Abstract
Objective
Salivary total protease and chitinase activities were measured by a broad-spectrum fluorescence resonance energy transfer approach as predictors of induction and resolution of gingival inflammation in healthy individuals by applying an experimental human gingivitis model.
Methods
Dental biofilm accumulated (21 days, Induction Phase) by omitting oral hygiene practices followed by a 2-week Resolution Phase to restore gingival health in an experimental gingivitis study. Plaque accumulation, as assessed by the Turesky Modification of the Quigley-Hein Plaque Index (TQHPI), and gingival inflammation, assessed using the Modified Gingival Index (MGI), scores were recorded and unstimulated saliva was collected weekly. Saliva was analysed for total protein, albumin, total protease activity and chitinase activity (n = 18).
Results
The TQHPI and MGI scores, as well as total protease activity, increased until day 21. After re-establishment of oral hygiene, gingival inflammation levels returned to values similar to baseline (day 0). Levels of protease activity decreased significantly, but not to baseline values. Furthermore, ‘fast’ responders, who responded immediately to plaque, exhibited significantly higher proteolytic activity throughout the experimental course than ‘slow’ responders, who showed a lagged inflammatory response.
Conclusion
The results indicate that differential inflammatory responses encompass inherent variations in total salivary proteolytic activities, which could be further utilised in contemporary diagnostic, prognostic and treatment modalities for periodontal diseases.
Key Points
| Total salivary protease activity was associated with gingival inflammation development among ‘fast’ responders in an experimental gingivitis study. |
| Salivary chitinase did not show any association with gingival inflammation scores, irrespective of the gingival inflammation development pattern. |
| Total salivary protease activity could be used to distinguish differential gingival inflammatory responses in saliva. |
Introduction
Periodontal diseases encompass the pathological inflammatory condition of the periodontal tissues, which is initiated by oral microbial biofilms. Without elimination of the biofilms, communities on the tooth surface will proceed towards subgingival growth. This leads to direct interactions with subgingival epithelium and immune cells, resulting in a cascade of inflammatory reactions and in chronic cases to destruction of gingival tissue [1]. During periodontal disease, both bacterial and host-derived proteins are released from the point of infection, i.e. the subgingival tissue, into the surrounding micro-environment: the gingival crevicular fluid (GCF) and saliva. In addition, as a result of bleeding, increased amounts of blood proteins, such as serum albumin and haemoglobin, are found in these oral fluids as well [2].
As sampling is relatively simple, GCF and saliva are considered ideal matrices for analysis of compounds involved in periodontal infection and inflammation. In this light, detailed molecular proteome analysis proved to be meaningful in characterising the comprehensive molecular composition of GCF and saliva and confirmed the nature of the immunological host response [3–7]. However, the technical complexity, time and costs of proteome analysis do not easily lead to practical applications for clinical chair-site usability. In this light and our experience though, enzyme activity in particular can be helpful to serve as molecular tool in bridging periodontal inflammatory processes to clinical chair-site practice [8–10]. A large advantage of monitoring enzyme activity is that their activity level directly mirrors the metabolic state of cells, rather than their presence alone [11, 12]. In addition, protease activity can specifically be measured by means of specific tailored substrates, monitoring either disappearance of the substrate or appearance of the cleavage product [12, 13]. The use of such peptide-based substrates enables proteolytic activity in complex matrices, and can chemically be adapted to hand-held applications. To exemplify, by combining magnetic nanoparticle technology with human and bacterial protease substrates, prototype chair-site tests have been developed for oral and non-oral applications [14–16].
The periodontal protease network, comprising both microbiological as well as host proteases, is highly complex and involved in a myriad of processes. Yet, the gingipains of Porphyromonas gingivalis commonly play a distinctive role in this process. Their protein levels are known to account for up to 85% of the total periodontal proteasome and are known to be involved in a number of processes, including bacterial co-aggregation, epithelial adherence through the haemagglutinin-adhesion domains and degradation of the extracellular matrix proteins [17–19]. Besides, their activity leads to expression of both proinflammatory and anti-inflammatory molecules [20–25]. Other well-described examples of bacterial proteases of the periodontal proteasome include karilysin from Tannerella forsythia and dentisilin from Treponema denticola. Karilysin cleaves fibrinogen, elastin and fibronectin and is known to inactivate the lectin pathway of the complement system by degrading mannose-binding lectin, ficolin-2 and ficolin-3. Furthermore, karilysin is able to detach tumour necrosis factor (TNF)-α from the surface of macrophages [26–28]. Dentilisin is known to be involved in circumventing the complement system by cleaving the negative regulator factor H [28, 29]. In addition, dentilisin degrades human fibrinogen at the inflamed site of periodontitis [30]. From the host side, collagenases and the metalloproteases (MMPs) are mainly involved during inflammation. In particular, MMP-1, MMP-3, MMP-8 and MMP-9 are found to be upregulated in case of periodontal disease and play a role in the regulation of cellular communication and immune function involving regulation of peptide hormones, cytokines and growth factors [31–37]. Indeed, in a parallel study, we recently showed that MMP-8 levels are associated with the extent and severity of gingival inflammation [35].
Besides, in previous studies it was reported that salivary chitinase levels also change in individuals with periodontal diseases. Chitinases, which are secreted by the palatal and three major salivary glands, hydrolyse chitin, a typical constituent of the yeast cell wall [31–34, 38, 39]. It was found that periodontal treatment for a period of 5 or 6 months resulted in a three- to four-fold decrease in chitinase activity. However, notably, the decrease in chitinase activity upon treatment did not correlate with clinical parameters such as attachment loss and bleeding on probing; only weak correlations were observed with the changes in probing pocket depth and plaque index [33]. Others found that levels of the acute-phase chitinase-like protein YKL-40 were found to be significantly higher in GCF of patients with gingivitis and periodontitis than in healthy controls [40]. Moreover, YKL-40 levels were found to be higher in patients with chronic periodontitis than in patients with gingivitis [41]. Experimental gingivitis studies have been used for evaluations of enquiries into gingivitis pathogenesis, as gingival inflammation is induced by plaque accumulation during abstention of oral hygiene, and resolved once oral hygiene is reinstituted. While several experimental gingivitis studies have reported variations in gingival inflammatory response patterns, few attempts have been made to characterise individual variability. In the experimental gingivitis study, conducted at Aarhus University where these data originate from, two different patterns of gingival inflammation response to plaque accumulation were identified and categorised as ‘slow’ and ‘fast’ responders [42]. ‘Slow’ responders had lagged inflammation response, whereas ‘fast’ responders responded immediately to plaque. As the pattern of gingival inflammation development is modulated by the host immune response, it is of relevance to investigate the immune components that may be associated with a more exacerbated clinical response. Accordingly, the aim of the present study was to assess the ability of total salivary protease and chitinase activity to monitor gingival disease progression and resolution using saliva, i.e. total oral fluid samples from the ‘fast’ and ‘slow’ responders. More specifically, it was investigated whether both total protease and chitinase activity would be discriminative in both groups as an attempt to explain the different stages of gingivitis development [42].
Materials and Methods
Design of the Study and Collection of Saliva Samples
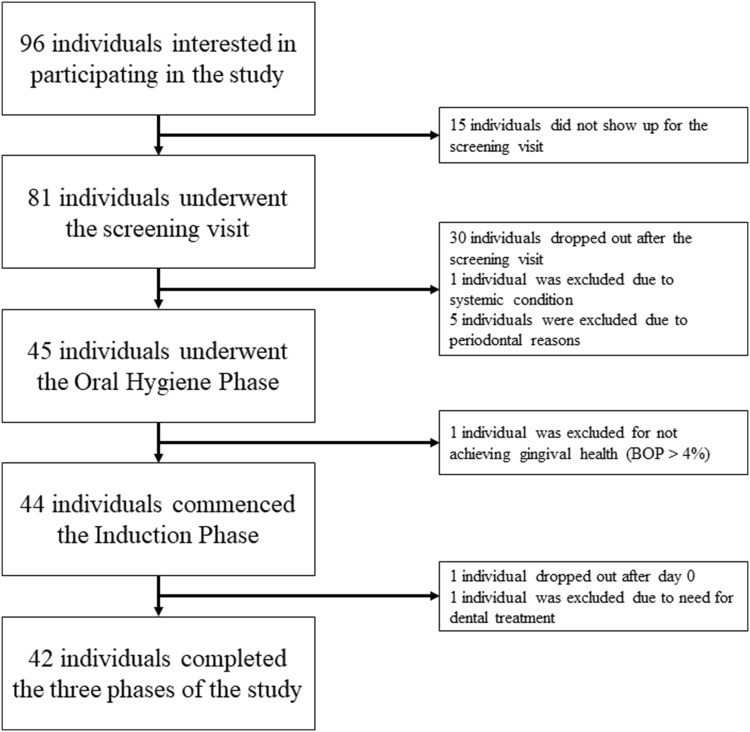
These analyses include a subset of participants from a larger experimental gingivitis study conducted at Aarhus University (Dataset II from Nascimento et al. [42]). Baseline characteristics are depicted in Table 1 and Fig. 1 provides a flowchart of patient recruitment and participation. This study protocol was approved by the Ethical Committee of Central Denmark Region (MOUTH: Reference 1-10-72-402-14). Before enrolment all participants were given oral and written information about the aim and design of the study, duration, demands, benefits and possible harm. Participation in this study was based on written informed consent. Exclusion criteria comprised use of or need for antibiotics (antibacterials) or anti-inflammatories within 6 weeks, systemic conditions with known effects on the immune system, probing pocket depths > 4 mm, interproximal attachment loss > 2 mm, cavitated caries, use of orthodontic appliances, presence of crowns or bridgework or inadequate restorations interfering with plaque control, pregnancy, planned pregnancy or breastfeeding, and motoric impairments that hinder effective personal oral hygiene [42].
Table 1.
Baseline characteristics of the 18 participants included in all three phases of the study
| Baseline characteristics | Mean (SD) |
|---|---|
| Age (years) | 25.2 (4.0) |
| TQHPI score—plaque | 0.18 (0.1) |
| MGI score—gingival inflammation | 0.02 (0.0) |
| Number of teeth | 29.1 (1.5) |
| Daily frequency of toothbrushing | 2.8 (0.4) |
MGI Modified Gingival Index, SD standard deviation, TQHPI Turesky Modification of the Quigley-Hein Plaque Index
Fig. 1.
Flowchart of patient recruitment and participation (see also Dataset II from Nascimento et al. [42]). In total, saliva samples from 18 individuals were available for analysis from the 42 individuals who completed the study
The study comprised three phases: (1) a pre-experimental Oral Hygiene Phase (day − 14 to day 0); (2) an Induction Phase (day 0–day 21); and (3) a Resolution Phase (day 21–day 35). The pre-experimental appointment (day − 14 to day 0) comprised clinical examinations, administration of questionnaires and measurement of cotinine by ELISA (enzyme-linked immunosorbent assay) in saliva to verify self-reported smoking status [42]. During the Oral Hygiene Phase, a professional prophylaxis (reduction of plaque, gingival inflammation and calculus) was performed by a dental hygienist. Individuals who achieved bleeding on probing < 4% and plaque < 20% were asked to proceed to the Induction Phase. Whole unstimulated saliva was collected as described previously [4]. At the beginning of the Induction Phase (day 0), the volunteers were asked to refrain from any kind of dental hygiene for 3 weeks. At day 7, 14 and 21 dental plaque and gingival scores, respectively, were registered and saliva was collected. After saliva collection and clinical examination at day 21, all participants were supplied with a new toothbrush and toothpaste in order to re-establish self-performed oral hygiene procedures (Resolution Phase). At day 35, after saliva collection and clinical examination, additional professional cleaning was provided for participants who required further care to restore gingival health. The clinical parameters recorded during the study included plaque accumulation, assessed using the Turesky Modification of the Quigley-Hein Plaque Index (TQHPI), and gingival inflammation, assessed by the Modified Gingival Index (MGI) [43, 44] (Table 1). The saliva samples were aliquoted for various aims, frozen down at − 80 °C and stored until further use. The saliva samples without protease inhibitors (n = 18) were available for further analysis.
Total Protein Content
Total protein content was measured using a Pierce™ BCA Protein Assay Kit (Thermo Scientific, West Palm Beach, FL, USA) in 96-well polystyrene microplates (Greiner Bio-One, Frieckenhausen, Germany), according to the manufacturer’s specifications, essentially as described previously [7]. Bovine serum albumin (BSA) was used as a standard. Optical readouts were obtained using a Multiskan™ FC Microplate Photometer (Thermo Scientific) in duplicate.
Albumin
Polypropylene microplates (Greiner Bio-One) were coated with a rabbit polyclonal antibody, anti-(human albumin) (Sigma-Aldrich, St Louis, MO, USA), overnight at room temperature. Saliva samples were subsequently serially diluted two-fold, and incubated for 2 h at 37 °C. Captured salivary albumin was detected using horseradish peroxidase (HRP)-conjugated rabbit anti-(human albumin) (GeneTex, Irvine, CA, USA). Human serum albumin (Sigma-Aldrich) was used as a standard.
Enzymatic Activities
Black 96-well polypropylene microplates (Greiner Bio-One) were used for all fluorescence-based enzymatic activity assays. Chitinase activity was quantified by adding 50 μL of saliva to a substrate solution of 4-methylumbelliferyl β-d-N,N0,N00-triacetylchitotrioside (Sigma-Aldrich) to a final reaction volume of 200 μL and a substrate concentration of 40 nM. The increase in fluorescence was subsequently acquired for 15 min at 37 °C [7, 33]. Total protease activity was measured based on the cleavage of a broad-spectrum fluorescence resonance energy transfer (FRET) substrate, designated PEK-054 ([FITC]-NleKKKKVLPIQLNAATDK-[KDbc]) [45]. Fluorescence was recorded at 2 min intervals in a BMG FLUOstar Galaxy microplate reader (MTX Lab Systems, Vienna, VA, USA). All enzymatic activity assays were performed in duplicate.
Data Analysis
Mean TQHPI and MGI scores were calculated for each individual. Two groups of individuals with different patterns of development in MGI scores during the Induction Phase were identified by group-based trajectory modelling (GBTM): ‘slow’ and ‘fast’ responders [42]. Accordingly, the ‘fast’ responders had higher mean MGI scores at the end of the first week of the Induction Phase, in contrast to the ‘slow’ responders, who needed an extra week to achieve similar mean MGI scores. Levels of protease and chitinase activity were log10 transformed due to non-Normal distribution and skewness. Enzymatic activity and clinical data were standardised in order to allow the comparability between estimates. Growth curve modelling, executed as multilevel mixed-effects regression analysis, was used to analyse the effect of time on TQHPI and MGI scores, as well as on salivary protease and chitinase. Such an approach was also used to analyse the association between the time-varying salivary activity level outcomes (protease and chitinase activity) and the clinical covariates (mean MGI, mean TQHPI, mean MGI at day − 14). Multilevel analyses were stratified by the patterns of gingival inflammation development. All analyses were performed using the software STATA® version 14.0 (StataCorp., College Station, TX, USA).
Results
Samples and clinical data from 18 healthy non-smoking volunteers, i.e. 13 females and five males (mean age: 25.33 ± 4.1 years, range 20–34), with at least 20 teeth were used in these analyses. Figure 2 displays the results from the growth curve modelling for the effect of time on MGI and TQHPI scores over the study period. Overall, both plaque and gingival scores increased steadily between day 0 and day 21 (p < 0.001). When oral hygiene was restored at day 21, both the plaque and gingival scores decreased, but the mean TQHPI score did not reach baseline levels at day 35 (p < 0.05) (Table 2). In a previous study in this population, we were able to identify individuals with different gingival development responses to plaque accumulation, dubbed ‘fast’ and ‘slow’ responders [42]. While the ‘slow’ responders showed a lagged gingival response to plaque accumulation, the fast responders seem to respond immediately. Even with a similar TQHPI, ‘fast’ responders had higher mean MGI scores, i.e. ~ 1.7 at the end of the first week of the Induction Phase, in contrast to the ‘slow’ responders, who showed MGI scores of ~ 0.7 and needed an extra week to achieve comparable mean MGI scores (p < 0.01).
Fig. 2.
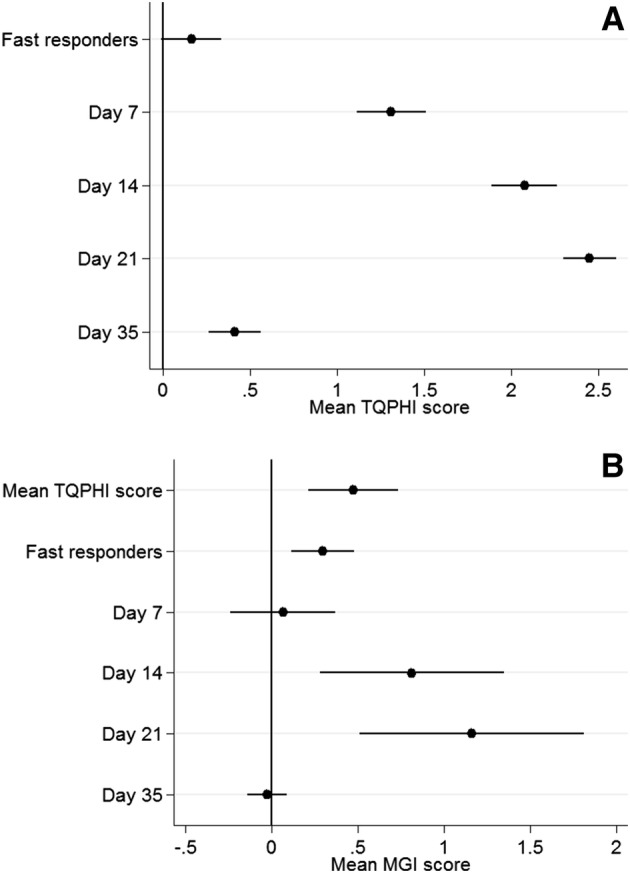
Results from mixed-effects multilevel models. a Mean TQHPI score as a function of day of experiment and pattern of gingival inflammation response. b Mean MGI score as a function of mean TQHPI, day of experiment and pattern of gingival inflammation response. Estimates given are the standardised regression coefficients (95% confidence interval). MGI modified gingival index, TQHPI Turesky Modification of the Quigley-Hein Plaque Index
Table 2.
Scoring of the plaque and gingival indices as well as the mean percentage with plaque and gingival index
| Index | Day | ||||
|---|---|---|---|---|---|
| 0 | 7 | 14 | 21 | 35 | |
| Median plaque index (range) | 0.11 (0–0.5)A | 1.44 (0.9–2.8)B | 2.44 (1.5–3.5)C | 3.0 (2.2–4.0)D | 0.5 (0.1–1.3)E |
| Mean plaque index (SD) | 0.14 (0.1)A | 1.59 (0.5)B | 2.52 (0.5)C | 3.01 (0.4)D | 0.57 (0.3)E |
| Median gingival index (range) | 0.0 (0–0.1)A | 0.38 (0.1–1.4)B | 1.28 (0.4–2.4)C | 1.9 (0.9–2.3)D | 0.12 (0–1.3)E |
| Mean gingival index (SD) | 0.01 (0.0)A | 0.49 (0.3)B | 1.34 (0.4)C | 1.85 (0.3)D | 0.17 (0.2)E |
Different capital letters in a row indicate statistical differences
SD standard deviation
Total salivary protein content, albumin concentration, protease activity and chitinase activity were determined at days 0 (baseline), 7, 14, 21 and 35 (Table 3). The distributions of all biochemical variables showed significant deviations from normal. Compared with baseline, overall protease activity was elevated at days 14, 21 and 35 but not at day 7 (Fig. 3a). At day 35 total protease activity was decreased compared with day 21 but did not reach baseline levels (Fig. 3a). Chitinase activity at baseline was 152.9 ± 85.6 ΔF/Δt, but no statistical differences in chitinase activity were found at later timepoints (Fig. 3b). Finally, the total protein content and albumin concentrations at baseline was 1361 ± 759 and 29.4 ± 11.2 μg/mL, respectively. No statistical differences were found for these two analyses at later timepoints (Table 3).
Table 3.
Saliva analysis
| Variable | Day | ||||
|---|---|---|---|---|---|
| 0 | 7 | 14 | 21 | 35 | |
| Total protein (μg/mL) | |||||
| Mean ± SD (na) | 1361 ± 758.6A (18) | 1188 ± 557.6A (18) | 1208 ± 680.1A (18) | 855.1 ± 474.3A (18) | 1286 ± 940.6A (15) |
| 95% CI for the mean | 984.4–1738 | 911.1–1466 | 869.5–1546 | 483.5–1227 | 765–1807 |
| Albumin (μg/mL) | |||||
| Mean ± SD (na) | 29.4 ± 11.2A (18) | 29.5 ± 13.2A (18) | 31.8 ± 14.4A (18) | 37.8 ± 11.2A (17) | 31.8 ± 13.6A (18) |
| 95% CI for the mean | 23.8–34.9 | 22.9–36 | 24.7–39 | 32.1–43.6 | 25–38.7 |
| Chitinase (ΔF/Δt) | |||||
| Mean ± SD (na) | 152.9 ± 85.6A (15) | 148.1 ± 99.7A (16) | 154.2 ± 105.2A (16) | 203.4 ± 115.2A (16) | 159.8 ± 98.6A (16) |
| 95% CI for the mean | 105.5–200.3 | 95.0–201.2 | 98.1–210.3 | 142.1–264.8 | 107.3–212.4 |
| Protease (ΔF/Δt) | |||||
| Mean ± SD (na) | 33.0 ± 32.0A (17) | 40.5 ± 32.2A (14) | 49.5 ± 34.2B (13) | 55.7 ± 29.0C (15) | 40.7 ± 39.7B (15) |
| 95% CI for the mean | 16.5–49.4 | 22–59.1 | 28.4–69.7 | 39.7–71.8 | 18.7–62.7 |
Different capital letters in a row indicate statistical differences. The p value was adjusted by the number of comparisons
CI confidence interval, SD standard deviation, ΔF/Δt protease and chitinase activity measured by increase of fluorescence in time
aNumber of samples assayed for the respective variable out of the total sample population of 18 participants
Fig. 3.
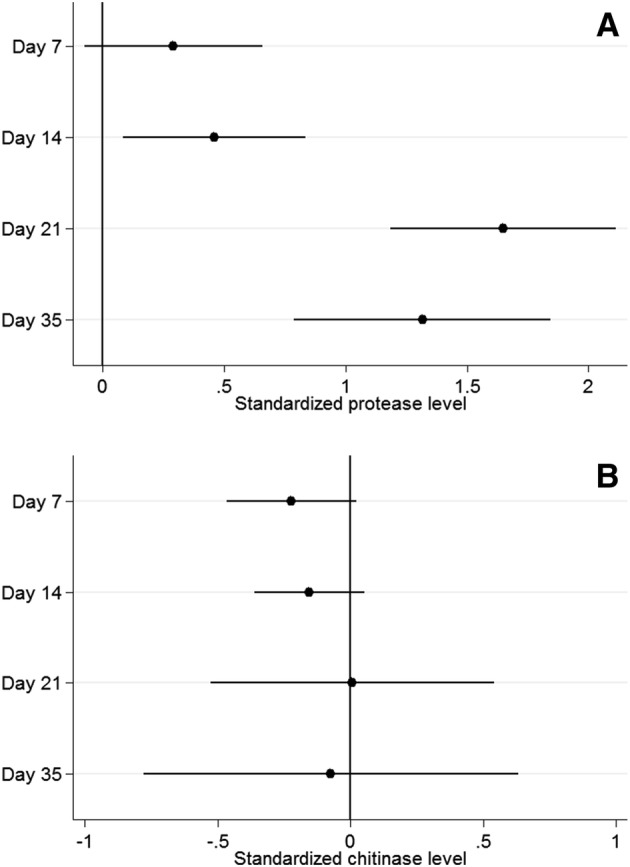
Results from mixed-effects multilevel models. a Salivary protease level and as a function of day of experiment. b Salivary chitinase level as a function of day of experiment. Estimates given are the standardised regression coefficients (95% confidence interval). Both analyses were adjusted for the standardised protein level
When investigating the association between the salivary markers and gingival inflammation development, results revealed that the protease activity increased over time, but reached statistical significance only at the end of the 3-week Induction Phase (day 21) (Fig. 3a). Stratified analysis based on the trajectories of gingival inflammation development (‘fast’ and ‘slow’ responders) was performed to investigate whether protease and chitinase activity would explain the different clinical response between the response groups. This analysis revealed that among ‘fast’ responders, the protease activity was significantly higher at the end of the first week of the Induction Phase (day 7) than at baseline, while ‘slow’ responders required an extra week to reach a significant increase in protease activity (Table 4). Additionally, ‘fast’ responders showed higher protease activity than ‘slow’ responders throughout the entire study period (Table 4). The stratified analysis also revealed that MGI was inversely associated with the protease activity among ‘fast’ responders. However, the chitinase activity was not associated with gingival inflammation development, irrespective of the response group (Table 5).
Table 4.
Results of mixed-effects linear regression of the clinical outcome ‘protease activity’ as a function of time, mean gingival inflammation over time and mean gingival inflammation at day − 14
| Explanatory variables | Protease activity | |||||
|---|---|---|---|---|---|---|
| Total sample (n = 18) | ‘Fast’ responders (n = 7) | ‘Slow’ responders (n = 11) | ||||
| Coefficient (95% CI) | p value | Coefficient (95% CI) | p value | Coefficient (95% CI) | p value | |
| Time (days) (reference: day 0) | < 0.001 | 0.017 | 0.020 | |||
| + 7 | 0.52 (0.14–1.17) | 1.40 (0.04–2.70) | 0.53 (0.00–1.05) | |||
| + 14 | 1.03 (0.41–2.48) | 2.21 (0.28–4.14) | 0.92 (0.04–0.21) | |||
| + 21 | 2.33 (0.54–4.12) | 3.50 (1.44–5.54) | 1.02 (0.09–1.95) | |||
| + 35 | 1.35 (0.85–1.87) | 1.04 (0.33–1.75) | 1.00 (0.46–1.54) | |||
| Gingival inflammation | − 0.33 (1.11–0.45) | 0.391 | − 0.73 (− 1.23 to − 0.25) | 0.031 | − 0.10 (− 0.41 to 0.21) | 0.443 |
| Gingival inflammation (day − 14) | − 0.05 (0.25–0.16) | 0.824 | 0.04 (− 0.20–0.30) | 0.662 | 0.01 (− 0.33 to 0.35) | 0.762 |
The coefficient denotes the mean difference of the protease activity outcome given the explanatory variables included in the model. The analysis was adjusted by protein level. Values in bold indicate statistical significance compared to baseline (p < 0.05)
CI confidence interval
Table 5.
Results of mixed-effects linear regression of the clinical outcome ‘chitinase activity’ as a function of time, mean gingival inflammation over time and mean gingival inflammation at day − 14
| Explanatory variables | Chitinase activity | |||||
|---|---|---|---|---|---|---|
| Total sample (n = 18) | ‘Fast’ responders (n = 7) | ‘Slow’ responders (n = 11) | ||||
| Coefficient (95% CI) | p value | Coefficient (95% CI) | p value | Coefficient (95% CI) | p value | |
| Time (days) (reference: day 0) | 0.938 | 0.663 | 0.701 | |||
| + 7 | − 0.27 (− 0.69 to 0.13) | − 0.35 (− 1.12 to 0.43) | − 0.27 (− 0.78 to 0.23) | |||
| + 14 | − 0.31 (− 1.21 to 0.60) | − 0.53 (− 1.75 to 0.70) | − 0.48 (− 1.64 to 0.67) | |||
| + 21 | − 0.17 (− 1.20 to 0.86) | 0.02 (− 1.12 to 1.16) | − 0.61 (− 2.20 to 0.97) | |||
| + 35 | − 0.07 (− 0.76 to 0.61) | 0.08 (− 0.52 to 0.68) | − 0.27 (− 1.28 to 0.75) | |||
| Gingival inflammation | 0.10 (− 0.47 to 0.66) | 0.974 | 0.26 (− 0.30 to 0.83) | 0.829 | 0.21 (− 0.65 to 1.08) | 0.715 |
| Gingival inflammation (day − 14) | − 0.05 (− 0.34 to 0.23) | 0.897 | − 0.01 (− 0.30 to 0.26) | 0.989 | − 0.19 (− 1.72 to 1.34) | 0.905 |
The coefficient denotes the mean difference of the chitinase activity outcome given the explanatory variables included in the model. The analysis was adjusted by protein level
CI confidence interval
Discussion
The aim of this study was to assess whether protease or chitinase activity could be used to monitor induction and resolution of gingivitis. For this, the association between total salivary protease activity, which includes both host and microbial proteases, and experimentally induced gingival inflammation were analysed using a broad-spectrum FRET approach. In addition, we assessed the chitinase activity in the same samples. Our results demonstrated that total salivary protease activity was associated with gingival inflammation development among ‘fast’ responders, while salivary chitinase did not show any association with gingival inflammation scores, irrespective of the gingival inflammation development pattern.
In the cohort where these data originate from, it was recently found that host MMP-8 levels are associated with the extent and severity of gingival inflammation [42]. In this study we further analysed total protease activity using a generic, fluorogenic protease substrate, the breakdown of which represents both total bacterial as well as total host protease activity [45]. Due to the fact that no albumin increase was detected throughout the investigation, leakage of proteases originating from serum could be excluded. In line with the clinical scores, i.e. on the plaque and gingival indexes, the protease activity, but not chitinase activity, increased until oral hygiene was restored. Proteolytic activity decreased between day 21 and day 35, but not to baseline levels, similar to the clinical scores. These findings are in line with earlier studies which showed that inflammation of the gingiva is characterised by protease activity of both bacterial and human origin [10, 24, 46–48], and with the earlier study on this expanded cohort that revealed that clinical ‘fast’ responders, compared with ‘slow’ responders, exhibit overall higher MMP-8 levels throughout the experimental course [42]. These observations collectively underline that differential inflammatory host responses of the periodontal tissues encompass variations in proteolytic activities, which are reflected in saliva, and strengthens the notion that therapeutic strategies on the resolution of inflammatory periodontal disease could also target the pharmacological inhibition of the proteolytic activity occurring in the oral milieu.
As mentioned earlier, the proteolytic activity was shown to be associated with the ‘fast’ but not with the ‘slow’ gingival response pattern (Table 4). We and others postulate that this might be a consequence of so-called priming of oral neutrophils, here case of the ‘fast’ responders. Previously, three distinct neutrophil subsets were identified: resting neutrophils, oral biofilm-primed parainflammatory neutrophils and proinflammatory neutrophils. These intermediately activated parainflammatory populations occur in the healthy oral cavity and may quickly shift to a proinflammatory phenotype during a periodontal unstable, i.e. diseased, environment. A next, subsequent exposure to the oral biofilm will rapidly activate the primed parainflammatory neutrophils to respond faster than the resting neutrophils [49–51]. Therefore, it can be envisaged that it was as a consequence of the activated neutrophils and proteases secreted by the biofilm that an increase in the total protease activity was detected. In addition, activation of human pro-MMPs by bacterial proteases from potent periopathogens, such as gingipains and dentisilin, will contribute to a magnification in total protease activity [52, 53].
One may consider the use of a generic protease substrate to be a limitation of the study. Firstly, by using PEK-054 it is not possible to identify the proteases involved and its source, i.e. whether its source is of human or bacterial origin. PEK-054 lacks specificity as it is hydrolysed by virtually all bacterial and human salivary proteases. Hence, to study the role of bacteria-specific proteases, and in particular gingipains of P. gingivalis, one might consider using recently developed bacteria-specific D-amino acid-containing protease substrates. While l-amino acids represent the vast majority of amino acids found in all natural proteins, the presence of d-amino acids is highly specific for bacteria [13]. Secondly, in case of periodontal disease, the full protease network is very complex. A variety of both human and bacterial proteases may reciprocally inhibit each other, or, in contrast, work synergistically to amplify their effects as stated earlier. To exemplify, it is known that bacterial proteases act synergistically in inactivating the complement system, with dentilisins, gingipains and karilysins all contributing by cleaving various molecules of the complement system [48, 54]. Accordingly, the gingipains of P. gingivalis are able to stimulate triggering receptor expressed on myeloid cells 1 (TREM-1) or membrane-bound MMPs, i.e. TNF-α-converting enzyme (TACE), which are considered to be amplifiers of the inflammatory response, expressed by immune and epithelial cells [21, 55–58]. Alternatively, bacterial proteases may manipulate the human-derived MMP activity in different ways, e.g. through upregulation by cleavage of the pro-MMPs or, in contrast, inactivation of metabolic active MMPs [47, 49, 52, 53, 59]. However, using PEK-054 quantification of total proteolytic activity in saliva is accessible and apparently mirrors the course of periodontal inflammation.
In contrast to protease activity levels, chitinase activity levels were relatively stable throughout the course of the study. Even though chitinase activity appeared highest at day 21, no statistical differences were found. Although chitinases have not been studied as extensively as proteases in periodontal disease, our findings support the data of studies pointing towards a lack of correlation between chitinase activity and gingivitis [33, 34, 40].
Conclusion
Total salivary protease activity, measured by a broad-spectrum FRET approach, seems to be a good tool to monitor induction and resolution of gingival inflammation in situ. Individuals with ‘fast’ gingival inflammation development present higher levels of total protease activity, which could be used to distinguish differential inflammatory responses.
Compliance with Ethical Standards
Funding
This work was supported by the author’s institutions and by the Aarhus University Research Foundation.
Conflict of interest
FJB, GGN, KN, AS, GNB, WEK, RL and NB declare no conflicts of interest related to this study.
Ethical approval and informed consent
This clinical study was approved by the Ethical Committee of Central Denmark Region (MOUTH: Reference 1-10-72-402-14). Participation in this study was based on informed consent. Before enrolment all participants were given oral and written information about the aim and design of the study, duration, demands, benefits and possible harm.
References
- 1.Schenkein HA. Host responses in maintaining periodontal health and determining periodontal disease. Periodontol. 2000;2006(40):77–93. doi: 10.1111/j.1600-0757.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- 2.Ebersole JL, Schuster JL, Stevens J, Dawson D, 3rd, Kryscio RJ, Lin Y, et al. Patterns of salivary analytes provide diagnostic capacity for distinguishing chronic adult periodontitis from health. J Clin Immunol. 2013;33:271–279. doi: 10.1007/s10875-012-9771-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bostanci N, Belibasakis GN. Gingival crevicular fluid and its immune mediators in theproteomic era. Periodontol. 2000;2018(76):68–84. doi: 10.1111/prd.12154. [DOI] [PubMed] [Google Scholar]
- 4.Bostanci N, Ramberg P, Wahlander A, Grossman J, Jonsson D, Barnes VM, et al. Label-free quantitative proteomics reveals differentially regulated proteins in experimental gingivitis. J Proteome Res. 2013;12:657–678. doi: 10.1021/pr300761e. [DOI] [PubMed] [Google Scholar]
- 5.Grant MM. What do’omic technologies have to offer periodontal clinical practice in the future? J Periodontal Res. 2012;47:2–14. doi: 10.1111/j.1600-0765.2011.01387.x. [DOI] [PubMed] [Google Scholar]
- 6.Lira-Junior R, Ozturk VO, Emingil G, Bostanci N, Bostrom EA. Salivary and serum markers related to innate immunity in generalized aggressive periodontitis. J Periodontol. 2017;88:3139–3147. doi: 10.1902/jop.2017.170287. [DOI] [PubMed] [Google Scholar]
- 7.Prodan A, Brand HS, Ligtenberg AJ, Imangaliyev S, Tsivtsivadze E, van der Weijden F, et al. Interindividual variation, correlations, and sex-related differences in the salivary biochemistry of young healthy adults. Eur J Oral Sci. 2015;123:149–157. doi: 10.1111/eos.12182. [DOI] [PubMed] [Google Scholar]
- 8.Afacan B, Ozturk VO, Emingil G, Kose T, Bostanci N. Alarm anti-protease trappin-2 negatively correlates with proinflammatory cytokines in patients with periodontitis. J Periodontol. 2018;89:58–66. doi: 10.1902/jop.2017.170245. [DOI] [PubMed] [Google Scholar]
- 9.Bostanci N, Emingil G, Afacan B, Han B, Ilgenli T, Atilla G, et al. Tumor necrosis factor-alpha-converting enzyme (TACE) levels in periodontal diseases. J Dent Res. 2008;87:273–277. doi: 10.1177/154405910808700311. [DOI] [PubMed] [Google Scholar]
- 10.Kaman WE, Galassi F, de Soet JJ, Bizzarro S, Loos BG, Veerman EC, et al. Highly specific protease-based approach for detection of Porphyromonas gingivalis in diagnosis of periodontitis. J Clin Microbiol. 2012;50:104–112. doi: 10.1128/JCM.05313-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galassi F, Kaman WE, Anssari Moin D, van der Horst J, Wismeijer D, Crielaard W, et al. Comparing culture, real-time PCR and fluorescence resonance energy transfer technology for detection of Porphyromonas gingivalis in patients with or without peri-implant infections. J Periodontal Res. 2012;47:616–625. doi: 10.1111/j.1600-0765.2012.01474.x. [DOI] [PubMed] [Google Scholar]
- 12.Kaman WE, Hays JP, Endtz HP, Bikker FJ. Bacterial proteases: targets for diagnostics and therapy. Eur J Clin Microbiol Infect Dis. 2014;33:1081–1087. doi: 10.1007/s10096-014-2075-1. [DOI] [PubMed] [Google Scholar]
- 13.Kaman WE, Voskamp-Visser I, de Jongh DM, Endtz HP, van Belkum A, Hays JP, et al. Evaluation of a D-amino-acid-containing fluorescence resonance energy transfer peptide library for profiling prokaryotic proteases. Anal Biochem. 2013;441:38–43. doi: 10.1016/j.ab.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 14.Alhogail S, Suaifan GARY, Bizzarro S, Kaman WE, Bikker FJ, Weber K, et al. On site visual detection of Porphyromonas gingivalis related periodontitis by using a magnetic-nanobead based assay for gingipains protease biomarkers. Microchim Acta. 2018;185(2):149. doi: 10.1007/s00604-018-2677-x. [DOI] [PubMed] [Google Scholar]
- 15.Chinnappan R, Al Attas S, Kaman WE, Bikker FJ, Zourob M. Development of magnetic nanoparticle based calorimetric assay for the detection of bovine mastitis in cow milk. Anal Biochem. 2017;523:58–64. doi: 10.1016/j.ab.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Wignarajah S, Suaifan GA, Bizzarro S, Bikker FJ, Kaman WE, Zourob M. Colorimetric assay for the detection of typical biomarkers for periodontitis using a magnetic nanoparticle biosensor. Anal Chem. 2015;87:12161–12168. doi: 10.1021/acs.analchem.5b03018. [DOI] [PubMed] [Google Scholar]
- 17.Abe N, Baba A, Takii R, Nakayama K, Kamaguchi A, Shibata Y, et al. Roles of Arg- and Lys-gingipains in coaggregation of Porphyromonas gingivalis: identification of its responsible molecules in translation products of rgpA, kgp, and hagA genes. Biol Chem. 2004;385:1041–1047. doi: 10.1515/BC.2004.135. [DOI] [PubMed] [Google Scholar]
- 18.Bengtsson T, Khalaf A, Khalaf H. Secreted gingipains from Porphyromonas gingivalis colonies exert potent immunomodulatory effects on human gingival fibroblasts. Microbiol Res. 2015;178:18–26. doi: 10.1016/j.micres.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Yamada M, Ikegami A, Kuramitsu HK. Synergistic biofilm formation by Treponema denticola and Porphyromonas gingivalis. FEMS Microbiol Lett. 2005;250:271–277. doi: 10.1016/j.femsle.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 20.Belibasakis GN, Bostanci N, Reddi D. Regulation of protease-activated receptor-2 expressionin gingival fibroblasts and Jurkat T cells by Porphyromonas gingivalis. Cell Biol Int. 2010;34:287–292. doi: 10.1042/CBI20090290. [DOI] [PubMed] [Google Scholar]
- 21.Bostanci N, Belibasakis GN. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 2012;333:1–9. doi: 10.1111/j.1574-6968.2012.02579.x. [DOI] [PubMed] [Google Scholar]
- 22.Furuta N, Takeuchi H, Amano A. Entry of Porphyromonas gingivalis outer membrane vesicles into epithelial cells causes cellular functional impairment. Infect Immun. 2009;77:4761–4770. doi: 10.1128/IAI.00841-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Brien-Simpson NM, Pathirana RD, Walker GD, Reynolds EC. Porphyromonas gingivalis RgpA-Kgp proteinase-adhesin complexes penetrate gingival tissue and induce proinflammatory cytokines or apoptosis in a concentration-dependent manner. Infect Immun. 2009;77:1246–1261. doi: 10.1128/IAI.01038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruggiero S, Cosgarea R, Potempa J, Potempa B, Eick S, Chiquet M. Cleavage of extracellular matrix in periodontitis: gingipains differentially affect cell adhesion activities of fibronectin and tenascin-C. Biochim Biophys Acta. 2013;1832:517–526. doi: 10.1016/j.bbadis.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stathopoulou PG, Galicia JC, Benakanakere MR, Garcia CA, Potempa J, Kinane DF. Porphyromonas gingivalis induce apoptosis in human gingival epithelial cells through a gingipain-dependent mechanism. BMC Microbiol. 2009;9:107. doi: 10.1186/1471-2180-9-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bryzek D, Ksiazek M, Bielecka E, Karim AY, Potempa B, Staniec D, et al. A pathogenic trace of Tannerella forsythia—shedding of soluble fully active tumor necrosis factor alpha from the macrophage surface by karilysin. Mol Oral Microbiol. 2014;29:294–306. doi: 10.1111/omi.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jusko M, Potempa J, Karim AY, Ksiazek M, Riesbeck K, Garred P, et al. A metalloproteinase karilysin present in the majority of Tannerella forsythia isolates inhibits all pathways of the complement system. J Immunol. 2012;188:2338–2349. doi: 10.4049/jimmunol.1101240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potempa J, Pike RN. Corruption of innate immunity by bacterial proteases. J Innate Immun. 2009;1:70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDowell JV, Huang B, Fenno JC, Marconi RT. Analysis of a unique interaction between the complement regulatory protein factor H and the periodontal pathogen Treponema denticola. Infect Immun. 2009;77:1417–1425. doi: 10.1128/IAI.01544-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bamford CV, Fenno JC, Jenkinson HF, Dymock D. The chymotrypsin-like protease complex of Treponema denticola ATCC 35405 mediates fibrinogen adherence and degradation. Infect Immun. 2007;75:4364–4372. doi: 10.1128/IAI.00258-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaetti-Jardim Junior E, Nakano V, Wahasugui TC, Cabral FC, Gamba R, Avila-Campos MJ. Occurrence of yeasts, enterococci and other enteric bacteria in subgingival biofilm of HIV-positive patients with chronic gingivitis and necrotizing periodontitis. Braz J Microbiol. 2008;39:257–261. doi: 10.1590/S1517-83822008000200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sardi Janaina C. O., Duque Cristiane, Mariano Flávia S., Peixoto Iza T. A., Höfling José F., Gonçalves Reginaldo B. Candida spp. in periodontal disease: a brief review. Journal of Oral Science. 2010;52(2):177–185. doi: 10.2334/josnusd.52.177. [DOI] [PubMed] [Google Scholar]
- 33.Van Steijn GJ, Nieuw Amerongen AV, Veerman E, Kasanmoentalib S, Overdijk B. Chitinase in whole and glandular human salivas and in whole saliva of patients with periodontal inflammation. Eur J Oral Sci. 1999;107:328–337. doi: 10.1046/j.0909-8836.1999.eos107503.x. [DOI] [PubMed] [Google Scholar]
- 34.Van Steijn GJ, Nieuw Amerongen AV, Veerman EC, Kasanmoentalib S, Overdijk B. Effect of periodontal treatment on the activity of chitinase in whole saliva of periodontitis patients. J Periodontal Res. 2002;37:245–249. doi: 10.1034/j.1600-0765.2002.00330.x. [DOI] [PubMed] [Google Scholar]
- 35.Nascimento GG, Baelum V, Sorsa T, Tervahartiala T, Skottrup PD, Lopez R. Salivary levels of MPO, MMP-8 and TIMP-1 are associated with gingival inflammation response patterns during experimental gingivitis. Cytokine. 2019;115:135–141. doi: 10.1016/j.cyto.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Sorsa T, Gursoy UK, Nwhator S, Hernandez M, Tervahartiala T, Leppilahti J, et al. Analysis of matrix metalloproteinases, especially MMP-8, in gingival creviculular fluid, mouthrinse and saliva for monitoring periodontal diseases. Periodontol. 2000;2016(70):142–163. doi: 10.1111/prd.12101. [DOI] [PubMed] [Google Scholar]
- 37.Uitto Veli-Jukka, Suomalainen Kimmo, Sorsa Timo. Salivary collagenase. Origin, characteristics and relationship to periodontal health. Journal of Periodontal Research. 1990;25(3):135–142. doi: 10.1111/j.1600-0765.1990.tb01035.x. [DOI] [PubMed] [Google Scholar]
- 38.Mushi MF, Bader O, Taverne-Ghadwal L, Bii C, Gross U, Mshana SE. Oral candidiasis among African human immunodeficiency virus-infected individuals: 10 years of systematic review and meta-analysis from sub-Saharan Africa. J Oral Microbiol. 2017;9:1317579. doi: 10.1080/20002297.2017.1317579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Overdijk B, Van Steijn GJ. Human Serum Contains a chitinase—identification of an enzyme, formerly described as 4-methylumbelliferyl-tetra-N-acetylchitotetraoside hydrolase (Mu-Tact Hydrolase) Glycobiology. 1994;4:797–803. doi: 10.1093/glycob/4.6.797. [DOI] [PubMed] [Google Scholar]
- 40.Roslind A, Johansen JS. YKL-40: a novel marker shared by chronic inflammation and oncogenic transformation. Methods Mol Biol. 2009;511:159–184. doi: 10.1007/978-1-59745-447-6_7. [DOI] [PubMed] [Google Scholar]
- 41.Keles ZP, Keles GC, Avci B, Cetinkaya BO, Emingil G. Analysis of YKL-40 acute-phase protein and interleukin-6 levels in periodontal disease. J Periodontol. 2014;85:1240–1246. doi: 10.1902/jop.2014.130631. [DOI] [PubMed] [Google Scholar]
- 42.Nascimento GG, Danielsen B, Baelum V, Lopez R. Identification of inflammatory response patterns in experimental gingivitis studies. Eur J Oral Sci. 2019;127:33–39. doi: 10.1111/eos.12588. [DOI] [PubMed] [Google Scholar]
- 43.Lobene RR, Weatherford T, Ross NM, Lamm RA, Menaker L. A modified gingival index for use in clinical trials. Clin Prev Dent. 1896;8:3–6. [PubMed] [Google Scholar]
- 44.Turesky S, Gilmore ND, Glickman I. Reduced plaque formation by the chloromethyl analogue of victamine C. J Periodontol. 1970;41:41–43. doi: 10.1902/jop.1970.41.41.41. [DOI] [PubMed] [Google Scholar]
- 45.Janus MM, Keijser BJ, Bikker FJ, Exterkate RA, Crielaard W, Krom BP. In vitro phenotypic differentiation towards commensal and pathogenic oral biofilms. Biofouling. 2015;31:503–510. doi: 10.1080/08927014.2015.1067887. [DOI] [PubMed] [Google Scholar]
- 46.Miao D, Godavikava V, Qian X, Seshadrinathan S, Kapila YL, Fenno JC. Treponema denticola upregulates MMP-2 activation in periodontal ligament cells: interplay between epigenetics and periodontal infection. Arch Oral Biol. 2014;59:1056–1064. doi: 10.1016/j.archoralbio.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva N, Abusleme L, Bravo D, Dutzan N, Garcia-Sesnich J, Vernal R, et al. Host response mechanisms in periodontal diseases. J Appl Oral Sci. 2015;23:329–355. doi: 10.1590/1678-775720140259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoneda M, Hirofuji T, Anan H, Matsumoto A, Hamachi T, Nakayama K, et al. Mixed infection of Porphyromonas gingivalis and Bacteroides forsythus in a murine abscess model: involvement of gingipains in a synergistic effect. J Periodontal Res. 2001;36:237–243. doi: 10.1034/j.1600-0765.2001.036004237.x. [DOI] [PubMed] [Google Scholar]
- 49.Rijkschroeff P, Jansen ID, van der Weijden FA, Keijser BJ, Loos BG, Nicu EA. Oral polymorphonuclear neutrophil characteristics in relation to oral health: a cross-sectional, observational clinical study. Int J Oral Sci. 2016;8:191–198. doi: 10.1038/ijos.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fine N, Hassanpour S, Borenstein A, Sima C, Oveisi M, Scholey J, et al. Distinct oral neutrophil subsets define health and periodontal disease states. J Dent Res. 2016;95:931–938. doi: 10.1177/0022034516645564. [DOI] [PubMed] [Google Scholar]
- 51.Fredriksson MI. Effect of priming in subpopulations ofperipheral neutrophils from patients with chronic periodontitis. J Periodontol. 2012;83:1192–1199. doi: 10.1902/jop.2012.110584. [DOI] [PubMed] [Google Scholar]
- 52.Sorsa T, Ingman T, Suomalainen K, Haapasalo M, Konttinen YT, Lindy O, et al. Identification of proteases from periodontopathogenic bacteria as activators of latent human neutrophil and fibroblast-type interstitial collagenases. Infect Immun. 1992;60:4491–4495. doi: 10.1128/iai.60.11.4491-4495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nieminen MT, Listyarifah D, Hagström J, Haglund C, Grenier D, Nordström D, et al. Treponema denticola chymotrypsin-like proteinase may contribute to orodigestive carcinogenesis through immunomodulation. Br J Cancer. 2018;118:428–434. doi: 10.1038/bjc.2017.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Potempa M, Potempa J. Protease-dependent mechanisms of complement evasion by bacterial pathogens. Biol Chem. 2012;393:873–888. doi: 10.1515/hsz-2012-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bostanci N, Bao K, Wahlander A, Grossmann J, Thurnheer T, Belibasakis GN. Secretome of gingival epithelium in response to subgingival biofilms. Mol Oral Microbiol. 2015;30:323–335. doi: 10.1111/omi.12096. [DOI] [PubMed] [Google Scholar]
- 56.Bostanci N, Thurnheer T, Belibasakis GN. Involvement of the TREM-1/DAP12 pathway in the innate immune responses to Porphyromonas gingivalis. Mol Immunol. 2011;49:387–394. doi: 10.1016/j.molimm.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 57.Willi M, Belibasakis GN, Bostanci N. Expression and regulation of triggering receptor expressed on myeloid cells 1 in periodontal diseases. Clin Exp Immunol. 2014;178:190–200. doi: 10.1111/cei.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bostanci N, Reddi D, Rangarajan M, Curtis MA, Belibasakis GN. Porphyromonas gingivalis stimulates TACE production by T cells. Oral Microbiol Immunol. 2009;24:146–151. doi: 10.1111/j.1399-302X.2008.00488.x. [DOI] [PubMed] [Google Scholar]
- 59.Jotwani R, Eswaran SVK, Moonga S, Cutler CW. MMP-9/TIMP-1 imbalance induced in human dendritic cells by Porphyromonas gingivalis. Fems Immunol Med Mic. 2010;58:314–321. doi: 10.1111/j.1574-695X.2009.00637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]