI1230β+×HKI163 were identified. The improved lines had good agronomic performance and possessed high lysine (0.294–0.332%), tryptophan (0.073–0.081%), and β-carotene (6.12–7.38 µg/g) content. These improved lines can be used as genetic resources for maize improvement.
Keywords: β-carotene, crtRB1, marker-assisted backcross breeding, opaque-2, quality protein maize
Introduction
Maize (Zea mays L.) is a staple food crop and currently grown in more than 150 countries, with a total harvest area of approximately 187 million hectares, producing 1138 million tonnes worldwide (FAOSTAT, 2018). It has good nutritional value, that is, 68.5% carbohydrates, 8% fat, 4% ash, 3% crude fiber, and 16.5% protein (Ullah et al., 2010). In addition, maize carotenoids contain both provitamin A (뗐α-carotene, β-carotene, and β-cryptoxanthin) and non-provitamin A (lutein and zeaxanthin) components. Maize, therefore, is of special importance for the nutrition of people from many countries in Africa, Asia, and Latin America, where protein-energy malnutrition (PEM) and vitamin A deficiency (VAD) affect more than a billion people. The demand for maize has steadily increased over the past decades and is expected to continue to rise in the forthcoming years, at least up until 2050 (Rosegrant et al., 2009). However, normal maize protein possesses low nutritional significance to humans because of very limited amounts of major amino acids, such as lysine (1.6–2.6%) and tryptophan (0.2–0.6%) (Moro et al., 1996), which is less than half of the recommended dose specified for human nutrition. Over the past three decades, many natural maize mutants associated to quality protein maize (QPM) with higher lysine and tryptophan content have been identified, that is, opaque-2 (o2) in chromosome 7 (Mertz et al., 1964), floury-2 (fl2) in chromosome 8 (Nelson et al., 1965), opaque-7 (o7) in chromosome 8 (Ma and Nelson, 1975), opaque-6 (o6) in chromosome 8 (McWhirter, 1971), and floury-3 (fl3) in chromosome 8 (Ma and Nelson, 1975). Among them, o2 mutant has been more popular and widely utilized in breeding programs for the improvement of protein quality. The recessive o2 allele improves the endosperm lysine and tryptophan levels by nearly two-fold. The gene-linked simple sequence repeat (SSR) markers umc1066, phi112, and phi057 have been used to identify the o2 gene (Yang et al., 2005; Gupta et al., 2013; Surender et al., 2017).
VAD is one of the serious health issues in developing and low-income countries and critically affects over 7 million pregnant women and 125 million children (Giuliano, 2017). β-Carotene is the best provitamin A (vitamin A precursor), and maize is a predominant source of β-carotene; however, very few maize varieties are rich in β-carotene, and many exhibiting varieties are inherently deficient in β-carotene (Muthusamy et al., 2014). Yan et al. (2010) revealed that crtRB1 is a major gene responsible for the β-carotene content in maize. This gene is positioned at chromosome 10 and encodes β-carotene hydroxylase, which is responsible for the biosynthesis of lycopene. Association mapping approach led to the identification of three polymorphisms, 5’TE (in the 5’-Untranslated Region), InDel4 (in the coding region), and 3’TE (spanning the sixth exon and 3’-Untranslated Region), in the crtRB1 gene that were significantly influencing the β-carotene content. Since then, polymerase chain reaction (PCR)-based codominant markers were developed based on these polymorphisms, and these markers aided breeders to identify and develop higher β-carotene content lines using marker-assisted selection (MAS). Moreover, Yan et al. (2010) reported the 3’TE favorable allele (allele 1, 543 bp) that is responsible for reduced transcript expression of the gene associated with higher β-carotene content, with an average increase of 6.50 μg/g in the maize endosperm in comparison with the unfavorable allelic class. Recently, this allele-based marker was successfully used to detect the crtRB1 gene in diverse maize lines (Muthusamy et al., 2014; Zunjare et al., 2018; Sagare et al., 2019).
To date, numerous maize hybrids with either provitamin A or QPM have been released and commercialized, but genotypes with both the nutritional traits are very limited. This situation necessitates developing maize genotypes with the combination of QPM and provitamin A. Our previous attempts have led to the development of two β-carotene-rich inbred lines viz., UMI1200β+ and UMI1230β+. In this study, our objective was aimed to introgress the o2 gene from HKI163 into UMI1200β+ and UMI1230β+. We, therefore, applied marker-assisted backcross (MAB) breeding using gene-specific markers for foreground selection and polymorphic SSRs for background selection. Our goal was to obtain innovative breeding materials with high β-carotene, lysine, and tryptophan contents.
Materials and Methods
Plant Genetic Materials
HKI163 is an inbred line containing the opaqueness gene (o2). Its grain lysine content is 0.340% in protein, and its tryptophan content is 0.082% in protein (Zunjare et al., 2018). It was obtained from Chaudhary Charan Singh Haryana Agricultural University, Uchani, India. UMI1200β+ and UMI1230β+ are improved inbred lines containing the β-carotene-associated gene crtRB1, with a grain lysine content of 0.130 and 0.150%, respectively, and tryptophan content of 0.024 and 0.029%, respectively. These β-carotene-rich inbred lines were developed by transferring crtRB1 gene from donor HP46715 (CIMMYT, Mexico) to local popular inbred lines viz., UMI1200 and UMI1230. The β-carotene contents of UMI1200β+ and UMI1230β+ were 9.073 and 9.232 µg/g, respectively.
Development of Backcross Progenies
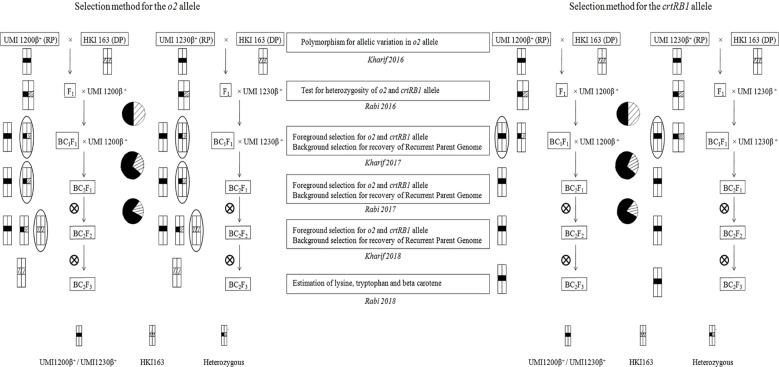
MAB breeding scheme that includes crossing, backcrossing, and selfing was undertaken as mentioned in Figure 1 . Backcross progenies were developed by crossing UMI1200β+ and UMI1230β+ (recurrent parents) with HKI163 (donor parent) following two cycles of backcrosses during 2016 to 2019. UMI1200β+ and UMI1230β+ were used as recurrent parents and crossed with HKI163 (donor) for developing F1 plants. Then, F1 plants were confirmed by foreground selection with crtRB1 and o2-linked markers. These F1 plants were used as the male parents to develop the BC1F1s. Likewise, another round of backcross was followed for UMI1200β+×HKI163 and UMI1230β+×HKI163 to develop BC2F1s using MAB breeding to reduce the linkage drag and to increase the recurrent parent genome percentage. Furthermore, selected BC2F1 plants that were heterozygous at the o2 loci and homozygous at the crtRB1 loci were self-pollinated to produce BC2F2 plants and BC2F3 plants.
Figure 1.
Scheme for the development of o2 and crtRB1 genes-derived improved lines using marker-assisted foreground and background selection.
Genomic DNA Isolation and PCR Analysis
Young leaf tissues from two-week-old plants were ground into powder using liquid nitrogen and stored at -80°C. Genomic DNA was isolated using the cetyl trimethylammonium bromide (CTAB) method (Murray and Thompson, 1980). The DNA was checked for its quantity and quality on a 0.8% agarose gel. The PCR for crtRB1 3′TE gene-specific and SSR primers and agarose gel electrophoresis were carried out following the method by Muthusamy et al. (2014) and Pukalenthy et al. (2019).
Foreground and Background Selection
o2 gene- and crtRB1 gene-linked markers were used for the foreground selection in backcross and selfed lines ( Table 1 ). Based on marker polymorphism between donor and recurrent parents, three SSR markers, umc1066, phi 112, and phi057, linked to the o2 gene and crtRB1 3′TE, which is linked to the crtRB1, were employed for foreground selection. For the background selection, a total of 236 SSR markers distributed on all 10 chromosomes of maize genome were used to identify polymorphic markers between the donor and recurrent parents. Furthermore, the SSR markers that showed polymorphism among the parents were used in the background selection to determine the recurrent parent genome (RPG) recovery percentage at each backcross generation. All of the SSR primer sequences used in background selection were obtained from the maize genome database (www.maizegdb.org) and synthesized by Eurofin Ltd, Bangalore, India.
Table 1.
Sequence information of the markers used for polymorphic studies and foreground screening.
| S. No. | Marker name | Forward sequence (5’-3’) | Reverse sequence (5’-3’) | Annealing |
|---|---|---|---|---|
| 1 | phi112 | TGCCCTGCAGGTTCACATTGAGT | AGGAGTACGCTTGGATGCTCTTC | 53°C |
| 2 | umc1066 | ATGGAGCACGTCATCTCAATGG | AGCAGCAGCAACGTCTATGACACT | 60°C |
| 3 | phi 057 | CTCATCAGTGCCGTCGTCCAT | CAGTCGCAAGAAACCGTTGCC | 63°C |
| 4 | crtRB1 | ACACCACATGGACAAGTTCG | ACACTCTGGCCCATGAACAC (R1) | 62°C -54°Ca |
| ACAGCAATACAGGGGACCAG (R2) | 54°C |
In total of 19 cycles, reduction of temperature by 0.5°C on each cycle starting from the initial 54–62°C.
Kernel Modification
The parents and heterozygous plant (O2/o2) seeds from backcrossed and selfed progenies (BC1F1, BC2F1, BC2F2, and BC2F3) were harvested and examined for the kernel modification using a standard light box screening method (Vasal et al., 1980). Maize kernels were categorized into five types viz., type 1, not opaque; type 2, 25% opaqueness; type 3, 50% opaqueness; type 4, 75% opaqueness; and type 5, 100% opaqueness (Vivek et al., 2008). In all of the generations, the kernels with 25% opaqueness were selected and forwarded to the next generation to fix the o2 allele in its homozygous recessive state and to reduce the undesirable traits caused by the modifier genes acting in the maize endosperm.
Investigation of Morphological Traits in Improved Lines
For the BC2F3 improved lines, observations for 15 morphological traits that were categorized and presented chronologically according to the plant stage data were taken using standard maize descriptors formulated by the International Board for Plant Genetic Resources (IBPGR) (Anonymous, 1991). Morphological traits viz., days to tasselling (days), days to silking (days), plant height (in centimeters), ear height (in centimeters), tassel length (in centimeters), number of tassel branches, leaf length (in centimeters) and leaf width (in centimeters), cob length (in centimeters), cob girth (in centimeters), number of kernel rows per cob, number of kernels per row, cob weight (in grams), single plant yield (in grams), and 100-kernel weight (in grams) were taken.
Estimation of Lysine, Tryptophan, and β-Carotene Contents
The lysine, tryptophan, and β-carotene contents were estimated in seeds of BC2F3 improved lines. The shelled seeds taken for estimation were shade dried and stored at 22–26°C before the analysis. Lysine and tryptophan contents in the endosperm were estimated according to the method described by Galicia et al. (2008). The estimations were done with two replications consisting of two blanks, four checks, and the samples using the spectrophotometer V- 770 UV-VIS-NIT (Japan). The absorbances of lysine and tryptophan were recorded at 390 and 560 nm, respectively. The estimated lysine and tryptophan values were measured with the unit (in percent) (Moro et al., 1996). β-Carotene extraction was done according to the method described by Kurilich and Juvik (1999). The β-carotene content was estimated by high-performance liquid chromatography (HPLC), and samples were eluted by C30 column (5 μm, 4.6 × 250 mm). The mobile phase was composed of acetonitrile:dichloromethane:methanol (75:20:5). The retention and the spectrum of the carotenoid compounds were found to have a flow rate of 0.4 ml/min and were compared to those of the standard (β-carotene standard-M/s Sigma Aldrich, India). Furthermore, it was reconstituted in the acetonitrile mixture in three different concentrations (1, 10, and 100 ppm).
Statistical Analysis
In BC1F1, BC2F1, and BC2F2 generations, the segregation distortion was studied by chi-square analysis for the deviation from the expected Mendelian ratio. In the background selection, the amplicons were scored as A for recurrent parent, B for donor parent, and H for heterozygous plants. The recovery percentage of the recurrent genome was calculated using the formula RPG (%) = [A + (0.5H)/(A + B + H)] × 100 (Benchimol et al., 2005).
Results
Development of Maize Inbred Lines With the O2 and crtRB1 Genes
Three SSR markers, umc1066, phi112, and phi057, located within the o2 gene were investigated for their polymorphisms among the donor HKI163 and the two recurrent parents viz., UMI1200β+ and UMI1230β+. Among them, umc1066 was found to be polymorphic between the donor and each of the two recurrent parents. This informative SSR marker was further used for the foreground selection. F1 progenies were produced from two independent crosses of UMI1200β+×HKI163 and UMI1230β+×HKI163. BC1F1 progenies were obtained by backcrossing the F1 plants with UMI1200β+ and UMI1230β+ as the recurrent parents. In the BC1F1 generation, individual plants homozygous at the crtRB1 and heterozygous at the o2 locus were identified using the crtRB1 and o2-gene specific markers and utilized for next backcrossing with the recurrent parent. Furthermore, BC2F1 progenies were obtained from the selected BC1F1 plants based on the dual-selection procedure involving foreground selection and light box screening. Applying similar selection procedures and selfing, progenies of BC2F1 generation were advanced to BC2F2 ( Figure 2 ) and BC2F3. Finally, from each cross, five BC2F3 lines with homozygous marker alleles for the CrtRB1 and o2 genes were developed ( Figure 3 ). The segregation patterns of backcross progenies are presented in Table 2 .
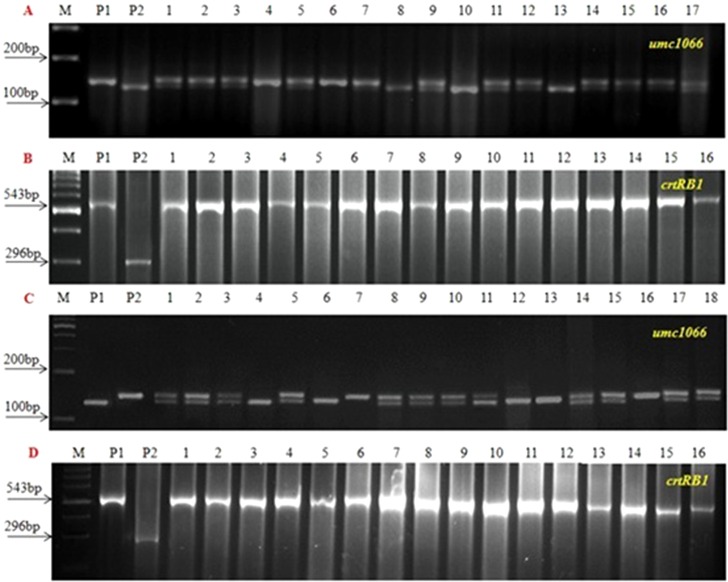
Figure 2.
Foreground screening in BC2F2 progenies. (A and B) UMI1200β+xHKI163 (C and D) UMI1230β+xHKI163, (M) Marker 100bp, (P1) UMI1200β+/UMI1230β+, (P2) HKI163 and (1-16) BC2F2 progenies.
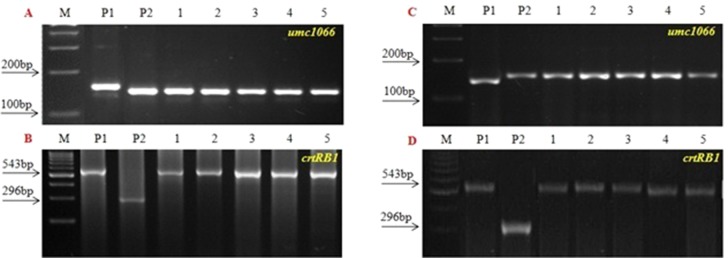
Figure 3.
PCR screening of five improved BC2F3 lines using o2 SSR and crtRB1 gene specific marker. (A and B) UMI1200β+ × HKI163 and (C and D) UMI1230β+ × HKI163. (M) Marker 100 bp, (P1) UMI1200β+/UMI1230β+, (P2) HKI163. (1-5) Improved lines from UMI1200β+×HKI163 (1-DBT6-1-5/25-8/25-4/25-4/25, 2- DBT6-1-5/25-8/25-9/25-9/25, 3- DBT6-1-5/25-10/25-15/25-15/25, 4- DBT6-1-5/25-10/25-17/25-17/25, 5- DBT6-1-5/25-14/25-11/25-11/25) and UMI1230β+×HKI163 (1- DBT7-1-6/25-9/25-37/25-37/25, 2- DBT7-1-6/25-9/25-57/25-57/25, 3- DBT7-1-6/25-12/25-23/25-23/25, 4- DBT7-1-6/25-27/25-3/25-3/25, 5- DBT7-1-6/25-27/25-67/25-67/25).
Table 2.
Segregation pattern of o2 allele in the backcross and selfed generation of UMI1200β+×HKI163 and UMI1230β+×HKI163.
| S. No. | Cross | Generation | No. of plants genotyped | Genotypic class | χ2 | P value | ||
|---|---|---|---|---|---|---|---|---|
| No. of A | No. of H | No. of B | ||||||
| 1 | UMI1200β+ × HKI163 | BC1F1 | 197 | 74 | 123 | 0 | 12.18782** | 0.000481 |
| BC2F1 | 194 | 84 | 110 | 0 | 03.484536ns | 0.061945 | ||
| BC2F2 | 126 | 33 | 082 | 11 | 19.143** | 0.000069 | ||
| 2 | UMI1230β+ × HKI163 | BC1F1 | 175 | 40 | 135 | 0 | 51.5714286** | 0.00006903 |
| BC2F1 | 100 | 43 | 057 | 0 | 01.96ns | 0.161513 | ||
| BC2F2 | 106 | 36 | 040 | 30 | 07.566ns | 0.0294 | ||
**Significant different (p < 0.01), the markers were significantly distorted from the expected segregation ratio (1:2:1); nsnon-significant (p ≥ 0.01), the markers were non-significant and fitted to the normal Mendalian ratio.
SSR-Based Genetic Background Analysis of Improved Lines
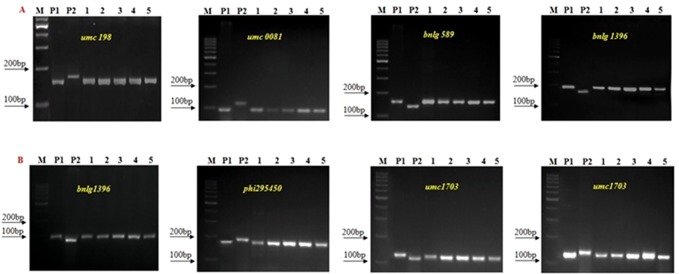
A set of 236 SSR markers distributed uniformly across the maize genome was used in polymorphism screening to select polymorphic markers between donor and recurrent parents. Among them, 104 and 107 SSR markers showed polymorphism between UMI1200β+ and HKI163 and UMI1230β+ and HKI163, respectively. The polymorphism percentage was recorded as 44.6 and 49.57%, respectively. Furthermore, these polymorphic markers were employed to screen the progenies derived from backcross and selfed generation for the recovery of RPG ( Figure 4 ). In BC1F1 generation, a total of 22 and 18 foreground positive plants from UMI1200β+×HKI163 and UMI1230β+×HKI163 were screened, which showed a recovery of RPG of 54.81% and 53.21%, respectively. Furthermore, the recovery of RPG increased in subsequent generations. The 22 and 31 selected positive plants from UMI1200β+×HKI163 and UMI1230β+×HKI163 in BC2F1 showed 82.21 and 79.81%, of RPG, respectively. Eight and six positive plants from UMI1200β+×HKI163 and UMI1230β+×HKI163 in BC2F2 showed 87.48 and 86.51% recovery of RPG, respectively. The highest recoveries of RPG 91.21% and 90.08% were obtained in each of five BC2F3 plants from UMI1200β+×HKI163 and UMI1230β+×HKI163.
Figure 4.
Background Screening of improved lines using SSR makers. (A) 1200β+xHKI163, (B) UMI1230β+xHKI163. (M) Marker 100bp, (P1) UMI 1200β+/UMI 1230β+, (P2) HKI163, (1-5) Improved lines from 1200β+xHKI163 (1-DBT6-1-5/25-8/25-4/25-4/25, 2-DBT6-1-5/25-8/25-9/25-9/25, 3- DBT6-1 5/25-10/25-15/25-15/25, 4-DBT6-1-5/25-10/25-17/25-17/25, 5- DBT6-1-5/25-14/25-11/25-11/-25) and 1230β+xHKI163(1- DBT7-1-6/25-9/25-37/25-37/25, 2-DBT6-1-6/25-9/25-57/25-57/25, 3- DBT7-1-6/25-12/25-23/25-23/25, 4- DBT7-1-6/24-27/25-3/25-3/25, 5-DBT7-1-6/25-27/25-67/25-67/25).
Kernel Modification
Opaqueness is the indicator for the presence of o2 allele, it is also tightly linked to the o2 gene, selecting the kernels along with the least opaqueness from generation to generation ensures that the o2 gene is fixed in its homozygous recessive state. Thus, we observed the opaqueness in selected foreground positive progenies from backcrossed and selfed progenies, along with HKI163, UMI1200β+, and UMI1230β+ for kernel modification. HKI163 kernels showed 25 and 50% opaqueness, whereas UMI1200β+ and UMI1230β+ exhibited 0% opaqueness. BC1F1, BC2F1, and BC2F2 progenies showed 0–100% opaqueness. Among them, progenies showing 25% were further selected and advanced to next generation, whereas the remainder were rejected. In maize, the endosperm modifier genes play a major role to produce undesirable characteristics, which affect the crop yield. Thus, we selected the progenies with 25% opaqueness to reduce the effect of the o2 modifier gene action. As a result, the recessive allele of o2/o2 was fixed in maize kernels and all of the BC2F3 lines showed 25% opaqueness ( Figure 5 ).
Figure 5.
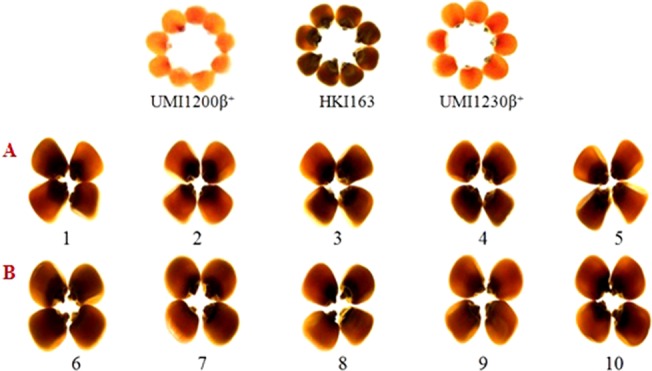
Endosperm modification of BC2F3 (A and B) of UMI1200β+xHKI163 and UMI1230β+xHKI163. (A) Five improved lines form the cross UMI1200β+xHKI163(1-DBT6-1-5/25-8/25-4/25-4/25, 2-DBT6-1-5/25-8/25-9/25-9/25, 3-DBT6-1-5/25-10/25-15/25-15/25, 4-DBT6-15/25-10/25-17/25-17/25, 5-DBT6-1-5/25-14/25-11/25-11/25) (B) Five improved lines from the cross UMI1230β+XHKI163(6-DBT7-1/25-9/25-37/25-37/25, 7-DBT7-1-6/25-9/25-57/25-57/25, 8-DBT7-1-6/25-12/25-23/25-23/25, 9-DBT7-1-6/25-27/25-3/25-3/25, 10-DBT7-1-6/25-27/25-67/25-67/25).
Morphological Characteristics of Improved Lines
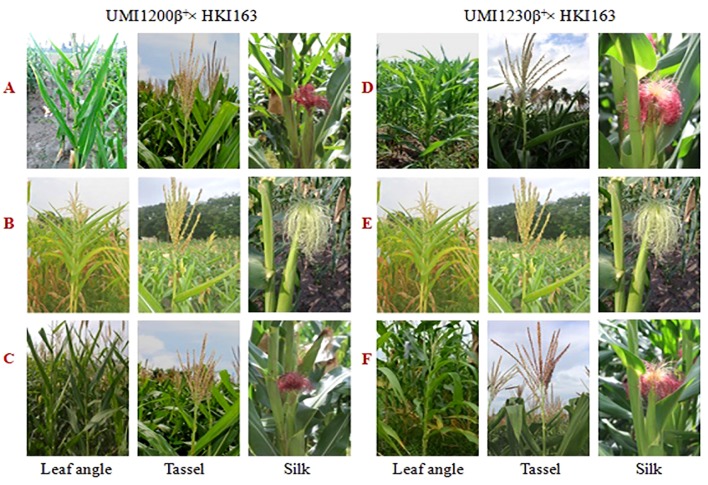
The morphological traits of the improved lines along with their donor and recurrent parents were presented in Table 3 . Five UMI1200β+×HKI163-based improved lines showed phenotypic resemblance ranging from 71.43% (number of kernels per row) to 98.38% (days to silking). Among them, DBT6-1-5/25-10/25-17/25-17/25 and DBT6-1-5/25-14/25-11/25-11/25 possessed high phenotypic resemblance with the recurrent parent. For instance, days to tasselling, days to silking, plant height, ear height, tassel length, cob girth, cob weight, 100-kernel weight, and single plant yield showed more than 90% similarity to UMI1200β+. Likewise, five UMI1230β+×HKI163-based improved lines showed phenotypic resemblance ranging from 82.43% (ear height) to 99.66% (100-kernel weight). DBT7-1-6/25-12/25-23/25-23/25 showed maximum similarity with the recurrent parent, followed by DBT7-1-6/25-27/25-67/25-67/25. These two lines exhibited more than 90% similarity to UMI1230β+ for the traits days to tasselling, days to silking, plant height, ear height, tassel length, leaf length, cob length, cob girth, cob weight, 100-kernel weight, and single plant yield ( Figure 6 ).
Table 3.
Comparison of BC2F3 improved lines from UMI1200β+×HKI163 and UMI1230β+×HKI163 with the parents for the recovery percentage of morphological traits.
| Morphological traits | UMI1200β+
(Recurrent parent) |
HKI163 (Donor parent) |
Identified positive lines | Recovery percentage (%) for morphological trait | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (UMI1200β+ × HKI163) | DBT6-1-5/25-8/25-4/25-4/25 | DBT6-1-5/25-8/25-9/25-9/25 | DBT6-1-5/25-10/25-15/25-15/25 | DBT6-1-5/25-10/25-17/25-17/25 | DBT6-1-5/25-14/25-11/25-11/25 | DBT6-1-5/25-8/25-4/25-4/25 | DBT6-1-5/25-8/25-9/25-9/25 | DBT6-1-5/25-10/25-15/25-15/25 | DBT6-1-5/25-10/25-17/25-17/25 | DBT6-1-5/25-14/25-11/25-11/25 | ||
| Days to tasseling (days) | 59.00 | 63.00 | 56.00 | 57.00 | 58.00 | 58.00 | 57.00 | 94.91 | 82.60 | 98.30 | 98.30 | 96.61 |
| Days to silking (days) | 62.00 | 65.00 | 58.00 | 60.00 | 61.00 | 60.00 | 59.00 | 93.54 | 96.77 | 98.38 | 96.77 | 95.16 |
| Plant height (cm) | 126.88 | 110.00 | 118.00 | 121.40 | 123.50 | 119.90 | 122.70 | 93.00 | 95.68 | 97.33 | 94.49 | 96.70 |
| Ear height (cm) | 71.00 | 62.00 | 68.00 | 67.70 | 69.30 | 68.30 | 68.00 | 95.77 | 95.35 | 97.60 | 96.19 | 95.77 |
| Tassel length (cm) | 21.00 | 26.20 | 18.00 | 19.20 | 18.10 | 19.50 | 20.30 | 85.71 | 91.42 | 86.19 | 92.38 | 96.66 |
| Number of tassel branches | 12.00 | 16.00 | 10.00 | 11.00 | 10.00 | 11.00 | 10.00 | 83.33 | 91.66 | 83.33 | 91.66 | 83.33 |
| Leaf length (cm) | 61.65 | 63.00 | 58.20 | 59.40 | 59.20 | 58.00 | 54.70 | 94.40 | 96.35 | 96.02 | 94.07 | 88.72 |
| Leaf width (cm) | 06.10 | 06.00 | 05.80 | 05.80 | 05.70 | 05.60 | 05.00 | 95.08 | 95.08 | 93.44 | 91.80 | 81.96 |
| Cob length (cm) | 15.20 | 17.00 | 13.00 | 13.80 | 14.00 | 13.00 | 12.40 | 85.52 | 90.78 | 92.10 | 85.52 | 81.57 |
| Cob girth(cm) | 13.80 | 13.00 | 12.00 | 11.50 | 13.00 | 12.70 | 13.10 | 86.95 | 83.33 | 94.20 | 92.02 | 94.92 |
| Number of kernel rows per cob | 14.00 | 12.00 | 12.00 | 10.00 | 12.00 | 10.00 | 10.00 | 85.71 | 85.71 | 71.43 | 85.71 | 71.43 |
| Number of kernels per row | 21.00 | 26.00 | 18.00 | 17.00 | 17.00 | 18.00 | 17.00 | 85.71 | 80.95 | 80.95 | 85.71 | 94.92 |
| Cob weight (g) | 108.00 | 112,00 | 98.00 | 92.80 | 94.30 | 101.00 | 97.90 | 90.74 | 85.92 | 87.31 | 93.51 | 90.64 |
| 100-kernel weight (g) | 24.00 | 22.00 | 23.00 | 22.30 | 22.80 | 23.10 | 22.80 | 95.83 | 92.91 | 95.00 | 96.25 | 95.00 |
| Single plant yield (g) | 90.67 | 57.00 | 82.11 | 83.07 | 84.11 | 86.21 | 87.32 | 90.55 | 91.61 | 92.76 | 95.08 | 96.31 |
| (UMI1200β+ × HKI163) | UMI1230β+ (Recurrent parent) | HKI163 (Donor parent) |
DBT7-1-6/25-9/25-37/25-37/25 | DBT7-1-6/25-9/25-57/25-57/25 | DBT7-1-6/25-12/25-23/25-23/25 | DBT7-1-6/25-27/25-3/25-3/25 | DBT7-1-6/25-27/25-67/25-67/25 | DBT7-1-6/25-9/25-37/25-37/25 | DBT7-1-6/25-9/25-57/25-57/25 | DBT7-1-6/25-12/25-23/25-23/25 | DBT7-1-6/25-27/25-3/25-3/25 | DBT7-1-6 /25-27/25-67/25-67/25 |
| Days to tasseling (days) | 61.00 | 63.00 | 58.00 | 58.00 | 59.00 | 60.00 | 58.00 | 95.08 | 95.08 | 96.72 | 98.36 | 95.08 |
| Days to silking (days) | 63.00 | 65.00 | 61.00 | 60.00 | 62.00 | 62.00 | 60.00 | 96.82 | 95.23 | 98.41 | 98.41 | 95.23 |
| Plant height (cm) | 112.00 | 110.00 | 110.50 | 109.70 | 104.60 | 101.00 | 105.20 | 98.66 | 97.94 | 93.39 | 90.17 | 93.92 |
| Ear height (cm) | 74.00 | 62.00 | 61.00 | 72.00 | 71.00 | 72.10 | 72.90 | 82.43 | 97.29 | 95.94 | 97.43 | 98.51 |
| Tassel length (cm) | 24.10 | 26.20 | 23.40 | 22.80 | 23.70 | 23.90 | 22.80 | 97.09 | 94.60 | 98.34 | 99.17 | 94.60 |
| Number of tassel branches | 14.00 | 16.00 | 13.00 | 12.00 | 13.00 | 12.00 | 12.00 | 92.85 | 85.71 | 92.85 | 85.71 | 85.71 |
| Leaf length (cm) | 61.00 | 63.00 | 58.40 | 60.20 | 59.00 | 59.20 | 58.90 | 95.73 | 98.68 | 96.72 | 97.04 | 96.55 |
| Leaf width (cm) | 06.50 | 06.00 | 06.30 | 6.10 | 05.70 | 05.80 | 05.90 | 96.92 | 93.84 | 87.69 | 89.23 | 90.76 |
| Cob length (cm) | 16.00 | 17.00 | 14.00 | 15.60 | 14.90 | 15.80 | 15.30 | 87.50 | 97.50 | 93.12 | 98.75 | 95.62 |
| Cob girth (cm) | 12.50 | 13.00 | 11.00 | 11.80 | 12.10 | 11.60 | 12.00 | 88.00 | 94.40 | 96.80 | 92.80 | 96.00 |
| Number of kernel rows per cob | 16.00 | 12.00 | 14.00 | 14.00 | 14.00 | 14.00 | 14.00 | 87.50 | 87.50 | 87.50 | 87.50 | 87.50 |
| Number of kernels per row | 23.00 | 26.00 | 22.00 | 22.00 | 22.00 | 21.00 | 19.00 | 95.65 | 95.65 | 95.65 | 91.30 | 82.60 |
| Cob weight (g) | 124.70 | 112.00 | 122.60 | 118.30 | 113.80 | 120.90 | 121.00 | 98.31 | 94.86 | 91.25 | 96.95 | 97.03 |
| 100-kernel weight (g) | 29.50 | 22.00 | 27.40 | 28.10 | 28.90 | 29.40 | 28.70 | 92.88 | 95.25 | 97.96 | 99.66 | 97.28 |
| Single plant yield (g) | 76.98 | 57.00 | 72.11 | 69.43 | 71.67 | 72.11 | 73.31 | 93.67 | 90.19 | 93.10 | 93.67 | 95.23 |
Figure 6.
Morphological traits of parents and improved lines of UMI1200β+xHKI163 and UMI1230β+xHKI163. (A) UMI1200β+, (B) HK163, (C) DBT6-1-5/25-10/25-17/25- 17/25, (D) UMI1230β+, (E) HKI163 (F) DBT7-1-6/25-12/25-23/25-23/25.
Analysis of Lysine, Tryptophan, and β-Carotene
All of the improved lines showed that lysine and tryptophan content increased many-fold over for both the β-carotene-rich parents viz., UMI1200β+ and UMI1230β+. Lysine and tryptophan contents varied from 0.294 to 0.332% and 0.073 to 0.081%, with an average of 0.314 and 0.077%. Among the improved lines, DBT 7-1-6/25-27/25-3/25-3/25 from UMI1230β+×HKI163 possessed higher levels of lysine (0.332%) and tryptophan (0.081%). Furthermore, accumulation of the β-carotene content was estimated in improved lines, which ranged from 6.127 to 7.387 µg/g with an average of 6.80 µg/g and higher than the QPM parent HKI163. DBT6-1-5/25-8/25-4/25-4/25 from UMI1200β+×HKI163 was found to have a high β-carotene content of 7.387 µg/g among improved lines. The lysine, tryptophan, and β-carotene contents of the improved lines are presented in Table 4 .
Table 4.
Lysine and tryptophan concentrations in parents and BC2F3 improved lines from UMI1200β+×HKI163 and UMI1230β+×HKI163.
| Carotenoid/Amino acids | UMI1200β+ | UMI1230β+ | HKI163 | (UMI1200β+ × HKI163) | (UMI1230β+ × HKI163) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DBT6-1-5/25-8/25-4/25-4/25 | DBT6-1-5/25-8/25-9/25-9/25 | DBT6-1-5/25-10/25-15/25-15/25 | DBT6-1-5/25-10/25-17/25-17/25 | DBT6-1-5/25-14/25-11/25-11/25 | DBT7-1-6/25-9/25-37/25-37/25 | DBT7-1-6/25-9/25-57/25-57/25 | DBT7-1-6/25-12/25-23/25-23/25 | DBT7-1-6/25-27/25-3/25-3/25 | DBT7-1-6/25-27/25-67/25-67/25 | ||||
| β- carotene (µg/g) | 9.073 | 9.232 | 0.800 | 7.387 | 6.127 | 6.665 | 7.187 | 6.865 | 7.123 | 7.265 | 6.321 | 6.812 | 6.312 |
| Lysine (%) | 0.130 | 0.150 | 0.340 | 0.294 | 0.312 | 0.299 | 0.322 | 0.331 | 0.298 | 0.317 | 0.308 | 0.332 | 0.327 |
| Tryptophan (%) | 0.024 | 0.029 | 0.082 | 0.077 | 0.079 | 0.080 | 0.073 | 0.080 | 0.073 | 0.077 | 0.079 | 0.081 | 0.075 |
Discussion
The Value of the Pyramiding O2 and crtRB1 Genes
Lysine, tryptophan, and β-carotene are the key nutritional traits in maize. The genetic nature and environmental factors have an influence on these traits. crtRB1 and o2 genes present on chromosomes 10 and 7 (Mertz et al., 1964; Vasal, 2000; Yang et al., 2004) provide increased β-carotene, lysine, and tryptophan contents. Molecular markers linked to these genes are available to facilitate direct selection in the breeding process. In this study, we successfully pyramided the o2 and crtRB1 genes in maize by MAS and several generations of backcrossing. The β-carotene content of the improved lines was increased by five- to six-fold for both crosses when compared to the QPM parent. The lysine and tryptophan contents of the improved lines were increased by two- and seven-fold for both crosses compared to the β-carotene parents. Thus, o2 and crtRB1 genes can work together in the same genetic background to control the content of lysine, tryptophan, and β-carotene.
Development of Improved Lines Through MAB Breeding
Parental polymorphism screening revealed that recurrent parents UMI1200β+ and UMI1230β+ were clearly distinguishable with o2 gene and CrtRB1 gene-linked markers umc1066 and crtRB1 3′TE from the donor line HKI163 and thus were used for foreground selection in the F1, BC1F1, BC2F1, BC2F2, and BC2F3 generations. In foreground selection, F1 and BC1F1 generations screening with crtRB1 allele indicated that all of the genotypes were heterozygous in the F1 generation and the segregation distortion in the BC1F1 generation (Babu et al., 2013). From the BC1F1 generation, the lines were fixed for the crtRB1 allele by selecting the plants with favorable allele (543bp) and rejecting the heterozygous plants with both allele (543bp+296bp). Therefore, no segregation existed for crtRB1 allele in the forwarded generations. Screening for the o2 gene revealed that BC2F1 of UMI1200β+×HKI163 and BC2F1 and BC2F2 of UMI1230β+×HKI163 showed approximately 50% of heterozygous plants with respect to the expected Mendalian ratio (1:1) in the backcross generations and (1:2:1) in the selfed generations. However, segregation distortion was observed in BC1F1 and BC2F2 of UMI1200β+×HKI163 and BC1F1 of UMI1230β+×HKI163. These results are in accordance with the previous reports (Liu et al., 2015; Tripathy et al., 2017; Goswami et al., 2019; Sagare et al., 2019). Furthermore, background analysis using genome-wide SSR markers revealed 91.21 and 90.08% recovery of RPG in each of the five BC2F3 plants from UMI1200β+×HKI163 and UMI1230β+×HKI163 and coupled with the earlier findings (Feng et al., 2015; Sarika et al., 2018).
Characteristics of Improved Lines
In addition to the background selection, phenotypic characterization is also useful to find the recovery percentage of recurrent parents (Manna et al., 2005; Gunjaca et al., 2008; Choudhary et al., 2014; Hossain et al., 2018). Phenotypic characterization among the parents and the improved lines showed more than 90% of recovery of the recurrent parents in morphological traits. Among them, DBT6-1-5/25-10/25-17/25-17/25 and DBT6-1-5/25-14/25-11/25-11/25 from UMI1200β+×HKI163 and DBT7-1-6/25-12/25-23/25-23/25 and DBT7-1-6/25-27/25-67/25-67/25 from UMI1230β+×HKI 163 possessed high phenotypic resemblance (90%) with their recurrent parents. Previously, several studies also reported more than 90% recovery of the recurrent parent characteristics in MAS-derived lines (Surender et al., 2017; Pukalenthy et al., 2019; Sagare et al., 2019). The lysine and tryptophan contents of the improved lines ranged from 0.294 to 0.331% and 0.073 to 0.080% for the cross UMI1200β+×HKI163 and 0.298 to 0.332% and 0.073 to 0.081% for the cross UMI1230β+×HKI163. On the average, lysine and tryptophan contents of the improved lines were 0.314 and 0.077%; they are at par with the QPM parent, three and seven-fold increases from the recurrent parents. Likewise, the average β-carotene contents of the improved lines for UMI1200β+×HKI163 and UMI1230β+×HKI163 were 6.846 and 6.766 µg/g, respectively, which were comparable to the β-carotene parents, six-fold higher than the QPM parent. Similar results were obtained by various studies (Muthusamy et al., 2014; Zunjare et al., 2018; Goswami et al., 2019). Overall, the improved inbred lines gained lysine and tryptophan contents but a slight reduction in β-carotene content (>2 ug) and grain yield. We followed the dual-selection procedure of molecular and light box screening to fix the o2 allele, which is the reason behind increasing lysine and tryptophan contents. We selected the progenies based on the good agronomic performance (>90%) and β-carotene content, even though some of the progenies recorded β-carotene contents at par with the recurrent parents with less agronomic performance. Thus, a slight reduction was observed in β-carotene content (>2 ug) of improved inbred lines. Moreover, introgression of o2 and crtRB1 genes caused a reduction in the grain yield. It is reported that QPM lines have some undesirable characteristics because of the modifier gene action in the endosperm. Thus, we used dual selection procedure to select the progenies and developed the improved inbred lines with less undesirable traits along with o2 and crtRB1 genes. However, it is not possible to stop the modifier gene (o2) activity and remove the undesirable traits completely. It might influence the yield attributing traits and reduces the yield performances. Thus, we obtained a reduction in the grain yield similar to a previous study (Lauderdale, 2000).
In the present study, using MAB breeding approach, we successfully pyramided the o2 and crtRB1 genes and developed the nutrition-rich inbreds, but introgression of multiple genes caused a slight reduction in the yield. To utilize these newly developed inbred lines effectively, our future research focus is on conducting multilocation trial (MLT) in various maize-growing regions and identifying the superior inbred lines to develop new hybrids. In addition, these inbred lines can be used as genetic resources for maize biofortification programs.
Data Availability
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
SN, FH, and VM designed the methods and experiments. SC, BP, DM, and RR developed backcross progenies and managed fieldwork. LJ and VC provided suggestions on experiments and monitored the work. SC, BP, DM, KAd, and KE conducted phenotype and genotype analysis. SC, VS, and KAr performed biochemical analysis. SC, BP, and DM analyzed the data. SC, KAd, and SN drafted the manuscript.
Funding
Financial support of Department of Biotechnology (DBT), Government of India (GOI), through project entitled ‘Enrichment of nutritional quality in maize through molecular breeding’ (BT/PR10922AGII/106/9442014 dt.25.3.2015) and Ministry of Science and Technology, Department of Biotechnology, Government of India DBT’s Twinning program for the NE for the project entitled ‘Marker assisted introgression of LyCE gene for enhanced ProA in maize’ (BT/166/NE/TBP/2011 dt 12.12.2011) is acknowledged. The funders had no role in the work design, data collection and analysis, or decision and preparation of the manuscript.
Conflict of Interest Statement
All authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Acknowledgments
The authors acknowledged the Centre of Innovation, Agricultural College and Research Institute, Tamil Nadu Agricultural University, Madurai, for providing lab and instrumental facilities.
References
- Anonymous (1991). “IBPGR. Descriptors for Maize,” in International Wheat and Maize Improvement Center (Mexico City: International Board for Plant Genetic; ). [Google Scholar]
- Babu R., Rojas N. P., Gao S., Yan J., Pixley K. (2013). Validation of the effects of molecular marker polymorphisms in LcyE and CrtRB1 on provitamin A concentrations for 26 tropical maize populations. Theor. Appl. Genet. 126, 389–399. 10.1007/s00122-012-1987-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchimol L., Souza C. L. D., Jr., Souza A. P. D. (2005). Microsatellite-assisted backcross selection in maize. Genet. Mol. Biol. 28, 789–797. 10.1590/S1415-47572005000500022 [DOI] [Google Scholar]
- Choudhary M., Muthusamy V., Hossain F., Thirunavukkarasu N., Pandey N., Jha S. K., et al. (2014). Characterization of β-carotene rich MAS-derived maize inbreds possessing rare genetic variation in β-carotene hydroxylase gene. Indian J. Genet. 74, 620–623. 10.5958/0975-6906.2014.00900.6 [DOI] [Google Scholar]
- Feng F., Wang Q., Liang C., Yang R., Li X. (2015). Enhancement of tocopherols in sweet corn by marker-assisted backcrossing of ZmVTE4. Euphytica 206, 513–521. 10.1007/s10681-015-1519-8 [DOI] [Google Scholar]
- FAOSTAT (2018). Crop prospects and food situation. Rome: Food and Agriculture Organization of the United Nation. Available from: http://faostat.fao.org (Accessed July 06, 2018).
- Galicia L., Nurit E., Rosales A., Palacios-Rojas N. (2008). Maize nutrition quality and plant tissue analysis laboratory. Lab. Protoc. 2008 10–17. [Google Scholar]
- Giuliano G. (2017). Provitamin A biofortification of crop plants: a gold rush with many miners. Curr. Opin. Biotechnol. 44, 169–180. 10.1016/j.copbio.2017.02.001 [DOI] [PubMed] [Google Scholar]
- Goswami R., Zunjare R. U., Khan S., Baveja A., Muthusamy V., Hossain F. (2019). Marker-assisted introgression of rare allele of β-carotene hydroxylase (crtRB1) gene into elite quality protein maize inbred for combining high lysine, tryptophan and provitamin A in maize. Plant Breed. 138, 174–183. 10.1111/pbr.12676 [DOI] [Google Scholar]
- Gunjaca J., Buhinicek I., Jukic M., Sarcevic H., Vrag olovic A., Kozic Z., et al. (2008). Discriminating maize inbred lines using molecular and DUS data. Euphytica 161, 165–172. 10.1016/j.copbio.2017.02.001 [DOI] [Google Scholar]
- Gupta H. S., Raman B., Agrawal P. K., Mahajan V., Hossain F., Thirunavukkarasu N. (2013). Accelerated development of quality protein maize hybrid through marker-assisted introgression of opaque-2 allele. Plant Breed. 132, 77–82. 10.1111/pbr.12009 [DOI] [Google Scholar]
- Hossain F., Muthusamy V., Pandey N., Vishwakarma A. K., Baveja A., Zunjare R. U., et al. (2018). Marker-assisted introgression of opaque2 allele for rapid conversion of elite hybrids into quality protein maize. J. Genet. 97, 287–298. 10.1007/s12041-018-0914-z [DOI] [PubMed] [Google Scholar]
- Kurilich A. C., Juvik J. A. (1999). Quantification of Carotenoid and Tocopherol Antioxidants in. Zea mays. J. Agric. Food Chem. 47, 1948–1955. 10.1021/jf981029d [DOI] [PubMed] [Google Scholar]
- Lauderdale J. (2000). Issues regarding targeting and adoption of quality protein maize (QPM) in CIMMYT Economics Working Paper 00–02. Mexico D.F. CIMMYT, pp. iv–31. [Google Scholar]
- Liu L., Jeffers D., Zhang Y., Ding M., Chen W., Kang M. S., et al. (2015). Introgression of the crtRB1 gene into quality protein maize inbred lines using molecular markers. Mol. Breed. 35, 154. 10.1007/s11032-015-0349-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Nelson O. E. (1975). Amino acid composition and storage proteins in two new high-lysine mutants in maize. Cereal Chem. 52, 412–419. [Google Scholar]
- Manna R., Okello O. K., Imanywoha J., Pixley K., Edema R. (2005). Enhancing introgression of the opaque-2 trait into elite maize lines using simple sequence repeats. Afr. Crop Sci. J. 13, 215–226. [Google Scholar]
- McWhirter K. S. (1971). A floury endosperm, high lysine locus on chromosome 10. Maize Genet. Coop. Newsletter 45, 184. [Google Scholar]
- Mertz E. T., Bates L. S., Nelson O. E. (1964). Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science 145, 279–280. 10.1126/science.145.3629.279 [DOI] [PubMed] [Google Scholar]
- Moro G. L., Habben J. E., Hamaker B. R., Larkins B. A. (1996). Characterization of the variability in lysine content for normal and opaque2 maize endosperm. Crop Sci. 36, 1651–1659. 10.2135/cropsci1996.0011183X003600060039x [DOI] [Google Scholar]
- Murray M. G., Thompson W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. 10.1093/nar/8.19.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy V., Hossain F., Thirunavukkarasu N., Choudhary M., Saha S., Bhat J. S., et al. (2014). Development of β-carotene rich maize hybrids through marker-assisted introgression of β-carotene hydroxylase allele. PLoS One 9, e113583. 10.1371/journal.pone.0113583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson O. E., Mertz E. T., Bates L. S. (1965). Second mutant gene affecting the amino acid pattern of maize endosperm proteins. Science 150, 1469–1470. 10.1126/science.150.3702.1469 [DOI] [PubMed] [Google Scholar]
- Pukalenthy B., Manickam D., Chandran S., Adhimoolam K., Sampathrajan V., Rajasekaran R., et al. (2019). Incorporation of opaque-2 into ‘UMI 1200’, an elite maize inbred line, through marker-assisted backcross breeding. Biotechnol. Biotechnol. Equip. 1–10. 10.1080/13102818.2018.1556121 [DOI]
- Rosegrant M. W., Ringler C., Sangi S. M., Sulser T. B., Zhu T., Cline S. A. (2009). Model description international model for policy analysis of agricultural commodities and trade. International food policy research institute; Washington. D.C. 12–13. [Google Scholar]
- Sagare D. B., Shetti P., Surender M., Reddy S. S. (2019). Marker-assisted backcross breeding for enhancing β-carotene of QPM inbreds. Mol. Breed. 39, 31. 10.1007/s11032-019-0939-x [DOI] [Google Scholar]
- Sarika K., Hossain F., Muthusamy V., Zunjare R. U., Baveja A., Goswami R., et al. (2018). Marker-assisted pyramiding of opaque2 and novel opaque16 genes for further enrichment of lysine and tryptophan in sub-tropical maize. Plant Sci. 272, 142–152. 10.1016/j.plantsci.2018.04.014 [DOI] [PubMed] [Google Scholar]
- Surender M., Shetti P., Sagare D. B., Durga Rani C. V., Jabeen F., Sudarshan M. R., et al. (2017). Development of QPM version of DHM117 maize hybrid using marker assisted selection. Int. J. Curr. Microbiol. App. Sci. 6, 3275–3289. 10.20546/ijcmas.2017.610.384 [DOI] [Google Scholar]
- Tripathy S. K., Lenka D., Ithape D. M., Maharana M. (2017). Development of opaque-2 introgression line in maize using marker assisted backcross breeding. Plant Gene 10, 26–30. 10.1016/j.plgene.2017.03.005 [DOI] [Google Scholar]
- Ullah I., Ali M., Farooqi A. (2010). Chemical and nutritional properties of some maize (Zea mays L.) varieties grown in NWFP, Pakistan. Pak. J. Nutr. 9, 1113–1117. 10.3923/pjn.2010.1113.1117 [DOI] [Google Scholar]
- Vasal S. K. (2000). The quality protein maize story. Food Nutr. Bull. 21, 445–450. 10.1177/156482650002100420 [DOI] [Google Scholar]
- Vasal S. K., Villegas E., Bjarnason M., Gelaw B., Goertz P. (1980). Genetic modifiers and breeding strategies in developing hard endosperm opaque-2 materials. CAB Direct. 33, 37–73. [Google Scholar]
- Vivek B. S., Krivanek A. F., Palacios Rojas N., Twumasi Afriyie S., Diallo A. O. (2008). Breeding Quality Protein Maize (QPM): Protocols for Developing QPM Cultivars. Mexico D.F. CIMMYT; 8-11 [Google Scholar]
- Yan J., Kandianis C. B., Harjes C. E., Bai L., Kim E. H., Yang X., et al. (2010). Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nat. Genet. 42, 322. 10.1038/ng.551 [DOI] [PubMed] [Google Scholar]
- Yang R., Yan Z., Wang Q., Li X., Feng F. (2018). Marker-assisted backcrossing of lcyE for enhancement of proA in sweet corn. Euphytica 214, 130. 10.1007/s10681-018-2212-5 [DOI] [Google Scholar]
- Yang W., Zheng Y., Ni S., Wu J. (2004). Recessive allelic variations of three microsatellite sites within the O2 gene in maize. Plant Mol. Boil. Rep. 22, 361–374. 10.1007/BF02772679 [DOI] [Google Scholar]
- Yang W., Zheng Y., Zheng W., Feng R. (2005). Molecular genetic mapping of a high-lysine mutant gene (opaque-16) and the double recessive effect with opaque-2 in maize. Mol. Breed. 15, 257–269. 10.1007/s11032-004-5947-8 [DOI] [Google Scholar]
- Zunjare R. U., Hossain F., Muthusamy V., Baveja A., Chauhan H. S., Bhat J. S., et al. (2018). Development of biofortified maize hybrids through marker-assisted stacking of β-carotene hydroxylase, lycopene-ε-cyclase and opaque2 genes. Front. Plant Sci. 9, 178. 10.3389/fpls.2018.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.