Abstract
Overexpression of AKR1B10 correlated with tumorigenesis of many human malignancies; however, the prognostic value of AKR1B10 expression in patients with hepatocellular carcinoma (HCC) still remains controversial. In this analysis, AKR1B10 expression in HCC tumors were evaluated in GEO, TCGA and Oncomine databases, and a survival analysis of AKR1B10 based on TCGA profile was performed. We found that AKR1B10 was significantly overexpressed in tumors compared with nontumors in 7 GEO series (GSE14520, GSE25097, GSE33006, GSE45436, GSE55092, GSE60502, GSE77314) and TCGA profile (all P < 0.05). Meta-analysis in Oncomine database revealed that AKR1B10 was significantly upregulated in cirrhosis, liver cell dysplasia and HCC compared with normal tissues (all P < 0.05). Kaplan-Meier analysis demonstrated that high AKR1B10 in tumors were significantly associated with worse overall survival (OS) in HCC patients (P < 0.05). Subgroup analysis showed that AKR1B10 overexpression were associated with poor 1-year, 3-year and 5-year OS (all P < 0.05). In addition, prognostic values of AKR1B10 upregulation for OS were more significant in HCC with hepatitis-virus-free (P = 0.00055), White race (P = 0.0029) and alcohol-free (P = 0.013), and both in male and female (P = 0.014 and P = 0.034, respectively). In conclusion: AKR1B10 was upregulated in tumors and correlated with worse OS in HCC patients.
Keywords: AKR1B10, hepatocellular carcinoma, survival, outcome
Introduction
Liver cancer, comprising 75%~85% cases of hepatocellular carcinoma (HCC), is predicted to be the sixth most commonly diagnosed cancer and the fourth leading cancer related deaths worldwide in 2018 1-4. In addition, the incidence of HCC will continue to rise over the next 10-20 years and to peak around 2030 based on a SEER registry projects study 3, 5. There has been a marked increase in HCC related annual death rates during the past two decades 3. Precise estimation of prognosis plays a critical role in treatment decision in HCC patients. Thus, to identify reliable prognostic biomarkers and to reveal HCC target for treatment is urgently required 6, 7.
Belongs to the aldo-keto reductase superfamily 8, 9, AKR1B10 is predominantly expressed in the small intestine, colon, liver, thymus and adrenal gland 8, 10. Upregulation of AKR1B10 has been observed in various human malignancies, including non-small cell lung carcinoma 11, pancreatic cancer 12, breast cancer 13, 14, oral squamous cell carcinoma 15, nasopharyngeal carcinoma 16 and HCC 17, 18. Conversely, underexpression of AKR1B10 has been observed in colon 19, gastric 20, endometrial cancer 21 and head and neck cancers 22. However, AKR1B10 in HCC has been controversially discussed. Some researches revealed that negatively correlated with serum alpha-fetoprotein (AFP) level, high AKR1B10 expression was found to be a favorable factor of overall survival and disease-free survival in patients with hepatitis B virus-related HCC 23-25. Another study found that high AKR1B10 expression was an independent risk factor for HCC development 26 and significantly correlated with liver damage25. Thus, to reevaluate the prognostic value of AKR1B10 in HCC patients is of great importance.
To help elucidate the possible relationship between AKR1B10 expression and HCC outcomes, we identified the AKR1B10 expression in GEO, Oncomine and TCGA databases and performed a survival analysis based on TCGA profile, in the hope of providing useful insights into hepatocarcinogenesis and aggressiveness.
Materials and Methods
Data resource and Description
Seven expression microarray series GSE14520, GSE25097, GSE33006, GSE45436, GSE55092, GSE60502 and GSE77314 containing HCC tumor and nontumor samples were downloaded from the Gene Expression Omnibus dataset (GEO, https://www.ncbi.nlm.nih.gov/geo/). Platforms and samples of GEO series were summarized in Table 1.
Table 1.
Details of GEO series included in this analysis
| GEO series | Contributor(s) | Tumor | Nontumor | Platform |
|---|---|---|---|---|
| GSE14520 | Roessler S et al, 2009 | 222 | 212 | Affymetrix Human Genome U133A 2.0 Array / Affymetrix HT Human Genome U133A Array |
| GSE25097 | Zhang C, 2010 | 268 | 243 | Rosetta/Merck Human RSTA Affymetrix 1.0 microarray, Custom CDF |
| GSE33006 | Huang Y et al, 2011 | 3 | 3 | Affymetrix Human Genome U133 Plus 2.0 Array |
| GSE45436 | Hsieh J, 2013 | 97 | 37 | Affymetrix Human Genome U133 Plus 2.0 Array |
| GSE55092 | Melis M et al, 2014 | 49 | 91 | Affymetrix Human Genome U133 Plus 2.0 Array |
| GSE60502 | Kao KJ, 2014 | 18 | 18 | Affymetrix Human Genome U133A Array |
| GSE77314 | Hou G et al, 2016 | 50 | 50 | Illumina Genome Analyzer (Homo sapiens) |
TCGA liver hepatocellular carcinoma mRNA normalized counts data derived from RNAseq Htseq platform were downloaded from Genomic Data Commons Data Portal (https://cancergenome.nih.gov/). TCGA RNAseq data contains 424 samples with 374 primary tumor and 50 solid tissue normal samples. RTCGAToolbox 27 and edgeR 28, 29 packages were used to identify AKR1B10 expression between tumor and normal samples.
Bioinformatics analysis for identifying AKR1B10 expression
Raw.CEL files of the microarray from each GEO dataset were normalized by quantile method of Robust Multichip Analysis (RMA) from R affy package 30. Gene expression of AKR1B10 was performed by tumor and normal comparison from R Limma package 31. Gene expression data of GSE77314 in XLSX form was downloaded from GEO dataset directly.
Studies compared AKR1B10 between cancer and normal samples in liver cancer were selected with threshold by p-Value ≤ 1E-4, fold change ≥ 2 and top 10% gene rank in Oncomine database (https://www.oncomine.org/).
Survival analysis
Liver Hepatocellular Carcinoma (TCGA, Provisional) database in cBioPortal for cancer genomics web service was used for identifying AKR1B10 for predicting the overall survival (OS) of HCC patients 32, 33. AKR1B10 Mrna expression levels calculated by log2 calculation were compared based on clinical attribute in HCC patients. To evaluate associations between candidate biomarkers and survival and linic-pathological features in HCC patients, gene data with Z scores and clinical data of HCC patients in Liver Hepatocellular Carcinoma (TCGA, Provisional) database were downloaded from cBioPortal and matched using VLOOKUP index in EXCEL. After excluding ten patients with liver histology of hepatocholangiocarcinoma (n = 7) and fibrolamellar carcinoma (n = 3) and six patients without gene expression levels, 361 HCC patients were included in the analysis. Additionally, the Kaplan Meier plotter online service (http://kmplot.com/analysis/) 34 was used for subgroup survival analysis of AKR1B10 with median cutoff in HCC patients.
Protein-protein interaction and pathway/biological process enrichment
Protein-Protein Interaction analysis (PPI) for AKR1B10 were performed by STRING database (https://string-db.org/). KEGG pathway and GO biological process enrichment analysis were conducted using Gene Set Enrichment Analysis (GSEA, http://software.broadinstitute.org/gsea/index.jsp). To investigate gene sets, AKR1B10 interactive genes were uploaded to Molecular Signatures Database in GSEA. A false discovery rate P value cut‑off of <0.05 was set as the screening condition.
Statistical analysis
Differences of gene expression between the individual groups were analyzed using student t test, Mann-Whitney U-test, Chi-square test and Ridit analysis based on variables types. PASW Statistics software version 23.0 from SPSS Inc. (Chicago, IL, USA) was used. A two-tailed P < 0.05 were considered significant for all tests.
Results
AKR1B10 expression comparison
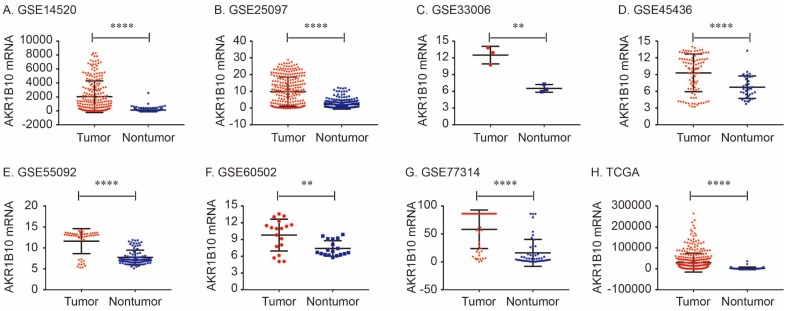
The details of GEO series included in this analysis were summarized in Table 1. As shown in Figure 1, AKR1B10 Mrna was significantly overexpressed in GSE14520, GSE25097, GSE33006, GSE45436, GSE55092, GSE60502. GSE77314 and TCGA datasets (all P < 0.01, Figure 1).
Figure 1.
AKR1B10 mRNA expression levels between tumor and nontumor tissues in HCC patients in GEO database series including GSE14520 (A), GSE25097 (B), GSE33006 (C), GSE45436 (D), GSE55092 (E), GSE60502 (F), GSE77314 (G) and TCGA database (H).
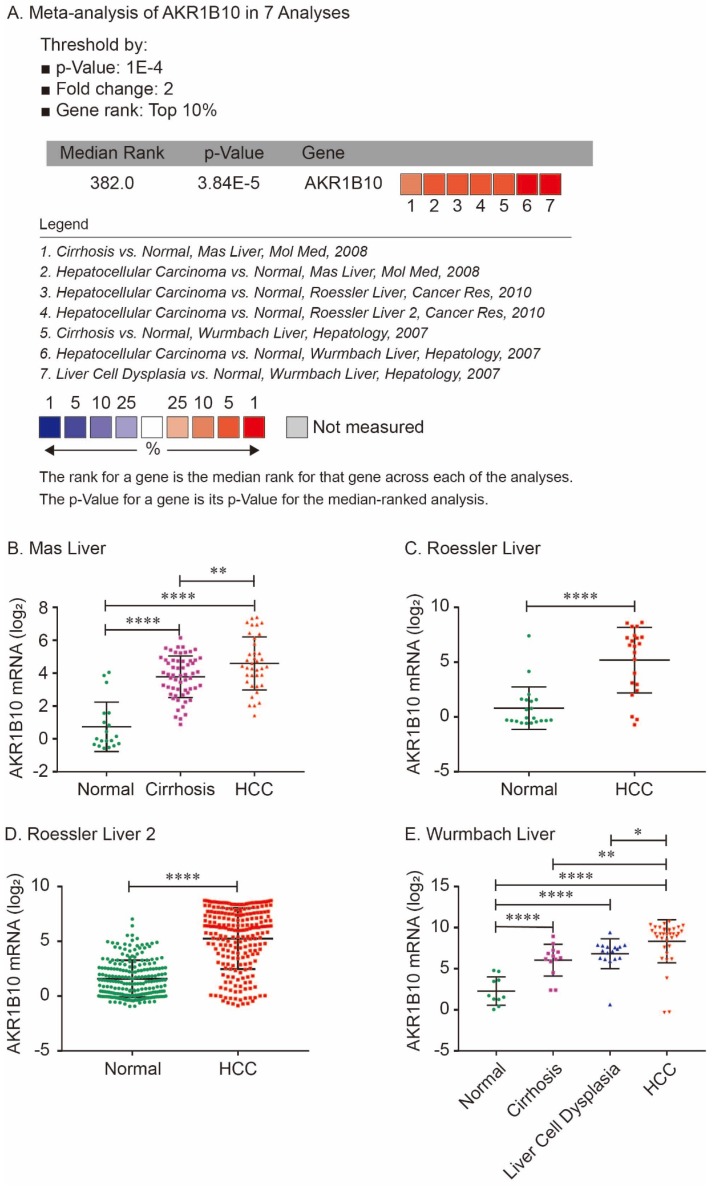
For validation, we performed meta-analysis of AKR1B10 expression in 7 analyses with threshold by p-Value ≤ 1E-4, fold change ≥ 2 and top 10% gene rank in Oncomine database. As shown in Figure 2, compared with that in normal liver tissues, AKR1B10 was significantly upregulated in cirrhosis (P < 0.0001, Figure 2A, 2B and 2E), liver cell dysplasia (P < 0.0001, Figure 2A and 2E) and HCC tumors (P < 0.0001, Figure 2A - 2E). Additionally, AKR1B10 was significantly higher in HCC tumor tissues than that in cirrhosis (P < 0.01, Figure 2B and 2E) and in liver cell dysplasia (P < 0.05, Figure 2E).
Figure 2.
Comparison of AKR1B10 mRNA expression levels across 7 analyses in Oncomine database. Meta-analysis of AKR1B10 expression in 7 analyses (A); AKR1B10 levels in normal, cirrhosis and HCC tissues in Mas Liver (B); AKR1B10 levels in normal and HCC tissues in Roessler Liver (C) and Roessler Liver 2 (D); AKR1B10 levels in normal, cirrhosis, liver cell dysplasia and HCC tissues in Wurmbach Liver (E).
Associations between AKR1B10 and OS in HCC patients
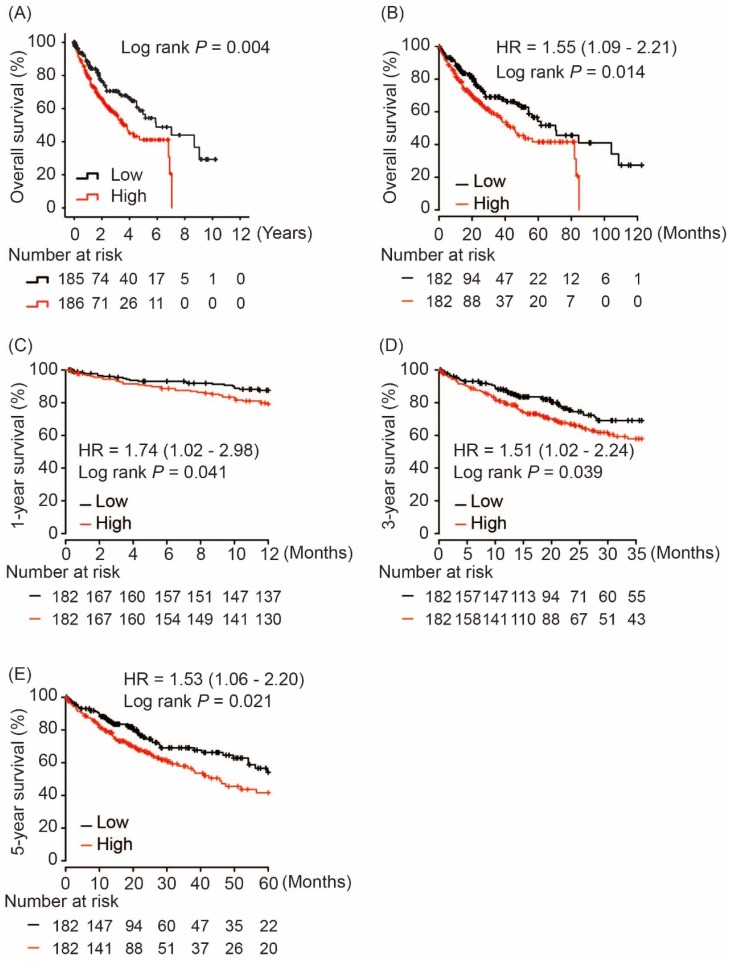
Using hash and survival packages in R program, we performed Kaplan-Meier analysis of AKR1B10 and OS in HCC patients, which showed that AKR1B10 overexpression in tumor tissues was significantly associated with poor OS in HCC patients (log rank P = 0.004, Figure 3A). A consistent result was validated in Kapan-Meier Plotter online service as shown in Figure 3B (log rank P = 0.014, Figure 3B). Moreover, subgroup analysis revealed that AKR1B10 upregulation in tumors was risk factor for 1-year, 3-year and 5-year OS in HCC patients (HR = 1.74, log rank P = 0.041; HR = 1.51, log rank P = 0.039 and HR = 1.53, log rank P = 0.021, respectively, Figure 3C - 3E).
Figure 3.
Overall survival of HCC patients grouped by AKR1B10 median cutoff in TCGA database (A, B); 1-year (C), 3-year (D) and 5-year overall survival comparison between AKR1B10 high and low groups.
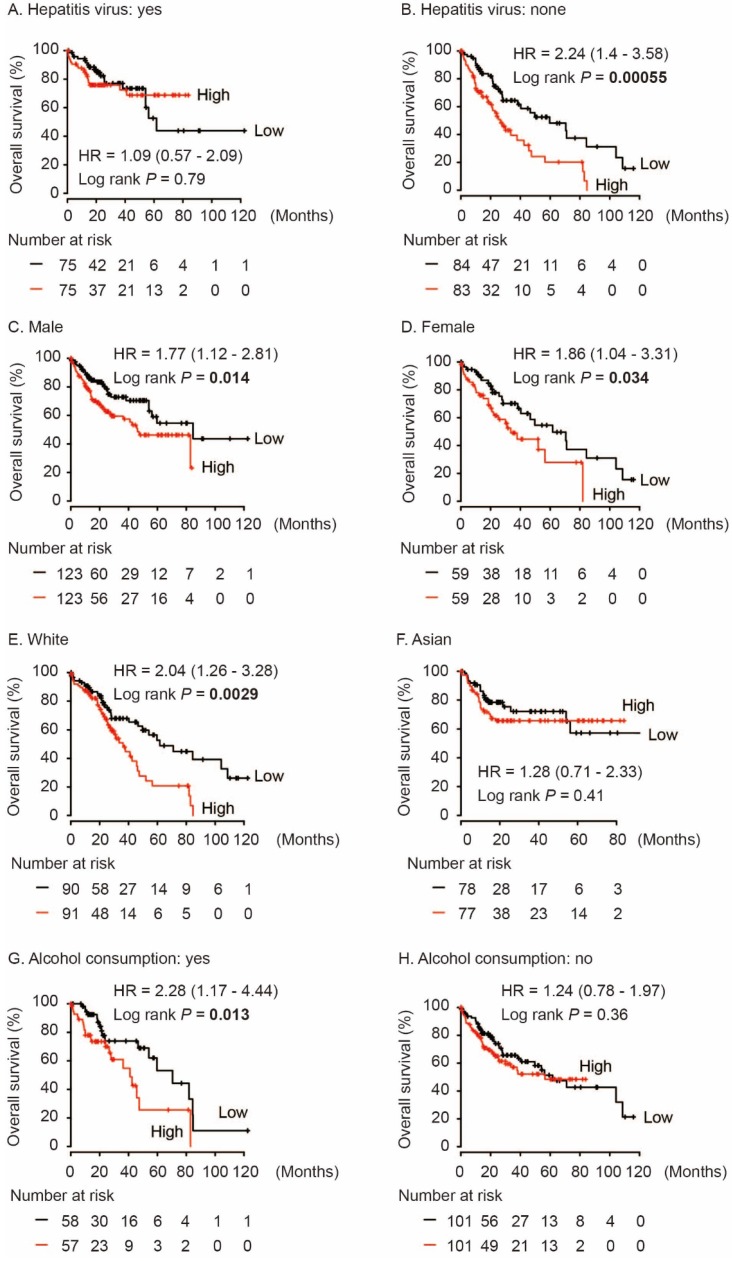
In addition, we performed subgroup survival analysis in different population. As shown in Figure 4, high AKR1B10 level significantly contributed to worse OS in HCC patients without hepatitis virus infection (HR = 2.24, log rank P = 0.00055, Figure 4B). And, AKR1B10 overexpression was significantly associated with OS both in male and female (HR = 1.77, log rank P = 0.014 and HR = 1.86, log rank P = 0.034, respectively, Figure 4C and 4D). AKR1B10 was risk factor for OS in White HCC (HR = 2.04, log rank P = 0.0029, Figure 4E) and in cases with alcohol consumption (HR = 2.28, log rank P = 0.013, Figure 4G).
Figure 4.
Subgroup analyses of overall survival comparison in different population [hepatitis virus infection status (A, B), gender (C, D), race (E, F) and alcohol consumption (G, H)] with AKR1B10 median cutoffs in HCC patients.
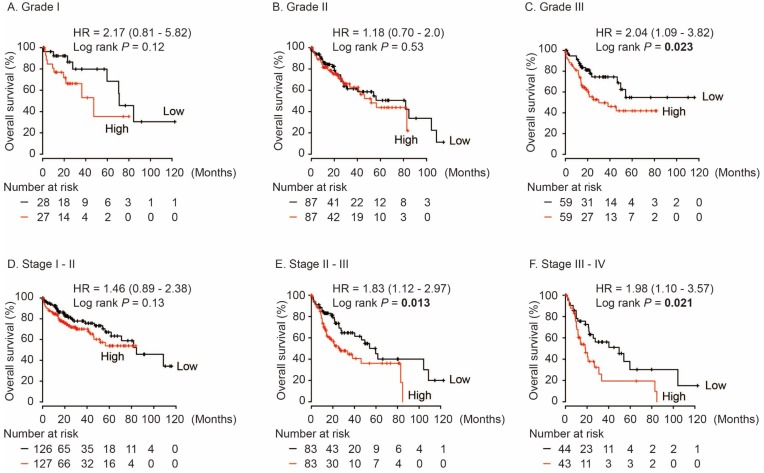
In HCC patients with neoplasm histologic Grade III, AKR1B10 was significantly associated with poor OS (HR = 2.04, log rank P = 0.023, Figure 5C), while no differences were found in HCC cases with Grade I and Grade II (log rank P > 0.05, Figure 5A and 5B). Moreover, AKR1B10 overexpression was significantly contributed to poor OS in HCC patients with stage II-III (HR = 1.83, log rank P = 0.013, Figure 5E) and stage III-IV (HR = 1.98, log rank P = 0.021, Figure 5F).
Figure 5.
Overall survival of HCC patients grouped by AKR1B10 median in different grades (A, B, C) and stages (D, E, F).
Association between AKR1B10 and clinico-pathological features in HCC patients
As shown in Table 2, more male cases in AKR1B10 high group (76.2% vs. 58.9%, P < 0.001) and patients in AKR1B10 high group were significantly older than those in AKR1B10 low group (62.0yr vs. 59.5yr, P = 0.031). More HCC cases had family history of cancer in AKR1B10 high group than those in AKR1B10 low group (43.1% vs. 30.0%, P = 0.01). More HCC cases had hepatocarcinoma risk factors (especially alcohol consumption) in AKR1B10 high group than those in AKR1B10 low group (P = 0.006). As we expected, HCC patients in AKR1B10 high group suffered from significantly advanced Ishak fibrosis status and advanced hepatic inflammation (P = 0.036 and P = 0.033, respectively).
Table 2.
Characteristics of HCC patients between AKR1B10 high and low groups
| Variables | AKR1B10 expression level | P value | |
|---|---|---|---|
| Low (n = 180) | High (n = 181) | ||
| Gender, male (%) | 106 (58.9) | 138 (76.2) | < 0.001 |
| Age, median (IQR), years | 59.5 (20) | 62 (15) | 0.031 |
| BMI, kg/m2, n (%) | 0.132 | ||
| <18.5 | 14 (7.8) | 7 (3.9) | |
| 18.5~24.99 | 79 (43.9) | 73 (40.3) | |
| 25~29.99 | 36 (20.0) | 52 (28.9) | |
| >30 | 35 (19.4) | 32 (17.7) | |
| NA | 16 (8.9) | 17 (9.4) | |
| Race, n (%) | 0.36 | ||
| Asian | 77 (42.8) | 79 (43.6) | |
| White | 91 (50.6) | 85 (47.0) | |
| Black or African American | 7 (3.9) | 10 (5.5) | |
| American Indian or Alaska Native | 2 (1.1) | 0 (0) | |
| NA | 3 (1.6) | 7 (3.9) | |
| Tumor status, n (%) | 0.923 | ||
| With tumor | 53 (29.4) | 56 (30.9) | |
| Tumor free | 115 (63.9) | 112 (61.9) | |
| NA | 12 (6.7) | 13 (7.2) | |
| Family history of cancer, n (%) | 54 (30.0) | 78 (43.1) | 0.01 |
| Hepatocarcinoma risk factors*, n (%) | 0.006 | ||
| Hepatitis virus infection | 55 (30.6) | 56 (30.9) | |
| Alcohol consumption | 48 (26.7) | 67 (37.0) | |
| Non-alcoholic fatty liver disease | 6 (3.3) | 6 (3.3) | |
| No risk factors | 54 (30.0) | 32 (17.7) | |
| Other | 8 (4.4) | 10 (5.5) | |
| NA | 9 (5.0) | 10 (5.5) | |
| Neoplasm histologic grade, n (%) | 0.054 | ||
| G1 | 35 (19.4) | 18 (9.9) | |
| G2 | 76 (42.2) | 95 (52.5) | |
| G3 | 60 (33.3) | 61 (33.7) | |
| G4 | 6 (3.3) | 5 (2.8) | |
| NA | 3 (1.7) | 2 (1.1) | |
| AJCC stage, n (%) | 0.203 | ||
| I | 84 (46.7) | 83 (45.9) | |
| II | 35 (19.4) | 47 (26.0) | |
| III | 48 (26.7) | 33 (18.2) | |
| IV | 4 (2.2) | 3 (1.7) | |
| NA | 9 (5.0) | 15 (8.3) | |
| Vascular invasion, n (%) | 0.999 | ||
| Macro | 8 (4.4) | 8 (4.4) | |
| Micro | 44 (24.4) | 45 (24.9) | |
| None | 99 (55.0) | 101 (55.8) | |
| NA | 29 (16.1) | 27 (14.9) | |
| Child-pugh classification, n (%) | 0.525 | ||
| A | 101 (56.1) | 111 (61.3) | |
| B | 9 (5.0) | 12 (6.6) | |
| C | 1 (0.6) | 0 (0) | |
| NA | 69 (38.3) | 58 (32.0) | |
| AFP > 400ng/ml, n (%) | 39 (21.7) | 25 (13.8) | 0.051 |
| Platelet, median (IQR), ×103/mm3 | 204.5 (182) | 178 (136) | 0.066 |
| New tumor event after initial treatment, n (%) | 46 (25.6) | 47 (26.0) | 0.929 |
| Ishak fibrosis status, n (%) | 0.036 | ||
| No fibrosis | 44 (24.4) | 28 (15.5) | |
| Portal fibrosis | 10 (5.6) | 21 (11.6) | |
| Fibrous speta | 11 (6.1) | 17 (9.4) | |
| Nodular Formation/Incomplete Cirrhosis | 3 (1.7) | 6 (3.3) | |
| Cirrhosis | 30 (16.7) | 40 (22.1) | |
| NA | 82 (45.6) | 69 (38.1) | |
| Hepatic inflammation, n (%) | 0.033 | ||
| None | 69 (38.3) | 45 (24.9) | |
| Mild | 43 (23.9) | 54 (29.8) | |
| Severe | 7 (3.9) | 11 (6.1) | |
| NA | 61 (33.9) | 71 (39.2) | |
| Follow up, median (IQR), years | 0.378 (1.8) | 0.334 (1.3) | 0.453 |
IQR, interquartile range; BMI, body mass index; AJCC, American Joint Committee on Cancer; AFP, alpha-fetoprotein; NA, not available.
* Sum of all risk factors compared with no risk factors.
PPI and KEGG/GO biological process enrichment
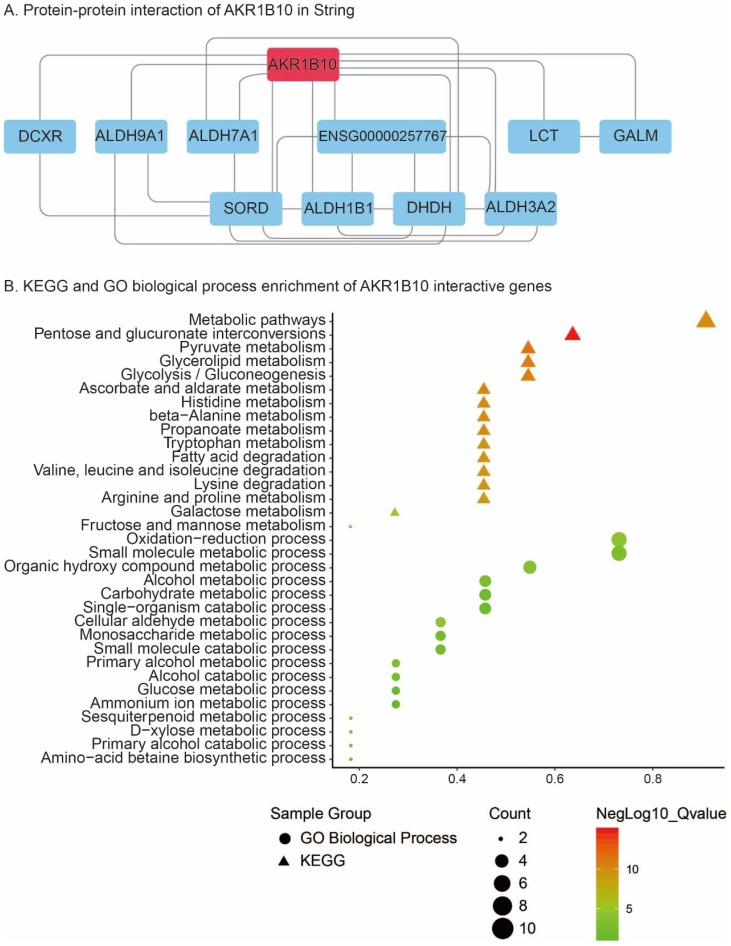
PPI analysis using STRING revealed that 10 genes including DCXR, ALDH9A1, ALDH7A1, ENSG00000257767, LCT, GALM, SORD, ALDH1B1, DHDH and ALDH3A2 were interacted with AKR1B10 (Figure 6A). KEGG pathway enrichment of AKR1B10 interactive genes showed that metabolic pathways, pentose and glucuronate interconversions, pyruvate metabolism, glycerolipid metabolism, glycolysis / gluconeogenesis, ascorbate and aldarate metabolism, histidine metabolism, beta-Alanine metabolism, propanoate metabolism, tryptophan metabolism, arginine and proline metabolism, fatty acid degradation, etc were the most enriched pathways (Figure 6B). Additionally, oxidation-reduction process, metabolic process including small molecular, organic hydroxy compound, alcohol, carbohydrate, single-organism, cellular aldehyde, glucose, ammonium ion, etc were most enriched GO biological process of AKR1B10 interactive genes (Figure 6B).
Figure 6.
Protein-protein interaction of AKR1B10 using String analysis (A); KEGG and GO biological process enrichment of its interactive genes (B)
Discussion
Over the past few years, new data have recently emerged that implicate AKR1B10 in tumor carcinogenesis in different systemic malignancies 35. AKR1B10 is expressed at lower levels in normal liver8, 10 and overexpressed in HCC tumors 22, 25. Matkowskyj K et al reported that AKR1B10 was overexpressed in 97% of HCC, with minimal to no expression in adjacent hepatic tissue, while hepatic adenomas and focal nodular hyperplasia did not exhibit expression of AKR1B10 17. A small sample study demonstrated that increased expression of AKR1B10 in moderately-differentiated HCC compared with well-differentiated HCC, poorly-differentiated HCC, and liver cirrhosis 18. Consistent with previous reports, we found that AKR1B10 was significantly upregulated in cirrhosis, liver cell dysplasia and HCC tumors compared with normal livers. Additionally, AKR1B10 was significantly higher in HCC tumor tissues than that in cirrhosis and in liver cell dysplasia. Hence, we assumed that AKR1B10 is emerging as a biomarker to distinguish hepatocellular carcinoma from benign liver lesions.
However, roles of AKR1B10 in hepatocarcinogenesis are still controversial. In a retrospective study of 168 cases, Schmitz K et al found that loss of AKR1B10 expression correlates with increased proliferative activity. A poorer prognosis in AKR1B10-negative HCCs was revealed compared with patients with strongly positive HCCs 36. In 110 patients with hepatitis B virus-related HCC, high AKR1B10 expression was negatively correlated with serum AFP level. Patients with high AKR1B10 expression had significantly higher DFS than those with low expression within 2 years after liver resection. High AKR1B10 expression was found to be a favorable factor of early recurrence and OS in HCC patients 23. In 255 HCC patients who underwent curative hepatectomy, high AKR1B10 expression was significantly associated with a lack of invasion of the major portal vein, a lack of intrahepatic metastasis, lower tumor stages, and lower AFP levels, which was found to be an independent predictor of both longer recurrence-free survival and longer disease-specific survival 24.
Conversely, our results demonstrated that high AKR1B10 in tumors were significantly associated with worse OS in HCC patients. AKR1B10 overexpression were associated with poor 1-year, 3-year and 5-year OS. A series publication enhanced our findings. Murata A et al reported that high AKR1B10 expression was an independent risk factor for HCC development in chronic hepatitis C patients. During the follow-up period after viral eradication, patients expressing high levels of AKR1B10 expressed markedly higher levels of alanine aminotransferase and AFP than did patients exhibiting low AKR1B10 expression 26. The oncogenetic functions of AKR1B10 in HCC tumor growth were validated in HCC cell lines SMMC-7721, HepG2 and Hep3B. Knockdown of AKR1B10 through shRNA in Hep3B cells showed significantly induced cell cycle arrest and inhibited cell growth 37. ShRNA-mediated silencing of AKR1B10 expression in HCC cells resulted in increased cell apoptosis, decreased colony formation and size, and enhanced cytoreductive response following exposure to doxorubicin chemotherapy 17. A recent study identified that AKR1B10 is a novel downstream target of interleukin-1 receptor-associated kinase 1 (IRAK1), which was found to be overexpressed in HCC and significantly correlated with IRAK1 expression. Knockdown of AKR1B10 negated IRAK1-induced tumor-initiating cells functions 38.
Functionally, AKR1B10 exerts a protective role through eliminating oxidative and carbonyl stresses and promoting epithelial proliferation for damage repair in inflammation. The KEGG pathway and GO biological process of AKR1B10 interactive genes in our study revealed that AKR1B10 should develop metabolic pathways and oxidation-reduction process. Unfortunately, we could not perform experimental research for probing potential oncogenic mechanisms of AKR1B10 in HCC development. And, no our own follow-up data of HCC patients were available. Considered the current controversial publications, we suggest a meta-analysis to evaluate the links between AKR1B10 and HCC prognosis. Even though, considered previous reports, we cautiously drew the hypothesis that AKR1B10 overexpression contributed to unfavorable prognosis in HCC patients.
Acknowledgments
This work was supported by National Natural Science Foundation of China (grant number 81803901 and 31601908) and Shanghai Sailing Program (grant number 17YF1416000).
References
- 1.Bray F, Ferlay J, Soerjomataram I. et al. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Heimbach JK, Kulik LM, Finn RS. et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358–80. doi: 10.1002/hep.29086. [DOI] [PubMed] [Google Scholar]
- 3.Omata M, Cheng AL, Kokudo N. et al. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int. 2017;11:317–70. doi: 10.1007/s12072-017-9799-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 5.Petrick JL, Kelly SP, Altekruse SF. et al. Future of Hepatocellular Carcinoma Incidence in the United States Forecast Through 2030. J Clin Oncol. 2016;34:1787–94. doi: 10.1200/JCO.2015.64.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo W, Tan HY, Wang N. et al. Deciphering hepatocellular carcinoma through metabolomics: from biomarker discovery to therapy evaluation. Cancer Manag Res. 2018;10:715–34. doi: 10.2147/CMAR.S156837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B, Finn RS. Personalized Clinical Trials in Hepatocellular Carcinoma Based on Biomarker Selection. Liver Cancer. 2016;5:221–32. doi: 10.1159/000367763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao D, Fan ST, Chung SS. Identification and characterization of a novel human aldose reductase-like gene. J Biol Chem. 1998;273:11429–35. doi: 10.1074/jbc.273.19.11429. [DOI] [PubMed] [Google Scholar]
- 9.Ma J, Cao D. Human aldo-keto reductases: structure, substrate specificity and roles in tumorigenesis. Biomol Concepts. 2011;2:115–26. doi: 10.1515/bmc.2011.010. [DOI] [PubMed] [Google Scholar]
- 10.Hyndman DJ, Flynn TG. Sequence and expression levels in human tissues of a new member of the aldo-keto reductase family. Biochim Biophys Acta. 1998;1399:198–202. doi: 10.1016/s0167-4781(98)00109-2. [DOI] [PubMed] [Google Scholar]
- 11.Kang MW, Lee ES, Yoon SY. et al. AKR1B10 is associated with smoking and smoking-related non-small-cell lung cancer. J Int Med Res. 2011;39:78–85. doi: 10.1177/147323001103900110. [DOI] [PubMed] [Google Scholar]
- 12.Chung YT, Matkowskyj KA, Li H. et al. Overexpression and oncogenic function of aldo-keto reductase family 1B10 (AKR1B10) in pancreatic carcinoma. Mod Pathol. 2012;25:758–66. doi: 10.1038/modpathol.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reddy KA, Kumar PU, Srinivasulu M. et al. Overexpression and enhanced specific activity of aldoketo reductases (AKR1B1 & AKR1B10) in human breast cancers. Breast. 2017;31:137–43. doi: 10.1016/j.breast.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Ma J, Luo DX, Huang C. et al. AKR1B10 overexpression in breast cancer: association with tumor size, lymph node metastasis and patient survival and its potential as a novel serum marker. Int J Cancer. 2012;131:E862–71. doi: 10.1002/ijc.27618. [DOI] [PubMed] [Google Scholar]
- 15.Ko HH, Peng HH, Cheng SJ. et al. Increased salivary AKR1B10 level: association with progression and poor prognosis of oral squamous cell carcinoma. Head Neck. 2018;40:2642–7. doi: 10.1002/hed.25370. [DOI] [PubMed] [Google Scholar]
- 16.He YC, Shen Y, Cao Y. et al. Overexpression of AKR1B10 in nasopharyngeal carcinoma as a potential biomarker. Cancer Biomark. 2016;16:127–35. doi: 10.3233/CBM-150548. [DOI] [PubMed] [Google Scholar]
- 17.Matkowskyj KA, Bai H, Liao J. et al. Aldoketoreductase family 1B10 (AKR1B10) as a biomarker to distinguish hepatocellular carcinoma from benign liver lesions. Hum Pathol. 2014;45:834–43. doi: 10.1016/j.humpath.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han C, Gao L, Zhao L. et al. Immunohistochemistry Detects Increased Expression of Aldo-Keto Reductase Family 1 Member B10 (AKR1B10) in Early-Stage Hepatocellular Carcinoma. Med Sci Monit. 2018;24:7414–23. doi: 10.12659/MSM.910738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohashi T, Idogawa M, Sasaki Y. et al. AKR1B10, a transcriptional target of p53, is downregulated in colorectal cancers associated with poor prognosis. Mol Cancer Res. 2013;11:1554–63. doi: 10.1158/1541-7786.MCR-13-0330-T. [DOI] [PubMed] [Google Scholar]
- 20.Yao HB, Xu Y, Chen LG. et al. AKR1B10, a good prognostic indicator in gastric cancer. Eur J Surg Oncol. 2014;40:318–24. doi: 10.1016/j.ejso.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 21.Hevir N, Sinkovec J, Lanisnik Rizner T. Decreased levels of AKR1B1 and AKR1B10 in cancerous endometrium compared to adjacent non-cancerous tissue. Chem Biol Interact. 2013;202:226–33. doi: 10.1016/j.cbi.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Laffin B, Petrash JM. Expression of the Aldo-Ketoreductases AKR1B1 and AKR1B10 in Human Cancers. Front Pharmacol. 2012;3:104. doi: 10.3389/fphar.2012.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang YY, Qi LN, Zhong JH. et al. High expression of AKR1B10 predicts low risk of early tumor recurrence in patients with hepatitis B virus-related hepatocellular carcinoma. Sci Rep. 2017;7:42199. doi: 10.1038/srep42199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ha SY, Song DH, Lee JJ. et al. High expression of aldo-keto reductase 1B10 is an independent predictor of favorable prognosis in patients with hepatocellular carcinoma. Gut Liver. 2014;8:648–54. doi: 10.5009/gnl13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonohara F, Inokawa Y, Hishida M. et al. Prognostic significance of AKR1B10 gene expression in hepatocellular carcinoma and surrounding non-tumorous liver tissue. Oncol Lett. 2016;12:4821–8. doi: 10.3892/ol.2016.5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murata A, Genda T, Ichida T. et al. Pretreatment AKR1B10 expression predicts the risk of hepatocellular carcinoma development after hepatitis C virus eradication. World J Gastroenterol. 2016;22:7569–78. doi: 10.3748/wjg.v22.i33.7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samur MK. RTCGAToolbox: a new tool for exporting TCGA Firehose data. PLoS One. 2014;9:e106397. doi: 10.1371/journal.pone.0106397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikolayeva O, Robinson MD. edgeR for differential RNA-seq and ChIP-seq analysis: an application to stem cell biology. Methods Mol Biol. 2014;1150:45–79. doi: 10.1007/978-1-4939-0512-6_3. [DOI] [PubMed] [Google Scholar]
- 29.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;2:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gautier L, Cope L, Bolstad BM. et al. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 31.Ritchie ME, Phipson B, Wu D. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerami E, Gao J, Dogrusoz U. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Aksoy BA, Dogrusoz U. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menyhárt O, Nagy A, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5:181006. doi: 10.1098/rsos.181006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kapoor S. AKR1B10 and its emerging role in tumor carcinogenesis and as a cancer biomarker. Int J Cancer. 2013;132:495. doi: 10.1002/ijc.27685. [DOI] [PubMed] [Google Scholar]
- 36.Schmitz KJ, Sotiropoulos GC, Baba HA. et al. AKR1B10 expression is associated with less aggressive hepatocellular carcinoma: a clinicopathological study of 168 cases. Liver Int. 2011;31:810–6. doi: 10.1111/j.1478-3231.2011.02511.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Zhou Y, Fei X. et al. Biostatistics mining associated method identifies AKR1B10 enhancing hepatocellular carcinoma cell growth and degenerated by miR-383-5p. Sci Rep. 2018;8:11094. doi: 10.1038/s41598-018-29271-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng BY, Lau EY, Leung HW. et al. IRAK1 Augments Cancer Stemness and Drug Resistance via the AP-1/AKR1B10 Signaling Cascade in Hepatocellular Carcinoma. Cancer Res. 2018;78:2332–42. doi: 10.1158/0008-5472.CAN-17-2445. [DOI] [PubMed] [Google Scholar]